Abstract
This work demonstrates in situ characterization of protein biomolecules in the aqueous solution using the System for Analysis at the Liquid Vacuum Interface (SALVI) and time-of-flight secondary ion mass spectrometry (ToF-SIMS). The fibronectin protein film was immobilized on the silicon nitride (SiN) membrane that forms the SALVI detection area. During ToF-SIMS analysis, three modes of analysis were conducted including high spatial resolution mass spectrometry, two-dimensional (2D) imaging, and depth profiling. Mass spectra were acquired in both positive and negative modes. Deionized water was also analyzed as a reference sample. Our results show that the fibronectin film in water has more distinct and stronger water cluster peaks compared to water alone. Characteristic peaks of amino acid fragments are also observable in the hydrated protein ToF-SIMS spectra. These results illustrate that protein molecule adsorption on a surface can be studied dynamically using SALVI and ToF-SIMS in the liquid environment for the first time.
Keywords: Chemistry, Issue 108, SALVI, ToF-SIMS, protein, water, in situ, molecular imaging, microfluidics
Introduction
Hydration is crucial to the structure,1 conformation,2 and biological activity3 of proteins. Proteins without water molecules surrounding them would not have viable biological activities. Specifically, water molecules interact with the surface and internal structure of proteins, and different hydration states of proteins make such interactions distinct.4 The interaction of proteins with solid surfaces is a fundamental phenomenon with implications in nanotechnology, biomaterials and tissue engineering processes. Studies have long indicated that conformational changes may occur as a protein encounters a surface. ToF-SIMS has been envisioned as the technique that has the potential to study the protein-solid interface.5-7 It is important to understand the hydration of proteins on solid surfaces, which potentially provides a fundamental understanding of the mechanism of their structure, conformation, and biological activity.
However, major surface analytical techniques are mostly vacuum-based and direct applications for volatile liquid studies are difficult due to the rapid evaporation of volatile liquid under the vacuum environment. We developed a vacuum compatible microfluidic interface, System for Analysis at the Liquid Vacuum Interface (SALVI), to enable direct observations of liquid surfaces and liquid-solid interactions using time-of-flight secondary ion mass spectrometry (ToF-SIMS).8-11 The unique aspects include the following: 1) the detection window is an aperture of 2-3 μm in diameter allowing direct imaging of the liquid surface, 2) surface tension is used to hold the liquid within the aperture, and 3) SALVI is portable among multiple analytical platforms.11,12
SALVI is composed of a silicon nitride (SiN) membrane as the detection area and a microchannel made of polydimethylsiloxane (PDMS). It is fabricated in the clean room, and the fabrication and key design factors have been detailed in previous papers and patents.8-12 The applications of ToF-SIMS as an analytical tool were demonstrated using a variety of aqueous solutions and complex liquid mixtures, some of which contained nanoparticles.13-17 Specifically, SALVI liquid ToF-SIMS allows dynamic probing of the liquid-solid interface of live biological systems (i.e., biofilms), single cells, and solid-electrolyte interface, opening new opportunities for in situ condensed phase studies including liquids using ToF-SIMS. However, the current design does not permit gas-liquid interactions yet. This is a direction for future development. SALVI has been used to study the hydrated protein film in this work for the first time.
Fibronectin is a commonly used protein dimer, consisting of two nearly identical monomers linked by a pair of disulfide bonds,18 which is well-known for its ability to bind cells.19,20 It was chosen as a model system to illustrate that the hydrated protein film could be dynamically probed using the SALVI liquid ToF-SIMS approach. The protein solution was introduced into the microchannel. After incubating for 12 hr, a hydrated protein film formed on the back side of the SiN membrane. Deionized (DI) water was used to rinse off the channel after protein introduction. Information was collected from hydrated fibronectin protein molecules in the SALVI microchannel using dynamic ToF-SIMS. DI water was also studied as a control to compare with results obtained from hydrated fibronectin thin film. Distinct differences were observed between the hydrated protein film and DI water. This work demonstrates that protein adsorption on surface in the liquid environment can be studied using the novel SALVI and liquid ToF-SIMS approach. The video protocol is intended to provide technical guidance for people who are interested in utilizing this new analytical tool for diverse applications of SALVI with ToF-SIMS and reduce unnecessary mistakes in liquid handling as well as ToF-SIMS data acquisition and analysis.
