Abstract
The RNA-binding protein Hfq is a global regulator which controls diverse cellular processes in bacteria. To begin understanding the role of Hfq in the Sinorhizobium meliloti–Medicago truncatula nitrogen-fixing symbiosis, we defined free-living and symbiotic phenotypes of an hfq mutant. Over 500 transcripts were differentially accumulated in the hfq mutant of S. meliloti Rm1021 when grown in a shaking culture. Consistent with transcriptome-wide changes, the hfq mutant displayed dramatic alterations in metabolism of nitrogen-containing compounds, even though its carbon source utilization profiles were nearly identical to the wild type. The hfq mutant had reduced motility and was impaired for growth at alkaline pH. A deletion of hfq resulted in a reduced symbiotic efficiency, although the mutant was still able to initiate nodule development and differentiate into bacteroids.
The soil-dwelling α-proteobacterium Sinorhizobium meliloti engages in a nitrogen-fixing symbiosis with legumes of the genera Medicago, Melilotus, and Trigonella. The establishment, stability, and productivity of this symbiosis depend on coordinated signal and nutrient exchanges between the host and its bacterial symbiont. In the earliest stages of the interaction, chemical signaling between the bacteria and plant initiates root nodule formation (Fisher and Long 1992). The bacteria invade the plant root by tunneling within a root hair and through subsequent cells via inward growth of the plant cell wall (termed an “infection thread”) into the developing nodule (Jones et al. 2007). During invasion, the bacteria continue to divide within infection threads. When an infection thread reaches a suitable plant cell, the bacteria pinch off from the tip of the infection thread, still surrounded by the plant membrane, and are released into the plant cell cytoplasm of the nodule cells. Here, they undergo terminal differentiation into nitrogen-fixing bacteroids able to convert, via nitrogenase, dinitrogen into ammonium that the plant can use for growth (Gibson et al. 2008). The microaerobic environment of the nodule supports expression of nif and fix genes required for nitrogen fixation and protects the activity of oxygen-labile nitrogenase (Fischer 1994). In exchange for fixed nitrogen (in the form of ammonium), the plant host provides its symbiont with fixed carbon in the form of C4-dicarboxylic acids (Yurgel and Kahn 2004).
Bacteria produce ammonia that crosses the symbiosome boundary to be assimilated in the plant. Recent evidence also shows that nitrogen-containing compounds are supplied by the host plant to their symbiotic partners. For example, amino acid transporters encoded by bra and aap were required for an effective symbiosis of Rhizobium leguminosarum with pea (Lodwig et al. 2003), because bacteroids become “symbiotic auxotrophs” for leucine, isoleucine, and valine (LIV) and must obtain these from the host plant (Prell et al. 2009). This reliance by rhizobia on host LIV amino acids was proposed to be an important checkpoint in nodule development: pea regulates R. leguminosarum bacteroid development and persistence by controlling the supply of LIV (Prell et al. 2009). A similar process likely occurs in the S. meliloti–Medicago spp. symbiosis, though it has not yet been experimentally determined.
In addition to the chemical and nutrient cues from both partners, development of a nodule depends on extensive remodeling of both bacterial and plant gene expression. Based on what is known about other bacterial–eukaryotic interactions, this remodeling of gene expression likely involves post-transcriptional regulatory cascades that are mediated by RNA-binding proteins like Hfq. The gene hfq appears to be evolutionarily conserved in many bacteria, including all α-, β-, and γ-proteobacteria, except those that have undergone genome reduction (Sun et al. 2002; Duan et al. 2009). In the best-studied models (Escherichia coli and Salmonella enterica), Hfq facilitates short and imperfect base pairing between a regulatory small (s)RNA and the 5′ untranslated region (UTR) of the regulated messenger (m)RNA (Brennan and Link 2007). This base pairing makes the ribosome binding site either more or less accessible (Brennan and Link 2007). A global role for Hfq in controlling post-transcriptional gene regulation in γ-proteobacteria is supported by high-throughput transcriptomic and proteomic studies, which demonstrated that 10 to 20% of Salmonella transcripts were under Hfq control (Sittka et al. 2007, 2008; Ansong et al. 2009). Stability of at least 30 sRNAs depended on Hfq (Zhang et al. 2003; Sittka et al. 2008). Similarly, in rhizobia, stability of at least two evolutionarily conserved sRNAs depends on Hfq (Voss et al. 2009).
Although hfq is better studied in γ-proteobacteria, fundamental information is starting to emerge on its importance in the α-subgroup as well. A Brucella hfq mutant was defective in its ability to invade and survive inside animal cells and was more sensitive to acid stress (Robertson and Roop 1999). A Caulobacter crescentus hfq mutant was impaired for survival under stress (Landt et al. 2008). Orthologs of Hfq exert post-transcriptional regulation over the nifA activator of nif and fix genes in Azorhizobium caulinodans, R. leguminosarum, and Rhodobacter capsulatus (Kaminski et al. 1994; Drepper et al. 2002; Zhang and Hong 2009). Overexpression of nifA was insufficient to fully rescue the phenotype of the hfq (nrfA) mutant in A. caulinodulans (Kaminski et al. 1994). These reports suggest that functions of hfq in the symbiosis are likely to be evolutionarily conserved and extend beyond its effects on nifA.
Although regulatory functions of hfq have been partly characterized in closely related α-proteobacteria, its roles in free-living and symbiotic Sinorhizobium meliloti have not been determined. Here, we investigate the role of S. meliloti hfq in bacterial gene regulation and symbiosis with its plant host.
RESULTS
Construction of the hfq mutant
S. meliloti hfq is chromosomally located upstream of hflX, an arrangement conserved in many bacteria (Supplementary Fig. S1). The hflX gene encodes a protein belonging to a family of phosphate-binding-loop triphosphatases (Polkinghorne et al. 2008; Jain et al. 2009). Upstream of hfq are genes encoding NtrYX, a two-component system postulated to be involved in nitrogen regulation and halotolerance (Pawlowski et al. 1991; Nogales et al. 2002); TrkA, a putative potassium transporter; and a putative aminotransferase. Global transcription analyses suggest that hfq and upstream genes are co-regulated (Barnett et al. 2004; Sauviac et al. 2007). To avoid polar effects on the downstream hflX and potential interference with expression of upstream genes, an in-frame deletion mutant of S. meliloti hfq, 1021Δhfq, was constructed. Affymetrix SymbiosisChip data (Supplementary Table S1) suggest that expression of upstream genes remains unchanged in 1021Δhfq and expression of downstream hflX increases only 1.6-fold. The hfq transcript was absent in the mutant, as expected. We also constructed a strain in which Δhfq was complemented by hfq carried on a pBBR plasmid.
Utilization of carbon and nitrogen sources by the hfq mutant
We used Phenotype microarrays (Biolog) to examine carbon and nitrogen metabolism of S. meliloti Rm1021, 1021Δhfq, and 1021Δhfq pHfq. Phenotype microarrays serve as a surrogate assay for the utilization of individual carbon or nitrogen sources (Biondi et al. 2009; Bochner 2009). The hfq mutant showed essentially the same carbon source utilization pattern as Rm1021 for the 182 carbon compounds of the PM1 and PM2 carbon utilization arrays: the exceptions were γ-aminobutyric acid (GABA) and pyruvic acid, on which 1021Δhfq respired more robustly than wild type (Supplementary Tables S2 and S3). Growth tests confirmed that the hfq mutant grew faster than Rm1021 in M9 medium with GABA or pyruvic acid as sole carbon sources (data not shown).
In contrast to carbon source utilization, utilization of nitrogen compounds differed substantially between the hfq mutant and wild-type strains (Supplementary Table S4). The wild type used 59 of the 96 tested nitrogen compounds. Respiration of the hfq mutant on 25 of these nitrogen sources decreased at least twofold (based on absorbance at 595 nm = 0.3 cutoff). In all, seven (L-valine, L-aspartic acid, D-asparagine, adenosine and the dipeptides Ala-Leu, Ala-Gly, and Ala-Thr) of the 25 compounds were not used by the mutant at all. The results of the phenotype arrays and growth tests indicate that the deletion of hfq has global effects on nitrogen metabolism in S. meliloti.
Microarrays reveal a global regulatory role for hfq
We compared transcriptomes of the wild type to 1021Δhfq grown to late exponential phase in a rich medium. For the data mining, we chose an arbitrary cutoff of signal log ratio (SLR) ≥0.96 (Barnett et al. 2004). Over 500 transcripts were differentially accumulated in the hfq mutant compared with the wild type. In total, 483 represented coding regions, accounting for approximately 7% of S. meliloti annotated open reading frames (ORF). Of the differentially accumulated transcripts, approximately 65% were decreased in the hfq mutant, whereas 35% were increased. Reverse-transcriptase quantitative polymerase chain reaction (qPCR) analyses confirmed Affymetrix SymbiosisChip data for selected genes (Table 1).
