Abstract
The fungal pathogen Histoplasma capsulatum causes respiratory and disseminated disease, even in immunocompetent hosts. In contrast to opportunistic pathogens, which are readily controlled by phagocytic cells, H. capsulatum yeasts are able to infect macrophages, survive antimicrobial defenses, and proliferate as an intracellular pathogen. In this review, we discuss some of the molecular mechanisms that enable H. capsulatum yeasts to overcome obstacles to intracellular pathogenesis. H. capsulatum yeasts gain refuge from extracellular obstacles such as antimicrobial lung surfactant proteins by engaging the β-integrin family of phagocytic receptors to promote entry into macrophages. In addition, H. capsulatum yeasts conceal immunostimulatory β-glucans to avoid triggering signaling receptors such as the β-glucan receptor Dectin-1. H. capsulatum yeasts counteract phagocyte-produced reactive oxygen species by expression of oxidative stress defense enzymes including an extracellular superoxide dismutase and an extracellular catalase. Within the phagosome, H. capsulatum yeasts block phagosome acidification, acquire essential metals such as iron and zinc, and utilize de novo biosynthesis pathways to overcome nutritional limitations. These mechanisms explain how H. capsulatum yeasts avoid and negate macrophage defense strategies and establish a hospitable intracellular niche, making H. capsulatum a successful intracellular pathogen of macrophages.
Keywords: cell wall, CR3, Dectin-1, fungal pathogenesis, glucan, iron, phagosome, ROS, zinc
Introduction
Histoplasma capsulatum is a member of a group of fungal pathogens that are characterized by thermal dimorphism and cause respiratory and disseminated disease in mammals [1–3]. In the environment, H. capsulatum grows as a saprobic conidia-producing mycelium. When H. capsulatum-contaminated soil is disturbed, the aerosolized mycelium fragments and conidia may be inhaled, and the small size of the conidia enables them to reach the lower respiratory tract. H. capsulatum cells respond to the elevated temperature in the mammalian host by activation of a transcriptional program that induces a switch to the yeast-phase morphology and expression of factors required for pathogenesis [4–7]. H. capsulatum is classified as a primary pathogen [8] due to its ability to cause disease in normal (i.e. immunocompetent) hosts, in contrast to opportunistic fungal pathogens that require some deficiency of host immune function (e.g. neutropenia, AIDS, etc.).
Factors produced by H. capsulatum yeast cells enable them to parasitize phagocytic immune cells. Pathogenic-phase yeasts are capable of invading phagocytic cells, including alveolar macrophages, polymorphonuclear leukocytes and dendritic cells [9–11]. These phagocytes serve as both the host cell and the vector by which infection dissemination is mediated. The innate immune response alone is insufficient to control H. capsulatum infection, in contrast to the ability of innate cells to control and eliminate some opportunistic fungal pathogens (e.g. Candida and Aspergillus species). Un-impaired by the innate immune system, continued proliferation of H. capsulatum yeasts exacerbates disease symptoms and leads to infection of extra-pulmonary sites. Upon activation of cell-mediated immunity, pro-inflammatory cytokines produced primarily by CD4+ T cells potentiate the antifungal properties of phagocytes and resolution of the H. capsulatum infection may occur. However, if an adequate T-cell response does not develop due to immuno-compromised conditions (e.g. HIV, anti-cytokine therapy, etc.), or as a result of larger inocula that establish high fungal burdens before activation of cell-mediated immunity, a life-threatening condition ensues [12–15]. Research over the past two decades has revealed a number of factors produced by H. capsulatum that subvert innate defenses and facilitate intracellular proliferation of the yeasts. In this review, we discuss findings that provide a molecular explanation of the mechanisms underlying H. capsulatum’s ability to surmount obstacles to infection and to survive within immune cells that are normally not conducive to fungal survival.
Extracellular obstacles to infection
Histoplasma capsulatum is almost exclusively found as an intracellular pathogen (Fig. 1). The species was so named by Samuel Darling because he observed it within phagocytic cells: ‘histo-’ because the microbe was found within histiocytes (a general term for tissue phagocytes), and ‘-plasma’ because he believed the microbe was a parasite-like creature. To secure intra-cellular residence, H. capsulatum must first overcome extracellular obstacles to infection (Fig. 2).
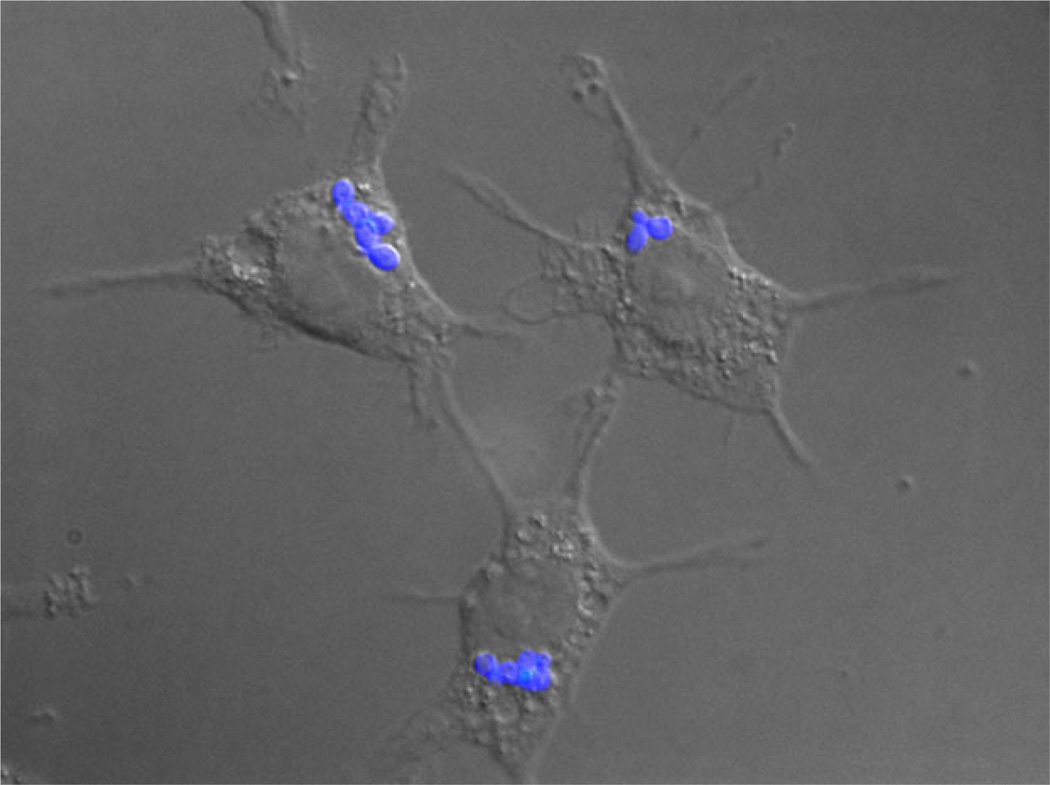
Fig. 1.
Histoplasma capsulatum is an intracellular pathogen of macrophages. The image shows H. capsulatum yeasts (blue fluorescence) within phagocytic P388D1 macrophage cells. Samples were fixed 2 h post-infection, and yeasts are stained using Uvitex (Polysciences, Inc., Warrington, PA).
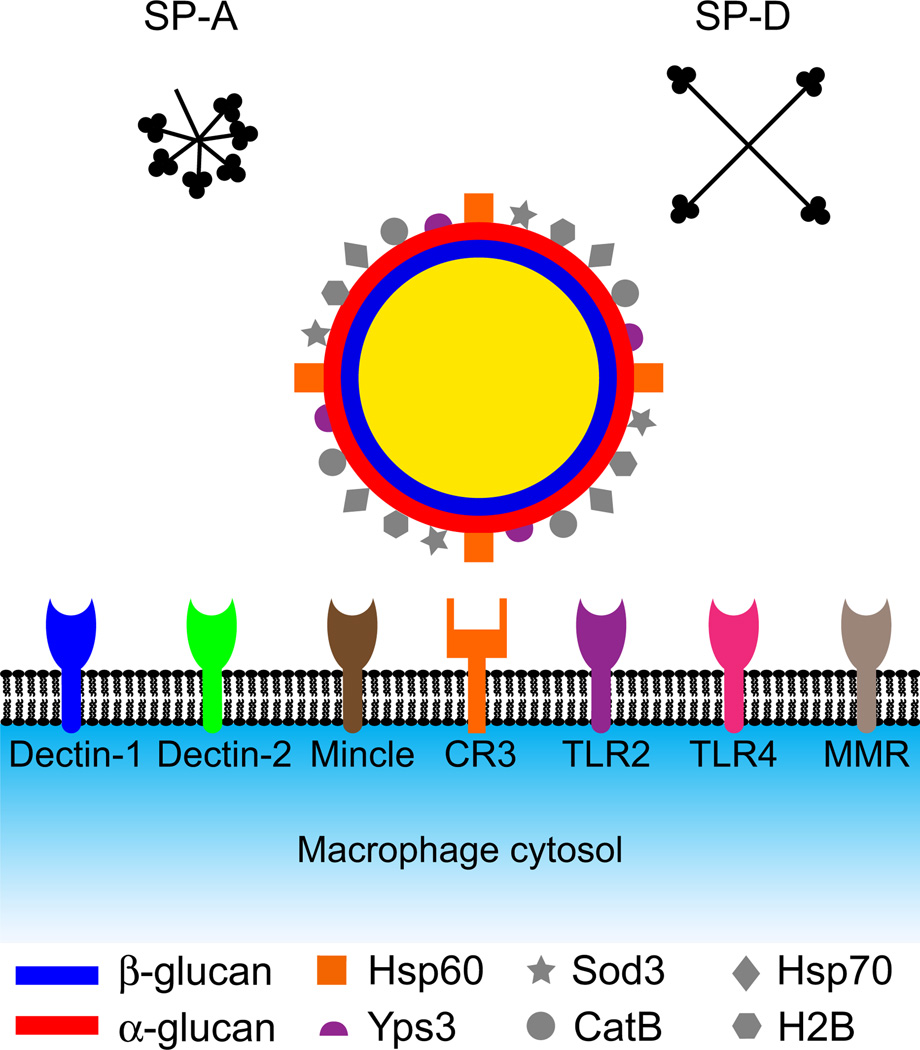
Fig. 2.
