Abstract
Context:
The widespread adoption of electronic health records (EHRs) offers significant opportunities to conduct research with clinical data from patients outside traditional academic research settings. Because EHRs are designed primarily for clinical care and billing, significant challenges are inherent in the use of EHR data for clinical and translational research. Efficient processes are needed for translational researchers to overcome these challenges. The Data QUEST Coordinating Center (DQCC), which oversees Data Query Extraction Standardization Translation (Data QUEST) – a primary-care, EHR data-sharing infrastructure – created processes that guide EHR data extraction for clinical and translational research across these diverse practices. We describe these processes and their application in a case example.
Case Description:
The DQCC process for developing EHR data extractions not only supports researchers’ access to EHR data, but supports this access for the purpose of answering scientific questions. This process requires complex coordination across multiple domains, including the following: (1) understanding the context of EHR data; (2) creating and maintaining a governance structure to support exchange of EHR data; and (3) defining data parameters that are used in order to extract data from the EHR. We use the Northwest-Alaska Pharmacogenomics Research Network (NWA-PGRN) as a case example that focuses on pharmacogenomic discovery and clinical applications to describe the DQCC process. The NWA-PGRN collaborates with Data QUEST to explore ways to leverage primary-care EHR data to support pharmacogenomics research.
Findings:
Preliminary analysis on the case example shows that initial decisions about how researchers define the study population can influence study outcomes.
Major Themes and Conclusions:
The experience of the DQCC demonstrates that coordinating centers provide expertise in helping researchers understand the context of EHR data, create and maintain governance structures, and guide the definition of parameters for data extractions. This expertise is critical to supporting research with EHR data. Replication of these strategies through coordinating centers may lead to more efficient translational research. Investigators must also consider the impact of initial decisions in defining study groups that may potentially affect outcomes.
Keywords: electronic health records, primary care, governance
Context
The widespread adoption of electronic health records (EHRs) offers significant opportunities to conduct research that addresses critical problems in clinical medicine.5,6 In contrast to claims-based data, which may capture only demographics, diagnoses, and procedures recorded for billing purposes, EHR systems provide a broader range of clinical data. These data include vital signs, diagnostic test results, social and family histories, prescriptions, and physical examination findings.7 EHRs are also a critical and effective source of data for studying small populations with rare conditions.8 For example, rural residing populations are less likely to participate in clinical research.9 Leveraging EHR data systems, which are widely adopted even in rural-serving primary care clinics, offers an important resource to address the paucity of rural subjects in clinical research studies.10 However, compared to large urban practices, rural-serving primary care practices, which are often independently owned and operated, may have fewer resources to support participation in EHR-based research.
EHRs are designed primarily for clinical care and billing. This leads to several potential problems inherent in the use of EHR data for clinical research.11,12 William Hersh and colleagues suggested that researchers consider five caveats related to data origin, or provenance, when planning to use EHR data for research: (1) Many EHR data have been transformed or coded for billing purposes; (2) Data entered in free text format may not be readily captured in easy-to-use formats; (3) EHRs may present multiple sources of data for a given measure or indicator of interest, resulting in potential inconsistencies in the data; (4) EHR data may not provide complete data; and (5) EHR data may contain inaccurate or incorrect data.3 Understanding the origin of information from EHR data is critical for accurately analyzing and interpreting EHR data for research.3 Researchers need efficient and feasible means of gathering this information.
There are several steps researchers may take to address problems encountered in the use of EHR data for research. When undertaking research with EHR data, researchers must consider both the technical specifications for creating data extractions and the impact of initial decisions that are needed to prepare EHR data for analysis. The DQCC team has developed an innovative three-step process to support development of EHR data extractions that includes helping investigators understand the context of EHR data, creating and maintaining data governance structures, and defining data parameters. The objectives of this paper are the following: (1) describe the process that the DQCC team uses; and (2) explore the impact of initial decisions about the study population that researchers make in preparing EHR data extractions for analysis. We suggest that the process that the DQCC has created is usable in other data sharing networks and will facilitate translational research with EHR data. We illustrate issues that arise when creating research data extractions and definitions of a patient population through a case example from pharmacogenomics research conducted in concert with the Northwest-Alaska Pharmacogenomics Research Network (NWA-PGRN).
