Abstract
Immune cell networks in tissues play a vital role in mediating local immunity and maintaining tissue homeostasis, yet little is known of the resident immune cell populations in the oral mucosa and gingiva. We have established a technique for the isolation and study of immune cells from murine gingival tissues, an area of constant microbial exposure and a vulnerable site to a common inflammatory disease, periodontitis. Our protocol allows for a detailed phenotypic characterization of the immune cell populations resident in the gingiva, even at steady state. Our procedure also yields sufficient cells with high viability for use in functional studies, such as the assessment of cytokine secretion ex vivo. This combination of phenotypic and functional characterization of the gingival immune cell network should aid towards investigating the mechanisms involved in oral immunity and periodontal homeostasis, but will also advance our understanding of the mechanisms involved in local immunopathology.
Keywords: Immunology, Issue 108, oral, gingival, periodontitis, oral immune cell network, tissue immunity, oral mucosal immunity
Introduction
Gingival tissues surround the human and murine dentition and are constantly exposed to the complex biofilm of the tooth 1. The immune cell network policing the gingival barrier is vital to maintaining tissue integrity, ensuring homeostasis with the local commensal microbes and, at the same time, providing effective immunity against pathogenic challenge 2. To achieve homeostasis, the immune system is carefully tailored to the gingival environment creating a highly-specialized immune cell network, yet little detail is known of the gingival immune cell populations and their role in maintaining tissue immunity 2.
When immune homeostasis is disrupted at the gingiva, either through increased host susceptibility and/or presence of dysbiotic microbial communities, an inflammatory condition, periodontitis arises 34. Periodontitis is a common inflammatory disease, leading to loss of tooth supporting structures. In its severe forms it is seen in approximately 10% of the general population 5. Dissecting the key factors involved in periodontitis susceptibility and progression has proven difficult 6. However, animal models have been extremely useful in understanding the mechanisms of periodontitis initiation and progression 7. Models can be employed to define the key cell populations and molecular mediators that are vital for maintaining immune homeostasis and drive development of periodontitis. Such insight will transform our understanding of gingiva-specific control of immune homeostasis and further our current understanding of disease pathogenesis.
Protocol
All experimental procedures described in this protocol followed required guidelines and were approved by the Institutional Animal Care and Use Committee, NIDCR/NIH.
1. Prepare in Advance
Prepare Complete media: RPMI supplemented with 2 mM L-glutamine, 100 units/ml penicillin, 100 ug/ml streptomycin and 10% FBS.
Prepare DNase media: 50 ml of RPMI media supplemented with 7.5 µg DNase (Make fresh, keep on ice at all times).
Prepare Collagenase-DNase media: 5 ml of DNase media plus 3.2 mg/ml of Collagenase Type IV (Make fresh, keep on ice at all times).
Pre-cool FACS buffer: 0.5 % FBS diluted in PBS.
Pre-cool centrifuge to 4 °C.
Warm shaker incubator to 37 °C.
2. Isolation, Dissection, and Cell Isolation from Gingival Blocks
Euthanize mice by exposing them to CO2 in an appropriate chamber, according to ARAC guidelines.
Open the thoracic and abdominal cavity to expose the heart and internal organs. To perfuse, make an incision in the inferior vena cava and perfuse 3 ml of PBS via the left ventricle using a 3 ml syringe with a 27 G needle.
Immobilize the body and head of the mouse on a pad with stomach facing up.
Cut each commissure of the lip towards the neck with scissors, separating the skin that covers mandible and neck region, exposing the underlying tissues and muscles. Dissect the lower lip to uncover the mandible.
Cut between the lower incisors to separate both sides of the mandible and immobilize each side to access the oral cavity.
Visualize and dissect the palate away from the nasal cavity.
Dissect areas using a 10 blade and include maxillary and mandibular molar areas with surrounding gingiva (Figure 1). Make vertical incisions just anterior to the first molar and posterior to the third molar and a horizontal incision at the border of the gingiva (white collar of tissue surrounding teeth).
Place the maxillary and mandibular blocks in a 50 ml conical tube with 5 ml Collagenase/DNase (keep on ice).
Incubate in a shaker incubator at 37 °C for 1 hr and during the last 5 min of incubation add 50 µl of 0.5 M EDTA.
