Abstract
Aims
We describe the development and interlaboratory study of modified Saccharomyces cerevisiae as a candidate material to evaluate a full detection workflow including DNA extraction and quantitative polymerase chain reaction (qPCR).
Methods and results
S. cerevisiae NE095 was prepared by stable insertion of DNA sequence External RNA Control Consortium-00095 into S. cerevisiae BY4739 to convey selectivity. For the interlaboratory study, a binomial regression model was used to select three cell concentrations, high (4 × 107 cells ml−1), intermediate (4 × 105 cells ml−1) and low (4 × 103 cells ml−1), and the number of samples per concentration. Seven participants, including potential end users, had combined rates of positive qPCR detection (quantification cycle <37) of 100%, 40%, and 0% for high, intermediate, and low concentrations, respectively.
Conclusions
The NE095 strain was successfully detected by all participants, with the high concentration indicating a potential target concentration for a reference material.
Significance and impact of the study
The engineered yeast has potential to support measurement assurance for the analytical process of qPCR, encompassing the method, equipment, and operator, to increase confidence in results and better inform decision-making in areas of applied microbiology. This material can also support process assessment for other DNA-based detection technologies.
Keywords: ERCC, Evaluation material, Interlaboratory study, Saccharomyces cerevisiae, qPCR, Reference material
1. Introduction
Data of high quality is essential in microbial detection, identification, and quantification because of the impact these organisms (pathogenic and beneficial) have on human life in areas including environmental monitoring, food safety, biothreat detection, and clinical outbreaks [1], [2], [3]. Despite the importance, analysis of microbial samples remains a practical and technological challenge when it comes to confidence in the measurements, especially for measurements made at the point of need or point of care where results are used to inform critical decision-making. In the biodefense field, for example, there are over 300 technologies marketed for use in biological detection claiming the ability to detect pathogens (biothreats) in suspicious materials [4], [5], [6]. However, there remains a scarcity of standards, reference materials, and third-party testing to demonstrate the reliability of these technologies in the hands of end users, despite considerable efforts by the stakeholder community and Federal government. Only a few commercially available biodetection technologies have been submitted to third-party validation, including the Razor™ EX BioDetection System [7], a qPCR-based assay.
qPCR is a well-established technique that provides selectivity and sensitivity in detecting nucleic acid markers [8], [9]. However, obtaining reliable data can be challenging because of factors that compromise nucleic acid amplification such as residue from the crude sample (matrix effect) [10]. In addition, measurement inaccuracy as a result of sample collection, processing, and nucleic acid extraction is often observed [10], [11], [12], [13]. The absence of method validation brings into question the reliability of the generated data [14]. Further, even with the use of a successful detection assay, such as the Biothreat Panel multiplexed PCR-based assay for the detection of Bacillus anthracis, Francisella tularensis and Yersinia pestis or the rapid, high-throughput, culture-based PCR methods to analyze samples for viable spores of B. anthracis and its surrogates [15], the final result in the field still depends upon the entire analytical process. The process includes the methods and protocols, the measurement workflow which encompasses all steps applied to obtain the final result, the performance of the equipment at the point of need, operator capabilities and skills, and proficiency testing. Without all of these components in place, results cannot be used with confidence to support decision-making.
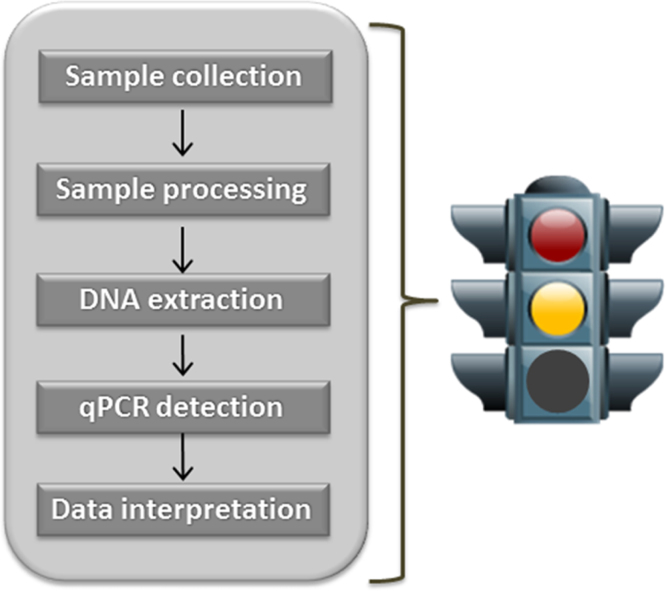
Control materials can serve to evaluate or validate analytical processes, establish reliable and comparable analytical results among laboratories or analysts through proficiency testing or competence assessment, and verify accuracy of measurement performance on a daily basis [16]. For example, human DNA Standard Reference Materials (SRMs) are used in the forensics community to reduce variability within and among laboratories [17], [18]. Control materials are typically thoroughly characterized using measurement methods with well understood biases and variability. Measurement controls such as reference materials can help provide confidence in the application of qPCR to microbial measurements, with the format of the material enabling performance evaluation at various workflow steps (Fig. 1). For instance, a reference material can be mixed into a matrix of interest to demonstrate a successful DNA extraction step, which is highly susceptible to matrix effects. The use of a reference material to demonstrate a successful qPCR analytical process can increase confidence when the measurement capability is applied in a real-case scenario, such as in a clinical setting, environmental monitoring, or biothreat scenario.
