Abstract
NG2 expressing cells (polydendrocytes, oligodendrocyte precursor cells) are the fourth major glial cell population in the central nervous system. During embryonic and postnatal development they actively proliferate and generate myelinating oligodendrocytes. These cells have commonly been studied in primary dissociated cultures, neuron cocultures, and in fixed tissue. Using newly available transgenic mouse lines slice culture systems can be used to investigate proliferation and differentiation of oligodendrocyte lineage cells in both gray and white matter regions of the forebrain and cerebellum. Slice cultures are prepared from early postnatal mice and are kept in culture for up to 1 month. These slices can be imaged multiple times over the culture period to investigate cellular behavior and interactions. This method allows visualization of NG2 cell division and the steps leading to oligodendrocyte differentiation while enabling detailed analysis of region-dependent NG2 cell and oligodendrocyte functional heterogeneity. This is a powerful technique that can be used to investigate the intrinsic and extrinsic signals influencing these cells over time in a cellular environment that closely resembles that found in vivo.
Keywords: Neuroscience, Issue 90, NG2, CSPG4, polydendrocyte, oligodendrocyte progenitor cell, oligodendrocyte, myelin, organotypic slice culture, time-lapse
Introduction
Organoytpic slice cultures of the central nervous system have proven to be extremely useful for studying neuron and glial cell biology in a semiintact system1–4. These cultures are relatively simple to adopt and retain many benefits of primary dissociated cell cultures such as manipulation of the extracellular environment and easy access for repeated long-term live cell imaging and electrophysiological recordings5–9. In addition, slice cultures maintain 3-dimensional tissue cytoarchitecture, regional neural connectivity, and most major cell types are present in the system. These properties make these cultures a unique and convenient system to investigate single cell behavior and physiology with cellular and environmental interactions.
NG2 cells are a population of glial cells in the mammalian central nervous system that continue to proliferate and generate myelinating oligodendrocytes during embryonic and postnatal development10. They have been extensively studied in dissociated primary cell cultures, and recent development of transgenic mouse lines with NG2 cell-specific expression of fluorescent proteins has facilitated in vivo fate mapping and electrophysiological recordings in acute slice preparations. Even with these studies, little is known about the temporal dynamics of NG2 cell proliferation and oligodendrocyte differentiation. Although dissociated cell culture is widely used for the relative facility in pharmacological and genetic manipulations, it is not suitable for interrogating functional differences of these cells in different brain regions particularly when it is desirable to maintain the context of the cellular microenvironment. Slice cultures provide a simple alternative that is amenable to pharmacological manipulations and have been used to investigate oligodendrocyte myelination11,12, cellular response after lysolecithin (LPC) or antibody induced demyelination13,14, and induction of remyelination via pharmacological treatment15.
A method to investigate and perform live imaging and fixed tissue (or postfixation) analysis of NG2 cell proliferation and oligodendrocyte differentiation in organotypic slice cultures taken from both the forebrain and cerebellum is described. This is a powerful method that can be used to study the cell fate of single NG2 cells after division16 and to discover region- and age-dependent differences in growth factor induced NG2 cell proliferation17. This relatively simple technique is widely accessible to further investigate cell intrinsic and/or environmental mechanisms regulating the physiology of these glial cells and their response to neuronal activity or myelin damage.
Protocol
NOTE: All animal procedures are following the guidelines and have been approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Connecticut.
NOTE: For these experiments constitutive NG2cre18 (JAX#008533) and inducible NG2creER16 (JAX#008538) transgenic mice crossed to Z/EG19 (JAX#003920) or gtRosa26:YFP reporter20 (JAX#006148) lines respectively were used to image NG2 cells and their progeny. To image mature oligodendrocytes, PLPDsRed transgenic mice21 were used. For consistent survival, slices can be isolated from mice up to postnatal day 10 from both the forebrain and cerebellum.
1. Slice Preparation
In a culture hood, place tissue culture inserts in six well plates and add 1 ml slice culture medium (recipe in reagents section). Put the plates in the cell culture incubator at 37 °C with 5% CO2 at least 2 hr prior to dissection.
Sterilize all tools and tissue chopper with 70% ethanol.
Bubble dissection buffer (recipe in reagents section) with 95% O2, 5% CO2 on ice for at least 15 min prior to the start of dissection.