Data Source
Data Query Extraction Standardization Translation (Data QUEST) is an infrastructure for sharing EHR data for research. Data QUEST was developed after an extensive needs and feasibility assessment with rural-serving primary care practices in the Washington, Wyoming, Alaska, Montana and Idaho (WWAMI) region Practice and Research Network (WPRN), The WPRN is a collaborative group of primary care practices across the five-state WWAMI region committed to research and quality improvement.16 The WPRN practices that partner with Data QUEST recognized data accessibility as a powerful way to facilitate research participation. Practices were also interested in supporting the including of rural populations in translational research. Our initial assessment identified local control of data management as a critical governance issue in developing the data sharing infrastructure.16 Our team created governance strategies to support data sharing, such as allowing partner organizations to store EHR data locally, requiring project-specific data queries to be approved by participating sites, and allowing partner organizations to physically terminate the links to the data sharing infrastructure at any time.13
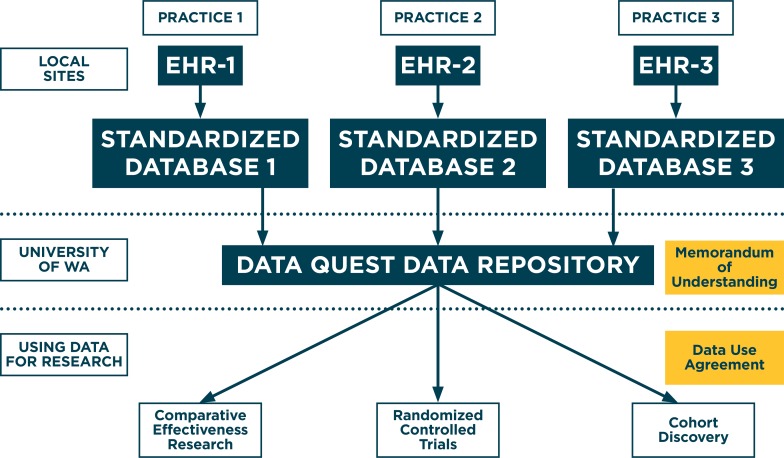
Data QUEST is implemented in over a dozen diverse primary-care practices serving primarily rural populations in the WPRN.14,15,16 Data QUEST (Figure 1) includes patient-level data stored securely within local practices’ firewalls and uses a federated data-sharing infrastructure to support regulation-compliant governance of data between primary care partners and researchers.16 Data QUEST includes a centralized data repository, hosted at the University of Washington (University of WA). Other existing distributed network solutions would have required larger digital infrastructures that are not feasible in small and rural-serving primary care practices. Data QUEST’s infrastructure and governance maintain compliance and are appropriate with the Health Insurance Portability and Accountability Act (HIPAA) and the Institutional Review Board (IRB). Data QUEST targets extraction from main data domains in the EHRs, including demographics, vital signs, diagnosis codes, diagnostic test results, social and family history, prescriptions, and physical examination findings.
Figure 1.
Structure of Data QUEST Data Flow and Management
While Data QUEST’s federated data sharing system provides participating organizations maximal local control of data, it may hinder the efficient use of data for research.17 Reports indicate that many data sharing infrastructures include some elements of local data control.18 Development of tools and processes to support data sharing can address some of these potential inefficiencies. The Scalable Architecture for Federated Translational Inquiries Network (SAFTINet) system addressed these challenges with the implementation of Master Consortium Agreements and Service Level Objective agreements, both of which are similar to processes used in the Data QUEST network.19 As trust and governance solidified, Data QUEST has expanded to include a central, de-identified data repository, improving speed and efficiency for cohort discovery.
Data QUEST Coordinating Center
The Data QUEST Coordinating Center (DQCC) is a team of researchers and scientists that provides consultation and support to those interested in working with Data QUEST. DQCC team member roles and responsibilities are described in Table 1. Members of the DQCC provide expertise in technical aspects of data sharing, developing and maintaining governance structures, and creating and sharing tools for efficient research. Consultations use a collaborative process to assess the feasibility of projects, ensure proper governance and regulatory compliance (e.g., HIPAA, IRB), create data extraction specifications, coordinate delivery of EHR data extractions, and advise on analyses.
Table 1.
Data QUEST Coordinating Center Roles and Responsibilities
| ROLE | RESPONSIBILITIES |
|---|---|
| Biomedical informatics experts |
|
| Practice-based research network clinical research experts |
|
| Research scientists |
|
| Program coordinators |
|
Case Description
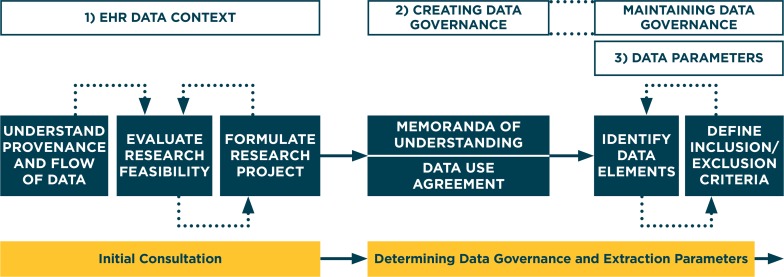
Through an iterative process, the DQCC has developed and adapted a process to successfully develop EHR data extractions that researchers can use to answer scientific questions. In this section, we describe the necessary steps and resources the DQCC uses in this process. We also describe a specific case from Data QUEST that includes a collaboration with pharmacogenomics researchers, and highlights the process the DQCC uses to develop EHR data extractions. The DQCC process of developing EHR data extractions, shown in Figure 2, provides researchers with access to the EHR data they need for answering scientific questions. This process requires coordination across multiple domains: (1) understanding the context of EHR data, (2) creating and maintaining a governance structure to support exchange of EHR data, and (3) defining data parameters that will be used to extract data from the EHR.1,2,3,4,20 We believe that this process can be implemented in similar practice-based research networks interested in building EHR data sharing capacity, and may be useful for supporting translational research with EHR data.