Subversion of extracellular obstacles to infection by Histoplasma capsulatum. Within alveolar spaces, H. capsulatum encounters the antifungal surfactant collectins SP-A and SP-D. To gain entry into macrophages for refuge from surfactant proteins, H. capsulatum yeasts interact with the phagocytic receptor CR3 (orange receptor with square binding pocket) using Hsp60 (orange squares) as the ligand. A number of signaling receptors (round binding pockets) may potentially interact with the yeast cell surface. The α-glucan polysaccharide layer (red) of the H. capsulatum cell wall conceals the immunostimulatory β-glucan layer (blue), preventing recognition of yeasts by the phagocyte β-glucan receptor Dectin-1 (blue receptor). Other cell-wall proteins that potentially interact with the macrophage include Yps3 (purple semi-circles), which may interact with TLR2 receptors, Hsp70 (diamonds), Sod3 (stars), CatB (circles) and H2B (hexagons). Mannose-type receptors on the macrophage include Dectin-2, Mincle, TLR2, TLR4 and the macrophage mannose receptor (MMR).
Surfactant proteins
The paucity of extracellular organisms suggests that intracellular residence is not only characteristic of, but also beneficial to, H. capsulatum yeasts. Indeed, the phagocyte may provide a refuge for H. capsulatum, limiting exposure to soluble extracellular antimicrobial factors. Within the alveolar space, the initial site of H. capsulatum infection, the extracellular environment composition includes surfactant fluid. The surfactant proteins A and D (SP-A and SP-D) present in this lipid-rich fluid have been shown to have host defense functions [16]. SP-A and SP-D are pulmonary col-lectins that comprise a collagen-like region and a C-type lectin domain that permits binding to polysaccharides of microbes. In vitro, both SP-A and SP-D have direct antifungal effects on H. capsulatum yeasts at physiological concentrations, with SP-D showing slightly more fungicidal activity in short-term assays [17]. Both SP-A and SP-D cause permeabilization of H. capsulatum yeast cells, which accounts for their fungicidal activity. Internalization of H. capsulatum yeasts by alveolar macrophages completely negates the fungicidal activity of SP-A and SP-D, indicating that alveolar macrophages shelter H. capsulatum from the antimicrobial effects of surfactant proteins. Interestingly, SP-A and SP-D are not active against Blastomyces dermatitidis, yeasts that are more commonly found extracellularly and probably have mechanisms to resist SP-A and SP-D [17]. Despite the sensitivity of H. capsulatum yeasts to surfactant in vitro, the loss of SP-A in outbred mice only slightly enhanced H. capsulatum infection and dissemination in vivo during acute stages (day 7), suggesting that H. capsulatum yeasts may minimize exposure to SP-A in vivo, presumably through refuge in alveolar macrophages [17].
Internalization by host phagocytes
Histoplasma capsulatum yeasts are internalized by host phagocytes through induction of phagocytosis. Blocking phagocytosis by inhibition of the actin cytoskeleton of phagocytes prevents yeast internalization [18,19]. Complement receptors appear to be the major phagocytic receptors for binding and uptake of H. capsulatum yeasts (Fig. 2), and this internalization does not require opsonization by complement [18,20,21]. These receptors include LFA-1 (CD11a/ CD18), CR3 (CD11b/CD18; Mac-1) and CR4 (CD11c/CD18), which are expressed on alveolar and interstitial macrophages as well as polymorphonuclear leukocytes and dendritic cells. Antibody blocking of the common CD18 subunit prevents 50–90% of binding and uptake of H. capsulatum yeast by human macrophages [18,20], indicating that the majority of H. capsulatum uptake is dependent on the complement receptors. Yeast binding to macrophages requires Ca2+ and Mg2+ ions, and is temperature-sensitive [20], consistent with the divalent cation-dependent structural dynamics of integrin binding properties [22]. Treatment of macrophages with soluble mannose or β-glucans did not prevent or minimally reduced binding, suggesting that CR3 interaction is sufficient for yeast adhesion [19,20]. On the other hand, binding of yeasts to stimulated murine macrophages was only partially reduced by antibodies to complement receptors, suggesting the possible existence of additional phagocytic receptors on murine cells [19].
The H. capsulatum Hsp60 protein was identified as the ligand for CD11/CD18 integrin-type receptors by purification of H. capsulatum proteins binding to CR3 [21]. Although Hsp60 is a canonical intracellular chaperone protein, it has also been found localized to the surface of H. capsulatum yeast cells; antibodies to H. capsulatum Hsp60 detect Hsp60 protein in cell-wall extracts and label non-permeabilized yeast cells [23], and vaccination with Hsp60 provides the host with a protective immune response [24]. In addition, recombinant Hsp60 may competitively block adhesion of yeasts to macrophages [21,25]. Thus, the Hsp60–complement receptor interaction appears to be a major ligand–receptor pairing mediating adhesion to and internalization of H. capsulatum yeasts by macrophages. How normally intracellular Hsp60 becomes localized to the cell surface and whether it serves additional non-adhesion functions is not understood at this time. Similar to H. capsulatum, the related dimorphic fungal pathogen Blastomyces dermatitidis expresses a CR3-interacting protein, Bad1. B. dermatitidis Bad1 may mediate adherence to macrophages, suggesting that CR3 may be a shared entry mechanism [26]. However, B. dermatitidis yeasts expressing Bad1, but lacking it on the yeast surface, retain virulence, indicating that Bad1-mediated adhesion of yeast to macrophages is not required for B. dermatitidis pathogenesis [27].
Host pattern recognition receptors
The close interaction of H. capsulatum yeasts with the macrophage surface to promote internalization risks engagement of pattern recognition receptors that may stimulate immune responses detrimental to H. capsulatum yeasts. Phagocytosis mediated by CR3 in the absence of other activating receptor interactions is generally non-inflammatory as there is a lack of additional danger signals. H. capsulatum’s success in becoming intracellular thus hinges on minimization of activation of macrophage signaling receptors while simultaneously triggering phagocytic receptors. Signaling receptors that recognize fungal pathogen-associated molecular patterns (PAMPs) include the Toll-like receptors (TLRs), the macrophage mannose receptor and C-type lectin receptors [28]. Most of these receptors contain a carbohydrate recognition domain that is capable of binding to molecules of the glycan-and glycoprotein-rich cell walls that comprise the surface of fungal cells.
Dectin-1 is the primary receptor for detection of fungal β-glucans [29]. β-glucans serve as a central component of the cell wall for many fungi [30], and, when bound to Dectin-1, induce an inflammatory response that may include increased production of reactive oxygen species and release of pro-inflammatory cytokines [31–34]. The H. capsulatum cell wall contains abundant β -glucans that have the potential to activate Dectin-1 [35,36]. However, mice lacking Dectin-1 do not have increased susceptibility to H. capsulatum infection [37]. Dectin-1-deficient mice have slightly higher, but not statistically increased, pulmonary fungal burdens after H. capsulatum infection, although this was measured after 13 days, corresponding to the adaptive immunity phase [38]. Together, these studies indicate little, if any, role for Dectin-1 in control of H. capsulatum infection, suggesting that H. capsulatum yeast avoids interaction with Dectin-1. Molecularly, most strains of H. capsulatum accomplish this by surrounding the yeast cell with non-immunostimulatory α-linked glucans that effectively conceal the β-glucans and drastically reduce yeast recognition by Dectin-1 [39]. Cell wall α-glucan is synthesized by an α- (1,3)-glucan synthase (Ags1) that is thought to extend β-glucan assembly from short α- (1,4)-linked glucan oligomers formed by the Amy1 α-amylase-like protein [40]. Ags1 and Amy1 are both required for H. capsulatum α-glucan synthesis [41,42]. In contrast to the avirulent mycelia, only H. capsulatum yeast cells produce α-glucan [36], supporting a pathogenesis-specific function. Consistent with this, loss of α-glucan by depletion of Ags1 or Amy1 functions markedly attenuates H. capsulatum virulence in vivo [41,42]. Curiously, H. capsulatum isolates belonging to the North American type 2 phylogenetic group naturally lack α-glucan (chemotype II strains) [43], and show variable recognition by Dectin-1 in vitro, depending on yeast growth conditions [35,38]. Depletion of Ags1 function has no effect on the virulence of these strains [35]. The molecular mechanism by which North American type 2 strains have circumvented the need for α-glucan remains unknown. Regardless of the mechanism, one strategy used by H. capsulatum yeasts is minimization of β-glucan exposure, which enables stealthy interaction of yeasts with macrophages without triggering Dectin-1 signaling (Fig. 2). B. dermatitidis and Paracoccidioides species also contain α-glucan in the yeast phase [44,45], presumably for the same β-glucan-masking purpose as H. capsulatum; however, the role of α-glucan has not been validated in these other dimorphic fungal pathogens.
TLR2, TLR4, Dectin-2, Mincle and the macrophage mannose receptor are pattern recognition receptors for various fungal mannans that potentially recognize mannan-containing PAMPs on the H. capsulatum surface. Early studies of H. capsulatum binding to human macrophages showed no reduction in yeast binding to macrophages in the presence of excess mannose competitor, suggesting a lack of mannan-type PAMP–pattern recognition receptor interactions [20]. However, recent studies have shown that Dectin-2, but not Mincle, recognizes H. capsulatum yeast cell surfaces, and initiates signal transduction in a heterologous signaling system [38]. The functional consequences of Dectin-2 detection of H. capsulatum mannans with regard to macrophage responses remain to be determined, as well as the specific mannan-type PAMP recognized.
TLR2 and TLR4 represent two additional signaling receptors that may detect H. capsulatum yeasts. TLR signaling is mediated through the MyD88 adaptor protein [46]. Consistent with some recognition of yeasts by TLRs, MyD88-deficient mice show increased susceptibility to H. capsulatum infection and show reduced cytokine production, which correlates with a reduced adaptive immune response [37]. At the macrophage level, phagocytes that express TLR2, compared to TLR2-deficient cells, contain more lipid bodies, which correlates with increased leukotriene production [47], suggesting TLR2-based recognition of H. capsulatum yeasts occurs. No similar effect was observed in TLR4-deficient cells, indicating the lack of a role for TLR4 recognition of yeasts. A stronger role for TLR2 recognition was found when using cell-wall biochemical fractions [47]. These findings suggest that the cell wall contains PAMPs that are recognized by TLR2, but that the spatial organization of the cell wall in live yeast limits TLR2 recognition by macrophages. Blocking TLR2 or TLR4 does not impair phagocytosis of H. capsulatum yeasts [19]; however, yeasts were still able to bind to CR3 in these experiments to mediate uptake.
