Abstract
Neuronal activity sculpts brain development by inducing the transcription of genes such as Brain-Derived Neurotrophic Factor (Bdnf) that modulate the function of synapses. Sensory experience is transduced into changes in gene transcription via the activation of calcium signaling pathways downstream of both L-type voltage gated calcium channels (L-VGCCs) and NMDA-type glutamate receptors (NMDARs). These signaling pathways converge on the regulation of transcription factors including Calcium-Response Factor (CaRF). Whereas CaRF is dispensable for the transcriptional induction of Bdnf following the activation of L-VGCCs, here we show that loss of CaRF leads to enhanced NMDAR-dependent transcription of Bdnf as well as Arc. We identify the NMDAR subunit-encoding gene Grin3a as a regulatory target of CaRF, and we show that expression of both Carf and Grin3a is depressed by the elevation of intracellular calcium, linking the function of this transcriptional regulatory pathway to neuronal activity. We find that light-dependent activation of Bdnf and Arc transcription is enhanced in the visual cortex of young CaRF knockout mice, suggesting a role for CaRF-dependent dampening of NMDAR-dependent transcription in the developing brain. Finally, we demonstrate that enhanced Bdnf expression in CaRF-lacking neurons increases inhibitory synapse formation. Taken together these data reveal a novel role for CaRF as an upstream regulator of NMDAR-dependent gene transcription and synapse formation in the developing brain.
Introduction
In the developing mammalian brain, transient sensory stimuli drive long-lasting changes in synapse development and neuronal function by inducing the transcription of activity-regulated genes (West and Greenberg 2011). Calcium plays a key role in this signal transduction by initiating intracellular signaling cascades that carry the signal to the nucleus where they activate multiple transcription factors that bind regulatory elements of activity-regulated genes (Lyons and West 2011). In addition to ubiquitous immediate early genes (IEGs) such as Fos and Jun, synaptic activity regulates a functionally important set of neuron-selective genes, which include the neurotrophin BDNF, the intracellular scaffolds Arc and Homer, the glutamate receptor binding protein Narp, and the transcription factor Npas4 (Leslie and Nedivi 2011; Hong et al. 2008; Bloodgood et al. 2013; Gu et al. 2013; Wang et al. 2006; Hu et al. 2010). Induced expression of these gene products couples synapse development and function with sensory-driven neuronal activity to adapt brain development to the environment.
Glutamate release at synapses drives calcium into neurons by opening both NMDA-type glutamate receptors (NMDARs) and L-type voltage-gated calcium channels (L-VGCCs). Calcium acts locally at these channels to induce the signaling cascades that regulate nuclear transcription factors, and the source of calcium entry can influence the specificity of downstream signaling (Bito et al. 1996; Dolmetsch et al. 2001; Karpova et al. 2013; Hardingham et al. 2002). We and others have used transcription of Bdnf as a model to discover the neuronal activity-regulated transcriptional mechanisms that are important for developmental synapse plasticity. BDNF is a key effector of activity-dependent synapse development and plasticity, and it is the precise temporal and spatial control of Bdnf transcription that permits this signaling molecule to regulate these dynamic processes (Leslie and Nedivi 2011; West et al. 2014).
The Bdnf gene contains nine alternative promoters, each of which is activity-inducible in neurons and most of which are regulated by multiple activity-dependent transcription factors (West et al. 2014). Bdnf exon IV-containing transcripts (Bdnf IV) are the most highly expressed and activity-regulated of the Bdnf splice variants in developing neurons, and the transcription factors that regulate Bdnf promoter IV have been well described. We first identified Calcium Response Factor (CaRF) as a transcription factor based on its ability to bind the L-VGCC-responsive calcium-response element 1 (CaRE1) in Bdnf promoter IV (Tao et al. 2002). Basal expression of Bdnf IV is reduced in the cortex of adult CaRF knockout mice, consistent with CaRF functioning as an activator of Bdnf promoter IV (McDowell et al. 2010). However L-VGCC-induced transcription of Bdnf IV is unaffected by the loss of CaRF and instead the transcription factor MEF2C mediates the actions of CaRE1 in response to this stimulus (Lyons et al. 2012).
