Abstract
The extracellular matrix (ECM) is a highly organized multimolecular structure, essential for life in higher organisms. Although substantial high-resolution structural information is available for relatively small fragments of ECM components, the inherent difficulty in preparing and analyzing samples of large, fibrous polymers impedes structural efforts. Here, we review recent advances in understanding the structure of three important ECM components: collagen, fibrillin and fibronectin. Emphasis is placed on the key role of intermolecular interactions in assembling larger, μm scale, structures.
Introduction
The extracellular matrix (ECM) is a complex network of glycoproteins and proteoglycans that originated with the advent of multicellular organisms [1]. Cells generate well-ordered ECM-complexes to surround and support themselves. The ECM then plays an essential role in the survival, migration and proliferation of these adjoining cells. Continuous ECM remodeling, catalyzed by various degradation enzymes, is common, and the arrangement and concentration of different macromolecules gives rise to a wide diversity of ECM forms in the various tissue types (skin, bone, cornea of the eye, etc.) [2]. The structure of the constituent polymers is rather well known at the domain or fragment level but is less well known at the levels of intact molecule or higher. The monomeric subunits are large, multi-domain and often inherently flexible, thus presenting problems to atomic resolution techniques such as single crystal diffraction or NMR. The polymers formed retain substantial heterogeneity and are cross-linked and difficult to extract from the matrix in undamaged form. Although progress is relatively slow, new approaches and tools are beginning to have an impact.
In this brief review, we have chosen to illustrate this field by discussing recent structural studies and current understanding of three archetypal ECM proteins: collagen (the most abundant protein in mammals), fibrillin and fibronectin. The first is a biopolymer of characteristic amino acid sequence, while the last two are modular proteins, constructed from repeating, autonomously folding domains with a high degree of structural similarity [3]. A major remaining structural problem is to define the various inter and intramolecular interactions made by molecules, especially in the context of structure at the μm level. We appreciate that this selection, with only three proteins, neglects many other important ECM molecules and provides only a part of the picture, however, space is limited. A particular area of neglect is polysaccharides, such as hyaluronan [4] which, with its receptors [5], plays a pivotal role in ECM hydration and elasticity.
Collagen
Collagen has a characteristic three residue repeat, Gly-Xaa-Yaa, in its primary structure, which results in a stable triple-helical conformation with the glycine residues at the core of the helix [6-8]. Proline and 4-hydroxyproline residues, usually found in positions Xaa and Yaa, function to stabilize the three individual polyproline II-like helices. After post-translational modification, secreted collagen helices self-assemble to cross-linked microfibrils and eventually μm-long fibrils. This spontaneous process creates large-scale molecular structures with properties of obvious interest to bioengineers [9]. Biology exploits the exceptional mechanical properties of collagen but it also uses it as a scaffold to attach a number of binding proteins to specific sites [10•].
The most abundant collagens are types I, II, and III, found in a range of tissues including tendon and skin; these form characteristic fibrils with identifiable repeat bands separated by 67 nm. These periodic patterns are still not very well understood but were early suggested to be related to the arrangement of triple-helices in fibrils and the inherent periodicity in the collagen primary structure, yielding five so-called D-periods [11-13]. The ability to generate recombinant collagens with defined composition is beginning to have an impact in structural studies; for example, there is evidence, from mutagenesis [14•] and thermostability experiments [15•], of unique domain-like characteristics in collagen type II and a recent study showed that some of these D-periods are in fact dispensable for banded fibril formation [16•].