Anesthetize mouse pups by placing them on ice for 5-10 min according to approved animal protocols. Ascertain the appropriate depth of anesthesia by confirming that the mice do not respond to tail and toe pinching.
Decapitate animals using sharp scissors following animal protocols. Quickly remove the skull over the forebrain and cerebellum by first making a sagittal cut followed by lateral incision, using sterilized tools. Cut cranial nerves from the ventral surface of the brain and cerebellum by carefully rolling the tissue to the side.
Place tissue in a sterile 35 mm dish containing ice-cold oxygenated dissection buffer.
Using a razor blade, sever the cerebellum and forebrain. Then make a mid-sagittal cut through the forebrain and cerebellum, separating the hemispheres.
Cut 300 μm-thick coronal and sagittal sections of the forebrain and cerebellum with a manual tissue chopper. NOTE: A manual tissue chopper allows rapid processing from dissection to culture incubation without the need for agarose embedding. A vibratome can also be used as described previously9.
Transfer separated slices into freshly bubbled cold dissection buffer on ice.
Use small dissecting or weighing spatulas to separate individual sections and transfer them to preincubated six well dishes with culture inserts.
Remove any excess dissection buffer from the surface of the culture insert using a disposable transfer pipette or a P 200 pipet tip. Two to three forebrain or three cerebellar slices can be placed on one culture insert. NOTE: For consistent results with forebrain cultures, it is ideal to take slices spanning the anterior one-third of the corpus callosum (Figure 1A) in order to study both gray and white matter regions in the same slice. Each animal usually yields 6 forebrain slices and 6 cerebellum slices.
Place the six well plates in the incubator and replace the slice medium with 1 ml of slice medium 1 day after slice preparation and then every other day until slice fixation.
Depending on the experiment, use slices after 5-7 days in culture. Use only slices that become transparent after the first few days and discard those that have uneven opaque regions. NOTE: Opaque regions within slices are visible by eye and appear as a clump of dark cells under phase contrast. After the technique has been mastered, dead opaque slices occur rarely, in less than 1-5% of slices.
2. Time-Lapse Imaging of NG2 cell Division and Oligodendrocyte Differentiation
NOTE: To perform time lapse imaging of NG2 cell proliferation and differentiation we use NG2cre:ZEG mice16 which express EGFP in NG2 cells and their progeny (Figure 1C-E). Reporter expression in this line is sufficiently robust to obtain live images of GFP+ cells in slices immediately after sectioning for a time period of at least four weeks. The clearest images are obtained after the initial thinning period of the slice, which occurs over the first 3-5 days in culture.
Throughout the experiment, keep the slices in a cell culture incubator at 37 °C and remove only for brief periods of time (less than 15 min per slice), keeping the lid on the six well plate while imaging.
Using an inverted fluorescence microscope equipped with a digital camera, capture images at the first time point using a 10X objective. NOTE: First time points will vary by experiment and can be the day when the slices were prepared or any time point after.
Capture images from multiple regions at time point one in both gray and white matter areas of the slice. Note the region imaged by specific landmarks within the slice and by mapping the image location on a schematic that can be used for subsequent imaging sessions. Useful landmarks can include edges of the slices and/or orientation of white matter tracts. Image four to eight locations on each slice at each time point.
Capture subsequent images at an interval of 4-6 hr if examining NG2 cell division and oligodendrocyte differentiation, ideally over 5-7 days.
Capture a final image of all regions and fix the slices immediately. Add 1 ml of fixative (4% PFA with 0.1M l-lysine 0.01 M sodium meta-periodate) to the bottom of the culture insert, then add another 1 ml on top of the slices. Take care not to squirt the fixative directly on the slice as it can detach from the membrane. Fix the tissue for 30 min and proceed with immunohistochemistry using developmental stage-specific markers as described in section 4.
Wash slices with 0.2 M sodium phosphate buffer 3 x 10 min. If necessary, store slices at 4 °C in 0.2 M sodium phosphate buffer for 1-3 days before immunostaining but ideally perform staining within 24 hr. Proceed with immunohistochemistry as described below in section 4 to determine the proportion of cells that have differentiated into oligodendrocytes (Figure 2D-F).
3. Fate Mapping of NG2 cell Progeny Using Inducible Reporter Transgenic Mice in Slice Cultures
NOTE: To track the fate of NG2 cells from a particular time point in the culture, inducible NG2creER:YFP transgenic mice can be used.
