Abstract
DNA methylation is a key epigenetic mechanism, needed for proper control over the expression of genetic information and silencing of repetitive elements. Exposure to ionizing radiation, aside from its strong genotoxic potential, may also affect the methylation of DNA, within the repetitive elements, in particular. In this study, we exposed C57BL/6J male mice to low absorbed mean doses of two types of space radiation – proton (0.1 Gy, 150 MeV, dose rate 0.53±0.08 Gy/min), and heavy iron ions (56Fe) (0.5 Gy, 600 MeV/n, dose rate 0.38±0.06 Gy/min). Radiation-induced changes in cardiac DNA methylation associated with repetitive elements were detected. Specifically, modest hypomethylation of retrotransposon LINE-1 was observed at day 7 after irradiation with either protons or 56Fe. This was followed by LINE-1, and other retrotransposons, ERV2 and SINE B1, as well as major satellite DNA hypermethylation at day 90 after irradiation with 56Fe. These changes in DNA methylation were accompanied by alterations in the expression of DNA methylation machinery and affected the one-carbon metabolism pathway. Furthermore, loss of transposable elements expression was detected in the cardiac tissue at the 90-day time-point, paralleled by substantial accumulation of mRNA transcripts, associated with major satellites. Given that the one-carbon metabolism pathway can be modulated by dietary modifications, these findings suggest a potential strategy for the mitigation and, possibly, prevention of the negative effects exerted by ionizing radiation on the cardiovascular system. Additionally, we show that the methylation status and expression of repetitive elements may serve as early biomarkers of exposure to space radiation.
Keywords: space radiation, heart, one-carbon metabolism pathway, DNA methylation, repetitive elements
1. Introduction
DNA methylation is a key epigenetic mechanism that plays a critical role during the development and in the maintenance of cellular homeostasis [1]. It is involved in the regulation of proper expression of genetic information in a sex-, tissue-, and cell type-dependent manner, as well as in the silencing of repetitive elements. In turn, alterations in methylation of DNA may lead to the development of pathological states, including heart disease. For instance, dilated cardiomyopathy has been characterized by genome-wide alterations in DNA methylation, including hypo- and hypermethylation of genes that play functional roles in the pathways linked to heart failure [2,3]. Changes in gene-specific methylation have also been described as molecular signatures of arrhythmia [4].
Alterations in DNA methylation are not limited to specific genes but can also be detected at repetitive elements, which comprise half to two-thirds of mammalian genomes [5,6]. They are the most highly methylated sequences in mammalian genomes, and are represented as transposable elements and satellite repeats [1,7]. Retrotransposons, such as Long Interspersed Nucleotide Element 1 (LINE-1), Endogenous Retroviruses 1 and 2 (ERV1 and ERV2) and Short Interspersed Nucleotide Elements B1 and B2 (SINE B1 and SINE B2), as well as the transposons Mariner and Charlie are among the most abundant transposable elements in mammalian genomes and together comprise ~40% of the mouse genome [8]. Satellite repeats are the centromere-associated repetitive sequences and are represented in the mouse as major satellites (6 Mb of 234 bp units located primarily at the pericentromeric regions) and minor satellites (~600 Kb of 120 bp units, located at centromeres) [9].
Previously considered as junk DNA, repetitive elements are now accepted as important regulators of stability and proper function of the genome, including expression of genetic information and chromatin structure [5,6]. Loss of epigenetic control over repetitive elements can result in unwanted alterations in their expression and the expression of genes within their regulatory network, and has been reported in numerous pathological states, including cardiac ischemic injury [10].
It is becoming increasingly evident that exposure to ionizing radiation, aside from its genotoxic potential, may also affect the cellular epigenome [11–13]. Of particular interest are the effects of space radiation, such as protons and heavy ions, since in some cases, epigenetic alterations represent the only long-term signatures of exposure. For instance, exposure to low mean absorbed doses of 56Fe (600 MeV, dose range 0.1 – 0.4 Gy), did not lead to increased production of reactive oxygen species, DNA damage or alterations in cellular senescence and apoptosis in the murine hematopoietic progenitor and stem cells (HPSCs). At the same time, exposure to the leukemogenic dose of 0.4 Gy of 56Fe has led to altered global and repetitive elements-associated methylation of DNA and DNA methylation machinery that were detectable in HSPCs for at least 5 months after irradiation [14]. Accumulating evidence indicates that alterations in DNA methylation is the general feature of space radiation and that these alterations are primary attributable to repetitive elements. Indeed, exposures to low absorbed mean doses of protons and 56Fe ions relevant to the space environment lead to significant alterations in DNA methylation and expression of repetitive elements in the bone marrow, liver and lung tissues [14–17]. As these changes in repetitive elements may persist for a long time post exposure, this suggests their potentially causative role in radiation-induced late tissue damage and disease development and progression. Furthermore, such epigenetic parameters may potentially serve as universal biomarkers of exposure.
Epidemiological studies indicate that exposures even to low doses of radiation may result in the development of heart diseases [18–20]. Recently, concerns were also raised in regards to the effects of space radiation on the cardiovascular system [21]. This particular study, the first of this kind to our knowledge, addressed the effects of protons and 56Fe ions on repetitive elements-associated DNA methylation and expression in the mouse heart.
2. Materials and Methods
2.1 Animals and irradiation
Male C57BL/6J mice, at 10 weeks of age (Jackson Laboratory, Bar Harbor, ME) were shipped to Brookhaven National Laboratories (BNL) in Upton, NY. After a one-week acclimation period, the mice were randomly assigned to experimental groups and were exposed to sham-irradiation (n=8), 0.1 Gy protons (150 MeV, dose rate 0.53±0.08 Gy/min) (n=9), or 0.5 Gy 56Fe (600 MeV/n, dose rate 0.38±0.06 Gy/min) (n=10). The dose of protons was chosen as likely during a solar particle event (SPE). The energy of 150 MeV is commonly used in a therapeutic setting and also represents energy near the maximum abundance of protons expected in most SPEs [21]. The dose of 56Fe was selected as likely to be encountered by astronauts due to the galactic cosmic rays during deep space exploration [22] and within the 56Fe dose range used in the previous studies [15,23,24]. At the selected energy of 600 MeV/n, thorough penetration of the animals with a relatively flat Bragg peak entrance region is expected. Dosimetry was performed by the NASA Space Radiation Laboratory (NSRL) physics dosimetry group at BNL to ensure the quality of exposure. For each exposure, animals were individually placed into clear Lucite cubes (3 in × 1½ in × 1½ in) with breathing holes. The focused beam of high-energy 56Fe particles was generated by the Booster accelerator at BNL and transferred to the experimental beam line at the NSRL facility. Dose calibration was performed with three parallel plate ion chambers that were positioned upstream of the target and a NIST traceable Far West thimble chamber. The values of the thimble chamber were then compared with the upstream ion chambers so that the desired dose could be delivered to the samples based on upstream ion chamber measurements. Sham irradiated mice served as controls and were placed into the same enclosures and for the same amount of time, since previous studies report no effect of sham irradiation on molecular end-points [14]. During the entire experiment, sham-irradiated mice were not housed together with irradiated mice. After irradiation, the mice were shipped to Loma Linda University (LLU) under climate-controlled conditions and were housed at LLU under a constant 12 h light:dark cycle. Regular chow and water were provided ad libitum. All procedures were approved by the Institutional Animal Care and Use Committees of LLU and BNL.