Cell-surface localized fungal molecules
As the cell wall is the primary surface that interacts with the macrophage, surface-localized factors of H. capsulatum yeasts are potential PAMPs that may be recognized or may influence the yeast–macrophage interaction. Cell wall-localized factors have often been investigated due to their role as immunologically dominant epitopes or vaccine substrates. These include the aforementioned Hsp60 protein, the extracellular catalase CatB/M-antigen [48], histone 2B [49] and Hsp70 [50] (Fig. 2). The yeast phase-specific protein Yps3, produced by some lineages of H. capsulatum [51–53], associates with the yeast cell surface via interactions with chitin [54]. Purified recombinant Yps3 was shown to bind to and activate TLR2 in microglial cells, leading to TLR2-dependent NF-κB stimulation and chemokine production [55]; however, no interaction of Yps3 in the normal context of a yeast cell was determined, and nor were the consequences of TLR2-dependent yeast interaction with macrophages. While pulmonary infection by H. capsulatum yeasts is not influenced by Yps3, H. capsulatum yeasts lacking Yps3 show reduced dissemination [56]. A speculative model based on these data suggests that Yps3-TLR2 interactions increase chemokine production, resulting in enhanced recruitment of phagocytes that serve as host cells for the yeast and vehicles for extra-pulmonary spread.
Intracellular challenges
Reactive oxygen species
The production of reactive oxygen species (ROS) is a central mechanism used by phagocytes to effect killing of fungal cells (Fig. 3). Phagocytic cells, particularly polymorphonuclear leukocytes and macrophages, produce reactive superoxide through assembly of the NADPH oxidase complex at the plasma or phagosomal membrane. Superoxide (O2−), and other reactive oxygen molecules derived from it [e.g. peroxide (H2O2) and hydroxyl radicals (•OH)] directly damage macromolecules, resulting in death of the microbe. Phagocyte-generated ROS differ from ROS generated by yeasts as by-products of aerobic respiration in that phagocyte-produced ROS are located extracellularly to the fungal cell. While most organisms possess various intracellular antioxidant enzymes to cope with metabolic-derived ROS, these enzymes are not positioned appropriately to deal with exogenous ROS, particularly superoxide, which does not cross membranes efficiently due to its charged nature. Thus, to survive, successful pathogens that interact with macrophages, and especially those that invade macrophages, must have mechanisms to avoid or rapidly neutralize any phagocyte-derived ROS. The ability to survive challenge with ROS-producing phagocytes specifically characterizes H. capsulatum yeast and not mycelia [11], consistent with the separation of the pathogenic and saprobic lifestyles of the two H. capsulatum forms.
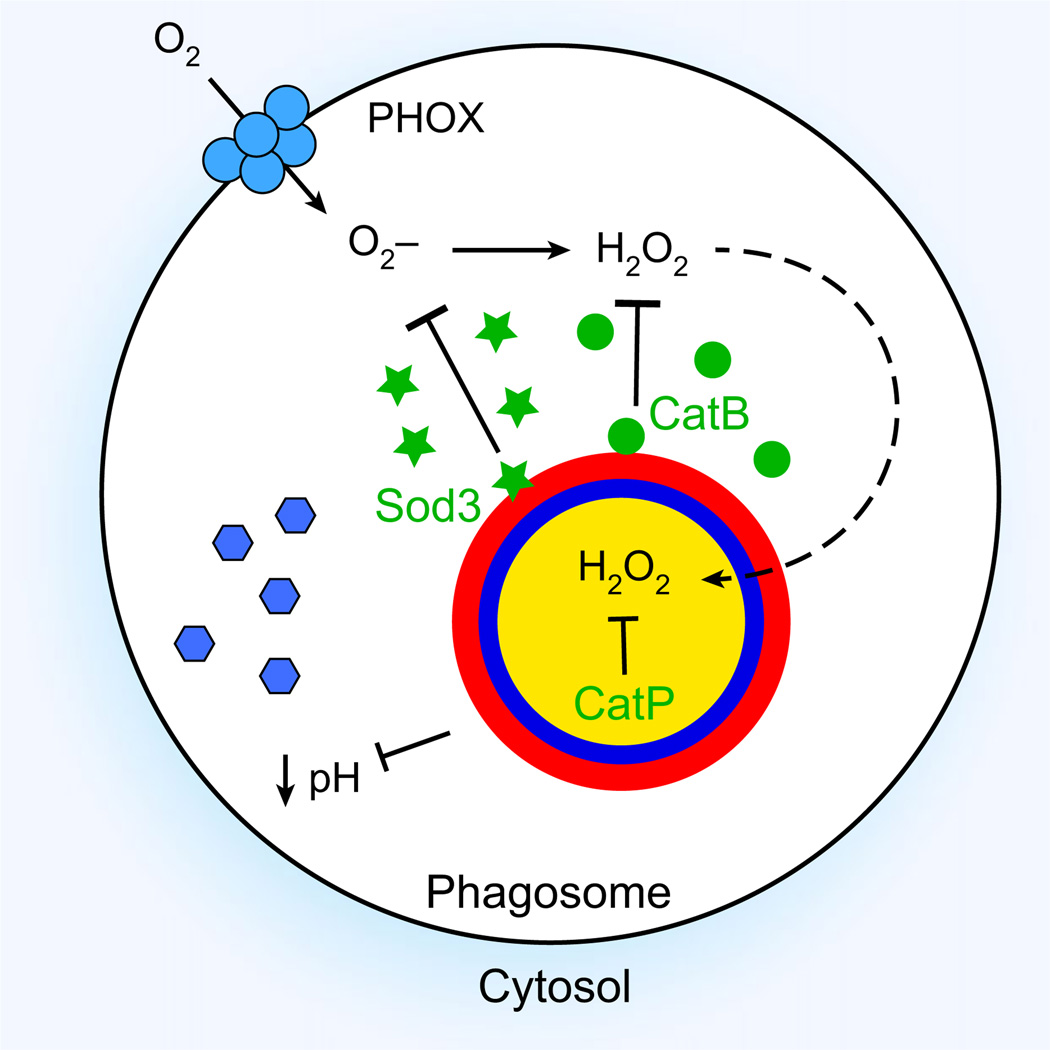
Fig. 3.
Histoplasma capsulatum mechanisms for survival in phagocytes. The schematic depicts a H. capsulatum yeast cell within a macrophage phagosome. The phagocyte NADPH oxidase complex (PHOX; blue membrane complex) produces superoxide (O2−) and peroxide (H2O2) within the phagosome. H. capsulatum eliminates these reactive oxygen molecules through production of extracellular superoxide dismutase Sod3 (green stars) and extracellular catalase CatB (green circles). If any H2O2 escapes destruction by CatB and enters the yeast cell (dashed arrow), further protection is provided by intracellular CatP. Acidification of the phagosome lumen presents an additional obstacle to intracellular survival by activation of acidic hydrolases (blue hexagons). H. capsulatum yeasts block acidification by an unknown mechanism, to maintain a near neutral pH.
Histoplasma capsulatum yeasts combat toxic ROS produced by phagocytes by expression of an efficient extracellular superoxide dismutase (Sod3) and catalase (CatB) (Fig. 3). Both H. capsulatum Sod3 and CatB are expressed specifically by H. capsulatum yeasts but not mycelia, indicating roles adapted to the pathogenic lifestyle of yeasts [57]. H. capsulatum Sod3 and CatB enzymes are secreted and associate with the yeast cell wall, positioning them to deal specifically with phagocyte-produced ROS but not yeast cytosolic or mitochondrial ROS [58,59]. Biochemically, extracellular Sod enzymes differ from intracellular Sod enzymes by functioning as monomers and lacking an electrostatic loop domain that guides superoxide to the catalytic metal co-factor [60]. As a consequence, the catalytic site is more accessible, resulting in rapid catalytic rates. H. capsulatum yeasts lacking Sod3 function are efficiently killed by phagocytes, and this killing is mediated by phagocyte-produced ROS [58]. H. capsulatum’s intracellular superoxide dismutase (Sod1) is unable to protect yeasts from exogenous superoxide or ROS-dependent killing by phagocytes, indicating spatial specificity of Sod3 and Sod1 for phagocyte-produced and intracellular ROS, respectively. Notably, Sod3 protects H. capsulatum yeasts from levels of superoxide produced by phagocytes that are sufficient to kill opportunistic fungal pathogens. In vivo, H. capsulatum yeasts lacking Sod3 function have attenuated virulence, and this attenuation is dependent on the ability of the host to produce superoxide, confirming the essential role of Sod3 in protecting H. capsulatum yeasts from phagocyte-derived ROS. Early studies of the yeast– macrophage interaction appeared to indicate that macrophages fail to produce ROS when stimulated with H. capsulatum yeasts [61–63], but no mechanism for how H. capsulatum may inhibit ROS generation was determined. However, the identification and analysis of Sod3 demonstrates that the lack of detectable oxidative burst in these studies results from highly efficient destruction of ROS by H. capsulatum yeasts [58].
Like Sod3, H. capsulatum’s CatB catalase is located extracellularly to the yeast cell in order to counter exogenous peroxide stress [59]. Loss of CatB reduced the ability of yeasts to eliminate extracellular peroxide, and slightly reduced survival against phagocytes [59]. Additional loss of the intracellular catalase (CatP) further reduced yeast virulence in vitro and in vivo [59]. The partial functional redundancy of CatP for CatB, despite its intracellular localization, probably stems from the ability of peroxide to pass through membranes due to lack of charge, unlike superoxide. Without the CatB/CatP defenses, H. capsulatum is killed by phagocyte-derived ROS. Together, these data indicate that H. capsulatum yeasts ensure their survival in macrophages through destruction of phagocyte-produced ROS by an extracellular ROS defense system consisting of Sod3 and CatB (Fig. 3).
