Abstract
JAM-A belongs to a family of immunoglobulin-like proteins called junctional adhesion molecules (JAMs) that localize at epithelial and endothelial intercellular tight junctions. JAM-A is also expressed on dendritic cells, neutrophils, and platelets. Homophilic JAM-A interactions play an important role in regulating paracellular permeability and leukocyte transmigration across epithelial monolayers and endothelial cell junctions, respectively. In addition, JAM-A is a receptor for the reovirus attachment protein, σ1. In this study, we used single molecular force spectroscopy to compare the kinetics of JAM-A interactions with itself and σ1. A chimeric murine JAM-A/Fc fusion protein and the purified σ1 head domain were used to probe murine L929 cells, which express JAM-A and are susceptible to reovirus infection. The bond half-life (t1/2) of homophilic JAM-A interactions was found to be shorter than that of σ1/JAM-A interactions . These results are in accordance with the physiological functions of JAM-A and σ1. A short bond lifetime imparts a highly dynamic nature to homophilic JAM-A interactions for regulating tight junction permeability while stable interactions between σ1 and JAM-A likely anchor the virus to the cell surface and facilitate viral entry.
Keywords: molecular force spectroscopy, atomic force microscopy (AFM), junctional adhesion molecule-A (JAM-A), reovirus attachment protein (sigma1)
INTRODUCTION
Junctional adhesion molecules (JAMs) are proteins that localize at intercellular tight junctions along with occludin and claudins (Martin-Padura et al., 1998). Apart from endothelial cells and epithelial cells, JAM family members are expressed on leukocytes and platelets (Cunningham et al., 2000; Sobocka et al., 2000). JAMs belong to the immunoglobulin (Ig) superfamily and are implicated in tight junction formation (Liu et al., 2000), monocyte transmigration (Martin-Padura et al., 1998), platelet activation (Sobocka et al., 2000), angiogenesis (Naik et al., 2003; Cooke et al., 2006), and attachment of mammalian reovirus (Barton et al., 2001b; Campbell et al., 2005). The JAM family includes JAM-A, JAM-B, JAM-C, JAM-4, and JAML proteins.
Structurally, all JAM proteins contain an N-terminal signal peptide, an extracellular region composed of two Ig-like domains (a membrane-distal, N-terminal D1 domain and a membrane-proximal, C-terminal D2 domain), a single membrane-spanning domain and a short cytoplasmic tail (Kostrewa et al., 2001). The cytoplasmic tail interacts with PDZ domain-containing scaffolding proteins including ZO-1, while the D1 domain interacts with the D1 domain of an opposing JAM-A molecule to form physiologically relevant homodimers (Ebnet et al., 2000; Kostrewa et al., 2001).
JAM-A was first discovered as an antigen on platelets for the F11 monoclonal antibody; engagement of platelets by F11 mediates granule release, fibrinogen binding, and aggregation (Kornecki et al., 1990). JAM-A was subsequently found to localize at regions of intercellular contact in epithelial and endothelial cell tight junctions (Martin-Padura et al., 1998). While JAM-A is capable of undergoing only homophilic interactions within the JAM family, JAM-B and JAM-C are capable of both homophilic and heterophilic interactions with each other (Liang et al., 2002). Support for JAM-A-mediated homophilic adhesion comes from the observation that transfected CHO cells show localization of JAM-A to regions of cell–cell contact formed between transfected cells (Naik et al., 2001). Inhibition of monocyte transmigration by monoclonal antibody BV11, which inhibits homophilic JAM-A interactions, strongly suggests that JAM-A-mediated interactions between monocytes and endothelial cells are important for transmigration (Martin-Padura et al., 1998). JAM-A can also undergo heterophilic interaction with leukocyte function associated antigen-1 (LFA-1, αLβ2 integrin) expressed on neutrophils and T lymphocytes (Ostermann et al., 2002). These findings suggest that a complex interaction of both heterophilic and homophilic interactions mediated by JAM-A facilitate the transmigration process. Furthermore, antibodies binding to the JAM-A homodimer interface have also been shown to delay the recovery of transepithelial resistance, highlighting the importance of homophilic JAM-A interactions in regulating the tight junction barrier (Liu et al., 2000; Mandell et al., 2004). Despite significant biochemical evidence for JAM-A homophilic interactions, neither the strength nor the kinetics of these interactions are well characterized.
