Abstract
This protocol describes a way to introduce topography to 3D biomaterials. The self-assembling behavior of magnetic nanoparticles can be exploited to form nanoscale to microscale fibers such that one can dissect the contribution of stiffness from topography from that due to tissue morphology. The magnetic particles are chemically cross-linked with several ECM proteins and then united using magnetic force-mediated assembly in a surrogate hydrogel to maintain the programmed topography. This process allows the creation of diverse topographic patterns in 3D, including isotropic, anisotropic (fibril), or interfaced architectures, without changing the bulk stiffness of the scaffold material. This anisotropic architecture guides the dendritic protrusions of cells, which can be compared to cells grown in an isotropic architecture lacking spatial guidance cues. Several cell types, such as fibroblasts and neurons, were cultured in this engineered 3D matrix. This technology provides an easy way to construct nano-bio interfaces for various biomedical engineering applications as well as dissect the role of topography in various cell behaviors.
Keywords: three-dimensional cell culture, extracellular matrix, topography, magnetic particles, magnetic field-directed self-assembly, biomaterial, nanocomposite material
INTRODUCTION
Tissue cells reside in a complex three-dimensional (3D) environment that is composed of heterogeneous biological building blocks in certain physical architectures where they interpret and process information about their surroundings received through myriads of biological and physicochemical signals (Chaudhuri et al., 2015; Mouw et al., 2014). Structurally, these architectures, i.e. topography, make up both the basement membrane and the interstitial matrix, thus playing a primary role in directing cell structure and function by providing physical and spatial cues to which cells respond in combination with biochemical components (Dalby et al., 2014). The biophysical interactions between cells and these architectures have a profound impact on cell processes such as differentiation, migration and proliferation (Griffith and Swartz, 2006; Hoffman et al., 2011; Hoffman-Kim et al., 2010; Ventre et al., 2014). Therefore, to recreate the topographical and chemical heterogeneity of the ECM architectures, 3D biomimetic scaffolds have approximated the in vivo architectural and signaling cues provided by the ECM for real time visualization of single cell dynamics and multicellular assembly (Tanner et al., 2012).
In this unit, the authors describe a protocol for in situ fabrication of nanoscale to microscale topography in 3D biomaterials to study the independent role of topography on cell processes (see Basic Protocol 1) (Kim et al., 2015). Magnetic beads coated with ECM proteins play the role of biological building blocks that comprise the topography and the magnetically directed assembly of these beads is used to create the topography. The authors also provide supplementary protocols for immobilizing the ECM proteins on the surface of magnetic beads (see Support Protocol 1) and for staining the cells cultured in the engineered 3D biomaterials (see Support Protocol 2).
STRATEGIC PLANNING
Magnetic field-directed self-assembly of magnetic particles
Magnetic particles used as topographic building blocks are superparamagnetic materials with magnetization that randomly changes direction under the influence of temperature. They are called ‘superparamagnetic’ because they show paramagnetic behavior but have much higher magnetic susceptibility than normal paramagnetic materials. In this protocol, the surfaces of these particles are functionalized to provide biological interfaces that mimic ECM materials. Because this functionalization is performed prior to the fabrication of the engineered matrix, there is no restriction on the chemistry and it is largely independent of matrix composition unless the functionalization disturbs the self-assembly of magnetic particles or severely enhances their aggregation. Here, authors coat the surface of carboxylated particles with ECM proteins such as fibronectins and laminins.
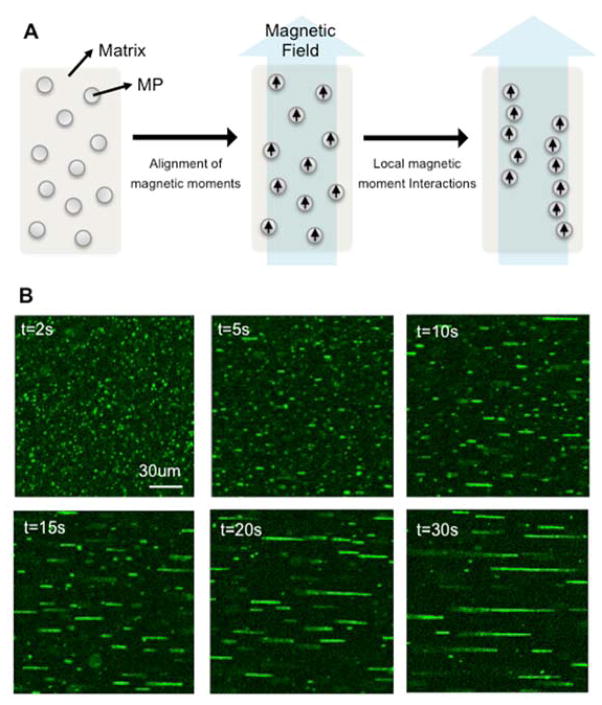
Briefly, in the absence of an external magnetic field, these particles are randomly dispersed in a liquid, as they have no net magnetic moments at room temperature. When an external magnetic field is applied, the interaction energy between the external magnetic field and the intrinsic magnetic moment of a particle overcomes the thermal fluctuation energy, thus fixing the magnetic moments parallel to the direction of the applied magnetic field line. Then the particles are arranged into chain-like aggregates, minimizing the interaction energy of all magnetic moments. In other words, the assembled particles form nanostructures akin to “fibers” along the magnetic field lines. Specifically, under the external magnetic field, the attractive magnetic force due to the magnetic dipoles of particles is balanced with the rheological resistance due to the fluid, and the particles in the fiber-like structure dynamically build or maintain their arrangement, establishing equilibrium among the many forces involved in the assembling process. This process is termed magnetic field-directed self-assembly. Particles immobilized in the fibers are released when we remove the external magnetic field. Therefore, to maintain the arrangement without the aid of the external magnetic field, we must solidify the liquid matrix to physically confine the self-assembled structures.