Induce Cre recombination and reporter expression by adding 100 nM 4OHT dissolved in ethanol to the culture medium. NOTE: Reporter positive cells should appear within 1-2 days at an induction efficiency of ~20-25% YFP+NG2+/NG2+ cells and at this time point will all be cells at the NG2 cell stage (Figure 2A-C)
Fix and wash the slices at different time points after various manipulations (described in sections 2.5 and 2.6).
4. Slice Immunohistochemistry
Cut the membranes with the slices attached out of the inserts using a scalpel.
Transfer slices to individual wells of a 24 well plate containing phosphate buffered saline (PBS) using a set of tweezers, taking care not to disrupt the composition of the slice when picking up the membrane.
Transfer the slices to a 24 well plate with blocking solution containing 5% normal goat serum (NGS) and 0.1% Triton-X100 in PBS for 1 hr.
Incubate the slices in primary antibody O/N in 5% NGS in PBS at 4 °C. For identifying cells in the NG2 cell stage, use antibodies to NG2 and PDGFRα. For identifying differentiated oligodendrocytes, use an antibody to the adenomatous polyposis coli antigen (APC; clone CC1).
On the second day wash slices in PBS 3 x15 min, then incubate in secondary antibodies in 5% NGS in PBS for 1 hr at RT. Wash slices in PBS 3 x15 min and mount on slides. Mount with slices up and membrane facing the slide so that the membrane will not be between the objective and the tissue.
Representative Results
Examples of representative data are given below that have been obtained using slice cultures from both the forebrain and cerebellum of P8 NG2cre:ZEG, NG2creER:YFP and PLPDsRed transgenic mouse lines. NG2 cells can be imaged over days in forebrain and cerebellum slices (Figure 1B-D, Video 1) and the phenotype of these cells can be determined after fixation and immunostaining with NG2 and CC1 antibodies (Figure 1E). In addition to live imaging, cell proliferation can also be assessed using immunohistochemical staining for cell proliferation markers such as Ki67 (Figure 1F). These data allow for detailed analysis of the temporal dynamics of oligodendrocyte differentiation after NG2 cell division on an individual cell basis. Fate mapping of NG2 cells can be carried out after induction of cre recombination in slices with 4OHT in NG2creER:YFP mice (Figure 2). YFP reporter-expressing NG2+ cells were found in the cultures two days after addition of 4OHT to the culture medium (Figure 2A). Six days after 4OHT induction, a proportion of the YFP+ cells had differentiated into CC1+ oligodendrocytes (Figure 2B). These data show the feasibility of performing fate mapping of NG2 cells in slices where the extracellular environment can be manipulated to test the effects of pharmacological agents or various insults on the fate of NG2 cells. Different concentrations of 4OHT can be used to achieve different degrees of cre recombination. For example, exposure to 100 nM 4OHT for 2 days causes reporter expression in approximately 20-25% of all NG2 expressing cells while 1µM 4OHT results in approximately 60-70% recombination efficiency. Finally, extensive myelin is present in these cultures and myelinating oligodendrocytes can be imaged in live and fixed slices using PLPDsRed transgenic mice (Figure 3, Video 2).
