Abstract
Huntington disease (HD) is a fatal progressive neurodegenerative disorder for which only symptomatic treatment is available. A better understanding of the pathology, and identification of biomarkers will facilitate the development of disease-modifying treatments. HD is potentially a good model of a neurodegenerative disease for development of biomarkers because it is an autosomal-dominant disease with complete penetrance, caused by a single gene mutation, in which the neurodegenerative process can be assessed many years before onset of signs and symptoms of manifest disease. Previous MRI studies have detected abnormalities in gray and white matter starting in premanifest stages. However, the understanding of how these abnormalities are related, both in time and space, is still incomplete. In this study, we combined deep gray matter shape diffeomorphometry and white matter DTI analysis in order to provide a better mapping of pathology in the deep gray matter and subcortical white matter in premanifest HD. We used 296 MRI scans from the PREDICT-HD database. Atrophy in the deep gray matter, thalamus, hippocampus, and nucleus accumbens was analyzed by surface based morphometry, and while white matter abnormalities were analyzed in (i) regions of interest surrounding these structures, using (ii) tractography-based analysis, and using (iii) whole brain atlas-based analysis. We detected atrophy in the deep gray matter, particularly in putamen, from early premanifest stages. The atrophy was greater both in extent and effect size in cases with longer exposure to the effects of the CAG expansion mutation (as assessed by greater CAP-scores), and preceded detectible abnormalities in the white matter. Near the predicted onset of manifest HD, the MD increase was widespread, with highest indices in the deep and posterior white matter. This type of in-vivo macroscopic mapping of HD brain abnormalities can potentially indicate when and where therapeutics could be targeted to delay the onset or slow the disease progression.
Keywords: Huntington, MRI, Shape, Diffeomorphometry, Atrophy, DTI
Highlights
-
•
Tracing temporal and spatial progression of HD is essential to design therapeutics.
-
•
We analyze subcortical gray matter shape and DTI in 296 premanifest HD individuals.
-
•
Changes in the shape of deep gray matter occur since the very early premanifest stages.
-
•
The in-vivo mapping of brain differences potentially indicate when and where therapeutics could be targeted.
1. Introduction
Huntington's disease (HD) is a devastating progressive neurodegenerative disorder that affects multiple domains, including motor, cognitive, and emotional, leading to incapacity for activities of daily living and, eventually, to death (Huntington, 1872, Folstein, 1991, Ross et al., 2014). It is caused by CAG repeat expansion in the gene Huntingtin (HTT), leading to polyglutamine expansion in the protein huntingtin (The Huntington's Disease Collaborative Research Group, 1993). Starting at the threshold of 36 CAGs, longer expansions lead to earlier ages of motor onset of HD. Diagnosis of “motor onset” of “manifest HD” is currently made in someone at risk, or tested genetically positive for the CAG expansion, on the basis of the clinical history and standardized Unified HD Rating Scale (UHDRS) motor exam yielding a clinical impression with “99% confidence” of the presence of HD in the context of the unequivocal presence of an otherwise unexplained extrapyramidal movement disorder (The Huntington's Disease Collaborative Research Group 1993, The Huntington's Disease Group 1996, Dorsey et al., 2013, Reilmann et al., 2014, Ross et al., 2014, Oster et al., 2015).
The course of HD can be divided into ‘premanifest’ and ‘manifest’ periods (Zhang et al., 2011, Dorsey et al., 2013, Tabrizi et al., 2013, Paulsen et al., 2014a, Paulsen et al., 2014b, Ross et al., 2014). The premanifest period can be further subdivided (Reilmann et al., 2014, Ross et al., 2014), and we use terminology from these references. First is a period when individuals are not distinguishable clinically (either by subjective symptoms or objective findings on exam) from controls (“presymptomatic”), usually up to 10–15 years before “motor onset.” Individuals may then enter the “prodromal” period, which is characterized by subtle motor, cognitive and behavioral changes, but not sufficient for a diagnosis of motor onset. Once motor and cognitive signs and symptoms begin, they progress inexorably over the course of the illness, which—with the exception of late-onset cases, who may die of other causes—is uniformly fatal (Ross et al., 1997, Dorsey et al., 2013).
A useful index of the length and severity of the individual's exposure to the effects of the mutant HTT gene is achieved by calculating a score based on the individual's age multiplied by (CAG – L), where L is a constant near the threshold of CAG repeat expansions for disease (Penney et al., 1997). This is termed the CAG-Age-Product (or CAP score). This score is useful for comparing data from cohorts of patients with a range of ages and CAG repeat lengths (Zhang et al., 2011, Ross et al., 2014).
There is growing consensus that intervention and treatment in HD should occur at the earliest stage possible. The development and testing of neuroprotective treatments will be facilitated by the identification of biomarkers in the manifest HD period and, optimally, premanifest stages. Neuroprotective treatments could include administration of gene silencing or other reagents directly into localized regions of the brain (Huntington's Disease Collaborative Research Group et al., 20, Kay et al., 2014, Ross et al., 2014). Because of the inverse relationship between the length of the CAG repeat expansion and age of onset, HD is a model disease that may provide proof of concept for neurodegenerative disease therapeutic modification. It also involves increasingly well-understood natural history, as demonstrated by large multicenter studies such as TRACK-HD (Tabrizi et al., 2009, Tabrizi et al., 2012, Tabrizi et al., 2013) and PREDICT-HD (Paulsen et al., 2006, Paulsen et al., 2008, Paulsen et al., 2014a, Paulsen et al., 2014b). These studies have identified white abnormalities in pre-manifest stage, in addition to the well-known atrophy of striatum and other subcortical gray matter structures.
Structural imaging from PREDICT, TRACK-HD, and other studies revealed striatal atrophy starting as early as 15 years before the onset, and continuing throughout the pre-manifest periods (Aylward et al., 1997, Aylward et al., 2000, Aylward et al., 2003, Aylward et al., 2011, Aylward et al., 2012, Paulsen et al., 2010, Paulsen et al., 2014a, Paulsen et al., 2014b, Tabrizi et al., 2012, Tabrizi et al., 2013). Extent of striatal atrophy adds predictive power for motor onset beyond age and CAG repeat length alone (Aylward et al., 2012, Aylward et al., 2013, Paulsen et al., 2014a, Paulsen et al., 2014b). In addition, the measurement of shape has enabled the delineation of regional atrophy in the striatum and other subcortical structures in the premanifest period (Younes et al., 2014a, Younes et al., 2014b). In parallel, diffusion tensor imaging (DTI) has revealed abnormalities in architecture and integrity in white matter and subcortical gray matter structures in both premanifest (Reading et al., 2005, Rosas et al., 2006, Kloppel et al., 2008, Stoffers et al., 2010) and manifest phases (Douaud et al., 2009, Vandenberghe et al., 2009, Della Nave et al., 2010, Bohanna et al., 2011, Delmaire et al., 2013, Sanchez-Castaneda et al., 2013).
In this study, we combine deep gray matter shape diffeomorphometry and white matter DTI analysis as means of better defining the topography of deep gray and white matter abnormalities in the premainifest stages of HD. Diffeomorphometry and geodesic shape analysis in computational anatomy (Van Camp et al., 2012, Miller et al., 2013, Miller et al., 2014, Younes et al., 2014a, Younes et al., 2014b) provide information about which structure subregions are affected. The association of this metric with DTI parameters in white matter regions through the pre-manifest course may provide additional details about the mapping of brain abnormalities. This could provide information about when, where, and how therapeutics could be administered in order to delay the onset or slow the progression of HD.
2. Methods
2.1. Participants
The data used here are from the PREDICT-HD study, in which subjects at risk for HD, but without motor signs and symptoms sufficient for diagnosis of manifest HD, had previously undergone elective predictive genetic testing. Premanifest and prodromal subjects are those that are found to be gene-expanded (CAG length greater than or equal to 36) but have not yet met traditional motor criteria for a diagnosis of HD. Diagnosis of HD is based on a Diagnostic Confidence Level (DCL) rating of “4” according to the motor assessment section of the Unified HD Rating Scale (UHDRS). Those that were found to negative for the CAG expansion (CAG at or below 30) were enrolled as comparison subjects. There were no subjects in with CAG between 30 and 36. Participants were recruited from 33 sites across the United States, Canada, Europe, and Australia, and had annual study visits consisting of a neurological motor examination, cognitive assessment, brain MRI (biennial), and psychiatric and functional assessment, with blood samples for genetic and biochemical analyses. Informed written consent was obtained from all subjects before participating in this study.
Subjects with CAG expansion were divided into three subgroups: low-CAP, medium-CAP, and high-CAP. Low-CAP individuals are presumably far from the symptomatic onset while the high-CAP individuals are the closest to the onset. The CAP score was computed as CAP = (age at entry) × (CAG − 33.66) (Zhang et al., 2011) and presumably indexes the cumulative exposure to the effects of mutant huntingtin. Cutoffs for groups were CAP < 290 (Low), 290 ≤ CAP ≤ 368 (Medium), and CAP > 368 (High).