Add 5 ml of cold DNase media to the 50 ml tube with tissues and gently swirl to mix.
Transfer the 4 blocks of tissue on a petri dish and cover with 500 µl of DNase media. Remove gingiva from each block of tissue, using a scalpel blade and pointing the blade into the gingival and interdental areas.
Transfer the tissues and the media from the petri dish as well as the previous DNase media used during dissection, to a new 50 ml tube through the cell strainer (70 µm size). Wash with DNase media (3-5 ml) all of the instruments used and filter.
Using the plunger of a sterile 3 ml syringe, mash the tissue against the strainer, filtering with cold DNase media (30 to 35 ml).
Wash the cell strainer with cold DNase media.
Centrifuge at 4 °C, 314 x g for 6 min.
Discard the supernatant, re-suspend in 1 ml of complete media.
Count cells (the expected amount should range between 8 x 105 to 1.2 x 106 cells with a viability of >80%) using an automated cell counter.
3. Stimulation and FACS Staining of Gingival Cells
Stimulate the single cell suspension with PMA (50 ng/ml) and Ionomycin (2.5 µg/ml) in the presence of 1 µl/ml of Brefeldin A.
Incubate for 3.5 hr at 37 °C and 5% CO2.
Spin down and wash with PBS and centrifuge at 4 °C; 314 x g for 6 min.
Stain the cells using a viability stain following the manufacturer's instructions.
Stain for extracellular markers (CD45, TCRγδ, TCRβ, CD4, CD8, TCRγδ or antibodies of choice) following the manufacturer's instructions.
Wash the cells with cold FACS buffer and centrifuge at 4 °C, 314 x g for 5 min.
Gently vortex to disrupt the cell pellet.
Fix the cells with 2% paraformaldehyde for 20 min at room temperature.
- Wash the cells with cold FACS buffer, centrifuge at 4 °C, 314 x g for 5 min and stain for intracellular cytokines.
- Fill the FACS tubes with a solution of cold 0.5% saponin diluted in FACS buffer and centrifuge at 4 °C, 314 x g for 5 min.
- Add 300 µl of 0.5% saponin solution to the FACS tubes, incubate for 15 min in the dark at 4 °C and then centrifuge at 4 °C, 314 x g for 5 min.
- Stain the cells for intracellular markers (IL-17A and IFNγ or cytokines of choice) following the manufacturer's instructions. Wash the cells with 0.5% saponin solution and centrifuge at 4 °C, 314 x g for 5 min.
- Wash again with FACS buffer and centrifuge at 4 °C, 314 x g for 5 min.
- Re-suspend the cells in 300 µl of FACS buffer.
Analyze results on a flow cytometer.
4. Flow Cytometry Analysis
Visualize cells to be analyzed on forward (FSC) and side scatter (SCC) and gate to exclude debris.
Select single cells with forward scatter area (FSC-A) vs. height (FSC-H) parameters.
Gate cells that are live (as indicated by viability staining) and positive for the hematopoietic marker CD45+ for further analysis (Figure 2A and 2C).
Continue analysis of specific markers as shown in Figure 2 and Figure 3.
Representative Results
To illustrate application of the protocol, we show representative results examining the immune cell network in the gingiva of mice with and without periodontitis (WT vs. LFA-/-, Figure 2A-C). Representative FACS plots show live CD45+ hematopoietic cells in the gingiva (Figure 2A, 2C). Isolation and processing of immune cells with this protocol yields sufficient cells to perform ex vivo stimulation and characterization of cytokine secretion patterns. Here we show IL-17 and IFN-γ staining in total CD45+ cells in the steady state (WT) and in mice exhibiting periodontitis (due to LFA deficiency) (Figure 2B-D). These data reveal the potential of this technique to capture cytokine profiles during tissue steady state (WT) and in the context of local periodontitis (LFA-/-). In mice susceptible to periodontitis we further characterized the gingival immune cell composition particularly of the T cell and Innate Lymphoid cell (ILC) compartments (Figure 3A-B). We were able to define the production of IL-17 and IFN-γ within the TCRβ+CD4+ T cell, TCRβ+CD8+ T cell, TCRγδ+ T cell, NK and ILC compartments (Figure 3C).