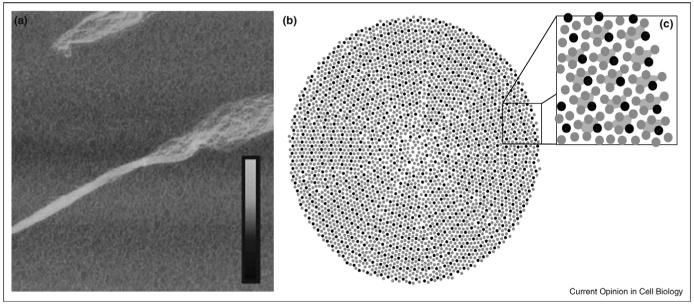
New structural observations are also helping to achieve a better idea of how collagen fibrils are assembled. Using contact-mode atomic force microscopy Bozec et al. [17••] proposed a model for collagen where individual triple-helices, or, possibly, microfibrils, twist around each other along the long axis of the fibril. This concept is comparable to a nanoscale rope composed of multiple twisted fibers, in agreement with past suggestions and observations (Figure 1a). Similar conclusions were reached in perhaps the most detailed study of fibrils so far [18••] with Orgel et al. investigating the structure of crystallite elements of collagen type I fibrils in situ using X-ray fiber diffraction techniques; a low resolution electron density map was obtained that allowed main chain tracking and some amino acid identification. Individual microfibrils were shown to adopt a right handed supertwist and to interdigitate with neighboring microfibrils. The overall packing is similar to the proposed quasihexagonally packed liquid-crystal collagen model (Figure 1b and c) [19] with intermolecular interactions involving the collagen N- and C-telopeptides critical in maintaining this arrangement, a result supported by computation [20,21].
Figure 1.
Supramolecular organization of collagen fibrils. (a) The superhelical twist of individual fibrillar elements can be seen in this atomic force microscopy image of a mechanically disrupted collagen fibril (ref. [51], reprinted with permission of Wiley-Liss, Inc., a subsidiary of John Wiley & Sons, Inc. Copyright Wiley-Liss, 2006). The box size is 5 μm × 5 μm and the inset height scale corresponds to 0-30 nm. (b-c) Cross-section model of molecular packing in collagen fibrils (adapted with permission from ref. [19]. Copyright Elsevier, 2002). Thousands of individual collagen triple-helices interact to form a single fibril with both ordered and disordered packing features. Collagen microfibrils are formed by five collagen molecules in a staggered arrangement, shown connected by trapezoids in (c).
Fibrillin
Fibrillin-1 is a large (350 kDa) multidomain glycoprotein that forms the major structural component of 10-12 nm elastic microfibrils in the ECM [22,23]. With elastin, it provides the necessary elasticity and resilience of a variety of tissues. A large number of matrix components that interact with fibrillin-1 have been identified, including integrins [24•], heparin [25•], latent transforming growth factor β-binding proteins and fibulin-2 [26]. These, together with homotypic fibrillin interactions, are likely to regulate microfibril assembly [27•]. Fibrillin-1 contains 47 epidermal growth factor-like (EGF) domains interspersed with seven transforming growth factor β-binding protein-like domains; 43 of the EGF domains are expected to bind calcium. Structural studies of fibrillin at the domain level are relatively advanced; a ‘dissection’ approach has been used to study recombinantly expressed domains a few at a time (e.g. [24•]). Ca2+-binding to EGF domains is believed to affect the large-scale fibrillin structure, a hypothesis extensively tested at the level of EGF domains [28] and fragments [29] using wildtype protein and mutations that are known to cause the Marfan syndrome.
Guanidine-extracted fibrillin microfibrils have a ‘beads on a string’ appearance, with approximate 57 nm periodicity when viewed by rotary shadowing electron microscopy (EM) [30] (Figure 2a). These periodic features of approximately 2.5 MDa mass are conserved between different tissue types [31] and, thus, can be mainly attributed to fibrillin molecules. The important issue of how microbrils are constructed from fibrillin molecules is not yet resolved. Several possible models have been proposed to explain the observed periodic pattern, including different orientations (parallel or antiparallel), globular (‘pleated’), non-globular, staggered or non-staggered arrangements of fibrillin molecules [30].
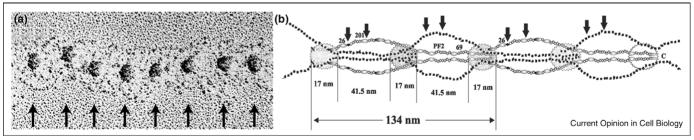
Figure 2.
Proposed model of fibrillin microfibrils (adapted with permission from reference [34••], Copyright The American Society for Biochemistry and Molecular Biology, 2007). (a) Rotary shadowed micrographs of guanidine-extracted microfibrils show characteristic repeats of approximately 57 nm (indicated by arrows). (b) A possible model of microfibril structure involves staggered, parallel molecules of fibrillin-1 with the N-terminal halves on the outside of the microfibril and C-terminal halves in the core. Individual fibrillin molecules extend over multiple sequential microfibril repeats. Antibody epitopes (mAb 26, mAb 201, and mAb 69) are labeled, and major collagenase cleavage sites are marked with arrows.