2.2. MRI
High-resolution structural 3T MRI scans (T1-weighted inversion recovery turboflash (MPRAGE), were used for the analyses of diffeomorphometry. Due to the multicentre and longitudinal nature of the PREDICT study, the image acquisition was heterogeneous, represented by multiple vendors (GE, Phillips, and Siemens and over 20 slightly different MR acquisition protocols (i.e., due to transmission and receive hardware). The main parameters of the data included in this study varied as follows: TR: 8–8.4, TE: 3.5–3.8, TI: 826–843, matrix: 256 × 256, voxel size: 1 × 1, slice thickness: 0.9–1.2. Similar data have been successfully used in previous publications authored by the Huntington's Study Group (HSG) PREDICT investigators (Paulsen et al., 2010, Paulsen et al., 2014a, Paulsen et al., 2014b, Muralidharan et al., 2014, Kim et al., 2015).
The DTIs were obtained in 3T scanners, B0 ≅ 1000, in axial orientation (matrix = 128 × 128, FOV = 256 × 256, slice thickness = 2 mm). The number of slices, gradients, and repetitions varied according to the center/scanner: 1) Phillips: 33 gradients, 70 slices, 2 repetitions; 2) Siemens (most common): 31 gradients, 70 slices, 2 repetitions, 3) Siemens (minority): 79 gradients, 50 slices, 3 repetitions. The repetitions were concatenated within a single section using DTIPrep (http://www.nitrc.org/projects/dtiprep/) (Oguz et al., 2014), followed by quality control inspection, and then converted to nifti format using DWIConvert. Details about the tensor calculation and quality control are fully described in our previous publications (Magnotta et al., 2012); similar data was used in (Matsui et al., 2015). The six elements of the diffusion tensor, the fractional anisotropy (FA), and the mean diffusivity (MD) were calculated using DTIStudio software (Laboratory of Brain Anatomical MRI and Center for Imaging Science at Johns Hopkins University), and visual quality control was performed to ensure that the values for scalar images were in proper range and that gross anatomical FA values were appropriate. Our previous publications showed that scalar measures such as FA and MD are stable in face of different gradient number and directions, minimizing the effect of different protocols in multicenter studies (Magnotta et al., 2012).
We used scans of 296 individuals enrolled in the PREDICT-HD study. When more than one session was available for an individual, the first visit was used. Table 1 summarizes the demographic information. Twenty-three DTI scans were not included because the original files were missing or incomplete, or because of rejections at the first step of the quality control, in the tensor and scalar calculations. For the shape diffeomorphometry, the sample size for each group varied by as many as five individuals in the comparison for each substructure due to the quality of the resulting triangulated mesh associated to each substructural segmentation forming the shape statistics. The demographic profile of each group was not significantly different among the comparison of shape in different structures and of the DTI.
Table 1.
Demographic information and sample size used in each comparison.
| Controls | Low-CAP | Medium-CAP | High-CAP | Total | ||
|---|---|---|---|---|---|---|
| Total | 84 | 58 | 76 | 78 | 296 | |
| DTI | 79 | 54 | 68 | 72 | 273 | |
| Diffeomorphometry | Caudate | 84 | 57 | 76 | 78 | 295 |
| Putamen | 81 | 57 | 76 | 73 | 287 | |
| Globus pallidus | 82 | 57 | 75 | 77 | 291 | |
| Thalamus | 84 | 58 | 76 | 74 | 292 | |
| Hippocampus | 79 | 56 | 75 | 73 | 283 | |
| Nc. accumbens | 84 | 58 | 76 | 78 | 296 | |
| Age | 46.3 ± 11.1 | 35.1 ± 9.9 | 42.8 ± 9.3 | 47.9 ± 9.8 | ||
| Gender | 53F/31M | 17F/41M | 23F/53M | 29F/49M | ||
2.3. Surface-based morphometry of subcortical structures
We extend the statistical analyses of diffeomorphometry followed in previous papers (Csernansky et al., 1998, Csernansky et al., 2000, Ashburner et al., 2003, Thompson et al., 2004, Wang et al., 2007, Qiu et al., 2009, Qiu et al., 2010, Miller et al., 2013, Miller et al., 2014, Younes et al., 2013). This procedure has three steps: (i) segmentation of the target structures, (ii) generation of a single template coordinate system from the population of baseline scans, and (iii) mapping of the template onto each of the target segmented structures represented via triangulated meshes.
The first step (segmentation of the target structures) was done using the BRAINS AutoWorkup processing pipeline published in (Pierson et al., 2011). After completion of AutoWorkup, all scans were individually inspected and manually corrected where necessary to ensure correct realignment and coregistration, tissue classification, and accuracy of brain and subcortical structures. The description of this process and its reliability is fully described in (Kim et al., 2014). The resulting pre-processed data included T1, T2, PD, and tissue classification volumetric images where the anterior commissure (AC point) was set to be the center of the image resampled to 1 mm resolution in a 256 × 256 × 256 matrix. Details of each step of MPRAGE pre-processing are found in our previous publication (Young Kim and Johnson, 2013).
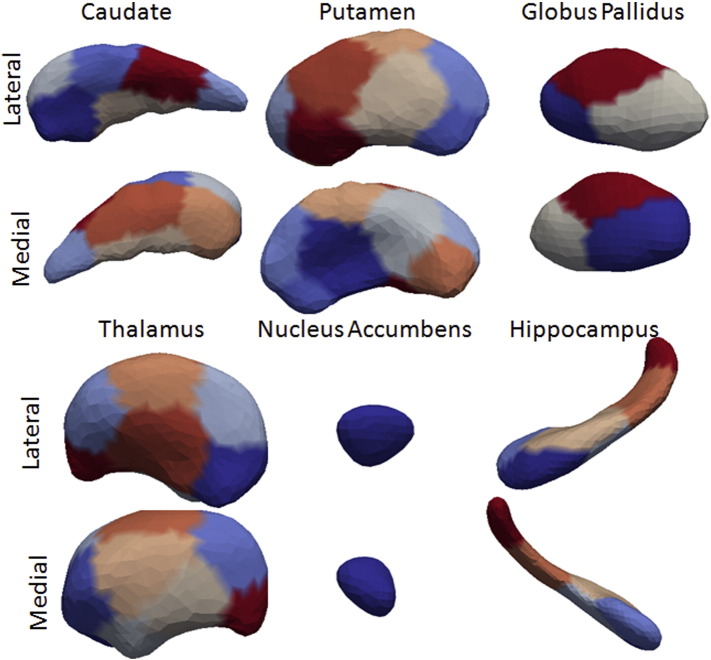
Binary images for the six substructures (caudate, globus pallidus, putamen, thalamus, hippocampus, and nucleus accumbens) were evaluated. To generate shape biomarkers indexed to a common coordinate system, we follow the procedure published in Younes et al. (2013) in which all surfaces are meshed and rigidly aligned via rotation and translation, with right subvolumes flipped before alignment to ensure that all structures could be compared. From rigidly aligned volumes, an average template triangulated mesh was generated based on a generative probability model over the entire population in which the observed surfaces are modeled as random deformations of the template (Ma et al., 2010). This template generation procedure solves a maximum-likelihood estimation problem for a statistical model in which the population is described as a random deformation of the template, itself defined as an unknown deformation of a fixed surface called hypertemplate. This procedure provides a coordinate system that represents the population via surface mapping onto the template. The resulting templates for the caudate, putamen, globus pallidus, thalamus, hippocampus, and nucleus accumbens (Fig. 1) become the coordinate systems, which are referenced for our p-value statistics and FWER (Family-wise Error Rate) calculations. These were computed by running the template generation algorithm on the population of baseline scans and are blind to group labels.
Fig. 1.
Template generated for the multiple brain structures. The colors represent the segments used for the surface based morphometry. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The high-dimensional diffeomorphometry shape statistics are generated indexed to each location of the template by computing the diffeomorphic correspondence between the template and each surface using large deformation diffeomorphic metric mapping (LDDMM) surface registration (Vaillant and Glaunes, 2005). The algorithm computes a smooth invertible mapping of the triangulated surface template Stemp onto the target surfaces Starget by minimizing the geodesic transformation energy and the matching cost that is the distance measurement between the mapped template (ϕ1 ⋅ Stemp) and the target surface.
| (1) |
The matching cost term δ(ϕ1 ⋅ Stemp, Starget) computes a norm between surfaces.
Shape coordinates are then deduced from the optimal diffeomorphism, resulting in one scalar measure per vertex on the template surfaces. For computational efficiency, we sub-discretized these measures by averaging them over small segments computed on the surface template (Younes et al., 2014a, Younes et al., 2014b). These segments are obtained by spectral clustering of the surface, a method that only relies on the surface geometry. This is achieved by computing the first k eigenvectors of the Laplace–Beltrami operator associated with the surface, where k is the intended number of segments, associating with each vertex a k-dimensional vector formed with the values of the eigenvectors evaluated at this point. These vectors are then used in a standard K-means algorithm to provide the k-desired segments. The number of segments was adjusted so that they cover an area of 150 mm2 on average, yielding 10 segments on the putamen, 8 on the caudate, 3 on the globus pallidus, 12 on the thalamus, 1 on the nucleus accumbens, and 6 on the hippocampus.