In addition to its physiological functions, JAM-A is a receptor for each of the three known serotypes of mammalian orthoreovirus (reovirus) (Barton et al., 2001b; Campbell et al., 2005), an important experimental model for studies of viral pathogenesis (Schiff et al., 2007). Both murine and human JAM-A proteins serve as receptors for reovirus (Barton et al., 2001b). Reovirus engages JAM-A via the σ1 protein, a filamentous trimer with an N-terminal tail and a C-terminal head (Chappell et al., 2002). The head domain of σ1 binds to JAM-A (Chappell et al., 2000; Barton et al., 2001b), while the tail domain recognizes cell-surface carbohydrate (Chappell et al., 2000; Barton et al., 2001a), which is sialic acid for serotype three strains (Barton et al., 2001a). The kinetic properties of σ1 interactions with JAM-A and several JAM-A mutants have been defined using surface plasmon resonance (Guglielmi et al., 2007). However, neither the interaction forces nor the kinetics of σ1/JAM-A interactions at the level of single molecules have been reported.
Atomic force microscopy (AFM) has permitted the characterization of interaction forces between ligands and receptors at the level of single molecules. This application of AFM, also called single molecule force spectroscopy, has significantly advanced an understanding of interactions between several types of cell-adhesion molecules such as E-cadherin, selectins, and some viral proteins (Baumgartner et al., 2000; Hanley et al., 2003; Chang et al., 2005). AFM also has an advantage over some other systems in that the receptors of interest can be probed in their natural state on the cell surface.
In this study, we applied single molecule force spectroscopy to define the interaction strength and kinetics of homophilic murine JAM-A (mJAM-A) interactions and mJAM-A/σ1 interactions. A chimeric mJAM-A/Fc fusion protein or purified σ1 head domain was chemically coupled to an AFM tip to probe adherent murine L929 (L) cells. L-cells express JAM-A and support reovirus infection (Barton et al., 2001a) but do not express JAM-B or JAM-C (Morris et al., 2006). We found that σ1/JAM-A interactions have a substantially longer bond half-life (t1/2) than JAM-A/JAM-A interactions and hence are kinetically more stable. These findings provide a biophysical basis for the different functions of JAM-A in regulating tight junction permeability and as a virus receptor.
MATERIALS AND METHODS
Cell lines and proteins
L-cells were cultured in DMEM (Sigma) supplemented to contain 10% fetal bovine serum (GIBCO), 100 units/ml penicillin and 100 mg/ml streptomycin (GIBCO). Cells were grown on 13 mm glass cover slips (Menzel-Glaser, Germany) in six-well culture plates (Nunc). The σ1 protein, containing the head domain plus a short region of the tail (residues 293–455), was expressed and purified as described (Schelling et al., 2007). mJAM-A/Fc fusion protein was purchased from R&D Systems, Inc.
Functionalization of tips
A one-step cross-linking procedure using EDC (Pierce) was employed to couple σ1 to the AFM tips (Figure 1) (Kunkel et al., 1981). Soft silicon nitride tips (Veeco, Santa Barbara, CA) were UV irradiated for 15 min and treated with a mixture of 30% H2O2/70% H2SO4 for 30 min. Tips were dried, treated with a 4% solution of 3-aminopropyltriethoxysilane (APTES, Sigma) in acetone for 3 min, rinsed three times in acetone, and incubated in a 2 mg/ml solution of BS3 (Pierce) for 30 min. Tips were incubated in 2 mg/ml mPEG-amine spacer (MW 2000, Nektar) for 2 h, followed by incubation with 1 M Tris buffer. The σ1 head domain (10 mg/ml) and freshly prepared EDC (1 mg/ml) were mixed in equal amounts. Tips were rinsed in PBS, incubated in this mixture for 2 h, rinsed, and blocked with 1% BSA.
Figure 1.
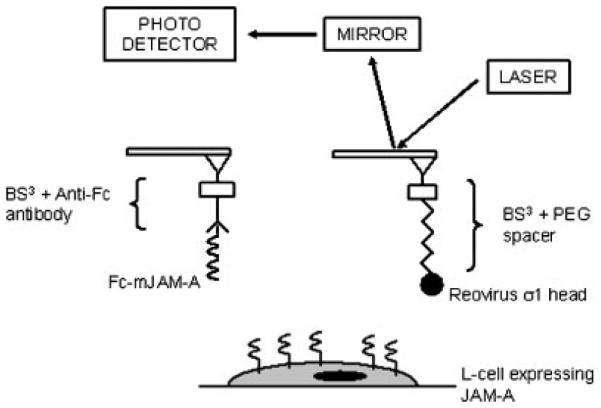
Schematic of the AFM procedure and functionalization methods used. The σ1 head domain was linked to the AFM tip via a PEG spacer, while the mJAM-A/Fc chimera was linked to the AFM tip using an anti-Fc antibody. L-cells, which express JAM-A, were probed using these functionalized tips.