A number of recent studies have addressed microfibril structure by different means. EM image analysis was used on samples extracted by collagenases [32••], giving data that supported a parallel, ‘pleated’ model, with fibrillin molecules occupying a single microfibril repeat. A similar model was assumed in a different study [33••] where recombinantly expressed fibrillin fragments were analyzed by small-angle X-ray scattering and EM single particle reconstruction. In contrast, a recent study of EM images of microfibrillar extracts analyzed by proteolytic degradation and antibody epitope mapping yielded data in support of a non-pleated, staggered model [34••], with individual fibrillin molecules extending over multiple repeats (Figure 2b). Although these published models still differ significantly, it is encouraging that they seem to be more convergent than previous ones. Some of the observed uncertainties and differences, for example the different extensibility of the extracted microfibril [34••], almost certainly arise from use of different extraction procedures.
Fibronectin
Fibronectin (FN) is a large dimeric plasma glycoprotein found only in vertebrates. FN is composed of three different domain types, FNI, FNII and FNIII, high-resolution structures of which have been available for some time [35]. A tightly controlled process transforms plasma FN to a fibrillar form within the ECM. Little is known about the fibrillogenesis process, or the structure of the fibrillar matrix formed. It is, however, believed that FN interdomain interactions [36-41,42•] and tension exerted through cell-surface receptors [43] are essential. Some of the association sites detected are cryptic [37,39], as interactions can only form after molecular rearrangement, possibly induced by FN stretching. Fibrillar FN has many binding partners including integrin cell-surface receptors, heparin, collagen, fibrin and fibulin [43]. It also has remarkable mechanical properties, including the ability to be stretched as much as fourfold by living cells [44].
Two different mechanisms have been proposed to explain this observed elasticity [45]. The first, postulates that multiple FNIII domains are reversibly unfolded under tension, hence the extension corresponds to stretching of the polypeptide chain [42•,46]. In contrast, the second mechanism assumes that FN extension is because of dissociation of weak inter-domain interactions, many of which involve the first two FNIII domains. In this case, FN extension involves FN transformation from a compact to an extended state. Although the domain-unfolding mechanism has been shown to be important in superfibronectin, a non-physiological FN aggregate [47•], two recent studies present data that favour the second mechanism.
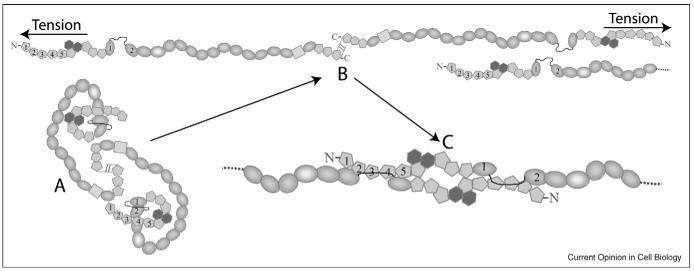
Abu-Lail et al. [48••] showed that FNIII and green fluorescent protein (GFP) domains have very similar mechanical unfolding strengths by single molecule force spectroscopy, and earlier studies showed that FN fibrils with embedded GFP domains maintain fluorescence under stretching in cell cultures [49••]. The fact that cell-tension does not unfold GFP domains or, by implication, FNIII domains, supports the domain-rearrangement mechanism. A separate study showed that disruption of a complex formed by the first two FNIII domains unveils a novel cryptic association site in the interdomain linker [50••]. This disrupted, ‘open’ state (1-2FNIIIopen) displays dramatically increased affinity to nM levels for the FN N-terminal domains (FN 30 kDa). Thus, a model was proposed where in vivo 1-2FNIIIopen is reproduced by cell-generated tension, leading to formation of a tight complex between two FN N-termini and the creation of a FN protofibril (Figure 3). This proposed mechanism could, in principle, be a general way of producing tension-induced signals.
Figure 3.
Proposed model of fibronectin fibrillogenesis [50••]. (A) FN exists in a globular, soluble state in plasma. The interdomain interactions defining this state are disrupted by cell-generated tension (B). (C) The N-termini of two extended FN molecules form a tight complex through the FN 30 kDa-1-2FNIIIopen interaction, thereby creating FN protofibrils.
