Abstract
Achieving high accuracy in orthology inference is essential for many comparative, evolutionary and functional genomic analyses, yet the true evolutionary history of genes is generally unknown and orthologs are used for very different applications across phyla, requiring different precision–recall trade-offs. As a result, it is difficult to assess the performance of orthology inference methods. Here, we present a community effort to establish standards and an automated web-based service to facilitate orthology benchmarking. Using this service, we characterize 15 well-established inference methods and resources on a battery of 20 different benchmarks. Standardized benchmarking provides a way for users to identify the most effective methods for the problem at hand, sets a minimum requirement for new tools and resources, and guides the development of more accurate orthology inference methods.
Supplementary information
The online version of this article (doi:10.1038/nmeth.3830) contains supplementary material, which is available to authorized users.
Subject terms: Standards, Sequence annotation, Protein function predictions, Phylogeny
Main
Evolutionarily related genes (homologs) across different species are often divided into gene pairs that originated through speciation events (orthologs) and gene pairs that originated through duplication events (paralogs)1. This distinction is useful in a broad range of contexts, including phylogenetic tree inference, genome annotation, comparative genomics and gene function prediction2,3,4. Accordingly, dozens of methods5 and resources6,7,8 for orthology inference have been developed.
Because the true evolutionary history of genes is typically unknown, assessing the performance of these orthology inference methods is not straightforward. Several indirect approaches have been proposed. Based on the notion that orthologs tend to be functionally more similar than paralogs (a notion now referred to as the ortholog conjecture9,10,11,12), Hulsen et al.13 used several measures of functional conservation (coexpression levels, protein–protein interactions and protein domain conservation) to benchmark orthology inference methods. Chen et al.14 proposed an unsupervised learning approach based on consensus among different orthology methods. Altenhoff and Dessimoz15 introduced a phylogenetic benchmark measuring the concordance between gene trees reconstructed from putative orthologs and undisputed species trees. More recently, several 'gold standard' reference sets, either manually curated16,17 or derived from trusted resources18, have been used as benchmarks. Finally, Dalquen et al.19 used simulated genomes to assess orthology inference in the presence of varying amounts of duplication, lateral gene transfer and sequencing artifacts.
This wide array of benchmarking approaches poses considerable challenges to developers and users of orthology methods. Conceptually, the choice of an appropriate benchmark strongly depends on the application at hand. Practically, most methods are not available as stand-alone programs and thus cannot easily be compared on a common set of data. Likewise, some benchmarks rely on complex pipelines that may be difficult to implement. If public results are available as part of a publication or a resource, inconsistent genome releases or identifiers severely complicate comparisons. Some methods or benchmarks can also be computationally costly to run. As a result, users cannot easily identify appropriate tools, and methodological progress is hampered.
Here, we report on a community effort to standardize and facilitate orthology benchmarking. For this effort, we established a shared reference data set and developed a web-based service for automatic orthology benchmarking (http://orthology.benchmarkservice.org). We then used these resources to run a community experiment to assess 15 well-established orthology inference methods and resources on a wide array of phylogenetic and functional benchmarks. By providing a way to automatically include new methods and disseminate results publicly, we hope to maintain an up-to-date and comprehensive assessment of state-of-the-art orthology tools.
Results
Here, we provide an overview of the benchmark service and orthology inference methods and then present benchmarking results in three categories: species discordance tests, reference gene trees and functional tests. The benchmark service alone required the evaluation of 70,390,701 orthologous relationships and the inference of 233,000 phylogenetic trees.
Benchmark service
To automate ortholog benchmarking on a broad range of tests (detailed below), we developed a publicly accessible web service (Fig. 1). Using this workflow, an orthology method developer first infers orthologs using the Quest for Orthologs (QfO) reference proteome data set. Orthology inference methods vary in the kind of output they provide—e.g., labeled gene trees and orthologous groups—but it is usually possible to reduce these to orthologous pairs, which thus constitute a natural 'common denominator' for benchmarking. The benchmark service accepts these pairwise orthologs predictions in OrthoXML20 or tab-delimited format. As the OrthoXML format also supports InParanoid-style clusters and hierarchical orthologous groups, the service can automatically convert these to pairwise relationships.
Figure 1. The Orthology Benchmark service facilitates assessment and comparison of orthology inference methods.
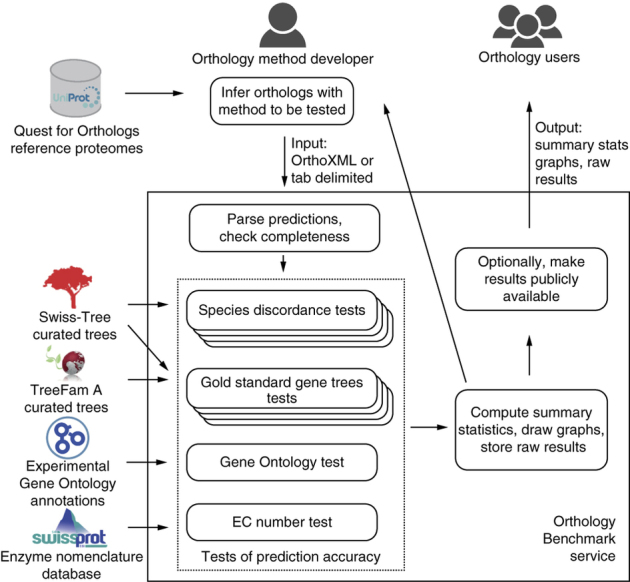
Orthology method developers run their methods on a reference proteome set and submit the inferred orthologs to the service. The predictions are subjected to a battery of phylogenetic and functional tests, and the results are returned to the method developer, who can choose to disclose them publicly.
Next, the service ensures that only predictions among valid reference proteomes are provided (with scoring implicitly assuming that the uploaded inferences are complete). Benchmarks are then selected and run in parallel; some may take up to several hours. Finally, statistical analyses determine the method's performance on each benchmark data set. Where possible, performance is measured in terms of precision (i.e., positive predictive value: the proportion of ortholog predictions that are correct) and recall (i.e., sensitivity, or true positive rate: the proportion of actual orthologs that are correctly predicted). Raw data and results are stored and provided to the submitter, who can choose to make the results publicly available. In order to achieve transparency and encourage improvements, we have released source code under an open source license (Mozilla Public License Version 2.0) at https://github.com/qfo/benchmark-webservice (also Supplementary Software).
