Abstract
Factor XI (FXI) is the zymogen of an enzyme (FXIa) that contributes to hemostasis by activating factor IX. Although bleeding associated with FXI deficiency is relatively mild, there has been resurgence of interest in FXI because of studies indicating it makes contributions to thrombosis and other processes associated with dysregulated coagulation. FXI is an unusual dimeric protease, with structural features that distinguish it from vitamin K–dependent coagulation proteases. The recent availability of crystal structures for zymogen FXI and the FXIa catalytic domain have enhanced our understanding of structure-function relationships for this molecule. FXI contains 4 “apple domains” that form a disk structure with extensive interfaces at the base of the catalytic domain. The characterization of the apple disk structure, and its relationship to the catalytic domain, have provided new insight into the mechanism of FXI activation, the interaction of FXIa with the substrate factor IX, and the binding of FXI to platelets. Analyses of missense mutations associated with FXI deficiency have provided additional clues to localization of ligand-binding sites on the protein surface. Together, these data will facilitate efforts to understand the physiology and pathology of this unusual protease, and development of therapeutics to treat thrombotic disorders.
Introduction
Factor XI (FXI) is the zymogen of a blood coagulation protease, factor XIa (FXIa), that contributes to hemostasis through activation of factor IX (FIX; Figure 1).1–3 The protein is a 160-kDa disulfide-linked dimer of identical 607 amino acid subunits, each containing 4 90- or 91-amino acid repeats called apple domains (A1 to A4 from the N-terminus) and a C-terminal trypsin-like catalytic domain.3–7 The structure is distinctly different from those of the well-characterized vitamin K–dependent coagulation proteases.1 FXI circulates in blood as a complex with high molecular weight kininogen (HK).8 Prekallikrein (PK), the zymogen of the protease α-kallikrein, is a monomeric homolog of FXI with the same domain structure9,10 that also circulates in complex with HK.11 A recent analysis of vertebrate genomes confirmed that FXI and PK are products of a duplication event during mammalian evolution.12 The ancestral predecessor is also a protease with 4 apple domains, but its functional properties have not been studied.12
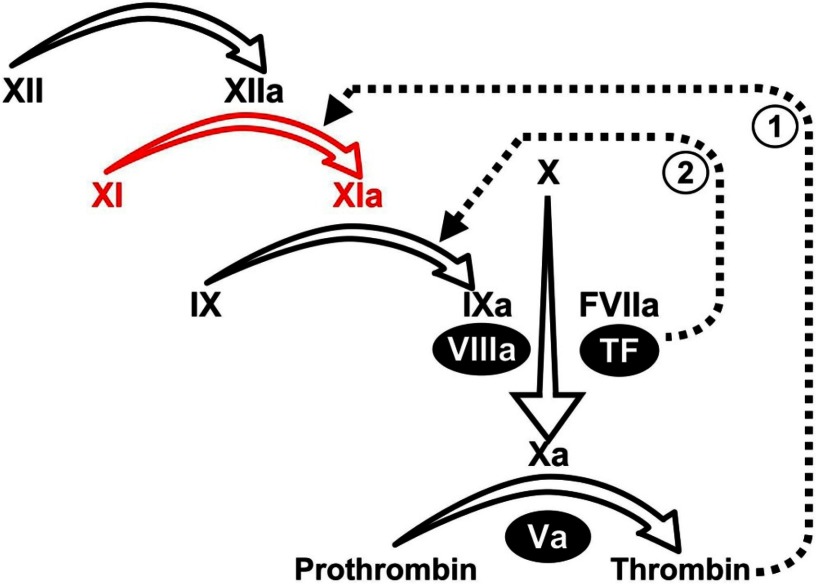
Figure 1.
Plasma coagulation protease reactions. Scheme for the cascade/waterfall model of thrombin generation triggered by activation of FXII. Conversion of FXI to FXIa is shown in red. A mechanism for initiation of coagulation by the FVIIa/TF complex through FX activation is also shown. Roman numerals indicate unactivated coagulation factors, and activated factors are indicated by “a.” Nonenzymatic cofactors are indicated by numerals in black ovals. The dotted line designated “1” indicates feedback activation of FXI by thrombin, whereas the dotted line indicated by “2” represents activation of FIX by FVIIa/TF.
In the cascade/waterfall models of coagulation that are the basis for the activated partial thromboplastin time (aPTT) assay, FXI activation by FXIIa initiates fibrin formation (Figure 1).1 However, newer schemes do not assign FXI a role in early fibrin generation, based largely on the observation that FXI deficiency causes relatively mild bleeding.1–3 FXIa is now postulated to be part of a feedback loop that sustains thrombin generation through FIX activation to consolidate coagulation (Figure 1).13–15 This appears to be particularly important in tissues with robust fibrinolytic activity, such as the oropharynx and urinary tract, which are common sites of bleeding in FXI-deficient patients.2,16–23
Congenital FXI deficiency is associated with a variable, mild to moderate bleeding disorder.2,16,17 Severe deficiency (< 15% of normal plasma activity) is prevalent in people of Jewish ancestry.2,16,17,24 The carrier rate is approximately 5% in Ashkenazi Jews, with severe (homozygous) deficiency found in 1 in 450 persons.16,24 Two mutations account for more than 90% of abnormal alleles in this population.24,25 The severe mutation Glu117Stop encodes a truncated protein,25 and homozygotes lack plasma FXI antigen. The more subtle missense mutation Phe283Leu causes a defect in FXI dimer formation.26–28 More than 180 other FXI gene mutations associated with FXI deficiency have been reported (http://www.factorxi.org, http://www.isth.org),29 including more than 100 single amino acid (missense) substitutions. Congenital deficiencies of vitamin K–dependent coagulation proteases are often associated with dysfunctional protein in plasma (cross-reactive material positive [CRM+] deficiency).30 In contrast, most cases of FXI deficiency are associated with low plasma levels of FXI protein (CRM− deficiency).31
There has been renewed interest in FXI as a result of population-based studies and work with animal models strongly indicating that this protein makes important contributions to arterial32–36 and venous37 thrombosis, ischemia-reperfusion injury in the central nervous system,38 and the pathology of sepsis.39 The observation that deficiency or inhibition of FXI interferes with platelet accumulation in growing thrombi33,35 has led to a reanalysis of the mechanisms involved in FXI activation and its interactions with platelets. In this review, we present a summary of recent structural data on FXI as it relates to current knowledge of protease function and to congenital FXI deficiency.
FXI structure
Crystal structures are available for full-length zymogen FXI,7 and the isolated active protease domain of FXIa in complex with natural and synthetic inhibitors.40–42 An NMR structure for the A4 domain dimer is also available.43 The apple domains of FXI and PK are members of the PAN (plasminogen, apple, nematode) module family, with homology to the N-terminal domains of hepatocyte growth factor and plasminogen.44 Each apple domain consists of 7 β-strands that fold into a curved antiparallel sheet cradling an α-helix (Figure 2A).7 Two disulfide bonds lock the helix onto the central β4 and β5 strands, whereas a third connects the N- and C-termini of the domain. This core topology is equivalent to PAN domains from hepatocyte growth factor,44 leech antiplatelet protein,45 and microneme antigen EtMIC5.46 The β4-β5 loop and β5-β6 crossover loop generate a small pocket on the opposite side of the sheet from the α-helix (Figure 2A).
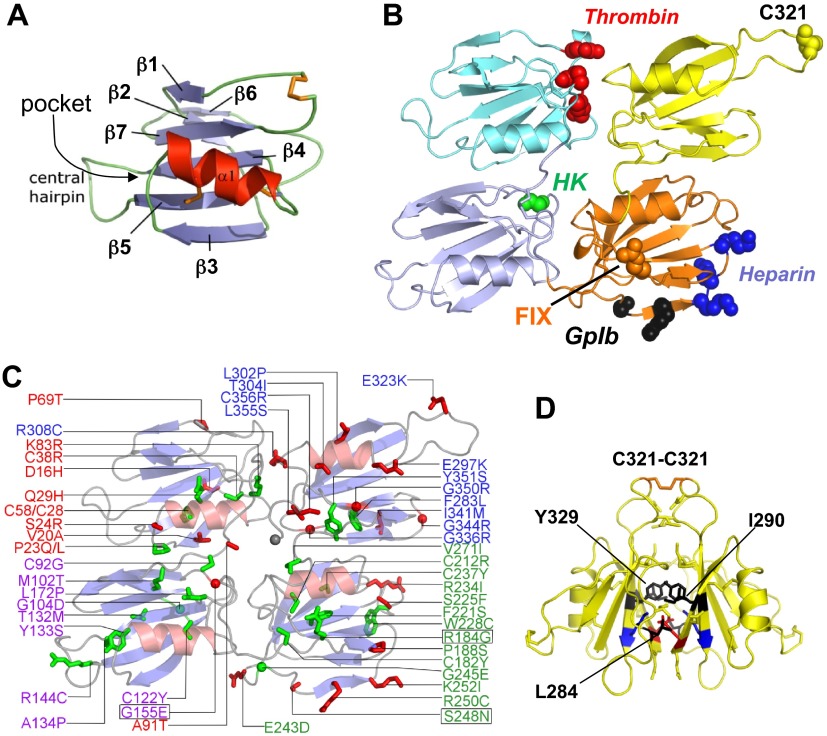
Figure 2.
Human FXI apple domains. Topology diagrams based on the FXI zymogen structure. For all diagrams, the first, second, third, and fourth apple domains (A1, A2, A3, and A4) are shown in cyan, light blue, orange, and yellow, respectively. (A) The apple domain consists of a 7-stranded β-sheet (blue) with a central α-helix positioned on top (red). Disulfide bonds are shown in orange. (B) Topology diagram showing the disk formed by the 4 apple domains, with the catalytic domain removed. Sites of residues implicated in ligand binding are red for thrombin, green for high-molecular-weight kininogen (HK), black for GPIb, blue for heparin, and orange for FIX. (C) Apple domain disk showing the positions of mutations identified in FXI-deficient patients that involve buried residues (green) or surface exposed residues (red). Mutations Gly155Glu, Arg184Gly, and Ser248Asn (boxed) are examples of rare CRM+ mutations. (D) Two A4 domains forming the FXI dimer interface. The Cys321 interchain disulfide bond is shown at the top in orange. Hydrophobic residues Leu284, Ile290, and Tyr329 are shown in black, and a salt bridge is formed between Lys331 (blue) and Glu287 (red).