Protocol
1. Cleaning and Sterilization of the SALVI Microchannel
- Sterilization of the Microchannel in SALVI
- Draw 2 ml of 70% ethanol aqueous solution into a syringe, connect the syringe with the inlet end of SALVI, and slowly inject 1 ml of the liquid in 10 min. Remove the syringe at the end of injection. Next, connect the inlet and outlet of SALVI using a polyetheretherketone (PEEK) union. Alternately, use a syringe pump to conduct the same procedure. For example, set the flow speed at 100 μl/min.
- Keep the SALVI microchannel filled with 70% ethanol solution at room temperature for 4 hr.
- Introduce Deionized (DI) Water into SALVI
- Draw 2 ml of sterilized DI water into a syringe, connect the syringe with the inlet of SALVI, and slowly inject 1 ml of the liquid in 10 min. Remove the syringe, and connect the inlet and outlet of SALVI using a PEEK union. Alternately, use a syringe pump as mentioned in step 1.1.1.
2. Immobilization of the Protein Film in SALVI
- Immobilize the Protein Film in SALVI
- Draw 2 ml of fibronectin (10 μg/ml in phosphate buffered saline, PBS) solution into a syringe, connect the syringe with the inlet of SALVI, slowly inject 1 ml of the liquid in 10 min, remove the syringe, and connect the inlet and outlet of SALVI using a PEEK union.
- Incubate the SALVI filled with the fibronectin solution in a clean culture dish, keep the dish in the hood at room temperature for 12 hr.
- Draw 2 ml of sterilized DI water into a syringe and connect the syringe with the inlet of SALVI. Slowly inject 1 ml of water in 10 min, remove the syringe, and connect the inlet and outlet of SALVI. Note: If equipped with a light microscope, check the SALVI under the microscope to ensure that the SiN membrane is intact before ToF-SIMS analysis. Now the SALVI device is ready for ToF-SIMS analysis.
3. Install SALVI into the ToF-SIMS Loadlock Chamber
Note: Gloves should be worn at all times when handling the SALVI device and installing it onto the ToF-SIMS stage to avoid potential contaminations during surface analysis.
- Mount SALVI onto the Stage
- Lay the ToF-SIMS stage on a flat and clean surface. Put the SALVI device onto the stage with the silicon wafer and SiN membrane facing upward. Fix the SALVI using four metal clamping pieces and screw the metal pieces holding the corner of the device into the metal plate by four screws.
- Gently roll up the PFTE tubing connected to the microfluidic channel. Use four metal pieces and four screws to hold the stiff part down. Make sure that the device is positioned in the open space on the SIMS stage so that it does not interfere with the primary ion beam during analysis. Additional image and illustration of the SALVI fabrication and installation are available in previous publications.8,9,11
- Mount the Faraday Cup onto the Stage
- Fix the Faraday cup on the stage by a screw.
- Load the Stage into the ToF-SIMS Loadlock Chamber
- Open the ToF-SIMS loadlock, hold and set the stage horizontally onto the loading platform, and close the loadlock door.
4. ToF-SIMS Data Acquisition
Turn on the vacuum pump and ensure suitable vacuum (e.g., on the order of 10-6 mbar or Torr) is reached in the loadlock chamber.
Move the SALVI into the main chamber once the vacuum is stabilized after about 30 min. Note: The vacuum should be lower than 5 x 10-6 mbar vacuum in the main chamber during data acquisition. Otherwise, higher vacuum is indicative of a leak in the SALVI system.
Adjust the primary ion beam and analyzer of ToF-SIMS with a spatial resolution of approximately 400 nm according to manufacturer recommendations. The current of primary beam is 1,200 pA while the cycle time is 30 μsec under DC mode. Note: This process largely follows manufacturer's recommendations.
Find the microfluidic channel using the optical microscope. Use the optical image center as a reference and align it to be consistent with the secondary ion image center.
Before each measurement, use a 1 KeV O2+ beam (~40 nA) to scan on the SiN window with a 500 x 500 µm2 area for ~15 sec to remove surface contamination. Also, use an electron flood gun to compensate surface charging during all measurements. Use the automatic function of the flood gun in the default mode for this step.
- Select positive or negative mode before data acquisition. Use the 25 keV Bi3+ beam as the primary ion beam in all measurements. Note: The steps are similar for positive and negative data acquisitions. For the interest of space, the positive mode is described in details here.
- Depth Profiling
- Scan the Bi3+ beam on a round area with a diameter of ~2 µm with 32 pixels by 32 pixels resolution.