Fig. 1.
Workflow steps for a downstream qPCR sample analysis. Analogous to a traffic light, the unsuccessful detection of a reference material provides a red light indicating the analytical process is unsuccessful and potentially helping identify the source of the problem. Successful detection of the material provides a yellow light indicating that the analytical process is working properly, that is the methods are appropriate, the equipment is functioning, and the user is proficient in the required skills. There is no green light since the material cannot be used to validate a specific detection assay for an organism of interest. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
One promising organism to meet these needs for a qPCR reference material is Saccharomyces cerevisiae (S. cerevisiae), a eukaryotic model system widely used in biology fields such as bioengineering [19]. Yeast can be genetically modified to enable specific detection and offers a low DNA extraction efficiency to challenge extraction protocols [11]. Its physiological resilience under low nutrient conditions and stability under various environmental conditions make yeast suitable for different formats, including a liquid or powder, for broader applicability. Moreover, S. cerevisiae has minimal health and security risks and can be handled without special precautions or training. It can therefore serve as a surrogate material for routine training and process evaluation in applications where the true agent of interest is a pathogen or threat agent and not easily used. Further, use of the yeast, and not the agent of interest, can essentially eliminate false positives during real microbial detection situations due to residual material on equipment.
The presence of a non-native target DNA sequence in the yeast genome can eliminate false positives from near-neighbor organisms. For specificity, the target sequence should be rare and not typically found in the environment of interest. NIST SRM 2374 contains a series of nucleic acid sequences selected by the External RNA Control Consortium (ERCC) as control sequences that are rarely (if at all) found in normal environmental conditions (temperature and pressure). One such sequence is ERCC-00095, derived from Methanocaldococcus jannaschii, a deep-sea vent archaeon found only in extremely harsh conditions (an extremophile).
The objective of this study was to develop a S. cerevisiae strain containing a non-yeast target DNA insert and evaluate the strain via interlaboratory study as a potential material for assessing the qPCR analytical process, in efforts toward a reference material for qPCR. Yeast cells were transformed by inserting the ERCC-00095 DNA sequence into the yeast genome. The modified yeast strain, termed NE095, was prepared at three different cell concentrations and evaluated using qPCR in an interlaboratory pilot study involving five public health laboratories, one mobile laboratory and one in-house laboratory. The NE095 was detectable at the expected concentrations in multiple laboratories and is suitable for a reference material.
2. Materials and methods
2.1. Preparation and characterization of engineered S. cerevisiae
2.1.1. cerevisiae transformation
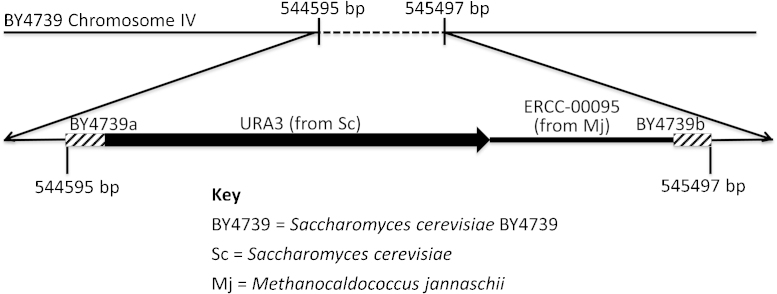
The parent organism was a URA3 deficient yeast strain: Saccharomyces cerevisiae BY4739 (MATalpha leu2Δ0 lys2Δ0 ura3Δ0) (procured from Open Biosystems) derived from S. cerevisiae S288C. The URA3 gene encodes for orotidine-5′-phosphate decarboxylase from S. cerevisiae S288C. A target DNA sequence and full-length URA3 gene were inserted via homologous recombination into chromosome IV of the yeast (Fig. 2, Supplemental material: S. cerevisiae NE095 transformation, Figs. S1 and S2). The DNA insert was prepared by ligating the target sequence, ERCC-00095 (from NIST SRM 2374, Genbank accession KC702204, without the polyA tail found in the SRM), to the URA3 gene by overlapping PCR. The full-length URA3 gene and URA3 promoter sequence were PCR amplified from a pYES2 vector (Life Technologies, part # V825-20). See Supplemental material for detailed methods describing the transformation process.