2.4. White matter analysis with DTI
The pipeline for the white matter DTI analysis consisted on mapping each subject's brain to a common brain template (JHU-MNI-SS) (Mori et al., 2008), using a sequence of linear transformations and then LDDMM (Miller et al., 2005, Wang et al., 2007, Ceritoglu et al., 2009). The JHU-MNI-SS is a multi-MRI contrast brain extensively segmented into more than two hundred structures (Oishi et al., 2008), allowing the automation of the atlas-based analysis (Faria et al., 2015). We used LDDMM because of the high accuracy on mapping, even for brains with large degrees of global or regional atrophy, as shown in previous studies (Oishi et al., 2009, Faria et al., 2010, Djamanakova et al., 2013). For the image post-processing, we used DTIStudio, DiffeoMap, and ROIEditor (Laboratory of Brain Anatomical MRI and Center for Imaging Science at Johns Hopkins University).
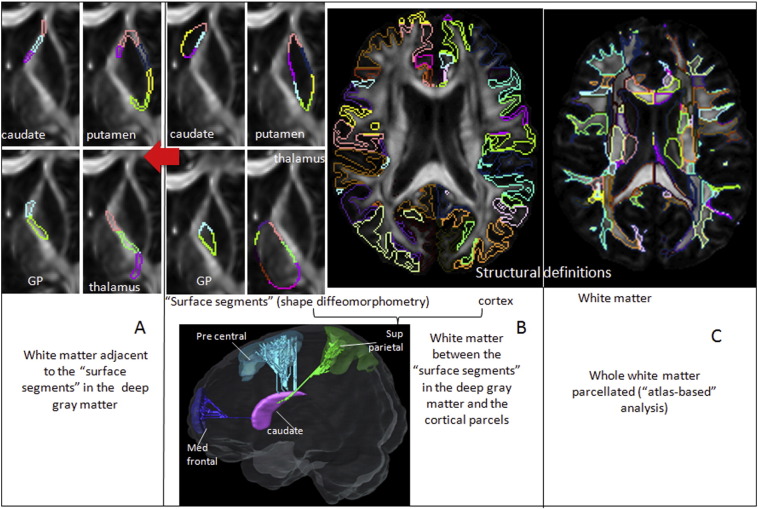
In order to identify the relationship between deep gray matter and deep white matter in HD, we used three approaches: A) the analysis of regions of interest (ROIs) in the white matter adjacent to the deep gray matter; B) the analysis of white matter pathways between the deep gray matter and cortex, using dynamic programming (DP); C) the analysis of the total white matter, regionally parcellated, using an atlas-based approach (ABA) (Fig. 2).
Fig. 2.
Schematic representation of the white matter analysis. A) ROIs in the white matter adjacent to the deep gray matter subregions used for shape diffeomorphometry (B, left panel). B) Examples of connections between deep gray matter and cortex; the cortical subregions in right panel are the parcels pre-defined in the template. C) Parcellation of the whole white matter in the atlas-based approach. GP = globus pallidus.
2.4.1. Analysis of the white matter adjacent to the deep gray matter
Each subregion defined in the surface of caudate, putamen, globus pallidus, and thalamus for the gray matter shape analysis (Fig. 2B, left panel) was expanded radially by 5 voxels (in a 1mm3 image resolution) through the adjacent white matter. Therefore, each segment in the shape analysis has the corresponding “white matter neighbor”, except where the surfaces are adjacent to the ventricles and in the putamen–globus pallidum interface.
2.4.2. Analysis of the white matter pathways between the striatum and the cortex
White matter pathways were traced using DP between each of the surface segments in striatum used for diffeomorphometry and the 38 cortical parcels in each hemisphere. The cortical parcels were previously defined in the template, as mentioned above. The DP algorithm applied is detailed in our previous paper (Li et al., 2014). Our choice of a probabilistic path-generation algorithm aims to minimize the false negatives that simple line propagation methods would generate, given that our seeds are located in the gray matter. Since high-angular resolution images are not available, DP is the natural option for tracing. Using a “not” operation we removed connections crossing a third seed or the ventricles. This minimized the double sampling of pathways and excluded implausible pathways. The streamlines traced in the template were eventually converted to regions of interest.
It is worth noting that algorithms such as DP trace the “shortest path” between chosen initial and terminal points that globally minimize a sequentially additive energy constraint defined by the tensor and, as happens to any other DTI-based tract-tracing method, does not necessarily reflect the existence of a biological connection. Therefore, they have to be interpreted as a macroscopic reconstruction of the white matter architecture or a region-growing tool, which can cluster anatomically related pixels based on DTI data, rather than a tool with which to investigate connectivity based on a cellular level structure.
2.4.3. Regional analysis of the whole white matter
Using our pre-segmented template and accurate image mapping, we automatically parceled the whole white matter of each individual into 98 ROIs. The segregation of cortex and peripheral white matter was threshold by FA > 0.2. This “atlas-based” analysis carries information about the structural anatomy, since each parcel is defined based on previous anatomical knowledge.
2.5. Group based linear effects statistical analysis
We have performed statistical analyses on each of the subcortical structures, making separate comparisons of the shape diffeomorphometry markers corresponding to the degree of atrophy relative to the template between the groups. We modeled the shape diffeomorphometry markers via linear effects. The analysis includes age, gender and log intracranial volumes as covariates, and computes statistics at each segment of the triangulated template surface returning p-values corrected for multiple comparisons using permutation testing.
The group analysis quantifies a mixed linear effects model of atrophy between the groups including controls and the low, medium and high-CAP score defined groups, and is given for each vertex v, scan j and subject s. For each control and patient subject, left and right structures are registered to the template, resulting in the computation of a normalized deformation marker Jv(s) that measures the amount of expansion/atrophy at vertex v of the template surface in registering it to subject s. The raw expansion/atrophy measure is defined as the logarithm of the local expansion/reduction in surface area around the vertex, and is interpreted mathematically as a log-jacobian on the template surface. We model the group variables as g(s) equaling 1 if subject s belongs to the group (either low, medium or high), and zero otherwise belonging to the controls. Our analysis uses gender (denoted d) and intracranial volume (denoted i) as covariates, resulting in the model
| (2) |
with ϵv(s) representing our noise model with ϵv(s) Gaussian distributed with variance σv2. We test for the null hypothesis with Hv0 : βv = 0 for all v, while correcting for multiple comparisons.
For volume testing the logarithm of the volume is used. The p-values of these models are computed using permutation sampling (using random permutations of residuals of the null hypothesis model). The test statistic is the log-likelihood difference between the null hypothesis and the Hv1 : βv ≠ 0 general hypothesis, which is equivalent to computing, for each coordinate v, the logarithm of ratio of the residual variance for the complete Hv1 hypothesis to the one obtained from the null hypothesis. Denoting this statistic by Sv, the family-wise error rates (FWER) are calculated by evaluating the maximum S* = maxvSv.
To compute p-values, the same statistic S⁎ is computed a large number of times with permuted residuals under the null, i.e., taking
| (3) |
where π is a random permutation of the subjects and the parameters and residuals are those obtained using maximum likelihood under the null hypothesis. The p-value is given by the fraction of times the values of S⁎ computed after permuting the residuals is larger than the value obtained with the true groups. The p-values that were observed via the linear effects modeling of deformation markers of Eq. (2) are provided for each structure including the left and right sides of the brain in the Supplementary Table 1. The volume statistics shown in the third column for the three groups provides p-values for the same linear effects model, also evaluated via permutations, in which J is replaced by the structure volume, for which no multiple testing correction is required.
For the DTI analysis, after mapping each subject brain scan to the common template, mean values of fractional anisotropy (FA), mean diffusivity (MD), radial diffusivity (RD), and axial diffusivity (AD) were obtained in each region of interest, in each of the three approaches mentioned before. Similarly, differences among groups (controls and each of the premanifest HD) were evaluated with permutation test and corrected for multiple comparisons with FWE. Monotonic linear fitting was used to describe the relationship between shape and DTI parameters versus CAP scores.