- To minimize the time required to punch-through the SiN membrane, use a long pulse width (i.e., 180 nsec, beam current ~1.0 pA at a cycle time of 100 μsec) Bi3+ beam. The intensities of secondary ions representative from the liquid surface increase when punch-through is reached. The punch-through time varies from 300 sec to 500 sec, depending on the batch of the SiN membrane. Note: This difference seems to be related to the batch of SiN membranes acquired according to our experience. However, the punch-through time does not really affect the data analysis.
- After the SiN membrane punch-through, keep the same current for about 200 sec to obtain sufficient data for 2D image reconstructions.
- Reduce the pulse width to 80 nsec for data collection to acquire spectra with better mass resolution. Continue this acquisition for about another 200 sec. The spectrum data shown in Figure 1 are obtained from this region.
- 2D Imaging and Mass Spectra
- Collect the 2D imaging and mass spectra data at the same time when measuring the depth profiling data. The ToF-SIMS instrument saves all the information of each spot during the experiment. The data are shown in different windows (FPanel - Spectra, FPanel - Profiles and FPanel - Images separately) of the digital control software. Check data in each panel judiciously during data acquisition.
5. ToF-SIMS Data Analysis
Note: Data analysis is initially performed using the IonToF ToF-SIMS instrumental software.
Open the analysis software of SIMS (Measurement Explorer) and then click the "profiles', 'spectra' and 'image' buttons, respectively, to process depth profile, m/z spectrum, and image data.
Open the data files that have the extension ".ita".
Select peaks of interest, type the species name in "ion" box, and label them with different colors in the spectra. Use the mouse scroll wheel to drag along the x-axis. Use Ctrl and scroll wheel to adjust peak heights and widths. Letter "L" switches between Linear and Log modes along the y-axis.
- Conduct data reconstruction before mass calibration. Otherwise, it is difficult to choose the spacing between peaks in the mass calibration step.
- Select the depth profiling data of interest and reconstruct the spectra according to the depth profile temporal series.
- Click the reconstruction button. In the "Start Reconstruction" window, type in a scan range of interest. Click "OK" to reconstruct the depth profile data.
- Perform a mass calibration. Press "F3" to open the "mass calibration" window. Choose peaks for calibration based on the specific sample.21 In this work, use the following peaks CH3+ (m/z 15), H3O+ (m/z 19), C2H5+ (m/z 29), H5O2+ (m/z 37), C3H7+ (m/z 43), H7O3+ (m/z 55), H9O4+ (m/z 71), H11O5+ (m/z 99) and H13O6+ (m/z 109) for the positive mode. Use the following peaks CH- (m/z 13), O- (m/z 16), OH- (m/z 17), C2H- (m/z 25), C3H- (m/z 37), and C4H- (m/z 49) for the negative mode.
- Select a peak, make sure that the downward arrow is pointed at the peak in the left bottom corner of the window. Click that peak and type the peak name in the "mass calibration" window, click "Add", and the ion of interest is included in the mass calibration.
- Finally, click the "Recalibrate" button. Check if the areas of peaks are in the right places according to their mass to charge ratios. In the "Spectrum" menu, select and click "Apply Mass Calibration" to "All Spectra" or "Selected spectra", depending on the specific analysis being conducted.
- As peak selection is necessary for sample analysis, determine characteristic peaks of the sample according to literature or other previous findings. Compare the intensity of peaks of interest in the m/z spectra. If necessary, add new peaks or delete useless peaks in the peak list.
- Add peaks. Click on a peak, find the red lines at the left bottom window. Hold the "Ctrl" key, scroll mouse horizontally to define peak width, which adds a peak to the peak list automatically. Another way to add a peak is to select a peak in the main spectrum window, and then to click the "add peak" button above the window. If there are many peaks to add, in the "Spectrum" menu, select "search peaks", check related specifications in the newly opened window, then add peaks as needed.
- To export a peak list, in the "Peak List" menu, select "Save…" the peak list in the "itmil" format. Use the Ctrl and left button to drag along the spectrum. In the spectra window, select a peak of interest and drag the two dashed lines to desirable positions.
- To import a peak list, in the "Mass interval List" menu, choose "Import…", locate an existing peak list file with the file extension ".ITPL". Click "Open".
- To use an "itmil" peak list, in the "Mass Interval List" menu, click "Open…", find the file, then click "Open".
- Apply a peak list to further analyze data. At the left-bottom of "Spectra" window, there is a window called "Mass Interval List". The imported peak list is shown up here.