Table 1.
Expression changes of selected genes in the 1021Δhfq mutanta
| Relative expression ratio 1021 Δ hfq mutant vs. Rm1021 |
||
|---|---|---|
| Gene | SymbiosisChipb | Real-time PCRc |
| SMc02118 (aapJ) | 2.2 | 4.1 ± 0.4 |
| SMc02119 (aapQ) | 4.1 | 11.0 ± 0.6 |
| SMc02120 (aapM) | 4.8 | 8.4 ± 0.4 |
| SMc01946 (livK) | 5.5 | 14.0 ± 0.3 |
| SMc01948 (livF) | 11.0 | 11.3 ± 0.3 |
| SMc01949 (livG) | 10 | 10.9 ± 0.4 |
| SMc01950 (livM) | 13.9 | 18.6 ± 0.4 |
| SMc03201 (bkdAa) | 5.4↓ | (15.0 ± 3.0)4↓ |
| SMc03202 (bkdAb) | 5.4↓ | (6.0 ± 1.0)4↓ |
| SMc03203 (bkdB) | 5.7↓ | (9.2 ± 0.6)4↓ |
| SMc00168 (sinI) | 1.8 | (2.6 ± 0.3) |
| SMc02584 (actR) | 3.1 | 4.8 ± 0.6 |
| SMc02585 (actS) | 3.5 | 4.7 ± 0.2 |
| SMc02484 (supA) | 6.34↓ | (13 ± 1.0)4↓ |
| Smc02485 (supB) | 4.34↓ | (16 ± 2.0)4↓ |
Downward arrows indicate decreased fold change in the 1021Δhfq mutant. Cultures for real-time polymerase chain reaction (PCR) were grown under the same conditions as, but independently of, those used for Symbiosis-Chip analysis.
Fold change is based on signal log ratio value from the SymbiosisChip data.
Fold change is calculated from cycle threshold (Ct) values obtained from real-time PCR experiments. The SMc00129 RNA was used an internal control. The Ct values of the internal control for the Rm1021 and 1021Δhfq were 28.64 and 28.67 for the aap and liv experiment, 31.43 and 31.77 for the sup experiment, 25.59 and 25.52 for the bkd qPCR experiment, and 25.81 and 26.46 for the sinI/act experiment, respectively. The standard errors of the means from qPCR experiments are shown (n = 3).
When grouped by predicted functional categories using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Ogata et al. 1999), the majority of hfq-dependent transcripts are predicted to be involved in transport, metabolism, motility, and membrane functions, and approximately 25% of hfq-dependent transcripts have unknown functions (Table 2). Transcripts encoding proteins related to DNA and protein synthesis were largely unaffected, suggesting that the global effects of hfq on gene regulation are probably not due to a general disruption of cellular functions.
Table 2.
Hfq-responsive functions identified by transcriptomic analysis
| Functional categoriesa | No. of differentially accumulated transcriptsb |
|---|---|
| Transport | 107 |
| Membrane proteins | 59 |
| Cellular processes | 48 |
| Central metabolism | 38 |
| Amino acid metabolism | 32 |
| Metabolism of cofactors and vitamins | 21 |
| Signal transduction | 15 |
| Regulatory functions | 14 |
| Energy metabolism | 14 |
| Fatty acid metabolism | 13 |
| Protein synthesis | 7 |
| DNA synthesis | 4 |
| Hypothetical proteins | 119 |
| Unknown functions | 61 |
Functional categories are defined according to KEGG (Ogata et al. 1999).
Some of these transcripts encode proteins predicted to have multiple functions.
Effect of hfq on transporters and metabolic genes
In the hfq mutant, transcripts corresponding to 140 transporter-related genes were differentially accumulated, representing approximately 25% of the total. Because of the high degree of amino acid sequence conservation among ABC transporters, predicting a transported solute is difficult. However, in S. meliloti and in closely related rhizobia, experimental evidence exists for substrates of many transporters. Transporter genes expressed in an hfq-dependent manner for which a transported solute has been predicted are shown in Supplementary Table S5. Increased expression in 1021Δhfq of livHMGFK and aapJQMP, encoding branched chain amino acid transporters, was confirmed by qPCR (Table 1).
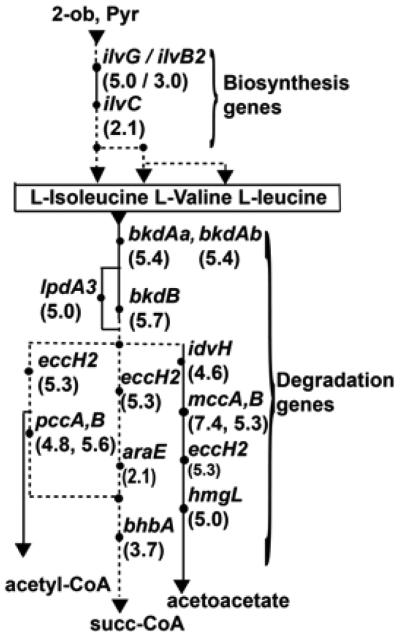
Over 100 genes predicted to be involved in cellular metabolism are differentially regulated in the hfq deletion mutant (Table 2). More than a dozen genes involved in metabolism of branched chain amino acids (LIV) were downregulated in the hfq mutant (Fig. 1). The decreased accumulation of the bkdAaAbB transcripts in 1021Δhfq was confirmed by qPCR (Table 1). The wild type and 1021Δhfq behaved similarly on leucine, isoleucine, or valine as sole carbon sources in the Phenotype microarrays; however, utilization of valine as a sole nitrogen source was severely reduced in the mutant. These results indicate that, even if transport and utilization of LIV are dependent on hfq, hfq is only essential when valine is used as a sole nitrogen source. Uptake and utilization of leucine and isoleucine may occur by both hfq-dependent and hfq-independent pathways.
Fig. 1.
Downregulated transcripts related to branched amino acid metabolism in the 1021Δhfq mutant. Reduced fold changes, calculated from SymbiosisChip signal log ratio values, are in parentheses. Abbreviations are 2-oxobutanoate = 2-ob, pyruvate = pyr, and succiny-CoA = succ-CoA. Assignment of genes and pathways are based on primary amino acid sequence similarity to known genes and the Kyoto Encyclopedia of Genes and Genomes pathway analysis (Ogata et al. 1999) and experimental evidence (Finan et al. 2001; Djordjevic et al. 2003; Prell et al. 2009). Solid circles indicate metabolic stages where Hfq-dependent regulation occurred. Dashed lines indicate that there are additional enzymatic steps in between the two circles. Solid lines indicate that there are no additional enzymatic steps between the two circles.
hutIH1GU and hutH2 mRNAs, encoding enzymes for conversion of L-histidine to glutamate and formamide, were downregulated in 1021Δhfq. Consistent with the predicted roles for the products of the hut genes (Finan et al. 2001), the hfq mutant’s ability to utilize histidine was reduced as determined by Phenotype microarrays. The utilization of L-glutamate and formamide by the hfq mutant remained unchanged, consistent with its regulatory effects on the hut genes.
Expression of PEP carboxykinase (pckA) and fructose-bisphosphate aldolase (fbaB), genes strongly induced during growth of S. meliloti on succinate (Barnett et al. 2004), was increased in 1021Δhfq. Gene pyc mRNA (encoding pyruvate carboxylase) was downregulated 2.3-fold in 1021Δhfq. Several genes required for metabolism of polyhydroxybutyrate and glycogen carbon storage compounds were expressed more strongly in the wild type than in 1021Δhfq. These include acetyl-CoA synthetase (acsA1), acetoacetyl-CoA reductase (phbB), glucose-1-phosphate adenylyltranferase (glcC), and methylmalonyl-CoA mutase (bhbA). The accumulation of transcripts relating to α-glucoside catabolism (aglA and thuB) (Jensen et al. 2005; Willis and Walker 1999) was strongly decreased in the hfq mutant. Expression of glpR, encoding a putative glycerol-3-phosphate regulon repressor, was increased 10-fold in the hfq mutant, with a concomitant decrease in expression of glycerol-3-phosphate uptake genes (SMb20416-20419, SMc02512, and 02514).
Motility and chemotaxis
Transcripts of most known flagellar, chemotaxis, and motility genes were decreased in the hfq mutant. Consistent with these changes, 1021Δhfq colonies were defective for swimming motility, as measured by colony diameter in soft agar (Fig. 2).
Fig. 2.
