Abstract
Hybridization is an important evolutionary force, because interspecific gene transfer can introduce more new genetic material than is directly generated by mutations. Pinus engelmannii Carr. is one of the nine most common pine species in the pine-oak forest ecoregion in the state of Durango, Mexico. This species is widely harvested for lumber and is also used in reforestation programmes. Interspecific hybrids between P.engelmannii and Pinus arizonica Engelm. have been detected by morphological analysis. The presence of hybrids in P. engelmannii seed stands may affect seed quality and reforestation success. Therefore, the goals of this research were to identify introgressive hybridization between P. engelmannii and other pine species in eight seed stands of this species in Durango, Mexico, and to examine how hybrid proportion is related to mean genetic dissimilarity between trees in these stands, using Amplified Fragment Length Polymorphism (AFLP) markers and morphological traits. Differences in the average current annual increment of putative hybrids and pure trees were also tested for statistical significance. Morphological and genetic analyses of 280 adult trees were carried out. Putative hybrids were found in all the seed stands studied. The hybrids did not differ from the pure trees in vigour or robustness. All stands with putative P. engelmannii hybrids detected by both AFLPs and morphological traits showed the highest average values of the Tanimoto distance, which indicates: i) more heterogeneous genetic material, ii) higher genetic variation and therefore iii) the higher evolutionary potential of these stands, and iv) that the morphological differentiation (hybrid/not hybrid) is strongly associated with the Tanimoto distance per stand. We conclude that natural pairwise hybrids are very common in the studied stands. Both morphological and molecular approaches are necessary to confirm the genetic identity of forest reproductive material.
Introduction
Hybridization and backcrossing to one or both of the parental types can lead to incorporation of alleles from one taxon into the gene pool of the other [1]. Interactions between environment and genetic structure can then lead to segregation of a novel taxon from parental types. Depending on the degree of differentiation, hybrid offspring of different taxa may be identified as species, subspecies, variants or races [2] [3]. Hybridization is an important evolutionary force, because interspecific gene transfer can introduce more new genetic material than is directly generated by mutation [4]. At least 30%, and possibly as much as 80%, of all species may have been originated by hybridization [5].
Plant hybridization often leads to the formation of hybrid zones [6]. As narrow geographic regions and “tension zones”, hybrid zones are active sites of evolutionary change and high levels of genetic variation [1] [3]. Stable hybrid zones are not suitable for the process of hybrid speciation [7]. The great majority of hybrid zones are maintained in equilibrium between dispersal and selection and may remain in balance for long periods [8]. In contrast, a hybrid species probably originates from a hybrid founder event, in which one or more early generation hybrids populate a new area and therefore become spatially or ecologically isolated from the parent species [7] [9].
In general, hybrids are unfit relative to their ancestors, in particular because of postmating reproductive barriers. These common barriers comprise hybrid weakness or inviability, hybrid sterility and hybrid breakdown. However, the exception applies that first generation (F1) hybrids, particularly between geographic races or closely related species, tend to exceed their parents in vegetative vigour or robustness [10].
Extensive variation in viability and fertility is observed both within and between interspecific hybrid generations. Some genotypic classes of hybrids possess lower, equivalent or higher levels of fitness relative to their parental taxa. Levels of variability tend to be highest in the second generation (F2) and first back-cross (BC) generations. Successive hybrid generations are characterized by lower levels of variability and increased fecundity and viability as a result of natural (differential) selection [10] [11].
Although most studies concerning hybridization are based on morphological evidence, introgression or hybridization are not necessarily indicated by the phenotypic expression of characters of one taxon in another. Identical characters in species may occur as a result of phenotypic plasticity, convergent evolution, or simply common ancestry [12]. Furthermore, individuals from hybrid swarms that have obtained most of their genes from one of the parental taxa are often morphologically indistinguishable from that taxon [13].
The use of molecular markers has shown that interspecific hybridization is even more common than assessed by morphological and cytogenetic evidence alone. The hybrid nature of many species has indeed been confirmed by molecular studies [14] [15] [16]. Some pronounced advantages of molecular markers over morphological markers are: i) absence of pleiotropic effects [17], ii) lack of deleterious or strong epistatic effects, iii) a high number of molecular markers can usually be identified, iv) most are multilocus, allowing many markers to be considered simultaneously, v) numerous alleles, per locus, may exist for some marker types [18], and that vi) genotypes can be identified at either the whole plant, tissue, or cellular levels [19]. Amongst other molecular markers, Amplified Fragment Length Polymorphism (AFLP) markers have been used effectively to detect both interspecific hybridization and introgression in animals, fungi and plants [20] [21] [22] [23] [24] [25].
Hybridization has been reported for very few Mexican pine species (e.g. [26] [27] [28] [29]). Recent diversification followed by secondary contact and hybridization may explain complex structures of intra- and interspecific morphological and genetic variation in the approximately 49 North American hard pines (Pinus section Trifoliae with subsections Australes, Contortae, and Ponderosae) [30]. Pinus subsection Ponderosae comprises approximately 17 species distributed from western Canada to Nicaragua. Although members of this group are of great ecological and economic importance, phylogenetic relationships between species are poorly understood [31] [32] [33]. Because of the very strong phylogenetic relationships among these species [34] [35] [30] and their very weak reproductive barriers, hybridization between Ponderosae species is quite possible [36][37][38][39]. Species divergence is recent and hybrid ancestry in some Ponderosae subsection species is inferred from an incomplete lineage sorting [40].
Apache pine (P. engelmannii Carr.) is, along with Pinus arizonica Engelm., Pinus arizonica var. cooperi (C.E. Blanco) Farjon and Pinus durangensis Martínez, one of the most commonly occurring Ponderosae species in the Mexican Sierra Madre Occidental [41] [31] and has a limited distribution in the mountains of Arizona and New Mexico. Interspecific hybrids between Apache pine, interior ponderosa pine (P. ponderosa var. scopulorum) and Arizona pine (P. arizonica) have been detected by morphological analysis [42] [43]. The presence of hybrids in seed stands may affect seed quality and reforestation success [9] [10] [44].