- Right-click the peak list file to be used, apply it to the "active spectrum" or "all spectra".
View the data profiles. In the "Profile" window, Use letter "M" to fit to window (full range) and letter "N" to fit to the y-axis. Use Ctrl and scroll wheel to adjust the profile width. Or use "/" and "*" on the key board to change the y-axis range and height.
- Export data for further plotting using other graphic software after the initial analysis.
- To export a depth profile, in the "Profile" window, click on the "File" menu, then select "Export", and then select "Save as" a ".txt" file.
- To export an m/z spectrum file, in the "Spectra" window, click on the "File" menu, then select "Export", and then select "Save as" a ".txt" file.
- To export an image file, in the "Image" window, print screen and save as the image file.
6. ToF-SIMS Data Plotting and Presentation
Note: Use a graphic tool to plot the data after the SIMS data are processed using the instrumental software.
Use a graphic tool to import the data.
Make a plot using the m/z as the x-axis and peak intensity as the y-axis to show the reconstructed spectrum. An example is given in Figure 1.
Make a plot using the sputter time as the x-axis and peak intensity as the y-axis to show the reconstructed depth profile temporal series. An example is given in Figure 2.
Combine reconstructed 2D images of different m/z and form a matrix to show the ion mapping. An example is given in Figure 2.
Representative Results
A couple of representative results are presented to demonstrate the advantages of the proposed protocol. By using the SALVI microfluidic interface, the primary ion beam (Bi3+) can directly bombard on the hydrated fibronectin film in DI water. Thus the molecular chemical mapping of the liquid surface can be successfully acquired.
Figure 1a and 1b show the positive ToF-SIMS mass spectra of the hydrated fibronectin film and DI water, respectively. The peaks of interest including water clusters (i.e., m/z 19, 37, 55, 73, 91 and 109) and amino acid fragments (i.e., m/z 30, 44, 61, 74, 81, 83, and 98) are indicated by the red arrows. In addition to the characteristic peaks of amino acid fragments, which have been widely reported using dry protein samples, the peaks of water clusters are observed in the mass spectra of hydrated fibronectin film in DI water. Moreover, the intensities of these water cluster peaks are very different from those in the mass spectra of DI water. This result provides the direct evidence of the property of water molecules surrounding and inside the hydrated protein molecules that play a role in their adsorption on a surface.
Figure 2a shows the depth profile time series of the hydrated fibronectin film and DI water. As the sample is under the SiN membrane, the intensities of selected second ions are negligible before the SiN membrane is punched through (i.e., 280 sec for fibronectin and 340 sec for DI water). Once the SiN membrane is punched through, the intensities of these secondary ions increase significantly. When using the long pulse width, the mass resolution is relatively low. Therefore, the short pulse width process is used to obtain data with better mass resolution for image and spectrum reconstructions as highlighted in Figure 2a. After a period of time (i.e., 60 sec for fibronectin and 220 sec for DI water), the reduced pulse width is used to obtain m/z spectra with better mass resolution. The final m/z spectra were reconstructed from 200 sec measurements highlighted in green shaded areas in the depth profile temporal series. Figure 2b shows the 2D images from the hydrated fibronectin film and DI water, which correspond to the green shaded area in Figure 2a. These 2D images represent the counts of selected secondary ions from the sample: the brighter the image, the higher the secondary ion count. Such intuitive images give a clear comparison of selected secondary ions between different samples, which is helpful in data analysis. According to our experiences, the difference in the SiN punch-through time varies from batch to batch of the SiN membranes. However, it does not affect the results of the dynamic ToF-SIMS analysis.
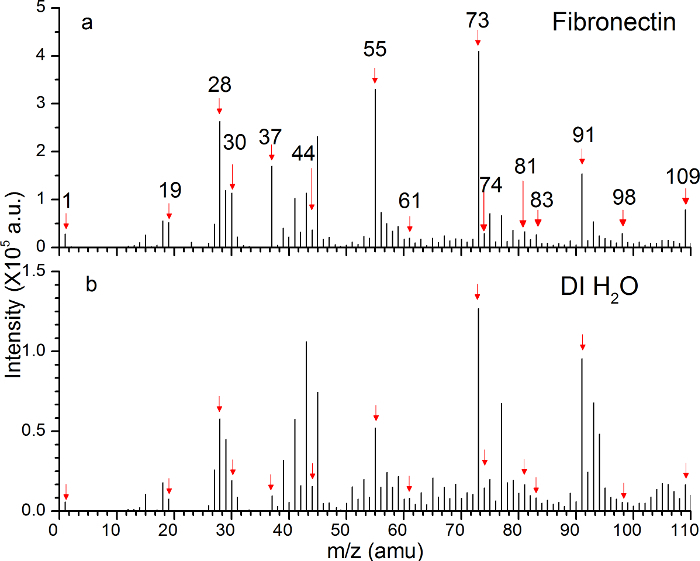 Figure 1: Comparisons of the ToF-SIMS positive spectra of the hydrated fibronectin film and DI water in the SALVI microchannel. (a) The reconstructed m/z spectrum of fibronectin. (b) The DI water spectrum.