Fig. 2.
Map of final linear construct inserted into BY4739 chromosome IV. BY4739a is a 50 bp region homologous to Open Biosystems strain S. cerevisiae BY4739 and corresponding to the non-coding region on chromosome IV between 544595 bp and 544644 bp. The full annotation of chromosome IV can be found at http://browse.yeastgenome.org/fgb2/gbrowse/scgenome/. URA3 is a 1106 bp sequence that encodes S. cerevisiae S288C orotidine-5′-phosphate decarboxylase, an enzyme that catalyzes the synthesis of pyrimidine ribonucleotides. The sequence was PCR amplified from a pYES2 vector (Life Technologies, part # V825-20) and includes a working copy of the 804 bp URA3 gene (Gene ID: 856692) as well as the URA3 promoter sequence. ERCC-00095 represents a 438 bp sequence from NIST SRM 2374: DNA Sequence Library for External RNA Controls. ERCC-00095 (GenBank accession number KC702204.1) is derived from donor microorganism M. jannaschii. The sequence was amplified from the ERCC-00095 plasmid using primers 95F and 95R (Table S1). BY4739b is 51 bp homologous to the non-coding region on chromosome IV between 545447 bp and 545497 bp in Open Biosystems strain S. cerevisiae BY4739.
2.1.2. NE095 characterization
The presence of the insert was demonstrated by PCR, and the sequence of the amplified insert, including the ERCC-00095 and URA3 gene, was confirmed by Sanger sequencing (analyzed by Eurofins MWG Operon, Alabama, USA) (Supplemental material: NE095 insert sequence confirmation, Figs. S3, S4, and S5). Once the insert was demonstrated, the yeast cells were cultured in SD/-Ura broth for subsequent experiments.
2.1.3. NE095 growth conditions and quantification via microscopy
NE095 cells were cultured in SD/-Ura broth overnight. Each culture was generated from one NE095 colony in 5 ml of broth (30 °C, ∼16 h,) followed by a 25-fold dilution in broth and subsequent growth (30 °C, ∼8 h). The number of replicate yeast cultures used for each set of experiments was as follows: binomial regression analysis (n = 8), stability study (n = 5) and interlaboratory study (n = 1). Samples for the interlaboratory study were collected from a single culture to assure that all participants obtained cells from the same batch. Variability in number of cells between batches under the selected experimental conditions was determined by cell counting while live/dead staining experiments were applied to estimate cell viability under different storage condition (see Supplemental material: S. cerevisiae NE095 material stability, Figs. S8, S9, and S10). Cells were counted using a cellometer (disposable hemocytometer) following the manufacturer’s instructions (Nexcelom, USA). Cellometers were loaded with yeast and imaged, and counts were performed using Image J software (version 1.47 v) [20]. Throughout this work, data are represented as the mean value ± one standard deviation, where the standard deviation serves as the estimate for the standard uncertainty.
2.1.4. DNA extraction and qPCR detection assay
The following protocol was used to analyze the genomic DNA (gDNA) for the yeast stability study and interlaboratory study, and to generate the regression model. gDNA from NE095 cells was extracted using 200 μl (800 cells, 8 × 104 cells, or 8 × 106 cells for low, intermediate, and high concentrations, respectively) aliquots of cells and a QIAamp DNA Blood Mini Kit (cat# 51104 QIAGEN, USA). The spin protocol for DNA purification from blood or body fluids was followed according to the manufacturer’s directions with a few modifications. DNA was incubated at 56 °C for 1 h following the addition of protease and lysis buffer. Extracted DNA was eluted in 200 μl elution buffer provided in the DNA extraction kit and stored at 4 °C prior to downstream analysis.