Tips with the mJAM-A/Fc chimera were generated by treatment with BS3, followed by incubation with mouse anti-human Fc antibody (5 μg/ml, Sigma) for 2 h. The reaction was quenched with Tris buffer, and the tips were incubated in a 5 μg/ml solution of mJAM-A/Fc chimera (R&D Systems) for 2 h. Tips were blocked with 1% BSA before experiments.
Mica was functionalized with mJAM-A/Fc chimera using an analogous procedure except that a higher concentration of mJAM-A/Fc protein (50 μg/ml) was used. For control experiments, mock functionalization of the tips and mica was performed using BSA. All the steps were similar except that BSA was used instead of σ1 or mJAM-A/Fc.
Data acquisition and analysis
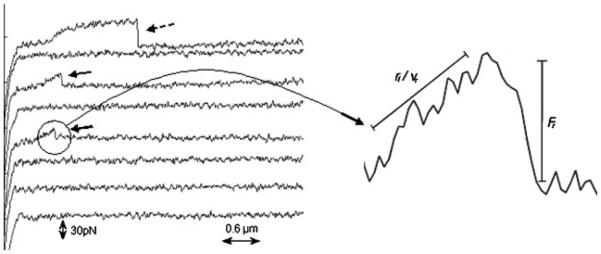
Force curves were acquired using a MultiMode™ Picoforce™ AFM (Veeco, Santa Barbara, CA) coupled to an upright microscope at room temperature using a fluid cell. Cell culture medium was injected into the fluid cell just before the experiments. The largest cantilever with a nominal spring constant of 0.01 N/m was used for obtaining force plots. Prior to obtaining force curves, the deflection sensitivity of the cantilever was obtained, and the spring constant was determined using the thermal tune module. The cantilever was positioned on an adherent cell to obtain force curves at different reproach velocities (1, 2.5, 5, and 10 μm/s). To minimize the number of adhesion events and maximize the probability of obtaining single-bond adhesion events, a contact force of 200 pN and contact time of 1 ms were used. The target adhesion frequency was 30%, which would provide >85% probability of the rupture forces being due to single bond rupture (Hanley et al., 2003). Force curves were analyzed for the magnitude of the rupture events and loading rate (defined as the slope of the retrace curve prior to the rupture event multiplied by the reproach velocity, Figure 2) using a code written in MATLAB version 6.5 (The MathWorks, Natick, MA).
Figure 2.
Typical force–distance curves with bond rupture events (bold arrows) and tether formation (dashed arrows). The loading rate for a given bond rupture event (Fr) is calculated by multiplying the slope (rf/vr) of the reproach curve prior to the rupture event by the reproach velocity of the cantilever.
Bell’s model parameter extraction
This model predicts a linear correlation between the magnitude of the bond rupture force and logarithm of the applied loading rate (Equation (1)) (Bell, 1978; Evans and Ritchie, 1997):
| (1) |
where f *=rupture force; rf=loading rate; xβ=reactive compliance; kB=Boltzmann constant; T=temperature; =unstressed off rate.
This model has been used previously to characterize binding interactions between homophilic N-cadherin and E-cadherin molecules on the surface of cells (Panorchan et al., 2006).
RESULTS
Overview of AFM strategy
The principle of molecular force spectroscopy using AFM is schematically shown in Figure 1. A cantilever with a sharp tip is functionalized with the protein of interest. A laser reflecting off the surface of the cantilever onto a photo detector monitors the deflections of the cantilever. Forces acting on the sharp silicon nitride tip deflect the cantilever causing a change in the position of the laser spot on the photo detector. The shift in position of the laser spot on the photo detector can be used to calculate the deflection of the cantilever. Since the spring constant of the cantilever can be determined by monitoring its thermal fluctuations, the deflection of the cantilever can be used to calculate the force acting on the tip.
The mJAM-A/Fc chimeric protein was linked to the AFM tip using anti-Fc antibody. This strategy was chosen to provide optimal accessibility of the N-terminal D1 domain of mJAM-A, which is involved in both JAM-A dimer formation (Kostrewa et al., 2001; Prota et al., 2003) and interactions with σ1 (Forrest et al., 2003; Guglielmi et al., 2007). For functionalizing tips with σ1 head domain, one end of a homo-bifunctional polyethylene glycol spacer (approximately 15 nm in length and containing amino groups at both ends) was attached to the silanized AFM tips using bis (Sulfosuccinimidyl) suberate (BS3) while the untagged σ1 head domain was linked to the other end of the spacer using 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC). EDC is a “zero-length” cross linker that covalently links an amino group to a carboxylic acid group (Kunkel et al., 1981). The PEG spacer was used to provide enhanced flexibility for the covalently attached σ1 head domain. The σ1 head domain has several surface-exposed aspartic and glutamic acid residues, which could engage the free amine group of the spacer in the presence of EDC via their carboxylic acid groups. The strategy chosen maximizes the probability that the JAM-A-binding site of the σ1 head domain will contact JAM-A.