Reactive nitrogen species
In addition to ROS production, activated phagocytes combat fungi by production of reactive nitrogen species (RNS). These species include nitric oxide (NO•), peroxynitrite (ONOO−) and nitrogen dioxide (•NO2). Inhibition of RNS production by phagocytes decreases their ability to control H. capsulatum [64,65], suggesting that H. capsulatum yeasts are susceptible to RNS. However, RNS is only fungistatic to H. capsulatum [65], indicating that H. capsulatum yeasts may contend with the lethal, but not the inhibitory, effects of RNS. The mechanism that underlies this ability is unknown, but transcriptional analysis of RNS-treated yeasts identified 59 genes that are up-regulated in response to nitrosative stress [66]. Cytokine activation of phagocytes stimulates RNS production, and reduced pro-inflammatory cytokine levels are correlated with increased susceptibility of mice to H. capsulatum infection [64,65,67]. These data suggest that RNS control H. capsulatum yeast, but that RNS-based control functions during the adaptive immunity stage of infection rather than the initial infection of phagocytes.
Phagosome acidification
Acidification of the phagosome/phagolysosome contributes to killing of microorganisms by activation of acidic lysosomal hydrolases. Conflicting reports exist regarding whether phagosome–lysosome fusion occurs in the H. capsulatum-infected macrophage, depending on the cell type and assay used [68–71]. Regardless, the lumen of the H. capsulatum-containing compartment does not become highly acidified. Live H. capsulatum yeasts, but not heat-killed yeasts, maintain a relatively neutral pH (~ pH 6.5) in the phagosome/ phagolysosome [69,71,72]. Maintenance of this more neutral phagosomal pH is essential for H. capsulatum infection of macrophages, as demonstrated by lack of survival of H. capsulatum yeast when the intracellular compartment is acidified [73]. A mutation in the 3-hydroxy-3-methyl-glutaryl CoA lyase gene (HCL1) results in acidification of the surrounding growth environment due to accumulation and secretion of acidic intermediates of leucine catabolism. This acidification does not impair in vitro growth, but severely compromises survival in macrophages, which is correlated with an acidified phagosomal pH [73]. In this study, phagosomal acidification was not dependent on the vacuolar ATPase, indicating that the mutant itself, not the macrophage, artificially acidified the compartment. While these data do not indicate that Hcl1 is part of H. capsulatum’s mechanism for neutralizing the phagosomal pH, they do provide genetic evidence that H. capsulatum must prevent acidification in order to survive in macrophages. The mechanisms behind this feature of H. capsulatum intracellular pathogenesis remain unknown.
Nutrient limitation
Intracellular pathogens must not only survive the defenses of the macrophage but also acquire resources for proliferation, and intracellular residence imposes nutritional challenges on H. capsulatum. In contrast to growth in rich media in vitro, the phagosomal compartment is generally considered to be nutrient-poor [74–77]. This is supported by studies of the intracellular lifestyle of bacteria and fungi, which show major transcriptional changes of nutritional genes in organisms within the phagosome compared to in culture [78–81]. In addition, removal of genes involved in nutrient synthesis or utilization may affect virulence without affecting in vitro growth of pathogenic bacteria and fungi [82–86]. To proliferate in macrophages, H. capsulatum yeast must catabolize whatever carbon, nitrogen and sulfur sources are available, as well as assimilating trace elements essential for life processes. Where nutrient gaps exist in the intracellular compartment, intracellular pathogens must possess sufficient biosynthetic capabilities to satisfy metabolic demands. Natural auxotrophies and derived auxotrophic H. capsulatum strains are enabling determination of which nutrient sources are and are not available, and which may be utilized in the intracellular compartment for yeast proliferation.
Limitation of essential metals
Biologically available iron is rare in the environment, and, within the macrophage, phagosome acquisition of iron is an even greater challenge. H. capsulatum yeasts have multiple strategies to acquire iron within host cells. Although H. capsulatum yeasts prevent acidification of the phagosomal compartment, the pH must still be lower than pH 7 in order to liberate iron from transferrin [87]. Thus, H. capsulatum must balance the need to avoid activation of acidic hydrolases of the macrophage with the need to acidify the compartment sufficiently to obtain iron. This balance is achieved by maintaining the pH at ~ 6.0–6.5, which is acidic enough to release one of the two iron atoms from transferrin but not sufficiently acidic for hydrolase activation. Consistent with this, chloroquine, which raises the phagosomal pH, decreases H. capsulatum survival in macrophages by reducing available intracellular iron [87].
A second method by which H. capsulatum acquires limited iron intracellularly is by secretion of iron-chelating siderophores (Fig. 4). H. capsulatum yeasts produce hydroxamate siderophores that scavenge iron from transferrin [88]. Iron limitation induces both transcriptional- and protein-level changes in H. capsulatum yeasts [89,90], including Sid1, a key enzyme in siderophore biosynthesis. Transcriptional changes are mediated by the iron-responsive GATA family transcription factor Sre1 [89]. Mutants of H. capsulatum that are unable to make siderophores have reduced proliferation in cultured macrophages [89,91]. In vivo, loss of siderophore production in North American type 2 strains drastically reduces pulmonary infection [91] at early stages. In contrast, a Panama strain that is deficient for siderophore synthesis did not show attenuation until day 15 after infection, a time point that coincides with adaptive immunity [89]. This discrepancy suggests that Panama strains have alternative iron acquisition strategies that operate in addition to siderophores.
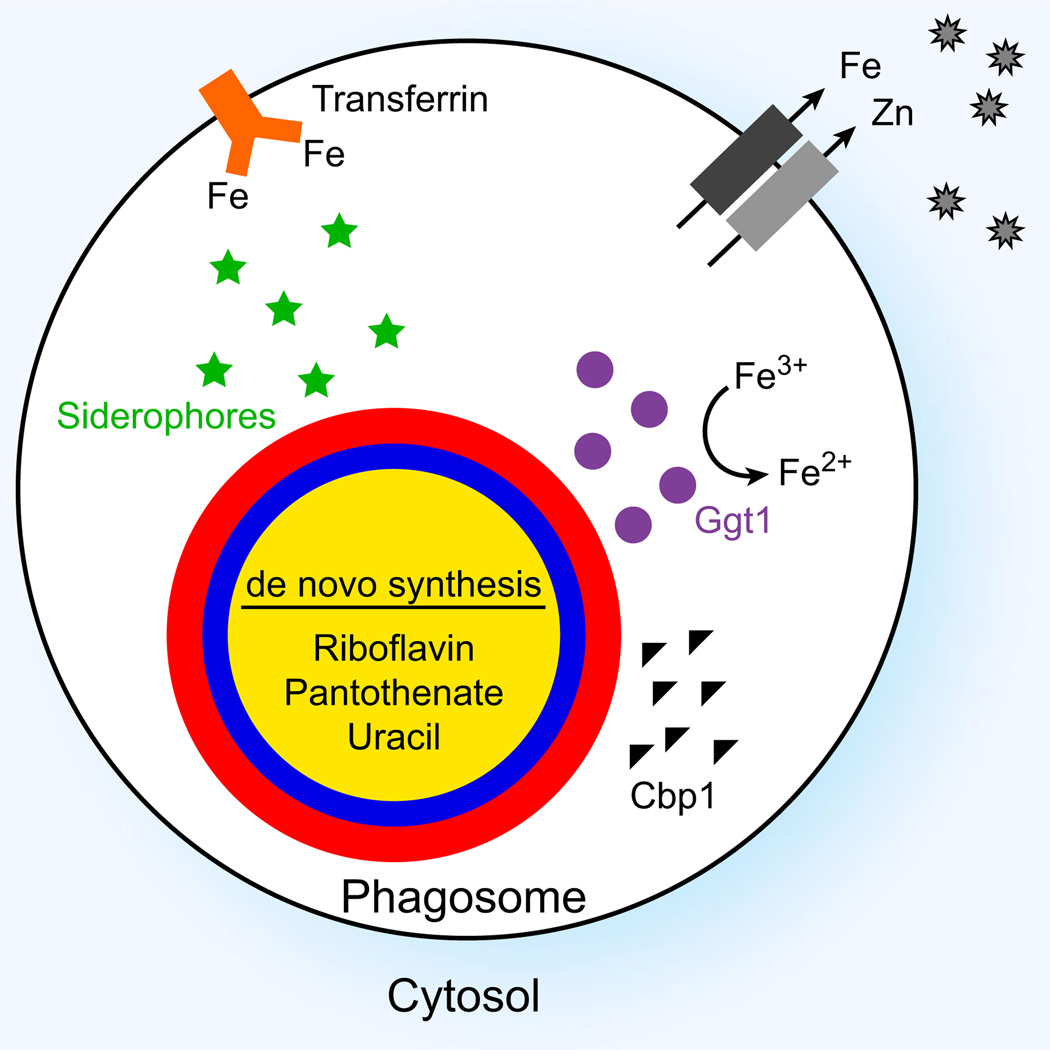
Fig. 4.
Histoplasma capsulatum mechanisms to cope with nutritional limitations and other obstacles to intracellular proliferation. H. capsulatum may acquire iron from transferrin (orange). Phagocytes restrict the availability of essential metals such as iron and zinc through membrane transporters (gray) and sequestering metallothioneins (gray star). H. capsulatum yeasts produce siderophores (green stars) with high affinity for iron to acquire iron within the phagosome. In addition, yeast cells produce γ-glutamyl transferase (Ggt1; purple circles) to reduce iron within the phagosome. To overcome nutrient limitations, H. capsulatum yeasts produce compounds such as uracil and the essential vitamins riboflavin and pantothenate using de novo synthesis pathways. The calcium-binding protein Cbp1 (black triangles) is highly expressed by yeasts, which facilitates intracellular pathogenesis through an unknown mechanism.