Conclusion
In all three ECM molecules visited, association interactions are the key to understand higher order structures. Intermolecular interactions define the packing order of collagen [18••], inter-domain interactions have a major role in models of fibrillin [27•] (whether ‘pleated’ or staggered) and interdomain association sites lead directly to FN fibrillogenesis models [50••]. Detailed structural descriptions of module interactions are, comparatively, fewer than structures of individual modules [3]. However, increasing attention to extraction procedures, new structural tools and an ability to express a variety of defined recombinant molecules can lead to a much better understanding of how the network of large, cross-linked molecules are laid down in the insoluble ECM by a series of precise association events.
Acknowledgement
Financial support was provided by the Wellcome Trust.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. edn 4. Garland; London: 2002. [Google Scholar]
- 2.Werb Z, Chin JR. Extracellular matrix remodeling during morphogenesis. Ann N Y Acad Sci. 1998;857:110–118. doi: 10.1111/j.1749-6632.1998.tb10111.x. [DOI] [PubMed] [Google Scholar]
- 3.Hohenester E, Engel J. Domain structure and organisation in extracellular matrix proteins. Matrix Biol. 2002;21:115–128. doi: 10.1016/s0945-053x(01)00191-3. [DOI] [PubMed] [Google Scholar]
- 4.Laurent TC, Laurent UB, Fraser JR. The structure and function of hyaluronan: An overview. Immunol Cell Biol. 1996;74:A1–A7. doi: 10.1038/icb.1996.32. [DOI] [PubMed] [Google Scholar]
- 5.Banerji S, Wright AJ, Noble M, Mahoney DJ, Campbell ID, Day AJ, Jackson DG. Structures of the Cd44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat Struct Mol Biol. 2007;14:234–239. doi: 10.1038/nsmb1201. [DOI] [PubMed] [Google Scholar]
- 6.Mayo KH. NMR and X-ray studies of collagen model peptides. Biopolymers. 1996;40:359–370. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C359::AID-BIP2%3E3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 7.Brodsky B, Persikov AV. Molecular structure of the collagen triple helix. Adv Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- 8.Okuyama K, Xu X, Iguchi M, Noguchi K. Revision of collagen molecular structure. Biopolymers. 2006;84:181–191. doi: 10.1002/bip.20381. [DOI] [PubMed] [Google Scholar]
- 9.Kotch FW, Raines RT. Self-assembly of synthetic collagen triple helices. Proc Natl Acad Sci U S A. 2006;103:3028–3033. doi: 10.1073/pnas.0508783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Leitinger B, Hohenester E. Mammalian collagen receptors. Matrix Biol. 2006;26:146–155. doi: 10.1016/j.matbio.2006.10.007. [An excellent recent review on collagen receptors.] [DOI] [PubMed] [Google Scholar]
- 11.Chapman JA, Hardcastle RA. The staining pattern of collagen fibrils. II. A comparison with patterns computer-generated from the amino acid sequence. Connect Tissue Res. 1974;2:151–159. doi: 10.3109/03008207409152100. [DOI] [PubMed] [Google Scholar]
- 12.Doyle BB, Hulmes DJ, Miller A, Parry DA, Piez KA, Woodhead-Galloway J. Axially projected collagen structures. Proc R Soc Lond B Biol Sci. 1974;187:37–46. doi: 10.1098/rspb.1974.0059. [DOI] [PubMed] [Google Scholar]
- 13.Hulmes DJ, Miller A, Parry DA, Piez KA, Woodhead-Galloway J. Analysis of the primary structure of collagen for the origins of molecular packing. J Mol Biol. 1973;79:137–148. doi: 10.1016/0022-2836(73)90275-1. [DOI] [PubMed] [Google Scholar]
- 14•.Steplewski A, Ito H, Rucker E, Brittingham RJ, Alabyeva T, Gandhi M, Ko FK, Birk DE, Jimenez SA, Fertala A. Position of single amino acid substitutions in the collagen triple helix determines their effect on structure of collagen fibrils. J Struct Biol. 2004;148:326–337. doi: 10.1016/j.jsb.2004.07.006. [See annotation to [16•].] [DOI] [PubMed] [Google Scholar]