Pinus engelmannii grows on dry to moderately moist canyon slopes, ridges, mesas, lower slopes, valleys and streamside terraces, at elevations of 1,500–2,700 m, in climates ranging from semiarid with bimodal precipitation to temperate-subhumid with most precipitation falling in summer [45] [46]. It is one of the nine most frequent pine species in the Sierra Madre Occidental in the state of Durango, Mexico [47]. It is widely harvested for lumber and is also used in reforestation programs in Durango [48].
The goals of this research were to identify introgressive hybridization between P. engelmannii and other pine species in eight seed stands of this species in the state of Durango, Mexico, and to examine how hybrid proportion is related to mean genetic dissimilarity between trees in these stands, using Amplified Fragment Length Polymorphism (AFLP) markers and morphological traits. We also hypothesized no differences in the average current annual increment of putative hybrids and pure trees, given that all the studied trees were healthy, dominant and superior phenotypes (plus trees).
Materials and Methods
We confirm that we provide the specific location of the field studies (Table 1). None vertebrate study was carried out. We confirm that the owners of the lands gave permission to conduct these studies on those sites.
Table 1. Details of the studied stands of Pinus engelmannii.
| Stand | No. | Code | Location | Property | Municipality | Latitude (N) | Longitude (W) | Elevation(m) | Age (years) |
|---|---|---|---|---|---|---|---|---|---|
| W | 1 | LM | La Mesa | Ejido Pueblo Nuevo | Pueblo Nuevo | 23° 26' 14.0" | 105° 03' 11.4" | 2,327 | 51–86 |
| W | 2 | CU | Cumbres | Predio Particular Cumbres | Durango | 23° 51' 55.3" | 105° 14' 27.3" | 2,441 | 38–103 |
| W | 3 | ARC | Adolfo Ruiz Cortinez | Ejido Adolfo Ruiz Cortinez | Durango | 23° 42' 46.7" | 105° 17' 13.7" | 2,330 | 48–87 |
| W | 4 | NP | Nueva Patria | Ejido Nueva Patria | San Dimas | 24° 03' 06.8" | 105° 29' 21.2" | 2,268 | 32–86 |
| E | 5 | RC | Rancho Los Castro | Ejido La Casita | Durango | 23° 44' 38.3" | 104° 45' 29.5" | 2,302 | 22–69 |
| E | 6 | R | El Río | Ejido La Casita | Durango | 23° 43' 26.8" | 104° 47' 27.1" | 2,335 | 22–55 |
| E | 7 | MC | Mesa Cebollas | Comunidad San Bernardino de Milpillas | Pueblo Nuevo | 23° 21' 28.6" | 104° 50' 27.2" | 2,543 | 51–86 |
| E | 8 | MCO | Mesa Coyotes | Comunidad San Bernardino de Milpillas | Pueblo Nuevo | 23° 25' 46.9" | 105° 01' 20.3" | 2,315 | 22–50 |
W = Western, E = Eastern.
Sampling sites
Eight Pinus engelmannii (Pe) seed stands in the state of Durango (NW Mexico) were analyzed: 1) La Mesa (LM), 2) Cumbres (CU), 3) Adolfo Ruiz Cortínez (ARC), 4) Nueva Patria (NP), 5) Rancho Los Castro (RC), 6) El Río (R), 7) Mesa Cebollas (MC), and 8) Mesa Coyotes (MCO). These uneven-aged seed stands are located in natural populations (Table 1 and Fig 1).
Fig 1. Location of the Pinus engelmannii, P. arizonica var cooperi, and P. durangensis stands.
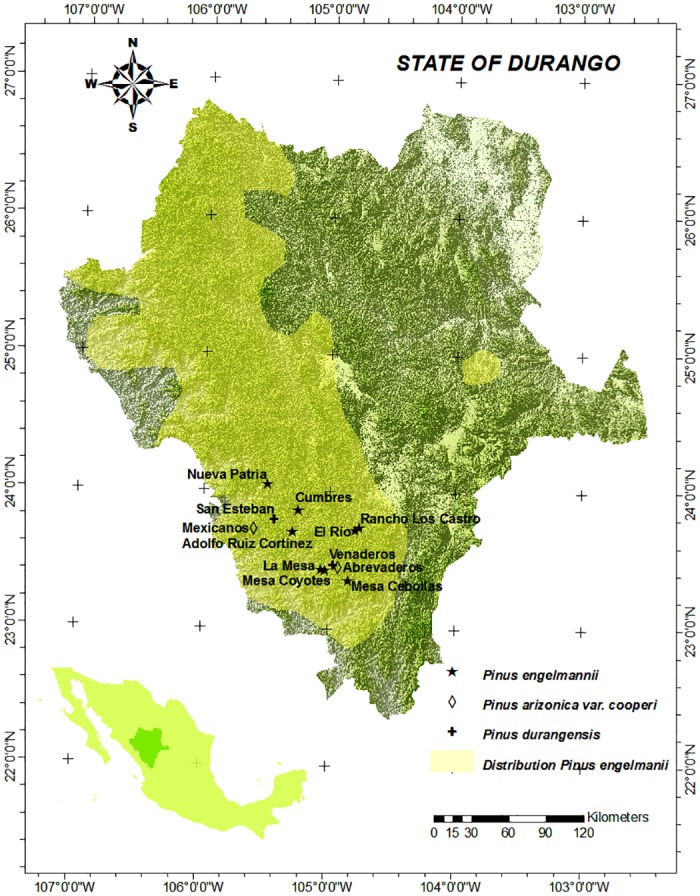
Location of the Pinus engelmannii, P. arizonica var cooperi, and P. durangensis stands in the state of Durango (NW Mexico). The distribution of P. engelmannii was taken from Little [78]. Data sources: Own compilation based on freely-accessible digital maps from INEGI, Mexico (http://www.inegi.org.mx/geo/contenidos/mapadigital/). The shp format of the distribution of P. engelmannii is freely available at: http://esp.cr.usgs.gov/data/little/pinuenge.pdf (accessed July 2015).