2.2. Tissue harvesting
At 7 days and 90 days after irradiation, groups of animals were sacrificed, and the heart was obtained and snap-frozen in liquid nitrogen. Frozen tissues were shipped on dry ice to the University of Arkansas for Medical Sciences for further molecular analyses and analysis of components of methionine metabolism with HPLC-EC. The researchers were blinded throughout all phases of the experiments; decoding only occurred after the final analyses were performed.
2.3. Nucleic Acids Extraction
RNA and DNA were extracted simultaneously from flash-frozen cardiac tissue using the AllPrep DNA/RNA extraction kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. DNA concentrations and integrity were analyzed by the Nanodrop 2000 (Thermo Scientific, Waltham, MA) and 1% agarose gel.
2.4. Analysis of methylation status of DNA repetitive elements by methylation-sensitive quantitative PCR
The methylation status of repetitive elements (LINE-1, ERV1, ERV2, SINE B1, SINE B2, Charlie, Mariner, major and minor satellites) was determined by methylation-sensitive McrBC-quantitative PCR (qPCR) as described before [14]. Primer sets are listed in Supplementary Table 2 of the Supplementary Material.
2.5. Analysis of methylation status of the long interspersed nuclear element-1 repetitive element by pyrosequencing
To determine the methylation status of LINE-1, total DNA was extracted, as described above. Genomic DNA (1 μg) underwent bisulfite conversion, and pyrosequencing analysis of LINE-1 ORF1 was performed on a PyroMark 96ID instrument.
2.6. Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total cardiac RNA was extracted as described above. Only RNA samples with the 260/280 ratios between 1.95 and 2.05 and the 260/230 ratios above 1.5 were considered for further molecular analyses. cDNA was synthesized using random primers and a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol (Life Technologies). The levels of gene transcripts were determined by quantitative Real Time PCR (qRT-PCR) using TaqMan Gene Expression Assays (Life Technologies). Assays for determination of mRNA abundance are provided in Supplementary Table 1. Assays for determination of expression of repetitive elements are provided in Supplementary Table 2. Each plate contained one experimental gene and a housekeeping gene. The cycle threshold (Ct) for each sample was determined from the linear region of the amplification plot. The ΔCt values for all genes were determined relative to the control gene Gapdh (Mm 99999915_g1, Life Technologies). The ΔΔCt were calculated using each exposed group means relative to control group means. The fold change data were calculated from the ΔΔCt values. All qRT-PCR reactions were conducted in triplicate and repeated twice.
2.7. Determination of analytical components of methionine metabolism
Snap-frozen heart samples were used to determine levels of S-adenosylhomocysteine (SAH), S-adenosylmethionine (SAM), methionine and adenosine using an HPLC-EC method. Details of this method were previously published elsewhere [25].
2.8. Statistical analysis
All data are presented as mean ± standard error of mean(s). All assessed parameters were measured within the same batch of animals. For each repetitive element and time-point combination, we analyzed DNA methylation and mRNA abundance in a one-way ANOVA comparing among types of radiation. We compared control to protons and 56Fe with a t-test in the ANOVA framework. We used Dunnett’s method to control Type I error 0.05 for the two comparisons. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software Inc. LaJolla, CA).
3. Results
3.1. Effects of protons and 56Fe irradiation on LINE-1 DNA methylation
Retrotransposon LINE-1 is the most abundant repetitive element in the mammalian genomes, comprising 20% and 21% of human and mouse genomes, respectively [8]. Its genomic abundance and presence of a large number of CpG sites (sites that may be potentially methylated) within its sequence have led to recognition of LINE-1 methylation status as a surrogate biomarker in the evaluation of global DNA methylation levels [26]. Previous studies clearly demonstrated that exposure to low doses of both low- and high-LET irradiation may affect the methylation status of LINE-1 in different cells and tissues [15,27–30]. Therefore, we first addressed the methylation of LINE-1 Open Reading Frame 2 (ORF2) that encodes the reverse transcriptase needed for LINE-1 retrotransposition. Methylation-sensitive McrBC-qPCR did not reveal significant alterations in DNA methylation of LINE-1 ORF2 7 days after exposure (Figure 1A). We then addressed the methylation status of LINE-1 ORF1 that encodes ORF1p, a nucleic acid chaperone, using pyrosequencing, the most sensitive and robust technique in the analyses of locus-specific DNA methylation. Analysis of 5 CpG sites within the ORF1 revealed minor, although significant, hypomethylation after exposure to protons and 56Fe at the 7-day time-point (−1.1% and −0.9%, respectively; p<0.01 for both exposure regimens).
Figure 1.
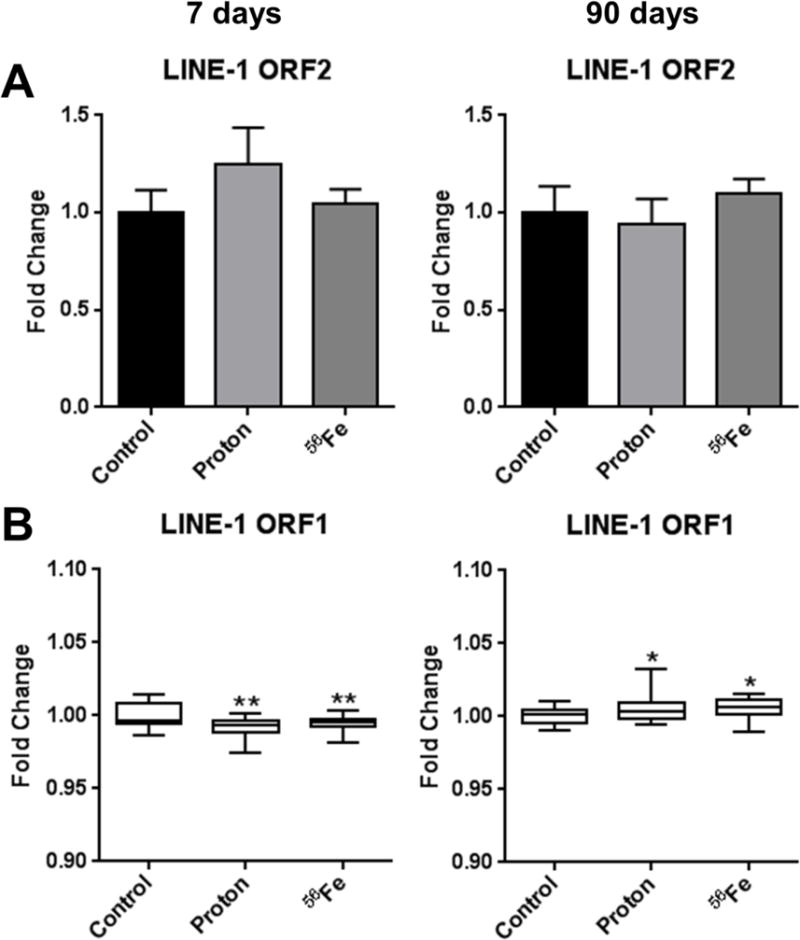
Effects of space radiation on the DNA methylation status of retrotransposon LINE-1 in the mouse heart. (A) Methylation of LINE-1 ORF2, as measured by methylation-sensitive real-time PCR. (B) Methylation of LINE-1 ORF1, as measured by pyrosequencing. Data are presented as mean ± SE. *p≤0.05, **p≤0.01, ANOVA with Dunnett’s test.