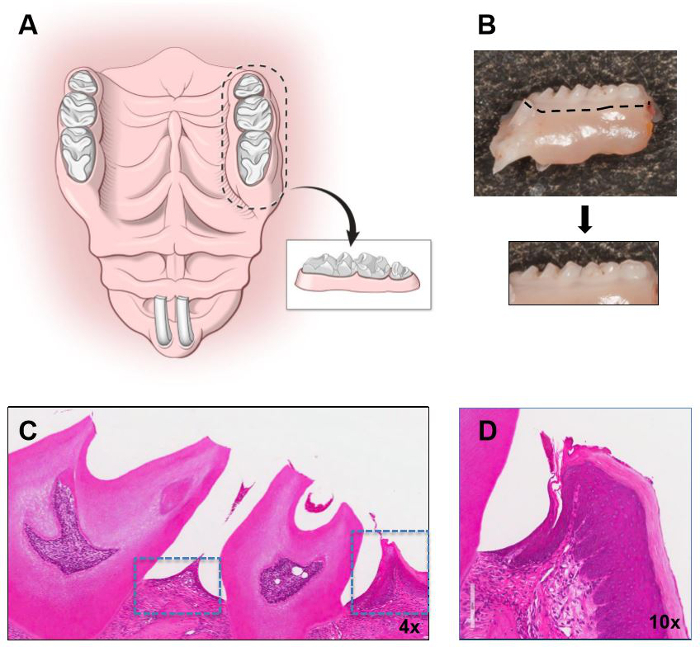 Figure 1: Isolation of murine gingiva. Illustration demonstrating the boarders of dissection in the maxilla and the isolated gingival segment to be used for gingival tissue dissection. (B) Segment of murine maxilla (molar area) with outline for block dissection and isolated block. Hematoxylin and Eosin stain (H&E) of a maxillary molar segment with gingival tissues outlined is shown in low (C) and higher magnification in (D). Original magnifications indicated. Please click here to view a larger version of this figure.
Figure 1: Isolation of murine gingiva. Illustration demonstrating the boarders of dissection in the maxilla and the isolated gingival segment to be used for gingival tissue dissection. (B) Segment of murine maxilla (molar area) with outline for block dissection and isolated block. Hematoxylin and Eosin stain (H&E) of a maxillary molar segment with gingival tissues outlined is shown in low (C) and higher magnification in (D). Original magnifications indicated. Please click here to view a larger version of this figure.
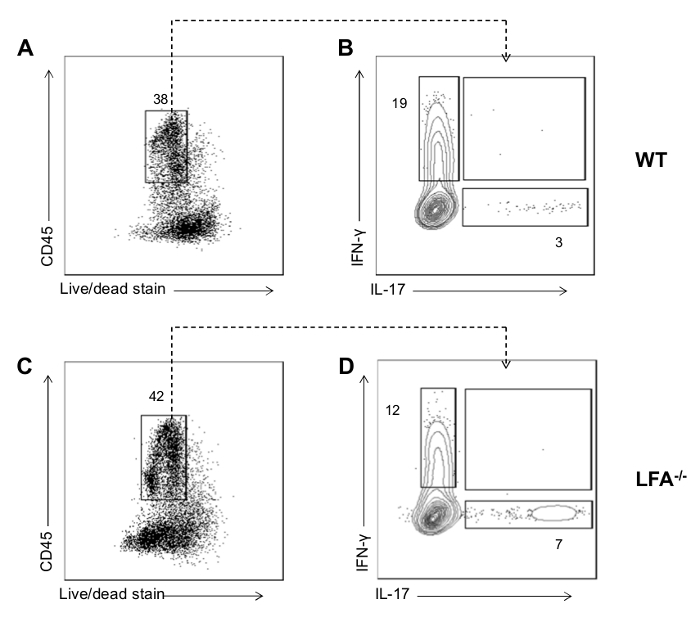 Figure 2: Representative FACS data for cytokine production from gingival cells. (A-C) FACS plot showing gate for live CD45+ cells in mouse gingival cell preparations following ex vivo re-stimulation with PMA and ionomycin in WT and LFA-/-. (B-D) Representative FACS plots showing IFN-γ and IL-17 staining in total CD45+ cells mouse from gingiva of mice with/without periodontitis (due to LFA-deficiency). Please click here to view a larger version of this figure.