Methods investigated
We investigated a broad array of well-established methods, including three tree-based methods: Ensembl Compara21, PANTHER 8.0 (ref. 22) and PhylomeDB23; seven graph-based methods (i.e., based on pairwise comparisons): Best Reciprocal Hits24, Reciprocal Smallest Distance (RSD)25, EggNOG26, Hieranoid27, InParanoid28, OMA29, OrthoInspector30 and a meta-method incorporating both tree- and graph-based methods, MetaPhOrs31. For some methods, multiple variants are included in the analysis (Online Methods). Each method inferred orthologs on the 754,149 protein sequences from 66 reference genomes except for MetaPhOrs, which inferred orthologs on all but three prokaryotes (Online Methods).
Generalized species tree discordance test
Orthology was first defined in the context of species tree inference, which requires genes related through speciation1. The species tree discordance test exploits this relationship by assessing the accuracy of orthologs in terms of the accuracy of the species tree that can be reconstructed from them15. The original protocol was limited to species tree 'comb' topology (a specific type of tree in which all bifurcations occur along a single path) and a small number of taxa (up to six). Here we overcome these two limitations by generalizing the orthology sampling procedure to any tree topology and employing larger reference trees from the SwissTree initiative. Furthermore, to minimize the possibility of gene–species tree discordance due to incomplete lineage sorting, we avoided sampling orthologs among species separated by branches shorter than 10 million years (myr) (Online Methods and Supplementary Fig. 1).
We observed different trade-offs between average discordance (Robinson–Foulds32 distance, as a proxy for the false discovery rate, the complement of precision) and the number of trees that can be sampled (proxy for recall) across all methods (Fig. 2). An ideal method would be placed in the lower right corner of Figure 2. When considering eukaryotes, results with the highest precision and lowest recall were obtained with OMA groups. At the other extreme, PANTHER 8.0 (all) tended to yield the highest recall and lowest precision results. Among the more balanced methods, no method consistently obtained a better balance than the other methods across all data sets, but Orthoinspector, InParanoid and PANTHER (LDO only) performed well overall. In terms of broad categories, there is no obvious systematic difference in performance between tree-based (Ensembl, PANTHER and PhylomeDB) and graph-based methods (the rest) or between methods relying on species tree (Ensembl, PANTHER, PhylomeDB, OMA GETHOGs, Hieranoid and EggNOG) and methods that do not. The latter point is perhaps unexpected, as one could expect knowledge of the species tree to provide an 'unfair' advantage in this particular benchmark. If there is any such effect, our results indicate that it is small.
Figure 2. The Generalized Species Tree Discordance test assesses the congruence of inferred orthologs with a trusted reference tree.

Benchmarking results are shown for eukaryotes. A trade-off between precision (measured in terms of tree error in the y-axis) and recall (measured in terms of completed tree samples in the x-axis; Online Methods) can be observed. Only high-confidence branches of the reference tree (L90, Online Methods), at least 10 myr long, are considered. Error bars indicate 95% confidence intervals and the line indicates the 'Pareto frontier'.
These trends persisted when we measured recall in terms of the number of inferred orthologs (Supplementary Fig. 2) or when we focused on other clades (Supplementary Figs. 3, 4, 5). Among vertebrates, the results were largely consistent, but we noted minor differences in the ranking of individual methods, with InParanoid Core yielding the highest precision and MetaPhOrs the highest recall (Supplementary Fig. 3). We also benchmarked the methods for their ability to recover ortholog relationships among 'universal' genes by applying the species discordance test on a tree spanning across archaea, bacteria and eukaryotes. Once again, there were slight variations in the precise ranking of methods, but the overall trends were very similar to what was observed for eukaryotes only (Supplementary Fig. 5). Finally, if we included (high-confidence) short branches as well, the average concordance of reconstructed trees substantially decreased, both because short branches tend to be harder to infer and because of potential incomplete lineage sorting around them; however, the relative position of the methods remained practically unchanged, which was a further indication of the robustness of the benchmark (Supplementary Fig. 6).
Reference gene trees
The second series of orthology benchmarks employs evolutionary relationships of gene pairs derived from annotated high-quality gene trees. Such reference trees are inferred through a careful combination of computational inference and expert curation: results obtained at each step of the tree inference pipeline (homolog identification, alignment, tree inference and gene–species tree reconciliation) are individually inspected, poor-quality sequences are excluded from the analysis and results are typically assessed using multiple models. This manual oversight is expected to yield gene phylogenies with high statistical support and topological consistency.
Concordance of orthology predictions was assessed with two sets of trees. The first was SwissTree16,33, a small collection of large- and high-confidence gene family phylogenies with different types of challenges for orthology prediction and species from all domains. The second, TreeFam-A34, consisted of a larger set of metazoan gene trees and thus covered a taxonomically restricted but wider range of protein families. Results obtained from the two benchmarks were quite similar (Fig. 3). On these benchmarks, virtually no trade-off between precision and recall appeared to be necessary. The best-performing methods were the ones that adopted a balanced precision–recall strategy, with MetaPhOrs doing particularly well. Methods with a more skewed precision–recall strategy (in particular, stringent OMA groups and permissive PANTHER (all)) fared poorly in comparison. This may be due in part to the nature of the reference gene tree data set, which focuses on gene families with a tractable evolutionary history. On ambiguous phylogenies, mistakes would become unavoidable and a skewed strategy could become preferable, depending on the application.
Figure 3. Benchmark results using sets of reference gene trees.

Evolutionary gene relationships are predicted for the QfO reference proteomes by 15 different methods. From the results, pairs of orthologous relationships are determined for each method and compared to those obtained from the reference gene trees of (a) SwissTree and (b) TreeFam-A. Error bars indicate 95% confidence intervals.