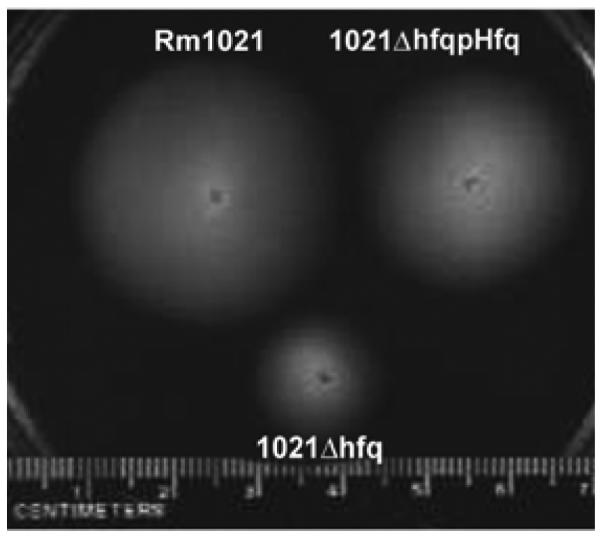
Swimming of Sinorhizobium meliloti A, Rm1021; B, 1021Δhfq; and C, 1021Δhfq pHfq. Plates containing 0.3% agar and 20-fold-diluted tryptone yeast (TY) medium with 3.4 mM CaCl2 were inoculated with 2 μl of washed log phase bacterial suspensions and incubated at 30°C for 3 days. Plates were imaged using a digital camera, with a ruler at the bottom of the photographed field. Scale bar = 1 mm.
Quorum-sensing genes
Gene hfq regulates quorum sensing (QS) in Vibrio spp. by destabilizing mRNA encoding QS regulators through multiple sRNAs (Lenz et al. 2004). Even though accumulation of the transcript corresponding to the LuxR-type AHL receptor SinR was unchanged in the hfq mutant, accumulation of the sinI mRNA, encoding acyl homoserine lactone (AHL) synthase, increased 1.8-fold. This was confirmed by qPCR (Table 1).
To determine if AHL or other QS compounds increased in the hfq mutant, we used bioassay-coupled thin-layer chromatography (TLC) to compare extracts of Rm1021, 1021Δhfq, and 1021Δhfq pHfq culture supernatants. The QS signal profile of 1021Δhfq was different from those of Rm1021 and 1021Δhfq pHfq (Fig. 3). These differences in the profiles of QS-active compounds may reflect differences in AHL synthesis via SinI, availability of substrates for SinI, or decreased degradation of QS compounds in the hfq mutant.
Fig. 3.
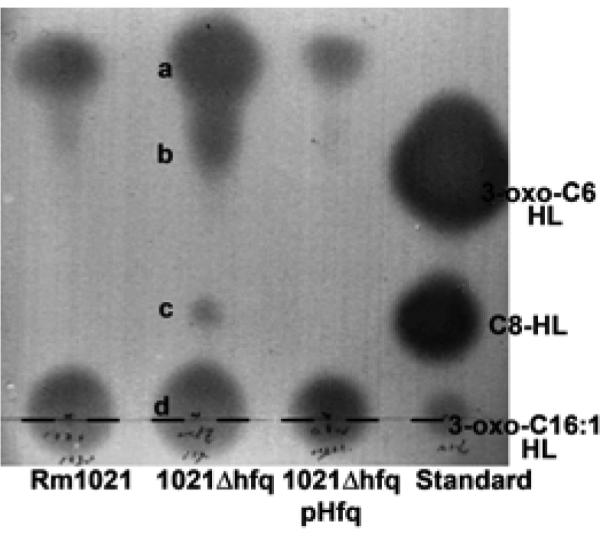
Thin-layer chromatography (TLC) analysis of acyl-homoserine lactones (AHLs) and active quorum-sensing (QS) compounds in dichloromethane supernatant extracts of tryptone yeast–grown Rm1021, 1021Δhfq, and 1021Δhfq pHfq. Each sample corresponding to 1 ml of culture supernatant was spotted on C18 TLC plates, shown by a dashed line. Active QS compounds were detected using an Agrobacterium tumefaciens reporter assay. Spots corresponding to putative AHLs or active QS compounds resolved by TLC in lane 2 are indicated by the letters a to d. AHL standards (right lane) were 500 pmol of 3-oxo-C6-homoserine lactone, 20 nmol of C8-HL (Sigma-Aldrich, Milwaukee, WI, U.S.A.), and 50 nmol of 3-oxo-C16:1-HL (gift from Anatol Eberhard, Cornell University, Ithaca, NY, U.S.A.). This experiment was repeated twice with results similar to those shown here.
Regulation of responses to pH stress
Accumulation of mRNA corresponding to the two-component ActSR regulatory system was increased in 1021Δhfq. This system is required for the adaptation to acidic conditions in closely related S. medicae (Tiwari et al. 1996) and is involved in detoxification, hydantoin utilization, microaerobic respiration, CO2 fixation, microaerobic induction of the nitrogen fixation regulators fixK and nifA, and nitrate assimilation (Fenner et al. 2004). In contrast, accumulation of RNA corresponding to SupAB (SMb20484-5) decreased in the hfq mutant. The SupAB systems participates in alkaline tolerance, possibly by controlling potassium uptake under high pH (Lin et al. 2009). Differential accumulation of actSR and supAB mRNAs was confirmed by qPCR (Table 1).
To test whether the resistance to acidic or alkaline conditions was altered in the hfq mutant, growth of wild-type, 1021Δhfq, and 1021Δhfq pHfq was compared in rich medium buffered to three different pHs. At pH 5.3, the hfq mutant grew similarly to the wild type (Fig. 4A), without even the modest growth defect that was observed at neutral pH (Fig. 4B). At pH 9.0, the growth defect of the hfq mutant was more pronounced (Fig. 4C). These observations indicate that the changes in accumulation of actSR and supAB, genes involved in pH stress resistance, are congruent with the observed phenotypes of the hfq mutant.
Fig. 4.
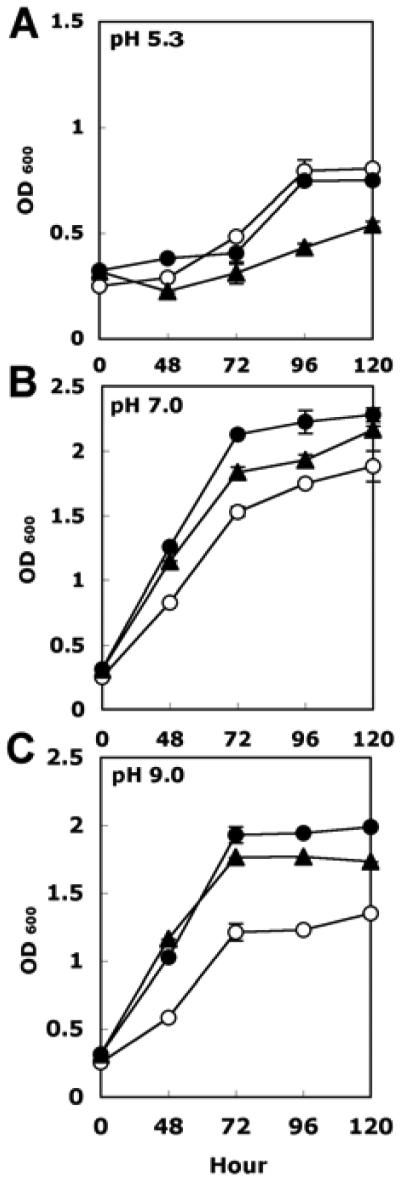
Comparison of growth of 1021Δhfq pBBR1-MCS3 (open circles) and the complemented strain 1021Δhfq pHfq (filled triangles) with wild-type vector control Rm1021 pBBR1-MCS3 (filled circles) as measured by optical density at 600 nm (OD600). Cells were grown in half-strength tryptone yeast (TY) liquid medium buffered with 40 mM morpholineethanesulfonic acid (MES) at A, pH 5.3 or B, pH 7.0 or with C, 40 mM TRIS at pH 9.0. Experiments were repeated three times with similar results. Representative data from one experiment are shown. Error bars denote the standard deviation of three technical replicates.
Noncoding sRNAs
Hfq modulates stability of complexes formed by regulatory sRNAs and their target transcripts. In S. meliloti, as well as in other bacteria, intracellular accumulation of sRNAs also depends on Hfq (Zhang et al. 2003; Valentin-Hansen et al. 2004; Brennan and Link 2007; Voss et al. 2009). The SymbiosisChip contains evenly tiled probe sets corresponding to S. meliloti intergenic regions (IG) >150 bp (Barnett et al. 2004), making it suitable for the identification of sRNAs (although certain limitations of this methodology are discussed below).