In situ programming of topography that mimics extracellular matrix architecture
In this unit, the authors describe a new method to provide topographical and chemical cues to cells in 3D scaffolds, using the self-assembling behavior of magnetic particles explained above. To achieve this, authors functionalize the superparamagnetic particles by immobilizing the proteins of interest, then mix these particles with cells in a liquid hydrogel, assemble these building blocks using magnetic field-directed self-assembly in a 3D matrix and solidify the matrix to maintain the programmed topography. Using this simple technique, one can fabricate diverse topographic patterns in many different types of hydrogels in 3D, and observe how cells behave in the programmed architecture. This process is independent of physico-chemical properties of the material, is biocompatible, and provides self-organized patterns at nanoscale to microscale resolution at low cost. It is also amenable to scaling to high-volume manufacturing.
Creating topography using the self-assembly of magnetic beads provides several advantages. First, it provides ease of anisotropy via nanoscale to microscale topography engineered into a 3D hydrogel. One way of modulating the topography is to control the dimension of chains. As the assembly simply follows in the same direction as that of the magnetic field line, any directions or curvatures of chains can be achieved by modulating the applied magnetic field line. The physical dimensions of these nano-chains, such as their length, width and inter-chain distance, can be controlled by either adjusting the duration of the applied magnetic field, the diameter of the nanoparticles or their initial concentration. For example, the chain length increases as the duration or the intensity of the applied magnetic field increases. Also, the width distribution of chains varies according to the diameter of the nanoparticles. Interestingly, the length and inter-chain distances are coupled. As the chain length increases, more nanoparticles are needed for elongation, thus fewer chains are formed. Another way of changing the topography is to use sequential polymerization of the matrix. This enables the fabrication of a hybrid structure composed of different topographic geometries. In this way, one can create a hybrid environment with stacked layers or form composites with different alignments and ECM proteins.
Secondly, this method allows one to create topography independent of other physicochemical or biological variables such as stiffness, porosity or molecular composition of the materials that comprise the matrix. In particular, the integration of chains in a matrix has little effect on the bulk stiffness or the molecular diffusion characteristics of pure matrices. Thus, one can create chains in various types of biological matrices, such as Matrigel and hyaluronic acid. Of course, the self-assembling profile of magnetic particles in a matrix is critically affected by the rheological properties of the matrix. For example, chains of particles in a matrix that has higher viscosity show greater elongation under the same conditions because the viscous resistance interferes with the magnetic attractions among the particles. Therefore, to obtain chains with certain desired dimensions, one has to define the assembling profile of particles in the matrix of interest to determine appropriate experimental conditions.
Topographical guidance on dendritic protrusion of cells
Tissue anisotropy provides spatial guidance cues for several cell types, such as fibroblasts and neurons in vivo. Similarly, the programmed topography in 3D biomaterials also plays a role in the physico-chemical guidance of cellular protrusions of dendritic cells. In a previous article, the authors fabricated an anisotropic (and isotropic) 3D matrix in which particles coated with fibronectin, laminin or BSA formed a fibril (and isotropic) architecture in Matrigel. They also showed that NIH 3T3 fibroblasts tend to extend their dendrites either parallel or perpendicular to the direction of chains in the matrix, independent of the protein composition of the chains, compared to cells grown in an isotropic 3D matrix. As a result, the authors concluded that either the topography alone or the topography along with a common integrin that facilitates binding to each of the ECM types is sufficient for guidance of dendritic extensions. This implies that the anisotropic topography formed by the self-assembly of magnetic particles in 3D matrix can guide cellular exploration and growth.
Fibril topography is composed of both nanoscale grooves made by aggregated particles on the surface of chains and microspaced patterns formed by the uniform distribution of chains. Several studies on the dendritic extensions of cells on 2D substrates have found that dendrites can extend in both parallel and perpendicular directions on hybrid patterns (Sur et al., 2012). In a similar way, the authors hypothesize that nanoscale grooves cause the perpendicular extension of cell dendrites, and the microspaced patterns guide parallel cellular protrusions. In other words, this magnetic self-assembly of particles creates a hybrid pattern that is composed of both microscale chain-like rails and perpendicular nanoscale grooves. A perpendicular combination of micro- and nano-patterns is able to guide both parallel and perpendicular extensions of dendrites.