Functional benchmarks
The third series of benchmarks evaluated orthology in terms of functional similarity. Although orthology is an evolutionary and not a functional relationship, we chose to include functional benchmarks for two reasons. First, for similar levels of sequence divergence, orthologs have been shown to be moderately (but significantly) more conserved than paralogs in terms of Gene Ontology (GO) annotation similarity11. For a given evolutionary distance, more accurate orthology inference is thus likely to be correlated with more functionally similar gene pairs. Second, many users are interested in using orthologs to identify functionally conserved genes; for this purpose, functional benchmarks are directly relevant.
We assessed functional similarity based on experimentally backed annotations from the UniProt–Gene Ontology Annotation (GOA) database35 and Enzyme Commission (EC) numbers from the ENZYME database36. Though the two benchmarks consider different aspects of gene function, the results were largely consistent. In both cases, orthology inference methods showed a clear trade-off between precision (measured as the average Schlicker semantic similarity37 of functional annotations associated with orthologs) and recall (measured as the number of ortholog relationships predicted; Fig. 4). The only exception was with the EC number benchmark, where MetaPhOrs falls beneath the 'Pareto frontier' (the frontier defined by the methods that are not outperformed by any other method in both precision and recall). However, MetaPhOrs is also the only method with missing taxa, and the three missing taxa contain a substantial number of genes with EC annotations (827 in total). This lack of EC annotations has a negative effect on the recall.
Figure 4. Benchmarks of functional similarity between inferred orthologous gene pairs.

Two different types of functional annotations are used: (a) experimentally supported GO annotations and (b) Enzyme Commission (EC) numbers. Error bars indicate 95% confidence intervals.
Discussion
The Orthology Benchmark service overcomes many of the practical complications previously associated with orthology benchmarking. It enables systematic comparison of a new method with state-of-the-art approaches on to a wide range of benchmarks. It replaces current practice, which typically includes fewer methods, fewer tests and less empirical data.
By relying on a common set of data for all methods, the benchmark service ensures that the results obtained by different methods are directly comparable. Previous benchmarking efforts required painstaking and error-prone mapping of proteins between different sources, releases and choice of alternative splicing variants. In contrast, by relying on a common set of data for all methods, the benchmark service ensures that the results obtained by different methods are directly comparable. The only caveat is that, since proteomes vary in quality and analytical difficulty, the results on the benchmark data set may not entirely reflect the quality of the orthology assignments otherwise provided by each resource. The choice of species included in the QfO reference proteomes (Online Methods) requires a compromise between (i) increasing the number of proteomes to make the benchmark set more representative of current resources and (ii) keeping the number of proteomes low to facilitate and encourage new submissions to the benchmark.
Submissions performed on a subset of the proteomes are discouraged, as all missing predictions are counted as false negatives. This provides an incentive for submitters to analyze the entire reference proteome data set. We considered alternative ways of handling submissions on partial data, but these approaches had major flaws. For example, one alternative was to extrapolate scores obtained on the subset of proteomes considered in a particular submission to all data. However, this approach could introduce a bias in the analyses (e.g., some methods only predict orthologs for 'easy' pairs of proteomes). Another alternative was to restrict comparisons to the intersection of proteomes analyzed by all methods. However, this approach results in an excessive waste of information, as the intersection can only decrease with each additional method.
Overall, results obtained across multiple phylogenetic and functional tests corroborated previous observations that the main difference among the established orthology inference methods lies in the trade-off they produce in terms of precision and recall13,15,17. However, this trade-off was not present in the reference gene tree test, perhaps because sequences with ambiguous location are typically excluded from these hand-curated trees. On these reference trees, the meta-method MetaPhOrs performed particularly well. The analysis also confirmed that the widely used reciprocal best hit approach has a relatively high precision but a relatively low recall38,39. Other methods fill different niches, with OMA group and PANTHER (all) often lying at the two extremes of the precision–recall trade-off. Among the more balanced approaches, InParanoid, Hieranoid and OrthoInspector showed solid performance in most benchmarks.
The decision of whether to favor a skewed or a balanced approach to the precision–recall trade-off strongly depends on the application. For instance, hypothesis-generating analyses may favor a high recall, while phylogenomic species tree inference typically requires high precision. Because of this, we refrained from computing a combined score, which would necessarily entail a statement of preference with respect to this trade-off.
To be deemed competitive, a method should ideally reach or exceed the Pareto frontier in at least a subset of the benchmarks. If it does not, the benchmark service may help uncover bugs or deeper flaws. Analogous to unit testing in software engineering, benchmarking can also provide quality control for new releases of established resources. In the course of the present community benchmarking effort, over a hundred sets of predictions were submitted to the service. Many submitters did not make their results publicly available, presumably after discovering poor outcome in some of the benchmarks. This clearly demonstrates the effectiveness of the benchmark service for quality control.
The bane of benchmarking is circularity. Despite our best efforts, not all circularity could be avoided. Some methods used knowledge of the species tree in their inference; however, this potentially unfair advantage produced a negligible difference in performance for these methods. More generally, many methods were trained or fine-tuned using some of the benchmarks considered here. For instance, parameters of the meta-method MetaPhOrs were in part trained using TreeFam-A31. Similarly, the latest versions of InParanoid28 and PhylomeDB23 used the benchmark service for parameter fine-tuning. As for the functional benchmarks, although GO annotations derived from sequence comparisons were excluded, experiments are often guided by sequence similarity to proteins with known function. Thus, even when restricting analyses to experimentally backed GO annotations, we cannot avoid circularity entirely. However, because the benchmarks are collectively underpinned by a large amount of data from a broad range of species (tens of thousands of trees and hundreds of thousands of pairs of functional annotations), the risk of overfitting seems low, and this potential risk will be monitored by the QfO benchmarking working group. New benchmarks may be introduced over time to detect and discourage overfitting.
Presently, the benchmark service uses orthologous gene pairs as 'common denominators' among all the methods. However, many resources provide richer outputs—such as reconciled gene trees or hierarchical orthologous groups—and may indeed be optimized for these. The performance on pairwise data is thus not entirely representative of what the data offer. In the future, however, the benchmark service could be extended to evaluate these richer, more specific orthology formats as well. Similarly, the benchmark service could also be extended to take into account confidence scores or posterior probabilities, which are particularly relevant to likelihood-based orthology inference methods40,41.