Hybridization to IG represented on the SymbiosisChip may result from several scenarios: i) 5′ and 3′ untranslated leader sequences of ORF; ii) read-through transcription of operon IG, or from a promoter distantly located from the IG; iii) bona fide genes that were not annotated prior to design of the SymbiosisChip (for example, cyoD, corresponding to IG SMb21489iSMb21688f1); iv) transcription from cryptic promoters that do not express functional genes; and v) noncoding sRNAs contained within an IG. These experiments identified 54 candidate hfq-dependent IG. Analyses of the genomic context of each IG ruled out scenarios i to iii, above, and identified 11 candidate sRNAs (Table 3). In all, 2 of these 11 were previously confirmed to be sRNAs by Northern hybridization and mapping and 3 had been predicted in silico (del Val et al. 2007; Ulve et al. 2007; Valverde et al. 2008). Transcripts for 6 of the 11 IG (Table 3, lines 1, 2, 4, 6, 8, and 9) were detectible in 5-week-old nodules (Barnett et al. 2004).
Table 3.
Selected intergenic regions (IGs) showing hfq-dependent differential hybridization
| IGa | Coordinatesb | Directionc | hfq Δ changed | Confirmed sRNAe | References |
|---|---|---|---|---|---|
| 1. SMa2167-SMa2165 | 1,220,730–1,221,104 | >>> | 3.0× ↑ | smA8 (in silico) | Valverde et al. 2008 |
| 2. SMb20539-SMb20540 | 566,661–567,238 | <<< | 2.8, 2.4× ↓ | N.D. | This study |
| 3. SMb20547-SMb20548 | 573,363–573,645 | >>> | 2.0× ↓ | N.D. | This study |
| 4. SMb21698-exoI2 | 667,960–668,302 | <<< | 3.0× ↓ | N.D. | This study |
| 5. SMc00290-SMc00291 | 1,860,431–1,860,626 | >>> | 2.4× ↓ | N.D. | This study |
| 6. SMc01209-SMc01210 | 1,718,586–1,719,313 | <<< | 3.3, 2.6× ↑ | N.D. | This study |
| 7. SMc01933-proS | 1,398,105–1,398,459 | <<< | 2.1× ↓ | SmrC9, sra32 (Northern) |
Ulve et al. 2007; del Val et al. 2008; Valverde et al. 2008 |
| 8. SMc02051-tig | 1,667,618–1,667,986 | <<< | 2.8× ↑ | sm7 (in silico) | Valverde et al. 2008 |
| 9. SMc02855-SMc02856 | 206,784–207,013 | <<< | 2.7× ↑ | N.D. | This study |
| 10. SMc03061-SMc03062 | 759,133–759,383 | <<< | 7.6× ↓ | Candidate 182 (in silico) | Valverde et al. 2008 |
| 11. SMc04453-SMc01885 | 2,320,931–2,321,247 | <<< | 2.2× ↑ | sm26 (Northern) | Valverde et al. 2008 |
IG is given in the format “left-to-right” with respect to the Sinorhizobium meliloti genome annotation. SMa = pSymA-located gene, SMb = pSymB, and SMc = chromosome.
Corresponds to the sequence coordinates for the probe set or sets that showed an hfq-dependent difference in transcript accumulation.
Transcription direction for the putative transcript detected in IG with respect to genome annotation (Galibert et al. 2001).
Magnitude and direction of fold change for the IG in 1021Δhfq versus wild-type Rm1021. Where two values are given, this indicates that a significant change was detected in two adjacent IG probe sets.
sRNA = regulatory small RNA and N.D. = not done.
Symbiotic phenotypes of the hfq mutant
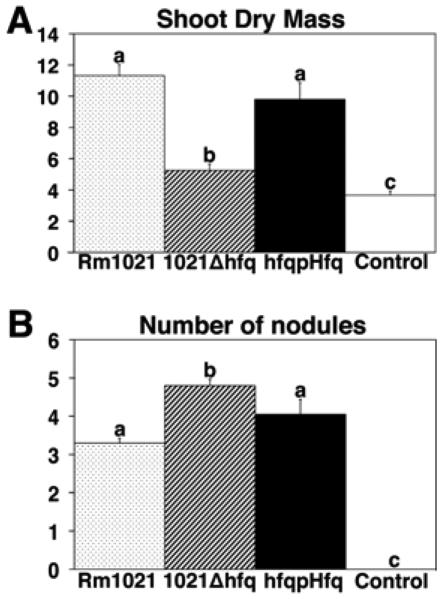
Medicago truncatula plants inoculated with the hfq mutant displayed a Fix− phenotype, characterized by nodules that were defective in nitrogen fixation. Normal nitrogen-fixing nodules are pink due to the presence of the oxygen-sequestering protein leghemoglobin. Nodules induced by 1021Δhfq were small, white, and cylindrical, similar to those induced by other nonfixing mutants (Fig. 5) (Griffitts and Long 2008). Plants inoculated with 1021Δhfq had significantly more nodules formed on their entire main roots than those inoculated with the wild type (Fig. 6), a phenotype commonly observed with Fix− mutants (Paau et al. 1985; Lodwig et al. 2003). However, there were no significant differences in nodule number between the wild type and 1021Δhfq above RT1 (the zone at the root tip at the time of inoculation) or between RT1 and RT2 (the section of the root susceptible to rhizobial infection during the following 24 h) (Caetano-Anolles et al. 1988). These regions correspond to sustained infections that initiated approximately 8 to 12 h after inoculation and 12 to 36 h after inoculation; therefore, the kinetics of nodulation initiation by the Rm1021 and the 1021Δhfq mutant were similar within 36 h after inoculations. Despite the hypernodulation, plants inoculated with the hfq mutant were stunted and their leaves were small (Fig. 5). Dry mass of 1021Δhfq-infected plant shoots was approximately half that of plants infected by wild-type Rm1021 but approximately 40% more than that of the control plants (Fig. 6). These results suggest the hfq mutant is providing its plant host with insufficient fixed nitrogen. Complementation of 1021Δhfq with pHfq restored the wild-type phenotypes (Figs. 5 and 6).
Fig. 5.
Symbiotic phenotype of Sinorhizobium meliloti 1021Δhfq. Top panels show 18-day-old shoots of Medicago truncatula seedlings inoculated with S. meliloti A, Rm1021; B, 1021Δhfq; and C, 1021Δhfq pHfq. D, E, and F, Representative nodules harvested from the plants in the upper panels. Nodules formed within the susceptible zones (RT1 and RT2) of main roots were harvested 18 days postinoculation. Images are on the same scale, which was assured by placing a ruler in the photographed field (left of panel A) and then cropping segments of the same size using Adobe Photoshop. Scale bar in F = 1 mm.
Fig. 6.
Shoot dry mass and number of nodules of Medicago truncatula plants infected by Rm1021, 1021Δhfq, and 1021Δhfq pHfq. A, Average shoot dry masses (n ≥ 7 plants). B, Average nodule numbers on main roots per plant (n ≥ 21 plants). Error bars represent standard error of the mean. Significant differences were determined using Student’s t test (P < 0.05) and are indicated by different letters.
To determine whether the Fix− phenotype of 1021Δhfq was due to an invasion defect, M. truncatula seedlings were inoculated with the wild type and 1021Δhfq expressing β-galactosidase from PhemA-lacZ (Fig. 7) (Leong et al. 1985). These observations indicated that the hfq mutant invaded nodule tissue successfully.
Fig. 7.
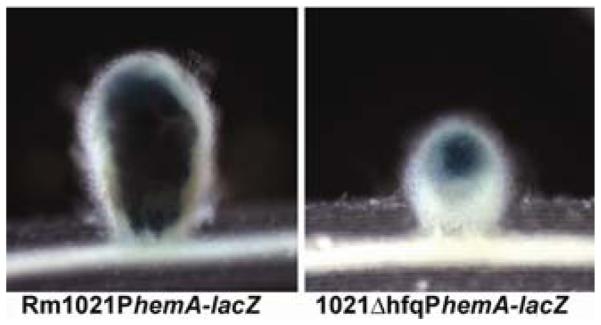
β-Galactosidase activity in nodules infected by Rm1021 (PhemA-lacZ) (left) and 1021Δhfq (PhemA-lacZ) (right). Eighteen-day-old nodules were harvested and stained as described (Griffitts et al. 2008).
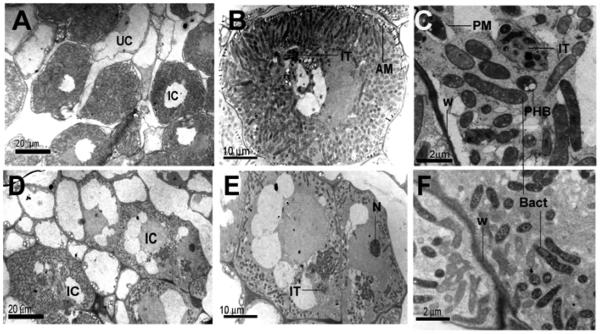
Transmission electron microscopy (TEM) was used to define the specific symbiotic block of the hfq mutant. Synchronous nodules harvested from the susceptible zone (between RT1 and RT2; discussed below) were used to minimize developmental variation. TEM of 3-week-old nodules indicates that hfq mutant bacteria were released from infection threads and differentiated into bacteroids (Fig. 8D through F). However, the number of bacteroids per host cell was lower for the hfq mutant than for the wild type, and their peribacteroid membranes (PM) were less visible (Fig. 8F compared with C). Both developed into elongated bacteroids similar in size to type 3 and type 4 bacteroids described by Vasse and associates (1990). The Fix− phenotype of the hfq mutant (Figs. 5 and 6) suggests that most, if not all, of the elongated bacteroids in the hfq mutant are type 3 bacteroids.