For analysis of the genetic structure, needles were sampled from 280 adult, dominant and superior phenotypes (plus trees) of Pinus engelmannii (35 individuals per stand), in 2013 (for selection criteria, see Wehenkel et al. [49]). The spacing was similar for all the sampling trees. The mean distance between these trees was 39 m. For purposes of comparison, further investigations were carried out on 70 adult trees of Pinus arizonica var. cooperi (Pco) from two seed stands (Abrevaderos (AB) and Mexicanos (MEX)) and 70 adult trees of Pinus durangensis (Pdu) from another two seed stands (Venaderos (VEN) and San Esteban (SE)), close to the P. engelmannii stands under study (Fig 1).
Morphological analysis of all trees was conducted in the field. Samples of cones and branchlets were collected for taxonomic determination in the laboratory, which was based on morphological characters from needles, branchlets and cones, supported with field data of the bark texture and branching pattern of the trees, comparing with descriptions of the species in [48] and [50]. Vouchers were deposited in the herbarium of the Centro Interdisciplinario de Investigación para el Desarrollo Integral Regional (CIIDIR) at Durango. Dasometric variables were recorded for each P. engelmannii tree, including age, diameter at breast height (DBH) and average current annual increment (CAI) at age 20 years, because the tree growth depends on the tree age (Table 1).
AFLP analysis
AFLP fingerprints were established according to the protocol described by Vos et al. [51]. DNA was extracted employing the DNeasy96 plant kit (QIAGEN) and digested with restriction enzymes EcoRI and MseI. Double-stranded MseI and EcoRI adaptors were ligated to the ends of the restriction fragments to generate template DNA. The restriction/ligation reaction was carried out at room temperature overnight. Subsequently, pre-selective amplification was conducted using the diluted ligation mix and primer combination E01/M03 (EcoRI-A/MseI-G). The final amplification reaction was initiated at 72°C for 2 min, followed by 20 cycles each consisting of 94°C for 10 sec, 56°C for 30 sec, and 72°C for 2 min, and a final step at 60°C for 30 min.
Selective amplification was carried out with the fluorescent-labelled (FAM) primer pair E35 (EcoRI-ACA) and M63+C (MseI-GAAC) for P. engelmannii, P. durangensis and P. arizonica var. cooperi. The fourth selective base was added to reduce the total number of peaks. Selective PCR cycling started at 94°C for 2 min, followed by 10 cycles, each consisting of 10 sec at 94°C, 30 sec at 65°C and 2 min at 72°C. The 65°C annealing temperature of the first cycle was subsequently reduced by 1°C for the next 10 cycles and continued at 56°C for 30 sec for the remaining 23 cycles, and was completed with a final extension step at 60°C for 30 min. All PCR reactions were carried out in a Peltier Thermal Cycler (MJ Research,Waltham, Massachusetts). The amplified restriction products were electrophoretically separated in a Genetic Analyzer (ABI 3100 16 capillaries), along with the internal size standard GeneScan 500 ROX (fluorescent dye ROX) from Applied Biosystems, Foster City, California, USA. The size of the AFLP fragments was resolved with the GeneScan 3.7 and Genotyper 3.7 software packages (Applied Biosystems) [52] [53] [54].
Scoring was fully automated and only strong and high quality fragments were considered. Only fragments above the signal threshold of 50 (minimum peak height) (according to ABI manual) and with a maximum peak width of 1.0, and fragment size ranging between 75 to 450 bp were considered. Two fragments were recorded only when the peak-peak distance between two signals was at least 0.5 bp [53].
Quality and reproducibility were tested by including reference samples in each plate and independent repetition (replicate PCRs) of at least 16 samples (i.e. a minimum of 16 individuals randomly chosen from each plate). All replicates presented the same AFLP pattern as in the first analyses (for further details, see [53]).
Finally, three binary AFLP matrices were created from the presence (code 1) or absence (code 0) at potential band positions. Each band detected corresponded to the presence of a dominant genetic variant (plus phenotype) with unknown mode of inheritance of this potential band position (detected fragment length) [54] [55]. The absence of a band reflected the presence of only recessive genetic (allelic) variants at the given position (locus).
Because Pe, Pco and Pdu are very closely related taxa [30] [34] [35] [40] and their physical and reproductive barriers are very weak [36] [37] [38], the degree of homoplasy was not expected to significantly increase [56] [57] [58].
To minimize the impact of size homoplasy [59] [60] and technical artifacts [55], only polymorphic loci with presence frequency between 5 and 95% (based across all 280 genotyped individuals) were selected for further study [61].
Hybrid identification
If a P. engelmannii stand includes hybrid trees, these should possess a genome that is a blend of alleles derived from both P. engelmannii and other pine species, caused by gene flow. These hybrids are detectable by genetic and/or stable morphological traits (whose expression is under exclusive genetic control), as stated in [62] and [63] regarding natural hybridization within seed sources and hybridization in sympatric populations of Pinus echinata Mill. and Pinus taeda L., respectively.
In order to test the hypothesis that genetic introgression exists between P. engelmannii and other pine species, 280 P. engelmannii trees were screened. The alternative hypothesis was that there is no hybridization among the studied species.
In order to determine the degree of hybridism, 204 AFLP markers were used in four separate STRUCTURE (version 2.3.4) analyses [64] [65] to compare i) P. engelmannii to P. arizonica var. cooperi, ii) P. engelmannii to P. durangensis separately for the western and the eastern stands.
The Bayesian clustering method, as implemented in STRUCTURE was used to test whether affiliation of individuals to species on the basis of morphology was congruent with the assignment based on the AFLP markers. The population number (K) = 2 was used for this purpose. If the probability of P. engelmannii affiliation of a putative P. engelmannii tree was less than 95% according to STRUCTURE, then that individual was recorded as a candidate hybrid. The affiliation probability was measured by the proportion of the dominant STRUCTURE populations in the eight P. engelmannii studied stands. Individuals were identified as first-generation (F1) hybrids when the probability of P. engelmannii affiliation of a putative P. engelmannii tree was in the range 45–55%.