Figure 1: Comparisons of the ToF-SIMS positive spectra of the hydrated fibronectin film and DI water in the SALVI microchannel. (a) The reconstructed m/z spectrum of fibronectin. (b) The DI water spectrum.
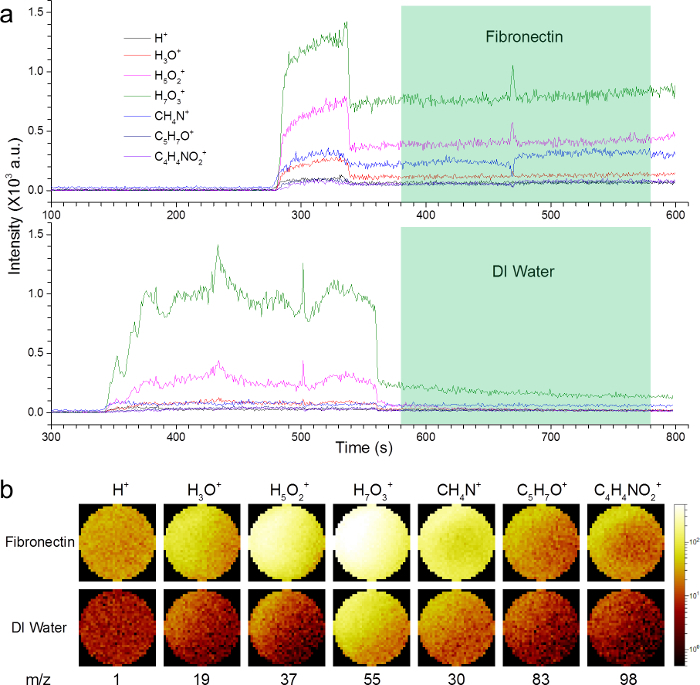 Figure 2:Depth profiles and 2D images of hydrated fibronectin and DI water. (a) The depth profiling time series of fibronectin and DI water in the positive mode. (b) 2D images reconstructed corresponding to the green shaded 200 sec time segments along the depth profile time series in (a). Several characteristic mass to charge ratios of relevance to water clusters (e.g., m/z 1, 19, 37, 55) are shown in addition to known amino acid fragments (e.g., m/z 30, 83, 98). Please click here to view a larger version of this figure.
Figure 2:Depth profiles and 2D images of hydrated fibronectin and DI water. (a) The depth profiling time series of fibronectin and DI water in the positive mode. (b) 2D images reconstructed corresponding to the green shaded 200 sec time segments along the depth profile time series in (a). Several characteristic mass to charge ratios of relevance to water clusters (e.g., m/z 1, 19, 37, 55) are shown in addition to known amino acid fragments (e.g., m/z 30, 83, 98). Please click here to view a larger version of this figure.
Discussion
SALVI is a microfluidic interface that allows dynamic liquid surface and liquid-solid interface analysis by vacuum based instruments, such as ToF-SIMS and scanning electron microscopy (SEM). Due to the usage of small apertures to expose liquid directly in vacuum, SALVI is suitable for many finely focused spectroscopy and imaging techniques without any modifications;22 the portability and versatility of microfluidics make it a true multimodal imaging platform. Distinct features and significance of SALVI compared to other existing techniques are summarized in several recent reviews.22-25 As the developer of the SALVI technology, our group has been actively applying it to a variety of studies involving dynamic liquid surfaces and liquid-solid interfaces. Some examples of new applications include mammalian cell membranes, bacteria biofilm attachment,15,16 or nanoparticle formation as a result of aqueous surface oxidations. In this paper, we present initial liquid ToF-SIMS results of the hydrated protein thin film adsorbed on the SiN surface.