3. Results
3.1. Shape diffeomorphometry
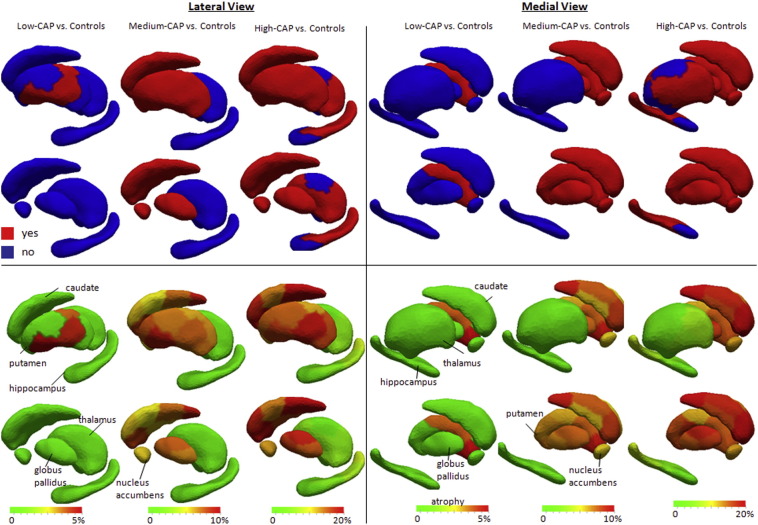
The shape diffeomorphometry analysis of subcortical gray matter change demonstrated differences in all three pre-manifest stages versus controls (Fig. 3). Modest differences, both in terms of location (restricted to central segments of putamen) and effect size were detected in the low-CAP group. In the medium-CAP, the differences extended to the whole surface of putamen, caudate, globus pallidus, and nucleus accumbens. In the high-CAP group, differences were also noticed in thalamus and hippocampus. The effect size was different in different regions of striatum and other subcortical structures. Especially large atrophy were noted in some regions, such as tail of caudate, showing up to 25% of atrophy.
Fig. 3.
Results from shape diffeomorphometry in both lateral (left panel) and medial (right panel) views. The top rows show whether there are significant differences between groups, and the bottom rows show the average degree of atrophy, if any. In the second and fourth rows, putamen (in the lateral view) and thalamus (in the medial view) were removed, to enable the visualization of structures lying behind them.
3.1.1. Analysis of the white matter adjacent to the deep gray matter
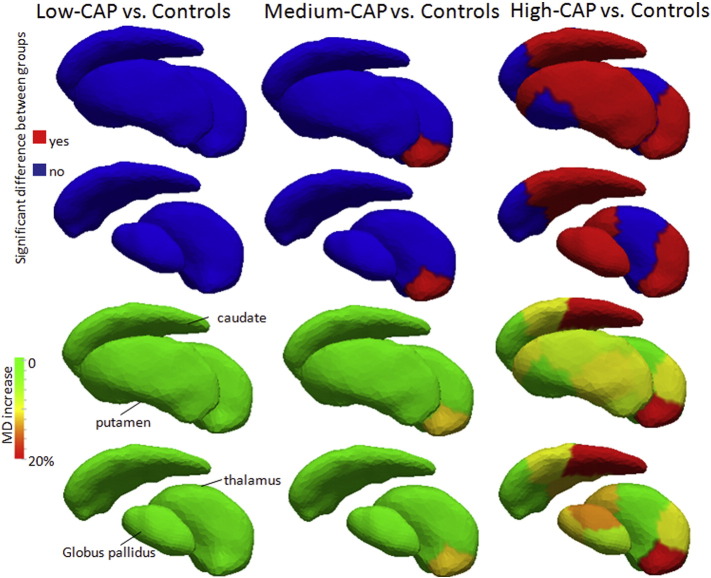
Medium Diffusivity (MD) was the most sensitive metric of white matter difference in HD. MD was increased in the white matter adjacent to the posterior portion of thalamus in the medium-CAP group. The increase was greater and widespread in the high-CAP group (Fig. 4). The MD values for each segment analyzed are reported in the Supplementary Table 2. The source of MD increase was the increase of both radial and axial diffusivity. Fractional anisotropy was not significantly different among groups.
Fig. 4.
Differences in MD between controls and each of the premanifest HD groups in the white matter adjacent to the segments of the deep gray matter and thalamus used for the diffeomorphometry analysis. Lateral view is shown. The top rows show whether there are significant differences between groups, and the bottom rows shows the respective MD increase of each groups compared to controls. In the second and fourth rows, putamen was removed, to enable the visualization of globus pallidum and thalamus.
3.1.2. Analysis of the white matter pathways between striatum and cortex
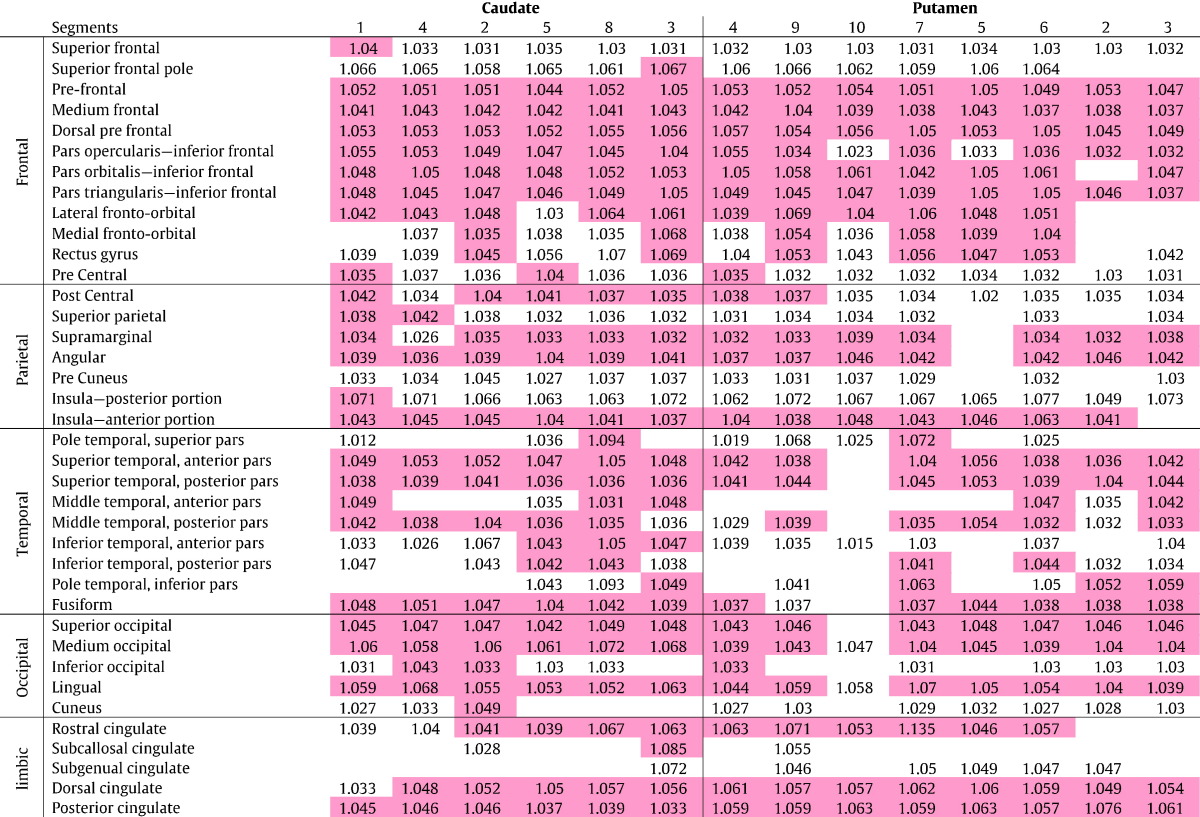
None of the pathways traced by DP between the striatum and cortical areas showed significant difference in DTI between controls and low-CAP or controls and medium-CAP group. Pathways between striatum and many cortical areas had significant MD increase in high-CAP group compared to controls, as shown in Table 2.
Table 2.
Differences in MD between high-CAP group and controls, in pathways between the striatum and the cortex. The numbers indicate the MD ratio of high-CAP/controls. Red cells are significant differences between groups, empty cells are pathways that couldn't be traced, i.e., DP resulted in zero streamlines. The subregions of striatum are ordered according to their anatomical position, from anterior to posterior, as much as possible.
3.1.3. Regional analysis of the whole white matter
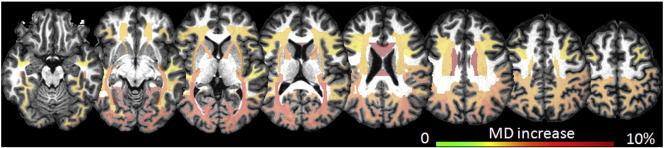
Once again, MD was the most sensitive DTI metric, showing differences between controls and high-CAP group as shown in Fig. 5. The effect size detected (maximum 8% of MD increase) was smaller than that detected in the analysis of the neighborhood of the deep gray matter (Fig. 4). The most affected areas were the posterior thalamic radiations, corpus callosum, and the peripheral occipital white matter. The MD increase was again due to increases in both radial and axial diffusivities. Although the comparison of MD between medium-CAP group and controls did not reveal significant differences, some areas showed increased MD close to the significance threshold, such as the posterior thalamic radiations. The MD values for each segment analyzed are reported in the Supplementary Table 3. The splenium of the corpus callosum had the lowest FA in the high-CAP group, although no area achieved the significance threshold for FA differences between groups.
Fig. 5.