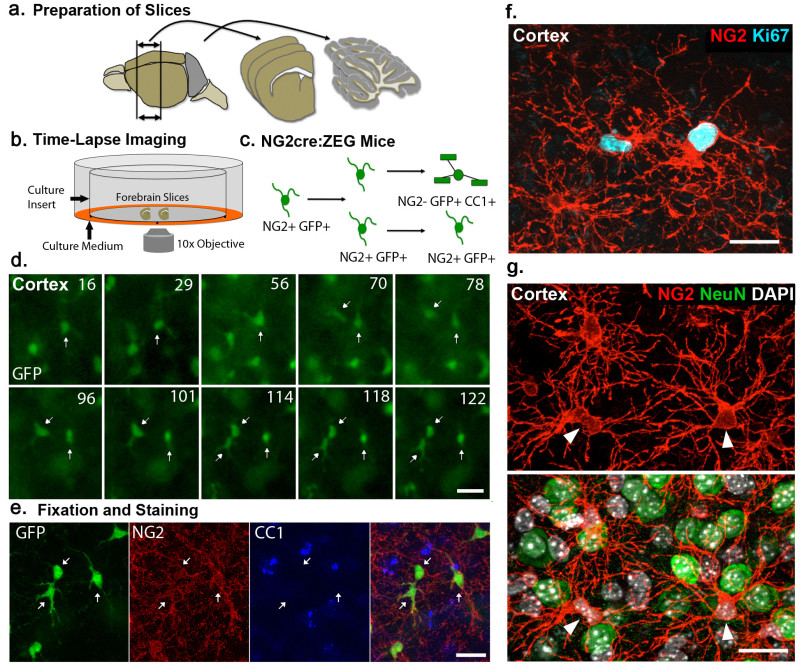 Figure 1. Slice culture method for long-term imaging of NG2 cell proliferation and differentiation(A-B) Schematic depicting the isolation of forebrain and cerebellum slices (A) and subsequent culturing and time-lapse imaging using slice culture inserts (B). (C) Depiction outlining NG2 cell division and subsequent oligodendrocyte differentiation in NG2cre:ZEG transgenic mice. (D) Example images acquired from cortical regions of NG2cre:ZEG mice showing a single GFP+ cell (arrows) dividing twice over a period of 122 hr, time depicted in top right corner as hours after the start of the imaging experiment. (E) Fixation and immunostaining with anti-NG2 and CC1 antibodies of the same slice reveals that the imaged dividing cells resulted in three NG2+ cells (arrows). (F) Example image showing two Ki67+NG2+ cells in a cortical slice culture. (G) Example image showing NG2+ cells (arrowheads) and their processes intertwined with NeuN+ neuronal nuclei in a cortical slice culture. Scale Bars 25 μm Please click here to view a larger version of this figure.
Figure 1. Slice culture method for long-term imaging of NG2 cell proliferation and differentiation(A-B) Schematic depicting the isolation of forebrain and cerebellum slices (A) and subsequent culturing and time-lapse imaging using slice culture inserts (B). (C) Depiction outlining NG2 cell division and subsequent oligodendrocyte differentiation in NG2cre:ZEG transgenic mice. (D) Example images acquired from cortical regions of NG2cre:ZEG mice showing a single GFP+ cell (arrows) dividing twice over a period of 122 hr, time depicted in top right corner as hours after the start of the imaging experiment. (E) Fixation and immunostaining with anti-NG2 and CC1 antibodies of the same slice reveals that the imaged dividing cells resulted in three NG2+ cells (arrows). (F) Example image showing two Ki67+NG2+ cells in a cortical slice culture. (G) Example image showing NG2+ cells (arrowheads) and their processes intertwined with NeuN+ neuronal nuclei in a cortical slice culture. Scale Bars 25 μm Please click here to view a larger version of this figure.
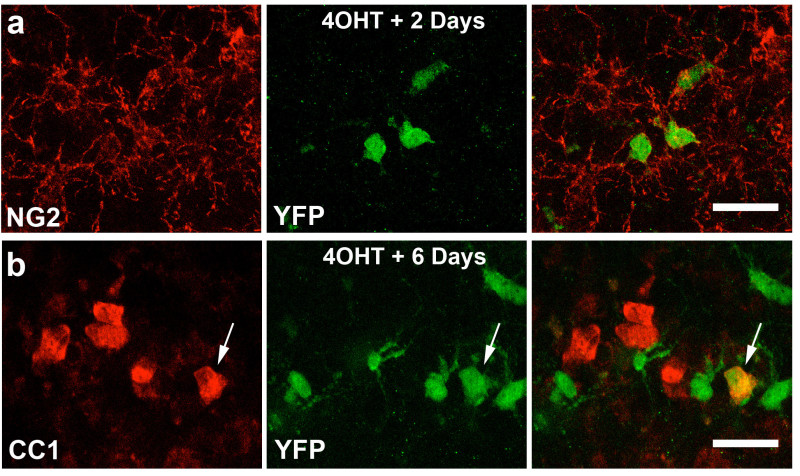 Figure 2. Induction of YFP expression to track the fate of NG2 cells in forebrain slice cultures(A-B) Images taken of the corpus callosum region of forebrain slice cultures from NG2creER:YFP mice treated with 4OHT for 24 hr and then left in culture for 2 days (A) or 6 days (B). Slices were fixed and immunostained for NG2 and YFP (A) or CC1 and YFP (B) demonstrating the differentiation of reporter positive NG2 cells into oligodendrocytes while in culture 6 days after cre recombination (arrow). Scale Bars 25 μm Please click here to view a larger version of this figure.