Histoplasma capsulatum yeasts utilize ferrous iron, and thus express multiple iron-reducing systems and iron transporters. Biochemical studies of extracellular iron reduction demonstrated that H. capsulatum yeasts produce three reductases, two soluble and one cell-associated [92,93]. Follow-up work identified one of these as a γ-glutamyl transferase (Fig. 4), which catalyzes a two-step reduction of ferric iron using reduced glutathione [94]. H. capsulatum yeasts lacking γ-glutamyl transferase function are impaired in the ability to proliferate within cultured macrophages, confirming acquisition of iron is a challenge faced by H. capsulatum. In addition, some strains (e.g. those of the Panama lineage) have genes encoding a high-affinity iron transport system (Fet3 and Ftr1) [91]. Paracoccidioides species and B. dermatitidis also produce hydroxymate siderophores and ferric reductases similar to those of H. capsulatum [95–97], suggesting common strategies for iron acquisition and utilization. However, the necessity for siderophores for fungal virulence in these organisms has not yet been established.
Intracellular nutrient availability is dynamic, as host responses to infection may cause limitation of certain nutrients, a concept termed ‘nutritional immunity’ [98]. Cytokine activation of macrophages may cause a further reduction in the free iron available to H. capsulatum yeasts in the phagosome, which provides one explanation of why Panama strains lacking siderophores are not impaired until adaptive immunity activates the macrophages [89]. Host cells actively remove iron when activated with interferon γ, which stimulates the sequestration of iron in an attempt to inhibit growth of the pathogen [99,100]. Zinc is another element that is central to the function of many metalloenzymes, and zinc availability is modulated by cytokine activation of macrophages. Treatment of H. capsulatum-infected macrophages with granulocyte-macrophage colony-stimulating factor decreased cellular levels of available iron and zinc [101]. This is correlated with the ability of granulocyte-macrophage colony-stimulating factor-treated macrophages to inhibit H. capsulatum growth and replication and to increase macrophage ROS production and phagosomal acidification [102]. In non-activated macrophages, H. capsulatum may effectively battle the host for iron and zinc, but activation of macrophages may restrict the availability of elements necessary for intracellular yeast proliferation (Fig. 4).
Pyrimidine limitation
The H. capsulatum-containing phagosomal compartment lacks available nucleic acids for H. capsulatum utilization. This was demonstrated by the isolation and characterization of a H. capsulatum uracil auxotroph. Mutants lacking Ura5 function grow in vitro as long as the medium is supplemented with uracil, indicating that H. capsulatum yeasts possess pyrimidine transport functions [103]. However, the uracil auxotroph is unable to grow in macrophages in culture, and lacks the ability to proliferate in vivo [104]. These results indicate that H. capsulatum’s pyrimidine biosynthetic pathway is essential for overcoming the lack of available uracil in the macrophage phagosome.
Scarcity of essential vitamins
To proliferate intracellularly, H. capsulatum yeasts use de novo biosynthesis of essential vitamins to overcome their absence in the phagosome. Loss of function of Rib2 (diaminohydroxyphosphoribosylaminopyrimidine deaminase) creates a riboflavin auxotrophy that may be rescued in vitro by supplementation with riboflavin [105]. However, there is essentially no replication of the rib2 mutant in cultured macrophages, and the mutant is severely attenuated in vivo. The intracellular yeasts are not killed during initial infection but are unable to proliferate intracellularly. Similar infection-related yeast growth inhibition results if H. capsulatum pantothenate biosynthesis is blocked, but not when biotin synthesis is impaired [105]. In vitro, wild-type H. capsulatum yeasts are able to synthesize all essential vitamins except for thiamine [105], which enables them to overcome the lack of riboflavin and pantothenate in the phagosome (Fig. 4). The growth of the biotin auxotroph in macrophages suggests that the phagosome contains sufficient biotin or that this co-factor is efficiently scavenged by intracellular yeasts.
Organic sulfur
Interestingly, H. capsulatum yeasts, but not mycelia, are auxotrophic for cysteine. This has been attributed to temperature-dependent lack of sulfite reductase expression, thereby preventing yeasts from incorporating inorganic sulfate into cysteine [106–109]. Despite the need for organic sulfur for yeast growth in vitro, wild-type H. capsulatum yeasts are able to proliferate within macrophages and establish infection in vivo, suggesting that the intracellular environment has ample organic sulfur to support H. capsulatum growth [110]. In addition, an undefined cysteine auxotroph strain derived by mutagenesis is fully virulent in vivo [111,112]. These results suggest that, while pyrimidines and essential vitamins are limiting in the macrophage, necessitating de novo synthesis by H. capsulatum yeasts, cysteine or an organic sulfur source is readily available to intracellular H. capsulatum yeasts.
Paracoccidioides brasiliensis shows a similar inorganic sulfur auxotrophy in the yeast phase, as organic sulfur is required for the transition from mycelia to yeasts [113–116] and many sulfur metabolism genes are differentially regulated between the yeast and mycelial phases [116–118]. The similar yeast requirement for organic sulfur in H. capsulatum and P. brasiliensis indicates that sufficient organic sulfur is available intracellularly [119]. Up-regulating the inorganic sulfur assimilatory pathway (by knockdown of the negative regulator Sulfur control protein) in P. brasiliensis rescues the dependence on organic sulfur for mycelial to yeast transition; however, the resulting ATP and redox imbalance impairs yeast growth [115]. These data for H. capsulatum and P. brasiliensis suggest that yeasts, but not mycelia, have streamlined expression of sulfur metabolism genes to exploit available organic sulfur in the phagosome. Interestingly, Legionella pneumophila, a bacterial pathogen that resides within the phagosome, is also naturally auxotrophic for cysteine [120]. It remains unknown what the actual organic sulfur compound(s) is (are) that are utilized by intracellular pathogens.
Undefined obstacles to intracellular proliferation
The first virulence factor identified for H. capsulatum was the small secreted calcium-binding protein Cbp1 [121]. Cbp1 has nanomolar affinity for calcium [122,123], but it remains unclear what role calcium may play in Cbp1 function or whether Cbp1 contributes to calcium homeostasis of intracellular yeasts. Cbp1 is abundantly produced specifically by H. capsulatum yeasts (not by mycelia), consistent with its function in H. capsulatum pathogenesis, and the protein is secreted into the phagosomal lumen [124]. The compact structure of Cbp1, created by intramolecular disulfide bonds, makes Cbp1 resistant to proteases that may be encountered in a phagolysosome. Despite the molecular and structural information on Cbp1, how this secreted protein contributes to H. capsulatum pathogenesis remains unknown, and consequently the obstacles that Cbp1 enables intracellular H. capsulatum yeast to overcome are undefined.
Conclusions
Histoplasma capsulatum is one of only a few microbial pathogens with a nearly exclusive intracellular lifestyle. Even though many human pathogenic fungi can exist within macrophages, they often have strategies for escape from the host cell [125–127]. In contrast, H. capsulatum tends to remain inside the macrophage for an extended time, proliferating to high numbers intracellularly. While opportunistic fungal pathogens are readily controlled by cells of the innate immune system, H. capsulatum, as a primary pathogen, survives and replicates within host phagocytes.
Histoplasma capsulatum’s success in establishing a permissive intracellular environment depends on multiple mechanisms to exploit the macrophage as its host cell. Two major obstacles facing intracellular pathogens are how to enter host cells without being recognized as a microbial invader and how to survive the onslaught of antimicrobial defense molecules (e.g. ROS and phagosome acidification). For the first task, H. capsulatum promotes its uptake by macrophages by interacting with phagocytic receptors while simultaneously minimizing recognition by signaling receptors. It achieves this by masking immunostimulatory molecules. H. capsulatum yeasts also effectively resist killing by macrophages by production of secreted defenses against fungicidal ROS and modulation of the phagosomal environment to prevent activation of lysosomal hydrolases. Production of external factors to specifically eliminate phagocyte-derived ROS is a common characteristic of pathogens that are able to infect and survive within phagocytes [128–132]. Thus, H. capsulatum accomplishes both stealth and survival, thereby preventing elimination and control by its host cell.
Successful intracellular pathogens need to do more than simply survive entry into phagocytes. Once established as an intracellular resident, H. capsulatum yeasts successfully acquire the nutrients necessary for proliferation. At least three mechanisms operate to capture limited iron, highlighting the challenge of nutritional immunity facing intracellular yeasts. Nevertheless, acquisition of essential metals may be H. capsulatum’s Achilles heel, as indicated by improved phagocyte control of H. capsulatum yeasts when macrophages are activated and metal sequestration is enhanced [87,99,102,133]. Although the primary carbon, nitrogen and sulfur sources utilized by H. capsulatum yeasts remain undefined, H. capsulatum yeasts have sufficient de novo synthesis pathways for biological molecules to provide for growth and replication, including biosynthesis of pyrimidines and essential vitamins. On the other hand, some nutrients, such as organic sulfur, appear to be sufficiently available in the phagosome to allow the proliferation of yeasts that are dependent on organic sulfur. These capabilities enable H. capsulatum to convert a challenging nutrient-poor phagosome into a permissive intracellular niche. Thus, a combination of survival strategies and nutrient acquisition mechanisms facilitate H. capsulatum’s success as an intracellular pathogen of macrophages.
Abbreviations
- Ags1
α-(1,3)-glucan synthase
- Amy1
α-amylase-like protein
- Cat
catalase
- Cbp1
calcium-binding protein
- PAMP
pathogen-associated molecular pattern
- ROS
reactive oxygen species
- Sod
superoxide dismutase
- SP
surfactant protein
- TLR
Toll-like receptor
Footnotes
Author Contributions
ALG and CAR analyzed the literature and co-wrote the review.