Figure 2.
Process that the Data QUEST Coordinating Center Uses to Create EHR Data Extractions
1. Understanding the Context of EHR Data
How EHR information is collected and entered is influenced by clinical workflow and variations in the processes of care.20,21 Knowledge of data provenance, the original context under which EHR information is collected, helps researchers understand advantages and limitations of EHR data and explore how the data can be used to answer a scientific question. The DQCC has worked closely with partner practices to understand the flow of data in their EHR systems. This knowledge, which grows with time and experience, is critical in guiding the design of scientific studies using EHR data.22 The DQCC helps researchers understand the context of EHR data through an initial consultation between the researcher and the DQCC. The DQCC meets with all potential researchers to understand their scientific questions and to provide advice and expertise about the feasibility of conducting studies using Data QUEST data. Additionally, the DQCC assesses whether data is available, complete, and valid.20 The DQCC also advises on how to formulate research projects that are acceptable and relevant to Data QUEST partner practices. If the DQCC determines that the scientific question is unfeasible using available data, researchers must consider whether the scientific question is modifiable, or whether a different source of data may be more appropriate.
2. Creating and Maintaining a Governance Structure to Support the Exchange of EHR Data
Data QUEST’s success is a direct result of its strong collaborative relationships with primary care practices, and its ongoing engagement with our stakeholders in partner practices.15 As research projects are proposed within the Data QUEST organizations, the DQCC works closely with partner practices to ensure that the practices understand the research and its procedures, as well as to address issues of data governance and security. Because Data QUEST works with diverse primary care practices, multiple health care systems may be involved in reviewing and approving projects. The DQCC has streamlined the governance process of working across diverse organizations by establishing the following: (1) a Memorandum of Understanding (MoU) that governs roles and responsibilities regarding Data QUEST for the DQCC and partner practices; and (2) Data Use Agreement (DUA) templates that can be easily adapted to specific research projects. To operationalize the research process, the DQCC asks researchers to create a one-page study overview following a specific template that the DQCC shares with partner practices to recruit practices to the study. The study overview describes the study protocol, potential benefits to project participation, and any potential impact on the practice that would result from participation. The DQCC and researchers then work collaboratively to create project specific DUAs. The DUAs outline the purpose of the study, list the necessary data elements, and describe how the data will be used to support this research. Data QUEST partner practices participating in a project must approve the DUA prior to the start of the project. The DQCC emphasizes the importance of compliance with HIPAA regulations and ensures that only data deemed essential to the conduct of the research study are collected. These tools and templates facilitate efficient communication between translational researchers and Data QUEST partner practices. Use of standard tools ensures that Data QUEST partner practices have all the information needed to quickly review and approve studies, and to authorize data extractions.
3. Defining Data Parameters
The process of defining data parameters that are used to create EHR data extractions is iterative between researchers and the DQCC. During this process, knowing the provenance and context of the EHR data, as well as understanding which data elements are available, is critical to producing EHR data extractions that answer the proposed scientific questions. The DQCC uses a template, completed for each study, that describes the available EHR data as a starting point for outlining and documenting data elements.
The researcher must identify which data elements are required for the project. Potential data elements may include the following: patient demographics (e.g., age, gender, race, ethnicity, ZIP code), patient insurance status, clinic characteristics (e.g., certain clinics within the practice organizations), provider identification (e.g., particular providers or types of providers), encounters (e.g., office visits with clinicians, telephone encounters, other types of encounters), vital signs, medications (e.g., type, dose, refills), International Classification of Diseases (ICD) diagnoses, laboratory results, past medical and surgical history, social history (e.g., tobacco, alcohol, drug use), family history, procedures (e.g., Current Procedural Terminology codes), referrals, and allergies. Researchers may prioritize comprehensiveness, and ask to include the type and maximum number of data elements. However, to ensure compliance with data governance issues, the DQCC requires researchers to limit data elements to only those that are necessary to answer the scientific question. For example, the DQCC encourages researchers to minimize protected health information (e.g., extract the year of birth rather than date of birth when age is needed) whenever possible.
The DQCC also works with the researcher to define patient inclusion and exclusion criteria for the data extract. For example, a researcher might choose to include patients of a certain gender, or those with a clinic visit within a certain period. The DQCC works with the researcher to finalize the data extract parameters, and contracts with a commercial vendor to obtain the EHR data extract.
Once a researcher receives an EHR data extraction, the researcher must make initial decisions to define the study population in preparation for data analysis. To some degree, this requires considerations similar to those described above for defining EHR data parameters; however, these considerations achieve a different purpose. For example, in order to reduce missing data, researchers may wish to exclude patients with infrequent visits. The DQCC provides expert consultation to researchers about defining inclusion and exclusion criteria, such as determining the time frame in which the patient must be seen in the clinic (e.g., ever seen versus seen in a discrete period), frequency of visits, or visits with specific provider types. The DQCC also consults with researchers to understand how data parameters or exclusion criteria may inadvertently select for certain types of patients, potentially biasing the results of analysis.