Then, in order to detect the degree of hybridization by morphological traits, vegetative and reproductive characters were assessed by using 14 qualitative and quantitative traits. Any tree displaying differences in these traits according to [48] and [50] was considered to be a putative hybrid. Character traits for morphologically "pure" P. engelmannii are shown in parenthesis: crown apex (rounded), crown density (open), twig diameter (1–3 cm), needle length (20–43 cm), needle width (1–2 mm), needle thickness (ca 1 mm), needle colour (dull pale green), number of stomata on dorsal face of needle (6–23), needle position (spreading-ascending to slightly drooping), young sheath length (2–4 cm); mature cone shape (ovoid to oblong), mature cone length (7–16 cm), mature cone width (6–12 cm), cone apophysis (elongate, strongly raised toward cone base). Characters not useful for distinguishing P. engelmannii from other related species (e.g. number of needles per fascicle) were not considered in this study.
Association between genetic distance (TD) and hybrid frequency per stand
To test the relationship between genetic distance and hybrid frequency (fhyp) in each P. engelmannii stand, the AFLP data were also used to calculate the mean genetic dissimilarity between the binary vectors of two individuals in each stand, a and b, using the Tanimoto distance (TD) [66]. The relationship between mean TDab and hybrid proportion per stand was then computed by the covariation (C) described by Gregorius et al. [67]. This method can detect types of covariation that are monotonous but not necessarily linear. C varies from -1 to 1, where C = 1 shows an entirely positive covariation and C = -1 a strictly negative covariation. If the denominator is zero, C is indeterminate [67]. In order to test the possibility that the observed degrees of covariation C[mean TDab x fhyp] were only produced by random events, rather than directed forces, a one-sided permutation test was performed (here 10,000 permutations; P(Z ≥ C) < 0.05) [68].
Differences in average current annual increment of putative hybrids and pure P. engelmannii trees
A permutation test based on randomly chosen reassignments was used to test whether the observed differences (Diff) in the mean values of the average current annual increment (CAI) of DBH at age 20 years of putative hybrids and pure trees in each P. engelmannii stand occur as random events, rather than by directed forces. If the P(Z ≥ Diff) is smaller than 0.05, we can expect statistically significant differences [68]. The average current annual increment (CAI) of DBH at age 20 years was determined from cores of wood taken with a Pressler borer.
Results
The AFLP primer combination resulted in 204 polymorphic bands of 75–450 base pairs across all individuals of Pinus engelmannii (Pe), P. arizonica var. cooperi (Pco) and P. durangensis (Pdu). Pe shared 87% of AFLP fragments with Pco and 90% with Pdu.
Fig 2 illustrates the STRUCTURE results for the eight Pe stands, two Pco stands and two Pdu stands, simulated with the population number (K) = 2. Pe individuals are those most clearly separated from the Pdu and Pco individuals.
Fig 2. STRUCTURE results simulated with K = 2.
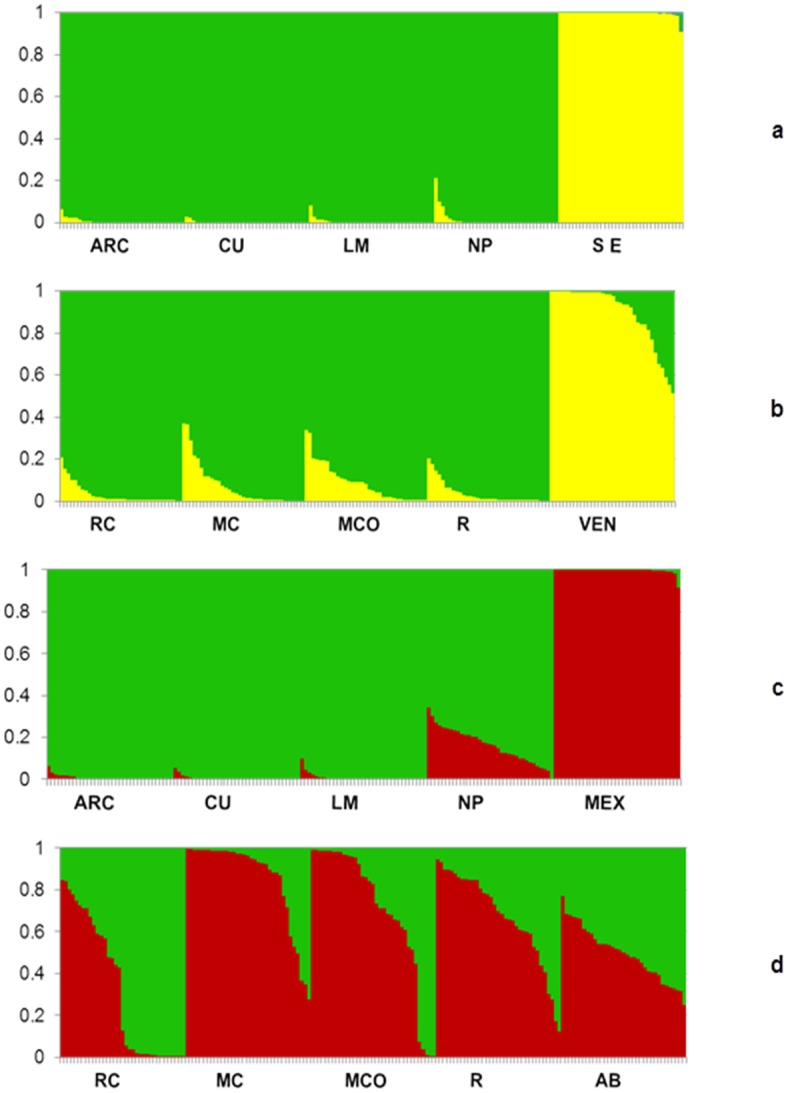
STRUCTURE results for the eight Pinus engelmannii stands (RC: Los Castro, MC: Mesa Cebollas, MCO: Mesa Coyotes, R: El Rio, ARC: Adolfo Ruiz Cortinez, CU: Cumbres, LM: La Mesa, NP: Nueva Patria), two Pinus durangensis stands (a) SE: San Esteban; (b) VEN: Venaderos, and two Pinus arizonica var. cooperi stands (c) MEX: Mexicanos, (d) AB: Abrevaderos), Durango, Mexico. Data were simulated with K = 2, as described in Material and Methods.