- 15•.Steplewski A, Majsterek I, McAdams E, Rucker E, Brittingham RJ, Ito H, Hirai K, Adachi E, Jimenez SA, Fertala A. Thermostability gradient in the collagen triple helix reveals its multi-domain structure. J Mol Biol. 2004;338:989–998. doi: 10.1016/j.jmb.2004.03.037. [See annotation to [16•].] [DOI] [PubMed] [Google Scholar]
- 16•.Steplewski A, Hintze V, Fertala A. Molecular basis of organization of collagen fibrils. J Struct Biol. 2007;157:297–307. doi: 10.1016/j.jsb.2006.10.009. [Collagen D-periods are shown to have distinguishable domain-like properties, such as thermal stability and tolerance for amino acid substitutions. Some of these domain-periods are, however, dispensable for fibril formation. Collectively, these three papers [14•,15•,16•] illustrate the power of recombinant methods in unraveling the structure of a complex system like the collagen fibril.] [DOI] [PubMed] [Google Scholar]
- 17••.Bozec L, van der Heijden G, Horton M. Collagen fibrils: nanoscale ropes. Biophys J. 2007;92:70–75. doi: 10.1529/biophysj.106.085704. [This study uses AFM on tendon collagen fibrils to show that the observed D-periods can be explained in terms of a spiral arrangement of fibrillar subcomponents, conceptually similar to rope construction.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Orgel JP, Irving TC, Miller A, Wess TJ. Microfibrillar structure of type I collagen in situ. Proc Natl Acad Sci U S A. 2006;103:9001–9005. doi: 10.1073/pnas.0502718103. [X-ray fiber diffraction experiments on tendon collagen produced an interpretable low resolution electron density map of crystalline elements in the fibril. Collagen microfibrils interdigitate and adopt a right-handed supertwist along the long axis of the fibril. Intermolecular interactions along the collagen termini maintain the overall packing arrangement.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulmes DJ. Building collagen molecules, fibrils, and suprafibrillar structures. J Struct Biol. 2002;137:2–10. doi: 10.1006/jsbi.2002.4450. [DOI] [PubMed] [Google Scholar]
- 20.Malone JP, Veis A. Heterotrimeric type I collagen C-telopeptide conformation as docked to its helix receptor. Biochemistry. 2004;43:15358–15366. doi: 10.1021/bi048304b. [DOI] [PubMed] [Google Scholar]
- 21.Malone JP, George A, Veis A. Type I collagen N-telopeptides adopt an ordered structure when docked to their helix receptor during fibrillogenesis. Proteins. 2004;54:206–215. doi: 10.1002/prot.10526. [DOI] [PubMed] [Google Scholar]
- 22.Jordan CD, Charbonneau NL, Sakai LY. Fibrillin microfibrils: connective tissue pathways that regulate shape and signaling. J Musculoskelet Neuronal Interact. 2006;6:366–367. [PubMed] [Google Scholar]
- 23.Whiteman P, Hutchinson S, Handford PA. Fibrillin-1 misfolding and disease. Antioxid Redox Signal. 2006;8:338–346. doi: 10.1089/ars.2006.8.338. [DOI] [PubMed] [Google Scholar]
- 24•.Lee SS, Knott V, Jovanovic J, Harlos K, Grimes JM, Choulier L, Mardon HJ, Stuart DI, Handford PA. Structure of the integrin binding fragment from fibrillin-1 gives new insights into microfibril organization. Structure. 2004;12:717–729. doi: 10.1016/j.str.2004.02.023. [The crystal structure of the EGF22-TB4-EGF23 fragment of fibrillin-1 shows substantial interdomain interactions mediated by the TB domain. This study suggests that other TB domains in fibrillin may function in a similar role.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Cain SA, Baldock C, Gallagher J, Morgan A, Bax DV, Weiss AS, Shuttleworth CA, Kielty CM. Fibrillin-1 interactions with heparin. Implications for microfibril and elastic fiber assembly. J Biol Chem. 2005;280:30526–30537. doi: 10.1074/jbc.M501390200. [Surface plasmon resonance experiments show the existence of four heparin-binding sites in fibrillin-1. Heparin competes for binding with other fibrillin partners, indicating a likely regulatory role in microfibril assembly.] [DOI] [PubMed] [Google Scholar]