Force spectroscopy of mJAM-A/L-cell interactions
Force–distance curves for homophilic mJAM-A interactions were obtained using cantilevers functionalized with recombinant mJAM-A/Fc chimeric protein. The functionalized cantilever was lowered onto a single L-cell. A contact force of 200 pN was applied for 1 ms by the cantilever before it was retracted at velocities of 1, 2.5, 5, or 10 μ/s. More than 500 force–distance curves were obtained for each velocity at an adhesion probability of <30%. Force–distance curves showing a clear single rupture event were analyzed for loading rate (defined as the slope of the retrace curve prior to the rupture event multiplied by the reproach velocity of the cantilever) and rupture force (Figure 2). Control experiments performed using anti-Fc antibody functionalized tips showed few adhesion events, indicating that the interactions being measured were specific to JAM-A (Figure 3). All the receptor/ligand pairs tested, corresponding to the histograms shown in Figure 3, are listed in Table 1.
Figure 3.
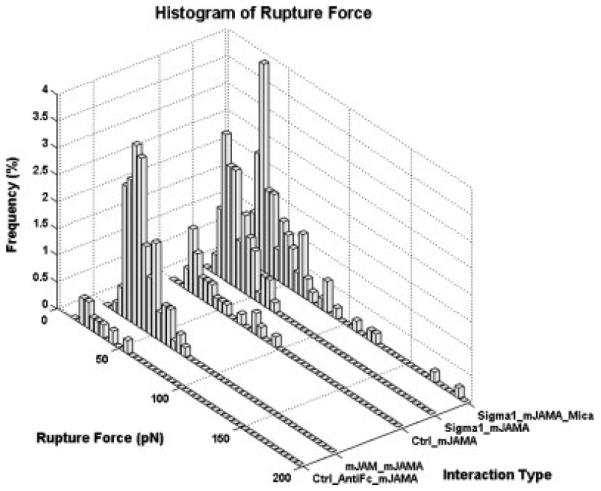
Histograms of bond rupture frequencies for different interaction types at reproach velocity of 5 μ/s. A contact force of 200 pN and a contact time of 1 ms were used for all experiments. Control experiments showed few adhesion events. Each histogram represents data analyzed from over 500 force curves.
Table 1.
Different interactions probed using AFM corresponding to the histograms depicted in Figure 3
| Interaction type | AFM tip | Substrate |
|---|---|---|
| 1. Ctrl_antiFc_mJAM-A | Anti-Fc only | L-cells |
| 2. mJAM-A_mJAM-A | Fc-mJAM-A | L-cells |
| 3. Ctrl_mJAMA | PEG + BSA only | L-cells |
| 4. σ1_mJAM-A | PEG + σ1 head | L-cells |
| 5. σ1_mJAM-A_mica | PEG + σ1 head | Fc-mJAM-A on mica |
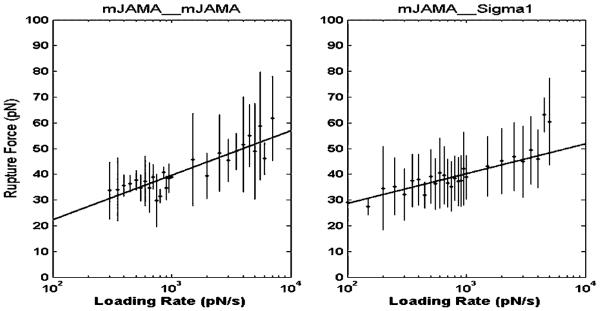
Following the method used by Hanley et al. (2004) and Panorchan et al. (2006), rupture force measurements were binned by increments of 50 pN/s for loading rates between 100 and 1000 pN/s and by increments of 500 pN/s for loading rates between 1000 and 10 000 pN/s. Each bin yielded a mean rupture force. By plotting the mean rupture force as a function of loading rate and extrapolating the fitted line, the unstressed dissociation rate and reactive compliance (xβ) were extracted using Bell’s model (Figure 4 and Equation (1) in Methods section).
Figure 4.
Loading rate curves for mJAM-A/mJAM-A and σ1 head/mJAM-A interactions. The loading rate and rupture force were obtained from the curves. The data points were binned as described in the text. The mean rupture force values for each bin were plotted against the logarithm of the corresponding loading rate.
Homophilic mJAM-A interactions showed an increase in bond rupture force with increasing loading rate. Fitting the data points to a line, the for mJAM-A interactions was found to be 0.688 ± 0.349 s−1 (Table 2). The bond dissociation time for mJAM-A interactions is very low when compared to that of σ1/mJAM-A interactions (see below). A low bond dissociation time is consistent with the physiological functions attributed to JAM-A in regulating tight junction permeability. The short bond lifetime imparts a highly dynamic nature to the interactions between JAM-A molecules necessary for the cell to regulate solute and solvent diffusion across the tight junction barriers.