Case Example
Pharmacogenomics Research Network
The NWA-PGRN is a collaborative research program supported by the National Institutes of General Medical Sciences. The program focuses specifically on pharmacogenomic discovery and application relevant to the health care of Northwest Native American, Alaska Native, and rural populations.23,24 The NWA-PGRN collaborates with Data QUEST to explore how primary care EHR data can be leveraged to support pharmacogenomics research and clinical applications. For example, EHR data facilitates pharmacogenomics research by helping to identify patients with rare or uncommon adverse drug events that may suggest pharmacogenomics markers.25,26 EHR systems also provide infrastructure that can help deploy pharmacogenomic tests, such as clinical decision support tools.27,28,29
As researchers within the NWA-PGRN, the DQCC worked with pharmacogenomic scientists who brought content expertise, while we provided expertise in working with EHR data from outpatient clinical settings. We were interested in determining whether primary care clinic EHR data could identify patients who might be candidates for potential pharmacogenomics testing, and focused on statins, a category of medications common in primary care.
Creating EHR Data Extractions to Identify Patients Using Statin Medications for Pharmacogenomics Investigations
The DQCC case example involves a research project that uses EHR data to identify and describe patients using statin medications. Statins are commonly prescribed medications that are effective in treating hyperlipidemia.30 Statin-induced myopathy is a clinical condition that includes myalgia, myositis, and rhabdomyolysis related to use of statin medications.31 The incidence of statin myopathy reported in clinical trials ranges from 0.44 to 5.34 per 10,000 years.32,33 Evidence suggests that some patients may be particularly susceptible to statin myopathy because of a genetic variation in a statin uptake protein (SLCO1B1/OATP1B1) or the cytochrome P450 enzyme system.34 The NWA-PGRN seeks to leverage primary care EHR systems to study patients who have been prescribed a statin medication. This case example is a detailed demonstration of the processes that the DQCC uses when collaborating with researchers to create EHR data extractions. We also include results of preliminary analyses comparing two populations of primary care patients to demonstrate the potential impact of initial decisions researchers make in their studies using EHR data.
1. Understanding the Context of the EHR Data
First, we explored whether the original scientific question—To what degree can primary care EHR data be used to identify patients who are taking statins and have evidence of statin-induced myopathy?—was feasible to answer with the data elements available in Data QUEST. When considering the study question, we found that a limitation that Data QUEST faces is its lack of specialty care and inpatient data. Severe statin myopathy that requires hospitalization, such as rhabdomyolysis, is unlikely to be found in primary care clinic EHRs. Thus the scientific question evolved to better fit the available data: How many patients in Data QUEST primary care practices are taking a statin medication, and who, based on available EHR data, have evidence of statin-based myopathy?
2. Governance
The DQCC facilitated communication with partner practices about the project, and compliance with all regulatory and governance issues. We created a one-page summary of the project proposal that was shared with potential partner practices. Once the DQCC developed the data parameters for the EHR extract, we drafted and distributed a DUA to each partner organization that agreed to participate. The DUA outlined the data elements that would be included in the extract, and how these elements would be used for the research. Each partner reviewed and signed the study-specific DUA. The DQCC then worked with a commercial vendor to obtain the EHR data extract.
3. Defining Data Parameters
We created a list of necessary data parameters for the project. Data elements requested included medications, diagnosis codes, specific laboratory results related to statin complications (e.g., liver, kidney, and thyroid function), family and social history variables such as history of cardiovascular disease and drug use, clinical encounter information (with anonymized provider data), height, weight, blood pressure, and patient demographics (gender, race or ethnicity, insurance, and ZIP code). To maximize patient privacy, only the month and year of birth were requested to calculate age. To limit the extract, we defined patient inclusion and exclusion criteria as adult patients aged 18 and older, since children are unlikely to use statin medications. In addition, we specified that the extract include only those patients who paid at least one visit to the clinic after July 1, 2007.
Findings
In this section, we describe the results from the process we used to define “active” patients, as well as the results from comparing the characteristics of groups of patients based on those definitions. We also describe the process of examining the medication data in the EHR data for different groups of active patients. These results highlight the impact of the initial decisions researchers make in preparing to analyze EHR data.