Figure 2: Representative FACS data for cytokine production from gingival cells. (A-C) FACS plot showing gate for live CD45+ cells in mouse gingival cell preparations following ex vivo re-stimulation with PMA and ionomycin in WT and LFA-/-. (B-D) Representative FACS plots showing IFN-γ and IL-17 staining in total CD45+ cells mouse from gingiva of mice with/without periodontitis (due to LFA-deficiency). Please click here to view a larger version of this figure.
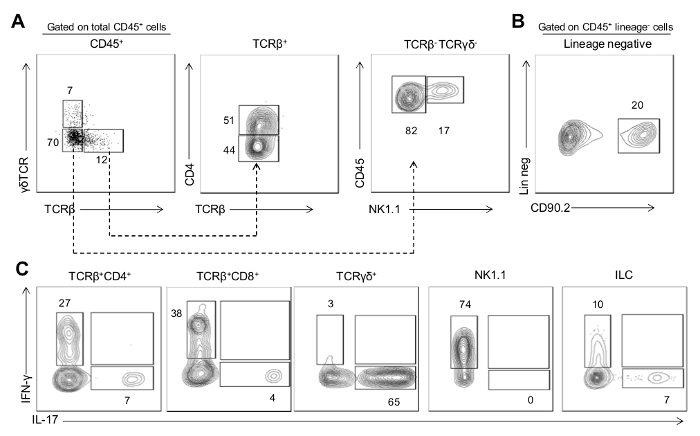 Figure 3: Cytokine production by immune populations in inflamed gingiva. (A) Representative FACS plot showing staining for TCRβ versus TCRγδ gated on CD45+ cells. Gating on TCRβ+ cells, middle plot shows CD4 versus TCRβ staining (allows for analysis of TCRβ+CD4+ T cells, TCRβ+CD8+ T cells and TCRγδ+ T cells). Gating on TCRβ- TCRγδ- cells, right plot shows CD45 versus NK1.1 staining (allows for analysis of NK cells). (B) Representative FACS plot showing staining for CD90.2 gated on CD45+ lineage negative cells (CD3−CD19−NK1.1−CD11c−CD11b−Ly6G−Ly6C−TCRγδ−TCRβ−) which identifies innate lymphoid cells. (C) Representative FACS plots showing IFN-γ and IL-17 staining in cell populations identified. Gingiva preparations are from 20 week old LFA-/- mice that develop periodontitis; data are representative of three independent experiments. Please click here to view a larger version of this figure.
Figure 3: Cytokine production by immune populations in inflamed gingiva. (A) Representative FACS plot showing staining for TCRβ versus TCRγδ gated on CD45+ cells. Gating on TCRβ+ cells, middle plot shows CD4 versus TCRβ staining (allows for analysis of TCRβ+CD4+ T cells, TCRβ+CD8+ T cells and TCRγδ+ T cells). Gating on TCRβ- TCRγδ- cells, right plot shows CD45 versus NK1.1 staining (allows for analysis of NK cells). (B) Representative FACS plot showing staining for CD90.2 gated on CD45+ lineage negative cells (CD3−CD19−NK1.1−CD11c−CD11b−Ly6G−Ly6C−TCRγδ−TCRβ−) which identifies innate lymphoid cells. (C) Representative FACS plots showing IFN-γ and IL-17 staining in cell populations identified. Gingiva preparations are from 20 week old LFA-/- mice that develop periodontitis; data are representative of three independent experiments. Please click here to view a larger version of this figure.
Discussion
The current technique successfully yields a large number of immune cells (from a single mouse) suitable, not only for phenotypic characterization, but also for functional studies ex vivo. Another group had previously published a protocol to isolate and characterize murine gingival immune cells and introduced the value of using multicolor flow cytometry in the study of periodontitis in animal models 9. Following considerable troubleshooting the key modification in this protocol was to include dissection of whole blocks of tissue in order to include both gingival collar areas and interdental gingiva (Figure 1). With gingival dissection alone, often full thickness flaps are not achieved and part of the gingiva is missed particularly in the interdental areas. Using this approach, numbers of immune cells harvested are significantly increased. This technique also allows for optimal isolation of gingival tissues both from maxilla and mandible.