Figure 2. Induction of YFP expression to track the fate of NG2 cells in forebrain slice cultures(A-B) Images taken of the corpus callosum region of forebrain slice cultures from NG2creER:YFP mice treated with 4OHT for 24 hr and then left in culture for 2 days (A) or 6 days (B). Slices were fixed and immunostained for NG2 and YFP (A) or CC1 and YFP (B) demonstrating the differentiation of reporter positive NG2 cells into oligodendrocytes while in culture 6 days after cre recombination (arrow). Scale Bars 25 μm Please click here to view a larger version of this figure.
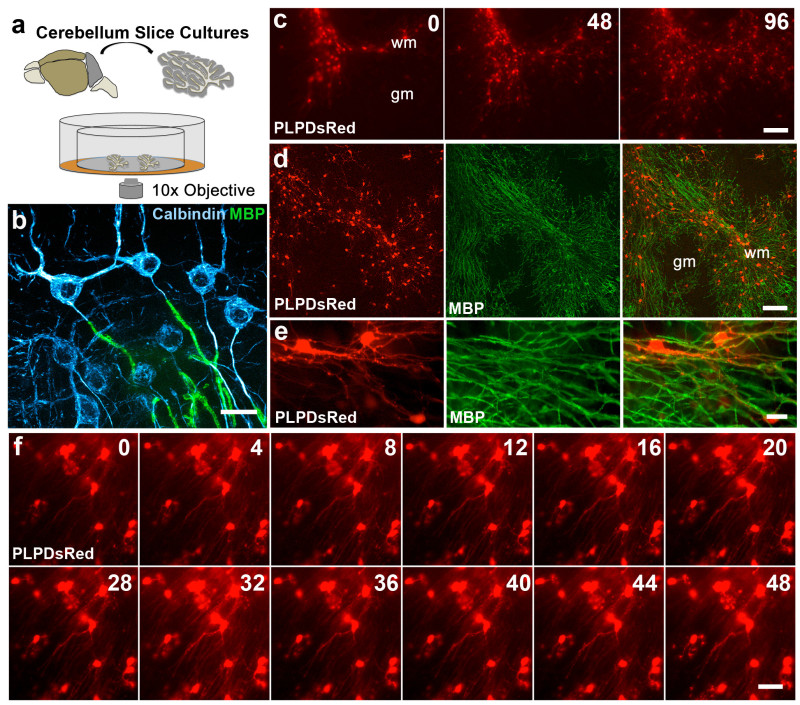 Figure 3. Imaging oligodendrocytes in slice cultures from PLPDsRed transgenic mice.(A) Schematic depicting the isolation, culturing and imaging of cerebellum slices from PLPDsRed transgenic mice. (B) Image taken from a fixed cerebellum slice after 5 days in vitro showing calbindin+ (cyan) purkinje neurons with myelinated myelin basic protein+ (MBP) (green) axons projecting into white matter regions of the slice. Scale Bar 25 μm. (C) Low magnification images captured from the same region every other day at the times indicated in hours in cerebellum slice cultures from PLPDsRed mice. Scale Bar 100 μm. (D) Low magnification image taken from a fixed PLPDsRed slice culture showing MBP expression in white matter regions where DsRed+ cells are concentrated. Scale Bar 100 μm. (E) High magnification image taken from a fixed PLPDsRed slice culture showing single DsRed+ oligodendrocytes with MBP+ processes. Scale Bar 20 μm. (F) Time-lapse sequence taken from a PLPDsRed cerebellum slice showing relatively stable cell bodies over the 48 hr imaging session, time indicated in upper right in hours. Scale Bar 25 μm. Please click here to view a larger version of this figure.