To illustrate differences between definitions of active patients in the NWA-PGRN study, we created two patient groups across two Data QUEST organizations that provided data from six primary care clinics. The overall group included patients with an office visit during a one-year study observation period, and was made up of two subgroups: Subgroup 1 consisted of patients with an office visit during the one-year study period; and Subgroup 2 consisted of patients with an office visit during the one-year study period, as well as both a visit in the year prior to and the year after the one-year study period. The overall group definition resulted in an additional 6,135 patients for the first organization, and an additional 2,675 patients for the second organization compared with the subgroup required to have three continuous years of visits (Table 2). We compared the characteristics of patients in the two independent subgroups at each organization. For both organizations, Subgroup 2 had a higher average age than did Subgroup 1. In the first organization, the proportion of female patients was higher among those required to have three continuous years of visits (69 percent in Subgroup 2 versus 63 percent in Subgroup 1, p<0.001), and in the second organization the reverse was true (65 percent in Subgroup 1 versus 59 percent in Subgroup 2, p<0.001). In both organizations, the mean number of prescriptions, defined as the number of prescriptions with evidence, was similar (0.1) in the two Subgroups. For both organizations, the proportions of patients with a statin prescription were higher in Subgroup 2 (required to have three years of visits). The decision regarding how to create the groups based on number of patient visits was made at the time of analysis rather than at the time that the original data parameters for creating the EHR data extract were defined. Enforcing these limitations at the time of EHR data extraction would have reduced the number of patients potentially available for analysis, and prevented us from detecting the differences in rates of statin prescriptions between the group definitions prior to undertaking the primary analysis.
Table 2.
Comparison of Selected Characteristics Across Different Patient Groups in Two Primary Care Organizations1
| CHARACTERISTICS | ORGANIZATION 1 | ORGANIZATION 2 | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| OVERALL | SUB-GROUP* 1 | SUB-GROUP* 2 | P VALUE2 | OVERALL | SUB-GROUP* 1 | SUB-GROUP* 2 | P VALUE2 | |
| Number of patients (all ages) | 9,365 | 6,135 | 3,230 | – | 4,665 | 2,675 | 1,990 | – |
| Age, mean years (SD)3 | 40.4 (16.6) | 37.8 (15.7) | 45.3 (17.0) | <0.001 | 49.1 (20.2) | 44.6 (19.6) | 55.2 (19.5) | <0.001 |
| Female, n (%)4 | 6,125 (65%) | 3,887 (63%) | 2,238 (69%) | <0.001 | 2,915 (62%) | 1,737 (65%) | 1,178 (59%) | <0.001 |
| Number of prescriptions documented during the 12 month study period, mean (SD) | 0.1 (0.4) | 0.1 (0.4) | 0.1 (0.5) | p>0.05 | 0.3 (0.9) | 0.2 (0.8) | 0.3 (1.1) | p>0.05 |
| Number of patients with a statin prescription documented during the 12-month study period, n(%) | 620 (7%) | 361 (6%) | 259 (8%) | <0.001 | 469 (10%) | 232 (9%) | 237 (12%) | <0.001 |
Notes:
Missing values are not included in this table.
p value based on Chi-square test or t-test comparing Subgroup 1 and Subgroup 2.
Age is defined as the patient age as of 2011.
Four patients at Organization 1 (in Subgroup 1) were missing information about gender.
Subgroup 1 defined as patients with an office visit during the 1-year study period, but not in all three years (before, during and after the 1-year study period). Subgroup 2 defined as patients with an office visit in the year prior, the year of, and the year after the 1-year study period.
Major Themes
The process the DQCC uses to work with researchers to develop EHR data extractions requires an understanding of the context of EHR data, knowledge of defining data parameters, and knowledge of developing and maintaining governance structures. In Table 3, we summarize the DQCC recommendations when considering the use of EHR data for research. While each of these recommendations outlines specific processes, there are common themes of collaboration and respect for the complexity of this process. Collaboration between academic researchers, community-based practices, and a multidisciplinary coordinating center team to foster successful research partnerships will ensure that researchers can successfully and efficiently leverage EHR data for clinical and translational research.
Table 3.
DQCC Recommendations when Considering use of EHR Data for Clinical Research
| CRITICAL COMPONENT | PROCESS |
|---|---|
| Create a multidisciplinary team. |
|
| Support collaborative relationships between practices and investigators. |
|
| Respect governance. |
|
| Understand and explore downstream consequences of data definitions and analytic steps. |
|
| Anticipate the complexity of the process. |
|
Initial decisions that researchers make in preparing to analyze EHR data extractions have an impact on the study populations. Preliminary analysis of EHR data extractions from the NWA PGRN case found that varied definitions of study populations resulted in differences in patient characteristics (age, gender). Incompleteness of EHR data remains a commonly cited concern when considering use of EHR data for research.1 In a recent systematic review of studies assessing EHR data quality, incompleteness of EHR data was the most commonly assessed dimension of EHR data quality.1 Requiring patients to be “active” in a clinic, as measured by requiring a clinical encounter before, during, and after the study observation period, ensures capture of outcomes that might have otherwise been missing or incomplete for patients who left the clinic. However, as evidenced in our findings, there may be important clinical differences between patients that meet these stricter inclusion criteria and those who do not—such as differences in average age and average number of prescriptions received. Researchers must weigh the risk of bias with the advantages of having more complete data. Working with experienced teams such as the DQCC can ensure receipt of comprehensive EHR data extractions that offer flexibility in defining sampling methods as researchers develop appropriate phenotypes for relevant research samples. Future research could explore analytic methods to address potential biases created by the varying approaches to identifying patient groups.