A remarkable feature of the FXI structure, which it probably shares with PK,10 is the intimate linkage of the apple domains into a disk-like platform around the base of the catalytic domain (Figure 2B).7,10 The disk has 2 sets of topologically equivalent interfaces. The side interfaces between A1 and A2 and between A3 and A4 are substantial, burying surface areas of 441 Å2 and 444 Å2, respectively. In comparison, the end interfaces between A1 and A4 (380 Å2) and between A2 and A3 (284 Å2) are smaller. The FXI subunit, therefore, is compact compared with the vitamin K–dependent proteases, whose elongated structures serve to position the catalytic domains above phospholipid membranes.1 Amino acid substitutions in the apple domains of FXI-deficient patients are shown in Figure 2C. With few exceptions, these mutations involve residues that are conserved in FXI, PK, and their ancestral predecessor (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Many involve buried residues (green in Figure 2C) that probably affect apple domain folding or disrupt formation of interdomain interfaces,29 which would explain why the majority of cases of FXI deficiency are CRM−.
FXI dimer
The 2 A4 domains form a substantial interface (886 Å2) between the dimer subunits, with the β-sheets packing against each other (Figure 2D).7 Cys321, which is located in a finger-like loop in A4, projects away from the body of the domain and forms the interchain disulfide bond (Figure 2B,D). However, FXI with substitutions for Cys321 still form stable dimers.28,47–51 Leu284, Ile290, and Tyr329, which form the hydrophobic core of the A4 domain interface, are required for dimer formation,7,50,51 as are salt bridges between Lys331 of one subunit and Glu287 of the other.7,48 The Cys321-Cys321 interchain bond forms poorly in FXI with a single alanine substitution of Leu284, Ile290, or Tyr329,51 indicating that the interface between A4 β-sheets forms before the disulfide bond. A number of A4 domain mutations associated with FXI deficiency interfere, or are predicted to interfere, with dimerization. The best characterized are Phe283Leu,26–28 which causes a partial dimerization defect, and Gly350Glu,27 which prevents dimer formation. Riley et al showed that the Phe283Leu substitution causes increased dimer dissociation and stabilizes the monomer through altered side-chain packing that is unfavorable for dimer formation.28
The dimeric structure of FXI, unique among coagulation proteases, has implications for inheritance of FXI deficiency. Although Glu117Stop and Phe283Leu are associated with a bleeding diathesis inherited as an autosomal recessive disorder (mutations in both alleles required for symptomatic deficiency),2,27 significant deficiency can occur in heterozygotes for some types of mutations, as follows. An amino acid substitution could prevent protein secretion (CRM− mutation) but not interfere with intracellular dimer formation. A FXI subunit containing such a mutation would exert a dominant-negative effect on normal FXI subunits by forming nonsecretable heterodimers with 1 normal and 1 abnormal subunit. The mutant subunit, in effect, traps wild-type subunits within the cell. This mechanism has been demonstrated in deficiencies of multimeric proteins, such as von Willebrand factor52 and fibrinogen.53 The FXI missense mutations Ser225Phe, Cys398Tyr, Gly400Val, and Trp569Ser exhibit a dominant negative effect when coexpressed with wild-type FXI and are associated with FXI deficiency that may be inherited in a dominant manner.27,54 It is probable that these mutations do not perturb apple domain disk structure significantly, allowing dimerization to occur. In this regard, it is interesting to note that 3 of 4 mutations known to operate by a dominant negative mechanism involve residues in the catalytic domain.
The structure of the FXI dimer is shown in Figure 3 with highlights on residues implicated in zymogen activation, substrate recognition, and receptor binding (also shown in Figure 2B). Many of the complex protein-protein interactions involving FXI and FXIa are mediated by its apple domains. Binding sites for HK,10,55,56 thrombin,7,57 heparin,58–61 FIX,62,63 and platelet receptors, such as glycoprotein Ib (GPIb),64 will be discussed in the following sections.
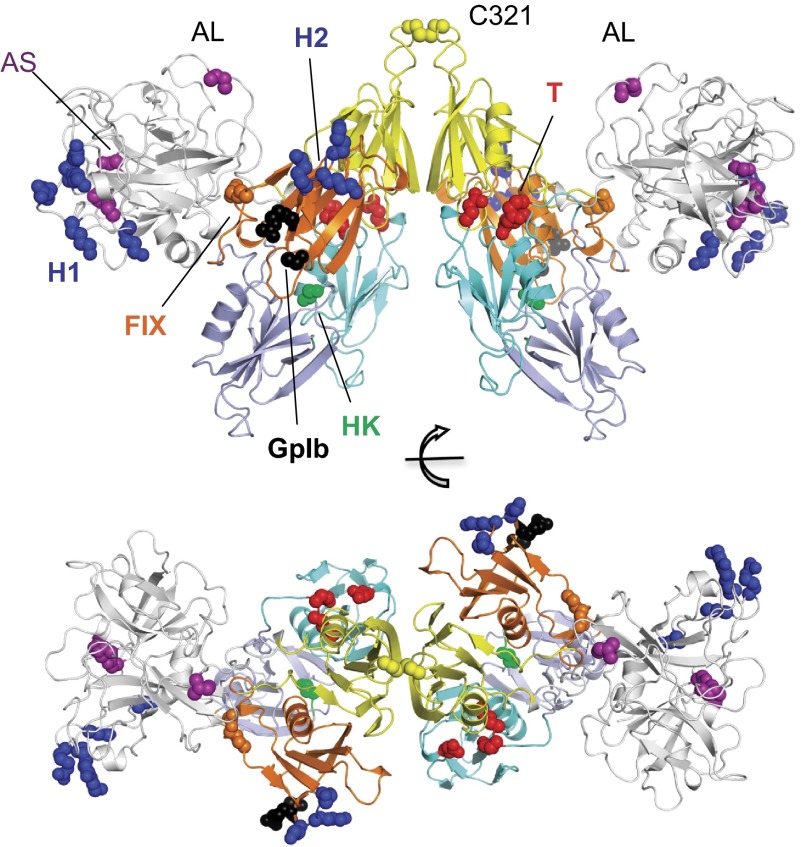
Figure 3.
The structure of zymogen FXI. Topology diagram of dimeric FXI viewed from 2 perspectives rotated 90 degrees. The catalytic domain is in white. Sites of residues implicated in ligand binding are red for thrombin (T), green for HK, black for GPIb, blue for heparin sites (H1 and H2), and orange for FIX. Positions for the activation loop (AL) cleavage site (Arg360-Ile370) residue Ile370 and active site (AS) residues Ser557, Asp462, and His413 are shown in purple.
FXI and PK binding to HK
HK is a 120-kDa multifunctional plasma protein with 6 domains, designated D1 to D6.65 Almost all FXI8 and 75% to 90% of PK11,66 circulate in a complex with HK. The D6 domain mediates binding to both proteins.65 The physiologic importance of the PK-HK interaction seems clear as cleavage of HK by α-kallikrein (activated PK) liberates the vasoactive peptide bradykinin.65 The importance of the FXI-HK interaction is not as evident, as HK deficiency is not associated with a bleeding disorder.30 In the presence of zinc ions, HK is required for optimal FXI binding to GP1b on activated platelets in suspension, enhancing the binding stoichiometry by approximately 2-fold.67–69 However, FXI binding to platelets assayed under flow (discussed in “FXI interactions with platelet receptors”) is not enhanced by HK.70
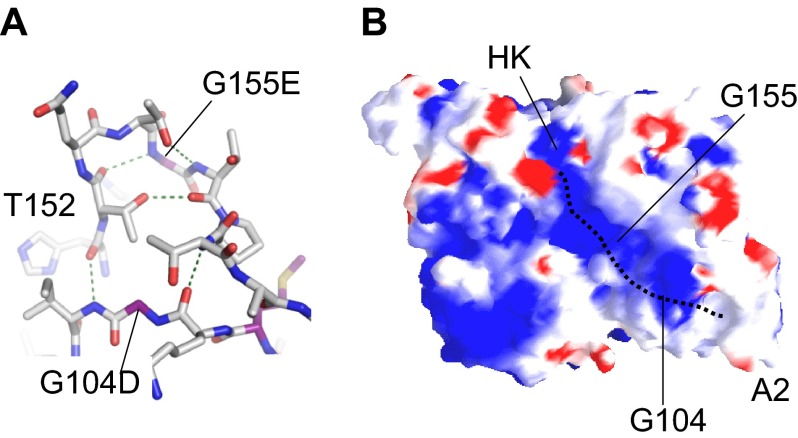
The sites on FXI and PK that bind HK are probably similar. Studies using FXI/PK chimeras, individual apple domains, and short peptide sequences derived from apple domains indicate the A2 domain is required for HK binding, with additional contributions from A1 and A4.55,56,71 Detailed mutagenesis to localize the binding site has not been performed; however, analysis of the natural FXI mutation Gly155Glu provides some information. Gly155Glu is a rare example of a mutation causing CRM+ FXI deficiency.72,73 Gly155 is located at the center of a loop connecting the β5 and β6 strands of A2. The Gly155 main chain nitrogen forms a hydrogen bond with the carbonyl group of Thr152 in a type I β-turn (Figure 4A). Gly155 points toward the center of a cavity in the apple domain disk and lies in a positively charged channel on the surface of A2 pointing away from the protease domain (Figure 4B).10 Recombinant FXI-Glu155 binds HK with approximately 10-fold lower affinity than wild-type FXI in a solid phase assay (D.G., unpublished observation, June 2008). The loop containing Gly155 forms hydrogen bonds with a second loop spanning residues 103 to 105 (Figure 4A) that contains the FXI mutation Gly104Asp and borders a pocket in the A2 domain at the end of the charged channel (Figure 4B). A Gly104Arg mutation in the PK A2 domain associated with CRM+ PK deficiency also causes decreased HK binding.74
Figure 4.