qPCR analysis was performed on a 7900HT PCR system (Life Technologies, Carlsbad, CA), except in the interlaboratory study where participants used their own qPCR technology. A TaqMan FAM-MGB primer and probe set was custom designed and synthesized by Life Technologies (Life Technologies, Carlsbad, CA) to amplify a 105 bp region of the ERCC-00095 sequence (Figs. S6 and S7). The reaction mixture contained 10 μl of Environmental MasterMix (Life Technologies, Carlsbad, CA), 4 μl of extracted genomic DNA, 1 μl of amplification primers and probe with the following sequences: CAGTCATCTTTAACCTCATCCCACAA (forward primer), CATTTGGCCCAAGAATTCATGGAAT (reverse primer), and CCCACTCAACAATCTT (probe), and 5 μl of sterilized water. Reaction conditions were: 5 min at 50 °C, 10 min at 95 °C for Taq polymerase activation, and 50 cycles of 15 s at 95 °C, 1 min at 60 °C for DNA amplification. A gDNA standard curve was prepared via 10-fold serial dilutions of the transformed yeast genomic DNA. The initial concentration was estimated by Nanodrop (Thermo Scientific, USA) and Qubit (Invitrogen, USA). The efficiency of amplification (E) for the transformed yeast using this customized qPCR assay (primers and probes) was 98.4% (Fig. S7). All chemicals were purchased from Fisher Scientific unless otherwise stated.
For the qPCR assay, primer and probe specificity was verified using Primer BLAST (results available at http://dx.doi.org/10.6084/m9.figshare.875419). Three primer combinations were compared to the GenBank nr database using Primer BLAST: forward-reverse, forward-probe, and probe-reverse. The only matches to the database for the primer-probe set were to M. jannaschii DSM 2661 (GenBank Accession: CP001781.1) and Methanocaldococcus vulcanius M7 complete genomes (L77117.1) as well as the ERCC-00095 (KC702204.1) and M. jannaschii spike-in control MJ-500-42 (DQ516759.1). The probe and reverse primer sequences had no mismatches to the Methanocaldococcus sequences or ERCC-00095, whereas the forward primer had two mismatches at the 5′ end (positions 1 and 3) to the M. vulcanius M7 genome, and three mismatches at the 5′ end (positions 1–3) for the M. jannaschii spike-in control MJ-500-42, ERCC-00095, and M. jannaschii DSM 2661 genome.
2.2. Interlaboratory study
2.2.1. Experimental design and statistical analysis approach
A logistic regression model was used to develop the experimental design for the interlaboratory study [21], specifically the cell concentrations and number of samples per concentration to include in the sample panel. To generate the regression model, four independent cultures were grown on two separate days with an average cell concentration of (6.3 ± 0.8) × 107 cells ml−1 (n = 8). A ten-fold cell dilution series was generated for each culture with cell concentrations ranging from 103 cells ml−1 to 107 cells ml−1. gDNA extracted from the cells was analyzed using the qPCR protocol described above. Samples were scored as positive if the quantification cycle (Cq) for detecting the ERCC-00095 insert was less than 37 for at least one of the triplicate reactions. The same Cq value criterion was applied to evaluate the raw data from the interlaboratory study. The qPCR data were analyzed using the statistical programming language R [22]. Source code and raw data for the binomial regression can be found at the following location: (http://dx.doi.org/10.6084/m9.figshare.875419).
2.2.2. Participants
Participants consisted of five public health laboratories, one mobile laboratory, and one in-house laboratory; six of these participants are laboratories currently active in the biosurveillance community. Each participant received the yeast sample panel, DNA extraction kit, primers and probes, and a detailed protocol (Supplemental material: Interlaboratory SOP). Participants used either the Applied Biosystems Instruments (ABI) 7500 or the Joint Biological Agent Identification and Diagnostic System (JBAIDS) platform for qPCR. To assure an even execution of the experiments, participants reported to the organizer laboratory the arrival date and condition of the sample panel, the platform and software used to process the samples and collect data, and any deviation from the recommended protocol. All participants submitted their results to the organizer laboratory within one week.
2.2.3. Yeast sample panel
Based upon the logistic regression model results, each sample panel was composed of ten randomized blinded samples: a 5% ethanol no template-control (blank), and nine yeast samples as follows: low (4.0 × 103 cells ml−1, n = 2), intermediate (4.0 × 105 cells ml−1, n = 5), and high (4.0 × 107 cells ml−1, n = 2). All samples in the panel came from a single overnight cell culture batch and were quantified by cell counting using a cellometer. Samples were shipped in 5% ethanol, a condition selected to maintain cell and DNA integrity for short term (2 weeks), liquid storage at room temperature. Stability of cells under the conditions used in the interlaboratory study was demonstrated by live/dead staining and qPCR as detailed in supporting information.