Critical steps for this protocol include a conservative dissection of gingival areas and an efficient rate of dissection. Specifically, blocks of tissue should be dissected to include only minimal tissue around teeth (gingiva) and not buccal tissues. Blocks should be dissected quickly and tissues and cells should remain on ice to ensure optimal cell viability.
For purposes of troubleshooting it is important to note that low yield of cells can be related to inadequate time of incubation with collagenase, inappropriate concentrations of collagenase and/or inadequate dissection of the gingiva from the tissue blocks. On that note, dissection of gingiva using magnifying loupes (2X-6X) can be helpful. Low viability of cells can be related to tardiness in the procedure and to media, buffers and cells not remaining continuously on ice.
Limitations of this protocol, inherent to the type of tissue dissected is the size of the gingival tissues, compared to other anatomical barrier sites studied (ex. Skin or gastrointestinal tract) in which larger tissues are available.
Nonetheless, the technique presented here has allowed us to characterize the dominant cell populations in the LFA-/- model which spontaneously developed periodontitis. In LFA-/- mice we identified a dominant IL-17 cytokine signature during the development of periodontitis 10. Using multicolor flow cytometric analysis we were able to define the cellular sources of IL-17 11. In this disease setting IL-17 was produced by TCRγδ T cells, CD4+ T cells and innate lymphoid cells (ILC) locally within the gingiva 10.
With the substantial number of cells isolated with this methodology and the ability to identify even small subpopulations, it will now be possible to develop a comprehensive understanding of the specialized immune cell network safeguarding gingival barrier integrity. This critical insight will allow us to evaluate immune responses in the gingival environment. By employing this technique we can now proceed to isolate specific constituents of the gingiva immune cell network by flow cytometric cell sorting, allowing additional analyses and characterizations to be undertaken.
Disclosures
Authors have nothing to disclose
Acknowledgments
Authors were funded in part by the intramural program of NIDCR (N.M.M) and supported by a Wellcome Trust Stepping Stones Fellowship (097820/Z/11/B to J.E.K) and by a Manchester Collaborative Centre for Inflammation Research grant (to J.E.K). The authors thank Teresa Wild for critically reviewing the manuscript.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining The Normal Bacterial Flora Of The Oral Cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Naik S. Compartmentalized And Systemic Control Of Tissue Immunity By Commensals. Nat Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: A Polymicrobial Disruption Of Host Homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. Immunomicrobial Pathogenesis Of Periodontitis: Keystones, Pathobionts, And Host Response. Trends Immunol. 2014;35:3–11. doi: 10.1016/j.it.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, et al. Prevalence Of Periodontitis In Adults In The United States: 2009 And 2010. J Dent Res. 2012;91:914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- Moutsopoulos NM, Lionakis MS, Hajishengallis G. Inborn Errors In Immunity: Unique Natural Models To Dissect Oral Immunity. J Dent Res. 2015. [DOI] [PMC free article] [PubMed]
- Hajishengallis G, Lamont RJ, Graves DT. The Enduring Importance Of Animal Modelsin Understanding Periodontal Disease. Virulence. 2015;6:229–235. doi: 10.4161/21505594.2014.990806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrow SO. Coligan JE, et al., editors. Analysis Of Flow Cytometry Data. Current Protocols In Immunology. 2001. Chapter 5, Unit 5 2. [DOI] [PubMed]
- Arizon M, et al. Langerhans Cells Down-Regulate Inflammation-Driven Alveolar Bone Loss. Proc Natl Acad Sci U S A. 2012;109:7043–7048. doi: 10.1073/pnas.1116770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos NM, et al. Defective Neutrophil Recruitment In Leukocyte Adhesion Deficiency Type I Disease Causes Local IL-17-Driven Inflammatory Bone Loss. Sci Transl Med. 2014;6:229–240. doi: 10.1126/scitranslmed.3007696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 Immune Axis: From Mechanisms To Therapeutic Testing. Nat Rev Immunol. 2014;14:585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]