Table 2.
Parameters extracted by extrapolating the loading rate curves shown in Figure 4
| Molecular pairs | xβ | Cell line | |
|---|---|---|---|
| mJAM-A/mJAM-A | 0.688 ± 0.349 | 0.547 ± 0.060 | L-cells |
| mJAM-A/σ1 head | 0.067 ± 0.041 | 0.817 ±0.073 | L-cells |
| *Cldn1/Cldn1 | 1.35 ± 1.31 | 0.36 ± 0.06 | Recombinant proteins |
Data presented for claudin-1 has been taken from previous work (Lim et al., 2008).
An interesting feature of the mJAM-A force–distance curves was the formation of occasional “tethers.” These force–distance curves are distinguished by the fact that the bond rupture is preceded by a long horizontal stretch of cantilever movement without any deflection (dashed arrow, Figure 2). This observation might be attributable to the interaction of mJAM-A on the tip with a free molecule of mJAM-A on the cell (not linked to the cytoskeleton). The interaction could have distorted the cell membrane as a long tube. Alternatively, this pattern could have been produced by dissociation of mJAM-A on the cell membrane from its cytoplasmic adaptor or cytoskeleton (Evans et al., 2005; Heinrich et al., 2005). Force–distance curves showing such tethers were excluded from the loading rate curve analysis.
Force spectroscopy of σ1/L-cell interactions
Force–distance curves for σ1/mJAM-A interactions were obtained on L-cells with tips functionalized with σ1 using an analogous approach. The data were analyzed using Bell’s model to extract the unstressed dissociation rate and reactive compliance (xβ) (Figure 4 and Table 2). The unstressed dissociation rate obtained for σ1/mJAM-A interactions (0.067 ± 0.041 s−1, Table 2) was much less than that obtained for homophilic mJAM-A interaction. These findings indicate that the bond half-life (t1/2) of the σ1/mJAM-A interaction is greater (by almost tenfold) than that of the homophilic mJAM-A interaction. A long bond t1/2 might facilitate stable adherence of the virus to the cell and help in its entry.
To ensure that the mJAM-A-binding domain of σ1 remained accessible after it was cross-linked to the AFM tip, control force curves were obtained using mica cover slips functionalized with mJAM-A. The force histogram profile obtained using these functionalized mica cover slips probed with σ1-functionalized tips was similar to that obtained using L-cells (Figure 3, Table 1). Furthermore, mica functionalized with BSA showed few adhesion events when probed with the σ1-functionalized tips (data not shown). Therefore, the method used to cross-link σ1 to the AFM tip did not adversely affect interactions of the σ1 head domain with mJAM-A.
DISCUSSION
In this study, we used single molecule force spectroscopy to elucidate the kinetics of homophilic JAM-A/JAM-A interactions and interactions between JAM-A and the reovirus σ1 protein. Although mJAM-A was used in this study, residues mediating interactions between JAM-A dimers are conserved between mJAM-A and hJAM-A, and both mJAM-A and hJAM-A bind reovirus σ1 (Kostrewa et al., 2001; Prota et al., 2003). Thus, our results are likely applicable for both human and murine homologs of JAM-A.
While both mJAM-A and σ1 interact specifically with L-cells, we find that σ1/L-cell interactions have a substantially longer half-life (t1/2) than mJAM-A/L-cell interactions. The lack of adhesion events in control experiments strongly suggests that these measurements describe specific interactions with mJAM-A on the surface of L-cells. The observed kinetic parameters of homophilic mJAM-A interactions closely resemble that of claudin-1 (Table 1), another tight junction protein that is also involved in regulating paracellular permeability of solutes. For claudin-1, rapid lateral association and dissociation between adjacent cells has also been observed using the corresponding GFP-tagged proteins (Sasaki et al., 2003). It is likely that such dynamic interactions provide cells with a platform to regulate solute flux.