Atlas-based white matter DTI analysis. The colors code the increase of MD in areas of significant differences between controls and high-CAP group. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Relationship between shape, DTI metrics, and CAP scores
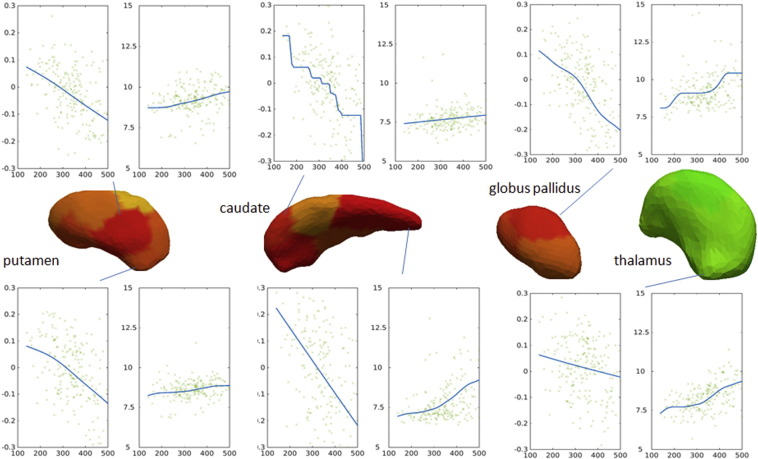
Fig. 6 shows the trends of shape and MD (y axis) abnormalities according to the CAP scores (x axis), fitted by monotonic curves. There is a positive correlation between the CAP scores and the MD, i.e., MD increases with higher CAP scores. There is a negative correlation between the CAP scores and the amount as each vertex dislocates to match the template (log-Jacobian), i.e., the atrophy increases with higher CAP scores.
Fig. 6.
Examples of plots of CAP scores (x-axis) vs. degree of atrophy (first plots, y-axis: log (Jacobian)) in putamen, caudate, globus pallidus, and thalamus; or vs. MD (second plots, y-axis: MD e10− 4, in mm2/s) in the adjacent white matter. For spatial localization, the various sub-segments of each structure are shown in the center. The colors represent the degree of atrophy in high-CAP versus vs. controls, as in Fig. 2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In this study, we combined deep gray matter shape diffeomorphometry and white matter DTI analysis as means of better defining the topography of deep gray and white matter abnormalities in the premanifest stages of HD. We used images from the PREDICT-HD study, which as noted above successfully includes sources of brain MRIs from multiple centers. Across-scanner variations might interfere with the detection of disease-specific structural abnormalities, thereby potentially limiting the use of group analysis collected at several centers. On the other hand, multicenter studies increased generalizability of results, and improved efficiency, particularly for rare or hard-to-recruit cases, such as HD (Paulsen et al., 2008, Tabrizi et al., 2009). Both the algorithm for segmenting subcortical structures and the scalar measures used for DTI analysis applied in this study has shown high multicenter reliability (Magnotta et al., 2012, Kim et al., 2015).
Volumetric structural imaging has identified imaging biomarkers for HD onset, such as the extent of caudate atrophy (Paulsen et al., 2014a, Paulsen et al., 2014b). The striatum has been shown to demonstrate strong cross-sectional and longitudinal atrophy in the pre-manifest phase (Aylward et al., 1997, Aylward et al., 2000, Rosas et al., 2001, Peinemann et al., 2005, Henley et al., 2009, Vandenberghe et al., 2009, Ginestroni et al., 2010, Georgiou-Karistianis et al., 2013, Sanchez-Castaneda et al., 2013). Other regions, such as the globus pallidus, thalamus and hippocampus, also undergo atrophy, though in smaller magnitude (Paulsen et al., 2010, Aylward et al., 2011, Younes et al., 2014a, Younes et al., 2014b) and mostly after motor the onset. In addition, the measurement of shape has enabled the delineation of regional abnormalities in the striatum and other subcortical structures in the premanifest period (Younes et al., 2014a, Younes et al., 2014b). Reduction of white matter volume was also reported (Tabrizi et al., 2009) in both premanifest and early HD cohorts (Crawford et al., 2013). DTI studies differ regarding specific areas of white matter abnormality in the premanifest stage (Reading et al., 2005, Rosas et al., 2006, Kloppel et al., 2008, Stoffers et al., 2010, Poudel et al., 2014).
In the present study, we integrated the characterization of deep gray matter shape with regional white matter DTI metrics in subjects with premanifest HD to improve the regional mapping of the abnormalities that precede HD onset. Regional abnormalities in the putamen shape, which can be anatomically interpreted as atrophy, occur in the very early stage, with differences being detected between the controls and the low-CAP group. In the medium-CAP group, shape is already extensively affected over the surface of caudate, putamen, and globus pallidus. The atrophy is greater in both location (involving thalamus and hippocampus) and degree in the high-CAP group compared to the other groups.
The MD in the white matter surrounding the deep gray matter was consistently increased close to the disease onset. In addition, higher MD was observed in the neighborhood of the posterior thalamus in the medium-CAP group. According the regional whole brain analysis of the white matter, MD was extensively larger in high-CAP compared to controls, particularly in posterior areas. A trend of increasing MD was seen even in the low-CAP group; in the medium-CAP, areas such as the splenium of the corpus callosum, posterior thalamic radiations, and occipital white matter had MD increase close to the threshold significance. Note that our method was conservative regarding the p-value cutoff, after the permutation and multiple comparisons corrections. This may explain the disagreement with other publications that showed white matter abnormalities occurring at least as early as subcortical gray matter abnormalities (Aylward et al., 2011, Tabrizi et al., 2013).
The tractography analysis revealed increased MD in the topography of streamlines traced between the striatum and multiple cortical areas in the high-CAP group, compared to controls, suggesting that pathology may follow paths of anatomic connections. This needs to be interpreted carefully, since DTI tractography may identify streamlines that imperfectly correspond to actual anatomic connections. Recently, Novak et al. (Novak et al., 2015) showed widespread differences in basal ganglia-cortical structural connectivity in early manifest HD subjects. Variations of DTI indices such as MD and FA may be interpreted as axonal disorganization or axonal degeneration, or defective myelination, or other effects. Our studies cannot define the cellular bases of the changes, but can help map their topography.
The sensitivity of MD or FA to detect group differences depends both on the regional microstructural architecture in question and on the disturbance to this architecture (Oh et al., 2009). In this study, MD was more sensitive than FA in detecting group differences, indicating that HD causes disturbances on the microstructural organization that are more sensitively detected by MD than by FA. Nevertheless, FA tends to be more sensitive to image noise (Pierpaoli and Basser, 1996) as demonstrated in our previous study about MultiCenter Reliability of Diffusion Tensor Imaging (Magnotta et al., 2012), in which among all the DTI scalar measures, FA was the one that had the largest coefficient of variance among all the protocols, although still quite small.
These results, while cross-sectional, rather than longitudinal, may be interpreted in relationship to CAP score(which is a genetic measure of the degree of exposure to the effects of the CAG repeat expansion in HTT), as the cross-sectional reflection of progressive changes in HD. Information from cross-sectional studies such as this one may be useful for guiding therapeutic trials, since subjects in clinical trials will be ascertained based on cross-sectional information and may be selected or stratified based on CAP score or comparable indices. Atrophy started in putamen and caudate, followed by globus pallidum, thalamus, and hippocampus. Our results are consistent with the hypothesis that detectible atrophy in the deep gray matter would precede detectible abnormalities in the white matter, and with a centripetal and posterior gradient of abnormalities in the white matter. There are, however, limitations on comparing white matter microstructural abnormalities and atrophy by imaging. The findings reported in this study may reflect a true biological phenomenon, or just the ability of MRI and of the image analysis employed to detect these abnormalities, or both. Differences in diffeomorphometry and diffusivity between groups were analyzed by the same statistical procedure, via linear effects corrected for multiple comparisons using permutation testing, as detailed in Section II.5. However, DTI is inherently noisier than T1-WIs, which may result on decreased power on detecting DTI abnormalities compared to atrophy. In this sense, our analysis may ensue very conservative results, particularly for DTI. Differences found at conservative group-level analysis, as in this study, can be interpreted with more confidence, and have greater chance to become features on predictive models, which is one of the PREDCIT-HD goals, than those found with a less conservative approach. The latter would better indicate small trends at group level and, indirectly, uncover underlying disease mechanisms, which are actually unlikely to be revealed by any MRI study in humans.
Our data suggest that atrophy is detected earliest in the deep gray matter, but that subjacent and closely connected white matter also becomes affected prior to predicted onset. A major current strategy for therapeutics involves lowering levels of mutant Htt, via RNA- or DNA-based strategies. One implementation currently in clinical trials involves infusion of antisense oligonucleotides into the lumbar CSF and thus, after CSF circulation, over the cortex. A concern based on our results is that these ASOs may not penetrate to deep gray and deep subcortical while matters structures we demonstrate to be affected during the pre-manifest period. A second potential implementation for clinical trials involves injection of viral vectors into the striatum. Our data suggest that this should be done early (as defined by CAP score), prior to the development of pathology in the deep white matter (thus perhaps even preventing the spread of pathology to the white matter). In addition, it may be necessary to inject the therapeutic agents into the white matter itself.