At the 90-day time-point, hypermethylation was observed in the LINE-1 ORF2 after exposure to 56Fe (although insignificant) and in LINE-1 ORF1 after exposure to protons and 56Fe (+0.8% and +0.82%, respectively; p<0.05 for both exposure regimens) (Figure 1B).
Studies indicate that high-LET radiation may affect the DNA methylation of selective LINE-1 elements (Lima et al., 2014). Taking this into consideration, we further assessed the DNA methylation in three LINE-1 elements that belong to A-type promoter-carriers – LINE-1 elements that maintain their retrotransposition activity in mice – L1MdA_I (0.21 MYR), L1MdA_II (1.62 MYR) and L1MdA_VI (4.62 MYR). While we did not identify significant changes in the DNA methylation in any of those LINE-1 elements at the 7-day time-point, significant DNA hypermethylation was detected in the evolutionary young L1MdA_I (2.4-fold increase; p<0.05 after 56Fe exposure) and L1MdA_II (7.1-fold increase; p<0.05 after protons or 56Fe) at the 90-day time-point (Figure 2A). No significant changes in DNA methylation of the evolutionary old L1MdA_VI LINE-1 element were identified at any time-point (Supplementary Figure 1).
Figure 2.
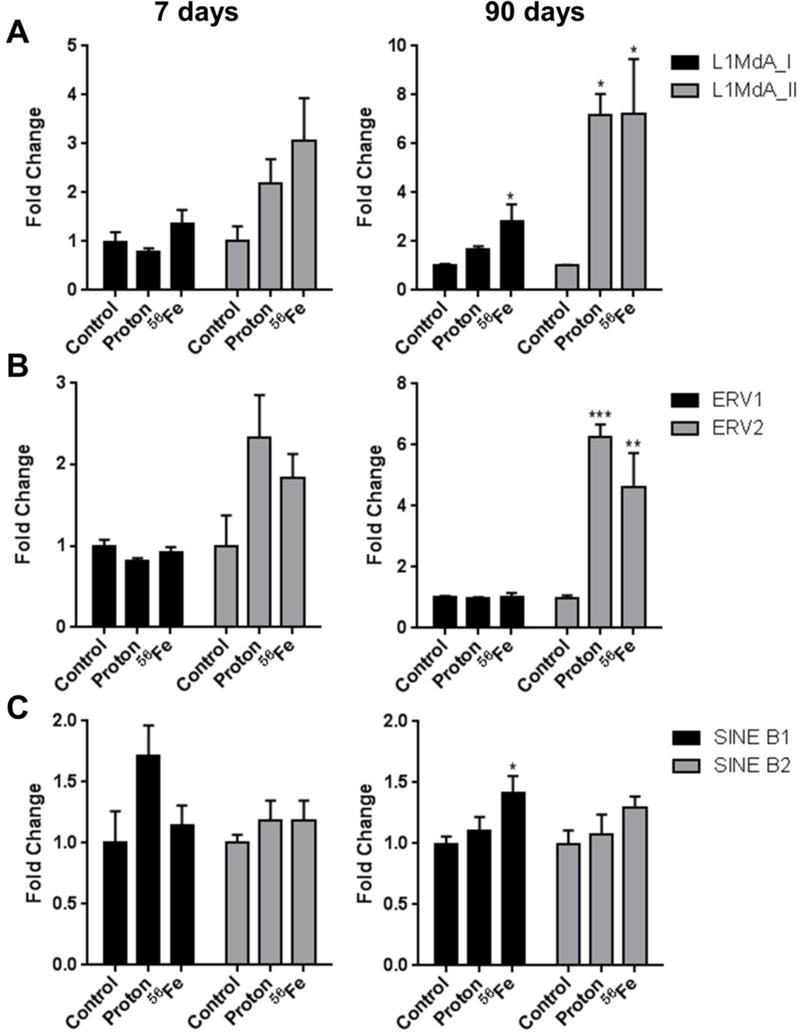
Effects of space radiation on the DNA methylation status of transposable elements in the mouse heart, as measured by methylation-sensitive real-time PCR. (A) LINE-1 elements L1MdA_I and L1MdA_VI; (B) autonomous retrotransposons ERV1 and ERV2; (C) non-autonomous retrotransposons SINE B1 and SINE B2. Data are presented as mean ± SE. *p≤0.05, **p≤0.01, ***p≤0.001, ANOVA with Dunnett’s test.
3.2. Effects of protons and 56Fe irradiation on other repetitive elements
Recent studies demonstrated that alterations in the methylation status of repetitive elements in response to environmental stressors are often non-unidirectional, and are rather characterized by loci of hypo- and hypermethylation (reviewed in [8]. Therefore, next we addressed the methylation of a panel of repetitive elements that belong to different classes – autonomous retrotransposons ERV1 and ERV2, non-autonomous retrotransposons SINE B1 and SINE B2, DNA transposons Charlie and Mariner and satellite DNA – major and minor satellites.
No significant changes were identified in the DNA methylation of ERV1 at both time-points and a strong DNA hypermethylation was observed in ERV2 elements 90 days after exposure (6.2-fold, p<0.001 and 4.3-fold, p<0.01 after protons and 56Fe, respectively) (Figure 2B).
The 7-day time-point was characterized by a 1.7-fold increase in the methylation of the SINE B1 retrotransposon in response to protons irradiation, although insignificant (p-0.061) (Figure 2C). Methylation of the other elements was affected to a lesser extent. At the 90-day time-point, a 1.4-fold (p<0.05) and 1.9-fold (p<0.01) hypermethylation in response to 56Fe irradiation was observed in SINE B1 and major satellites, respectively (Figure 2C and Figure 3B, respectively). Exposure to protons did not affect methylation of repetitive elements substantially.
Figure 3.
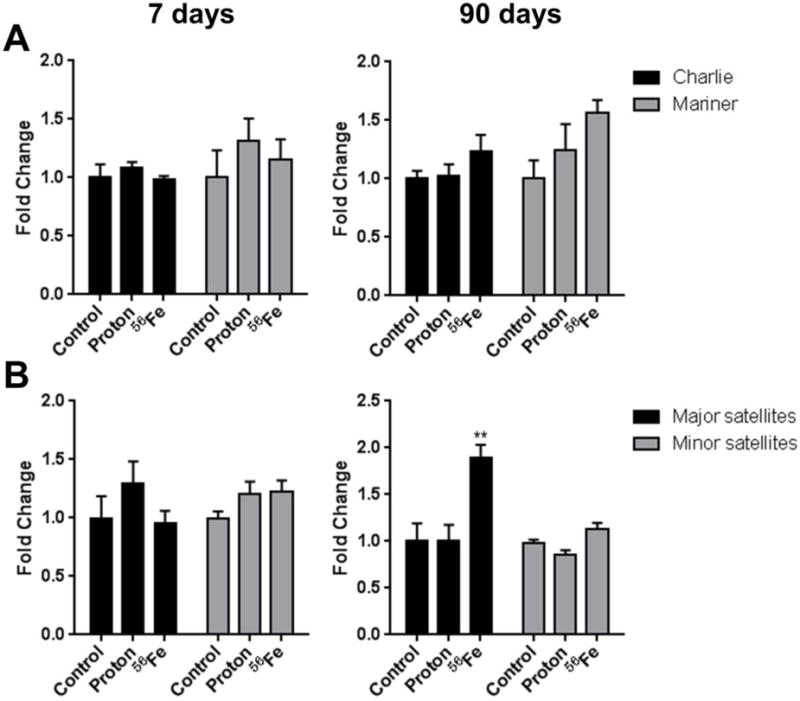
Effects of space radiation on the DNA methylation status of transposable elements and satellite DNA in the mouse heart, as measured by methylation-sensitive real-time PCR. (A) DNA transposons Charlie and Mariner; (B) major and minor satellites. Data are presented as mean ± SE. *p≤0.05, **p≤0.01, ANOVA with Dunnett’s test.