Methods
Quest for orthologs reference proteomes and species tree.
The QfO consortium has defined a consensus data set of proteomes and common file formats6,7 to be used by diverse orthology inference methods, allowing for standardized benchmarks and aiding integration of multiple ortholog sources. The QfO Reference Proteomes data sets were created as a collection of data providing a representative protein for each gene in the genome of selected species. Such data sets have been generated annually from the UniProt Knowledgebase (UniProtKB) database42 for the past five years. To this end, a gene-centric pipeline has been developed and enhanced over these years by UniProt. The QfO Reference Proteomes are a manually compiled subset of the UniProt reference proteomes, comprising well-annotated model organisms and organisms of interest for biomedical research and phylogeny, with the intention to provide broad coverage of the tree of life. These complete, nonredundant reference proteomes are publicly available at ftp://ftp.ebi.ac.uk/pub/databases/reference_proteomes/QfO. The data sets are provided either in SeqXML20 format or as a collection of FASTA files.
The benchmarking effort reported here uses the reference proteomes data set released in 2011, which comprises 754,149 nonredundant protein sequences from 66 species (40 eukaryotes and 26 bacteria–archaea).
The reference species tree used in this study was produced by the QfO species tree working group, which surveyed the literature to establish a well-supported tree topology for the 66 species43 (Supplementary Fig. 1). The internal nodes of this reference species tree have assigned confidence levels based on the agreement among the resources surveyed (L90: congruent, significant branch support; L70: congruent; L50: one alternative species tree topology; L30: default level; L10: two or more alternative species tree topologies have been reported; for more detail, see Boeckmann et al.43). The latest version of the tree can be retrieved from http://swisstree.vital-it.ch/species_tree. To minimize the chance of including cases of incomplete lineage sorting in the species tree discordance benchmark, we estimated the evolutionary times of all internal branches using the timetree resource44 and collapsed branches that were shorter than 10 myr.
Orthology databases and methods.
EggNOG26 (http://eggnogdb.embl.de) is a database of Orthologous Groups (OGs) and functional annotation covering prokaryotic and eukaryotic species. Since version 4.1, the EggNOG method is also capable of producing fine-grained (for example, pairwise) orthology predictions based on the automated analysis of phylogenetic trees. For this study, the complete set of 66 reference proteomes was independently analyzed using the EggNOG pipeline, which involved 1) joining proteins into inparalogous groups from closely related species and 2) de novo reconstruction of 38,513 OGs by clustering the obtained inparalogous groups based on triangles of their reciprocal best hits45. Phylogenetic analysis and automated tree interpretation for each OG was subsequently performed using the workflow described in PhylomeDB22 as implemented in the ETE Toolkit v2.3 (ref. 46). The phylogenetic approach used included testing three aligners (MAFFT47 v6.861b, Muscle48 v3.8.31 and Clustal Omega49 v1.2.1) and five evolutionary models (LG, WAG, JTT, VT and MtREV); applying alignment consensus and soft trimming techniques (M-Coffee50 v10, trimAl51 v1.3); and using maximum likelihood tree inference (PhyML52 v3). This workflow is labeled as eggnog41 when using the ETE-build command and was applied in a per-OG basis. Pairwise orthology predictions were derived from each tree using the species overlap algorithm53 after rooting trees to midpoint. The predictions were submitted to the benchmark service in July 2015.
Ensembl Compara21 uses a gene–species tree reconciliation pipeline. The predictions were run using the code released in version 81 of the Ensembl (July 2015). However, Treebest (the software used to build phylogenetic trees) had to be adapted to accept alignments of protein sequences. Treebest makes a consensus out of trees built with various phylogenetic methods and some of them required nucleotide sequences, which were not provided in the QfO data set. The list of maximum-likelihood models and distance methods (used for neighbor joining) was thus updated to: WAG, JTT and Dayhoff instead of WAG and HKY (for maximum likelihood), and JTT, Kimura and mixed amino acid models instead of dN, dS and mixed nucleotide models (for neighbor joining). The predictions were submitted to the benchmark service in June 2015. An older submission based on version 66 of the Ensembl code (June 2011) is also present on the benchmark service.
Hieranoid27 performs pairwise orthology analysis using InParanoid at each node in a guide (species) tree as it progresses from its leaves to the root. This concept reduces the total runtime complexity from a quadratic to a linear function of the number of species. We ran Hieranoid 2.0. Hieranoid outputs ortholog groups structured as species trees with orthologs at all levels, hence there can be many outparalogs within an ortholog group. The trees were therefore parsed to extract ortholog pairs only at the last common ancestor of two species, for all species pairs. The predictions were submitted to the benchmark service in April 2015.
InParanoid28 is a graph-based algorithm that aims to generate orthologous groups that include all inparalogs but no outparalogs between species pairs. Version 4.1 of the algorithm was run with default parameters. Two variants were tested in this study: the regular InParanoid output containing all predicted pairs of orthologs (labeled InParanoid in the plots) and a high-confidence set including only orthologs with InParanoid's maximum confidence score of 1.0 (labeled Inparanoid (core)). The predictions were submitted to the benchmark service in June 2011.
MetaPhOrs31 (Meta Phylogeny-based Orthologs) is a repository of orthologs and paralogs that were computed using phylogenetic trees available in several databases or computed from graph-based orthologous groups. For each orthology–paralogy prediction, MetaPhOrs (http://orthology.phylomedb.org/) provides two reliability scores: Evidence Level (informing about number of repositories from which prediction is retrieved) and Consistency Score (defining overall agreement of source databases about given prediction). MetaPhOrs does not include predictions for the three reference genomes Streptomyces coelicolor, Thermotoga maritima and Pyrococcus kodakaraensis (strain KOD1). The predictions were submitted to the benchmark service in February 2013.