FXI-binding site for HK. (A) The relative positions of Gly104 and Gly155 in the A2 domain are shown. Gly155 is involved in a tight β-turn forming a hydrogen bond with Thr152. The compact hydrogen bond network extends through Thr152 and Thr158 to form contacts to the main chain through residues Lys103 and Ile105. Changes at Gly104 or Gly155 will probably disrupt this network. (B) Charged surface (blue represents positive; red, negative) representation of the underside of the FXI apple domain disk, showing positions of Gly104 and Gly155. These residues are required for proper formation of a charged channel that is a probable binding site for HK. The dotted line represents the predicted binding site of HK that terminates in a pocket at the base of the A2 domain.
FXI zymogen activation
In vitro, known plasma activators of FXI include FXIIa,4 α-thrombin,13,14,75 the prothrombin activation intermediate meizothrombin,76 and FXIa (autoactivation),13,15 all of which cleave FXI at the Arg369-Ile370 bond.5,6 Although FXIIa activates FXI in the cascade/waterfall model (Figure 1), the absence of a bleeding disorder in FXII-deficient patients indicates that alternative mechanisms for FXI activation exist. This is demonstrated clearly by the work of Spronk et al using mice lacking FIX, FXI, or FXII on a background of low tissue factor (TF) expression.77 Low TF mice have a severe bleeding disorder but are viable.78 Superimposing FIX or FXI deficiency on the low TF background results in death in utero from bleeding, implying FIX activation by FXIa is required for viability in the setting of a hemostatic system crippled by low TF levels. Mice with deficiency of FXII and low TF, on the other hand, are viable and phenotypically similar to low TF mice with normal FXII expression. Although these data confirm the impression that FXIIa is not required for FXI activation, they should not be interpreted as indicating that FXIIa does not activate FXI in vivo. Indeed, FXIIa-mediated activation of FXI appears to play a central role in formation of pathologic thrombi in murine thrombosis models.33,38
Naito and Fujikawa13 and von dem Borne et al15 were the first to present results indicating that FXI is activated by thrombin in plasma. In purified protein systems, FXI activation by thrombin is enhanced by charged substances, such as dextran sulfate or heparin,13,14 but the relevance to activation in vivo is not clear. Thrombin interacts with most of its substrates through 2 electropositive anion-binding exosites (ABE I and ABE II).79 Using a panel of thrombins with site-specific mutations, Yun et al showed that ABE I and ABE II are required for efficient FXI activation in the presence of dextran sulfate.80 Here ABE II interacts with dextran sulfate, whereas ABE I probably binds FXI. Recent experiments in plasma, however, did not identify a role for ABE I in FXI activation, as wild-type thrombin and thrombin with mutations in ABE I initiated FXI-dependent thrombin generation similarly.81
Available information indicates that thrombin binds to the A1 domain of FXI, with residues Glu66, Lys83, and Gln84 forming part of the binding site.7,57,75 These residues cluster near the interface with the A4 domain (Figures 2B, 3A) and are suitably positioned in proximity to the activation loop containing the Arg369-Ile370 cleavage site. The FXI residues involved in binding FXIIa have not been definitively determined, and the interaction may extend beyond a single apple domain surface. For example, although studies using peptide mimicry point to a binding site for FXIIa on the A4 domain,82 a monoclonal antibody that recognizes A2 selectively interferes with FXI activation by FXIIa.76,81
A role for the FXI dimer in promoting zymogen activation by thrombin in trans has been proposed.7 Here the activating protease would bind one subunit of the dimer and cleave Arg369-Ile370 on the other. Consistent with this, Wu et al used a series of recombinant monomeric FXI proteins in a purified system to show that the dimer is required for optimal FXI activation by thrombin, FXIIa, and FXIa.50 Some of these studies used dextran sulfate to facilitate FXI activation, which may not suitably model the physiologic environment. A second study with similar FXI monomers gave somewhat contradictory results, with monomers proving more susceptible than dimers to spontaneous autoactivation by FXIa, but here experiments were conducted in conditioned media (unpurified system).51
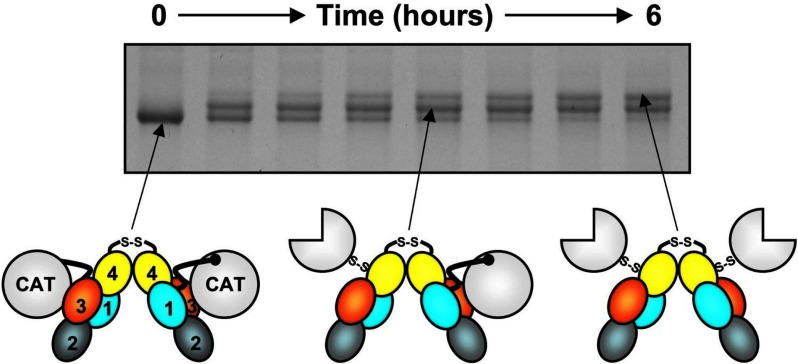
The term FXIa describes the protease form of FXI that has been cleaved at the Arg369-Ile370 peptide bonds on both subunits of the dimer. Recently, FXI activation by thrombin or FXIIa was shown to proceed through an intermediate with 1 activated subunit, referred to as 1/2-FXIa.3,83 The intermediate can be observed on nonreducing sodium dodecyl sulfate-polyacryalmide gels because FXI, 1/2-FXIa, and FXIa migrate at slightly different rates (Figure 5). 1/2-FXIa can be detected in plasma induced to undergo contact activation.83 Given that conversion of FXI to fully activated FXIa is a slow process, 1/2-FXIa may be a major species of activated FXI. The functional properties of this protease are discussed further in “Recognition and cleavage of FIX by FXIa.”
Figure 5.
FXI activation. Each FXI subunit is activated by cleavage between Arg369 and Ile370. FXI (0-hour time point) migrates slightly faster than FXIa (top band, 6 hours) on nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Activation of FXI (schematic at left) by α-thrombin (shown) or FXIIa proceeds through an intermediate with 1 cleaved subunit (center), designated 1/2-FXIa. Subsequent conversion of 1/2-FXIa to fully activated FXIa (right) appears to be a slower process. In the diagrams, the A1, A2, A3, and A4 domains are shown in light blue, cyan, orange, and yellow, respectively, and the catalytic domains (CAT) are in white. Circles represent unactivated catalytic domains; three-fourths circles, activated catalytic domains. In these diagrams, the catalytic domain moves relative to the apple domain disk after cleavage of Arg369-Ile370, exposing a surface on A3 thought to contain a FIX-binding site.
FXIa protease domain structure and interactions with inhibitors
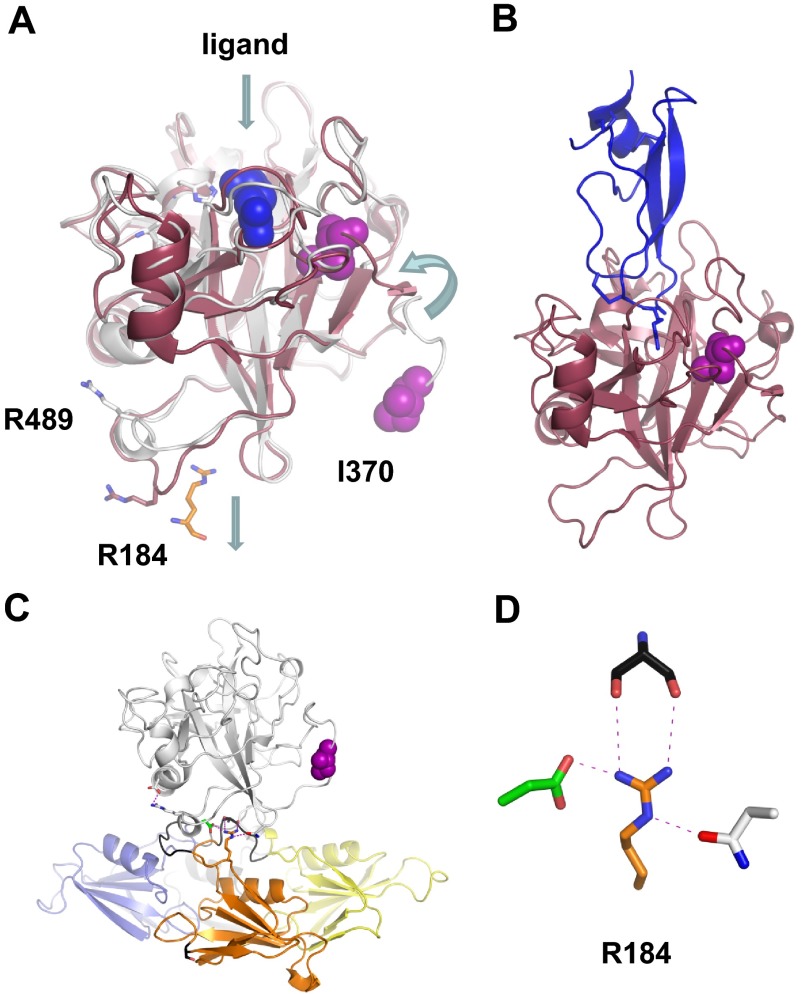
There are no structures available for full-length FXIa or 1/2-FXIa, but structures for isolated FXIa catalytic domain in complex with inhibitors have been reported.40–42 Comparison with the zymogen structure reveals conformational changes expected with activation of a serine protease, including a 12Å movement of Ile370 (the N-terminus of the catalytic domain, Figure 6A) into the protease domain core. Trypsin-like proteases, including FXIa, cleave their substrates after basic amino acids (arginine in the case of FXIa) that are designated the P1 residue. The site on the protease that interacts with the substrate P1 residue is the S1 site. Changes in the catalytic domain that occur during FXI activation result in a salt bridge forming between the Ile370 N-terminal amine and the Asp554 side chain. This allows formation of the S1 site that will be occupied by the side chain of the P1 arginine from a substrate or inhibitor. A distinct conformational change is also observed in loops on the side of the catalytic domain opposite the active site cleft. This involves unraveling of a short α-helix in the zymogen and a closing together of the loop containing Arg489 and a β-hairpin to fill a pocket left empty by removal of the A3 domain, which forms an interface with the base of the catalytic domain in the zymogen (Figure 6A,C). The potential importance of this conformational change to substrate recognition is discussed in “Recognition and cleavage of FIX by FXIa.”
Figure 6.