- 26.El-Hallous E, Sasaki T, Hubmacher D, Getie M, Tiedemann K, Brinckmann J, Batge B, Davis EC, Reinhardt DP. Fibrillin-1 interactions with fibulins depend on the first hybrid domain and provide an adaptor function to tropoelastin. J Biol Chem. 2007;282:8935–8946. doi: 10.1074/jbc.M608204200. [DOI] [PubMed] [Google Scholar]
- 27•.Marson A, Rock MJ, Cain SA, Freeman LJ, Morgan A, Mellody K, Shuttleworth CA, Baldock C, Kielty CM. Homotypic fibrillin-1 interactions in microfibril assembly. J Biol Chem. 2005;280:5013–5021. doi: 10.1074/jbc.M409029200. [A solid phase binding assay revealed three homotypic interactions in fibrillin-1, involving the N- and C-termini of the protein. A regulatory role in microfibril assembly is suggested.] [DOI] [PubMed] [Google Scholar]
- 28.Whiteman P, Willis AC, Warner A, Brown J, Redfield C, Handford PA. Cellular and molecular studies of Marfan syndrome mutations identify co-operative protein folding in the cbEGF12-13 region of fibrillin-1. Hum Mol Genet. 2007;16:907–918. doi: 10.1093/hmg/ddm035. [DOI] [PubMed] [Google Scholar]
- 29.Mellody KT, Freeman LJ, Baldock C, Jowitt TA, Siegler V, Raynal BD, Cain SA, Wess TJ, Shuttleworth CA, Kielty CM. Marfan syndrome-causing mutations in fibrillin-1 result in gross morphological alterations and highlight the structural importance of the second hybrid domain. J Biol Chem. 2006;281:31854–31862. doi: 10.1074/jbc.M602743200. [DOI] [PubMed] [Google Scholar]
- 30.Handford PA, Downing AK, Reinhardt DP, Sakai LY. Fibrillin: from domain structure to supramolecular assembly. Matrix Biol. 2000;19:457–470. doi: 10.1016/s0945-053x(00)00100-1. [DOI] [PubMed] [Google Scholar]
- 31.Lu Y, Sherratt MJ, Wang MC, Baldock C. Tissue specific differences in fibrillin microfibrils analysed using single particle image analysis. J Struct Biol. 2006;155:285–293. doi: 10.1016/j.jsb.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 32••.Lu Y, Holmes DF, Baldock C. Evidence for the intramolecular pleating model of fibrillin microfibril organisation from single particle image analysis. J Mol Biol. 2005;349:73–85. doi: 10.1016/j.jmb.2005.03.066. [This study shows that the intramolecular ‘pleating’ model of fibrillin-1 microfibril organization provides the best fit for stain exclusion patterns in EM micrographs. This model was then used to arrange in space single particle reconstructions of fibrillin-1 fragments in [33••].] [DOI] [PubMed] [Google Scholar]
- 33••.Baldock C, Siegler V, Bax DV, Cain SA, Mellody KT, Marson A, Haston JL, Berry R, Wang MC, Grossmann JG, et al. Nanostructure of fibrillin-1 reveals compact conformation of EGF arrays and mechanism for extensibility. Proc Natl Acad Sci U S A. 2006;103:11922–11927. doi: 10.1073/pnas.0601609103. [See annotation to [32••].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Kuo CL, Isogai Z, Keene DR, Hazeki N, Ono RN, Sengle G, Peter Bachinger H, Sakai LY. Effects of fibrillin-1 degradation on microfibril ultrastructure. J Biol Chem. 2007;282:4007–4020. doi: 10.1074/jbc.M606370200. [Analysis of antibody epitopes and collagenase cleavage sites in the fibrillin-1 microfibril monitored by EM supports a staggered model of microfibril organization. Significant differences in the properties of the microfibril were observed depending on the extraction method employed.] [DOI] [PubMed] [Google Scholar]
- 35.Potts JR, Campbell ID. Structure and function of fibronectin modules. Matrix Biol. 1996;15:313–320. doi: 10.1016/s0945-053x(96)90133-x. discussion 321. [DOI] [PubMed] [Google Scholar]
- 36.Morla A, Ruoslahti E. A fibronectin self-assembly site involved in fibronectin matrix assembly: reconstruction in a synthetic peptide. J Cell Biol. 1992;118:421–429. doi: 10.1083/jcb.118.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingham KC, Brew SA, Huff S, Litvinovich SV. Cryptic self-association sites in type III modules of fibronectin. J Biol Chem. 1997;272:1718–1724. doi: 10.1074/jbc.272.3.1718. [DOI] [PubMed] [Google Scholar]