It is quite critical to carefully handle the highlighted steps in the protocol. For example, when drawing liquid into the syringe in steps 1 and 2, make sure that there are not any bubbles inside the syringe. Because once the bubbles are injected into SALVI, they may induce leakage in vacuum.9 Thus, if any bubbles are observed, it is prudent to get rid of them by setting the syringe upright and gently pushing the bubbles out. In addition, it is very important to make the SiN window in SALVI flat for optimized ToF-SIMS analysis, as described in step 3. This levelness of the detection area directly affects the data quality according to the design of ToF-SIMS.26 It is necessary to visually inspect the device after securing everything onto the SIMS stage. If the SiN window is not horizontal, adjust the screws and change the height of the SALVI device to ensure that everything is at the same plane as much as possible. These delicate points need experience and carefulness, which determine the success and reliability of the whole experiment.
As shown in Figure 1b, the peaks with m/z 19, 37, 55, 73, 91 and 109 have high counts. This indicates that there are a good number of water clusters (H3O+, H5O2+, H7O3+, H9O4+, H11O5+ and H13O6+) in the positive secondary ions. The other water clusters (data not shown) with higher m/z values also have relatively strong peaks compared with their neighboring peaks. These large amounts of water clusters are emitted from the DI water surface after the SiN punch-through. Water cluster signals in the positive mode have not been observable in the previous work; and this was attributed to the low counts of secondary ions and non-optimized ToF-SIMS conditions in the past.8 With the continuous effort in optimizing the SALVI and ToF-SIMS acquisition conditions such as the primary ion beam (i.e., Bi+ vs. Bi3+), pulse width and current conditions during depth profiling, it is now possible to observe up to 44 water clusters in the positive mode using the conditions presented in this paper. Results shown here are promising to further investigate the role of water in dynamics of various biological molecules and biological systems.
Here as an example to illustrate the advantages of SALVI and ToF-SIMS in studying biological molecular dynamics, the hydration of fibronectin protein was presented. The peaks of m/z 30, 44, 61, 74, 81, 83, and 98 belong to the amino acid fragments according to the previous report,27,28 which are much stronger than those of DI water shown in Figure 1b. This difference in the increased intensity of the amino acid fragments in the fibronectin sample suggests that these secondary ions are indeed from the proteins themselves. In addition, the peaks of water clusters in Figure 1a are much stronger than those of DI water in Figure 1b, which implies that the properties of water molecules in the protein adsorption surface and interfacial solution could be very different from those in pure water.
Figure 2a shows representative depth profiling temporal series of two different samples: fibronectin and DI water in the SALVI microchannel. The depth profiling data were collected as the SiN window was being bombarded by the Bi3+ primary ion beam with the long pulse width. As seen in Figure 2a, the SiN window is punched through at around 300 sec. Before the SiN window in SALVI is punched through, the intensities of selected secondary ions are pretty low. Once the SiN window is punched through, the intensities of the secondary ions immediately increase as the liquid surface starts being bombarded by the primary ion beam. For example, the intensities of H+ and H3O+ show a large increase, indicating the observation of the water surface.8,9 Although the intensities are unstable right after the punch-through or at the interface, they may reach relatively stable values after a period of time (e.g., 60 sec for fibronectin and 220 sec for DI water, respectively, in Figure 2a). The difference in the long pulse width drilling does not really affect the analysis results illustrated here when reconstructing m/z spectra (e.g., Figure 1), because the effective m/z spectra are acquired after the SiN punch-through. However, a researcher may choose to use the same conditions for all the samples for consistency and ease in choosing data sections during data analysis. In order to obtain better mass resolution, the pulse width was subsequently reduced and the intensities were decreased accordingly in the latter portion of the depth profiling experiment. The depth profile time series were continuously collected for another 200 sec to provide ample data for m/z spectral analysis. The segments of time that m/z spectra were reconstructed are highlighted in green shaded areas.
Fibronectin has been widely used for surface adsorption prior to cell culturing; many studies have shown fibronectin is responsible for cell adhesion and cell-biomaterial interactions.19,20,29 A recent study characterized the thickness of fibronectin adsorbed on poly(hydroxylethyl methacrylate) brushes using ellipsometry. The fibronectin thin film was determined to be 3-12 nm thick.30 Such thickness is reasonable for the depth profiling analysis in an aqueous solution.8,9 With the development of SALVI, direct analysis of the hydrated protein adsorbed on the solid surface is now possible. This approach potentially can have wide applications in studying the interactions of proteins with solid surfaces dynamically.