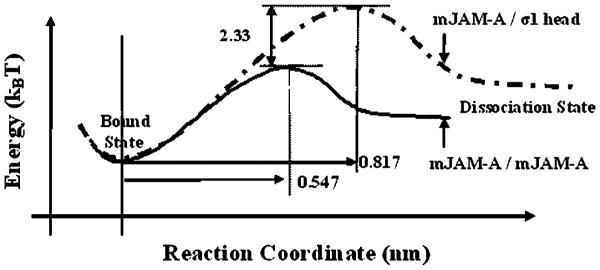
Dissociation of mJAM-A/mJAM-A and σ1/mJAM-A complexes was found to follow a single step energy activation barrier process (Figure 5). The topography of the energy landscapes of the dissociation of mJAM-A/mJAM-A and mJAM-A/σ1 complexes can be compared by plotting the geometric locations of their bound states on the same reactive coordinates (Figure 5). The unstressed off rate for the two interactions can be used to estimate the energy difference (ΔG) between the activation barriers of mJAM-A/mJAM-A and mJAM-A/σ1 complex dissociation and is given by (Bell, 1978; Merkel et al., 1999; Evans, 2001):
| (2) |
Figure 5.
Energy landscape for dissociation of σ1/mJAM-A and mJAM-A/mJAM-A constructed based on the kinetic parameters obtained from single molecule force spectroscopy experiments.
Here and denote the unstressed dissociation rate constants of the mJAM-A/mJAM-A and σ1/mJAM-A interactions, respectively. The analysis reveals that the activation barrier of the σ1/mJAM-A complex is 2.33kBT higher than that of mJAM-A/mJAM-A complex (Figure 5), indicating that σ1/mJAM-A forms a more stable bond when compared to the mJAM-A/mJAM-A interaction.
The differences in half-life for homophilic JAM-A and σ1/JAM-A interactions are consistent with published findings. When JAM-A dimers are mixed with σ1 in solution, σ1 displaces JAM-A from the dimer interface, presumably by binding to a region near this interface (Guglielmi et al., 2007). Therefore, once formed, the σ1/JAM-A complex is more stable in solution than JAM-A/JAM-A homodimers. Surface plasmon resonance studies have found that the σ1 head domain binds to hJAM-A with a KD of ~5 × 10−9 M (Guglielmi et al., 2007; Schelling et al., 2007). However, hJAM-A homophilic interactions are not detected on a biosensor surface (E. S. Barton, J. C. Forrest, and T.S. Dermody, unpublished observations). The lack of detection of homophilic JAM-A interactions in these studies may result from several factors, including steric interference from the tag appended to JAM-A for capture and the rigidity of the biosensor surface. However, our findings suggest that a short half-life in comparison to σ1/mJAM-A interactions contributes to the inability to detect JAM-A homophilic interactions using surface plasmon resonance. At this juncture it is not possible to ascertain the exact cause of the difference in unstressed off rate for mJAM-A/mJAM-A and σ1/mJAM-A interactions. However, while there is significant overlap such as Glu121, are not required for σ1 binding (Guglielmi et al., 2007). Therefore, we speculate that the differences in off-rate could arise from slight differences in binding sites or additional contacts between σ1 and JAM-A that could strengthen the σ1/JAM-A interaction after the complex is formed. Another possible explanation for the observed differences is the presence of a, as yet unknown, receptor for the σ1 head domain that is recruited following σ1 binding to JAM-A. It has been shown previously that the tail portion of σ1 can interact with sialic acid residues present on L-cells (Barton et al., 2001a). We used purified σ1 head domain in our experiments that did not possess the tail portion and hence it is unlikely that the observed difference was due to sialic acid residues. However, it is possible that the head domain has other binding partners that could strengthen the interaction following the initial complex formation with JAM-A.
Our results show how the kinetic differences of protein interactions, probed using single molecule force spectroscopy experiments, bear correlation to their physiological functions. Further experiments using JAM-A with mutated residues could help in clarifying the observed differences in JAM-A/JAM-A and σ1/JAM-A interactions.
Acknowledgements
We thank members of our laboratories for many useful discussions during the conduct of this research. This work was supported by the Biomedical Research Council (Agency for Science, Technology and Research [A*STAR], Singapore), United States Public Health Service awards T32 GM08554 (K.M.G.) and R01 GM67853 (T.S.D. and T.S.), the Elizabeth B. Lamb Center for Pediatric Research, and United States Public Health Service awards CA68485 for the Vanderbilt-lngram Cancer Center and DK20593 for the Vanderbilt Diabetes Research and Training Center.