References
- 1.Ajello L. The medical mycological iceberg. HSMHA Health Rep. 1971;86:437–448. [PMC free article] [PubMed] [Google Scholar]
- 2.Rippon JW. Dimorphism in pathogenic fungi. Crit Rev Microbiol. 1980;8:49–97. doi: 10.3109/10408418009085078. [DOI] [PubMed] [Google Scholar]
- 3.Kauffman CA. Histoplasmosis: a clinical and laboratory update. Clin Microbiol Rev. 2007;20:115–132. doi: 10.1128/CMR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci USA. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster RH, Sil A. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc Natl Acad Sci USA. 2008;105:14573–14578. doi: 10.1073/pnas.0806221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemecek JC, Wüthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 7.Medoff G, Sacco M, Maresca B, Schlessinger D, Painter A, Kobayashi GS, Carratu L. Irreversible block of the mycelial-to-yeast phase transition of Histoplasma capsulatum. Science. 1986;231:476–479. doi: 10.1126/science.3001938. [DOI] [PubMed] [Google Scholar]
- 8.Walsh TJ, Dixon DM. Spectrum of mycoses. In: Baron S, editor. In Medical Microbiology. 4th. Galveston, TX: University of Texas Medical Branch at Galveston; 1996. [PubMed] [Google Scholar]
- 9.De Sanchez SB, Carbonell LM. Immunological studies on Histoplasma capsulatum. Infect Immun. 1975;11:387–394. doi: 10.1128/iai.11.2.387-394.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gildea LA, Morris RE, Newman SL. Histoplasma capsulatum yeasts are phagocytosed via very late antigen-5, killed, and processed for antigen presentation by human dendritic cells. J Immunol. 2001;166:1049–1056. doi: 10.4049/jimmunol.166.2.1049. [DOI] [PubMed] [Google Scholar]
- 11.Schaffner A, Davis CE, Schaffner T, Markert M, Douglas H, Braude AI. In vitro susceptibility of fungi to killing by neutrophil granulocytes discriminates between primary pathogenicity and opportunism. J Clin Invest. 1986;78:511–524. doi: 10.1172/JCI112603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wüthrich M, Filutowicz HI, Warner T, Deepe GS, Klein BS. Vaccine immunity to pathogenic fungi overcomes the requirement for CD4 help in exogenous antigen presentation to CD8+ T cells: implications for vaccine development in immune-deficient hosts. J Exp Med. 2003;197:1405–1416. doi: 10.1084/jem.20030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deepe GS. Modulation of infection with Histoplasma capsulatum by inhibition of tumor necrosis factor-α activity. Clin Infect Dis. 2005;41(Suppl. 3):S204–S207. doi: 10.1086/429999. [DOI] [PubMed] [Google Scholar]
- 14.Deepe GS. Role of CD8+ T cells in host resistance to systemic infection with Histoplasma capsulatum in mice. J Immunol. 1994;152:3491–3500. [PubMed] [Google Scholar]
- 15.Zhou P, Freidag BL, Caldwell CC, Seder RA. Perforin is required for primary immunity to Histoplasma capsulatum. J Immunol. 2001;166:1968–1974. doi: 10.4049/jimmunol.166.3.1968. [DOI] [PubMed] [Google Scholar]
- 16.Brummer E, Stevens DA. Collectins and fungal pathogens: roles of surfactant proteins and mannose binding lectin in host resistance. Med Mycol. 2010;48:16–28. doi: 10.3109/13693780903117473. [DOI] [PubMed] [Google Scholar]
- 17.McCormack FX, Gibbons R, Ward SR, Kuzmenko A, Wu H, Deepe GS. Macrophage-independent fungicidal action of the pulmonary collectins. J Biol Chem. 2003;278:36250–36256. doi: 10.1074/jbc.M303086200. [DOI] [PubMed] [Google Scholar]
- 18.Newman SL, Bucher C, Rhodes J, Bullock WE. Phagocytosis of Histoplasma capsulatum yeasts and microconidia by human cultured macrophages and alveolar macrophages. Cellular cytoskeleton requirement for attachment and ingestion. J Clin Invest. 1990;85:223–230. doi: 10.1172/JCI114416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J-S, Huang J-H, Hung L-Y, Wu S-Y, Wu-Hsieh BA. Distinct roles of complement receptor 3, Dectin-1, and sialic acids in murine macrophage interaction with Histoplasma yeast. J Leukoc Biol. 2010;88:95–106. doi: 10.1189/jlb.1109717. [DOI] [PubMed] [Google Scholar]
- 20.Bullock WE, Wright SD. Role of the adherence-promoting receptors, CR3, LFA-1, and p150,95, in binding of Histoplasma capsulatum by human macrophages. J Exp Med. 1987;165:195–210. doi: 10.1084/jem.165.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long KH, Gomez FJ, Morris RE, Newman SL. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J Immunol. 2003;170:487–494. doi: 10.4049/jimmunol.170.1.487. [DOI] [PubMed] [Google Scholar]
- 22.Springer TA, Wang J-H. The three-dimensional structure of integrins and their ligands, and conformational regulation of cell adhesion. Adv Protein Chem. 2004;68:29–63. doi: 10.1016/S0065-3233(04)68002-8. [DOI] [PubMed] [Google Scholar]
- 23.Guimarães AJ, Frases S, Gomez FJ, Zancopé -Oliveira RM, Nosanchuk JD. Monoclonal antibodies to heat shock protein 60 alter the pathogenesis of Histoplasma capsulatum. Infect Immun. 2009;77:1357–1367. doi: 10.1128/IAI.01443-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez FJ, Allendoerfer R, Deepe GS. Vaccination with recombinant heat shock protein 60 from Histoplasma capsulatum protects mice against pulmonary histoplasmosis. Infect Immun. 1995;63:2587–2595. doi: 10.1128/iai.63.7.2587-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habich C, Kempe K, Gomez FJ, Lillicrap M, Gaston H, van der Zee R, Kolb H, Burkart V. Heat shock protein 60: identification of specific epitopes for binding to primary macrophages. FEBS Lett. 2006;580:115–120. doi: 10.1016/j.febslet.2005.11.060. [DOI] [PubMed] [Google Scholar]
- 26.Brandhorst TT, Wüthrich M, Finkel-Jimenez B, Warner T, Klein BS. Exploiting type 3 complement receptor for TNF-α suppression, immune evasion, and progressive pulmonary fungal infection. J Immunol. 2004;173:7444–7453. doi: 10.4049/jimmunol.173.12.7444. [DOI] [PubMed] [Google Scholar]
- 27.Brandhorst T, Wüthrich M, Finkel-Jimenez B, Klein B. A C-terminal EGF-like domain governs BAD1 localization to the yeast surface and fungal adherence to phagocytes, but is dispensable in immune modulation and pathogenicity of Blastomyces dermatitidis. Mol Microbiol. 2003;48:53–65. doi: 10.1046/j.1365-2958.2003.03415.x. [DOI] [PubMed] [Google Scholar]
- 28.Willment JA, Brown GD. C-type lectin receptors in antifungal immunity. Trends Microbiol. 2008;16:27–32. doi: 10.1016/j.tim.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Brown GD, Gordon S. Immune recognition: a new receptor for β-glucans. Nature. 2001;413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 30.Latgé J-P. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol. 2007;66:279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- 31.Underhill DM, Rossnagle E, Lowell CA, Simmons RM. Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood. 2005;106:2543–2550. doi: 10.1182/blood-2005-03-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheeler RT, Fink GR. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2006;2:e35. doi: 10.1371/journal.ppat.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by Dectin-1 and Toll-like receptor 2. J Exp Med. 2003;197:1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodridge HS, Shimada T, Wolf AJ, Hsu Y-MS, Becker CA, Lin X, Underhill DM. Differential use of CARD9 by Dectin-1 in macrophages and dendritic cells. J Immunol. 2009;182:1146–1154. doi: 10.4049/jimmunol.182.2.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards JA, Alore EA, Rappleye CA. The yeast-phase virulence requirement for α-glucan synthase differs among Histoplasma capsulatum chemotypes. Eukaryot Cell. 2011;10:87–97. doi: 10.1128/EC.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanetsuna F, Carbonell LM, Gil F, Azuma I. Chemical and ultrastructural studies on the cell walls of the yeast-like and mycelial forms of Histoplasma capsulatum. Mycopathol Mycol Appl. 1974;54:1–13. doi: 10.1007/BF02055967. [DOI] [PubMed] [Google Scholar]
- 37.Coady A, Sil A. MyD88-dependent signaling drives host survival and early cytokine production during Histoplasma infection. Infect Immun. 2015;83:1265–1275. doi: 10.1128/IAI.02619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, LeBert V, Hung CY, Galles K, Saijo S, Lin X, Cole GT, Klein BS, Wüthrich M. C-type lectin receptors differentially induce Th17 cells and vaccine immunity to the endemic mycosis of North America. J Immunol. 2014;192:1107–1119. doi: 10.4049/jimmunol.1302314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum α-(1,3) -glucan blocks innate immune recognition by the β-glucan receptor. Proc Natl Acad Sci. 2007;104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grün CH, Hochstenbach F, Humbel BM, Verkleij AJ, Sietsma JH, Klis FM, Kamerling JP, Vliegenthart JFG. The structure of cell wall α-glucan from fission yeast. Glycobiology. 2005;15:245–257. doi: 10.1093/glycob/cwi002. [DOI] [PubMed] [Google Scholar]
- 41.Marion CL, Rappleye CA, Engle JT, Goldman WE. An α-(1,4)-amylase is essential for α-(1,3) -glucan production and virulence in Histoplasma capsulatum. Mol Microbiol. 2006;62:970–983. doi: 10.1111/j.1365-2958.2006.05436.x. [DOI] [PubMed] [Google Scholar]
- 42.Rappleye CA, Engle JT, Goldman WE. RNA interference in Histoplasma capsulatum demonstrates a role for α-(1,3) -glucan in virulence. Mol Microbiol. 2004;53:153–165. doi: 10.1111/j.1365-2958.2004.04131.x. [DOI] [PubMed] [Google Scholar]
- 43.Domer JE. Monosaccharide and chitin content of cell walls of Histoplasma capsulatum and Blastomyces dermatitidis. J Bacteriol. 1971;107:870–877. doi: 10.1128/jb.107.3.870-877.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanetsuna F, Carbonell LM. Cell wall composition of the yeast-like and mycelial forms of Blastomyces dermatitidis. J Bacteriol. 1971;106:946–948. doi: 10.1128/jb.106.3.946-948.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanetsuna F, Carbonell LM, Azuma I, Yamamura Y. Biochemical studies on the thermal dimorphism of Paracoccidioides brasiliensis. J Bacteriol. 1972;110:208–218. doi: 10.1128/jb.110.1.208-218.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA., Jr MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 47.Sorgi CA, Secatto A, Fontanari C, Turato WM, Belangér C, de Medeiros AI, Kashima S, Marleau S, Covas DT, Bozza PT, et al. Histoplasma capsulatum cell wall β-glucan induces lipid body formation through CD18, TLR2, and Dectin-1 receptors: correlation with leukotriene B4 generation and role in HIV-1 infection. J Immunol. 2009;182:4025–4035. doi: 10.4049/jimmunol.0801795. [DOI] [PubMed] [Google Scholar]