3.3. Effects of protons and 56Fe irradiation on the expression of repetitive elements
Ionizing radiation has been shown to affect the expression of repetitive elements, including exposure to low absorbed mean doses of protons and 56Fe ions (Miousse et al., 2014b; Nzabarushimana et al., 2014; Nzabarushimana et al., 2015). At the 7-day time-point, a significant 2.2-fold loss of LINE-1 ORF2 expression was detected after exposure to 56Fe (p<0.05), in congruence with its modest hypomethylation (Figure 4A). Exposure to protons resulted in an insignificant 1.8-fold decrease in the expression of LINE-1 (p-0.069). At the 90-day time-point, a 1.45-fold insignificant decrease in the expression of LINE-1 ORF2 was observed in the heart after exposure to 56Fe (p=0.074).
Figure 4.
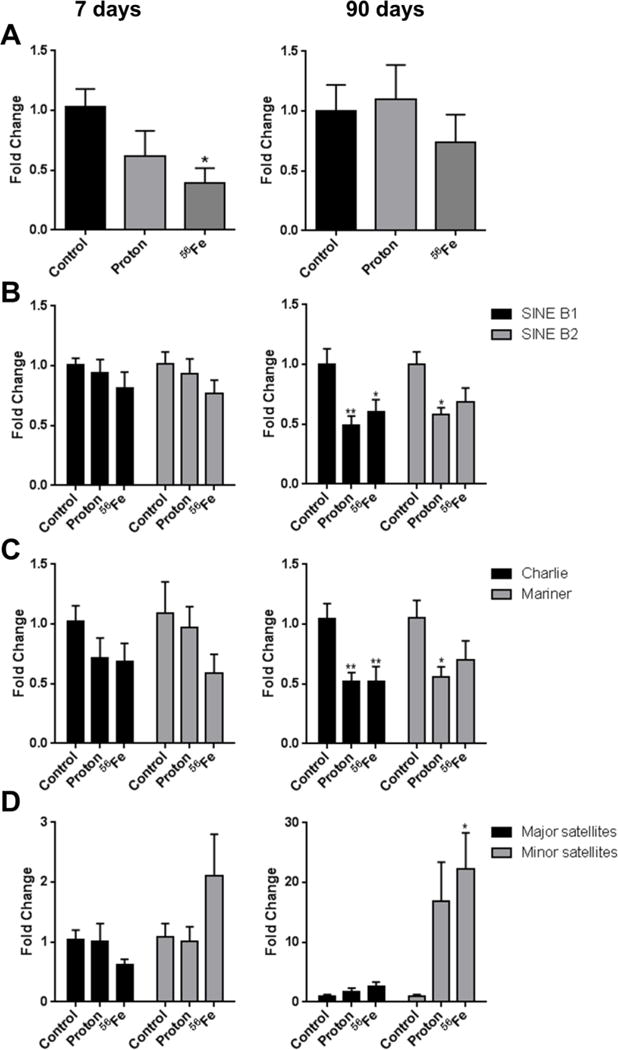
Effects of space radiation on the expression of repetitive elements in the mouse heart, as measured by quantitative real-time PCR. (A) Retrotransposons LINE-1; (B) SINE B1, and SINE B2; (C) DNA transposons Charlie and Mariner; (D) major and minor satellites. Data are presented as mean ± SE. *p≤0.05, **p≤0.01, ANOVA with Dunnett’s test.
Modest decreases in the expression of SINE B1, SINE B2, Mariner and Charlie were observed at the 7-day time-point with somewhat more pronounced effects observed after exposure to 56Fe (Figure 4B–D). At the same time, a nearly 2-fold reactivation of centromeric minor satellite repeats was detected, although insignificant (p=0.062). The 90-day time-point was characterized by even more diverse changes in the expression of repetitive elements. At this time, expression of all retrotransposons (SINE B1 and SINE B2) and transposons (Mariner and Charlie) was significantly decreased in the heart tissue, independently of radiation type. While the expression of major satellites was not affected significantly, dramatic accumulation of mRNA transcripts associated with minor satellites was observed (15.6- and 22.4-fold for proton and 56Fe, respectively; p=0.058 for protons and p<0.05 for 56Fe).
3.4. Effects of 56Fe and protons on DNA methylation machinery
Several mechanisms may contribute to radiation-induced alterations in DNA methylation and expression of repetitive elements. Earlier studies reported that irradiation may affect the mRNA levels and enzymatic activity of DNA methyltransferases, enzymes involved in the regulation of normal patterns of DNA methylation [31]. In our study, at the 7-day time-point very, subtle overexpression of Dnmt1 and Dnmt3a DNA methyltransferases was observed (Figure 5). A more pronounced overexpression of the Uhrf1 gene, involved in recruiting Dnmt1 to hemimethylated sites during replication, in response to protons exposure was observed, although it was not statistically significant (2.94-fold, p=0.059). At the 90-day time-point, a significant loss of Dnmt3a and Dnmt3b was observed after exposure to protons (−1.7- and −1.8-fold, respectively, p<0.05), and 56Fe (−1.76- and −1.8-fold, with p<0.01 and <0.05, respectively).
Figure 5.
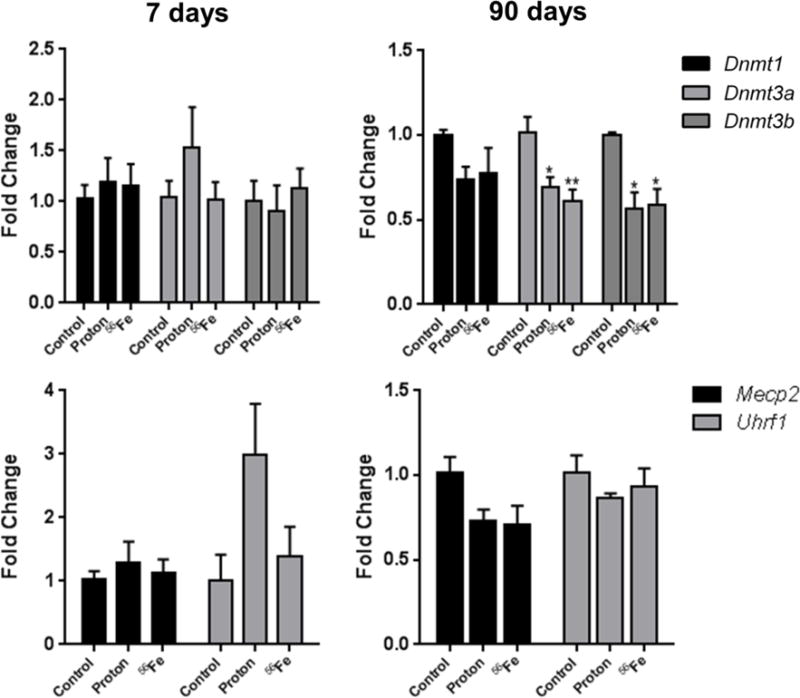
Effects of space radiation on the expression of DNA methylation machinery, as measured by quantitative real-time PCR. Data are presented as mean ± SE. *p≤0.05, **p≤0.01, ANOVA with Dunnett’s test.