OMA29 (Pairs, Groups, HOGs) is a publicly available resource (http://omabrowser.org/) that provides orthology predictions among thousands of proteomes from all domains of life. OMA uses evolutionary distance estimates from Smith–Waterman alignments to infer orthologs. A distinct feature among graph-based methods is the witness of nonorthology step in its pipeline, where cases of differential gene losses get detected. OMA provides three different groupings of orthologs: (i) the raw pairwise ortholog relationships form the OMA Pairs, a gene-centric view that lists all the orthologs for a given gene. (ii) OMA Groups, a very stringent type of grouping where all member proteins are orthologous to one another within a group. OMA Groups have been designed mainly for species tree inference purposes, as gene trees built from them should be congruent with the species tree. (iii) Lastly, we constructed hierarchical orthologous groups (OMA HOGs). These are nested groups that contain genes that descend from a single common ancestral gene within a given taxonomic range using the GETHOGs algorithm54. The predictions were submitted to the benchmark service in June 2011 (OMA pairs and groups) and in March 2013 (OMA HOGs).
OrthoInspector30 is a database of precomputed orthology and inparalogy relationships and a stand-alone package allowing large-scale predictions of orthology between thousands of proteomes (http://lbgi.fr/orthoinspector/). The resource has recently undergone a major new release, with improved speed and visualisation tools, but the inference algorithm is unchanged from the initial graph-based method described in Linard et al.55. The predictions were submitted to the benchmark service in June 2011.
PANTHER 8.022 is based on version 8.0 of the PANTHER database (http://pantherdb.org), released in 2012 (the current version is 10.0, released in 2015). Family membership of each sequence is based on HMM scoring to the PANTHER 'library' of HMMs (at both the family and subfamily levels). Sequences were aligned with MAFFT56 and the resulting alignment was used to construct phylogenetic trees with the GIGA program57. GIGA (version 1.1 was used for PANTHER version 8.0) uses a species tree to guide tree construction, and all nodes in the tree are labeled as speciation or gene duplication events; these labeled nodes are used to infer orthologs (pairs of genes with a speciation event as their common ancestor). PANTHER predicts two types of orthologs: least-diverged orthologs (LDO) and other orthologs (O). LDO pairs can be simplistically thought of as 'the same gene' in two different species. Formally, the two genes created by each gene duplication event in the tree are treated asymmetrically: the least diverged duplicate (the one with the shortest branch immediately following the duplication) remains in the same LDO group as its ancestor, while the other duplicate founds a new LDO group. The benchmarking was performed on either LDO only, or all orthologs (including both LDO and O). The predictions were submitted to the benchmark service in February 2013.
PhylomeDB23 (http://phylomedb.org/) is a publicly available repository of phylomes, i.e., the complete collection of phylogenies for all genes of a given species in a predefined evolutionary context. PhylomeDB is unique among other repositories in that it follows an approach that is both gene centric and genome wide. PhylomeDB uses its phylogenetic trees to infer orthology and paralogy relationships. For the Quest for Orthologs project, 42 phylomes were reconstructed using different combinations of the 66 species in the benchmark. A total of 458,108 phylogenetic trees were generated, which were later combined to provide orthology predictions for all proteins included in the benchmark. Briefly, each tree was scanned and only the partition of up to 30 sequences, including the seed protein, was kept. Then, evolutionary relationships were computed for those protein sequences based on a species overlap approach. Redundant predictions across the 42 phylomes were unified using the Consistency Score (CS) as implemented in MetaPhOrs (see above). Only those predictions having a Consistency Score greater or equal to 0.5 across the whole data set were called orthologs. The predictions were submitted to the benchmark service in June 2013.
RBH24 (Reciprocal best hit) is a classic method consisting of identifying the pairs of genes with mutually highest alignment score between every pair of species. Here, we use reciprocal blastp hits as orthologs, with minimum E-value of 1e–2, and we keep all hits that are ≥99% of the highest score. The predictions were submitted to the benchmark service in January 2016.
RSD25 infers orthology relationships by finding pairs of genes whose nearest gene, computed using PAML, is the other gene in the pair. Candidates genes are also filtered using BLAST E-value and multiple-sequence alignment divergence thresholds. This method is implemented in the database RoundUp58, a large-scale orthology database developed by the Wall Lab. The database is no longer maintained, but the source code is still available at https://github.com/todddeluca/reciprocal_smallest_distance/. To identify orthologs, we ran the algorithm with divergence and E-value cutoffs of 0.8 and 1e–5, respectively. The predictions were submitted to the benchmark service in February 2012.
Benchmarks.
Generalized species tree discordance. The idea behind the species tree discordance test is simple. Two genes are orthologous if they started diverging through a speciation event. Therefore, if we sample putative orthologous genes such that all resulting genes are related through speciation events, the resulting tree should be congruent with the species tree. Previously, we presented a sampling strategy for fully imbalanced tree topologies15. Here, we extend this idea to arbitrary reference trees, including those with soft polytomies (unresolved nodes).
The following procedure is repeated a large number of times. We start with a random gene in a random genome. We then attempt to sample a maximal path along the tree by selecting an orthologous gene in the 'next' species in the tour from the list of reported orthologs (Supplementary Fig. 7a). If there are multiple possibilities in the choice of the 'next' species due to soft polytomies, or in the choice of the orthologous counterparts due to one-to-many or many-to-many orthology, a choice is made at random. If there is no predicted ortholog at any step along the path, the sample is deemed unsuccessful. Alternatively, if at least one orthologous counterpart is predicted at each step, this results in a set of n sequences. Assuming that i) the reference tree is correct, ii) the retrieved orthologs are all correct and iii) all within-species variation is fixed (i.e., no incomplete lineage sorting), it is easy to prove that the unrooted evolutionary tree relating these sequences should only contain speciation nodes and should therefore be congruent with the reference species tree.
Proof: The n sequences sampled through the circular tour are sampled by starting from a random sequence and retrieving n − 1 pairs of orthologs. By construction, these n − 1 pairs of orthologs belong to pairs of species that have distinct last common ancestors and thus coalesce in different speciation nodes in the phylogenetic tree of these sequences. Therefore, that tree contains at least n − 1 distinct speciation nodes. However, the rooted, fully-resolved evolutionary tree of n species has exactly n − 1 internal nodes. Thus, all the internal nodes of the gene tree are speciation nodes. Since we assume that there is no incomplete lineage sorting, as long as the input orthologs are correct, the tree relating these sequences should be congruent with the species tree.