2.2.4. Statistical analysis for intermediate samples
The experimental design generated 35 intermediate concentration samples, which were prepared and distributed among seven labs (five samples to each lab; samples are nested within lab). In most cases, three replicate Cq measurements were taken for each sample. Measurements with undetermined runs (no detection at all) were modeled as being right-censored at 40 (i.e., the corresponding observed Cq was assumed to be greater than 40, but otherwise unknown). Let denote the natural log of the value observed in replicate measurement from sample at lab. We analyzed the observed data with the following random effects or hierarchical model:
where denotes a global average ln(Cq) across all labs and samples, is a random effect for lab i , is a random effect for sample , and is a random error corresponding to replicate measurement from sample at lab . Each variance component is assigned a prior distribution such that and follow a uniform distribution between 0 and 0.5. The overall mean was assigned a uniform prior distribution from 3 to 3.9, corresponding to a range of mean Cq values given by (20.1, 49.4). The posterior distributions of the model parameters and given the observed data were evaluated using Markov chain Monte Carlo simulation via the R package rjags. The simulation included 5 chains, each recording 10,000 observations following a 50,000 iteration burn in.
3. Results
3.1. NE095 characterization
The DNA insert was successfully prepared and incorporated into chromosome IV to produce the NE095 strain. NE095 has a functional URA3 gene and otherwise is not anticipated to have any other changes in phenotype or traits. The modified yeast strain grows under typical conditions in uracil-deficient medium, as expected. Sequencing confirmed the presence of the expected 1,645 bp insert, including the functional URA3 gene and the ERCC-00095 sequence (Fig. S5). In addition, the insert was successfully amplified by the selected qPCR primers and probes. An alignment of the qPCR primers with the de-novo assembly of the insert sequence revealed insertions 4 bp from the 5′ end of the forward and reverse primers (Fig. S6). Additionally, positions 1–3 from the 5′ end of the forward primer that were in disagreement with the GenBank sequences, due to the previously identified insertions, as identified with Primer BLAST agree with the Sanger sequence data. Primer mismatches can decrease assay efficiency; however the mismatches are at the 5′ end and less likely to impact the qPCR assay efficiency [23]. Even with the mismatches the qPCR assay efficiency was over 98%, which is suitable for quantitative assays.
The stability of the NE095 was monitored at room temperature in three different storage conditions, where it was determined that 5% ethanol was a suitable storage condition for the time frame required in the interlaboratory study (see supporting information). In 5% ethanol, no change in cell concentration was observed after two weeks of storage (p > 0.05) (Table S4). The stability of the yeast (membrane integrity) in the medium was also confirmed by live/dead staining while the integrity of the DNA was confirmed using qPCR (Figs. S8, S9, S10).
3.2. Experimental design for the interlaboratory study
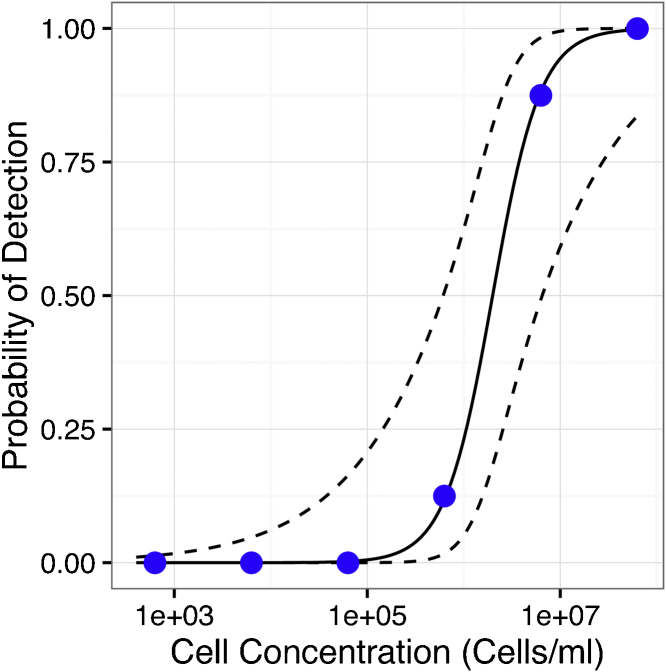
The appropriate sample concentrations for the interlaboratory study were determined by fitting the qPCR results from a dilution series of yeast cells to a binomial regression model (Fig. 3) The probability of detection for a sample with 107 cells ml−1 was 99.7% with a 95% confidence interval of 83.6–99.9%. The probability of detection for a sample with a concentration of 103 cells ml−1 was 4.2 × 10−3% with a 95% confidence interval of 3.6 × 10−6% to 4.7%. Based on these probabilities of detection, 103 cells ml−1 and 107 cells ml−1 were chosen as the negative and positive control samples, respectively. A concentration of 1.9 × 106 cells ml−1 had a 50% probability of detection with a 95% confidence interval for detection of 18–81% based on the generated regression model. However, because of an observed increase in DNA recovery with storage time (Fig. S10), a lower sample concentration (105 cells ml−1) was chosen as the intermediate value to represent samples near the limit of detection. That concentration had a 0.0052% probability of detection with a 95% confidence interval of 0.0001–21%. Five samples were distributed to the participants for the intermediate concentration, compared to 2 samples for the high and low concentration samples, to increase the statistical power to assess probability of detection at the intermediate concentration.