Fig. 8.
Electron micrographs of Medicago truncatula cells from 3-week-old nodules infected by A, B, and C, Rm1021 or D, E, and F, the 1021Δhfq mutant. Nodules were harvested from main roots between the position of the root tip at the time of inoculation (RT1) and the position of the root tip 24 h after inoculation (RT2). IC, Infected cells; UC, uninfected cells; IT, infection threads; W, plant cell wall; N, nucleolus; Bact, bacteroids; AM, amyloplasts; PHB, polyhydroxybutyrate granules. Brightness and contrast of images were adjusted using default settings of Adobe Photoshop version 8.0.
Wild-type bacteroids may store carbon more efficiently than the hfq mutant. Recently released wild-type bacteroids harbored large polyhydroxybutyrate (PHB) granules, whereas most hfq cells did not (Fig. 8C compared with F). It was previously observed that plant amyloplasts are located near the intercellular spaces of host cells in a normal symbiosis but not in Fix− nodules (Vasse et al. 1990). Consistent with this report, amyloplasts were present in wild-type nodules but were rare in hfq mutant nodules (Fig. 8B compared with E).
DISCUSSION
Influence of Hfq on carbon and nitrogen metabolism
Under nonstress conditions in half-strength tryptone yeast (TY) medium, the hfq mutant grew somewhat more slowly than the wild type, with a growth rate of 0.0251 U h–1 (compared with 0.0271 U h–1 for Rm1021 and 0.0252 U h–1 for 1021Δhfq pHfq) (Fig. 4B). The Phenotype microarray and Affymetrix SymbiosisChip experiments suggest that slower growth is not a result of a general growth defect but, rather, a consequence of very specific changes in uptake of nutrients and metabolism. The most striking free-living phenotype observed in our studies was the large number of ABC transporter transcripts that showed differential accumulation. This suggests that S. meliloti Hfq controls either i) stability or translation of many different ABC transporter transcripts directly through sRNA interactions with ABC transporter mRNAs; ii) stability or translation of a single or small number of transcripts via sRNA, with consequent effects on ABC transporter gene expression; or iii) some combination of i and ii.
Our results showing increased accumulation of transcripts corresponding to amino acid uptake systems in the hfq mutant (livHMGFK, aapJQMP, SMc02356-02358) suggest that Hfq reduces uptake of branched chain and polar amino acids in free-living cells. In Rhizobium leguminosarum, BraGDEFC (ortholog of LivHMGFK) has a very broad substrate specificity: affinity for polar and branched-chain amino acids such as L-Glu, 2-aminoisobutyric acid, L-His, GABA, L-Leu, L-Ile, and L-Val; and lesser affinity for D-amino acids (Hosie et al. 2002). R. leguminosarum AapJQMP transports L-amino acids, primarily Glu, Asp, His, and Pro, and can affect amino acid efflux (Walshaw and Poole 1996). SMc02356 is 47% identical to LivK but its transported solute is unknown. These results are interesting in light of reports that plant-supplied branchedchain amino acids (Leu, Ile, and Val) regulate bacteroid development and persistence, and that bra and aap are required for their bacterial uptake (Prell et al. 2009). Similar regulation may occur in S. meliloti but, because substrate specificity of LivK and AapJ is predicted to be broad and has not been experimentally determined, we cannot rule out a role for other amino acids during the symbiosis. In fact, similar transporters in the plant pathogen Agrobacterium tumefaciens (which is a close relative of S. meliloti) transport GABA, not LIV (Haudecoeur et al. 2009). Thus, it is possible that LivK and SMc02356 participate in GABA transport in S. meliloti. In addition to livK and SMc02356, mRNA corresponding to a putative GtsA GABA solute-binding protein also increased in 1021Δhfq. If one or more of these are, indeed, used for GABA transport in S. meliloti, their upregulation could explain why 1021Δhfq appeared to utilize GABA as sole carbon source better than the wild type in Phenotype microarrays.
Our transporter data have important implications for symbiosis because S. meliloti transports a broad range of growthlimiting nutrients from the soil, rhizosphere (Mauchline et al. 2006), and, perhaps, from nodules (Lodwig et al. 2003; Prell and Poole 2006). Unregulated synthesis of ABC transporters could explain why, in two cases, transport was not well coordinated with metabolism. For example, mRNA of pckA and fbaB, involved in C4-dicarboxylate catabolism, increased in the mutant, but dctA, encoding a C4-dicarboxylate transporter, did not. Also, whereas synthesis of branched-chain amino acid transporters apparently increased in the hfq mutant, corresponding utilization genes were downregulated (Fig. 1).
α-Glucosides (trehalose, sucrose, and maltose), present in root exudates and decaying plant matter, are likely important carbon sources for soil-dwelling rhizobia. mRNAs encoding α-glucoside transport and utilization functions were strongly downregulated in the hfq mutant. The thu genes are primarily involved in trehalose utilization (Jensen et al. 2002) and the agl genes in sucrose utilization (Willis and Walker 1999), though there is some cross-specificity. It was reported that an aglE thuE double mutation reduced import of α-glucosides 90% (Jensen et al. 2002) but, in our Phenotype microarray assays, 1021Δhfq respired as well as the wild type on trehalose, sucrose, or maltose. This suggests that either decreased expression of agl and thu genes in the hfq mutant is not rate-limiting for utilization or it may simply reflect that different growth conditions were used for SymbiosisChips (complex TY medium) and Phenotype microarrays (α-glucoside is the sole carbon source).
Hfq modulates responses to pH stress
In α- and γ-proteobacteria, hfq mutants are defective in resistance to pH and envelope stress, reactive oxygen species, osmotic and heat shock, and stationary phase growth (Robertson and Roop 1999; Sittka et al. 2007; Ansong et al. 2009). In the present study, we did not observe differential regulation of large numbers of stress-related genes, probably because cells were grown in rich medium under nonstress conditions. However, we detected differential accumulation of mRNA for actSR, a global regulator important for acid tolerance, and for supAB, implicated in response to alkaline stress. Although Hfq has been implicated in acid tolerance in other species, including Brucella and Salmonella (Ansong et al. 2009; Robertson and Roop 1999), we found that deletion of hfq was more deleterious for growth at high pH than at low pH. Further characterization of the role of Hfq in S. meliloti stress adaptation will expand our knowledge of its role in stress response and provide clues as to its function in nodule bacteria.
Hfq affects accumulation of sRNA and transcripts encoding regulatory proteins
Hfq is known to control synthesis of regulatory proteins. Thus, it may indirectly affect expression of large sets of downstream genes. We found hfq-dependent changes in expression of genes encoding transcriptional regulators actR, glpR, cheY, and seven others with unknown functions. One or more of these may be responsible for Hfq-dependent but indirect alterations in S. meliloti gene expression.
We expected that, if there are sRNAs whose stability or expression depend on Hfq, then we would be able to detect differential accumulation of these sRNAs via the tiled intergenic probes on the SymbiosisChip. These experiments provided proof of concept (Table 3): five of the sRNAs we detected were previously confirmed or predicted in S. meliloti (del Val et al. 2007; Ulve et al. 2007; Valverde et al. 2008). A recent study demonstrated that S. meliloti Hfq likely interacts with SmrC15 and SmrC16 sRNAs and controls expression of Sra33 sRNA (Voss et al. 2009). Although we saw hybridization corresponding to these sRNAs, our experiments did not detect statistically significant hfq-dependent differences in their accumulation. There are probably other Hfq-dependent sRNAs that we did not identify in study, because we did not optimize our methods for sRNA detection. For example, the RNeasy columns (Qiagen) we used for purification of RNA do not efficiently retain RNA smaller than 200 nucleotides and we did not size fractionate RNA to increase identification efficiency (Landt et al. 2008; Valverde et al. 2008). Moreover, we used Affymetrix software with default settings for analysis: employing software specifically designed for analysis of evenly tiled probes would likely improve detection sensitivity.
The hfq mutant is severely impaired for symbiosis with M. truncatula.