Figure 2b shows the selected 2D images of different secondary ions from fibronectin and DI water samples, respectively. 2D images are obtained from the depth profiling temporal data in green shaded areas in Figure 2a, which correspond to the liquid surface after the SiN punch-through as described earlier. The images of selected amino acid fragment secondary ions CH4N+, C5H7O+ and C4H4NO2+ (i.e., m/z 30, 83 and 98) for the fibronectin thin film were much brighter than those for DI water. This indicates that the amino acid fragment secondary ions are observed from the fibronectin molecules inside the 2 μm diameter aperture. Furthermore, the images of selected peaks (e.g., m/z 1, 19, 37, and 55) corresponding to several water clusters of fibronectin are remarkably brighter than those of DI water. This result demonstrates that the hydration of fibronectin makes the water molecules surrounding the biomolecule distinct from the water molecules in the DI water.
In summary, this work has provided striking evidence on protein hydration induced water molecule property alterations. Such results could provide an improved fundamental understanding on the structure, conformation, and biological activity of proteins adsorbed on a solid surface in the liquid microenvironment.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We are grateful to the Pacific Northwest National Laboratory (PNNL) Chemical Imaging Initiative-Laboratory Directed Research and Development (CII-LDRD) and Materials Synthesis and Simulation across Scales (MS3) Initiative LDRD fund for support. Instrumental access was provided through a W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL) Science Themed Proposal. EMSL is a national scientific user facility sponsored by the Office of Biological and Environmental Research (BER) at PNNL. The authors thank Mr. Xiao Sui, Mr. Yuanzhao Ding, and Ms. Juan Yao for proof reading the manuscript and providing useful feedback. PNNL is operated by Battelle for the DOE under Contract DE-AC05-76RL01830.
References
- Tompa K, Bokor M, Verebelyi T, Tompa P. Water rotation barriers on protein molecular surfaces. Chem. Phys. 2015;448:15–25. [Google Scholar]
- Maruyama Y, Harano Y. Does water drive protein folding? Chem. Phys. Lett. 2013;581:85–90. [Google Scholar]
- Chaplin M. Opinion - Do we underestimate the importance of water in cell biology. Nat. Rev. Mol. Cell Biol. 2006;7(11):861–866. doi: 10.1038/nrm2021. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. Mapping hydration dynamics around a protein surface. Proc. Natl. Acad. Sci. U. S. A. 2007;104(47):18461–18466. doi: 10.1073/pnas.0707647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia N, May CJ, McArthur SL, Castner DG. Time-of-flight secondary ion mass spectrometry analysis of conformational changes in adsorbed protein films. Langmuir. 2002;18(10):4090–4097. [Google Scholar]
- Gray JJ. The interaction of proteins with solid surfaces. Curr. Opin. Struct. Biol. 2004;14(1):110–115. doi: 10.1016/j.sbi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Wagner MS, Horbett TA, Castner DG. Characterization of the structure of binary and ternary adsorbed protein films using electron spectroscopy for chemical analysis, time-of-flight secondary ion mass spectrometry, and radiolabeling. Langmuir. 2003;19(5):1708–1715. doi: 10.1016/s0142-9612(02)00612-9. [DOI] [PubMed] [Google Scholar]
- Yang L, Yu X-Y, Zhu Z, Iedema MJ, Cowin JP. Probing liquid surfaces under vacuum using SEM and and ToF-SIMS. Lab Chip. 2011;11(15):2481–2484. doi: 10.1039/c0lc00676a. [DOI] [PubMed] [Google Scholar]
- Yang L, Yu X-Y, Zhu ZH, Thevuthasan T, Cowin JP. Making a hybrid microfluidic platform compatible for in situ imaging by vacuum-based techniques. J. Vac. Sci. Technol. A. 2011;29(6):061101. [Google Scholar]
- Yu X-Y, Yang L, Zhu ZH, Cowin JP, Iedema MJ. Probing aqueous surfaces by ToF-SIMS. LC GC N. Am. 2011. pp. 34–38. [DOI] [PubMed]
- Yu X-Y, Yang L, Cowin J, Iedema M, Zhu Z. Systems and methods for analyzing liquids under vacuum. 8,555,710 B2. US patent. 2013