Fig. 3.
Logistic regression model. The model was generated based on the number of positive qPCR reactions (at least 1 of 3 reactions with a Cq value <37) using a cell dilution series from 8 independent cultures. The filled circles represent the experimental data. The solid and dashed lines indicate the regression model and the 95% pointwise confidence interval, respectively.
3.3. Interlaboratory study
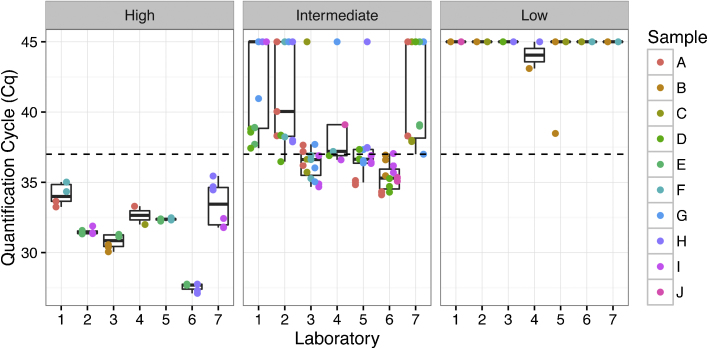
A total of 190 qPCR data points were collected from the interlaboratory study (Fig. 4). Laboratory 4 ran single qPCR reactions instead of the triplicate reactions requested in the provided protocol. Despite the single run, the data were otherwise suitable to be included in the analysis. Laboratory 6 extracted DNA from 400 μl of cells, as opposed to 200 μl as described in the protocol, and obtained the lowest quantification cycle (Cq) values for the high concentration samples. Thus, data from laboratory 6 were excluded from further analysis because of deviations from the provided protocol. Potential contamination was monitored by blank samples (data not shown) and no detection was observed as expected. High concentration (107 cells ml−1) samples were positive (detection) across all laboratories while low samples (103 cells ml−1) scored negative since any detectable Cq values (43.1 and 38.5) were above the threshold of 37. For intermediate samples (105 cells ml−1), 40% of the samples (12 out of 30) were scored as positive, suggesting that this concentration is approaching the yeast concentration required to produce a positive qPCR signal in essentially all measurements.
Fig. 4.
Boxplots describing the quantification cycle (Cq) as a function of laboratory for the 10 samples analyzed in the interlaboratory study. The concentration of samples was individually randomized and blinded for each participant. The circles (●) in the plots represent individual datapoints, with different colors indicating the randomization of the samples. A value of 45 was assigned for undetectable Cq values in order to visualize the generated data. The dashed line indicates the threshold (Cq = 37) used to indicate positive detection. Note that data from laboratory 6 were not included in the statistical analysis because of deviation from the provided protocol.
According to the posterior distributions of their respective variance parameters, the probability that the variability due to differences between labs exceeds the variability due to differences between samples is 80%. That is, in 80% of the simulated iterations the standard deviation parameter for the labs, σlab, had a larger value than the standard deviation parameter for the samples, σsample. The posterior median of is 1.54, with a 95% posterior credible interval given by (0.50, 5.44) (Fig. S11).
4. Discussion
In this study we aimed to demonstrate the use of yeast as a candidate reference material in a workflow composed of various steps that are not necessarily measurable individually, and we envision the yeast having broader application, such as to assess performance parameters related to quantitative methods. We successfully developed S. cerevisiae NE095, a yeast strain with a stable DNA insertion for qPCR measurements, and demonstrated its detection in the hands of potential users via interlaboratory study. This work demonstrates the potential of the yeast material to be further developed into a reference material to evaluate processes associated with nucleic acid-based detection technologies. The insert incorporated into the yeast genome attributes specificity by eliminating the risk of false positive amplification from environmental background, near-neighbor organisms, or contaminants. Because the yeast is a versatile organism that can be manipulated in a broad range of environments, the authors foresee its use as a reference material in multiple areas of microbial detection including environmental monitoring, food safety, biothreat detection, and clinical outbreaks.
The modified yeast strain was developed to evaluate analytical processes used for microbial measurements and to instill confidence in measurement capabilities, particularly for applications where critical decision-making depends upon the measurement results. NE095 can challenge nucleic acid detection technologies and processes through using a pre-determined amount of cells in a typical qPCR workflow (e.g., sample collection, DNA extraction, qPCR with a custom assay for the target DNA sequence) with the expectation of positive detection. Qualitative assessment would provide a yes/no answer to detection, whereas lack of detection may indicate error in sample processing or defective equipment. When used for quantitative assessment, NE095 can evaluate the efficiency of the analytical process, track changes in efficiency over time, or determine the effects of modifications in the process or operator skills on efficiency. Additionally, NE095 can be used as a confidence checker (to assure correct equipment function), in proficiency testing, training, and other applications. It is noteworthy to mention that NE095 is not designed to validate detection assays/technologies for a specific organism of interest (e.g., pathogen or biothreat) but rather the analytical process in place to assure that it is working properly.