- 48.Guimarães AJ, Hamilton AJ, de M, Guedes HL, Nosanchuk JD, Zancopé-Oliveira RM. Biological function and molecular mapping of M antigen in yeast phase of Histoplasma capsulatum. PLoS One. 2008;3:e3449. doi: 10.1371/journal.pone.0003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nosanchuk JD, Steenbergen JN, Shi L, Deepe GS, Casadevall A. Antibodies to a cell surface histone-like protein protect against Histoplasma capsulatum. J Clin Invest. 2003;112:1164–1175. doi: 10.1172/JCI19361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomez FJ, Gomez AM, Deepe GS. An 80-kilodalton antigen from Histoplasma capsulatum that has homology to heat shock protein 70 induces cell-mediated immune responses and protection in mice. Infect Immun. 1992;60:2565–2571. doi: 10.1128/iai.60.7.2565-2571.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keath EJ, Painter AA, Kobayashi GS, Medoff G. Variable expression of a yeast-phase-specific gene in Histoplasma capsulatum strains differing in thermotolerance and virulence. Infect Immun. 1989;57:1384–1390. doi: 10.1128/iai.57.5.1384-1390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keath EJ, Abidi FE. Molecular cloning and sequence analysis of yps-3, a yeast-phase-specific gene in the dimorphic fungal pathogen Histoplasma capsulatum. Microbiology. 1994;140:759–767. doi: 10.1099/00221287-140-4-759. [DOI] [PubMed] [Google Scholar]
- 53.Bohse ML, Woods JP. Expression and interstrain variability of the YPS3 gene of Histoplasma capsulatum. Eukaryot Cell. 2007;6:609–615. doi: 10.1128/EC.00010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bohse ML, Woods JP. Surface localization of the Yps3p protein of Histoplasma capsulatum. Eukaryot Cell. 2005;4:685–693. doi: 10.1128/EC.4.4.685-693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aravalli RN, Hu S, Woods JP, Lokensgard JR. Histoplasma capsulatum yeast phase-specific protein Yps3p induces Toll-like receptor 2 signaling. J Neuroinflammation. 2008;5:30. doi: 10.1186/1742-2094-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bohse ML, Woods JP. RNA interference-mediated silencing of the YPS3 gene of Histoplasma capsulatum reveals virulence defects. Infect Immun. 2007;75:2811–2817. doi: 10.1128/IAI.00304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holbrook ED, Edwards JA, Youseff BH, Rappleye CA. Definition of the extracellular proteome of pathogenic-phase Histoplasma capsulatum. J Proteome Res. 2011;10:1929–1943. doi: 10.1021/pr1011697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Youseff BH, Holbrook ED, Smolnycki KA, Rappleye CA. Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 2012;8:e1002713. doi: 10.1371/journal.ppat.1002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holbrook ED, Smolnycki KA, Youseff BH, Rappleye CA. Redundant catalases detoxify phagocyte reactive oxygen and facilitate Histoplasma pathogenesis. Infect Immun. 2013;81:2334–2346. doi: 10.1128/IAI.00173-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gleason JE, Galaleldeen A, Peterson RL, Taylor AB, Holloway SP, Waninger-Saroni J, Cormack BP, Cabelli DE, Hart PJ, Culotta VC. Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc Natl Acad Sci USA. 2014;111:5866–5871. doi: 10.1073/pnas.1400137111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fleischmann J, Wu-Hsieh B, Howard DH. The intracellular fate of Histoplasma capsulatum in human macrophages is unaffected by recombinant human interferon-gamma. J Infect Dis. 1990;161:143–145. doi: 10.1093/infdis/161.1.143. [DOI] [PubMed] [Google Scholar]
- 62.Wolf JE, Kerchberger V, Kobayashi GS, Little JR. Modulation of the macrophage oxidative burst by Histoplasma capsulatum. J Immunol. 1987;138:582–586. [PubMed] [Google Scholar]
- 63.Eissenberg LG, Goldman WE. Histoplasma capsulatum fails to trigger release of superoxide from macrophages. Infect Immun. 1987;55:29–34. doi: 10.1128/iai.55.1.29-34.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lane TE, Wu-Hsieh BA, Howard DH. Anti-Histoplasma effect of activated mouse splenic macrophages involves production of reactive nitrogen intermediates. Infect Immun. 1994;62:1940–1945. doi: 10.1128/iai.62.5.1940-1945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakamura LT, Wu-Hsieh BA, Howard DH. Recombinant murine γ-interferon stimulates macrophages of the RAW cell line to inhibit intracellular growth of Histoplasma capsulatum. Infect Immun. 1994;62:680–684. doi: 10.1128/iai.62.2.680-684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nittler MP, Hocking-Murray D, Foo CK, Sil A. Identification of Histoplasma capsulatum transcripts induced in response to reactive nitrogen species. Mol Biol Cell. 2005;16:4792–4813. doi: 10.1091/mbc.E05-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allendoerfer R, Deepe GS. Intrapulmonary response to Histoplasma capsulatum in γ-interferon knockout mice. Infect Immun. 1997;65:2564–2569. doi: 10.1128/iai.65.7.2564-2569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eissenberg LG, Schlesinger PH, Goldman WE. Phagosome-lysosome fusion in P388D1 macrophages infected with Histoplasma capsulatum. J Leukoc Biol. 1988;43:483–491. doi: 10.1002/jlb.43.6.483. [DOI] [PubMed] [Google Scholar]
- 69.Eissenberg LG, Goldman WE, Schlesinger PH. Histoplasma capsulatum modulates the acidification of phagolysosomes. J Exp Med. 1993;177:1605–1611. doi: 10.1084/jem.177.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newman SL, Gootee L, Kidd C, Ciraolo GM, Morris R. Activation of human macrophage fungistatic activity against Histoplasma capsulatum upon adherence to type 1 collagen matrices. J Immunol. 1997;158:1779–1786. [PubMed] [Google Scholar]
- 71.Strasser JE, Newman SL, Ciraolo GM, Morris RE, Howell ML, Dean GE. Regulation of the macrophage vacuolar ATPase and phagosome–lysosome fusion by Histoplasma capsulatum. J Immunol. 1999;162:6148–6154. [PubMed] [Google Scholar]
- 72.Taylor ML, Espinosa-Schoelly ME, Iturbe R, Rico B, Casasola J, Goodsaid F. Evaluation of phagolysosome fusion in acridine orange stained macrophages infected with Histoplasma capsulatum. Clin Exp Immunol. 1989;75:466–470. [PMC free article] [PubMed] [Google Scholar]
- 73.Isaac DT, Coady A, Van Prooyen N, Sil A. The 3-hydroxy-methylglutaryl coenzyme A lyase HCL1 is required for macrophage colonization by human fungal pathogen Histoplasma capsulatum. Infect Immun. 2013;81:411–420. doi: 10.1128/IAI.00833-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lorenz MC, Fink GR. Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryot Cell. 2002;1:657–662. doi: 10.1128/EC.1.5.657-662.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naderer T, McConville MJ. The Leishmania–macrophage interaction: a metabolic perspective. Cell Microbiol. 2008;10:301–308. doi: 10.1111/j.1462-5822.2007.01096.x. [DOI] [PubMed] [Google Scholar]
- 76.Muñoz-Elías EJ, McKinney JD. Carbon metabolism of intracellular bacteria. Cell Microbiol. 2006;8:10–22. doi: 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- 77.Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 78.Fan W, Kraus PR, Boily M-J, Heitman J. Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot Cell. 2005;4:1420–1433. doi: 10.1128/EC.4.8.1420-1433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Faucher SP, Mueller CA, Shuman HA. Legionella pneumophila transcriptome during intracellular multiplication in human macrophages. Front Microbiol. 2011;2:60. doi: 10.3389/fmicb.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 83.Hu G, Cheng P-Y, Sham A, Perfect JR, Kronstad JW. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol. 2008;69:1456–1475. doi: 10.1111/j.1365-2958.2008.06374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, Hu G, Kronstad JW, Perfect JR. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio. 2011;2:e00103–e00111. doi: 10.1128/mBio.00103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barelle CJ, Priest CL, Maccallum DM, Gow NAR, Odds FC, Brown AJP. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 87.Newman SL, Gootee L, Brunner G, Deepe GS. Chloroquine induces human macrophage killing of Histoplasma capsulatum by limiting the availability of intracellular iron and is therapeutic in a murine model of histoplasmosis. J Clin Invest. 1994;93:1422–1429. doi: 10.1172/JCI117119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Howard DH, Rafie R, Tiwari A, Faull KF. Hydroxamate siderophores of Histoplasma capsulatum. Infect Immun. 2000;68:2338–2343. doi: 10.1128/iai.68.4.2338-2343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hwang LH, Mayfield JA, Rine J, Sil A. Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathog. 2008;4:e1000044. doi: 10.1371/journal.ppat.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Winters MS, Spellman DS, Chan Q, Gomez FJ, Hernandez M, Catron B, Smulian AG, Neubert TA, Deepe GS. Histoplasma capsulatum proteome response to decreased iron availability. Proteome Sci. 2008;6:36. doi: 10.1186/1477-5956-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hilty J, George Smulian A, Newman SL. Histoplasma capsulatum utilizes siderophores for intracellular iron acquisition in macrophages. Med Mycol. 2011;49:633–642. doi: 10.3109/13693786.2011.558930. [DOI] [PubMed] [Google Scholar]