The data available in EHR systems are not always exactly what researchers want or need for analyses. Medication information critical to pharmacogenomics research is available in the primary care based EHRs, but is often limited to the dates when medications were prescribed, making it difficult to measure medication adherence.35 Despite this limitation, we were able to identify two groups of patients who had active prescriptions for a statin medication, building toward a usable phenotype algorithm for future medication adverse-event studies. Our results demonstrate the capacity of EHR data from Data QUEST to identify patients who have been prescribed a statin medication. This cohort identification process is critical for pursuing further research to study genetic aspects of statin induced myopathy. For example, future research could leverage Data QUEST to identify and contact patients for enrollment in a study to test for genetic markers that may predispose patients to statin induced myopathy. Growing work to link EHR-extracted data with additional types of data, such as claims and patient-reported outcomes, also has the potential to address this shortfall.36,37
EHR data extractions can offer unprecedented opportunities to examine critical outcomes among large numbers of patients in real-world settings. Creating research-ready data sets from EHR data is complicated, however, and needs expert consultation to guide researchers in defining usable and reliable data sets. Leveraging our lessons learned in the DQCC, we identified three key components necessary for creating data extractions from EHR data for clinical research: (1) understanding the context of EHR data, (2) creating and maintaining a strong governance structure to support exchange of EHR data, and (3) defining data parameters for extracting data from the EHR. Critical among these strategies is infusing expert consultation (see Table 1) in the process to assist researchers in navigating the complexities inherent in using EHR data for research.
Given the rapidly growing adoption of EHR systems across diverse health care systems, there will be increasing opportunities for translational researchers to conduct research with EHR data. The development of efficient and scalable processes that support collaborative research between translational researchers and health care systems is needed. The DQCC process described here potentially addresses this existing gap. This process promotes efficient translational research with EHR data across diverse, independent practice organizations that do not share an overarching governance structure. The DQCC process is generally applicable to any clinical research network that includes distributed EHR data, regardless of the chosen data model. To our knowledge, the DQCC process has only been used with Data QUEST in the WWAMI region Practice and Research Network (WPRN), and not in other networks. This process provides general guidance for new and existing networks to facilitate data sharing across diverse partners.
We are unaware of a comprehensive report of governance processes or tools in similar networks, but such a report would be helpful to identify shared themes related to governance. These themes could be used to define a generalizable process of governance that could be disseminated more widely. As described by Paolino and colleagues, innovations in data sharing governance, specifically around DUAs and IRB approvals, can increase the efficiency and the flexibility of using EHR data sets for research.38 Future research by the Data QUEST team to test the effect of the DQCC governance and operational model on translational research process outcomes is needed.
Conclusion
Development and implementation of Data QUEST infrastructure, which supports the sharing of EHR data for research, faces unique challenges in response to the diversity of participating primary care organizations. In this report, we demonstrate the unique capacity of Data QUEST to provide robust EHR data from across diverse, rural-serving primary care organizations, which supports essential clinical and translational research across diverse, rural-serving primary care organizations. Our process for creating EHR data extractions—which includes understanding the context of EHR data, supporting and maintaining governance structures, and defining data parameters—can be used as a guide for other distributed data networks. Collaboration with an experienced team that is familiar with working with EHR data and has existing partnerships with the practices where EHR data are collected is critical to success. Without the knowledge of data provenance and strong partnerships, it is impossible to understand the context of EHR data and to develop and maintain functional governance strategies to work with it. Sustaining efforts to maintain expertise and relationships that support data sharing infrastructures is a challenge on a national scale. Coordinating centers, such as the DQCC, play a critical role in providing this expertise and supporting these relationships.39 With these strategies and collaborators in place, researchers can successfully use EHR data for research within and across diverse health care organizations.