The probability of detection at a given concentration was modeled using binomial regression with a logistic link function (Fig. 3). Binomial regression has been used traditionally in toxicology studies [24] but has gained application in microbiology in the last 10 years. For instance, Janse et al. [25] used binomial regression analysis to support the development of a rapid qPCR assay to distinguish Burkholderia mallei and Burkholderia pseudomallei strains, two closely related, highly virulent bacteria. Results from the regression model were used to select the concentration levels, including a positive control (high concentration), negative control (low concentration), and intermediate concentration samples, for the interlaboratory study. We believe the logistic regression model was adequate for this study, where the main goal was to determine the probability of detection at various concentration levels, e.g., probit [26].
The interlaboratory study revealed that all participants succeeded in detecting the yeast as predicted by the organizer laboratory. Laboratories 4 and 5 observed Cq’s for samples at the low concentration level (103 cells ml−1); however those values did not meet the established criteria for positive scoring (Cq < 37) and were therefore scored as negative in agreement with predictions. Although a common cycle threshold for qPCR analysis is 40, a threshold of 37 was selected to reduce the chance of false positives due to low levels of contamination or instrument noise. Statistical analysis also showed that variability between samples (within labs) is about the same scale as the variability between repeated measurements made on a single sample (within sample) (Fig. S11). The estimated variability among laboratories is larger than the estimated variability between samples and between repeated measurements for 80% of the iterations. Variations in Cq values observed across all laboratories might be due to differences in arrival date, sample processing date, calibration of the qPCR instrument, adherence to the distributed protocol, or other factors.
This work is the initial phase of a larger project with the end goal of providing a material for process evaluation in qPCR and other nucleic acid-based techniques as a means to improve confidence in the analytical process and ultimately the measurement results. This study established cell concentrations that can be expected to produce a positive or negative qPCR result using this protocol and gives insight into an appropriate cell concentration for the development of a reference material. The next step is to preserve the yeast in a format that provides stability over longer time periods, such as a dry format (e.g., via lyophilization). Using the system described herein, the results indicate that a yeast concentration of 107 cells ml−1 should result consistently in positive detection. Dry yeast cells could be rehydrated to challenge the qPCR workflow or crushed into a powder to challenge additional steps involved in field microbial risk assessment, such as sampling [27]. In addition to challenging the qPCR analytical process, a quantitative reference material composed of intact yeast cells has potential application for a broad range of other processes where microbial cells or cell components are measured.
5. Conclusions
The need for stable, well-characterized biological materials to aid in reproducible and reliable analytical processes and improve confidence in microbial measurements is relevant to nearly any activity involving detection or quantification of microorganisms. In this study, we describe the development and assessment of a candidate material based on S. cerevisiae to evaluate nucleic-acid based detection technologies (e.g., qPCR). We successfully incorporated the ERCC-00095 DNA sequence into the S. cerevisiae genome to confer selectivity, and demonstrated via interlaboratory study the detection of NE095 in blinded samples in the hands of potential future users. In addition to a qualitative (yes/no) detection of the yeast, quantitative detection was also achieved by challenging the capability of qPCR to detect predetermined cell concentrations. These results support the further development of NE095 as a reference material for qPCR technologies involved in microbial detection and risk assessment.
Disclaimers
Opinions expressed in this paper are the authors’ and do not necessarily reflect the policies and views of the Department of Homeland and Security (DHS), National Institute of Standards and Technology (NIST), or affiliated venues. Certain commercial equipment, instruments, or materials are identified in this paper only to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the NIST, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose. Official contribution of NIST; not subject to copyrights in USA.
Conflicts of interest
None declared.
Acknowledgements
The Department of Homeland Security (DHS) Science and Technology Directorate sponsored the production of this material under Interagency Agreement HSHQPM-12-X-00078 with the National Institute of Standards and Technology (NIST). We thank the Florida Department of Health, Michigan Department of Community Health, Minnesota Department of Health, New York Department of Health, Washington Department of Health and 4th Civil Support Team, Georgia Army National Guard for participating in the interlaboratory study.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bdq.2016.01.001.
Contributor Information
S.M. Da Silva, Email: sdasilva@nist.gov.