According to the results of STRUCTURE analyses, hybrids were found in all the studied stands. On the basis of morphological features, hybrids occurred in 37.5% of the stands. These hybrids were probably Pe x P. arizonica var. arizonica and in one case, Pe x P. maximinoi. Forty-two Pe x Pinus ssp. hybrid individuals were detected both by AFLPs and morphological traits (15% of all putative Pe), 163 putative hybrids by AFLPs (58% of all putative Pe) and 42 by morphological traits (15%) (Table 2). The morphological traits did not discover hybridization in 121 trees (74% of all detected hybrids using AFLPs). The current annual increment (CAI) did not significantly differ between the Pe trees and their hybrids.
Table 2. Mean Tanimoto distance and minimum number of hybrids per Pinus engelmannii stand.
| Stand | Mean Tanimoto distance | AFLPs (No.) Pe x Pinus ssp. | AFLPs (No.) Pe x Pco | AFLPs (No.) Pe x Pdu | Morphological (No.) |
|---|---|---|---|---|---|
| ARC | 0.40 | 1 | 1 | 1 | 0 |
| CU | 0.32 | 1 | 1 | 0 | 0 |
| LM | 0.53 | 1 | 1 | 1 | 0 |
| NP | 0.33 | 35 | 32 | 3 | 0 |
| RC | 0.48 | 23 | 19 | 7 | 0 |
| MC | 0.60 | 35 | 35 | 14 | 2 (Pe x Par, Pe x Pmax) |
| MCO | 0.53 | 32 | 30 | 19 | 6 (Pe x Par) |
| R | 0.62 | 35 | 35 | 8 | 34 (Pe x Par) |
Mean Tanimoto distance and minimum number of hybrids detected by AFLPs (probability < 95% of affiliation of a putative P. engelmannii tree to the species, STUCTURE analysis with K = 2) and morphological traits per P. engelmannii seed stand. Pe = Pinus engelmannii, Pco = P. arizonica var. cooperi, Pdu = P. durangensis, Par = P. arizonica var. arizonica, Pmax = P. maximinoi, RC = Pe seed stand in Rancho Los Castro, MC = Pe seed stand in Mesa Cebollas, MCO = Pe seed stand in Mesa Coyotes, R = Pe seed stand in El Río, ARC = Pe seed stand in Adolfo Ruiz Cortinez, CU = Pe seed stand in Cumbres, LM = Pe seed stand in La Mesa, NP = Pe seed stand in Nueva Patria.
STRUCTURE analysis indicated that 55% of all Pe individuals had genetic introgression from Pco, and 19% from Pdu (Table 2). In addition, 3% of Pco in the MEX stand showed introgression from Pe, 100% of Pco from Pe in AB, 3% of Pdu from Pe in SE stand and 46% of Pdu from Pe in the VEN stand.
By the results of STRUCTURE analysis only eight of the putative Pe trees were identified as first-generation (F1) hybrids Pe x Pco (in each of these four stands: RC, R, MCO, and MC).
Statistically significant covariations (C) of the mean Tanimoto distance (TD) with hybrid frequency per P. engelmannii stand were found for the combination TD and frequency of Pe x Pdu (C = 0.86, P = 0.035), Pe x Pco x Pdu (C = 0.92, P = 0.020) and morphological traits (C = 0.99, P = 0.015).
Discussion and Conclusions
The Pinus engelmannii trees share many AFLP fragments with P. arizonica var. cooperi and with P. durangensis, due to the relatively recent diversification of Ponderosae species and the very weak physical and reproductive barriers between them [34][35] [36][37][38][40] followed by introgression [30]. This interspecific gene transfer is maintained by: i) wind pollination, ii) weak reproductive isolating barriers, iii) longevity, iv) overlapping generations, v) large effective population sizes [30] and vi) the overlapping geographical extension (sympatric distribution) of these three Ponderosae species.
The STRUCTURE analysis showed that many individuals from several P. engelmannii seed stands were hybridized with P. arizonica and P. durangensis. Only eight first-generation (F1) hybrids were found and a relatively high frequency of introgressed individuals was observed, as [69] recently found for Salix. This appears to indicate that F1 hybrids are not stable and backcrosses are the rule, so the alternative hypothesis referred to lack of hybridization is discarded. These results are consistent with reports of interspecific hybridization between Apache pine and Arizona pine (e.g. [42] [43]). Morphological analysis confirmed hybridization in two stands (R and MC) in which P. engelmannii and P. arizonica var. arizonica grow together; however, other cases of hybridization evidenced by AFLPs were not morphologically detected, as found also by [69] for Siberian willows. Natural pairwise and triple hybrids have also been found in some Mexican Quercus [70]. The similar current annual increment (CAI) observed for hybrids and pure trees in the eight P. engelmannii stands under study suggests that hybrids and pure trees did not differ in terms of vigour or robustness [10]. Fitness advantages of pure vs introgressed and F1 individuals were not found and were not expected, because all the studied individuals were healthy, dominant and superior phenotypes (plus trees).
All the stands with putative P. engelmannii hybrids detected both by AFLPs as by morphological traits (R, MC, MCO) also showed the highest average values of the Tanimoto distance (Table 2), indicating: i) more heterogeneous genetic material [4], ii) higher genetic variation and therefore iii) the higher evolutionary potential in these stands [1] [71], and iv) that the morphological differentiation (hybrid/not hybrid) is strongly associated with the Tanimoto distance per stand.
The isolated El Rio stand included the highest degree of hybridization (morphologically almost 100%). This stand appears to represent a stable hybrid zone [8]. As the hybrids are not spatially or ecologically isolated from the parental species [9] [7], and the morphological traits are combinations of traits of those species (no novel characteristics were found), hybrid speciation is not plausible in the population.