The FXIa protease domain. (A) Topologic diagram showing the superposition of FXI (white) and FXIa (red) protease domain crystal structures. Conformational changes include a 12 Å shift in the position of Ile370 into the protease core after cleavage of the Arg369-Ile370 bond, and an unraveling of an α-helix containing Arg489 to fill a pocket left empty by removal of the A3 domain and Arg184. The Arg side chain from the FXIa ligand in panel B is shown in blue. (B) Structure of FXIa in complex with PN2-KPI domain inhibitor (blue). (C) The side chain of Arg184 is buried in the FXI zymogen through contacts with the protease domain. This residue is thought to act as a switch, which is released on zymogen activation to engage substrate FIX. (D) Stick diagram showing the side chain of Arg184 that forms 3 noncovalent interactions with the side chains of Ser268 (gray) from the A3 domain, and Asp488 (green) and Asn566 (white) from the protease domain in the FXI zymogen structure.
Activated FXIa is subject to negative regulation by plasma serpins, including antithrombin, C1-inhibitor, protease nexin 1, and protein Z–dependent protease inhibitor.59–61,84–87 Basic amino acids in the autolysis loop of the catalytic domain (Arg504, Lys505, Arg507, and Lys509) are determinants of serpin specificity.87 Heparin enhances serpin-mediated FXIa inhibition through a mechanism involving heparin-binding sites on the A3 (Lys252, Lys253, and Lys 255; Figures 2B, 3)58,59 and catalytic (Lys529, Arg530, Arg532, Lys536, and Lys540)61,62 domains (Figure 3). The A3-binding site facilitates inhibition through a template mechanism in which both protease and serpin bind to heparin.61 In contrast, heparin binding to the catalytic domain facilitates inhibition in a manner not completely explained by a template mechanism, and probably involves charge neutralization or an allosteric effect that overcomes repulsive interactions between the serpin and catalytic domain.61
FXIa is also regulated by protease nexin 2 (PN2),41,88,89 which is released from activated platelets.88 FXIa inhibition requires the Kunitz-type protease inhibitor (KPI) domain of PN2. A crystal structure of the FXIa catalytic domain complex with PN2-KPI shows that the inhibitor has a characteristic disulfide-stabilized double-loop structure that fits into the FXIa active site, with the P1 arginine occupying the S1-binding pocket (Figure 6B).41
Recognition and cleavage of FIX by FXIa
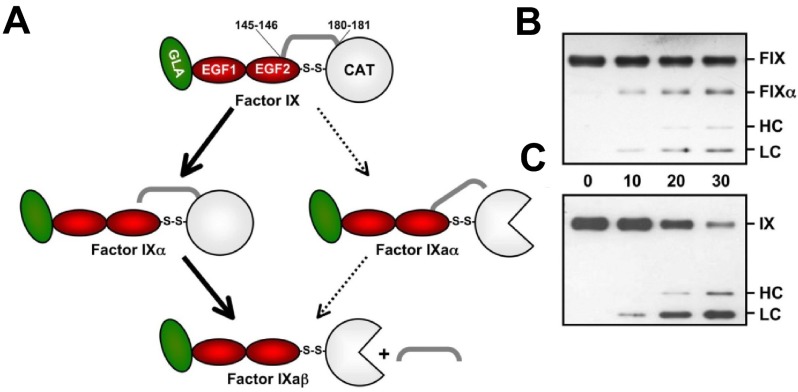
FXIa's major substrate, FIX, has an elongated structure that includes (from the N-terminus) a calcium-binding Gla domain, 2 EGF repeats, a 55-amino acid activation peptide, and a C-terminal serine protease domain (Figure 7A).90 Conversion of FIX to the protease FIXaβ requires cleavages of the Arg145-Ala146 and Arg180-Val181 bonds releasing the activation peptide. When FIX is activated by the factor VIIa (FVIIa)/TF complex (Figure 1), there is preference for initial cleavage after Arg145, resulting in accumulation of the intermediate FIXα, followed by cleavage after Arg180 (Figure 7B).91–93 In contrast, relatively little or no FIXα accumulates during FIX activation by FXIa (Figure 7C).50,94–96 More than 1 mechanism is compatible with this observation. Cleavage of the FIX activation sites by FXIa could be sequential, with the rate of the second cleavage greater than the first. Indeed, FXIa, like FVIIa/TF, cleaves FIX initially at the Arg145-Ala146 bond.83,96 Alternatively, the protease may cleave sequentially but not release the intermediate before the second cleavage. This is consistent with data showing that cleavage rates of FIX and FIXα by FXIa are roughly similar (ie, Arg180-Val181 is not cleaved more rapidly than Arg145-Ala146).95 The possibility that FXIa requires its 2 catalytic domains to cleave the FIX activation sites simultaneously has been refuted, as 1/2-FXIa83 and monomeric variants of FXIa50 activate FIX similarly to dimeric FXIa.
Figure 7.
FIX activation. (A) FIX contains (from the N-terminus) a Gla domain (green), 2 epidermal growth factor (EGF) domains (red), an activation peptide (AP; gray bar), and a catalytic (CAT) domain (white). FIX is converted to FIXaβ by cleavage after Arg145 and Arg180, releasing the AP. FVIIa/TF initially cleaves FIX after Arg145 to form the intermediate FIXα, with subsequent cleavage after Arg180 to form FIXaβ (bold arrows). White three-fourths circle represents the active catalytic domain of FIXaβ. During this reaction, FIXα accumulates before formation of FIXaβ. FXIa also cleaves FIX initially after Arg145; however, no FIXα accumulates. Initial cleavage of zymogen FIX after Arg180 to generate the intermediate FIXaα appears to be a minor reaction (thin arrows). (B-C) Reducing Western blots of FIX (100nM) activated by 1nM FVIIa/TF (B) or FXIa (C) over 30 minutes (time indicated between panels). Markers at the right indicate migration of standards for zymogen FIX (FIX), the large fragment of FIXα (FIXα), the heavy chain (HC) of FIXaβ, and the light chain (LC) of FIXα or FIXaβ. No FIXα accumulates during activation by FXIa.
Initial recognition of FIX involves exosites on FXIa that are distinct from the catalytic active site.96–98 FIX does not bind FXI,99 indicating that conformational changes during conversion of FXI to FXIa expose the exosites. An antibody that recognizes an epitope on the FXIa apple domain disk competitively inhibited FIX activation by FXIa.98 Subsequent work with recombinant proteins localized a FIX-binding site to the A3 domain (Figures 2B, 3).62,63 A critical residue for FIX activation is Arg184,63 located near the N-terminus of A3. An Arg184Gly mutation was recently reported in a CRM+ FXI-deficient patient, and recombinant FXIa with this mutation activates FIX poorly.100 In the zymogen structure, Arg184 occupies a central location in a loop connecting A2 and A3 that is buried under the protease domain (Figure 6A,C). The long Arg side chain extends such that the guanidinium group fills the small pocket formed by the interface between the apple domains and catalytic domain. Here, Arg184 interacts with 3 residues: 2 from the catalytic domain (Asp488, Asn566) and 1 from the A3 domain (Ser268; Figure 6D).
Arg184 is conserved in FXI across species12 but is not present in PK, consistent with its role in interactions with FIX. The Arg184 side chain must be exposed in FXIa by conformational changes that occur after cleavage of the Arg369-Ile370 bond in the activation loop. Full-length FXI and FXIa have been analyzed by small-angle x-ray scattering and electron microscopy, which revealed a change in the shape of the molecule on activation, consistent with a shift in the position of the apple domain disk relative to the catalytic domain.43 This could break interactions between Arg184 and Asp488, Asn566, and Ser268 (Figure 6D). Arg184 may, therefore, be part of a switch that holds FXI in an inactive conformation in the zymogen and, when the switch is thrown, facilitates engagement of FIX.
It appears that residues in the FIX Gla domain are required for binding to FXIa,99 explaining the enhancing effect of calcium ions on the reaction.90 There is also evidence that it is the Gla domain that binds to the A3 domain.101 Interestingly, loss of the A3 exosite has a more deleterious effect on cleavage of FIX at Arg180-Val181 than at Arg145-Ala146.83,96 This suggests that the A3 exosite primarily facilitates cleavage of FIXα to FIXaβ96 and that a distinct exosite is involved in the initial interaction with FIX that facilitates conversion to FIXα. Recent work by Sinha et al indicates that this exosite is located on the FXIa catalytic domain.96 Interestingly, unlike binding to the A3 exosite, the interaction with the catalytic domain exosite does not required calcium, suggesting that the interaction does not involve the FIX Gla domain.
FXI interactions with platelet receptors
Although platelets do not appear to enhance FXI activation in a static system,75 there is growing evidence that platelets affect FXI and FXIa behavior under flow.35,70 When human blood lacking FXII activity is perfused over collagen at arterial shear rates, platelets deposit in aggregates and fibrin strands form.70 Addition of an antibody that blocks FXIa activation of FIX prevents fibrin formation. Interestingly, adding inhibitors of FVIIa/TF does not affect fibrin formation, suggesting that platelets may play a role in FXI activation. Platelets may also localize FXIa to the site of fibrin formation, as fibrin does not form when blood lacking platelets is perfused over collagen.