The following are the supplementary data related to this article.
P-values for deformation markers on each structure corrected for multiple comparisons via permutation tests. *p-value < 0.000025.
MD values in the white matter ROIs defined in the neighborhood of the deep gray matter.
MD values in the white matter (wm) ROIs defined by the automated whole-brain analysis.
Funding
A. V. Faria is supported by NIH/NIBIB R03-EB014357 and AHA 12SDG12080169. C. A. Ross was supported by P50 NS16375 (NINDS Baltimore HD Center) and NS40068 (PREDICT-HD). M. I. Miller, J. T. Ratnanather and L. Younes are partially supported by NIH P41-EB015909, R01-EB000975, R01-EB008171, U01-NS082085. PREDICT-HD and J. S. Paulsen are supported by the NIH/NINDS NS40068 and CHDI Foundation, Inc. A3917.
Acknowledgments
The authors thank the PREDICT-HD sites, the study participants, and the National Research Roster for Huntington Disease Patients and Families.
References
- Ashburner J., Csernansky J.G., Davatzikos C., Fox N.C., Frisoni G.B., Thompson P.M. Computer-assisted imaging to assess brain structure in healthy and diseased brains. Lancet Neurol. 2003;2(2):79–88. doi: 10.1016/s1474-4422(03)00304-1. [DOI] [PubMed] [Google Scholar]
- Aylward E.H., Codori A.M., Rosenblatt A., Sherr M., Brandt J., Stine O.C., Barta P.E., Pearlson G.D., Ross C.A. Rate of caudate atrophy in presymptomatic and symptomatic stages of Huntington's disease. Mov. Disord. 2000;15(3):552–560. doi: 10.1002/1531-8257(200005)15:3<552::AID-MDS1020>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Aylward E.H., Harrington D.L., Mills J.A., Nopoulos P.C., Ross C.A., Long J.D., Liu D., Westervelt H.K., Paulsen J.S. Regional atrophy associated with cognitive and motor function in prodromal Huntington disease. J. Huntingtons Dis. 2013;2(4):477–489. doi: 10.3233/JHD-130076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylward E.H., Li Q., Stine O.C., Ranen N., Sherr M., Barta P.E., Bylsma F.W., Pearlson G.D., Ross C.A. Longitudinal change in basal ganglia volume in patients with Huntington's disease. Neurology. 1997;48(2):394–399. doi: 10.1212/wnl.48.2.394. [DOI] [PubMed] [Google Scholar]
- Aylward E.H., Liu D., Nopoulos P.C., Ross C.A., Pierson R.K., Mills J.A., Long J.D., Paulsen J.S. Striatal volume contributes to the prediction of onset of Huntington disease in incident cases. Biol. Psychiatry. 2012;71(9):822–828. doi: 10.1016/j.biopsych.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylward E.H., Nopoulos P.C., Ross C.A., Langbehn D.R., Pierson R.K., Mills J.A., Johnson H.J., Magnotta V.A., Juhl A.R., Paulsen J.S. Longitudinal change in regional brain volumes in prodromal Huntington disease. J. Neurol. Neurosurg. Psychiatry. 2011;82(4):405–410. doi: 10.1136/jnnp.2010.208264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylward E.H., Rosenblatt A., Field K., Yallapragada V., Kieburtz K., McDermott M., Raymond L.A., Almqvist E.W., Hayden M., Ross C.A. Caudate volume as an outcome measure in clinical trials for Huntington's disease: a pilot study. Brain Res. Bull. 2003;62(2):137–141. doi: 10.1016/j.brainresbull.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Bohanna I., Georgiou-Karistianis N., Sritharan A., Asadi H., Johnston L., Churchyard A., Egan G. Diffusion tensor imaging in Huntington's disease reveals distinct patterns of white matter degeneration associated with motor and cognitive deficits. Brain Imaging Behav. 2011;5(3):171–180. doi: 10.1007/s11682-011-9121-8. [DOI] [PubMed] [Google Scholar]
- Ceritoglu C., Oishi K., Li X., Chou M.C., Younes L., Albert M., Lyketsos C., van Zijl P.C., Miller M.I., Mori S. Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. NeuroImage. 2009;47(2):618–627. doi: 10.1016/j.neuroimage.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford H.E., Hobbs N.Z., Keogh R., Langbehn D.R., Frost C., Johnson H., Landwehrmeyer B., Reilmann R., Craufurd D., Stout J.C., Durr A., Leavitt B.R., Roos R.A., Tabrizi S.J., Scahill R.I. Corpus callosal atrophy in premanifest and early Huntington's disease. J. Huntingtons Dis. 2013;2(4):517–526. doi: 10.3233/JHD-130077. [DOI] [PubMed] [Google Scholar]
- Csernansky J.C., Joshi S., Wang L., Gado M., Miller J.P., Grenander U., Miller M.I. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. Proc. Natl. Acad. Sci. 1998;95:11406–11411. doi: 10.1073/pnas.95.19.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky J.G., Wang L., Joshi S., Miller J.P., Gado M., Kido D., McKeel D., Morris J.C., Miller M.I. Early DAT is distinguished from aging by high-dimensional mapping of the hippocampus. Neurology. 2000;55(11):1636–1643. doi: 10.1212/wnl.55.11.1636. [DOI] [PubMed] [Google Scholar]
- Della Nave R., Ginestroni A., Tessa C., Giannelli M., Piacentini S., Filippi M., Mascalchi M. Regional distribution and clinical correlates of white matter structural damage in Huntington disease: a tract-based spatial statistics study. AJNR Am. J. Neuroradiol. 2010;31(9):1675–1681. doi: 10.3174/ajnr.A2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmaire C., Dumas E.M., Sharman M.A., van den Bogaard S.J., Valabregue R., Jauffret C., Justo D., Reilmann R., Stout J.C., Craufurd D., Tabrizi S.J., Roos R.A., Durr A., Lehericy S. The structural correlates of functional deficits in early huntington's disease. Hum. Brain Mapp. 2013;34(9):2141–2153. doi: 10.1002/hbm.22055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamanakova A., Faria A.V., Hsu J., Ceritoglu C., Oishi K., Miller M.I., Hillis A.E., Mori S. Diffeomorphic brain mapping based on T1-weighted images: improvement of registration accuracy by multichannel mapping. J. Magn. Reson. Imaging. 2013;37(1):76–84. doi: 10.1002/jmri.23790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey E.R., Beck C.A., Darwin K., Nichols P., Brocht A.F., Biglan K.M., Shoulson I. Natural history of Huntington disease. JAMA Neurol. 2013;70(12):1520–1530. doi: 10.1001/jamaneurol.2013.4408. [DOI] [PubMed] [Google Scholar]
- Douaud G., Behrens T.E., Poupon C., Cointepas Y., Jbabdi S., Gaura V., Golestani N., Krystkowiak P., Verny C., Damier P., Bachoud-Levi A.C., Hantraye P., Remy P. In vivo evidence for the selective subcortical degeneration in Huntington's disease. NeuroImage. 2009;46(4):958–966. doi: 10.1016/j.neuroimage.2009.03.044. [DOI] [PubMed] [Google Scholar]
- Faria A.V., Oishi K., Yoshida S., Hillis A., Miller M.I., Mori S. Content-based image retrieval for brain MRI: An image-searching engine and population-based analysis to utilize past clinical data for future diagnosis. NeuroImage Clin. 2015;7:367–376. doi: 10.1016/j.nicl.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria A.V., Zhang J., Oishi K., Li X., Jiang H., Akhter K., Hermoye L., Lee S.K., Hoon A., Stashinko E., Miller M.I., van Zijl P.C., Mori S. Atlas-based analysis of neurodevelopment from infancy to adulthood using diffusion tensor imaging and applications for automated abnormality detection. NeuroImage. 2010;52(2):415–428. doi: 10.1016/j.neuroimage.2010.04.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein S.E. The psychopathology of Huntington's disease. Res. Publ. Assoc. Res. Nerv. Ment. dis. 1991;69:181–191. [PubMed] [Google Scholar]
- Georgiou-Karistianis N., Scahill R., Tabrizi S.J., Squitieri F., Aylward E. Structural MRI in Huntington's disease and recommendations for its potential use in clinical trials. Neurosci. Biobehav. Rev. 2013;37(3):480–490. doi: 10.1016/j.neubiorev.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Ginestroni A., Battaglini M., Diciotti S., Della Nave R., Mazzoni L.N., Tessa C., Giannelli M., Piacentini S., De Stefano N., Mascalchi M. Magnetization transfer MR imaging demonstrates degeneration of the subcortical and cortical gray matter in Huntington disease. AJNR Am. J. Neuroradiol. 2010;31(10):1807–1812. doi: 10.3174/ajnr.A2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley S.M., Wild E.J., Hobbs N.Z., Scahill R.I., Ridgway G.R., Macmanus D.G., Barker R.A., Fox N.C., Tabrizi S.J. Relationship between CAG repeat length and brain volume in premanifest and early Huntington's disease. J. Neurol. 2009;256(2):203–212. doi: 10.1007/s00415-009-0052-x. [DOI] [PubMed] [Google Scholar]
- Huntington G. On chorea. Med. Surg. Report. 1872;26:320–321. [Google Scholar]
- Huntington's Disease Collaborative Research Group. Aronin N., DiFiglia M. Huntingtin-lowering strategies in Huntington's disease: antisense oligonucleotides, small RNAs, and gene editing. Mov. Disord. 2014;29(11):1455–1461. doi: 10.1002/mds.26020. [DOI] [PubMed] [Google Scholar]
- Kay C., Skotte N.H., Southwell A.L., Hayden M.R. Personalized gene silencing therapeutics for Huntington disease. Clin. Genet. 2014;86(1):29–36. doi: 10.1111/cge.12385. [DOI] [PubMed] [Google Scholar]
- Kim R.E., Lourens S., Long J.D., Paulsen J.S., Johnson H.J. Preliminary analysis using multi-atlas labeling algorithms for tracing longitudinal change. Front. Neurosci. 2015;9:242. doi: 10.3389/fnins.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.Y., Magnotta V.A., Liu D., Johnson H.J. Stable Atlas-based Mapped Prior (STAMP) machine-learning segmentation for multicenter large-scale MRI data. Magn. Reson. Imaging. 2014;32(7):832–844. doi: 10.1016/j.mri.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppel S., Draganski B., Golding C.V., Chu C., Nagy Z., Cook P.A., Hicks S.L., Kennard C., Alexander D.C., Parker G.J., Tabrizi S.J., Frackowiak R.S. White matter connections reflect changes in voluntary-guided saccades in pre-symptomatic Huntington's disease. Brain. 2008;131(Pt 1):196–204. doi: 10.1093/brain/awm275. [DOI] [PubMed] [Google Scholar]
- Laboratory of Brain Anatomical MRI and Center for Imaging Science at Johns Hopkins University. "MRI Studio: an image processing program." from http://www.mristudio.org.