Figure 3. Imaging oligodendrocytes in slice cultures from PLPDsRed transgenic mice.(A) Schematic depicting the isolation, culturing and imaging of cerebellum slices from PLPDsRed transgenic mice. (B) Image taken from a fixed cerebellum slice after 5 days in vitro showing calbindin+ (cyan) purkinje neurons with myelinated myelin basic protein+ (MBP) (green) axons projecting into white matter regions of the slice. Scale Bar 25 μm. (C) Low magnification images captured from the same region every other day at the times indicated in hours in cerebellum slice cultures from PLPDsRed mice. Scale Bar 100 μm. (D) Low magnification image taken from a fixed PLPDsRed slice culture showing MBP expression in white matter regions where DsRed+ cells are concentrated. Scale Bar 100 μm. (E) High magnification image taken from a fixed PLPDsRed slice culture showing single DsRed+ oligodendrocytes with MBP+ processes. Scale Bar 20 μm. (F) Time-lapse sequence taken from a PLPDsRed cerebellum slice showing relatively stable cell bodies over the 48 hr imaging session, time indicated in upper right in hours. Scale Bar 25 μm. Please click here to view a larger version of this figure.
Video 1. Live Imaging of NG2 cell division in a cortical slice culture. Representative time lapse-sequence showing multiple cell divisions in a cortical slice culture taken from an NG2cre:ZEG transgenic mouse. Video displayed at 5 frames per second, montage of images shown in Figure 1. (See "Video_1.mov" under Downloads)
Video 2. Live imaging of oligodendrocytes in cerebellum slice cultures Representative time lapse-sequence showing small changes in oligodendrocyte morphology (arrow) imaged over 48 hr in a cerebellum slice taken from a PLPDsRed transgenic mouse. Video displayed at 3 frames per second , montage of images shown in Figure 3. (See "Video_2.mov" under Downloads)
Discussion
Myelination in the central nervous system is essential for efficient neuronal communication and axonal survival22. NG2 cells continuously generate myelinating oligodendrocytes into adulthood while maintaining a resident population in most brain regions16,23–25. Some genetic and molecular mechanisms regulating the differentiation of these cells have been described but much remains to be discovered. Organotypic slice cultures are a convenient tool to investigate these mechanisms due to their unique characteristics of maintaining anatomical regions, easy manipulation of the extracellular environment, robust myelination, and the presence of all major cell types. These characteristics facilitate investigation of short and long-term interactions between NG2 cells, oligodendrocytes and axons11,26. In addition, cell transplantation is relatively easy to perform and can be used to investigate region-dependent differences in cell behavior17. Furthermore, pharmacological treatments can be added to the culture medium to investigate molecular mechanisms influencing NG2 cell proliferation and/or differentiation in normal17,27,28 and demyelinated cultures15,29. Finally, it is technically feasible to use slice cultures to perform screens for high throughput analysis of compounds that direct NG2 cells to proliferate or differentiate, possibly even after a demyelinating insult30.
Current methods to investigate oligodendrocyte lineage cells and their interactions with axons in a controlled culture setting include co-cultures with dissociated dorsal root ganglion (DRG) or embryonic cortical neurons and NG2 cells31,32, which were based on original preparations developed to investigate DRG-Schwann cell interactions33. These cultures have been used to investigate fundamental properties of axon and oligodendrocyte lineage cell interactions including neuronal activity-dependent signaling to induce differentiation and myelin production32,34–36 in addition to other questions such as the dependence of axon diameter and NG2 cell density controlling proliferation and the onset of differentiation37. While these coculture systems are ideal to address such questions, direct correlation and application to the in vivo situation is not always clear. As mentioned previously, organotypic slice cultures provide a unique condition of maintaining many of the local neuronal connections and three dimensional cytoarchitecture that are lost in co-culture preparations of dissociated cells. Furthermore unlike organotypic slice cultures, co-culture systems of dissociated cells often do not include other glial cells and extracellular matrix molecules which have shown to play key roles in the development, maintenance and physiology of NG2 cells, oligodendrocytes, and myelin. With the dramatic growth in the variety of tools available for specific labeling and manipulation of distinct cell populations with viral vectors and transgenic animals, long-term imaging and fate mapping in organotypic slice cultures should prove to be a powerful technique for the investigation of NG2 cell proliferation, differentiation and oligodendrocyte myelination.