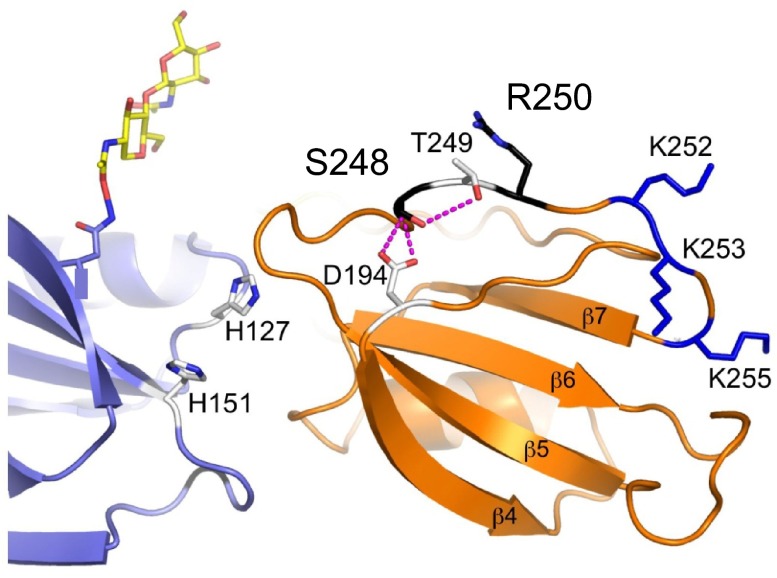
FXI lacks the Gla domain that facilitates binding of vitamin K-dependent coagulation proteases to phospholipid surfaces.5–7 Yet, there is solid evidence that FXI binds to platelets. Greengard et al measured approximately 1500 binding sites for FXI per activated platelet.67 Subsequent work determined that platelet GP1b mediates binding and that FXI competes with von Willebrand factor, but not thrombin, for binding to GP1b.102 Optimal binding of FXI to platelets requires residues in the FXI A3 domain (Figures 2B, 3),64,68,69 the N-terminal domain of the GP1bα chain,102 HK, and zinc ions.67–69 The A3 residues involved, Ser248 and Arg250, are near to, but distinct from, the A3 heparin-binding site.58,59 A CRM+ missense mutation in the A3 domain, Ser248Asn, was identified in a family with a bleeding disorder.103 FXI-Asn248 binds platelets with 5-fold reduced affinity compared with wild-type FXI,103 and other Ser248 substitutions (Gln or Ala) showed similar defects.104 Figure 8 shows the local environment around Ser248. The overall structure of the A3 domain is probably not affected significantly by the Ser248Asn mutation, as FXI-Asn248 is (1) secreted, (2) activated by FXIIa, and (3) activates FIX similar to wild-type FXIa. Cumulatively, the findings make it probable that Ser248 and Arg250 are central to a site for interactions with platelet GPIb.
Figure 8.
Platelet-binding site on FXI. The locations of 2 residues (Ser248 and Arg250 in black), that probably form a GPIb platelet-binding site on the FXI A3 domain (orange), are shown relative to residues that form the heparin-binding site (Ly252, Lys253, and Lys255 in blue). Ser248 forms hydrogen bonds with Asp194 and Thr249, which are probably disrupted in the hereditary FXI mutation Ser248Gln. The adjacent A2 domain is shown in light blue. The position of an N-linked glycan moiety attached to residue Asn108 of the A2 domain is shown in yellow and red.
FXIa also binds to platelets (∼ 250 sites per platelet) but does so by a mechanism that appears to differ from that of FXI.105 FXIa binding to platelets does not require the A3 domain or HK, and involves residues in the catalytic domain between Cys527 and Cys542 in the vicinity of the heparin-binding site (Figure 3).106 The binding site on platelets for FXIa has not been identified but appears to be distinct from that of FXI, as FXI does not compete with FXIa for binding.105
The observation that each platelet contains 1500 FXI-binding sites was somewhat perplexing, considering that there are approximately 25 000 copies of GP1b per platelet.107 New data from White-Adams et al may offer a solution to this conundrum; this group reported that FXI binds to a platelet receptor for apolipoprotein E called ApoER2′ or LRP8.70 FXI binds to soluble ApoER2′ with an affinity of approximately 60nM. Human and murine platelets bind and spread on glass coated with FXI or fibrinogen. Binding to FXI, but not fibrinogen, was abrogated by receptor-associated protein, which blocks ligand binding to members of the low-density lipoprotein receptor family. Platelet binding to FXI was also blocked by soluble ApoER2′, and platelets from ApoER2-deficient mice did not bind FXI. Under conditions of physiologic shear, soluble ApoER2′ or its low-density lipoprotein-binding domain prevented platelet binding to immobilized FXI.
There are an estimated 2000 copies of ApoER2′ per platelet, which agrees well with the number of FXI-binding sites.67 Furthermore, ApoER2′ colocalizes with GP1b on platelets,108 suggesting that FXI binding involves both receptors. Consistent with this, antibodies to GP1b block platelet binding to immobilized FXI under physiologic shear. It is not clear whether the platelet-binding site on the FXI A3 domain is required for interactions with ApoER2′ or whether a different site is involved.
Future directions
Several important aspects of FXI pathophysiology require additional study. Over the past 50 years, many investigators have commented on the weak correlation between bleeding and FXI activity measured by conventional assays. This is reflected in 2 studies involving 49 FXI-deficient kindreds (242 persons), which determined that severity of bleeding was similar in patients with severe and mild deficiency.21,22 In contrast, work with patients primarily with the Glu117Stop and/or Phe283Leu mutations showed bleeding risk to be much greater in severe than mild deficiency (odds ratio = 13.0 and 2.6, respectively).109 Diverse factors could be contributing to this disagreement, including differences in clinical criteria for bleeding, variation in genetic backgrounds or FXI mutations, or coinheritance of other bleeding disorders. That being said, some patients with severe FXI deficiency do not bleed abnormally, even with trauma to tissues that typically cause problems for FXI-deficient patients. We suspect that the manner in which we measure FXI activity in clinical laboratories does not accurately assess the protein's contribution to hemostasis.
FXI-deficient plasma exhibits a prolonged aPTT clotting time, and FXI deficiency is currently defined as low activity in an aPTT-based assay in which patient and FXI-deficient plasma are mixed. However, the aPTT only requires FXI to function as a component of contact activation initiated fibrin formation, and does not assess FXI functions such as feedback activation by thrombin, inhibition of fibrinolysis, and interactions with platelets. The FXI Ser248Asn variant illustrates the limitations of the aPTT in assessing FXI function. This mutation is associated with bleeding and defective FXI binding to platelets but does not affect the aPTT assay, which does not contain platelets.103 Assays that capture the importance of FXI to sustained (rather than initial) thrombin generation, probably in platelet-rich plasma under flow, will be required to address the deficiencies of the aPTT. Allen et al made the interesting observation that the contribution of FXI to thrombin generation varied substantially when platelets from different normal donors were tested in a cell-based model of TF-initiated coagulation.110 Platelets, therefore, may turn out to be key for accurately assessing FXI function.
As presented in this review, FXI has features that distinguish it from vitamin K–dependent coagulation proteases, making it difficult to extrapolate from the lessons learned from the large body of work on these enzymes to predict structure-function relationships for FXI. There remain many important unanswered questions regarding FXI and FXIa from a structural and enzymologic perspective. Areas of active investigation include determining the functional significance of the dimeric structure of FXI/FXIa, elucidating the manner in which FXI/FXIa engages platelets, and identifying the functional consequences of platelet binding.
New evidence indicates that the dimeric structure of FXI facilitates its activation by FXIIa and thrombin.50 However, the FXI homolog PK (a monomer) is activated rapidly by FXIIa, as is a monomeric FXI chimera containing the PK A4 domain.62 It will be important to determine whether dimerization is critical for FXI activation under physiologic conditions, probably using flow-based systems, or perhaps in vivo.
It has been proposed that one subunit of a FXIa dimer could bind to a platelet receptor, leaving the other free to interact with the substrate FIX.105 An intriguing possibility that remains to be tested is that 1/2-FXIa binds to GP1b, or perhaps ApoER2, through the A3 domain of its unactivated subunit while binding to FIX through A3 on the activated subunit. Note that in Figure 3B the A3 domains point in opposite directions from the longitudinal plane of the molecule. As FXI lacks a Gla domain, the dimeric structure may represent an alternative solution to the problem of tethering the protease to a surface at a wound site while allowing interaction with its substrate. Such a hypothesis is also best tested in flow-based systems, where the FXI-platelet interaction is probably important to protease function.
Recent studies associating FXIa activity and pathologic coagulation in humans and animal models have stimulated considerable interest in this protease as a target for clinical intervention, with potential advantages over targeting thrombin or FXa in terms of side effects and therapeutic window. Work on FXI structure is at an early stage, compared with the vitamin K-dependent coagulation factors, with crystal structural data becoming available only within the past 5 years. Structures defining full-length FXIa and ligand complexes with substrate, receptors, and activating enzymes will be required to completely understand the molecular basis of processes such as zymogen activation, platelet binding, and substrate cleavage. This will be critical to understanding the role of this protease in disease processes and to the development of specific inhibitors for clinical purposes.
Acknowledgments
This work was supported by the British Heart Foundation (grants PG/04/129/18095 and RG/07/002/23132; J.E.) and the National Heart, Lung, and Blood Institute (awards HL58837 and HL81326; D.G.).
Footnotes
The online version of this article contains a data supplement.
Authorship
Contribution: J.E., P.A.M., and D.G. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonas Emsley, Centre for Biomolecular Sciences, School of Pharmacy, University of Nottingham, Nottingham, NG7 2RD, United Kingdom; e-mail: jonas.emsley@nottingham.ac.uk; or David Gailani, Hematology/Oncology Division, Vanderbilt University School of Medicine, Nashville, TN 37232; e-mail: dave.gailani@vanderbilt.edu.