- Yu X-Y, Liu B, Yang L, Zhu Z, Marshall MJ. Microfluidic electrochemical device and process for chemical imaging and electrochemical analysis at the electrode-liquid interface in situ. 20,140,038,224 A1. US patent. 2014
- Yang L, Zhu Z, Yu X-Y, Thevuthasan S, Cowin JP. Performance of a microfluidic device for in situ ToF-SIMS analysis of selected organic molecules at aqueous surfaces. Anal. Methods. 2013;5(10):2515–2522. [Google Scholar]
- Yang L, et al. In situ SEM and ToF-SIMS analysis of IgG conjugated gold nanoparticles at aqueous surfaces. Surf. Interface Anal. 2014;46(4):224–228. [Google Scholar]
- Hua X, et al. In situ molecular imaging of hydrated biofilm in a microfluidic reactor by ToF-SIMS. Analyst. 2014;139(7):1609–1613. doi: 10.1039/c3an02262e. [DOI] [PubMed] [Google Scholar]
- Hua X, et al. Two-dimensional and three-dimensional dynamic imaging of live biofilms in a microchannel by time-of-flight secondary ion mass spectrometry. Biomicrofluidics. 2015;9(3):031101. doi: 10.1063/1.4919807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, et al. In situ chemical probing of the electrode-electrolyte interface by ToF-SIMS. Lab Chip. 2014;14(5):855–859. doi: 10.1039/c3lc50971k. [DOI] [PubMed] [Google Scholar]
- Pankov R, Yamada KM. Fibronectin at a glance. J. Cell Sci. 2002;115(20):3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- Pierschbacher MD, Hayman EG, Ruoslahti E. Location of the cell-attachment site in fibronectin with monoclonal antibodies and proteolytic fragments of the molecule. Cell. 1981;26(2):259–267. doi: 10.1016/0092-8674(81)90308-1. [DOI] [PubMed] [Google Scholar]
- Engvall E, Ruoslahti E. Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int. J. Cancer. 1977;20(1):1–5. doi: 10.1002/ijc.2910200102. [DOI] [PubMed] [Google Scholar]
- Green FM, Gilmore IS, Seah MP. TOF-SIMS: Accurate mass scale calibration. J. Am. Soc. Mass Spectrom. 2006;17(4):514–523. doi: 10.1016/j.jasms.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Yu X-Y, Liu B, Yang L. Imaging liquids using microfluidic cells. Microfluid. Nanofluid. 2013;15(6):725–744. [Google Scholar]
- Shi H, Lercher JA, Yu X-Y. Sailing into uncharted waters: recent advances in the in situ monitoring of catalytic processes in aqueous environments. Catal. Sci. Technol. 2015;5(6):3035–3060. [Google Scholar]
- Deleu M, Crowet JM, Nasir MN, Lins L. Complementary biophysical tools to investigate lipid specificity in the interaction between bioactive molecules and the plasma membrane: A review. Biochim. Biophys. Acta-Biomembr. 2014;1838(12):3171–3190. doi: 10.1016/j.bbamem.2014.08.023. [DOI] [PubMed] [Google Scholar]
- Kraft ML, Klitzing HA. Imaging lipids with secondary ion mass spectrometry. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2014;1841(8):1108–1119. doi: 10.1016/j.bbalip.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Gilmore IS. SIMS of organics-Advances in 2D and 3D imaging and future outlook. J. Vac. Sci. Technol. A. 2013;31(5):050819. [Google Scholar]
- Muramoto S, et al. ToF-SIMS Analysis of Adsorbed Proteins: Principal Component Analysis of the Primary Ion Species Effect on the Protein Fragmentation Patterns. J. Phys. Chem. C. 2011;115(49):24247–24255. doi: 10.1021/jp208035x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüning C, Hellweg S, Dambach S, Lipinsky D, Arlinghaus HF. Improving the interpretation of ToF-SIMS measurements on adsorbed proteins using PCA. Surf. Interface Anal. 2006;38(4):191–193. [Google Scholar]
- Gustavsson J, et al. Surface modifications of silicon nitride for cellular biosensor applications. J. Mater. Sci.-Mater. Med. 2008;19(4):1839–1850. doi: 10.1007/s10856-008-3384-7. [DOI] [PubMed] [Google Scholar]
- Deng J, Ren TC, Zhu JY, Mao ZW, Gao CY. Adsorption of plasma proteins and fibronectin on poly(hydroxylethyl methacrylate) brushes of different thickness and their relationship with adhesion and migration of vascular smooth muscle cells. Regen Biomater. 2014. pp. 17–25. [DOI] [PMC free article] [PubMed]