Several critical steps should be carried out when performing these experiments to ensure slice survival and data reproducibility. First, in order to generate healthy slices, sectioning and isolation of slices should be performed as quickly as possible and in ice cold dissection buffer. Second, while automated image position acquisition with a motorized stage on the fluorescence microscope can be useful, there are many factors that can disrupt the precise relocation of the regions imaged and manual repeated acquisition is necessary to get consistently reliable images over multiple days. This is particularly necessary if the slices are kept in a cell culture incubator between imaging sessions and not in a stage-top incubator. Third, during slice fixation, staining, and mounting it is extremely important to handle the slices gently as any disruption of the tissue will result in the inability to relocate live imaged regions and cells. There are no specific modifications to traditional methods used for slice culture preparations that are used for studies of neurons and astrocytes.
There are several limitations to the slice culture method that must be considered. Astrocytes, microglia and NG2 cells are known to respond to CNS damage with increased proliferation at the site of injury and the formation of a glial scar. The process of preparing the slices results in some reorganization and an initial reactive glial response from these cells. NG2 cell proliferation rates return to levels similar to those reported in vivo for matched stages of development after several days in culture however the increased proliferation rates and initial reactive phenotype of these cells must be considered when interpreting the data. The presence of horse serum in the culture medium could alter NG2 cell proliferation and should be considered when interpreting the findings of any experiment. In addition to this, while there is spontaneous neuronal activity, sensory input and long range connections between neurons are lacking, which could lead to altered neuronal activity within the slice. Finally, without the use of specific promoter driven viral vectors or transgenic mice, cell type-specific genetic manipulations within slice culture systems is challenging. While these limitations should be considered when designing experiments, slice cultures are a unique system which can be utilized to gain novel insight into questions that cannot be answered with dissociated cell cultures or in the intact animal.
Disclosures
The authors declare that they have no competing financial interests
Acknowledgments
This work was funded by grants from the National Multiple Sclerosis Society (RG4179 to A.N.), the National Institutes of Health (NIH R01NS073425 and R01NS074870 to A.N.) and the National Science Foundation (A.N.). We thank Dr. Frank Kirchhoff (University of Saarland, Homburg Germany) for providing PLPDsRed transgenic mice. We thank Youfen Sun for her assistance in maintaining the transgenic mouse colony.
References
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. Journal of Neuroscience Methods. 1991;37(2) doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Gogolla N, Galimberti I, DePaola V, Caroni P. Preparation of organotypic hippocampal slice cultures for long-term live imaging. Nature Protocols. 2006;1(3):1165–1171. doi: 10.1038/nprot.2006.168. [DOI] [PubMed] [Google Scholar]
- Gähwiler BH, Capogna M, Debanne D, McKinney RA, Thompson SM. Organotypic slice cultures: a technique has come of age. Trends in Neurosciences. 1997;20(10):471–477. doi: 10.1016/s0166-2236(97)01122-3. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Kurotani T, Toyama K. Neural connections between the lateral geniculate nucleus and visual cortex in vitro. Science. 1989;245(4914):192–194. doi: 10.1126/science.2749258. [DOI] [PubMed] [Google Scholar]
- Bahr BA, Kessler M, et al. Stable maintenance of glutamate receptors and other synaptic components in long-term hippocampal slices. Hippocampus. 1995;5(5):425–439. doi: 10.1002/hipo.450050505. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature Neuroscience. 2004;7(2) doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Kapfhammer JP, Gugger OS. The analysis of purkinje cell dendritic morphology in organotypic slice cultures. Journal of Visualized Experiments JoVE. 2012. [DOI] [PMC free article] [PubMed]
- Elias L, Kriegstein A. Organotypic slice culture of E18 rat brains. Journal of Visualized Experiments JoVE. 2007. p. e235. [DOI] [PMC free article] [PubMed]
- Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nature Reviews Neuroscience. 2009;10(1):9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- Haber M, Vautrin S, Fry EJ, Murai KK. Subtype-specific oligodendrocyte dynamics in organotypic culture. Glia. 2009;57(9):1000–1013. doi: 10.1002/glia.20824. [DOI] [PubMed] [Google Scholar]