In Mexican Arbutus, hybrids are common and backcrossing occasionally occurs, particularly in disturbed areas [72] [73]. Recent data [74] [75] indicate that hybridization can be accelerated by climate change. Global climate change is resulting in wide-scale habitat modification that will result in increasing opportunities for hybridization, in an analogous manner to other forms of anthropogenic disturbance [76]. The consequences of hybridization on the evolution of a species will depend, both on the relative fitness of hybrid offspring compared with offspring of pure species, as on the frequency of hybrid matings [76]. New research lines are necessary to explore how introgressive hybridization in P. engelmannii would influence its adaptation to environmental changes.
We conclude that natural pairwise hybrids are very common in the P. engelmannii stands under study and that these hybrids are sometimes not morphologically obvious. Considering that the AFLPs results are less biased in the estimation of hybrids, morphological traits are not a good proxy to estimate hybridization in this species.Thus, both morphological and molecular approaches are necessary to confirm the genetic identity of forest reproductive material. Molecular approaches should involve a combination of plastid data, nuclear and mitochondrial DNA sequences and crossing experiments. The genetic identity of forest reproductive material is essential for a complete understanding of tree species phylogeny, for developing effective breeding programs, and for seed quality and reforestation success [30] [44] [77].
Supporting Information
(XLSX)
Acknowledgments
We thank the Unión de Permisionarios Forestales Unidad de Administración Forestal La Flor Durango A.C. and Agrupación de Silvicultores Región El Salto S.C. for their helpful assistance and for the allocation of the data set.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1.Harrison RG. Hybrids and hybrid zones: historical perspective In Harrison RG, editors. Hybrid zones and the evolutionary process. Oxford UK: Oxford University Press; 1993. pp. 3–12. [Google Scholar]
- 2.Futuyma DJ. Evolutionary biology Massachusetts: Sinauer, Sunderland; 1998. [Google Scholar]
- 3.Tovar-Sánchez E, Oyama K. Natural hybridization and hybrid zones between Quercus crassifolia and Quercus crassipes (Fagaceae) in Mexico: morphological and molecular evidence. Am. J. Bot. 2004; 91: 1352–1363. 10.3732/ajb.91.9.1352 [DOI] [PubMed] [Google Scholar]
- 4.Anderson E. Introgressive hybridization. New York: John Wiley and Sons, Inc.; 1949. [Google Scholar]
- 5.Wendel JF, Stewart J. Molecular evidence for homoploid reticulate evolution among Australian species of Gossypium. Evol. 1991; 45: 694–711. [DOI] [PubMed] [Google Scholar]
- 6.Zhou R, Shi S, Wu CI. Molecular criteria for determining new hybrid species–an application to the Sonneratia hybrids. Molec. Phy. Evol. 2005; 35: 595–601. [DOI] [PubMed] [Google Scholar]
- 7.Ungerer MC, Baird SJ, Pan J, Rieseberg LH. Rapid hybrid speciation in wild sunflowers. Proceedings of the National Academy of Sciences. 1998; 95: 11757–11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton NH, Hewitt GM. Analysis of hybrid zones. Ann. R. Ecol. System.1985; 113–148. [Google Scholar]
- 9.Rieseberg LH. Hybrid origins of plant species. Ann. R. Ecol. System. 1997; 359–389. [Google Scholar]
- 10.Rieseberg LH, Carney SE. Plant hybridization. New Phytol. 1998; 140: 599–624. [DOI] [PubMed] [Google Scholar]
- 11.Arnold ML, Hodges SA. Are natural hybrids fit or unfit relative to their parents? Tren. Ecol. Evol. 1995; 10: 67–71. [DOI] [PubMed] [Google Scholar]
- 12.Linder CR, Taha I, Seiler GS, Snow AA, Rieseberg LH. Long-term introgression of crop genes into wild sunflower populations. Theo. App. Genet. 1998; 96: 339–347. [DOI] [PubMed] [Google Scholar]
- 13.Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: setting conservation guidelines. Tren. Ecol. Evol. 2001; 16: 613–622. [Google Scholar]
- 14.Arnold ML. Natural hybridization and evolution Oxford: Oxford Series in ecology and evolution: Oxford University Press; 1997. [Google Scholar]
- 15.Mallet J. Hybrid speciation. Nature. 2007; 446: 279–283. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan Z, Fehrer J. Molecular evidence for a natural primary triple hybrid in plants revealed from direct sequencing. Ann. Bot. 2007; 99: 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havey MJ, Muehlbauer FJ. Linkages between restriction fragment length, isozyme, and morphological markers in lentil. Theoretical and Applied Genetics, 1989; 77: 395–401. 10.1007/BF00305835 [DOI] [PubMed] [Google Scholar]
- 18.Michelmore RW, Hulbert SH. Molecular markers for genetic analysis of phytopathogenic fungi. Annual Review of Phytopathology.1987; 25: 383–404. [Google Scholar]
- 19.Stuber Charles W. Biochemical and molecular markers in plant breeding. Plant Breed. Rev. 1992; 9: 37–61. [Google Scholar]
- 20.Liu Z, Karsi A, Li P, Cao D, Dunham R. An AFLP-based genetic linkage map of channel catfish (Ictalurus punctatus) constructed by using an interspecific hybrid resource family. Genetics. 2003; 165: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo YP, Saukel J, Mittermayr R, Ehrendorfer F. AFLP analyses demonstrate genetic divergence, hybridization, and multiple polyploidization in the evolution of Achillea (Asteraceae Anthemideae). New Phytol. 2005; 166: 273–290. [DOI] [PubMed] [Google Scholar]