- 38.Aguirre KM, McCormick RJ, Schwarzbauer JE. Fibronectin self-association is mediated by complementary sites within the amino-terminal one-third of the molecule. J Biol Chem. 1994;269:27863–27868. [PubMed] [Google Scholar]
- 39.Hocking DC, Sottile J, McKeown-Longo PJ. Fibronectin’s III-1 module contains a conformation-dependent binding site for the amino-terminal region of fibronectin. J Biol Chem. 1994;269:19183–19187. [PubMed] [Google Scholar]
- 40.Litvinovich SV, Ingham KC. Interactions between type III domains in the 110 kDa cell-binding fragment of fibronectin. J Mol Biol. 1995;248:611–626. doi: 10.1006/jmbi.1995.0246. [DOI] [PubMed] [Google Scholar]
- 41.Sechler JL, Rao H, Cumiskey AM, Vega-Colon I, Smith MS, Murata T, Schwarzbauer JE. A novel fibronectin binding site required for fibronectin fibril growth during matrix assembly. J Cell Biol. 2001;154:1081–1088. doi: 10.1083/jcb.200102034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J Mol Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [The stability of different FN type III domains to mechanical unfolding was shown to be different by more that two fold in multidomain protein constructs. Constructs involving the first two type III domains indicated the presence of a stabilizing interaction between them.] [DOI] [PubMed] [Google Scholar]
- 43.Mao Y, Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24:389–399. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix-cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 45.Erickson HP. Stretching fibronectin. J Muscle Res Cell Motil. 2002;23:575–580. doi: 10.1023/a:1023427026818. [DOI] [PubMed] [Google Scholar]
- 46.Erickson HP. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc Natl Acad Sci U S A. 1994;91:10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Ohashi T, Erickson HP. Domain unfolding plays a role in superfibronectin formation. J Biol Chem. 2005;280:39143–39151. doi: 10.1074/jbc.M509082200. [Formation of superfibronectin catalyzed by anastellin is shown to involve a conformational rearrangement of the third type III domain of FN. An engineered disulphide crosslink preventing unfolding in this domain inhibited this catalysis.] [DOI] [PubMed] [Google Scholar]
- 48••.Abu-Lail NI, Ohashi T, Clark RL, Erickson HP, Zauscher S. Understanding the elasticity of fibronectin fibrils: unfolding strengths of FN-III and GFP domains measured by single molecule force spectroscopy. Matrix Biol. 2006;25:175–184. doi: 10.1016/j.matbio.2005.10.007. [Single molecule AFM showed that the mechanical unfolding strengths of GFP and FN type III domains are very similar over a large range of pulling speeds. Previously [49••], GFP-labeled FN was shown to retain fluorescence under tension in cell-stretching assays, indicating that type III domains are not mechanically unfolded under these conditions.] [DOI] [PubMed] [Google Scholar]
- 49••.Ohashi T, Kiehart DP, Erickson HP. Dual labeling of the fibronectin matrix and actin cytoskeleton with green fluorescent protein variants. J Cell Sci. 2002;115:1221–1229. doi: 10.1242/jcs.115.6.1221. [See annotation for [48••].] [DOI] [PubMed] [Google Scholar]
- 50••.Vakonakis I, Staunton D, Rooney LM, Campbell ID. Interdomain association in fibronectin: insight into cryptic sites and fibrillogenesis. EMBO J. 2007;26:2575–2583. doi: 10.1038/sj.emboj.7601694. [The first two type III domains of FN form a complex as shown by NMR. Disruption of this complex unveils a high-affinity association site for the FN N-terminus. This study suggests that a similar event in vivo could lead to FN protofibrils.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen CK, Goh MC. Fibrous long spacing type collagen fibrils have a hierarchical internal structure. Proteins. 2006;64:227–233. doi: 10.1002/prot.20949. [DOI] [PubMed] [Google Scholar]