References
- 1.Furie B, Furie BC. Molecular basis of blood coagulation. In: Hoffman H, Benz EJ, Shattil SJ, et al., editors. Hematology, Basic Principles and Practice. 5th ed. Philadelphia, PA: Churchill Livingstone-Elsevier; 2009. pp. 1819–1836. [Google Scholar]
- 2.Seligsohn U. Factor XI in haemostasis and thrombosis: past, present and future. Thromb Haemost. 2007;98(1):84–89. [PubMed] [Google Scholar]
- 3.Gailani D, Smith SB. Structural and functional features of factor XI. J Thromb Haemost. 2009;7(suppl 1):75–78. doi: 10.1111/j.1538-7836.2009.03414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouma BN, Griffin JH. Human blood coagulation factor XI: purification, properties, and mechanism of activation by activated factor XII. J Biol Chem. 1977;252(18):6432–6437. [PubMed] [Google Scholar]
- 5.Fujikawa K, Chung DW, Hendrickson LE, Davie EW. Amino acid sequence of human factor XI, a blood coagulation factor with four tandem repeats that are highly homologous with plasma prekallikrein. Biochemistry. 1986;25(9):2417–2424. doi: 10.1021/bi00357a018. [DOI] [PubMed] [Google Scholar]
- 6.McMullen BA, Fujikawa K, Davie EW. Location of the disulfide bonds in human coagulation factor XI: the presence of tandem apple domains. Biochemistry. 1991;30(8):2056–2060. doi: 10.1021/bi00222a008. [DOI] [PubMed] [Google Scholar]
- 7.Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006;13(6):557–558. doi: 10.1038/nsmb1095. [DOI] [PubMed] [Google Scholar]
- 8.Thompson RE, Mandle R, Jr, Kaplan AP. Association of factor XI and high molecular weight kininogen in human plasma. J Clin Invest. 1977;60(6):1376–1380. doi: 10.1172/JCI108898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMullen BA, Fujikawa K, Davie EW. Location of the disulfide bonds in human plasma prekallikrein: the presence of four novel apple domains in the amino-terminal portion of the molecule. Biochemistry. 1991;30(8):2050–2056. doi: 10.1021/bi00222a007. [DOI] [PubMed] [Google Scholar]
- 10.Hooley E, McEwan PA, Emsley J. Molecular modeling of the prekallikrein structure provides insights into high-molecular-weight kininogen binding and zymogen activation. J Thromb Haemost. 2007;5(12):2461–2466. doi: 10.1111/j.1538-7836.2007.02792.x. [DOI] [PubMed] [Google Scholar]
- 11.Mandle RJ, Colman RW, Kaplan AP. Identification of prekallikrein and high-molecular-weight kininogen as a complex in human plasma. Proc Natl Acad Sci U S A. 1976;73(11):4179–4183. doi: 10.1073/pnas.73.11.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponczek MB, Gailani D, Doolittle RF. Evolution of the contact phase of vertebrate blood coagulation. J Thromb Haemost. 2008;6(11):1876–1883. doi: 10.1111/j.1538-7836.2008.03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naito K, Fujikawa K. Activation of human blood coagulation factor XI independent of factor XII: factor XI is activated by thrombin and factor XIa in the presence of negatively charged surfaces. J Biol Chem. 1991;266(12):7353–7358. [PubMed] [Google Scholar]
- 14.Gailani D, Broze GJ., Jr Factor XI activation in a revised model of blood coagulation. Science. 1991;253(5022):909–912. doi: 10.1126/science.1652157. [DOI] [PubMed] [Google Scholar]
- 15.von dem Borne PA, Meijers JC, Bouma BN. Feedback activation of factor XI by thrombin in plasma results in additional formation of thrombin that protects fibrin clots from fibrinolysis. Blood. 1995;86(8):3035–3042. [PubMed] [Google Scholar]
- 16.Asakai R, Chung DW, Davie EW, Seligsohn U. Factor XI deficiency in Ashkenazi Jews in Israel. N Engl J Med. 1991;325(3):153–158. doi: 10.1056/NEJM199107183250303. [DOI] [PubMed] [Google Scholar]
- 17.Seligsohn U. Factor XI deficiency in humans. J Thromb Haemost. 2009;7(suppl 1):84–87. doi: 10.1111/j.1538-7836.2009.03395.x. [DOI] [PubMed] [Google Scholar]
- 18.Bouma BN, Meijers JC. New insights into factors affecting clot stability: a role for thrombin activatable fibrinolysis inhibitor (TAFI; plasma procarboxypeptidase B, plasma procarboxypeptidase U, procarboxypeptidase R). Semin Hematol. 2004;41(suppl 1):13–19. doi: 10.1053/j.seminhematol.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Nesheim M, Bajzar L. The discovery of TAFI. J Thromb Haemost. 2005;3(10):2139–2146. doi: 10.1111/j.1538-7836.2005.01280.x. [DOI] [PubMed] [Google Scholar]
- 20.Mosnier LO, Bouma BN. Regulation of fibrinolysis by thrombin activatable fibrinolysis inhibitor, an unstable carboxypeptidase B that unites the pathways of coagulation and fibrinolysis. Arterioscler Thromb Vasc Biol. 2006;26(11):2445–2453. doi: 10.1161/01.ATV.0000244680.14653.9a. [DOI] [PubMed] [Google Scholar]
- 21.Ragni MV, Sinha D, Seaman F, Lewis JH, Spero JA, Walsh PN. Comparison of bleeding tendency, factor XI coagulant activity, and factor XI antigen in 25 factor XI-deficient kindreds. Blood. 1985;65(3):719–724. [PubMed] [Google Scholar]
- 22.Bolton-Maggs PH, Young Wan-Yin B, McCraw AH, Slack J, Kernoff PB. Inheritance and bleeding in factor XI deficiency. Br J Haematol. 1988;69(4):521–528. doi: 10.1111/j.1365-2141.1988.tb02409.x. [DOI] [PubMed] [Google Scholar]
- 23.Bolton-Maggs PH, Patterson DA, Wensley RT, Tuddenham EG. Definition of the bleeding tendency in factor XI-deficient kindreds: a clinical and laboratory study. Thromb Haemost. 1995;73(2):194–202. [PubMed] [Google Scholar]
- 24.Peretz H, Mulai A, Usher S, et al. The two common mutations causing factor XI deficiency in Jews stem from distinct founders: one of ancient Middle Eastern origin and another of more recent European origin. Blood. 1997;90(7):2654–2659. [PubMed] [Google Scholar]
- 25.Asakai R, Chung DW, Ratnoff OD, Davie EW. Factor XI (plasma thromboplastin antecedent) deficiency in Ashkenazi Jews is a bleeding disorder that can result from three types of point mutations. Proc Natl Acad Sci U S A. 1989;86:7667–7671. doi: 10.1073/pnas.86.20.7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meijers JC, Davie EW, Chung DW. Expression of human blood coagulation factor XI: characterization of the defect in factor XI type III deficiency. Blood. 1992;79(6):1435–1440. [PubMed] [Google Scholar]
- 27.Kravtsov DV, Wu W, Meijers JC, et al. Dominant factor XI deficiency caused by mutations in the factor XI catalytic domain. Blood. 2004;104(1):128–134. doi: 10.1182/blood-2003-10-3530. [DOI] [PubMed] [Google Scholar]
- 28.Riley PW, Cheng H, Samuel D, Roder H, Walsh PN. Dimer dissociation and unfolding mechanism of coagulation factor XI apple 4 domain: spectroscopic and mutational analysis. J Mol Biol. 2007;367(2):558–573. doi: 10.1016/j.jmb.2006.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders RE, Shiltagh N, Gomez K, et al. Structural analysis of eight novel and 112 previously reported missense mutations in the interactive FXI mutation database reveals new insight on FXI deficiency. Thromb Haemost. 2009;102(2):287–301. doi: 10.1160/TH09-01-0044. [DOI] [PubMed] [Google Scholar]
- 30.Gailani D, Neff AT. Rare coagulation factor deficiencies. In: Hoffman H, Benz EJ, Shattil SJ, et al., editors. Hematology, Basic Principles and Practice. 5th ed. Philadelphia, PA: Churchill Livingstone-Elsevier; 2009. pp. 1939–1952. [Google Scholar]
- 31.Saito H, Ratnoff OD, Bouma BN, Seligsohn U. Failure to detect variant (CRM+) plasma thromboplastin antecedent (factor XI) molecules in hereditary plasma thromboplastin antecedent deficiency: a study of 125 patients of several ethnic backgrounds. J Lab Clin Med. 1985;106(6):718–722. [PubMed] [Google Scholar]
- 32.Wang X, Cheng Q, Xu L, et al. Effects of factor IX or factor XI deficiency on ferric chloride-induced carotid artery occlusion in mice. J Thromb Haemost. 2005;3(4):695–702. doi: 10.1111/j.1538-7836.2005.01236.x. [DOI] [PubMed] [Google Scholar]
- 33.Renné T, Pozgajová M, Grüner S, et al. Defective thrombus formation in mice lacking coagulation factor XII. J Exp Med. 2005;202(2):271–281. doi: 10.1084/jem.20050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doggen CJ, Rosendaal FR, Meijers JC. Levels of intrinsic coagulation factors and the risk of myocardial infarction among men: opposite and synergistic effects of factors XI and XII. Blood. 2006;108(13):4045–4051. doi: 10.1182/blood-2005-12-023697. [DOI] [PubMed] [Google Scholar]
- 35.Tucker EI, Marzec UM, White TC, et al. Prevention of vascular graft occlusion and thrombus-associated thrombin generation by inhibition of factor XI. Blood. 2009;113(4):936–944. doi: 10.1182/blood-2008-06-163675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salomon O, Steinberg DM, Koren-Morag N, Tanne D, Seligsohn U. Reduced incidence of ischemic stroke in patients with severe factor XI deficiency. Blood. 2008;111(8):4113–4117. doi: 10.1182/blood-2007-10-120139. [DOI] [PubMed] [Google Scholar]
- 37.Meijers JC, Tekelenburg WL, Bouma BN, Bertina RM, Rosendaal FR. High levels of coagulation factor XI as a risk factor for venous thrombosis. N Engl J Med. 2000;342(10):696–701. doi: 10.1056/NEJM200003093421004. [DOI] [PubMed] [Google Scholar]