The hfq mutant showed severe symbiotic defects consistent with a nonfixing phenotype. This phenotype may be due at least in part to an evolutionarily conserved effect of Hfq on nifA expression (Kaminski et al. 1994; Drepper et al. 2002; Zhang and Hong 2009). In shake cultures, we failed to observe differential accumulation of nifA in the hfq mutant; however, because nifA is known to be expressed at low levels under these conditions, this hypothesis remains to be tested.
Hfq-dependent changes in nitrogen metabolism may be deleterious to symbiosis. As discussed above, uptake of branchedchain amino acids by the bacterium is required for an effective symbiosis (Prell et al. 2009). However, the SymbiosisChip data for free-living bacteria suggest increased, not decreased, uptake of branched-chain amino acids by the hfq mutant. It is possible that regulation of Liv/Aap transporter synthesis is different in free-living rhizobia and bacteroids. It is also possible that changes in branched-chain amino acid catabolism (Fig. 1) contribute, in part, to the symbiotic phenotype of the hfq mutant. These changes in accumulation were not dramatic; however, the hfq mutant failed to utilize valine as a sole nitrogen source in Phenotype microarrays. If valine is the sole branched-chain amino acid transported to the bacteria within the nodule, a failure to utilize valine could result in symbiotic defects.
TEM suggests that 1021Δhfq does not accumulate as much PHB as does the wild type. Although regulation of PHB utilization in planta was not tested in this study, and PHB biosynthesis is not essential for an effective symbiosis (Wang et al. 2007), we note that PHB utilization transcripts (acsA1, phbB, and bhbA) were differentially regulated by hfq during growth in culture, perhaps at least partially explaining the observed lack of PHB granules in bacteroids formed by 1021Δhfq.
Comparing global gene expression in wild-type nodules to hfq mutant nodules, in coordination with assays of symbiotic phenotypes of putative Hfq-dependent genes, will help us unravel the role of Hfq in symbiosis.
MATERIALS AND METHODS
Growth of bacterial strains
Bacteria were grown in Luria-Bertani medium with NaCl at 5 g/liter (Fisher Scientific, Pittsburgh) or TY (Beringer 1974) rich medium or in M9 sucrose minimal medium (Meade and Signer 1977) supplemented with antibiotics at the following concentrations: streptomycin at 500 μg/ml, neomycin at 50 to 200 μg/ml, tetracycline at 2.5 to 5 μg/ml, gentamicin at 30 μg/ml, ampicillin at 100 μg/ml, and kanamycin at 50 μg/ml. E. coli and S. meliloti were grown at 37 and 30°C, respectively. Plasmids were mobilized into S. meliloti by triparental conjugation using helper plasmid pRK600 (Finan et al. 1986). For pH growth assays, cultures were grown in half-strength TY supplemented with 1.7 mM CaCl2 and buffered with either 40 mM morpholineethanesulfonic acid (MES) to pH 5.3 or 40 mM TRIS to pH 9.0 (Fenner et al. 2004). The final pH of spent cultures was monitored and did not vary by more than 0.2 units from the initial pH.
Construction of strains and plasmids
To create an in-frame deletion of hfq, regions flanking hfq (SMc01048) were PCR amplified using primer pairs MG201hfqF1/MG202hfqR1 and MG203hfqF2/MG204hfqR2 (all oligonucleotide sequences are in Supplementary Table S6). The resulting PCR products were digested with BamHI and ligated to each other, and the entire ligation reaction was used as a template for a second round of PCR using MG201hfqF1 and MG204hfqR2. The resulting PCR product containing the hfq deletion, with start and stop codons in frame (junction sequence ATGTCCTGA), was cloned into pCR2.1 (Invitrogen, Carlsbad, CA, U.S.A.) and sequenced to confirm absence of PCR-introduced mutations. The fragment containing the hfq deletion was excised from pCR2.1 as a 1.4-kb SpeI-XbaI fragment, recloned into suicide vector pJQ200SK (Quandt and Hynes 1993) to yield plasmid pJQ200SK-hfq9, and then mobilized into S. meliloti Rm1021 by triparental mating. Gentamicin-resistant transconjugants containing a single crossover in hfq were selected. Sucrose counterselection for double recombinants was performed using 5% sucrose as previously described (Barnett et al. 2001). Both presence of the in-frame hfq deletion and absence of the hfq ORF in 1021Δhfq was confirmed by PCR and sequencing.
To construct an hfq plasmid for complementation experiments, a wild-type copy of the gene was amplified using primers MG201hfqF1 and MG204hfqR2 and Rm1021 DNA as a template. The resulting PCR product, containing the entire hfq ORF plus 580 bp of upstream and 778 bp of downstream DNA, was cloned into pCR2.1, sequenced, and recloned into the SpeI-XbaI of pBBR1-MCS3 (Kovach et al. 1995), resulting in pHfq.
Affymetrix SymbiosisChip analysis
Three biological replicates were used for each strain, wildtype S. meliloti Rm1021 or its isogenic mutant 1021Δhfq. For each replicate, a single bacterial colony was inoculated into 20 ml of TY with streptomycin at 500 μg/ml and grown with shaking at 200 rpm and 30°C in 250-ml baffled flasks. Cells were harvested during late exponential phase (optical density at 600 nm [OD600] = 0.84 to 0.89). RNA extraction, cDNA synthesis, cDNA fragmentation and labeling, and hybridization to Affymetrix SymbiosisChips were performed as previously described (Barnett et al. 2004). The dual-genome Affymetrix SymbiosisChip contains “probe sets” for all S. meliloti ORF annotated in 2001 (Galibert et al. 2001), intergenic regions over 150 bp, and approximately 10,000 probe sets corresponding to expressed sequence tags of the plant host, M. truncatula.
cDNA (4 μg) was hybridized to each chip. Affymetrix software (GCOS v1.4 and DMT v3.1) was used for data analysis and mining as previously described (Barnett et al. 2004). Affymetrix comparison analysis was performed on six arrays (three Rm1021 and three 1021Δhfq) to yield nine pairwise comparisons. An average SLR for the increase or decrease change was judged to be significant only if at least eight of the nine pairwise comparisons were deemed by the software to be significantly changed (P value ≤ 0.05). SLR is expressed as the log2 ratio of the change (i.e., a SLR of 1 equals a twofold change). The data discussed in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (Edgar et al. 2002) and are accessible through GEO Series accession number GSE19346.
qPCR
Cultures were grown under the same conditions but independently of those used for SymbiosisChip analysis. RNA was extracted using Total RNA Isolation and DNA-free kits (Ambion, Austin, TX, U.S.A.). Quality of RNA preparations was assessed using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, U.S.A.). First-strand cDNA was synthesized from 2 μg of RNA using a SuperScript VILO cDNA synthesis kit (Invitrogen). Gene-specific primers for qPCR were designed using Primer Express 3.0 (Applied Biosystems, Foster City, CA, U.S.A.). SMc02461 and SMc00129 were used as internal controls because cycle threshold values of these genes were similar under a number of different conditions (Becker et al. 2004). qPCR reactions were performed using SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7500 FAST real-time PCR system. Data from the experiments were processed using ABI 7500 software V2.0.1. Each sample contained pooled RNA preparations from three independent cultures. Each average value was from three independent cDNA and qPCR reactions.
Phenotype microarray (Biolog) assays
Stationary-phase cultures grown in TY were centrifuged, washed three times, and resuspended in inoculation fluid (IF-0; Biolog, Inc., Hayward, CA, U.S.A. ) to an OD600 = 0.2 as per the manufacturer’s instructions. IF-0 buffer was supplemented with 40 nM D-biotin and 120 nM cyanocobalamin for carbon utilization assays with PM1 and PM2 MicroPlates, or with 2.5 mM MgCl2 and 2.5 mM CaCl2 for the nitrogen utilization assays with the PM3 MicroPlate. Cell suspensions (100 μl) were added to each well and plates were incubated at 30°C. Absorbance at 590 nm in three independently inoculated experiments was measured using a Wallac 1420 plate reader (Perkin Elmer, Waltham, MA, U.S.A.) immediately following inoculation and after 3 days of inoculation. For growth confirmation tests, either 0.2% GABA or 0.2% pyruvic acid was used as a sole carbon source in M9 minimal medium with ammonium as the nitrogen source. In M9 minimal medium, 4.5 mM L-Val was used as sole nitrogen sources, with sucrose as the carbon source.
Motility assays
Swimming motility was assayed on soft agar (0.3%) plates with either 20-fold diluted or full-strength TY medium with CaCl2 at 3.4 mM. Plates were spot inoculated with 2 μl of washed log phase (0.5 to 0.8. at OD600) bacterial suspensions.