BASIC PROTOCOL 1: CREATING AN ANISOTROPIC ECM MICROENVIRONMENT BY NANOPARTICLE ASSEMBLY
In this protocol, the authors describe how to form topographical patterns in in vitro three-dimensional hydrogel matrices. They applied this method to observe cell behavior in a topographically anisotropic architecture, using the 3T3 fibroblast cell line. The authors analyzed the topography formed by self-assembled magnetic beads and observed enhanced protrusions of fibroblast dendrites in this engineered matrix.
This method produced various bio-interfaces by immobilizing different types of ECM proteins on the surface of magnetic beads. The authors used fibronectin and laminin as representative structural proteins comprising the ECM and bovine serum albumin (BSA) as a control interface.
This protocol was developed for observing cell behavior in 3D engineered matrix in LabTek 8-well chambered coverglass systems that require only small volumes of sample (100~200 μl). This protocol can also be used to make samples in Corning 12-well cell culture plates by scaling up all the reagents to volumes in the range of 500 μl to 1 ml.
Materials
Matrigel (Corning Matrigel Growth Factor Reduced Basement Membrane Matrix, product number 354230)
Hyaluronic Acid (HyStem™ Hydrogel Kit 7.5ml, catalog number GS311)
ECM protein-coated superparamagnetic particles (Support Protocol 1)
Cell culture medium (10% FBS, 1% Penicilin/Spreptomycin and 1% L-glutamine in DMEM (1×))
Serum-free culture medium (1% p/s and 1% L-glutamine in DMEM (1×))
Cell suspension (NIH 3T3 Fibroblast cell line) at 1 × 106 cells/ml in cell culture medium
LabTek 8-well chambered coverglass
Corning 12-well Plate (Corning 3512 Costar Cell Culture Plates, Product#3512)
37°C, 10% CO2, cell culture incubator
Microscope with 10× objective
NdFeB magnet (K&J Magnetics, Inc., catalog number BX8X8X8, 25.4 mm X 25.4 mm X 25.4 mm)
Prepare the chamber
-
1
Place LabTek 8-well chambered coverglass on ice and allow it to chill.
-
2
Keep Matrigel on ice.
Matrigel remains at liquid-phase from approximately 2–8°C and begins to gell at 37°C. Therefore, all the chambers, Matrigel and reagents should be kept on ice during the experimental process from sample preparation to topography fabrication. -
3
Drop 20 μl of liquid Matrigel on the glass surface of the well of the LabTek chambered coverglass, and, with gentle strokes, spread the Matrigel on the glass surface using a pipet tip. Keep the chambered coverglass on ice while spreading Matrigel.
Any type of polymer capable of solidification without an interfering magnetic force can be used to create topography. For example, hyaluronic acid can also be used as a matrix material. Several studies have demonstrated that the aligned magnetic chains are incorporated in a polymeric material through photopolymerization. Thus, this could be a universal tool to program topographies in 3D biomaterials. -
4
Place the LabTek coverglass in a 37°C, 10% CO2 cell culture incubator for 4 min to allow the Matrigel to polymerize.
It is crucial to cover the bottom of the chamber with Matrigel to prevent the cells from reaching the bottom of the chamber. If cells attach to the glass substrate, their behavior is affected by the glass substrate, which has much higher stiffness than the 3D Matrigel environment.If the gelled Matrigel is left in the incubator too long, the thin layer of Matrigel covering the bottom will dry out.
Prepare the sample
-
5
During, or before the time of Matrigel polymerization, detach cells, count them, and prepare a cell suspension of 1 × 106 cells/ml in culture medium.
In this process, one can determine cell type and cell concentration.In the case of using NIH/3T3 fibroblasts, the authors detach the cells as follows: First, wash the 75 cm2 flask with 13 ml of PBS. Apply 1 ml of 0.25 % Trypsin-EDTA to the cells and incubate them in a 37°C, 10% CO2 cell culture incubator for 2 min. Then, move them into a 15 ml tube and spin down cells at 1200 rpm for 4 min. Finally, re-suspend the cells in 1 ml of cell culture media. When you use other cell lines, the conditions for detaching cells should be empirically determined. -
6
Thoroughly mix 20 μl of cell suspension (2 × 104 cells) and 10 μl of ECM-coated magnetic particles (10 mg/ml) in 100 μl of Matrigel on ice.
-
7
Spread this mixture on the Matrigel-coated coverglass.
Induce topography in the sample
-
8
Keep the magnet on ice and allow it to chill.
-
9
Place the coverglass with the sample on the cold magnet and wait 6 min to allow the magnetic beads to assemble into fiber-like structures.
The magnetic field application time varies according to other parameters: the matrix material (which has different rheological properties), the magnetic field intensity, the magnetization of magnetic particles, and so on. For the purpose of changing the dimensions of the topography, you can also control these parameters. -
10
Gently remove the coverglass from the magnet and briefly observe the sample using a 10X microscope to confirm that the self-assembled chains are properly formed.
-
11
Incubate the sample in a 37°C, 10% CO2 cell culture incubator for 30 min to allow the Matrigel to polymerize and the chains to be confined.