- 38.Kleinschnitz C, Stoll G, Bendszus M, et al. Targeting coagulation factor XII provides protection from pathological thrombosis in cerebral ischemia without interfering with hemostasis. J Exp Med. 2006;203(3):513–518. doi: 10.1084/jem.20052458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker EI, Gailani D, Hurst S, Cheng Q, Hanson SR, Gruber A. Survival advantage of coagulation factor XI-deficient mice during peritoneal sepsis. J Infect Dis. 2008;198(2):271–274. doi: 10.1086/589514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin L, Pandey P, Babine RE, et al. Crystal structures of the FXIa catalytic domain in complex with ecotin mutants reveal substrate-like interactions. J Biol Chem. 2005;280(6):4704–4712. doi: 10.1074/jbc.M411309200. [DOI] [PubMed] [Google Scholar]
- 41.Navaneetham D, Jin L, Pandey P, et al. Structural and mutational analyses of the molecular interactions between the catalytic domain of factor XIa and the Kunitz protease inhibitor domain of protease nexin 2. J Biol Chem. 2005;280(43):36165–36175. doi: 10.1074/jbc.M504990200. [DOI] [PubMed] [Google Scholar]
- 42.Jin L, Pandey P, Babine RE, Weaver DT, Abdel-Meguid SS, Strickler JE. Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors. Acta Crystallogr D Biol Crystallogr. 2005;61(10):1418–1425. doi: 10.1107/S0907444905024340. [DOI] [PubMed] [Google Scholar]
- 43.Samuel D, Cheng H, Riley PW, et al. Solution structure of the A4 domain of factor XI sheds light on the mechanism of zymogen activation. Proc Natl Acad Sci U S A. 2007;104(10):15693–15698. doi: 10.1073/pnas.0703080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tordai H, Bányai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999;461(1):63–67. doi: 10.1016/s0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- 45.Huizinga EG, Schouten A, Connolly TM, Kroon J, Sixma JJ, Gros P. The structure of leech anti-platelet protein, an inhibitor of haemostasis. Acta Crystallogr D Biol Crystallogr. 2001;57(8):1071–1078. doi: 10.1107/s0907444901007405. [DOI] [PubMed] [Google Scholar]
- 46.Brown PJ, Gill AC, Nugent PG, McVey JH, Tomley FM. Domains of invasion organelle proteins from apicomplexan parasites are homologous with the Apple domains of blood coagulation factor XI and plasma pre-kallikrein and are members of the PAN module superfamily. FEBS Lett. 2001;497(1):31–38. doi: 10.1016/s0014-5793(01)02424-3. [DOI] [PubMed] [Google Scholar]
- 47.Meijers JC, Mulvihill ER, Davie EW, Chung DW. Apple four in human blood coagulation factor XI mediates dimer formation. Biochemistry. 1992;31(19):4680–4684. doi: 10.1021/bi00134a021. [DOI] [PubMed] [Google Scholar]
- 48.Dorfman R, Walsh PN. Noncovalent interactions of the Apple 4 domain that mediate coagulation factor XI homodimerization. J Biol Chem. 2001;276(9):6429–6438. doi: 10.1074/jbc.M010340200. [DOI] [PubMed] [Google Scholar]
- 49.Cheng Q, Sun MF, Kravtsov DV, Aktimur A, Gailani D. Factor XI apple domains and protein dimerization. J Thromb Haemost. 2003;1(11):2340–2347. doi: 10.1046/j.1538-7836.2003.00418.x. [DOI] [PubMed] [Google Scholar]
- 50.Wu W, Sinha D, Shikov S, et al. Factor XI homodimer structure is essential for normal proteolytic activation by factor XIIa, thrombin, and factor XIa. J Biol Chem. 2008;283(27):18655–18664. doi: 10.1074/jbc.M802275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zucker M, Zivelin A, Landau M, Rosenberg N, Seligsohn U. Three residues at the interface of factor XI (FXI) monomers augment covalent dimerization of FXI. J Thromb Haemost. 2009;7(6):97097–97105. doi: 10.1111/j.1538-7836.2009.03353.x. [DOI] [PubMed] [Google Scholar]
- 52.Bodó I, Katsumi A, Tuley E, Eikenboom J, Dong Z, Sadler J. Type I von Willebrand disease mutation Cys1149Arg causes intracellular retention and degradation of heterodimers: a possible mechanism for dominant mutations in oligomeric proteins. Blood. 2001;98(10):2973–2979. doi: 10.1182/blood.v98.10.2973. [DOI] [PubMed] [Google Scholar]
- 53.Brennan S, Wyatt J, Medicina D, Callea F, George P. Fibrinogen brescia: hepatic endoplasmic reticulum storage and hypofibrinogenemia because of gamma 284 Gly → Arg mutation. Am J Pathol. 2000;157(1):189–196. doi: 10.1016/s0002-9440(10)64530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kravtsov DV, Monahan PE, Gailani D. A classification system for cross-reactive material-negative factor XI deficiency. Blood. 2005;105(12):4671–4673. doi: 10.1182/blood-2004-05-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renné T, Gailani D, Meijers JC, Müller-Esterl W. Characterization of the H-kininogen-binding site on factor XI: a comparison of factor XI and plasma prekallikrein. J Biol Chem. 2002;277(7):4892–4899. doi: 10.1074/jbc.M105221200. [DOI] [PubMed] [Google Scholar]
- 56.Renné T, Sugiyama A, Gailani D, Jahnen-Dechent W, Walter U, Müller-Esterl W. Fine mapping of the H-kininogen binding site in plasma prekallikrein apple domain 2. Int Immunopharmacol. 2002;2(13):1867–1873. doi: 10.1016/s1567-5769(02)00170-4. [DOI] [PubMed] [Google Scholar]
- 57.Baglia FA, Walsh PN. A binding site for thrombin in the apple 1 domain of factor XI. J Biol Chem. 1996;271(7):3652–3658. doi: 10.1074/jbc.271.7.3652. [DOI] [PubMed] [Google Scholar]
- 58.Ho DH, Badellino K, Baglia FA, Walsh PN. A binding site for heparin in the apple 3 domain of factor XI. J Biol Chem. 1998;273(26):16382–16390. doi: 10.1074/jbc.273.26.16382. [DOI] [PubMed] [Google Scholar]
- 59.Zhao M, Abdel-Razek T, Sun MF, Gailani D. Characterization of a heparin-binding site on the heavy chain of factor XI. J Biol Chem. 1998;273(47):31153–31159. doi: 10.1074/jbc.273.47.31153. [DOI] [PubMed] [Google Scholar]
- 60.Badellino KO, Walsh PN. Localization of a heparin-binding site in the catalytic domain of factor XIa. Biochemistry. 2001;40:7569–7580. doi: 10.1021/bi0027433. [DOI] [PubMed] [Google Scholar]
- 61.Yang L, Sun MF, Gailani D, Rezaie AR. Characterization of a heparin-binding site on the catalytic domain of factor XIa: mechanism of heparin acceleration of factor XIa inhibition by the serpins antithrombin and C1-inhibitor. Biochemistry. 2009;48:1517–1524. doi: 10.1021/bi802298r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Y, Gailani D. Identification of a factor IX binding site on the third apple domain of activated factor XI. J Biol Chem. 1996;271(46):29023–29028. doi: 10.1074/jbc.271.46.29023. [DOI] [PubMed] [Google Scholar]
- 63.Sun MF, Zhao M, Gailani D. Identification of amino acids in the factor XI apple 3 domain required for activation of factor IX. J Biol Chem. 1999;274(51):36373–36378. doi: 10.1074/jbc.274.51.36373. [DOI] [PubMed] [Google Scholar]
- 64.Baglia FA, Gailani D, López JA, Walsh PN. Identification of a binding site for glycoprotein Ib-alpha in the Apple 3 domain of factor XI. J Biol Chem. 2004;279(44):45470–45476. doi: 10.1074/jbc.M406727200. [DOI] [PubMed] [Google Scholar]
- 65.Sainz IM, Pixley RA, Colman RW. Fifty years of research on the plasma kallikrein-kinin system: from protein structure and function to cell biology and in-vivo pathophysiology. Thromb Haemost. 2007;98(1):77–83. [PubMed] [Google Scholar]
- 66.Scott CF, Colman RW. Function and immunochemistry of prekallikrein-high molecular weight kininogen complex in plasma. J Clin Invest. 1980;65(2):413–421. doi: 10.1172/JCI109684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greengard JS, Heeb MJ, Ersdal E, Walsh PN, Griffin JH. Binding of coagulation factor XI to washed human platelets. Biochemistry. 1986;25(13):3884–3890. doi: 10.1021/bi00361a022. [DOI] [PubMed] [Google Scholar]
- 68.Baglia FA, Jameson BA, Walsh PN. Identification and characterization of a binding site for platelets in the Apple 3 domain of coagulation factor XI. J Biol Chem. 1995;270(12):6734–6740. doi: 10.1074/jbc.270.12.6734. [DOI] [PubMed] [Google Scholar]
- 69.Baglia FA, Gailani D, Lopez JA, Walsh PN. Identification of a binding site for glycoprotein Ibα in the apple 3 domain of factor XI. J Biol Chem. 2004;279(44):45470–45476. doi: 10.1074/jbc.M406727200. [DOI] [PubMed] [Google Scholar]
- 70.White-Adams TC, Berny MA, Tucker EI, et al. Identification of coagulation factor XI as a ligand for platelet apolipoprotein E receptor 2 (ApoER2). Arterioscler Thromb Vasc Biol. 2009;29(10):1602–1607. doi: 10.1161/ATVBAHA.109.187393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baglia FA, Jameson BA, Walsh PN. Fine mapping of the high molecular weight kininogen binding site on blood coagulation factor XI through the use of rationally designed synthetic analogs. J Biol Chem. 1992;267(6):4247–4252. [PubMed] [Google Scholar]
- 72.Mitchell M, Harrington P, Cutler J, Rangarajan S, Savidge G, Alhaq A. Eighteen unrelated patients with factor XI deficiency, four novel mutations and a 100% detection rate by denaturing high-performance liquid chromatography. Br J Haematol. 2003;121(3):500–502. doi: 10.1046/j.1365-2141.2003.04302.x. [DOI] [PubMed] [Google Scholar]
- 73.O'Connell NM, Saunders RE, Lee CA, Perry DJ, Perkins SJ. Structural interpretation of 42 mutations causing factor XI deficiency using homology modeling. J Thromb Haemost. 2005;3(1):127–138. doi: 10.1111/j.1538-7836.2004.01088.x. [DOI] [PubMed] [Google Scholar]
- 74.Katsuda I, Maruyama F, Ezaki K, Sawamura T, Ichihara Y. A new type of plasma prekallikrein deficiency associated with homozygosity for Gly104Arg and Asn124Ser in apple domain 2 of the heavy-chain region. Eur J Haematol. 2007;79(1):59–68. doi: 10.1111/j.1600-0609.2007.00871.x. [DOI] [PubMed] [Google Scholar]