N.J. Lin, Email: nancy.lin@nist.gov.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Butler T. Plague gives surprises in the first decade of the 21st century in the United States and worldwide. Am. J. Trop. Med. Hyg. 2013;89(4):788–793. doi: 10.4269/ajtmh.13-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gandhi M., Chikindas M.L. Listeria: a foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 2007;113(1):1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Gueimonde M. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013;4:202. doi: 10.3389/fmicb.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emanuel P., Caples M. US Army RDECOM; 2011. Chemical, Biological, Radiological Technology Survey. [Google Scholar]
- 5.U.S. Department of Homeland Security, Framework for a Biothreat Field Response Mission Capability. Washington, D.C., 2011.
- 6.U.S. Department of Homeland Security, Guide for the Selection of Biological Agent Detection Equipment for Emergency First Responders, 2nd ed. Whashington, D.C., 2007.
- 7.Hadfield T. RAZOR EX anthrax air detection system for detection of Bacillus anthracis spores from aerosol collection samples: collaborative study. J. AOAC Int. 2013;96(2):392–398. doi: 10.5740/jaoacint.cs2012-06. [DOI] [PubMed] [Google Scholar]
- 8.Kim J., Lim J., Lee C. Quantitative real-time PCR approaches for microbial community studies in wastewater treatment systems: applications and considerations. Biotechnol. Adv. 2013;31(8):1358–1373. doi: 10.1016/j.biotechadv.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Botes M., de Kwaadsteniet M., Cloete T.E. Application of quantitative PCR for the detection of microorganisms in water. Anal. Bioanal. Chem. 2013;405(1):91–108. doi: 10.1007/s00216-012-6399-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997;63(10):3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson N.D., Morrow J.B. DNA extract characterization process for microbial detection methods development and validation. BMC Res. Notes. 2012:668. doi: 10.1186/1756-0500-5-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossen L. Inhibition of PCR by components of food samples: microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 1992;17(1):37–45. doi: 10.1016/0168-1605(92)90017-w. [DOI] [PubMed] [Google Scholar]
- 13.Bustin S.A. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 14.U.S. Government Accoutability Office, Anthrax Detection: Agencies need to validate sampling activities in order to increase confidence in negative results. GAO: Washington, D.C., 2005.
- 15.Kane S.R. Rapid: high-throughput, culture-based PCR methods to analyze samples for viable spores of Bacillus anthracis and its surrogates. J. Microbiol. Methods. 2009;76(3):278–284. doi: 10.1016/j.mimet.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Bipm, International vocabulary of metrology and general concepts and associated terms (VIM) JCGM. 2008;200:2008. doi: 10.1016/j.clinbiochem.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Kline M.C. Production and certification of NIST Standard Reference Material 2372 human DNA Quantitation Standard. Anal. Bioanal. Chem. 2009;394(4):1183–1192. doi: 10.1007/s00216-009-2782-0. [DOI] [PubMed] [Google Scholar]
- 18.Kline M.C. Results from the NIST 2004 DNA quantitation study. J. Forensic Sci. 2005;50(3):571–578. [PubMed] [Google Scholar]
- 19.Akada R. Genetically modified industrial yeast ready for application. J. Biosci. Bioeng. 2002;94(6):536–544. doi: 10.1016/s1389-1723(02)80192-x. [DOI] [PubMed] [Google Scholar]
- 20.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smieja M. Replicate PCR testing and probit analysis for detection and quantitation of Chlamydia pneumoniae in clinical specimens. J. Clin. Microbiol. 2001;39(5):1796–1801. doi: 10.1128/JCM.39.5.1796-1801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Team, R.C., R: A language and environment for statistical computing. R foundation for Statistical computing, Vienna, Austria. http://www.R-project.org. 2013.
- 23.Stadhouders R. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J. Mol. Diagn. 2010;12(1):109–117. doi: 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altshuler B. Modeling of dose-response relationships. Environ. Health Perspect. 1981;42:23–27. doi: 10.1289/ehp.814223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janse I. Multiplex qPCR for reliable detection and differentiation of Burkholderia mallei and Burkholderia pseudomallei. BMC Infect. Dis. 2013;13:86. doi: 10.1186/1471-2334-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardini F. Suitability of log-linear models to evaluate the microbiological quality of baby clams (Chamelea gallina L.) harvested in the Adriatic Sea. Int. J. Food Microbiol. 2000;54(1–2):63–74. doi: 10.1016/s0168-1605(99)00170-1. [DOI] [PubMed] [Google Scholar]
- 27.ASTM . ASTM International; West Conshohocken, PA, USA: 2010. Standard Guide for Operational Guidelines for Initial Response to a Suspected Biothreat Agent. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.