Gelation is required to physically confine the self-assembled topography made of magnetic beads.Allow the sample to gel as rapidly as possible after removing it from the magnet. The magnetic particles will erse again if the external magnetic field is removed from the material system. When the external magnetic field is not applied to the matrix, the sample slowly loses its self-assembled topography. -
12
Add 300 μl of serum-free culture medium per well.
Media can be customized according to the purpose of the experiment. One can use different types of media that contain drugs or other reagents to observe the effects of those on cell behavior in an engineered matrix. -
13
Incubate the sample for 12–18 hours to allow the cells to respond.
The incubation time can vary according to the cell type. If more time is required, replace the media each day.
SUPPORT PROTOCOL 1: CHEMICAL CROSS-LINKING OF THE ECM PROTEINS ON THE SURFACE OF MAGNETIC NANOPARTICLES
Various protein conjugation protocols can be used, and this process should be adjusted for the purpose of the experiment. Ideally, surface functionalization of nanoparticles is performed prior to hydrogel polymerization. Hence, there is no restriction on the chemistry and it is largely independent of matrix composition.
Materials
300 nm of carboxyl-superparamagnetic beads (Adem Tech, catalog number 20131)
Diluted activation buffer (1X) (Adem Tech, catalog number 20820)
Diluted storage buffer (1X) (Adem Tech, catalog number 20820)
1-ethyl-3-(3-dimethylaminopropyl carbodiimide hydrochloride) (Adem Tech, catalog number 20820)
Bovine Albumin Serum (BSA)
Fluorescent fibronectin (Cytoskeleton Inc., catalog number FNR02, labeled with HiLyte 488)
Fluorescent laminin (Cytoskeleton Inc., catalog number LMN01, labeled with Rhodamine)
1.5 ml tubes
Shaker (mixer)
Activation of beads
-
1
Add 0.5 mg of carboxyl-superparamagnetic beads (300nm) to a 1.5 ml tube.
-
2
Wash the beads with activation buffer (1X, diluted in D.I. water) 3 times.
To wash the beads, place the tube on the magnet for 20 sec and pipette off the supernatant carefully, leaving beads undisturbed. Then, remove the tube from the magnet, resuspend the beads carefully in 100 μl of activation buffer, and mix them well. The separation time of the magnetic beads can vary according to the size of the particles or the intensity of the magnetic field produced by the magnet. -
3
Dissolve 4 mg of 1-ethyl-3-(3-dimethylaminopropyl carbodiimide hydrochloride) (EDC) in 1 ml activation buffer.
-
4
Add 100 μl of the EDC solution for each 100 μl of particle solution to the tube and vortex to mix properly.
-
5
Incubate this solution for 1 hour at room temperature with shaking.
Protein conjugation
-
6
During incubation, prepare an ECM protein solution at 1 mg/ml and dilute the solution in activation buffer (1X).
-
7
Add 20 μl of protein solution to the activated sample per tube.
-
8
Incubate this solution overnight (at least 4 hours) at room temperature with shaking.
-
9
During incubation, prepare the bovine serum albumin (BSA) solution in the activation buffer (0.5 mg/ml).
-
10
Add 200 μl of BSA solution per tube to block nonspecific binding.
-
11
Incubate this solution for 1 hour at room temperature with shaking.
-
12
Wash the beads with the storage buffer 3 times.
-
13
Resuspend the beads to yield a final concentration of 1 mg/ml in the tube.
The final concentration of the beads can be controlled. -
14
Store this solution at 2–8°C.
SUPPORT PROTOCOL 2: STAINING AND IMAGING CELLS IN A 3D ANISOTROPIC MATRIX
Here, authors provide a simple process to stain the nucleus and the actins. One can observe other intracellular proteins through appropriate fixation, permeabilization and immunostaining processes.
Materials
4% (w/v) paraformaldehyde in D.I. water
Sample of cells in an anisotropic 3D matrix in LabTek 8-well chambered coverglass (Basic Protocol 1)
Phosphate-buffered saline (PBS, pH 7.2, KD Medical, Cat# RGE-3190)
Blocking solution (1% BSA in PBS)
Phalloidin conjugated to Alexa Fluor 633 (Life Technologies, catalog nuber A22284)
Hoechst 33342 (Invitrogen, catalog number H3570)
Confocal microscopy
Fix the sample
-
1
After the cell incubation, gently aspirate the culture media from the LabTek coverglass and wash the sample with 200 μl of PBS twice.
-
2
Apply 300 μl of 4 % paraformaldehyde to the sample per well and incubate the sample overnight at room temperature.
The authors increase the incubation time to make sure the sample is fixed, but 3 hours of incubation is probably sufficient. -
3
Gently aspirate the 4 % paraformaldehyde solution from the sample, and wash it with 200 μl PBS three times.
-
4
Add 300 μl of blocking buffer to the well to block the sample and prevent nonspecific binding of staining proteins, and allow the sample to rest 30 min at room temperature.
-
5
Aspirate the blocking buffer and wash the sample with 200 μl of PBS twice.
Stain the cells
-
6
Prepare 200 μl of cell staining solution composed of 1:50 Phalloidin conjugated to Alexa Fluor 633 and 1:500 Hoechst 33342 in PBS.