- 75.Kravtsov DV, Matafonov A, Tucker EI, et al. Factor XI contributes to thrombin generation in the absence of factor XII. Blood. 2009;114(2):452–458. doi: 10.1182/blood-2009-02-203604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von dem Borne PA, Mosnier LO, Tans G, Meijers JC, Bouma BN. Factor XI activation by meizothrombin: stimulation by phospholipid vesicles containing both phosphatidylserine and phosphatidylethanolamine. Thromb Haemost. 1997;78(2):834–839. [PubMed] [Google Scholar]
- 77.Spronk HMH, Wilhelm S, Heemskerk H, et al. Feedback activation of factor XI by thrombin is essential for haemostasis in vivo. J Thromb Haemost. 2009;7(suppl 2) Abstract PL-TU-003. [Google Scholar]
- 78.Pawlinski R, Pedersen B, Erlich J, Mackman N. Role of tissue factor in haemostasis, thrombosis, angiogenesis and inflammation: lessons from low tissue factor mice. Thromb Haemost. 2004;92(3):444–450. doi: 10.1160/TH04-05-0309. [DOI] [PubMed] [Google Scholar]
- 79.Crawley JT, Zanardelli S, Chion CK, Lane DA. The central role of thrombin in hemostasis. J Thromb Haemost. 2007;5(suppl 1):95–101. doi: 10.1111/j.1538-7836.2007.02500.x. [DOI] [PubMed] [Google Scholar]
- 80.Yun TH, Baglia FA, Myles T, et al. Thrombin activation of factor XI on activated platelets requires the interaction of factor XI and platelet glycoprotein Ib alpha with thrombin anion-binding exosites I and II, respectively. J Biol Chem. 2003;278(48):48112–48119. doi: 10.1074/jbc.M306925200. [DOI] [PubMed] [Google Scholar]
- 81.Matafonov A, Kravtsov DV, Tucker EI, et al. Factor XI contributes to thrombin generation in the absence of factor XIIa. Blood. 2008;112(11):1058a–1059a. doi: 10.1182/blood-2009-02-203604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baglia FA, Jameson BA, Walsh PN. Identification and characterization of a binding site for factor XIIa in the Apple 4 domain of coagulation factor XI. J Biol Chem. 1993;268(6):3838–3844. [PubMed] [Google Scholar]
- 83.Smith SB, Verhamme IM, Sun MF, Bock PE, Gailani D. Characterization of novel forms of coagulation factor XIa: independence of factor XIa subunits in factor IX activation. J Biol Chem. 2008;283(11):6696–6705. doi: 10.1074/jbc.M707234200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wuillemin WA, Hack CE, Bleeker WK, Biemond BJ, Levi M, ten Cate H. Inactivation of factor XIa in vivo: studies in chimpanzees and in humans. Thromb Haemost. 1996;76(4):549–555. [PubMed] [Google Scholar]
- 85.Knauer DJ, Majumdar D, Fong PC, Knauer MF. SERPIN regulation of factor XIa: the novel observation that protease nexin 1 in the presence of heparin is a more potent inhibitor of factor XIa than C1 inhibitor. J Biol Chem. 2000;275(48):37340–37346. doi: 10.1074/jbc.M003909200. [DOI] [PubMed] [Google Scholar]
- 86.Rezaie AR, Sun MF, Gailani D. Contributions of basic amino acids in the autolysis loop of factor XIa to serpin specificity. Biochemistry. 2006;45(31):9427–9433. doi: 10.1021/bi060820+. [DOI] [PubMed] [Google Scholar]
- 87.Zhang J, Tu Y, Lu L, Lasky N, Broze GJ. Protein Z-dependent protease inhibitor deficiency produces a more severe murine phenotype than protein Z deficiency. Blood. 2008;111(10):4973–4978. doi: 10.1182/blood-2007-12-126391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith RP, Higuchi DA, Broze GJ. Platelet coagulation factor XIa-inhibitor, a form of Alzheimer amyloid precursor protein. Science. 1990;248(4959):1126–1128. doi: 10.1126/science.2111585. [DOI] [PubMed] [Google Scholar]
- 89.Scandura JM, Zhang Y, Van Nostrand WE, Walsh PN. Progress curve analysis of the kinetics with which blood coagulation factor XIa is inhibited by protease nexin-2. Biochemistry. 1997;36(2):412–420. doi: 10.1021/bi9612576. [DOI] [PubMed] [Google Scholar]
- 90.Schmidt AE, Bajaj SP. Structure-function relationships in factor IX and factor IXa. Trends Cardiovasc Med. 2003;13(1):39–45. doi: 10.1016/s1050-1738(02)00210-4. [DOI] [PubMed] [Google Scholar]
- 91.Osterud B, Rapaport SI. Activation of factor IX by the reaction product of tissue factor and factor VII: additional pathway for initiating blood coagulation. Proc Natl Acad Sci U S A. 1977;74(12):5260–5264. doi: 10.1073/pnas.74.12.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bajaj SP, Rapaport SI, Russell WA. Redetermination of the rate-limiting step in the activation of factor IX by factor XIa and by factor VIIa/tissue factor: explanation for different electrophoretic radioactivity profiles obtained on activation of 3H- and 125I-labeled factor IX. Biochemistry. 1983;22(17):4047–4053. doi: 10.1021/bi00286a009. [DOI] [PubMed] [Google Scholar]
- 93.Lawson JH, Mann KG. Cooperative activation of human factor IX by the human extrinsic pathway of blood coagulation. J Biol Chem. 1991;266(17):11317–11327. [PubMed] [Google Scholar]
- 94.Osterud B, Bouma BN, Griffin JH. Human blood coagulation factor IX: purification, properties, and mechanism of activation by activated factor XI. J Biol Chem. 1978;253(17):5946–5951. [PubMed] [Google Scholar]
- 95.Wolberg AS, Morris DP, Stafford DW. Factor IX activation by factor XIa proceeds without release of a free intermediate. Biochemistry. 1997;36(14):4074–4079. doi: 10.1021/bi962274y. [DOI] [PubMed] [Google Scholar]
- 96.Sinha D, Marcinkiewicz M, Navaneetham D, Walsh PN. Macromolecular substrate-binding exosites on both the heavy and light chains of factor XIa mediate the formation of the Michaelis complex required for factor IX-activation. Biochemistry. 2007;46(34):9830–9839. doi: 10.1021/bi062296c. [DOI] [PubMed] [Google Scholar]
- 97.Ogawa T, Verhamme IM, Sun MF, Bock PE, Gailani D. Exosite-mediated substrate recognition of factor IX by factor XIa: the factor XIa heavy chain is required for initial recognition of factor IX. J Biol Chem. 2005;280(25):23523–23530. doi: 10.1074/jbc.M500894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sinha D, Seaman FS, Walsh PN. Role of calcium ions and the heavy chain of factor XIa in the activation of human coagulation factor IX. Biochemistry. 1987;26(13):3768–3775. doi: 10.1021/bi00387a005. [DOI] [PubMed] [Google Scholar]
- 99.Aktimur A, Gabriel MA, Gailani D, Toomey JR. The factor IX gamma-carboxyglutamic acid (Gla) domain is involved in interactions between factor IX and factor XIa. J Biol Chem. 2003;278(10):7981–7987. doi: 10.1074/jbc.M212748200. [DOI] [PubMed] [Google Scholar]
- 100.Guella I, Soldà G, Spena S, et al. Molecular characterization of two novel mutations causing factor XI deficiency: a splicing defect and a missense mutation responsible for a CRM+ defect. Thromb Haemost. 2008;99(3):523–530. doi: 10.1160/TH07-12-0723. [DOI] [PubMed] [Google Scholar]
- 101.Gailani D, Smith SB, Agah S, Bajaj SP. An analysis of cleavage of the factor IX activation sites by factor XIa. Blood. 2008;112(11):1060a. [Google Scholar]
- 102.Baglia FA, Shrimpton CN, Emsley J, et al. Factor XI interacts with the leucine-rich repeats of glycoprotein Ibalpha on the activated platelet. J Biol Chem. 2004;279(47):49323–49329. doi: 10.1074/jbc.M407889200. [DOI] [PubMed] [Google Scholar]
- 103.Sun MF, Baglia FA, Ho D, et al. Defective binding of factor XI-N248 to activated human platelets. Blood. 2001;98(1):125–129. doi: 10.1182/blood.v98.1.125. [DOI] [PubMed] [Google Scholar]
- 104.Ho DH, Badellino K, Baglia FA, et al. The role of high molecular weight kininogen and prothrombin as cofactors in the binding of factor XI A3 domain to the platelet surface. J Biol Chem. 2000;275(33):25139–25145. doi: 10.1074/jbc.M001890200. [DOI] [PubMed] [Google Scholar]
- 105.Sinha D, Seaman FS, Koshy A, Knight LC, Walsh PN. Blood coagulation factor XIa binds specifically to a site on activated human platelets distinct from that for factor XI. J Clin Invest. 1984;73(6):1550–1556. doi: 10.1172/JCI111361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller TN, Sinha D, Baird TR, Walsh PN. A catalytic domain exosite (Cys527-542) in factor XIa mediates binding to a site on activated platelets. Biochemistry. 2007;46(50):14450–14460. doi: 10.1021/bi701310x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bergmeier W, Chauhan AK, Wagner DD. Glycoprotein Ibalpha and von Willebrand factor in primary platelet adhesion and thrombus formation: lessons from mutant mice. Thromb Haemost. 2008;99(2):264–270. doi: 10.1160/TH07-10-0638. [DOI] [PubMed] [Google Scholar]
- 108.Pennings MT, Derksen RH, van Lummel M, et al. Platelet adhesion to dimeric beta-glycoprotein I under conditions of flow is mediated by at least two receptors: glycoprotein Ibalpha and apolipoprotein E receptor 2′. J Thromb Haemost. 2007;5(2):369–377. doi: 10.1111/j.1538-7836.2007.02310.x. [DOI] [PubMed] [Google Scholar]
- 109.Brenner B, Laor A, Lupo H, Zivelin A, Lanir N, Seligsohn U. Bleeding predictors in factor XI deficient patients. Blood Coagul Fibrinolysis. 1997;8(8):511–515. doi: 10.1097/00001721-199711000-00005. [DOI] [PubMed] [Google Scholar]
- 110.Allen GA, Wolberg AS, Oliver JA, Hoffman M, Roberts HR, Monroe DM. Impact of procoagulant concentration on rate, peak, and total thrombin generation in a model system. J Thromb Haemost. 2004;2(3):402–413. doi: 10.1111/j.1538-7933.2003.00617.x. [DOI] [PubMed] [Google Scholar]