3.5. Effects of 56Fe and protons on the one-carbon metabolism pathway
Given that the aberrant expression of DNA methyltransferases could not explain the observed alterations in the methylation of repetitive elements, next we addressed the tissue levels of components involved in the one-carbon metabolism pathway, and metabolism of methionine, in particular. The latter is needed for the synthesis of proteins and polyamines, serves as a precursor for glutathione, and is needed for the synthesis of S-adenosylmethionine (SAM) – a major donor of methyl groups for DNA methylation. Alterations in the one-carbon metabolism pathway may significantly affect DNA methylation [32].
At the 7-day time point, we did not identify substantial changes in the tissue levels of methionine or its down-stream product SAM. At the same time, exposure to both 56Fe and protons were associated with modestly reduced levels of SAH and, subsequently, increased SAM/SAH ratios (Table 1). These finding are in good agreement with the lack of changes in DNA methylation at the 7-day time-point.
Table 1.
Methionine metabolism in hearts of mice exposed to 0.1 Gy of protons, 0.5 Gy of 56Fe, or control treatment (nmol/mg protein, mean ± SEM).
| Metabolite | 7 days post-irradiation | 90 days post-irradiation | ||||
|---|---|---|---|---|---|---|
| Control | Protons | 56Fe | Control | Protons | 56Fe | |
| Methionine | 0.45 ± 0.01 | 0.44 ± 0.027 | 0.52 ± 0.05 | 0.35 ± 0.11 | 0.48 ± 0.05 | 0.50 ± 0.04 |
| SAM | 1.02 ± 0.07 | 1.09 ± 0.07 | 1.14 ± 0.13 | 0.68 ± 0.09 | 0.85 ± 0.07* | 0.93 ± 0.04* |
| SAH | 0.43 ± 0.04 | 0.33 ± 0.01 | 0.30 ± 0.026* | 0.30 ± 0.01 | 0.27 ± 0.01 | 0.30 ± 0.05 |
| SAM/SAH | 2.38 ± 0.16 | 3.30 ± 0.17* | 3.82 ± 0.25* | 2.30 ± 0.23 | 3.20 ± 0.34 | 3.40 ± 0.57 |
| Adenosine | 2.90 ± 0.49 | 1.76 ± 0.17 | 2.30 ± 0.36 | 0.71 ± 0.10 | 0.40 ± 0.05 | 0.51 ± 0.12 |
p<0.05 compared to sham-irradiated control
At the 90-day time-point, the levels of both methionine and SAM were substantially increased in cardiac tissue, suggesting that alterations in one-carbon metabolism pathway are, at least in part, attributable to repetitive elements-associated DNA hypermethylation.
4. Discussion
A limited number of studies have examined the effects of space radiation on cardiac function and structure in animal models. Exposure to protons (0.5 Gy, 1 GeV) and 56Fe (0.15 Gy, 1 GeV/n) in a mouse model induced cardiac infiltration of CD68-positive cells, increased DNA oxidation, myocardial fibrosis, and modified cardiac function, both at baseline and in response to myocardial infarction, in a radiation-type specific manner [33,34]. Exposure to 28Si (0.1–0.5 Gy, 300 MeV/n) caused prolonged apoptosis and increased expression of the common pro-inflammatory cytokines IL-1β, IL-6, or TNF-α in a mouse model [35]. These studies have convincingly demonstrated that exposure to low mean absorbed doses of space radiation may significantly impact the cardiovascular system and lead to the development of heart diseases.
Epigenetic alterations are becoming increasingly recognized among the driving forces of disease development and progression, including heart diseases. Genome-wide alterations in DNA methylation are observed in dilated cardiomyopathy [2], and heart failure is accompanied by altered methylation of genes important for cardiovascular function [3]. Alterations in the promoter methylation occurs in genes that are up-regulated in failing hearts [36,37], providing evidence that epigenetic changes are involved in gene regulation in cardiovascular disease.
Ionizing radiation is a potent epigenotoxic stressor that may significantly, and often for a long-term or even permanently, affect the major epigenetic mechanisms of regulation – methylation of DNA, histone modification and non-coding RNAs. In this study, we aimed to examine the effects of low mean absorbed doses of two common types of space radiation (0.1 Gy of protons, 150 MeV and 0.5 Gy of 56Fe, 600 MeV/n) on cardiac DNA methylation in a mouse model.
In this analysis, we included retrotransposons (capable of autonomous propagation – LINE-1, ERV1, and ERV2 and non-autonomous elements SINE B1 and SINE B2), DNA transposons (Charlie and Mariner), and satellite repeats (major and minor satellites). These repetitive elements comprise about one third of the mouse genome and are usually heavily methylated in order to prevent their aberrant activity [8].
First, we analyzed the methylation of LINE-1 ORF2 using the conventional methylation-sensitive RT-PCR. We did not identify significant changes in the methylation of LINE-1 after exposure to protons or 56Fe at either time-point. Utilization of a more robust and accurate approach for locus-specific DNA methylation – pyrosequencing – allowed us to identify modest hypomethylation of the cardiac LINE-1 ORF1 at the 7-day time-point, independently of the type of exposure. These findings are in good agreement with previous studies that reported global and LINE-1-specific hypomethylation early after exposure to ionizing radiation [11,29].
This hypomethylation at the early time-point, was followed by hypermethylation of LINE-1 ORF1 at the 90-day time-point. This DNA hypermethylation stemmed from the evolutionary young LINE-1 elements, while DNA methylation in evolutionary old elements remained unchanged. Similar patterns were also observed in the recent study by Lima et al, where time-dependent fluctuations in LINE-1 methylation were observed after exposure to 56Fe within the same tissue [15]. Together, these studies provide evidence that space-radiation induced changes in DNA methylation are dynamic and may vary depending on the time after irradiation. At the same time, based on accumulating evidence [16] and the results of the current study, it seems that relatively early post-irradiation time-points are characterized by global and repetitive elements-associated DNA hypomethylation, while later time points are characterized by DNA hypermethylation.