- 92.Timmerman MM, Woods JP. Ferric reduction is a potential iron acquisition mechanism for Histoplasma capsulatum. Infect Immun. 1999;67:6403–6408. doi: 10.1128/iai.67.12.6403-6408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Timmerman MM, Woods JP. Potential role for extracellular glutathione-dependent ferric reductase in utilization of environmental and host ferric compounds by Histoplasma capsulatum. Infect Immun. 2001;69:7671–7678. doi: 10.1128/IAI.69.12.7671-7678.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zarnowski R, Cooper KG, Brunold LS, Calaycay J, Woods JP. Histoplasma capsulatum secreted γ–glutamyltransferase reduces iron by generating an efficient ferric reductant. Mol Microbiol. 2008;70:352–368. doi: 10.1111/j.1365-2958.2008.06410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Silva-Bailão MG, Bailão EFLC, Lechner BE, Gauthier GM, Lindner H, Bailão AM, Haas H, de Almeida Soares CM. Hydroxamate production as a high affinity iron acquisition mechanism in Paracoccidioides spp. PLoS One. 2014;9:e105805. doi: 10.1371/journal.pone.0105805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zarnowski R, Woods JP. Glutathione-dependent extracellular ferric reductase activities in dimorphic zoopathogenic fungi. Microbiology. 2005;151:2233–2240. doi: 10.1099/mic.0.27918-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holzberg M, Artis WM. Hydroxamate siderophore production by opportunistic and systemic fungal pathogens. Infect Immun. 1983;40:1134–1139. doi: 10.1128/iai.40.3.1134-1139.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lane TE, Wu-Hsieh BA, Howard DH. Iron limitation and the γ-interferon-mediated anti-Histoplasma state of murine macrophages. Infect Immun. 1991;59:2274–2278. doi: 10.1128/iai.59.7.2274-2278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Byrd TF, Horwitz MA. Interferon γ-activated human monocytes downregulate transferrin receptors and inhibit the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. J Clin Invest. 1989;83:1457–1465. doi: 10.1172/JCI114038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Winters MS, Chan Q, Caruso JA, Deepe GS. Metallomic analysis of macrophages infected with Histoplasma capsulatum reveals a fundamental role for zinc in host defenses. J Infect Dis. 2010;202:1136–1145. doi: 10.1086/656191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Subramanian Vignesh K, Landero Figueroa JA, Porollo A, Caruso JA, Deepe GS. Granulocyte macrophage-colony stimulating factor induced Zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity. 2013;39:697–710. doi: 10.1016/j.immuni.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Worsham PL, Goldman WE. Development of a genetic transformation system for Histoplasma capsulatum: complementation of uracil auxotrophy. Mol Gen Genet. 1990;221:358–362. doi: 10.1007/BF00259400. [DOI] [PubMed] [Google Scholar]
- 104.Retallack DM, Heinecke EL, Gibbons R, Deepe GS, Woods JP. The URA5 gene is necessary for Histoplasma capsulatum growth during infection of mouse and human cells. Infect Immun. 1999;67:624–629. doi: 10.1128/iai.67.2.624-629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garfoot AL, Zemska O, Rappleye CA. Histoplasma capsulatum depends on de novo vitamin biosynthesis for intraphagosomal proliferation. Infect Immun. 2014;82:393–404. doi: 10.1128/IAI.00824-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Salvin SB. Cysteine and related compounds in the growth of the yeast like phase of Histoplasma capsulatum. J Infect Dis. 1949;84:275–283. doi: 10.1093/infdis/84.3.275. [DOI] [PubMed] [Google Scholar]
- 107.Howard DH, Dabrowa N, Otto V, Rhodes J. Cysteine transport and sulfite reductase activity in a germination-defective mutant of Histoplasma capsulatum. J Bacteriol. 1980;141:417–421. doi: 10.1128/jb.141.1.417-421.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Boguslawski G, Akagi JM, Ward LG. Possible role for cysteine biosynthesis in conversion from mycelial to yeast form of Histoplasma capsulatum. Nature. 1976;261:336–338. doi: 10.1038/261336a0. [DOI] [PubMed] [Google Scholar]
- 109.Stetler DA, Boguslawski G. Cysteine biosynthesis in a fungus, Histoplasma capsulatum. Sabouraudia. 1979;17:23–34. doi: 10.1080/00362177985380041. [DOI] [PubMed] [Google Scholar]
- 110.Wu-Hsieh BA, Howard DH. Intracellular growth inhibition of Histoplasma capsulatum induced in murine macrophages by recombinant γ-interferon is not due to a limitation of the supply of methionine or cysteine to the fungus. Infect Immun. 1992;60:698–700. doi: 10.1128/iai.60.2.698-700.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jacobson ES, Harrell AC. Cysteine-independent and cysteine-requiring yeast strains of Histoplasma capsulatum. Mycopathologia. 1982;77:69–73. doi: 10.1007/BF00437386. [DOI] [PubMed] [Google Scholar]
- 112.Jacobson ES, Harrell AC. A prototrophic yeast strain of Histoplasma capsulatum. Mycopathologia. 1982;77:65–68. doi: 10.1007/BF00437385. [DOI] [PubMed] [Google Scholar]
- 113.Medoff G, Painter A, Kobayashi GS. Mycelial- to yeast-phase transitions of the dimorphic fungi Blastomyces dermatitidis and Paracoccidioides brasiliensis. J Bacteriol. 1987;169:4055–4060. doi: 10.1128/jb.169.9.4055-4060.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Andrade RV, Paes HC, Nicola AM, de Carvalho MJA, Fachin AL, Cardoso RS, Silva SS, Fernandes L, Silva SP, Donadi EA, et al. Cell organisation, sulphur metabolism and ion transport-related genes are differentially expressed in Paracoccidioides brasiliensis mycelium and yeast cells. BMC Genom. 2006;7:208. doi: 10.1186/1471-2164-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Menino JF, Saraiva M, Gomes-Rezende J, Sturme M, Pedrosa J, Castro AG, Ludovico P, Goldman GH, Rodrigues F. P. brasiliensis virulence is affected by SconC, the negative regulator of inorganic sulfur assimilation. PLoS One. 2013;8:e74725. doi: 10.1371/journal.pone.0074725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ferreira ME, Marques Edos R, Malavazi I, Torres I, Restrepo A, Nunes LR, de Oliveira RC, Goldman MHS, Goldman GH. Transcriptome analysis and molecular studies on sulfur metabolism in the human pathogenic fungus Paracoccidioides brasiliensis. Mol Genet Genomics. 2006;276:450–463. doi: 10.1007/s00438-006-0154-4. [DOI] [PubMed] [Google Scholar]
- 117.Paris S, Duran-Gonzalez S, Mariat F. Nutritional studies on Paracoccidioides brasiliensis: the role of organic sulfur in dimorphism. Sabouraudia. 1985;23:85–92. doi: 10.1080/00362178585380151. [DOI] [PubMed] [Google Scholar]
- 118.Marques ER, Ferreira MES, Drummond RD, Felix JM, Menossi M, Savoldi M, Travassos LR, Puccia R, Batista WL, Carvalho KC, et al. Identification of genes preferentially expressed in the pathogenic yeast phase of Paracoccidioides brasiliensis, using suppression subtraction hybridization and differential macroarray analysis. Mol Genet Genomics. 2004;271:667–677. doi: 10.1007/s00438-004-1016-6. [DOI] [PubMed] [Google Scholar]
- 119.Brummer E, Hanson LH, Restrepo A, Stevens DA. Intracellular multiplication of Paracoccidioides brasiliensis in macrophages: killing and restriction of multiplication by activated macrophages. Infect Immun. 1989;57:2289–2294. doi: 10.1128/iai.57.8.2289-2294.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Feeley JC, Gorman GW, Weaver RE, Mackel DC, Smith HW. Primary isolation media for Legionnaires disease bacterium. J Clin Microbiol. 1978;8:320–325. doi: 10.1128/jcm.8.3.320-325.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sebghati TS, Engle JT, Goldman WE. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science. 2000;290:1368–1372. doi: 10.1126/science.290.5495.1368. [DOI] [PubMed] [Google Scholar]
- 122.Beck MR, DeKoster GT, Hambly DM, Gross ML, Cistola DP, Goldman WE. Structural features responsible for the biological stability of Histoplasma’s virulence factor CBP. Biochemistry. 2008;47:4427–4438. doi: 10.1021/bi701495v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Batanghari JW, Deepe GS, Di Cera E, Goldman WE. Histoplasma acquisition of calcium and expression of CBP1 during intracellular parasitism. Mol Microbiol. 1998;27:531–539. doi: 10.1046/j.1365-2958.1998.00697.x. [DOI] [PubMed] [Google Scholar]
- 124.Kügler S, Young B, Miller VL, Goldman WE. Monitoring phase-specific gene expression in Histoplasma capsulatum with telomeric GFP fusion plasmids. Cell Microbiol. 2000;2:537–547. doi: 10.1046/j.1462-5822.2000.00078.x. [DOI] [PubMed] [Google Scholar]
- 125.Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol. 2006;16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 126.Ma H, Croudace JE, Lammas DA, May RC. Expulsion of live pathogenic yeast by macrophages. Curr Biol. 2006;16:2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 127.Uwamahoro N, Verma-Gaur J, Shen H-H, Qu Y, Lewis R, Lu J, Bambery K, Masters SL, Vince JE, Naderer T, et al. The pathogen Candida albicans hijacks pyroptosis for escape from macrophages. mBio. 2014;5:e00003–e00014. doi: 10.1128/mBio.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Piddington DL, Fang FC, Laessig T, Cooper AM, Orme IM, Buchmeier NA. Cu, Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect Immun. 2001;69:4980–4987. doi: 10.1128/IAI.69.8.4980-4987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.De Groote MA, Ochsner UA, Shiloh MU, Nathan C, McCord JM, Dinauer MC, Libby SJ, Vazquez-Torres A, Xu Y, Fang FC. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fang FC, DeGroote MA, Foster JW, Bäumler AJ, Ochsner U, Testerman T, Bearson S, Giárd JC, Xu Y, Campbell G, et al. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc Natl Acad Sci USA. 1999;96:7502–7507. doi: 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Melillo AA, Mahawar M, Sellati TJ, Malik M, Metzger DW, Melendez JA, Bakshi CS. Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J Bacteriol. 2009;191:6447–6456. doi: 10.1128/JB.00534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol Microbiol. 2009;71:240–252. doi: 10.1111/j.1365-2958.2008.06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brummer E, Stevens DA. Antifungal mechanisms of activated murine bronchoalveolar or peritoneal macrophages for Histoplasma capsultum. Coin Exp Immunol. 1995;102:65–70. doi: 10.1111/j.1365-2249.1995.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]