Given our evidence for the stimulus-specific contributions of CaRF to Bdnf promoter IV regulation, here we set out to determine the requirement for CaRF in NMDAR-induced transcription of Bdnf. Here we report that loss of CaRF leads to enhanced NMDAR-dependent induction of both Bdnf IV and the activity-regulated gene Arc, and we investigate potential mechanisms as well as biological consequences of this change in transcriptional regulation. These data reveal a novel role for CaRF as an upstream regulator of NMDAR-dependent processes in the developing brain.
Materials and Methods
Dissociated Neuron Cultures
Neuron-enriched cultures were generated from cortex of male and female E16.5 CD1 mouse embryos (Charles River Laboratories) and cultured as previously described (McDowell et al. 2010; Tao et al. 2002). Withdrawal from tetrodotoxin (TTX WD) was done by treating neurons for 48 hrs with 1μM TTX (Tocris) prior either to harvesting cells (for the control condition) or washing out the TTX with Neurobasal medium (Invitrogen). Isotonic membrane depolarization with 55mM extracellular KCl was done as previously described (Lyons et al. 2012). APV (Tocris) was used at a concentration of 100μM. Nimodipine (Sigma) was used at a concentration of 5μM. EGTA (Sigma) was used at 2.5mM. Actinomycin D (Sigma) was used at 30nM. Pharmacological blockers were added 2 min prior to TTX WD or KCl addition and maintained throughout the period of stimulation. All experiments were conducted in accordance with an animal protocol approved by the Duke University Institutional Animal Care and Use Committee.
Quantitative PCR
RNA was harvested from cultured mouse cortical neurons on DIV7 following 90min or 6hrs of KCl-mediated membrane depolarization or TTX WD as described in the text. RNA was harvested using the Absolutely RNA Miniprep Kit (Agilent) and cDNA was synthesized by Superscript II (Invitrogen). Quantitative SYBR green PCR was performed on an ABI 7300 real-time PCR machine (Applied Biosystems) using intron-spanning primers (IDT): Mouse Arc F: 5′- GAGCCTACAGAGCAGGAGA -3′, R: 5′- TGCCTTGAAAGTGTCTTGGA -3′; Mouse Bdnf exon I F: 5′- AGTCTCCAGGACAGCAAAGC -3′, R: 5′- GCCTTCATGCAACCGAAGTA -3′; Mouse Bdnf exon IV: F: 5′- CGCCATGCAATTTCCACTATCAATAA -3′, R: 5′- GCCTTCATGCAACCGAAGTATG -3′; Mouse Carf F: 5′- GCATTGACAAATGGGATTCCGTC -3′, R: 5′- GTTGAAGAACCTTTGCTGGCTC -3′; Mouse/Rat Carf F: 5′-TGCCGTCTTAGGAGTTGTGA-3′, R: 5′-CTCGACTTCCTGCATTGACA-3′. Mouse cFos F: 5′- TTTATCCCCACGGTGACAGC -3′, R: 5′- CTGCTCTACTTTGCCCCTTCT -3′; Mouse Gapdh F: 5′- CATGGCCTTCCGTGTTCCT -3, R: 5′- TGATGTCATCATACTTGGCAGGTT-3′; Mouse Grin1 F: 5′- GCTCAGAAACCCCTCAGACA -3′, R: 5′- GGCATCCTTGTGTCGCTTGTAG -3′; Mouse Grin2a F: 5′- TACTCCAGCGCTGAACATTG -3′, R: 5′- TCAGCTGGACCTGTGTCTTG -3′; Mouse Grin2b F: 5′- GAGCATAATCACCCGCATCT -3′, R: 5′- AAGGCACCGTGTCCGTATCC -3′; Mouse Grin2c F: 5′- GCAGAACTTCCTGGACTTGC -3′, R: 5′- CTCTTCACGGGAGCAGTAGG -3′; Mouse Grin2d F: 5′- TTTTGAGGTGCTGGAGGAGT -3′, R: 5′- GTCTCGGTTATCCCAGGTGA -3′; Mouse Grin3a F: 5′- AAA GCC ATT TGC CAT TGA AG -3′, R: 5′- GAA TCC TAT GCA CAG CAG CA -3′; Mouse Grin3b F: 5′- CTACATCAAGGCGAGCTTCC -3′, R: 5′- AGCTTGCAGTCCGCATCTAT -3′. Data were all normalized to expression of the housekeeping gene Gapdh to control for sample size and processing. For CaRF luciferase assay, RNA was harvested from HEK293T cells at 2 days after transfection. cCaRE-Luc plasmid was previously described (Pfenning et al. 2010; Lyons et al. 2012) and TK-renilla luciferase is from Promega. The firefly luciferase mRNA levels were normalized for each well to cotransfected renilla luciferase mRNA levels. The primer sequences for firefly luciferase were 5′-GAGGTGAACATCACGTACGCG-3′ and 5′-AAGAGAGTTTTCACTGCATACGACG-3 and for renilla luciferase were 5′-GAAACTTCTTGGCACCTTCAACA-3′ and 5′-GCTTATCTACGTGCAAGTGATGATTT-3′
RNAi and Lentiviral Infection
Four independent shRNAs were used to knockdown mouse Carf, and were paired with a vector-matched control (Ctrl). Carf shRNA1 (5′-GAAGACAGCACCAGCAATTAC-3′) and Ctrl1(5′-AAACAAGCCCATTCGCGGATT-3), which is a scrambled version of Carf shRNA1, were cloned into vector pLLx3.8 (Zhou et al. 2007). Carf shRNA2 (TRCN0000086260; 5′-GCAGATGAACATAGCCCTCAA-3′), Carf shRNA3 (TRCN0000086262; 5′-GACGATGGTGAGAAGTCAGAA-3′), Carf shRNA4 (TRCN0000086261; 5′-CCAGCCAGGATATACATTAAA-3′) were purchased from Thermo Scientific and the empty pLKO.1 vector was used as Ctrl2. Bdnf shRNA (TRCN0000065384; 5′-CAAGGCTGTTAGAGAGATAATTGGA-3′) were purchased from Thermo Scientific and the empty pLKO.1 vector was used as Ctrl. The rat GluN3A and rat CaRF-myc expression plasmids were constructed by placing the rat Grin3a or Carf-myc coding sequences under control of the ubiquitin promoter in the lentiviral expression vector pFUIGW (Lois et al. 2002). The C-terminally truncated rat CaRF(1-268) expression plasmid was cloned by PCR from rat brain cDNA generating a fragment expressing amino acids 1-268, which corresponds to the shortest truncated version of mouse CaRF (AAs 1-251, Fig. S4). The construct was placed under control of the ubiquitin promoter in the lentiviral expression vector pFUIGW (Lois et al. 2002). For viral infection of neurons, shRNA or viral expression constructs were packaged as lentiviral particles in HEK 293T cells following standard procedures. Concentrated viruses were titered on mouse cortical neurons which were infected for 6 hrs on day in vitro (DIV) 1 at a multiplicity of infection of 1 in BME medium (Sigma) with 0.4μg/mL added polybrene (Sigma).