A least-squares distance tree is reconstructed for each set of putative orthologous sequences. After aligning the sequences with MAFFT47, maximum likelihood distances and their variances (using the inverse Fisher information) are estimated using the EstimatePam() function in the Darwin programming environment59 for each pair of sequences. Next, the gene tree is estimated using Darwin's MinSquareTree() function, which is a fast implementation of the weighted least-squares trees60 constrained to non-negative branch lengths61. We have previously shown that orthology benchmarking results obtained with such distance trees are consistent with more computationally demanding Maximum likelihood trees15. The Robinson–Foulds32 distance between this gene tree and the reference tree measures the false discovery rate, while the total number of trees is used as a proxy of recall. Due to the stochastic nature of the algorithm, repeated runs of the benchmark may lead to slightly (albeit nonsignificantly) different results.
Reference gene trees. Reference gene trees labeled with speciation and duplication events were downloaded from SwissTree on March 23, 2015 (http://swisstree.vital-it.ch/) and Treefam-A version 7 (http://www.treefam.org/). As sequences analyzed in these two resources can differ from those of the QfO reference proteomes, sequences were mapped based on gene identifiers or sequence identifiers. After mapping, for each family the n(n − 1)/2 induced pairwise evolutionary relationships were extracted and compared with the orthologous predictions from each orthology prediction method as follows. Let G = {gi} be the set of all genes in the reference gene tree and RO = {(gi, gj) | gi ∈ G, gj ∈ G, gi ≠ gj, label(gi,gj) = speciation} the set of true orthologs according to the reference tree. Likewise, let RP be the set of nonorthologous relations in that family and P = {(gi, gj)}, be the set of all predictions made by the orthology method. With PF = {(gi, gj) | (gi, gj) ∈ P ∩ gi ∈ G ∩ gj ∈ G}, we denote the set of orthologs where both members are part of the reference gene family. Now, the true/false positives/negatives are simply TP = PF ∩ RO, FP = PF ∩ RP, FN = RO − PF and TN = RP − PF. From these values we can compute positive predictive values (PPV) and true positive rate (TPR): PPV = |TP|/(|TP| + |FP|), TPR = |TP|/(|TP| + |FN|).
We can further estimate the uncertainties of these rates by treating them as binomially distributed random variables, for example, σ2(PPV) = PPV(1 − PPV)/(|TP| + |FP|). Finally, we combine all the families by building averages of the rates. As an example, for the positive predictive value this results in,
Functional tests.We downloaded the Gene Ontology annotations62 for all the genes in the reference genomes from the November 2014 release of UniProt-GOA35 and excluded any annotation with a 'NOT' qualifier from this set. For the analysis shown here, we only use annotations with experimental evidence codes (EXP, IPI, IDA, IMP, IGI and IEP). Likewise, we collected the hierarchical EC number assignments of the ENZYME database36 maintained by Swiss-Prot. The computation of the functional similarities between gene pairs is done in the same way for both types of data, using the approach of Schlicker et al.37: the semantic similarity between annotations sim(i,j) is measured using Lin's metric63; between any two genes, the most similar pairs of annotations are identified and averaged, i.e.,
where pi is the set of function annotations associated with protein i.
Code availability.
The source code is available under an open source license (Mozilla Public License Version 2.0) at https://github.com/qfo/benchmark-webservice.
Supplementary Information
Integrated supplementary information
Internal branches are coloured according to their confidence levels: L90+≥10myr: green, L90+<10myr: purple, L70: blue, L50: orange, L30: pink and L10: red. For the species tree discordance test we collapse all branches below L90 or having shorter branches as 10myr based on estimates from the timetree resource44. The tree can be retrieved in electronic format from http://swisstree.vital-it.ch/species_tree.
Same as main Figure 2, but with number of orthologous relationships on the x-axis instead of number of complete tree samplings: the Generalised Species Tree Discordance test assesses the congruence of inferred orthologs with a trusted reference tree, focusing on eukaryotes. A trade-off between precision (measured in terms of tree error in the y-axis) and recall (measured in number of orthologous relationships in the x-axis) can be observed. Only high-confidence branches of the reference tree (L90, see Methods), at least 10 myr long, are considered. Results for other clades are provided in Supplementary Figure 3, 4, 5. Error bars indicate 95% confidence intervals.
Same as main Figure 2, but covering vertebrate genomes only.
Same as Figure 2, but covering fungal genomes only.
Same as Figure 2, but covering all genomes in the reference genome dataset, including prokaryotes.
Generalised Species Tree Discordance test on eukaryotes from Figure 2, but also including short (L90+) branches—and therefore including potentially ILS (left-hand figure). The right hand figure depicts Figure 2 for comparison. The relative position of the methods does not substantially change, but the average discordance (y-axis) is considerably higher when including the short branches—due to a combination of stochastic error in the reconstruction of the gene trees and potential incomplete lineage sorting around short branches.
Generalised species discordance test: (a) illustration and (b) pseudocode of the ortholog sampling strategy, with two potential paths along two possible bifurcating trees (orange and blue dashed lines).
Supplementary information
Supplementary Figures 1–7 (PDF 4244 kb)
Orthology benchmark service. (ZIP 405 kb)
Acknowledgements
This work was supported by Swiss National Science Foundation grant PP00P3_150654 (to C.D.), UK Biotechnology and Biological Sciences Research Council grant BB/L018241/1 (to C.D.), Spanish Ministry of Economy and Competitiveness grant BIO2012-37161 (to T.G.), Qatar National Research Fund NPRP 5-298-3-086 (to T.G.), European Research Council grant ERC-2012-StG-310325 (to T.G.), National Institutes of Health (NIH) grant R24 OD011883 (to S.E.L.), U41 HG002273 (to S.E.L. and P.D.T.), U41 HG007822 (to M.J.M. and I.X.), Swiss State Secretariat for Education, Research and Innovation (SERI) funding (to I.X. and C.D.), US National Science Foundation EAGER Award #1355632 (to K.S.) and ANR project BIP-BIP ANR-10-BINF-03-02 (to O.L.). Furthermore, A.S.d.S., J.H.-C., M.J.M., M.M. and P.B. acknowledge support from the European Molecular Biology Laboratory, M.M. acknowledges support from the Wellcome Trust (WT095908), S.E.L. acknowledges support from Lawrence Berkeley National Laboratory core funds (Office of Basic Energy Sciences and US Department of Energy Contract No. DE-AC02-05CH11231), L.J.J. acknowledges support from the Novo Nordisk Foundation (Grant No. NNF14CC0001) and L.P.P. acknowledges support from the La Caixa–CRG International Fellowship Program.