REFERENCES
- Barton ES, Connolly JL, Forrest JC, Chappell JD, Dermody TS. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 2001a;276:2200–2211. doi: 10.1074/jbc.M004680200. [DOI] [PubMed] [Google Scholar]
- Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell FJ, Nusrat A, Parkos CA, Dermody TS. Junction adhesion molecule is a receptor for reovirus. Cell. 2001b;104:441–451. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- Baumgartner W, Hinterdorfer P, Ness W, Raab A, Vestweber D, Schindler H, Drenckhahn D. Cadherin interaction probed by atomic force microscopy. Proc. Natl Acad. Sci. U.S.A. 2000;97:4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Campbell JA, Schelling P, Wetzel JD, Johnson EM, Forrest JC, Wilson GA, Aurrand-Lions M, Imhof BA, Stehle T, Dermody TS. Junctional adhesion molecule a serves as a receptor for prototype and field-isolate strains of mammalian reovirus. J. Virol. 2005;79:7967–7978. doi: 10.1128/JVI.79.13.7967-7978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MI, Panorchan P, Dobrowsky TM, Tseng Y, Wirtz D. Single-molecule analysis of human immunodeficiency virus type 1 gp120-receptor interactions in living cells. J. Virol. 2005;79:14748–14755. doi: 10.1128/JVI.79.23.14748-14755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell JD, Duong JL, Wright BW, Dermody TS. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 2000;74:8472–8479. doi: 10.1128/jvi.74.18.8472-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell JD, Prota AE, Dermody TS, Stehle T. Crystal structure of reovirus attachment protein sigma1 reveals evolutionary relationship to adenovirus fiber. Embo. J. 2002;21:1–11. doi: 10.1093/emboj/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke VG, Naik MU, Naik UP. Fibroblast growth factor-2 failed to induce angiogenesis in junctional adhesion molecule-A-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2006;26:2005–2011. doi: 10.1161/01.ATV.0000234923.79173.99. [DOI] [PubMed] [Google Scholar]
- Cunningham SA, Arrate MP, Rodriguez JM, Bjercke RJ, Vanderslice P, Morris AP, Brock TA. A. novel protein with homology to the junctional adhesion molecule. Characterization of leukocyte interactions. J. Biol. Chem. 2000;275:34750–34756. doi: 10.1074/jbc.M002718200. [DOI] [PubMed] [Google Scholar]
- Ebnet K, Schulz CU, Meyer Zu, Brickwedde MK, Pendl GG, Vestweber D. Junctional adhesion molecule interacts with the PDZ domain- containing proteins AF-6 and ZO-1. J. Biol. Chem. 2000;275:27979–27988. doi: 10.1074/jbc.M002363200. [DOI] [PubMed] [Google Scholar]
- Evans E. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- Evans E, Heinrich V, Leung A, Kinoshita K. Nano- to microscale dynamics of P-selectin detachment from leukocyte interfaces. I. Membrane separation from the cytoskeleton. Biophys. J. 2005;88:2288–2298. doi: 10.1529/biophysj.104.051698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest JC, Campbell JA, Schelling P, Stehle T, Dermody TS. Structure-function analysis of reovirus binding to junctional adhesion molecule 1. Implications for the mechanism of reovirus attachment. J. Biol. Chem. 2003;278:48434–48444. doi: 10.1074/jbc.M305649200. [DOI] [PubMed] [Google Scholar]
- Guglielmi KM, Kirchner E, Holm GH, Stehle T, Dermody TS. Reovirus binding determinants in junctional adhesion molecule-A. J. Biol. Chem. 2007;282:17930–17940. doi: 10.1074/jbc.M702180200. [DOI] [PubMed] [Google Scholar]
- Hanley W, McCarty O, Jadhav S, Tseng Y, Wirtz D, Konstantopoulos K. Single molecule characterization of P-selectin/ligand binding. J. Biol. Chem. 2003;278:10556–10561. doi: 10.1074/jbc.M213233200. [DOI] [PubMed] [Google Scholar]
- Hanley WD, Wirtz D, Konstantopoulos K. Distinct kinetic and mechanical properties govern selectin-leukocyte interactions. J. Cell Sci. 2004;117:2503–2511. doi: 10.1242/jcs.01088. [DOI] [PubMed] [Google Scholar]
- Heinrich V, Leung A, Evans E. Nano- to microscale dynamics of P-selectin detachment from leukocyte interfaces. II. Tether flow terminated by P-selectin dissociation from PSGL-1. Biophys. J. 2005;88:2299–2308. doi: 10.1529/biophysj.104.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornecki E, Walkowiak B, Naik UP, Ehrlich YH. Activation of human platelets by a stimulatory monoclonal antibody. J. Biol. Chem. 1990;265:10042–10048. [PubMed] [Google Scholar]