Recent studies clearly indicate that alterations in DNA methylation induced by environmental stressors are not unidirectional and can be characterized by loci of both hypo- and hypermethylation [38]. Therefore, next we addressed the DNA methylation status of ERV1, ERV2, SINE B1, SINE B2, Charlie, Mariner, major and minor satellites in the murine hearts. While minor and non-significant alterations were observed at the 7-day time-point, modest hypermethylation, similar to hypermethylation of LINE-1, was observed in the methylation of repetitive elements at the 90-day time-point. This DNA hypermethylation was more pronounced after exposure to 56Fe, and affected primarily ERV2, SINE B1, Mariner and major satellites. This finding is in good agreement with our previous observation of delayed DNA hypermethylation in repetitive elements in the murine lung after exposure to low mean absorbed doses of 56Fe [16].
To investigate the mechanisms of radiation-induced changes in DNA methylation, we first addressed the expression of a panel of genes, involved in the regulation and maintenance of normal DNA methylation patterns. Although the 7-day time-point was characterized by hypomethylation of repetitive elements, minor increases in the expression of Dnmt1 and Dnmt3a methyltransferases, as well as Uhrf1 were observed. DNA methyltransferases and Uhrf1 are involved in radiation-induced DNA repair [39] and may hence be up-regulated in response to DNA damage. Interestingly, a recent study indicates that overexpression of Uhrf1 drives DNA hypomethylation, suggesting another possible mechanism for the observed LINE-1 hypomethylation at an early time point [40]. While significant hypermethylation was observed in repetitive elements at the 90-day time-point, DNA methyltransferases exhibited unidirectional trends towards their decreased expression. Because similar trends were observed in the expression of Mecp2, the enzyme involved in binding methyl groups to DNA, this may be potentially perceived as a compensatory mechanism in response to DNA hypermethylation. Further studies will be clearly needed to investigate this effect and its consequences.
Because methionine is required for the biosynthesis of SAM, the major methyl donor used by methyltransferases for DNA methylation, the methionine metabolism pathway is closely linked to DNA methylation status. In our study, at the 7-day time-point, only a modest decrease in the cardiac tissue levels of SAH were detected, in congruence with the lack of substantial changes in DNA methylation and DNA methylation machinery. The 90-day time-point was characterized by substantially increased levels of methionine and SAM, and, with that, an associated increase in SAM/SAH ratio. These findings suggest that the delayed effects of space irradiation are associated with the increased synthesis of the donors of methyl groups which may explain the observed hypermethylation within the repetitive elements.
At the same time, other mechanisms of the observed radiation-induced alterations in DNA methylation cannot be excluded. For instance, recent advances in understanding the role of 5-hydroxymethylation and methyl-deoxygenases may provide further knowledge in the process of DNA demethylation. In this study, we addressed the expression of Tet1, the major deoxygenase involved in the conversion of 5-methylcytosine into 5-hydroxymethylcytosine. While at the 7-day time-point non-significant increases in the expression of Tet1 were observed in cardiac tissue, congruent with weak hypomethylation at this time, significant decreases were observed at the 90-day time-point (−2.2-fold, p<0.01 for both, protons and 56Fe) (Figure 6A), which was associated with hypermethylation of repetitive elements. This finding clearly indicates that the mechanisms of radiation-induced alterations in DNA methylation are complex and may involve multiple pathways.
Figure 6.
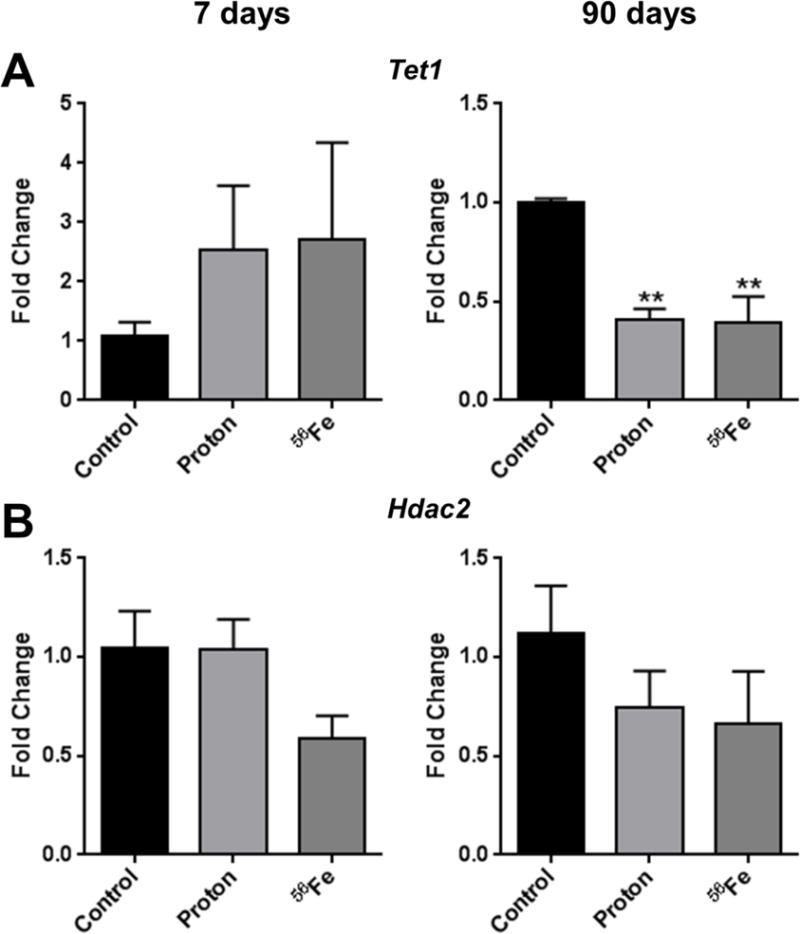
Effects of space radiation on the expression of (A) methyl-deoxygenase Tet1 and (B) histone deacetylase Hdac2, as measured by quantitative real-time PCR. Data are presented as mean ± SE. *p≤0.05, **p≤0.01, ANOVA with Dunnett’s test.
DNA methylation plays a critical role in the silencing of repetitive elements [1] and observed alterations in their methylation status may result in the aberrant expression of repetitive elements. In this study, loss of LINE-1 expression was detected after exposure to protons or 56Fe despite its modest hypomethylation at the 7-day time-point and was still detectable at the 90-day time-point. Furthermore, the mRNA levels of other transposable elements, including SINE B1, SINE B2, Charlie, and Mariner were also decreased after irradiation. This indicates that other mechanisms, such as histone modifications, may be involved in their silencing. Indeed, the expression of the histone deacetylase Hdac2, one of the major regulators of transposable elements expression [41], was decreased in response to protons or 56Fe exposure, although insignificantly (Fig. 6B). The studies that will explore further the interplay between DNA methylation and histone modifications in the regulation of transposable elements expression are warranted.
Observed alteration in the methylation and expression of transposable elements may significantly affect the cellular epigenome, alter the expression of genetic information and potentially lead to disease development. It has been shown that retrotransposons like LINE-1 and SINE can influence the expression of adjacent gene promoters [42–44]. Furthermore, SINEs have been shown to function as insulators, facilitating the expression of numerous genes [45]. Decreased expression of LINE-1 in cardiomyocytes is associated with reduced oxidative stress [10].