Source data
Author Contributions
A.M.A. and C.D. conceived the project, with contributions from the rest of the QfO benchmarking working group (A.S.d.S., B.B., C.P., E.S., F.S., K.F., K.S., J.H.-C., M.J.M., M.M., P.D.T., S.E.L. and T.G.); A.S.d.S. and M.J.M. contributed the reference proteomes; A.M.A., B.B., F.S. and K.F. contributed benchmarks; A.M.A., B.L., D.S., E.S., K.F., J.H.-C., L.P.P., M.M., S.C.-G., P.D.T., T.D. and T.G. contributed predictions; P.B., O.L., C.v.M., I.X. and L.J.J. supervised contributions of benchmarks or predictions. A.M.A. and C.-M.T. designed and implemented the benchmark service; D.A.D. and A.M.A. assessed the impact of incomplete lineage sorting on orthology inference; A.M.A. and J.H.-C. generated plots; A.M.A., B.B., C.D., E.S., F.S., K.F., K.S., M.M., P.D.T., S.E.L. and T.G. analyzed the results; A.M.A., B.B., C.D., E.S. and L.J.J. wrote the manuscript, with feedback from all other coauthors; C.D. coordinated the project.
Competing interests
The authors declare no competing financial interests.
Footnotes
Christophe Dessimoz: A list of authors who are consortium members is provided at the end of the manuscript.
Contributor Information
Christophe Dessimoz, Email: christophe.dessimoz@unil.ch.
Quest for Orthologs consortium:
Adrian M Altenhoff, Brigitte Boeckmann, Salvador Capella-Gutierrez, Todd DeLuca, Kristoffer Forslund, Jaime Huerta-Cepas, Benjamin Linard, Cécile Pereira, Leszek P Pryszcz, Fabian Schreiber, Alan Sousa da Silva, Damian Szklarczyk, Clément-Marie Train, Odile Lecompte, Ioannis Xenarios, Kimmen Sjölander, Maria J Martin, Matthieu Muffato, Toni Gabaldón, Suzanna E Lewis, Paul D Thomas, Erik Sonnhammer, and Christophe Dessimoz
References
- 1.Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. doi: 10.2307/2412448. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 3.Gabaldón T, Koonin EV. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dessimoz C. Editorial: orthology and applications. Brief. Bioinform. 2011;12:375–376. doi: 10.1093/bib/bbr057. [DOI] [PubMed] [Google Scholar]
- 5.Altenhoff, A.M. & Dessimoz, C. in Evolutionary Genomics (ed. Anisimova, M.) Ch. 9 (Humana Press, 2012).
- 6.Gabaldón T, et al. Joining forces in the quest for orthologs. Genome Biol. 2009;10:403. doi: 10.1186/gb-2009-10-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dessimoz C, et al. Toward community standards in the quest for orthologs. Bioinformatics. 2012;28:900–904. doi: 10.1093/bioinformatics/bts050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonnhammer ELL, et al. Big data and other challenges in the quest for orthologs. Bioinformatics. 2014;30:2993–2998. doi: 10.1093/bioinformatics/btu492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nehrt NL, Clark WT, Radivojac P, Hahn MW. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput. Biol. 2011;7:e1002073. doi: 10.1371/journal.pcbi.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas PD, Wood V, Mungall CJ, Lewis SE, Blake JA. On the use of gene ontology annotations to assess functional similarity among orthologs and paralogs: a short report. PLoS Comput. Biol. 2012;8:e1002386. doi: 10.1371/journal.pcbi.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altenhoff AM, Studer RA, Robinson-Rechavi M, Dessimoz C. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput. Biol. 2012;8:e1002514. doi: 10.1371/journal.pcbi.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Zhang J. The ortholog conjecture is untestable by the current gene ontology but is supported by RNA sequencing data. PLoS Comput. Biol. 2012;8:e1002784. doi: 10.1371/journal.pcbi.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulsen T, Huynen MA, de Vlieg J, Groenen PM. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006;7:R31. doi: 10.1186/gb-2006-7-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeckmann B, Robinson-Rechavi M, Xenarios I, Dessimoz C. Conceptual framework and pilot study to benchmark phylogenomic databases based on reference gene trees. Brief. Bioinform. 2011;12:423–435. doi: 10.1093/bib/bbr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trachana K, et al. Orthology prediction methods: a quality assessment using curated protein families. Bioessays. 2011;33:769–780. doi: 10.1002/bies.201100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salichos L, Rokas A. Evaluating ortholog prediction algorithms in a yeast model clade. PLoS ONE. 2011;6:e18755. doi: 10.1371/journal.pone.0018755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalquen DA, Altenhoff AM, Gonnet GH, Dessimoz C. The impact of gene duplication, insertion, deletion, lateral gene transfer and sequencing error on orthology inference: a simulation study. PLoS ONE. 2013;8:e56925. doi: 10.1371/journal.pone.0056925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt T, Messina DN, Schreiber F, Sonnhammer ELL. SeqXML and OrthoXML: standards for sequence and orthology information. Brief. Bioinform. 2011;12:485–488. doi: 10.1093/bib/bbr025. [DOI] [PubMed] [Google Scholar]
- 21.Vilella AJ, et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 2014;42:D897–D902. doi: 10.1093/nar/gkt1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- 26.Powell S, et al. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber F, Sonnhammer ELL. Hieranoid: hierarchical orthology inference. J. Mol. Biol. 2013;425:2072–2081. doi: 10.1016/j.jmb.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Sonnhammer ELL, Östlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43:D234–D239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altenhoff AM, et al. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;43:D240–D249. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linard B, et al. OrthoInspector 2.0: software and database updates. Bioinformatics. 2015;31:447–448. doi: 10.1093/bioinformatics/btu642. [DOI] [PubMed] [Google Scholar]
- 31.Pryszcz LP, Huerta-Cepas J, Gabaldón T. MetaPhOrs: orthology and paralogy predictions from multiple phylogenetic evidence using a consistency-based confidence score. Nucleic Acids Res. 2011;39:e32. doi: 10.1093/nar/gkq953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson DF, Foulds LR. Comparison of phylogenetic trees. Math. Biosci. 1981;53:131–147. doi: 10.1016/0025-5564(81)90043-2. [DOI] [Google Scholar]