- Kostrewa D, Brockhaus M, D’Arcy A, Dale GE, Nelboeck P, Schmid G, Mueller F, Bazzoni G, Dejana E, Bartfai T, Winkler FK, Hennig M. X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. Embo. J. 2001;20:4391–4398. doi: 10.1093/emboj/20.16.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel GR, Mehrabian M, Martinson HG. Contact-site cross-linking agents. Mol. Cell Biochem. 1981;34:3–13. doi: 10.1007/BF02354846. [DOI] [PubMed] [Google Scholar]
- Liang TW, Chiu HH, Gurney A, Sidle A, Tumas DB, Schow P, Foster J, Klassen T, Dennis K, DeMarco RA, Pham T, Frantz G, Fong S. Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM 2 interacts with T, NK, and dendritic cells through JAM 3. J. Immunol. 2002;168:1618–1626. doi: 10.4049/jimmunol.168.4.1618. [DOI] [PubMed] [Google Scholar]
- Lim TS, Vedula SR, Kausalya PJ, Hunziker W, Lim CT. Single- molecular-level study of claudin-1-mediated adhesion. Langmuir. 2008;24:490–495. doi: 10.1021/la702436x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Nusrat A, Schnell FJ, Reaves TA, Walsh S, Pochet M, Parkos CA. Human junction adhesion molecule regulates tight junction resealing in epithelia. J. Cell Sci. 2000;113(Pt 13):2363–2374. doi: 10.1242/jcs.113.13.2363. [DOI] [PubMed] [Google Scholar]
- Mandell KJ, McCall IC, Parkos CA. Involvement of the junctional adhesion molecule-1 (JAM1) homodimer interface in regulation of epithelial barrier function. J. Biol. Chem. 2004;279:16254–16262. doi: 10.1074/jbc.M309483200. [DOI] [PubMed] [Google Scholar]
- Martin-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M, Fruscella P, Panzeri C, Stoppacciaro A, Ruco L, Villa A, Simmons D, Dejana E. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- Morris AP, Tawil A, Berkova Z, Wible L, Smith CW, Cunningham SA. Junctional adhesion molecules (JAMs) are differentially expressed in fibroblasts and co-localize with ZO-1 to adherens-like junctions. Cell Commun. Adhes. 2006;13:233–247. doi: 10.1080/15419060600877978. [DOI] [PubMed] [Google Scholar]
- Naik MU, Mousa SA, Parkos CA, Naik UP. Signaling through JAM-1 and alphavbeta3 is required for the angiogenic action of bFGF: dissociation of the JAM-1 and alphavbeta3 complex. Blood. 2003;102:2108–2114. doi: 10.1182/blood-2003-04-1114. [DOI] [PubMed] [Google Scholar]
- Naik UP, Naik MU, Eckfeld K, Martin-DeLeon P, Spychala J. Characterization and chromosomal localization of JAM-1, a platelet receptor for a stimulatory monoclonal antibody. J. Cell Sci. 2001;114:539–547. doi: 10.1242/jcs.114.3.539. [DOI] [PubMed] [Google Scholar]
- Ostermann G, Weber KS, Zernecke A, Schroder A, Weber C. JAM-1 is a ligand of the beta(2) integrin LFA-1 involved in transendothelial migration of leukocytes. Nat. Immunol. 2002;3:151–158. doi: 10.1038/ni755. [DOI] [PubMed] [Google Scholar]
- Panorchan P, Thompson MS, Davis KJ, Tseng Y, Konstantopoulos K, Wirtz D. Single-molecule analysis of cadherin-mediated cell-cell adhesion. J. Cell Sci. 2006;119:66–74. doi: 10.1242/jcs.02719. [DOI] [PubMed] [Google Scholar]
- Prota AE, Campbell JA, Schelling P, Forrest JC, Watson MJ, Peters TR, Aurrand-Lions M, Imhof BA, Dermody TS, Stehle T. Crystal structure of human junctional adhesion molecule 1: implications for reovirus binding. Proc. Natl Acad. Sci. U.S.A. 2003;100:5366–5371. doi: 10.1073/pnas.0937718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui C, Furuse K, Mimori-Kiyosue Y, Furuse M, Tsukita S. Dynamic behavior of paired claudin strands within apposing plasma membranes. Proc. Natl Acad. Sci. U.S.A. 2003;100:3971–3976. doi: 10.1073/pnas.0630649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelling P, Guglielmi KM, Kirchner E, Paetzold B, Dermody TS, Stehle T. The reovirus sigma1 aspartic acid sandwich: a trimerization motif poised for conformational change. J. Biol. Chem. 2007;282:11582–11589. doi: 10.1074/jbc.M610805200. [DOI] [PubMed] [Google Scholar]
- Schiff LA, Nibert ML, Tyler KL. Orthoreoviruses and their replication. In: Knipe DM, Howley PM, editors. In Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2007. 1853–1915. [Google Scholar]
- Sobocka MB, Sobocki T, Banerjee P, Weiss C, Rushbrook JI, Norin AJ, Hartwig J, Salifu MO, Markell MS, Babinska A, Ehrlich YH, Kornecki E. Cloning of the human platelet F11 receptor: a cell adhesion molecule member of the immunoglobulin superfamily involved in platelet aggregation. Blood. 2000;95:2600–2609. [PubMed] [Google Scholar]