The most pronounced changes, however, were detected in the expression of repetitive elements that are transcribed from the satellite repeats in centromeric (minor satellites) and pericentromeric (major satellites) regions and are involved in the formation of heterochromatin [46–48]. Increases in their expression may significantly impact the chromatin status, centromeric cohesion and dissociation during chromosome segregation [46,47]. A recent study has shown that a pronounced (up to 27-fold) increase in satellite repeat transcripts was a common feature of all examined human failing hearts when compared to normal hearts [49]. It remains unknown whether this satellite-associated aberrant transcription is one of the mechanisms or consequences of the disease. Our data indicate that transcriptional activation of satellite repeats may occur as an early response to irradiation (7-day time-point), and may persist over time (90-day time-point), suggesting that the accumulation of satellite DNA mRNA transcripts may serve as one of the driving forces of the disease, rather than being merely a consequence.
In conclusions, we report that exposure to low absorbed mean doses of protons and 56Fe ions lead to non-unidirectional alterations in the methylation and expression of repetitive elements. We further show that space radiation affects the one-carbon metabolism pathway, and these changes, at least in part, contribute to alterations in DNA methylation. Given that the one-carbon metabolism pathway can be modulated by dietary modifications, these findings suggest a potential strategy for the mitigation and, possibly, prevention of the negative effects exerted by space radiation on the cardiovascular system. Additionally, we show that the methylation status and expression of repetitive elements may serve as early biomarkers of exposure to space radiation. Further studies, especially those that investigate the effects of combined exposures to low mean absorbed doses of protons and heavy ions (conditions most relevant to the space environment) as well as time- and dose-dependent correlations between space radiation and DNA methylation endpoints are clearly needed and are being currently performed at our laboratories.
Supplementary Material
Highlights.
Radiation-induced dynamic changes in cardiac DNA methylation were detected
Early LINE-1 hypomethylation was followed by hypermethylation at a later time-point
Radiation affected one-carbon metabolism in the heart tissue
Irradiation resulted in accumulation of satellite DNA mRNA transcripts
Acknowledgments
We are thankful to Dr. Kristy Kutanzi for the critical reading and editing of the manuscript. This work was supported by National Aeronautics and Space Administration [NNX10AD59G]; National Space Biomedical Research Institute [RE03701 through NCC 9-58]; the National Institutes of Health [CA148679 to MB, R37 CA71382 to MHJ, 1P20GM109005, and Clinical and Translational Science Award UL1TR000039 and KL2TR000063], US Veterans Administration, the Arkansas Biosciences Institute (ABI), ABI Grant for Core Metabolomics Laboratory, and ACH Foundation Grants for studies of metabolic changes in children with Autism and patients with Down Syndrome.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no conflict of interests.
References
- 1.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 2.Haas J, Frese KS, Park YJ, Keller A, Vogel B, Lindroth AM, Weichenhan D, Franke J, Fischer S, Bauer A, Marquart S, Sedaghat-Hamedani F, Kayvanpour E, Kohler D, Wolf NM, Hassel S, Nietsch R, Wieland T, Ehlermann P, Schultz JH, Dosch A, Mereles D, Hardt S, Backs J, Hoheisel JD, Plass C, Katus HA, Meder B. Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol Med. 2013;5:413–429. doi: 10.1002/emmm.201201553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Movassagh M, Choy M, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PloS One. 2010;5:e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duygu B, Poels EM, da Costa Martins Paula A. Genetics and epigenetics of arrhythmia and heart failure. Front Genet. 2013;4:219. doi: 10.3389/fgene.2013.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowley M, Oakey RJ. Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet. 2013;9:e1003234. doi: 10.1371/journal.pgen.1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedoroff NV. Presidential address. transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- 7.Ross JP, Rand KN, Molloy PL. Hypomethylation of repeated DNA sequences in cancer. Epigenomics. 2010;2:245–269. doi: 10.2217/epi.10.2. [DOI] [PubMed] [Google Scholar]
- 8.Miousse IR, Chalbot MC, Lumen A, Ferguson A, Kavouras IG, Koturbash I. Response of transposable elements to environmental stressors. Mutat Res Rev Mutat Res. 2015;765:19–39. doi: 10.1016/j.mrrev.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ugarkovic D. Functional elements residing within satellite DNAs. EMBO Rep. 2005;6:1035–1039. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucchinetti E, Feng J, Silva R, Tolstonog GV, Schaub MC, Schumann GG, Zaugg M. Inhibition of LINE-1 expression in the heart decreases ischemic damage by activation of akt/PKB signaling. Physiol Genomics. 2006;25:314–324. doi: 10.1152/physiolgenomics.00251.2005. [DOI] [PubMed] [Google Scholar]
- 11.Koturbash I, Pogribny I, Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue–a possible mechanism contributing to radiation carcinogenesis? Biochem Biophys Res Commun. 2005;337:526–533. doi: 10.1016/j.bbrc.2005.09.084. [DOI] [PubMed] [Google Scholar]
- 12.Baulch JE, Aypar U, Waters KM, Yang AJ, Morgan WF. Genetic and epigenetic changes in chromosomally stable and unstable progeny of irradiated cells. PloS One. 2014;9:e107722. doi: 10.1371/journal.pone.0107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jangiam W, Tungjai M, Rithidech KN. Induction of chronic oxidative stress, chronic inflammation and aberrant patterns of DNA methylation in the liver of titanium-exposed CBA/CaJ mice. Int J Radiat Biol. 2015;91:389–398. doi: 10.3109/09553002.2015.1001882. [DOI] [PubMed] [Google Scholar]
- 14.Miousse IR, Shao L, Chang J, Feng W, Wang Y, Allen AR, Turner J, Stewart B, Raber J, Zhou D, Koturbash I. Exposure to low-dose 56Fe-ion radiation induces long-term epigenetic alterations in mouse bone marrow hematopoietic progenitor and stem cells. Radiat Res. 2014;182:92–101. doi: 10.1667/RR13580.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lima F, Ding D, Goetz W, Yang AJ, Baulch JE. High LET 56Fe ion irradiation induces tissue-specific changes in DNA methylation in the mouse. Environ Mol Mutagen. 2014;55:266–277. doi: 10.1002/em.21832. [DOI] [PubMed] [Google Scholar]
- 16.Nzabarushimana E, Miousse IR, Shao L, Chang J, Allen AR, Turner J, Stewart B, Raber J, Koturbash I. Long-term epigenetic effects of exposure to low doses of 56Fe in the mouse lung. J Radiat Res. 2014;55:823–828. doi: 10.1093/jrr/rru010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nzabarushimana E, Prior S, Miousse IR, Pathak R, Allen AR, Latendresse J, Olsen RH, Raber J, Hauer-Jensen M, Nelson GA. Combined exposure to protons and 56Fe leads to overexpression of Il13 and reactivation of repetitive elements in the mouse lung. Life Sci Space Res (Amst) 2015;7:1–8. doi: 10.1016/j.lssr.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azizova T, Muirhead C, Druzhinina M, Grigoryeva E, Vlasenko E, Sumina M, O’Hagan J, Zhang W, Haylock R, Hunter N. Cardiovascular diseases in the cohort of workers first employed at mayak PA in 1948–1958. Radiat Res. 2010;174:155–168. doi: 10.1667/RR1789.1. [DOI] [PubMed] [Google Scholar]