- 33.Zhang X, Krause K-H, Xenarios I, Soldati T, Boeckmann B. Evolution of the ferric reductase domain (FRD) superfamily: modularity, functional diversification, and signature motifs. PLoS ONE. 2013;8:e58126. doi: 10.1371/journal.pone.0058126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiber F, Patricio M, Muffato M, Pignatelli M, Bateman A. TreeFam v9: a new website, more species and orthology-on-the-fly. Nucleic Acids Res. 2014;42:D922–D925. doi: 10.1093/nar/gkt1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimmer EC, et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2012;40:D565–D570. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlicker A, Domingues FS, Rahnenführer J, Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf YI, Koonin EV. A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes. Genome Biol. Evol. 2012;4:1286–1294. doi: 10.1093/gbe/evs100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dalquen DA, Dessimoz C. Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals. Genome Biol. Evol. 2013;5:1800–1806. doi: 10.1093/gbe/evt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sennblad B, Lagergren J. Probabilistic orthology analysis. Syst. Biol. 2009;58:411–424. doi: 10.1093/sysbio/syp046. [DOI] [PubMed] [Google Scholar]
- 41.Akerborg O, Sennblad B, Arvestad L, Lagergren J. Simultaneous Bayesian gene tree reconstruction and reconciliation analysis. Proc. Natl. Acad. Sci. USA. 2009;106:5714–5719. doi: 10.1073/pnas.0806251106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res.40, D71–D75 (2012). [DOI] [PMC free article] [PubMed]
- 43.Boeckmann B, et al. Quest for Orthologs (QfO) entails Quest for Tree of Life (QfToL): in search of the gene stream. Genome Biol. Evol. 2015;7:1988–1999. doi: 10.1093/gbe/evv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen LJ, et al. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huerta-Cepas J, Dopazo J, Gabaldón T. ETE: a python environment for tree exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 48.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 53.Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldón T. The human phylome. Genome Biol. 2007;8:R109. doi: 10.1186/gb-2007-8-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altenhoff AM, Gil M, Gonnet GH, Dessimoz C. Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS ONE. 2013;8:e53786. doi: 10.1371/journal.pone.0053786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Linard B, Thompson JD, Poch O, Lecompte O. OrthoInspector: comprehensive orthology analysis and visual exploration. BMC Bioinformatics. 2011;12:11. doi: 10.1186/1471-2105-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas PD. GIGA: a simple, efficient algorithm for gene tree inference in the genomic age. BMC Bioinformatics. 2010;11:312. doi: 10.1186/1471-2105-11-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeLuca TF, Cui J, Jung J-Y, St Gabriel KC, Wall DP. Roundup 2.0: enabling comparative genomics for over 1800 genomes. Bioinformatics. 2012;28:715–716. doi: 10.1093/bioinformatics/bts006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonnet GH, Hallett MT, Korostensky C, Bernardin L. Darwin v. 2.0: an interpreted computer language for the biosciences. Bioinformatics. 2000;16:101–103. doi: 10.1093/bioinformatics/16.2.101. [DOI] [PubMed] [Google Scholar]
- 60.Wikipedia contributors. Least squares inference in phylogeny. Wikipedia, The Free Encyclopediahttps://en.wikipedia.org/w/index.php?title=Least_squares_inference_in_phylogeny&oldid=552325441 (2013) (last accessed 7 March 2016).
- 61.Felsenstein, J. Inferring Phylogenies (Palgrave Macmillan, 2004).
- 62.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res.43, D1049–D1056 (2015). [DOI] [PMC free article] [PubMed]
- 63.Lin, D. An information-theoretic definition of similarity. in Proceedings of the 15th International Conference on Machine Learning (ed. Shavlik, J.W.) 296–304 (Morgan Kaufmann, San Francisco, California, USA, 1998).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Internal branches are coloured according to their confidence levels: L90+≥10myr: green, L90+<10myr: purple, L70: blue, L50: orange, L30: pink and L10: red. For the species tree discordance test we collapse all branches below L90 or having shorter branches as 10myr based on estimates from the timetree resource44. The tree can be retrieved in electronic format from http://swisstree.vital-it.ch/species_tree.
Same as main Figure 2, but with number of orthologous relationships on the x-axis instead of number of complete tree samplings: the Generalised Species Tree Discordance test assesses the congruence of inferred orthologs with a trusted reference tree, focusing on eukaryotes. A trade-off between precision (measured in terms of tree error in the y-axis) and recall (measured in number of orthologous relationships in the x-axis) can be observed. Only high-confidence branches of the reference tree (L90, see Methods), at least 10 myr long, are considered. Results for other clades are provided in Supplementary Figure 3, 4, 5. Error bars indicate 95% confidence intervals.
Same as main Figure 2, but covering vertebrate genomes only.
Same as Figure 2, but covering fungal genomes only.
Same as Figure 2, but covering all genomes in the reference genome dataset, including prokaryotes.
Generalised Species Tree Discordance test on eukaryotes from Figure 2, but also including short (L90+) branches—and therefore including potentially ILS (left-hand figure). The right hand figure depicts Figure 2 for comparison. The relative position of the methods does not substantially change, but the average discordance (y-axis) is considerably higher when including the short branches—due to a combination of stochastic error in the reconstruction of the gene trees and potential incomplete lineage sorting around short branches.
Generalised species discordance test: (a) illustration and (b) pseudocode of the ortholog sampling strategy, with two potential paths along two possible bifurcating trees (orange and blue dashed lines).
Supplementary Figures 1–7 (PDF 4244 kb)
Orthology benchmark service. (ZIP 405 kb)