-
7
Allow the sample to rest for 3 hours at room temperature
-
8
Gently aspirate the staining solution and wash the sample with 200 μl PBS three times.
Visualize the sample
-
9
Examine the sample by confocal microscopy.
Some samples may be stored before they are examined, but storage conditions vary according to the fluorophore used for staining. If the sample is liquid and is stored, it will lose fluorescence due to molecular diffusion and photobleaching. Therefore, it is recommended that one proceed promptly to confocal microscopy after staining is complete.
REAGENTS AND SOLUTIONS
Blocking buffer
Prepare a fresh solution of 1% BSA (w/w) in DI water. BSA was purchased from Sigma-Aldrich. This buffer can be stored at 2–8°C for one month.
Matrigel
Corning Matrigel Growth Factor Reduced (GFR) Basement Membrane Matrix, 10 ml, product number 354230
Paraformaldehyde, 4% (w/v)
Dilute the 16% paraformaldehyde (Electron Microscopy Science, Paraformaldehyde 16% solution, EM grade, Cat. #15710) in DI water to a 4% solution.
PBS
PBS, pH 7.2, KD Medical, Cat# RGE-3190.
Staining material
Phalloidin conjugated to Alexa Fluor 633 (Life Technologies, catalog nuber A22284)
Hoechst 33342 (Invitrogen, catalog number H3570)
EDC solution
Dissolve 2 mg of powder-type EDC in 500 μl of activation buffer to make a final concentration of 4 mg/ml. Always prepare a fresh solution per experiment.
Dry stock powdered EDC should be stored at −20°C and avoided placing it in humid conditions, as EDC is spoilt when it comes in contact with water.
COMMENTARY
Background Information
Many hydrogel materials, such as alginate and reconstituted laminin-rich ECM (e.g. Matrigel) have been used to provide in vitro ECM environments for real time visualization of single cell dynamics or multicellular assembly (Doyle et al., 2013; Wang et al., 2013). However, they are not able to recreate the topographical or biochemical heterogeneity of ECM architectures because amorphous gels usually form, with no fibrils or spatial heterogeneities. In contrast, 3D scaffolds to approximate the in vivo architectural and signaling cues provided by the ECM, such as collagen type I or synthetic polymers cross-linked with ECM proteins have been tailored to form fibrillar architectures by tuning polymerization conditions including protein concentration, temperature and pH (Raub et al., 2007; Raub et al., 2008). However, the physical characteristics of the scaffold, especially the matrix rigidity, are unfortunately strongly correlated with protein concentration (Levental et al., 2009).
To decouple the topography with other physico-chemical properties of the 3D scaffold, techniques such as photolithographic patterning, electrospinning, and molecular self-assembly have been employed (Dvir et al., 2011; Santos et al., 2012). Lithographic methods such as multiphoton chemical patterning and photodegradation selectively expose functional groups one at a time to multiphoton excitation to immobilize the desired oligopeptides for each site of ligand binding in a surrogate ECM hydrogel (DeForest et al., 2009; DeForest and Tirrell, 2015). These methods produce very precise patterns of adhesion ligands in a hydrogel but can only be used with certain types of materials and they are largely low-throughput. Additionally, the cells are exposed to chemicals and irradiation during the fabrication process. A low cost alternative is the method of electrospinning. Briefly, this process involves “jetting” to transform an electrically charged polymer solution into nano-fibers with a distinct chemistry to achieve scaffolds with tunable mechanical properties (Dong et al., 2009; Ionescu et al., 2010). Cells cannot be incorporated in situ using this method, due to the deleterious effects of the solvents and the electric fields applied during fabrication. In addition, the jetting process employed to generate nano-fibers can only achieve specific 3D geometries. As a bottom-up approach, molecular self-assembly of amphiphile peptides by their non-covalent molecular interactions generates nano-fibrillar supermolecules that are similar in architecture to that of native ECMs such as type I collagens and fibronectin (Ahn et al., 2015; Hartgerink et al., 2001). However, tuning the mechanical properties or geometrical arrangement of the nano-fibers used in the construction of 3D structures is not readily achieved with this method. Due to the limitations of these currently employed technologies, despite our extensive understanding of the biochemical reactions that control cellular fates, dissecting the role of physical cues that also modulate or may potentially override signals by biochemical cues remains elusive.
The protocol introduced in this unit allows one to recreate topography in 3D using magnetic field-directed self-assembly of surface-functionalized magnetic particles that serve as biological building blocks. Because a magnetic force can penetrate most types of organic materials with minimal attenuation, this method can provide a simple way of fabricating biomimetic 3D scaffolds that incorporate topographical features independent of other physico-chemical parameters of the matrix material. This approach provides a tissue-mimetic platform to dissect the role of topographical cues received from fibrillar geometries from that due to the chemical composition of the ECM in surrogate 3D hydrogels.
Critical Parameters and Troubleshooting
Engineered matrix polymerization
In Basic Protocol 1, the assembling of particle chains takes approximately 10 min in Matrigel and the sample should be placed on the magnet during this time. While the particles are assembled, cells descend to the bottom of the well plate due to gravity. Therefore, when one fabricates an isotropic matrix that does not require the self-assembling of magnetic particles, the researcher should allow the cells to settle for the same amount of time to ensure that the sample has approximately the same concentration of cells at the bottom.