- Bin JM, Leong SY, Bull S-J, Antel JP, Kennedy TE. Oligodendrocyte precursor cell transplantation into organotypic cerebellar shiverer slices: a model to study myelination and myelin maintenance. PloS one. 2012;7(7) doi: 10.1371/journal.pone.0041237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgbauer E, Rao TS, Webb M. Lysolecithin induces demyelination in vitro in a cerebellar slice culture system. Journal of Neuroscience Research. 2004;78(2):157–166. doi: 10.1002/jnr.20248. [DOI] [PubMed] [Google Scholar]
- Harrer MD, von Budingen HC, et al. Live imaging of remyelination after antibody-mediated demyelination in an ex model for immune mediated CNS damage. Experimental Neurology. 2009;216(2):431–438. doi: 10.1016/j.expneurol.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Fancy SPJ, Harrington EP, et al. Axin2 as regulatory and therapeutic target in newborn brain injury and remyelination. Nature Neuroscience. 2011;14(8):1009–1016. doi: 10.1038/nn.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Hill RA, Dietrich D, Komitova M, Suzuki R, Nishiyama A. Age-dependent fate and lineage restriction of single NG2 cells. Development. 2011;138(4):745–753. doi: 10.1242/dev.047951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RA, Patel KD, Medved J, Reiss AM, Nishiyama A. NG2 Cells in White Matter But Not Gray Matter Proliferate in Response to PDGF. Journal of Neuroscience. 2013;33(36):14558–14566. doi: 10.1523/JNEUROSCI.2001-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135(1):145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28(3-4):147–155. [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genetics. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Hirrlinger PG, Scheller A, et al. Expression of reef coral fluorescent proteins in the central nervous system of transgenic mice. Molecular and Cellular Neuroscience. 2005;30(3):291–303. doi: 10.1016/j.mcn.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Nave K-A. Myelination and support of axonal integrity by glia. Nature. 2010;468(7321):244–252. doi: 10.1038/nature09614. [DOI] [PubMed] [Google Scholar]
- Rivers LE, Young KM, et al. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nature Neuroscience. 2008;11(12):1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimou L, Simon C, Kirchhoff F, Takebayashi H, Gotz M. Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. The Journal of Neuroscience. 2008;28(41):10434–10442. doi: 10.1523/JNEUROSCI.2831-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68(4):668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidou K, Anderson KI, Strachan D, Edgar JM, Barnett SC. Time-lapse imaging of the dynamics of CNS glial-axonal interactions in vitro and ex vivo. PloS one. 2012;7(1):e30775. doi: 10.1371/journal.pone.0030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoumari AM, Baulieu EE, Schumacher M. Progesterone increases oligodendroglial cell proliferation in rat cerebellar slice cultures. Neuroscience. 2005;135(1):47–58. doi: 10.1016/j.neuroscience.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Azim K, Butt AM. GSK3β negatively regulates oligodendrocyte differentiation and myelination in vivo. Glia. 2011;59(4):540–553. doi: 10.1002/glia.21122. [DOI] [PubMed] [Google Scholar]
- Hussain R, El-Etr M, et al. Progesterone and nestorone facilitate axon remyelination: a role for progesterone receptors. Endocrinology. 2011;152(10):3820–3831. doi: 10.1210/en.2011-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nature Reviews. Molecular Cell Biology. 2007;8(10):839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- Lubetzki C, Demerens C, et al. Even in culture, oligodendrocytes myelinate solely axons. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(14):6820–6824. doi: 10.1073/pnas.90.14.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerens C, Stankoff B, et al. Induction of myelination in the central nervous system by electrical activity. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9887–9892. doi: 10.1073/pnas.93.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood PM, Bunge RP. Evidence that sensory axons are mitogenic for Schwann cells. Nature. 1975;256(5519):662–664. doi: 10.1038/256662a0. [DOI] [PubMed] [Google Scholar]
- Stevens B, Tanner S, Fields RD. Control of myelination by specific patterns of neural impulses. The Journal of Neuroscience. 1998;18(22):9303–9311. doi: 10.1523/JNEUROSCI.18-22-09303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Porta S, Haak LL, Gallo V, Fields RD. Adenosine: a neuron-glial transmitter promoting myelination in the CNS in response to action potentials. Neuron. 2002;36(5):855–868. doi: 10.1016/s0896-6273(02)01067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H, Lee PR, Fields RD. Control of local protein synthesis and initial events in myelination by action potentials. Science. 2011;333(6049):1647–1651. doi: 10.1126/science.1206998. New York, N.Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SS, Kelland EE, Tokar E, De la Torre AR, Chan JR. The geometric and spatial constraints of the microenvironment induce oligodendrocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(38):14662–14667. doi: 10.1073/pnas.0805640105. [DOI] [PMC free article] [PubMed] [Google Scholar]