- Li M., Ratnanather J.T., Miller M.I., Mori S. Knowledge-based automated reconstruction of human brain white matter tracts using a path-finding approach with dynamic programming. NeuroImage. 2014;88:271–281. doi: 10.1016/j.neuroimage.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Miller M.I., Younes L. A bayesian generative model for surface template estimation. Int. J. Biomed. Imaging. 2010;2010 doi: 10.1155/2010/974957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnotta V.A., Matsui J.T., Liu D., Johnson H.J., Long J.D., Bolster B.D., Jr., Mueller B.A., Lim K., Mori S., Helmer K.G., Turner J.A., Reading S., Lowe M.J., Aylward E., Flashman L.A., Bonett G., Paulsen J.S. Multicenter reliability of diffusion tensor imaging. Brain Connect. 2012;2(6):345–355. doi: 10.1089/brain.2012.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui J.T., Vaidya J.G., Wassermann D., Kim R.E., Magnotta V.A., Johnson H.J., Paulsen J.S. Prefrontal cortex white matter tracts in prodromal Huntington disease. Hum. Brain Mapp. 2015;36(10):3717–3732. doi: 10.1002/hbm.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.I., Beg M.F., Ceritoglu C., Stark C. Increasing the power of functional maps of the medial temporal lobe by using large deformation diffeomorphic metric mapping. Proc. Natl. Acad. Sci. U. S. A. 2005;102(27):9685–9690. doi: 10.1073/pnas.0503892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.I., Younes L., Ratnanather J.T., Brown T., Trinh H., Postell E., Lee D.S., Wang M.C., Mori S., O'Brien R., Albert M. The diffeomorphometry of temporal lobe structures in preclinical Alzheimer's disease. Neuroimage Clin. 2013;3:352–360. doi: 10.1016/j.nicl.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.I., Younes L., Trouve A. Diffeomorphometry and geodesic positioning systems for human anatomy. Technol. Singap. World Sci. 2014;2(1):36. doi: 10.1142/S2339547814500010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Oishi K., Jiang H., Jiang L., Li X., Akhter K., Hua K., Faria A.V., Mahmood A., Woods R., Toga A.W., Pike G.B., Neto P.R., Evans A., Zhang J., Huang H., Miller M.I., van Zijl P., Mazziotta J. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage. 2008;40(2):570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan P., Fishbaugh J., Johnson H.J., Durrleman S., Paulsen J.S., Gerig G., Fletcher P.T. Diffeomorphic shape trajectories for improved longitudinal segmentation and statistics. Med. Image Comput. Comput. Assist. Interv. 2014;17(Pt 3):49–56. doi: 10.1007/978-3-319-10443-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak M.J., Seunarine K.K., Gibbard C.R., McColgan P., Draganski B., Friston K., Clark C.A., Tabrizi S.J. Basal ganglia-cortical structural connectivity in Huntington's disease. Hum. Brain Mapp. 2015 doi: 10.1002/hbm.22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguz I., Farzinfar M., Matsui J., Budin F., Liu Z., Gerig G., Johnson H.J., Styner M. DTIPrep: quality control of diffusion-weighted images. Front. Neuroinform. 2014;8:4. doi: 10.3389/fninf.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.S., Kubicki M., Rosenberger G., Bouix S., Levitt J.J., McCarley R.W., Westin C.F., Shenton M.E. Thalamo-frontal white matter alterations in chronic schizophrenia: a quantitative diffusion tractography study. Hum. Brain Mapp. 2009;30(11):3812–3825. doi: 10.1002/hbm.20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Faria A., Jiang H., Li X., Akhter K., Zhang J., Hsu J.T., Miller M.I., van Zijl P.C., Albert M., Lyketsos C.G., Woods R., Toga A.W., Pike G.B., Rosa-Neto P., Evans A., Mazziotta J., Mori S. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer's disease participants. NeuroImage. 2009;46(2):486–499. doi: 10.1016/j.neuroimage.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Zilles K., Amunts K., Faria A., Jiang H., Li X., Akhter K., Hua K., Woods R., Toga A.W., Pike G.B., Rosa-Neto P., Evans A., Zhang J., Huang H., Miller M.I., van Zijl P.C., Mazziotta J., Mori S. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. NeuroImage. 2008;43(3):447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster E., Eberly S.W., Dorsey E.R., Kayson-Rubin E., Oakes D., Shoulson I. Informativeness of early Huntington disease signs about gene status. J. Huntingtons Dis. 2015;4(3):271–277. doi: 10.3233/JHD-150156. [DOI] [PubMed] [Google Scholar]
- Paulsen J.S., Long J.D., Johnson H.J., Aylward E.H., Ross C.A., Williams J.K., Nance M.A., Erwin C.J., Westervelt H.J., Harrington D.L., Bockholt H.J., Zhang Y., McCusker E.A., Chiu E.M., Panegyres P.K. Clinical and biomarker changes in premanifest Huntington disease show trial feasibility: a decade of the PREDICT-HD study. Front. Aging Neurosci. 2014;6:78. doi: 10.3389/fnagi.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J.S., Long J.D., Ross C.A., Harrington D.L., Erwin C.J., Williams J.K., Westervelt H.J., Johnson H.J., Aylward E.H., Zhang Y., Bockholt H.J., Barker R.A. Prediction of manifest Huntington's disease with clinical and imaging measures: a prospective observational study. Lancet Neurol. 2014;13(12):1193–1201. doi: 10.1016/S1474-4422(14)70238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J.S., Hayden M., Stout J.C., Langbehn D.R., Aylward E., Ross C.A., Guttman M., Nance M., Kieburtz K., Oakes D., Shoulson I., Kayson E., Johnson S., Penziner E. Preparing for preventive clinical trials: the Predict-HD study. Arch. Neurol. 2006;63(6):883–890. doi: 10.1001/archneur.63.6.883. [DOI] [PubMed] [Google Scholar]
- Paulsen J.S., Langbehn D.R., Stout J.C., Aylward E., Ross C.A., Nance M., Guttman M., Johnson S., MacDonald M., Beglinger L.J., Duff K., Kayson E., Biglan K., Shoulson I., Oakes D., Hayden M. Detection of Huntington's disease decades before diagnosis: the Predict-HD study. J. Neurol. Neurosurg. Psychiatry. 2008;79(8):874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J.S., Nopoulos P.C., Aylward E., Ross C.A., Johnson H., Magnotta V.A., Juhl A., Pierson R.K., Mills J., Langbehn D., Nance M. Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res. Bull. 2010;82(3–4):201–207. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinemann A., Schuller S., Pohl C., Jahn T., Weindl A., Kassubek J. Executive dysfunction in early stages of Huntington's disease is associated with striatal and insular atrophy: a neuropsychological and voxel-based morphometric study. J. Neurol. Sci. 2005;239(1):11–19. doi: 10.1016/j.jns.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Penney J.B., Jr., Vonsattel J.P., MacDonald M.E., Gusella J.F., Myers R.H. CAG repeat number governs the development rate of pathology in Huntington's disease. Ann. Neurol. 1997;41(5):689–692. doi: 10.1002/ana.410410521. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C., Basser P.J. Toward a quantitative assessment of diffusion anisotropy. Magn. Reson. Med. 1996;36(6):893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- Pierson R., Johnson H., Harris G., Keefe H., Paulsen J.S., Andreasen N.C., Magnotta V.A. Fully automated analysis using BRAINS: AutoWorkup. NeuroImage. 2011;54(1):328–336. doi: 10.1016/j.neuroimage.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel G.R., Stout J.C., Dominguez D.J., Salmon L., Churchyard A., Chua P., Georgiou-Karistianis N., Egan G.F. White matter connectivity reflects clinical and cognitive status in Huntington's disease. Neurobiol. Dis. 2014;65:180–187. doi: 10.1016/j.nbd.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Qiu A., Adler M., Crocetti D., Miller M., Mostofsky S. Basal ganglia shapes predict social, communication, and motor dysfunctions in boys with autism spectrum disorder. J. Am. Acad. Child Adolesc. Psychiatry. 2010;49:539–551. doi: 10.1016/j.jaac.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Qiu A., Crocetti D., Adler M., Mahone E., D. MB, Miller M., Mostofsky S. Basal ganglia volume and shape in children with attention deficit hyperactivity disorder. Am. J. Psychiatr. 2009;166:74–82. doi: 10.1176/appi.ajp.2008.08030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading S.A., Yassa M.A., Bakker A., Dziorny A.C., Gourley L.M., Yallapragada V., Rosenblatt A., Margolis R.L., Aylward E.H., Brandt J., Mori S., van Zijl P., Bassett S.S., Ross C.A. Regional white matter change in pre-symptomatic Huntington's disease: a diffusion tensor imaging study. Psychiatry Res. 2005;140(1):55–62. doi: 10.1016/j.pscychresns.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Reilmann R., Leavitt B.R., Ross C.A. Diagnostic criteria for Huntington's disease based on natural history. Mov. Disord. 2014 doi: 10.1002/mds.26011. [DOI] [PubMed] [Google Scholar]