- 22.Shasany AK, Darokar MP, Dhawan S, Gupta AK, Gupta S, Shukla AK, Khanuja SP. Use of RAPD and AFLP markers to identify inter-and intraspecific hybrids of Mentha. J. of Heredity. 2005; 96: 542–549. [DOI] [PubMed] [Google Scholar]
- 23.Gompert Z, Fordyce JA, Forister ML, Shapiro AM, Nice CC. Homoploid hybrid speciation in an extreme habitat. Science. 2006; 314: 1923–1925. [DOI] [PubMed] [Google Scholar]
- 24.Skrede I, Carlsen T, Stensrud Ø, Kauserud H. Genome wide AFLP markers support cryptic species in Coniophora (Boletales). Fun. Biol. 2012; 116: 778–784. [DOI] [PubMed] [Google Scholar]
- 25.Koerber GR, Anderson PA, Seekamp JV. Morphology, physiology and AFLP markers validate that green box is a hybrid of Eucalyptus largiflorens and E. gracilis (Myrtaceae). Austral. System. Bot. 2013; 26: 156–166. [Google Scholar]
- 26.Critchfield WB. Crossability and relationships of the closed-cone pines. Silvae Genet. 1967; 16: 89–97. [Google Scholar]
- 27.Conkle MT, Critchfield WB. Genetic variation and hybridization of ponderosa pine. Ponderosa pine: the species and its management.1988; 27: 43. [Google Scholar]
- 28.Quijada A, Liston A, Robinson W, Álvarez BE. The ribosomal ITS region as a marker to detect hybridization in pines. Mol. Ecol. 1997; 6: 995–996. [Google Scholar]
- 29.Delgado P, Salas LR, Vázquez LA, Wegier A, Anzidei M, Alvarez B, et al. Introgressive hybridization in Pinus montezumae Lamb and Pinus pseudostrobus Lindl. (Pinaceae): Morphological and molecular (cpSSR) evidence. J. Plant Scien. 2007; 168: 861–875. [Google Scholar]
- 30.Hernández LS, Gernandt DS, Pérez JA, Jardón BL. Phylogenetic relationships and species delimitation in Pinus Section Trifoliae Inferred from Plastid DNA. PLOS ONE. 2013; 8: e70501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Styles BT. Genus Pinus: a Mexican purview In: Ramamoorthy TP, Bye R, Lot A, Fa J. editors. Biological diversity of Mexico, origins and distribution. 1993. pp. 397–420. [Google Scholar]
- 32.Rehfeldt GE. Genetic variation in the Ponderosae of the Southwest. Am. J. Bot. 1993; 330–343. [Google Scholar]
- 33.Gernandt DS, Hernández LS, Salgado HE, Pérez de la Rosa J. Phylogenetic relationships of Pinus subsection Ponderosae inferred from rapidly evolving cpDNA regions. Syst. Bot. 2009; 34: 481–491. [Google Scholar]
- 34.Gernandt DS, López GG, García SO, Liston A. Phylogeny and classification of Pinus. Taxon. 2005; 54: 29–42. [Google Scholar]
- 35.Vargas-Mendoza CF, Rodríguez-Banderas A, Ibarra-Sánchez CL, Romero-Salas EA, Medina-Jaritz NB, and Alcalde-Vázquez R. Phylogenetic Analysis of Mexican Pine Species Based on Three Loci from Different Genomes (Nuclear, Mitochondrial and Chloroplast). INTECH Open Access Publisher, 2011. [Google Scholar]
- 36.Little EL, Francis IR. Botanical descriptions of forty artificial pine hybrids. No. 1345. US Dept. of Agriculture, 1965.
- 37.Garrett Peter W. Species hybridization in the genus Pinus. 1979.
- 38.Critchfield WB. Crossability and relationships of Washoe pine. Madroño. 1984; 144–170. [Google Scholar]
- 39.Epperson BK, Frank WT, Willyard Ann. Chloroplast diversity in a putative hybrid swarm of Ponderosae (Pinaceae). American Journal of Botany. 2009; 96: 707–712. 10.3732/ajb.0800005 [DOI] [PubMed] [Google Scholar]
- 40.Willyard A, Cronn R, Liston A. Reticulate evolution and incomplete lineage sorting among the ponderosa pines. Molecular Phylogenetics and Evolution. 2009; 52: 498–511. 10.1016/j.ympev.2009.02.011 [DOI] [PubMed] [Google Scholar]
- 41.González-Elizondo MS, González-Elizondo M, Tena Flores JA, Ruacho GL. López EL. Vegetación de la Sierra Madre Occidental, México: Una Síntesis. Acta Bot. Mex; 2012; 100: 351–403. [Google Scholar]
- 42.Peloquin RL. Variation and hybridization patterns in Pinus ponderosa and Pinus engelmannii Dissertation, University of California at Santa Barbara; 1971; p 196. [Google Scholar]
- 43.Peloquin RL. The identification of three-species hybrids in the ponderosa pine complex. Southwestern Naturalist. 1984; 29: 115–122. [Google Scholar]
- 44.Roe AD, MacQuarrie CJ, Gros-Louis MC, Simpson JD, Lamarche J, Beardmore T, et al. Fitness dynamics within a poplar hybrid zone: I. Prezygotic and postzygotic barriers impacting a native poplar hybrid stand. Ecol. and Evol. 2014; 4: 1629–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villa-Salas AB, Manon G, Cecilia A. Multiresource management research in northern Sonora. In: IUFRO/MAB conference: research on multiple use of forest resources: Proceedings; 1980 May 18–23; Flagstaff, AZ Gen. Tech. Rep. WO-25. Washington, DC: U.S. Department of Agriculture. Forest Service; 1980: 20–25 [Google Scholar]
- 46.Barton AM, Teeri JA. The ecology of elevational positions in plants: drought resistance in five montane pine species in southeastern Arizona. Am. J. Bot. 1993; 15–25. [Google Scholar]
- 47.Silva-Flores R, Pérez-Verdín G, Wehenkel C. Patterns of tree species diversity in relation to climatic factors on the Sierra Madre Occidental, Mexico. PLOS ONE. 2014; 9: e105034 10.1371/journal.pone.0105034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.García AA, González-Elizondo MS. Pináceas de Durango. 2a. Ed Xalapa, Veracruz: CONAFOR-Instituto de Ecología A.C.; 2003. [Google Scholar]
- 49.Wehenkel C, Simental-Rodríguez LS, Silva-Flores R, Hernández-Díaz C, López-Sánchez CA, Antúnez P. Discrimination of 59 seed stands of various Mexican pine species based on 43 dendrometric, climatic, edaphic and genetic traits. Forstarchiv. 2015; 86:194–201. [Google Scholar]
- 50.Debreczy Z, Rácz I. Conifers around the World. Vols. 1–2 DendroPress Ltd., Budapest: 20111089 pp. [Google Scholar]