TLC analysis of QS signals
Two independent 10-ml cultures for each strain were grown in TY liquid medium buffered to pH 6.5 with 10 mM MES to prevent alkaline inactivation of AHL. Supernatants were collected when cultures reached OD600 = 0.98 to 1.08 and extracted with dichloromethane or acidified ethyl acetate as described (Teplitski et al. 2003; Gao et al. 2005). The extracts were dried down and then resuspended in 100 μl of 70:30 methanol/water. Suspensions of two independent extracts of the same strain were pooled together. Then, 10 μl from each pooled sample (corresponding to 1 ml of the original supernatant) were spotted onto Whatman KC18F TLC plates (Whatman, Maidstone, U.K.). Plates were developed with 60:40 methanol/water and dried. To detect AHL, the plates were overlaid with an agar suspension of A. tumefaciens NT1 pZLR4 and the substrate 5-bromo-4-chloro-3-indolyl b-D-galactopyranoside as previously described (Cha et al. 1998; Gao et al. 2005). pZLR4 carries a traR receptor gene and a traG::lacZ fusion whose expression is induced in the presence of AHL and other active QS compounds.
Nodulation assays
Seed of M. truncatula A17 were surface sterilized and germinated as previously described (Gao et al. 2005). Approximately 160 seedlings were grown in 30 pouches (Mega International, St. Paul, MN, U.S.A.), incubated in walk-in plant growth chambers (at 24°C under a cycle of 16 h of light and 8 h of darkness). Cultures of Rm1021 and 1021Δhfq, each containing the PhemA-lacZ plasmid (for LacZ staining) (Leong et al. 1985), or Rm1021 and 1021Δhfq (for transmission electron microscopy and nodulation assays), were grown to mid-log phase in TY medium. Cells were washed, resuspended in equal volumes of water, and spot inoculated on the primary roots of plants (100 μl/plant). The position of the primary root tips was marked at the time of inoculation (RT1) on the pouch face for each seedling, as was the position of the root 24 h later (RT2) to indicate the developmental zones that are transiently susceptible to infection as defined by Caetano-Anolles and associates (1988). We can infer that nodules formed above RT1 were initiated within approximately 12 h after inoculation, whereas nodules formed between RT1 and RT2 were initiated between 12 and 36 h after inoculation. The kinetics of nodule formation within the RT1-RT2 region, above RT1, and below RT2 was monitored throughout the duration of the experiment. Dry mass of individual seedlings was measured as an indicator of symbiotic nitrogen fixation efficiency.
Microscopy
For visualization of nodule bacteria by light microscopy, a plasmid expressing lacZ from the S. meliloti hemA promoter (Leong et al. 1985) was introduced into Rm1021 and 1021Δhfq strains. These bacteria were used to infect plants as described above. Eighteen days after inoculation, at least 20 Rm1021- and 1021Δhfq-induced nodules were harvested from susceptible zones. Nodules were stained for β-galactosidase activity as previously described (Griffitts et al. 2008).
For TEM, nodules were harvested from susceptible zones 3 weeks postinoculation. Before harvesting for TEM experiments, several nodules from roots that had been inoculated in parallel by PhemA-lacZ-labeled Rm1021 and 1021Δhfq were collected from susceptible zones and stained to monitor synchronous nodule development. Eighteen nodules from each time point (six nodules each of Rm1021, 1021Δhfq, and 1021Δhfq pHfq) were processed as previously described: tissues were fixed in 4% formaldehyde and 0.1 M sodium cacodylate buffer, dehydrated, and embedded in LR White hard grade (London Resin, Reading, U.K.) (Gao et al. 2001). Two of six processed nodules of each kind were sectioned longitudinally, first by semithin sections until infected host tissue was visible, and then by 60-nm-thin sections between the semithin sections until halfway through the nodules to cover the entire infected area. Discontinuous thin sections were stained with uranyl acetate and lead citrate. Ultrathin sections were examined using a Hitachi H-7500EM electron microscope.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Bauer and R. Fisher for critically reading the manuscript, A. Eberhard for the gift of 3-oxo-C16:1-HSL, H. J. Hao for assistance with electron microscopy, and A. Al-Agely and B. Creary for technical support of this project. This work was supported by United States Department of Agriculture NRI 2007-35319-18158 (to M. Teplitski), NIH GM069342 (to S. R. Long), and Hoover Circle Fund (to S. R. Long).
Footnotes
*The e-Xtra logo stands for “electronic extra” and indicates that six supplementary tables and one supplementary figure are available online.
AUTHOR-RECOMMENDED INTERNET RESOURCES
NCBI Gene Expression Omnibus series: www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19346 RhizoGATE Sinorhizobium meliloti portal: www.rhizogate.de Sinorhizobium meliloti/Medicago truncatula symbiosisChip: cmgm.stanford.edu/biology/long/genechip.htm
LITERATURE CITED
- Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, Adkins JN, McClelland M, Heffron F, Smith RD. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: Implications for virulence and global protein translation. PLoS ONE. 2009;4:e4809. doi: 10.1371/journal.pone.0004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett MJ, Hung DY, Reisenauer A, Shapiro L, Long SR. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J. Bacteriol. 2001b;183:3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett MJ, Toman CJ, Fisher RF, Long SR. A dual-genome Symbiosis Chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16636–16641. doi: 10.1073/pnas.0407269101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Berges H, Krol E, Bruand C, Ruberg S, Capela D, Lauber E, Meilhoc E, Ampe F, de Bruijn FJ, Fourment J, Francez-Charlot A, Kahn D, Kuster H, Liebe C, Puhler A, Weidner S, Batut J. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant-Microbe Interact. 2004;17:292–303. doi: 10.1094/MPMI.2004.17.3.292. [DOI] [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Tatti E, Comparini D, Giuntini E, Mocali S, Giovannetti L, Bazzicalupo M, Mengoni A, Viti C. Metabolic capacity of Sinorhizobium (Ensifer) meliloti strains detected by phenotype microarray. Appl. Environ. Microbiol. 2009;75:5396–5404. doi: 10.1128/AEM.00196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR. Global phenotypic characterization of bacteria. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2009;33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Caetano-Anolles G, Crist-Estes DK, Bauer WD. Chemotaxis of Rhizobium meliloti to the plant flavone luteolin requires functional nodulation genes. J. Bacteriol. 1988;170:3164–3169. doi: 10.1128/jb.170.7.3164-3169.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha C, Gao P, Chen YC, Shaw PD, Farrand SK. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- del Val C, Rivas E, Torres-Quesada O, Toro N, Jimenez-Zurdo JI. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol. Microbiol. 2007;66:1080–1091. doi: 10.1111/j.1365-2958.2007.05978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic MA, Chen HC, Natera S, Van Noorden G, Menzel C, Taylor S, Renard C, Geiger O, Weiller GF, Sinorhizobium DNA Sequencing Consortium A global analysis of protein expression profiles in Sinorhizobium meliloti: Discovery of new genes for nodule occupancy and stress adaptation. vMol. Plant-Microbe Interact. 2003;16:508–524. doi: 10.1094/MPMI.2003.16.6.508. [DOI] [PubMed] [Google Scholar]
- Drepper T, Raabe K, Giaourakis D, Gendrullis M, Masepohl B, Klipp W. The Hfq-like protein NrfA of the phototrophic purple bacterium Rhodobacter capsulatus controls nitrogen fixation via regulation of nifA and anfA expression. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2002;215:221–227. doi: 10.1111/j.1574-6968.2002.tb11394.x. [DOI] [PubMed] [Google Scholar]
- Duan Y, Zhou L, Hall DG, Li W, Doddapaneni H, Lin H, Liu L, Vahling CM, Gabriel DW, Williams KP, Dickerman A, Sun Y, Gottwald T. Complete genome sequence of citrus huang-longbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant-Microbe Interact. 2009;22:1011–1020. doi: 10.1094/MPMI-22-8-1011. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner BJ, Tiwari RP, Reeve WG, Dilworth MJ, Glenn AR. Sinorhizobium medicae genes whose regulation involves the ActS and/or ActR signal transduction proteins. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2004;236:21–31. doi: 10.1016/j.femsle.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Finan TM, Kunkel B, De Vos GF, Signer ER. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM, Weidner S, Wong K, Buhrmester J, Chain P, Vorholter FJ, Hernandez-Lucas I, Becker A, Cowie A, Gouzy J, Golding B, Puhler A. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9889–9894. doi: 10.1073/pnas.161294698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer HM. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RF, Long SR. Rhizobium--plant signal exchange. Nature. 1992;357:655–660. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dreano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernandez-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thebault P, Vandenbol M, Vorholter FJ, Weidner S, Wells DH, Wong K, Yeh KC, Batut J. The composite genome of the legume symbiont Sino-rhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- Gao M, D’Haeze W, De Rycke R, Wolucka B, Holsters M. Knockout of an azorhizobial dTDP-L-rhamnose synthase affects lipopolysaccharide and extracellular polysaccharide production and disables symbiosis with Sesbania rostrata. Mol. Plant-Microbe Interact. 2001;14:857–866. doi: 10.1094/MPMI.2001.14.7.857. [DOI] [PubMed] [Google Scholar]