Acknowledgments
We acknowledge the Northwest Alaska Pharmacogenomics Research Network group for supporting the infrastructure and data collection, and Imara West for her assistance in data cleaning and analysis. This project was funded by the National Institute of General Medical Science (U01 GM092676) and the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1TR000423). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disciplines
Health Information Technology | Primary Care
References
- 1.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. 2013 Jan 1;20(1):144–51. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012 May 2;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 3.Hersh WR, Weiner MG, Embi PJ, Logan JR, Payne PR, Bernstam EV, Lehmann HP, Hripcsak G, Hartzog TH, Cimino JJ, Saltz JH. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care. 2013 Aug;51(8 Suppl 3):S30–7. doi: 10.1097/MLR.0b013e31829b1dbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell J, Buchan I. Electronic health records should support clinical research. J Med Internet Res. 2005 Jan-Mar;7(1):e4. doi: 10.2196/jmir.7.1.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prokosch HU, Ganslandt T. Perspectives for medical informatics. Reusing the electronic medical record for clinical research. Methods Inf Med. 2009;48(1):38–44. [PubMed] [Google Scholar]
- 6.Hsiao CJ, Hing E. Use and characteristics of electronic health record systems among office-based physician practices: United States, 2001–2013. NCHS Data Brief. 2014;(143):1–8. [PubMed] [Google Scholar]
- 7.Weiner MG, Lyman JA, Murphy S, Weiner M. Electronic health records: high-quality electronic data for higher-quality clinical research. Inform Prim Care. 2007;15(2):121–7. doi: 10.14236/jhi.v15i2.650. [DOI] [PubMed] [Google Scholar]
- 8.Devers K, Grey B, Ramos C, Shaw A, Blavin F, Waidmann T. Assistant Secretary for Planning and Evaluation; 2013. Sep, The feasibility of using electronic health records and other electronic health data for research on small populations [Internet]. [place unknown] [cited 2014 July, 27]; Available from: http://aspe.hhs.gov/sp/reports/2013/ElectronicHealthData/rpt_ehealthdata.pdf. [Google Scholar]
- 9.Baquet CR, Commiskey P, Mullins CD, Mishra SI. Recruitment and participation in clinical trials: socio-demographic, rural/urban, and health care access predictors. Cancer Detect Prev. 2006;30(1):24–33. doi: 10.1016/j.cdp.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack D, Zhang S, Douglas M, Sow C, Strothers H, Rust G. Disparities in Primary Care EHR Adoption Rates. Journal of Health Care for the Poor and Underserved. 2016;27(1):327–38. doi: 10.1353/hpu.2016.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick RS, Steen EB, Detmer DE. The Computer-Based Patient Record: An Essential Technology for Health Care [Internet] Washington D.C: National Academy Press; 1997. [cited 2015 June 26]; Available from: http://www.nap.edu/openbook.php?record_id=5306. [PubMed] [Google Scholar]
- 12.Crossing the Quality Chasm [Internet] Washington D.C: National Academy Press; 2001. [cited 2015 June 26]; Available from: https://www.iom.edu/~/media/Files/Report%20Files/2001/Crossing-the-Quality-Chasm/Quality%20Chasm%202001%20%20report%20brief.pdf. [Google Scholar]
- 13.Lin CP, Stephens KA, Baldwin LM, Keppel GA, Whitener RJ, Echo-Hawk A, Korngiebel D. Developing governance for federated community-based EHR data sharing. AMIA Jt Summits Transl Sci Proc. 2014 Apr 7;2014:71–6. [PMC free article] [PubMed] [Google Scholar]
- 14.Cole AM, Stephens KA, Keppel GA, Lin CP, Baldwin LM. Implementation of a health data-sharing infrastructure across diverse primary care organizations. J Ambul Care Manage. 2014;37(2):164–70. doi: 10.1097/JAC.0000000000000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin CP, Black RA, Laplante J, Keppel GA, Tuzzio L, Berg AO, Whitener RJ, Buchwald DS, Baldwin LM, Fishman PA, Greene SM, Gennari JH, Tarczy-Hornoch P, Stephens KA. Facilitating health data sharing across diverse practices and communities. AMIA Jt Summits Transl Sci Proc. 2010 Mar 1;2010:16–20. [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens KA, Lin CP, Baldwin LM, Echo-Hawk A, Keppel GA, Buchwald D, Whitener RJ, Korngiebel DM, Berg AO, Black RA, Tarczy-Hornoch P. LC Data QUEST: A Technical Architecture for Community Federated Clinical Data Sharing. AMIA Jt Summits Transl Sci Proc. 2012;2012:57–62. [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy DB, Propp K, Cohen A, Sabharwal R, Schachter AA, Rein AL. Learning from Health Information Exchange Technical Architecture and Implementation in Seven Beacon Communities. EGEMS (Wash DC) 2014 May 5;2(1):1060. doi: 10.13063/2327-9214.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sittig DF, Hazlehurst BL, Brown J, Murphy S, Rosenman M, Tarczy-Hornoch P, Wilcox AB. A survey of informatics platforms that enable distributed comparative effectiveness research using multi-institutional heterogeneous clinical data. Medical care. 2012 Jul;50(Suppl):S49. doi: 10.1097/MLR.0b013e318259c02b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schilling LM, Kwan BM, Drolshagen CT, Hosokawa PW, Brandt E, Pace WD, Uhrich C, Kamerick M, Bunting A, Payne PR, Stephens WE, George JM, Vance M, Giacomini K, Braddy J, Green MK, Kahn MG. Scalable Architecture for Federated Translational Inquiries Network (SAFTINet) Technology Infrastructure for a Distributed Data Network. EGEMS (Wash DC) 2013 Oct 7;1(1):1027. doi: 10.13063/2327-9214.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hersh WR, Cimino J, Payne PR, Embi P, Logan J, Weiner M, Bernstam EV, Lehmann H, Hripcsak G, Hartzog T, Saltz J. Recommendations for the use of operational electronic health record data in comparative effectiveness research. EGEMS (Wash DC) 2013;1(1):1018. doi: 10.13063/2327-9214.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spence D. Data, data everywhere. BMJ. 2013 Feb 4;346:f725. doi: 10.1136/bmj.f725. [DOI] [PubMed] [Google Scholar]
- 22.Johnson KE, Kamineni A, Fuller S, Olmstead D, Wernli KJ. How the provenance of electronic health record data matters for research: a case example using system mapping. EGEMS (Wash DC) 2014;2(1):1058. doi: 10.13063/2327-9214.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.PGRN [Internet] [cited 2015 June, 26]; Available from: http://www.nigms.nih.gov/Research/specificareas/PGRN/background/Pages/mission2010.aspx.