Magnet
The authors use a commercial neodymium magnet to create magnetic field lines along which the particles align. For any particular magnet, the researcher should find a specific region that provides a relatively uniform magnetic field in which to place the sample. If even part of the sample is in a region that has a large field gradient, the magnetic particles will aggregate or the topographic architecture will be distorted. The researcher can use micro-patterned magnets fabricated by MEMS technology to produce a specific magnetic field pattern or increase the throughput of this platform.
Magnetic particles
The characteristics of magnetic particles critically affect the self-assembling behavior in a matrix and protein immobilization process. Here, the authors use commercialized magnetic beads and a protein crosslinking kit from Ademtech Inc., and modify the protein crosslinking process from the basic protocol that this company provides. If the researcher directly synthesizes the magnetic particles or uses other types of commercialized magnetic particles, Support Protocol 2 should be modified according to the type of magnetic particles.
Anticipated Results
Basic Protocol 1
Time-lapse imaging with confocal microscopy of the 3D cell sample should show exploration and protrusion of cell dendrites.
Support Protocol 1
If the proteins are labeled with fluorescence, the protein immobilization on the surface of magnetic particles should result in a sample that is ready for fluorescence confocal imaging of the magnetic particles and their chain formation process.
Support Protocol 2
Staining of the cells in a 3D engineered matrix should result in a sample that is ready for fluorescence confocal imaging of the cell body and nucleus being studied. Z-stacks collected with confocal microscopy will allow for 3D image reconstruction.
Time Considerations
Basic Protocol 1
It takes approximately 2 hr to prepare a sample of cells in 3D engineered matrix using the technique described in Basic Protocol 2. In addition, it takes at least 12 hr of incubation at 37°C to allow cells to explore and extend dendrites in the 3D engineered matrix. The total incubation time for each cell line being studied should be determined by the researcher.
Support Protocol 1
It takes approximately 15 hr to crosslink the proteins on the surface of carboxylated magnetic particles at room temperature, as described in Basic Protocol 2. The time for crosslinking proteins on the activated particles takes most of the reaction time, and it should take at least 4 hr of incubation to immobilize the proteins. This process should be optimized according to the proteins that the researcher wants to immobilize.
Support Protocol 2
It takes approximately 4 hr to prepare a sample of stained cells in 3D matrix described in Support Protocol 2.
Figure 1.
Magnetic field-directed assembly of magnetic particles as an in situ programming method of topography. (A) Self-assembling principle of magnetic particles under the application of an external magnetic field. (B) Time-lapse images of magnetic particle self-assembly. Green objects represent magnetic particles coated with HiLyte488-fibronectins. The self-assembly was performed in culture media.
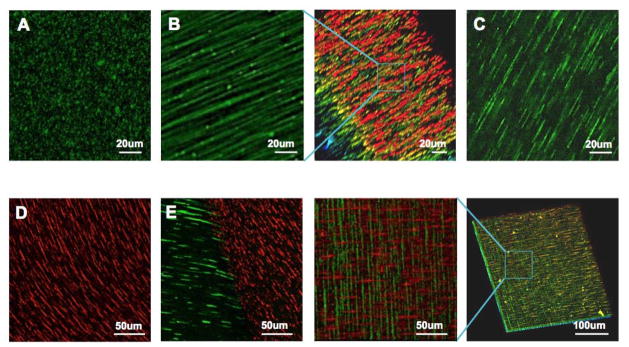
Figure 2.
Topographical patterns that can be achieved using this technology. (A) Isotropic topography with random dispersion of magnetic particles. (B) Anisotropic topography with fibril chains and 3D distribution in gelled Matrigel. (C) Anisotropic topography in a gelled hyaluronic acid. (D) Curved chains fabricated by the application of a curved magnetic field. (E) Two adjoining topographies with different surface proteins and orientations. (F) Two stacked topographies with different surface proteins and orientations. Green and red objects represent magnetic particles coated with HiLyte488-fibronectins and Rhodamine-laminins, respectively.
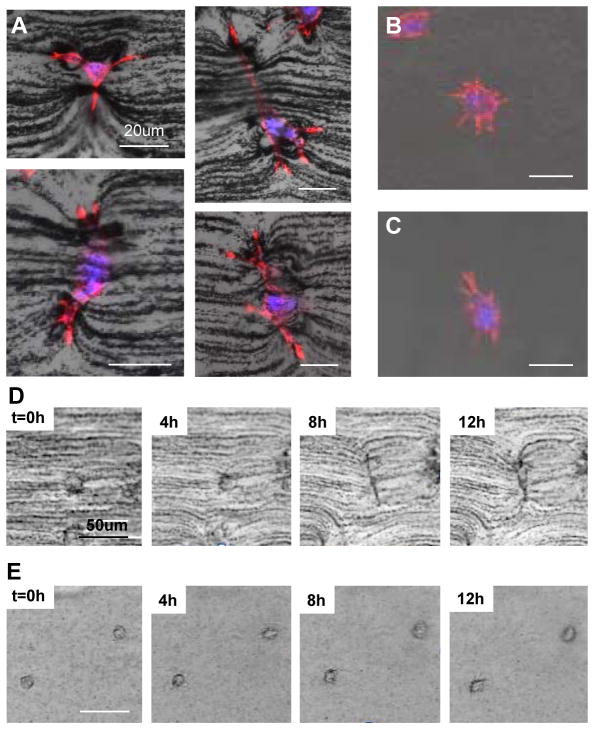
Figure 3.