- 51.Vos P, Hogers R, Bleeker M. AFLP: A new concept for DNA fingerprinting. Nucleic Acids Res. 1995; 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuchma O, Vornam B, Finkeldey R. Mutation rates in Scots pine (Pinus sylvestris L.) from the Chernobyl exclusion zone evaluated with AFLP and microsatellite markers. Mutat. Res. 2011; 725: 29–35. 10.1016/j.mrgentox.2011.07.003 [DOI] [PubMed] [Google Scholar]
- 53.Simental-Rodríguez SL, Quiñones-Pérez CZ, Moya D, Hernández TE, López SC, Wehenkel C. The relationship between species diversity and genetic structure in the rare Picea chihuahuana Tree Species Community, Mexico. PLOS ONE. 2014; 9 e111623 10.1371/journal.pone.0111623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vuylsteke M, Mank R, Antonise R, Bastiaans E, Senior ML, Stuber CW, Melchinger AE et al. Two high-density AFLP® linkage maps of Zea mays L.: analysis of distribution of AFLP markers. Theor. Appl. Genet. 1999; 99: 6, 921–935. [Google Scholar]
- 55.Krauss SL. Accurate gene diversity estimates from amplified fragment length polymorphism (AFLP) markers. Mol. Ecol. 2000; 9: 1241–1245. [DOI] [PubMed] [Google Scholar]
- 56.O’Hanlon PC, Peakall RA simple method for the detection of size homoplasy among amplified fragment length polymorphism fragments. Mol Ecol. 2000; 9:815–816. [DOI] [PubMed] [Google Scholar]
- 57.Althoff DM, Gitzendanner MA, Segraves KA The Utility of amplified fragment length polymorphisms in phylogenetics: a comparison of homology within and between genomes. Syst Bio. 2007; 56: 477–484. [DOI] [PubMed] [Google Scholar]
- 58.Gort G, van Eeuwijk FA. Review and simulation of homoplasy and collision in AFLP. Euphytica. 2012; 183: 389–400. [Google Scholar]
- 59.Vekemans X, Hardy OJ. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol. 2004; 13: 921–935. [DOI] [PubMed] [Google Scholar]
- 60.Caballero A, Quesada H, Rolán AE. Impact of amplified fragment length polymorphism size homoplasy on the estimation of population genetic diversity and the detection of selective loci. Genetics. 2008; 179: 539–554. 10.1534/genetics.107.083246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.San Cristobal M, Chevalet C, Peleman J, Heuven H, Brugmans B, Van Schriek, et al. Genetic diversity in European pigs utilizing amplified fragment length polymorphism markers. Animal Genetics. 2006; 37: 232–238. [DOI] [PubMed] [Google Scholar]
- 62.Xu S, Tauer CG, Nelson CD. Natural hybridization within seed sources of shortleaf pine (Pinus echinata Mill.) and loblolly pine (Pinus taeda L.). Tree Genetics & Genomes. 2008; 4: 849–858. [Google Scholar]
- 63.Edwards Burke M, Hamrick J, Price R. Frequency and direction of hybridization in sympatric populations of Pinus taeda and P. echinata (Pinaceae). American Journal of Botany. 1997; 84: 879–879. [PubMed] [Google Scholar]
- 64.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000; 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. 2007; 7:574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deichsel G, Trampisch HJ. Clusteranalyse und Diskriminanzanalyse. Stuttgart: Ed. Fischer G.; 1985.
- 67.Gregorius HR, Degen B, König A. Problems in the analysis of genetic differentiation among populations–a case study in Quercus robur. Silvae Gen. 2007; 56: 190–199. [Google Scholar]
- 68.Manly BFJ. Randomization, Bootstrap and Monte Carlo methods in biology. Chapman and Hall, London, 1997. [Google Scholar]
- 69.Fogelqvist J, Verkhozina AV, Katyshev AI, Pucholt P, Dixelius Ch, Rönnberg-Wästljung AC, Lascoux M, Berlin S. Genetic and morphological evidence for introgression between three species of willows. BMC Evolutionary Biology. 2015; 15:193 10.1186/s12862-015-0461-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peñaloza-Ramírez JM, González-Rodríguez A, Mendoza-Cuenca L, Caron H, Kremer A, Oyama K. Interspecific gene flow in a multispecies oak hybrid zone in the Sierra Tarahumara of Mexico. Ann. Bot. 2010; 105 (3): 389–399. 10.1093/aob/mcp301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tovar-Sánchez E, Oyama K. Effect of hybridization of the Quercus crassifolia x Quercus crassipes complex on the community structure of endophagous insects. Oecologia. 2006; 147: 702–713. [DOI] [PubMed] [Google Scholar]
- 72.González-Elizondo MS, González-Elizondo M, Sørensen PD. Arbutus bicolor (Ericaceae), a New Species from Mexico. Acta Bot. Mex. 2012; 99: 55–72. [Google Scholar]
- 73.González-Elizondo MS, González-Elizondo M, Zamudio-Ruiz S. Delimitación taxonómica de los complejos Arbutus mollis y A. occidentalis (Ericaceae). Acta Bot. Mex. 2012; 101: 49–81. [Google Scholar]
- 74.Gómez JM, González-Megías A, Lorite J, Abdelaziz M, Perfectti F. The silent extinction: climate change and the potential hybridization-mediated extinction of endemic high-mountain plants. Biodiversity and Conservation. 2015; 24: 1843–1857. [Google Scholar]
- 75.Muhlfeld CC, Kovach RP, Jones LA, Al-Chokhachy R, Boyer MC, Leary RF, Lowe WH, Luikart G, Allendorf FW. Invasive hybridization in a threatened species is accelerated by climate change. Nature Climate Change. 2014; 4: 620–624. [Google Scholar]
- 76.Chunco AJ. Hybridization in a warmer world. Ecology and Evolution 2014; 4: 2019–2031. 10.1002/ece3.1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ehrenberg C, Gustafsson Å, Forshell C, Simak M. Seed quality and the principles of forest genetics. Hereditas. 1955; 41: 291–366. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