- 19.Little M, Tawn E, Tzoulaki I, Wakeford R, Hildebrandt G, Paris F, Tapio S, Elliott P. A systematic review of epidemiological associations between low and moderate doses of ionizing radiation and late cardiovascular effects, and their possible mechanisms. Radiat Res. 2008;169:99–109. doi: 10.1667/RR1070.1. [DOI] [PubMed] [Google Scholar]
- 20.Schultz-Hector S, Trott K. Radiation-induced cardiovascular diseases: Is the epidemiologic evidence compatible with the radiobiologic data? Int J Radiat Oncol Biol Phys. 2007;67:10–18. doi: 10.1016/j.ijrobp.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 21.NCRP. (NCRP Report No. 153).Information Needed to make Radiation Protection Recommendations for Space Missions Beyond Low-Earth Orbit. 2006 [Google Scholar]
- 22.Cucinotta FA. Review of NASA approach to space radiation risk assessments for mars exploration. Health Phys. 2015;108:131–142. doi: 10.1097/HP.0000000000000255. [DOI] [PubMed] [Google Scholar]
- 23.Christofidou-Solomidou M, Pietrofesa RA, Arguiri E, Schweitzer KS, Berdyshev EV, McCarthy M, Corbitt A, Alwood JS, Yu Y, Globus RK, Solomides CC, Ullrich RL, Petrache I. Space radiation-associated lung injury in a murine model. Am J Physiol Lung Cell Mol Physiol. 2015;308:L416–L428. doi: 10.1152/ajplung.00260.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farin AM, Manzo ND, Kirsch DG, Stripp BR. Low- and high-LET radiation drives clonal expansion of lung progenitor cells in vivo. Radiat Res. 2015;183:124–132. doi: 10.1667/RR13878.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melnyk S, Pogribna M, Pogribny IP, Yi P, James SJ. Measurement of plasma and intracellular S-adenosylmethionine and S-adenosylhomocysteine utilizing coulometric electrochemical detection: Alterations with plasma homocysteine and pyridoxal 5′-phosphate concentrations. Clin Chem. 2000;46:265–272. [PubMed] [Google Scholar]
- 26.Miousse IR, Koturbash I. The fine LINE: Methylation drawing the cancer landscape. Biomed Res Int. 2015;2015:131547. doi: 10.1155/2015/131547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aypar U, Morgan WF, Baulch JE. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutat Res. 2011;707:24–33. doi: 10.1016/j.mrfmmm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Goetz W, Morgan MN, Baulch JE. The effect of radiation quality on genomic DNA methylation profiles in irradiated human cell lines. Radiat Res. 2011;175:575–587. doi: 10.1667/RR2390.1. [DOI] [PubMed] [Google Scholar]
- 29.Koturbash I, Boyko A, Rodriguez-Juarez R, McDonald RJ, Tryndyak VP, Kovalchuk I, Pogribny IP, Kovalchuk O. Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis. 2007;28:1831–1838. doi: 10.1093/carcin/bgm053. [DOI] [PubMed] [Google Scholar]
- 30.Newman MR, Sykes PJ, Blyth BJ, Bezak E, Lawrence MD, Morel KL, Ormsby RJ. The methylation of DNA repeat elements is sex-dependent and temporally different in response to X radiation in radiosensitive and radioresistant mouse strains. Radiat Res. 2014;181:65–75. doi: 10.1667/RR13460.1. [DOI] [PubMed] [Google Scholar]
- 31.Koturbash I, Baker M, Loree J, Kutanzi K, Hudson D, Pogribny I, Sedelnikova O, Bonner W, Kovalchuk O. Epigenetic dysregulation underlies radiation-induced transgenerational genome instability in vivo. Int J Radiat Oncol Biol Phys. 2006;66:327–330. doi: 10.1016/j.ijrobp.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Waterland RA. Assessing the effects of high methionine intake on DNA methylation. J Nutr. 2006;136:1706S–1710S. doi: 10.1093/jn/136.6.1706S. [DOI] [PubMed] [Google Scholar]
- 33.Sasi SP, Yan X, Lee J, Sisakyan H, Carrozza J, Goukassian DA. Radiation-associated degenerative cardiovascular risks during normal aging and after adverse CV event 10 months post-initial exposure. J Radiat Res. 2014;55:i111–i112. [Google Scholar]
- 34.Yan X, Sasi SP, Gee H, Lee J, Yang Y, Mehrzad R, Onufrak J, Song J, Enderling H, Agarwal A. Cardiovascular risks associated with low dose ionizing particle radiation. PloS One. 2014;9:e110269. doi: 10.1371/journal.pone.0110269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tungjai M, Whorton EB, Rithidech KN. Persistence of apoptosis and inflammatory responses in the heart and bone marrow of mice following whole-body exposure to 28Silicon (28Si) ions. Radiat Environ Biophys. 2013;52:339–350. doi: 10.1007/s00411-013-0479-4. [DOI] [PubMed] [Google Scholar]
- 36.Movassagh M, Choy MK, Knowles DA, Cordeddu L, Haider S, Down T, Siggens L, Vujic A, Simeoni I, Penkett C, Goddard M, Lio P, Bennett MR, Foo RS. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–2422. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Movassagh M, Vujic A, Foo R. Genome-wide DNA methylation in human heart failure. Epigenomics. 2011;3:103–109. doi: 10.2217/epi.10.70. [DOI] [PubMed] [Google Scholar]
- 38.Miousse IR, Chalbot MC, Aykin-Burns N, Wang X, Basnakian A, Kavouras IG, Koturbash I. Epigenetic alterations induced by ambient particulate matter in mouse macrophages. Environ Mol Mutagen. 2014;55:428–435. doi: 10.1002/em.21855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mistry H, Tamblyn L, Butt H, Sisgoreo D, Gracias A, Larin M, Gopalakrishnan K, Hande MP, McPherson JP. UHRF1 is a genome caretaker that facilitates the DNA damage response to gamma-irradiation. Genome Integr. 2010;1:7. doi: 10.1186/2041-9414-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montoya-Durango DE, Liu Y, Teneng I, Kalbfleisch T, Lacy ME, Steffen MC, Ramos KS. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat Res. 2009;665:20–28. doi: 10.1016/j.mrfmmm.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Estecio MR, Gallegos J, Dekmezian M, Lu Y, Liang S, Issa JP. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol Cancer Res. 2012;10:1332–1342. doi: 10.1158/1541-7786.MCR-12-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, Goel A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63:635–646. doi: 10.1136/gutjnl-2012-304219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev. 2010;20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roman AC, Gonzalez-Rico FJ, Fernandez-Salguero PM. B1-SINE retrotransposons: Establishing genomic insulatory networks. Mob Genet Elements. 2011;1:66–70. doi: 10.4161/mge.1.1.15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci U S A. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferri F, Bouzinba-Segard H, Velasco G, Hube F, Francastel C. Non-coding murine centromeric transcripts associate with and potentiate aurora B kinase. Nucleic Acids Res. 2009;37:5071–5080. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haider S, Cordeddu L, Robinson E, Movassagh M, Siggens L, Vujic A, Choy MK, Goddard M, Lio P, Foo R. The landscape of DNA repeat elements in human heart failure. Genome Biol. 2012;13:R90. doi: 10.1186/gb-2012-13-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