Cell behavior in an engineered matrix. (A) Dendritic protrusion of fibroblasts in an anisotropic matrix, (B) in an isotropic matrix, (C) and in pure Matrigel without topography. (D) Time-lapse images of fibroblasts grown in anisotropic, (E) and in isotropic matrices. In the anisotropic matrix, a single cell elongates its body and extends dendrites, deforming the fibril architecture composed of chains. On the other hand, cells in an isotropic matrix explore the architecture of the matrix by extending their filopodia in all directions.
Figure 4.
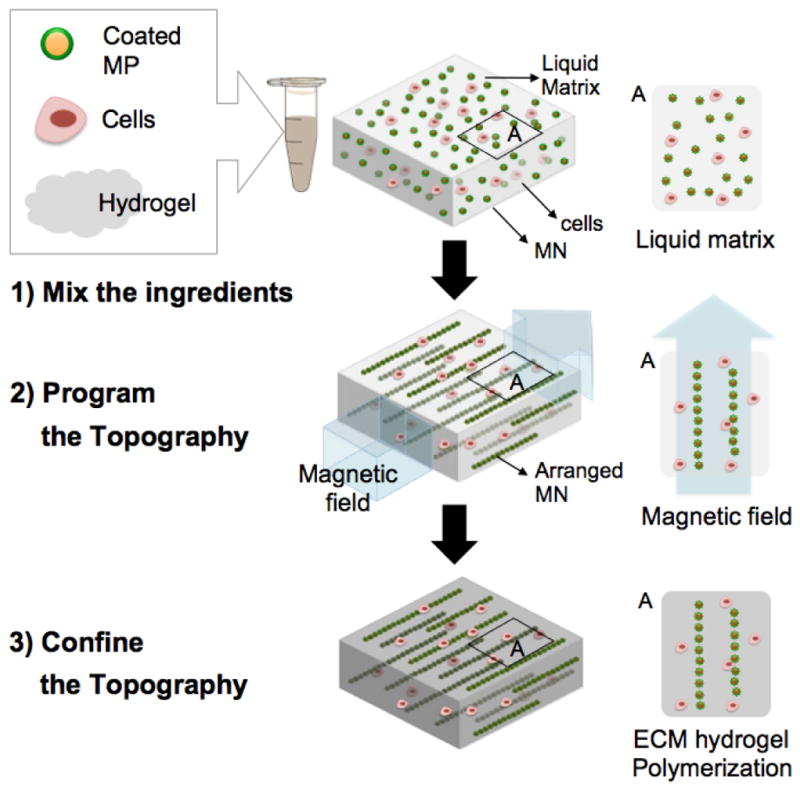
A schematic of the fabrication process. Functionalized magnetic nanoparticles are mixed with cells in liquid ECM or mixed in liquid ECM alone. A magnetic field is then applied to this mixture before polymerization of the hydrogel, to facilitate assembly of long chain-like structures (12–14). The applied magnetic field should be sufficient to overcome the rheological resistance within the hydrogel. Self-organization into specific geometric patterns with defined anisotropy is achieved by simply controlling the external field arrangement. After chains are formed with the desired dimensions, the matrix is polymerized to preserve the programmed topography. The magnetic field is then removed. A control sample where no magnetic field was applied was also fabricated and the nanoparticles are randomly distributed throughout the matrix, for comparison.
Figure 5.
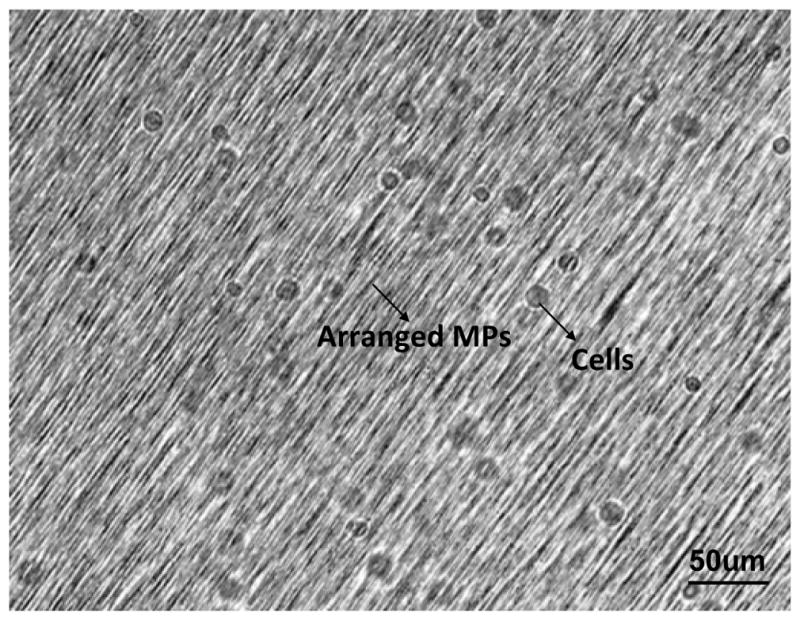
NIH/3T3 fibroblasts seeded in anisotropic matrix directly following matrix solidification.