- Rosas H.D., Goodman J., Chen Y.I., Jenkins B.G., Kennedy D.N., Makris N., Patti M., Seidman L.J., Beal M.F., Koroshetz W.J. Striatal volume loss in HD as measured by MRI and the influence of CAG repeat. Neurology. 2001;57(6):1025–1028. doi: 10.1212/wnl.57.6.1025. [DOI] [PubMed] [Google Scholar]
- Rosas H.D., Tuch D.S., Hevelone N.D., Zaleta A.K., Vangel M., Hersch S.M., Salat D.H. Diffusion tensor imaging in presymptomatic and early Huntington's disease: selective white matter pathology and its relationship to clinical measures. Mov. Disord. 2006;21(9):1317–1325. doi: 10.1002/mds.20979. [DOI] [PubMed] [Google Scholar]
- Ross C.A., Aylward E.H., Wild E.J., Langbehn D.R., Long J.D., Warner J.H., Scahill R.I., Leavitt B.R., Stout J.C., Paulsen J.S., Reilmann R., Unschuld P.G., Wexler A., Margolis R.L., Tabrizi S.J. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol. 2014;10(4):204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- Ross C.A., Becher M.W., Colomer V., Engelender S., Wood J.D., Sharp A.H. Huntington's disease and dentatorubral-pallidoluysian atrophy: proteins, pathogenesis and pathology. Brain Pathol. 1997;7(3):1003–1016. doi: 10.1111/j.1750-3639.1997.tb00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Castaneda C., Cherubini A., Elifani F., Peran P., Orobello S., Capelli G., Sabatini U., Squitieri F. Seeking Huntington disease biomarkers by multimodal, cross-sectional basal ganglia imaging. Hum. Brain Mapp. 2013;34(7):1625–1635. doi: 10.1002/hbm.22019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers D., Sheldon S., Kuperman J.M., Goldstein J., Corey-Bloom J., Aron A.R. Contrasting gray and white matter changes in preclinical Huntington disease: an MRI study. Neurology. 2010;74(15):1208–1216. doi: 10.1212/WNL.0b013e3181d8c20a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi S.J., Langbehn D.R., Leavitt B.R., Roos R.A., Durr A., Craufurd D., Kennard C., Hicks S.L., Fox N.C., Scahill R.I., Borowsky B., Tobin A.J., Rosas H.D., Johnson H., Reilmann R., Landwehrmeyer B., Stout J.C. Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8(9):791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi S.J., Reilmann R., Roos R.A., Durr A., Leavitt B., Owen G., Jones R., Johnson H., Craufurd D., Hicks S.L., Kennard C., Landwehrmeyer B., Stout J.C., Borowsky B., Scahill R.I., Frost C., Langbehn D.R. Potential endpoints for clinical trials in premanifest and early Huntington's disease in the TRACK-HD study: analysis of 24 month observational data. Lancet Neurol. 2012;11(1):42–53. doi: 10.1016/S1474-4422(11)70263-0. [DOI] [PubMed] [Google Scholar]
- Tabrizi S.J., Scahill R.I., Owen G., Durr A., Leavitt B.R., Roos R.A., Borowsky B., Landwehrmeyer B., Frost C., Johnson H., Craufurd D., Reilmann R., Stout J.C., Langbehn D.R. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington's disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013;12(7):637–649. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- The Huntington's Disease Group Unified Huntington's disease rating scale: reliability and consistency. Mov. Disord. 1996;11(2):136–142. doi: 10.1002/mds.870110204. [DOI] [PubMed] [Google Scholar]
- Thompson P.M., Hayashi K.M., De Zubicaray G.I., Janke A.L., Rose S.E., Semple J., Hong M.S., Herman D.H., Gravano D., Doddrell D.M., Toga A.W. Mapping hippocampal and ventricular change in Alzheimer disease. NeuroImage. 2004;22(4):1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Vaillant M., Glaunes J. Surface matching via currents. LNCS. 2005;3565:381–392. doi: 10.1007/11505730_32. [DOI] [PubMed] [Google Scholar]
- Van Camp N., Blockx I., Camon L., de Vera N., Verhoye M., Veraart J., Van Hecke W., Martinez E., Soria G., Sijbers J., Planas A.M., Van der Linden A. A complementary diffusion tensor imaging (DTI)-histological study in a model of Huntington's disease. Neurobiol. Aging. 2012;33(5):945–959. doi: 10.1016/j.neurobiolaging.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Vandenberghe W., Demaerel P., Dom R., Maes F. Diffusion-weighted versus volumetric imaging of the striatum in early symptomatic Huntington disease. J. Neurol. 2009;256(1):109–114. doi: 10.1007/s00415-009-0086-0. [DOI] [PubMed] [Google Scholar]
- Wang L., Beg F., Ratnanather T., Ceritoglu C., Younes L., Morris J.C., Csernansky J.G., Miller M.I. Large deformation diffeomorphism and momentum based hippocampal shape discrimination in dementia of the Alzheimer type. IEEE Trans. Med. Imaging. 2007;26(4):462–470. doi: 10.1109/TMI.2005.853923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes L., Albert M., Miller M.I. Inferring changepoint times of medial temporal lobe morphometric change in preclinical Alzheimer's disease. Neuroimage Clin. 2014;5:178–187. doi: 10.1016/j.nicl.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes L., Ratnanather J.T., Brown T., Aylward E., Nopoulos P., Johnson H., Magnotta V.A., Paulsen J.S., Margolis R.L., Albin R.L., Miller M.I., Ross C.A. Regionally selective atrophy of subcortical structures in prodromal HD as revealed by statistical shape analysis. Hum. Brain Mapp. 2013 doi: 10.1002/hbm.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes L., Ratnanather J.T., Brown T., Aylward E., Nopoulos P., Johnson H., Magnotta V.A., Paulsen J.S., Margolis R.L., Albin R.L., Miller M.I., Ross C.A. Regionally selective atrophy of subcortical structures in prodromal HD as revealed by statistical shape analysis. Hum. Brain Mapp. 2014;35(3):792–809. doi: 10.1002/hbm.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young Kim E., Johnson H.J. Robust multi-site MR data processing: iterative optimization of bias correction, tissue classification, and registration. Front. Neuroinform. 2013;7:29. doi: 10.3389/fninf.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Long J.D., Mills J.A., Warner J.H., Lu W., Paulsen J.S. Indexing disease progression at study entry with individuals at-risk for Huntington disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011;156b(7):751–763. doi: 10.1002/ajmg.b.31232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P-values for deformation markers on each structure corrected for multiple comparisons via permutation tests. *p-value < 0.000025.
MD values in the white matter ROIs defined in the neighborhood of the deep gray matter.
MD values in the white matter (wm) ROIs defined by the automated whole-brain analysis.