- 24.Pharmacogenomics Research Network (PGRN) [Internet] [cited 2015 June, 26]; Available from: http://www.pgrn.org/display/pgrnweborganization/NWAP+Profile.
- 25.Vilar S, Harpaz R, Santana L, Uriarte E, Friedman C. Enhancing adverse drug event detection in electronic health records using molecular structure similarity: application to pancreatitis. PLoS One. 2012;7(7):e41471. doi: 10.1371/journal.pone.0041471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haerian K, Varn D, Vaidya S, Ena L, Chase HS, Friedman C. Detection of pharmacovigilance-related adverse events using electronic health records and automated methods. Clin Pharmacol Ther. 2012;92(2):228–34. doi: 10.1038/clpt.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano MJ, Stafford RS. Electronic health records and clinical decision support systems: impact on national ambulatory care quality. Arch Intern Med. 2011;171(10):897–903. doi: 10.1001/archinternmed.2010.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawamoto K, Lobach DF, Willard HF, Ginsburg GS. A national clinical decision support infrastructure to enable the widespread and consistent practice of genomic and personalized medicine. BMC Med Inform Decis Mak. 2009;9:17. doi: 10.1186/1472-6947-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch BM, Kawamoto K. Clinical decision support for genetically guided personalized medicine: a systematic review. J Am Med Inform Assoc. 2013 Mar-Apr;20(2):388–400. doi: 10.1136/amiajnl-2012-000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeWilde S, Carey IM, Bremner SA, Richards N, Hilton SR, Cook DG. Evolution of statin prescribing 1994–2001: a case of agism but not of sexism? Heart. 2003 Apr;89(4):417–21. doi: 10.1136/heart.89.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sathasivam S, Lecky B. Statin induced myopathy. BMJ. 2008;337:a2286. doi: 10.1136/bmj.a2286. [DOI] [PubMed] [Google Scholar]
- 32.Gaist D, Rodriguez LA, Huerta C, Hallas J, Sindrup SH. Lipid-lowering drugs and risk of myopathy: a population-based follow-up study. Epidemiology. 2001;12(5):565–9. doi: 10.1097/00001648-200109000-00017. [DOI] [PubMed] [Google Scholar]
- 33.Graham DJ, Staffa JA, Shatin D, Andrade SE, Schech SD, La Grenade L, Gurwitz JH, Chan KA, Goodman MJ, Platt R. Incidence of hospitalized rhabdomyolysis in patients treated with lipid-lowering drugs. JAMA. 2004 Dec 1;292(21):2585–90. doi: 10.1001/jama.292.21.2585. [DOI] [PubMed] [Google Scholar]
- 34.Ghatak A, Faheem O, Thompson PD. The genetics of statin-induced myopathy. Atherosclerosis. 2010 Jun;210(2):337–43. doi: 10.1016/j.atherosclerosis.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 35.Bayley KB, Belnap T, Savitz L, Masica AL, Shah N, Fleming NS. Challenges in using electronic health record data for CER: experience of 4 learning organizations and solutions applied. Med Care. 2013;51(8 Suppl 3):S80–6. doi: 10.1097/MLR.0b013e31829b1d48. [DOI] [PubMed] [Google Scholar]
- 36.Estabrooks PA, Boyle M, Emmons KM, Glasgow RE, Hesse BW, Kaplan RM, Krist AH, Moser RP, Taylor MV. Harmonized patient-reported data elements in the electronic health record: supporting meaningful use by primary care action on health behaviors and key psychosocial factors. J Am Med Inform Assoc. 2012 Jul-Aug;19(4):575–82. doi: 10.1136/amiajnl-2011-000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howie L, Hirsch B, Locklear T, Abernethy AP. Assessing the value of patient-generated data to comparative effectiveness research. Health Aff (Millwood) 2014;33(7):1220–8. doi: 10.1377/hlthaff.2014.0225. [DOI] [PubMed] [Google Scholar]
- 38.Paolino AR, Lauf SL, Pieper LE, Rowe J, Vargas IM, Goff MA, Daley MF, Tuzzio L, Steiner JF. Accelerating Regulatory Progress in Multi-Institutional Research. EGEMS (Wash DC) 2014 Jul 10;2(1):1076. doi: 10.13063/2327-9214.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilcox A, Holve E. Sustaining the effective use of health care data: a message from the editors. EGEMS (Wash DC) 2014;2(2):1141. doi: 10.13063/2327-9214.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]