Significance Statement.
Biomimetic extracellular matrix (ECM) topographies driven by the magnetic field-directed self-assembly of ECM protein-coated magnetic beads were fabricated. This novel bottom-up method allows researchers to program isotropic, anisotropic and diverse hybrid ECM patterns without changing other physico-chemical properties of the scaffold material. A 3D anisotropic matrix fabricated using this technology is able to guide the dendritic protrusions of cells.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute. The work was also supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1161). We thank George Leiman for editorial assistance.
LITERATURE CITED
Uncategorized References
- Ahn S, Deravi LF, Park SJ, Dabiri BE, Kim JS, Parker KK, Shin K. Self-Organizing Large-Scale Extracellular-Matrix Protein Networks. Advanced Materials. 2015;27:2838. doi: 10.1002/adma.201405556. [DOI] [PubMed] [Google Scholar]
- Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ. Substrate stress relaxation regulates cell spreading. Nature Communications. 2015;6 doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby MJ, Gadegaard N, Oreffo ROC. Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nature Materials. 2014;13:558–569. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- DeForest CA, Polizzotti BD, Anseth KS. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nature Materials. 2009;8:659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest CA, Tirrell DA. A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nature Materials. 2015;14:523–531. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- Dong B, Smith ME, Wnek GE. Encapsulation of Multiple Biological Compounds Within a Single Electrospun Fiber. Small. 2009;5:1508–1512. doi: 10.1002/smll.200801750. [DOI] [PubMed] [Google Scholar]
- Doyle AD, Petrie RJ, Kutys ML, Yamada KM. Dimensions in cell migration. Curr Opin Cell Biol. 2013;25:642–649. doi: 10.1016/j.ceb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir T, Timko BP, Kohane DS, Langer R. Nanotechnological strategies for engineering complex tissues. Nature Nanotechnology. 2011;6:13–22. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman-Kim D, Mitchel JA, Bellamkonda RV. Topography, Cell Response, and Nerve Regeneration. Annual Review of Biomedical Engineering. 2010;12:203–231. doi: 10.1146/annurev-bioeng-070909-105351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu LC, Lee GC, Sennett BJ, Burdick JA, Mauck RL. An anisotropic nanofiber/microsphere composite with controlled release of biomolecules for fibrous tissue engineering. Biomaterials. 2010;31:4113–4120. doi: 10.1016/j.biomaterials.2010.01.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Staunton JR, Tanner K. Independent Control of Topography for 3D Patterning of the ECM Microenvironment. Adv Mater. 2015 doi: 10.1002/adma.201503950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu HM, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw JK, Ou GQ, Weaver VM. Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Bio. 2014;15:771–785. doi: 10.1038/nrm3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raub CB, Suresh V, Krasieva T, Lyubovitsky J, Mih JD, Putnam AJ, Tromberg BJ, George SC. Noninvasive assessment of collagen gel microstructure and mechanics using multiphoton Microscopy. Biophys J. 2007;92:2212–2222. doi: 10.1529/biophysj.106.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raub CB, Unruh J, Suresh V, Krasieva T, Lindmo T, Gratton E, Tromberg BJ, George SC. Image correlation spectroscopy of multiphoton images correlates with collagen mechanical properties. Biophys J. 2008;94:2361–2373. doi: 10.1529/biophysj.107.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos E, Hernandez RM, Pedraz JL, Orive G. Novel advances in the design of three-dimensional bio-scaffolds to control cell fate: translation from 2D to 3D. Trends in Biotechnology. 2012;30:331–341. doi: 10.1016/j.tibtech.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Sur S, Pashuck ET, Guler MO, Ito M, Stupp SI, Launey T. A hybrid nanofiber matrix to control the survival and maturation of brain neurons. Biomaterials. 2012;33:545–555. doi: 10.1016/j.biomaterials.2011.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner K, Mori H, Mroue R, Bruni-Cardoso A, Bissell MJ. Coherent angular motion in the establishment of multicellular architecture of glandular tissues. Proc Natl Acad Sci U S A. 2012;109:1973–1978. doi: 10.1073/pnas.1119578109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventre M, Natale CF, Rianna C, Netti PA. Topographic cell instructive patterns to control cell adhesion, polarization and migration. Journal of the Royal Society Interface. 2014;11 doi: 10.1098/rsif.2014.0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lacoche S, Huang L, Xue B, Muthuswamy SK. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. P Natl Acad Sci USA. 2013;110:163–168. doi: 10.1073/pnas.1201141110. [DOI] [PMC free article] [PubMed] [Google Scholar]