- Gao M, Chen H, Eberhard A, Gronquist MR, Robinson JB, Rolfe BG, Bauer WD. sinI- and expR-dependent quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 2005;187:7931–7944. doi: 10.1128/JB.187.23.7931-7944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffitts JS, Long SR. A symbiotic mutant of Sinorhizobium meliloti reveals a novel genetic pathway involving succinoglycan biosynthetic functions. Mol. Microbiol. 2008;67:1292–1306. doi: 10.1111/j.1365-2958.2008.06123.x. [DOI] [PubMed] [Google Scholar]
- Griffitts JS, Carlyon RE, Erickson JH, Moulton JL, Barnett MJ, Toman CJ, Long SR. A Sinorhizobium meliloti osmosensory two-component system required for cyclic glucan export and symbiosis. Mol. Microbiol. 2008;69:479–490. doi: 10.1111/j.1365-2958.2008.06304.x. [DOI] [PubMed] [Google Scholar]
- Haudecoeur E, Planamente S, Cirou A, Tannieres M, Shelp BJ, Morera S, Faure D. Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14587–14592. doi: 10.1073/pnas.0808005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie AH, Allaway D, Galloway CS, Dunsby HA, Poole PS. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J. Bacteriol. 2002;184:4071–4080. doi: 10.1128/JB.184.15.4071-4080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Dhimole N, Khan AR, De D, Tomar SK, Sajish M, Dutta D, Parrack P, Prakash B. E. coli HflX interacts with 50S ribosomal subunits in presence of nucleotides. Biochem. Biophys. Res. Commun. 2009;379:201–205. doi: 10.1016/j.bbrc.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JB, Peters NK, Bhuvaneswari TV. Redundancy in periplasmic binding protein-dependent transport systems for trehalose, sucrose, and maltose in Sinorhizobium meliloti. J. Bacteriol. 2002;184:2978–2986. doi: 10.1128/JB.184.11.2978-2986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JB, Ampomah OY, Darrah R, Peters NK, Bhuvaneswari TV. Role of trehalose transport and utilization in Sinorhizobium meliloti–alfalfa interactions. Mol. Plant-Microbe Interact. 2005;18:694–702. doi: 10.1094/MPMI-18-0694. [DOI] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: The Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski PA, Desnoues N, Elmerich C. The expression of nifA in Azorhizobium caulinodans requires a gene product homologous to Escherichia coli HF-I, an RNA-binding protein involved in the replication of phage Q beta RNA. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4663–4667. doi: 10.1073/pnas.91.11.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L. Small non-coding RNAs in Caulobacter crescentus. Mol. Microbiol. 2008;68:600–614. doi: 10.1111/j.1365-2958.2008.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Leong SA, Williams PH, Ditta GS. Analysis of the 5′ regulatory region of the gene for delta-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res. 1985;13:5965–5976. doi: 10.1093/nar/13.16.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DX, Tang H, Wang ET, Chen WX. An ABC transporter is required for alkaline stress and potassium transport regulation in Sinorhizobium meliloti. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2009;293:35–41. doi: 10.1111/j.1574-6968.2009.01500.x. [DOI] [PubMed] [Google Scholar]
- Lodwig EM, Hosie AH, Bourdes A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature. 2003;422:722–726. doi: 10.1038/nature01527. [DOI] [PubMed] [Google Scholar]
- Mauchline TH, Fowler JE, East AK, Sartor AL, Zaheer R, Hosie AH, Poole PS, Finan TM. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17933–17938. doi: 10.1073/pnas.0606673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade HM, Signer ER. Genetic mapping of Rhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 1977;74:2076–2078. doi: 10.1073/pnas.74.5.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J, Campos R, BenAbdelkhalek H, Olivares J, Lluch C, Sanjuan J. Rhizobium tropici genes involved in free-living salt tolerance are required for the establishment of efficient nitrogen-fixing symbiosis with Phaseolus vulgaris. Mol. Plant-Microbe Interact. 2002;15:225–232. doi: 10.1094/MPMI.2002.15.3.225. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paau AS, Leps WT, Brill WJ. Regulation of nodulation by Rhizobium meliloti 102F15 on its mutant which forms an unusually high number of nodules on alfalfa. Appl. Environ. Microbiol. 1985;50:1118–1122. doi: 10.1128/aem.50.4.1118-1122.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski K, Klosse U, de Bruijn FJ. Characterization of a novel Azorhizobium caulinodans ORS571 two-component regulatory system, NtrY/NtrX, involved in nitrogen-fixation and metabolism. Mol. Gen. Genet. 1991;231:124–138. doi: 10.1007/BF00293830. [DOI] [PubMed] [Google Scholar]
- Polkinghorne A, Ziegler U, Gonzalez-Hernandez Y, Pospischil A, Timms P, Vaughan L. Chlamydophila pneumoniae HflX belongs to an uncharacterized family of conserved GTPases and associates with the Escherichia coli 50S large ribosomal subunit. Microbiology. 2008;154:3537–3546. doi: 10.1099/mic.0.2008/022137-0. [DOI] [PubMed] [Google Scholar]
- Prell J, Poole P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006;14:161–168. doi: 10.1016/j.tim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Prell J, White JP, Bourdes A, Bunnewell S, Bongaerts RJ, Poole PS. Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12477–12482. doi: 10.1073/pnas.0903653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- Robertson GT, Roop RM., Jr The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 1999;34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x. [DOI] [PubMed] [Google Scholar]
- Sauviac L, Philippe H, Phok K, Bruand C. An extracyto-plasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J. Bacteriol. 2007;189:4204–4216. doi: 10.1128/JB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Eberhard A, Gronquist M, Gao M, Robinson JB, Bauer WD. Chemical identification of N-acyl homoserine lactone quorum sensing signals produced by Sinorhizobium meliloti strains in defined medium. Arch. Microbiol. 2003;180:494–497. doi: 10.1007/s00203-003-0612-x. [DOI] [PubMed] [Google Scholar]
- Tiwari RP, Reeve WG, Dilworth MJ, Glenn AR. Acid tolerance in Rhizobium meliloti strain WSM419 involves a two-component sensor-regulator system. Microbiology. 1996;142:1693–1704. doi: 10.1099/13500872-142-7-1693. [DOI] [PubMed] [Google Scholar]
- Ulve VM, Sevin EW, Cheron A, Barloy-Hubler F. Identification of chromosomal alpha-proteobacterial small RNAs by comparative genome analysis and detection in Sinorhizobium meliloti strain 1021. BMC Genomics. 2007;8:467. doi: 10.1186/1471-2164-8-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Valverde C, Livny J, Schluter JP, Reinkensmeier J, Becker A, Parisi G. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics. 2008;9:416. doi: 10.1186/1471-2164-9-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Camut S, Truchet G. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 1990;172:4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss B, Hölscher M, Baumgarth B, Kalbfleisch A, Kaya C, Hess WR, Becker A, Evguenieva-Hackenberg E. Expression of small RNAs in Rhizobiales and protection of a small RNA and its degradation products by Hfq in Sinorhizobium meliloti. Biochem. Biophys. Res. Commun. 2009;390:331–336. doi: 10.1016/j.bbrc.2009.09.125. [DOI] [PubMed] [Google Scholar]
- Walshaw DL, Poole PS. The general L-amino acid permease of Rhizobium leguminosarum is an ABC uptake system that also influences efflux of solutes. Mol. Microbiol. 1996;21:1239–1252. doi: 10.1046/j.1365-2958.1996.00078.x. [DOI] [PubMed] [Google Scholar]
- Wang C, Saldanha M, Sheng X, Shelswell KJ, Walsh KT, Sobral BW, Charles TC. Roles of poly-3-hydroxybutyrate (PHB) and glycogen in symbiosis of Sinorhizobium meliloti with Medicago sp. Microbiology. 2007;153:388–398. doi: 10.1099/mic.0.29214-0. [DOI] [PubMed] [Google Scholar]
- Willis LB, Walker GC. A novel Sinorhizobium meliloti operon encodes an alpha-glucosidase and a periplasmic-binding-protein-dependent transport system for alpha-glucosides. J. Bacteriol. 1999;181:4176–4184. doi: 10.1128/jb.181.14.4176-4184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurgel SN, Kahn ML. Dicarboxylate transport by rhizobia. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2004;28:489–501. doi: 10.1016/j.femsre.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hong G. Post-transcriptional regulation of NifA expression by Hfq and RNase E complex in Rhizobium leguminosarum bv. viciae. Acta Biochim. Biophys. Sin. (Shanghai) 2009;41:719–730. doi: 10.1093/abbs/gmp060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.