Immunofluorescence
Embryonic mouse cortical neurons were cultured on PDL/laminin coated glass coverslips (Bellco) and fixed in 4% paraformaldehyde at room temperature for 10mins. Neurons were blocked in 10% normal goat serum and permeabilized in 0.3% Triton X-100 prior to antibody incubation. Coverslips were incubated in primary antibodies overnight at 4°C. Secondary antibodies were incubated at room temperature for 1 hr. Hoechst dye (0.1μg/ml, Sigma) was used to label nuclei. Primary antibodies used in this study for immunocytochemistry were mouse anti-MEF2D, 1:1000 (BD Biosciences; #610774). mouse anti-Gephyrin, 1:500 (Synaptic Systems; 147021), rabbit anti-GAD65, 1:500 (Millpore; AB5082), chicken anti-GFP, 1:2000 (Millpore; AB16901).
Western Blotting
Cells were homogenized in homogenization buffer (320 mM Sucrose, 10mM HEPES pH 7.4, 2mM EDTA, 1mM DTT and protease inhibitors). Nuclear pellet was centrifuged out at 1500xg for 15min. Cytosolic and membrane fractions were separated by centrifuging at 200000xg for 20min. Membrane, cytoplasmic, and nuclear extracts were run for SDS-PAGE and transferred to Nitrocellulose for western blotting following standard procedures. Primary antibodies used in this study for western blotting were mouse anti-Actin, 1:5000 (Millipore; MAB1501); rabbit anti-CaRF, 1:500 (#4510,McDowell et al., 2010); rabbit anti-Histone H3, 1:5000 (Millipore; catalog #070-690); rabbit anti-GluN3A, 1:1000 (Millipore; #07-356); mouse anti-Transferrin Receptor, 1:2000 (Invitrogen; #13-6800).
Electrophoretic Mobility Shift Assays (EMSA)
Nuclear extracts from cultured mouse cortical neurons infected on DIV1 with viruses expressing the shRNAs targeting Carf or their respective controls were harvested on DIV7 and used for EMSA as previously described (McDowell et al. 2010). Probes were end labeled with 32P and EMSA images were captured on a Storm phosphorimager (Molecular Dynamics). The high affinity CaRF-binding cCaRE probe, CaRE1 and mCaRE1 have been previously described (Pfenning et al. 2010; Lyons et al. 2012). The probes of two potential CaRF binding sites near Grin3a promoter are listed here (gCaRE1: 5′-TCATTATGAGACAGA, gCaRE2: 5′-TCTCGAGGAGACAGA).
BDNF ELISA
Two-site BDNF ELISA was performed as previously described (Hong et al. 2008; McDowell et al. 2010). Total protein concentration in the lysate was measured by BCA protein assay kit (Pierce) and BDNF protein concentrations were measured by the BDNF Emax ImmunoAssay System (Promega).
Pilocarpine-induced Seizure
Adult (8-12 week old) male C57BL6/J mice (Charles River Labs) were weighed and injected with 1 mg/kg methyl scopolamine nitrate (i.p.). 30 min later mice were injected i.p. with either 337 mg/kg pilocarpine HCl or saline (control mice) and monitored for status epilepticus as previously described (Wijayatunge et al. 2014). 1, 3, or 6 hrs following the onset of status epilepticus, mice were deeply anesthetized with isofluorane and decapitated for brain harvesting. Brains of control mice were harvested 3 hrs after the saline injection. 5-6 mice were used for each time point. The hippocampus was rapidly dissected bilaterally, flash frozen on dry ice/ethanol, and stored at −80°C prior to RNA harvesting, cDNA synthesis, and quantitative PCR as described above.
Dark Adaptation and Light Exposure
Male and female Carf wildtype (WT) and knockout (KO) adult mice (McDowell et al. 2010) or pups with their mothers at postnatal day 14 (P14) were transferred from their normal housing room with a 12 hr:12 hr light:dark cycle into a light-tight dark housing room to maintain constant darkness for 7 days. Animals in the unstimulated (dark) condition were killed and their eyes enucleated in the dark prior to bringing the body into the light to dissect the brain. Animals in the stimulated (light-exposed) condition were removed from the dark room and exposed to normal lighting for 6 hrs prior to tissue harvesting. For MK-801 (Sigma) experiments, 6-month-old adult were injected intraperitoneally (i.p.) in the dark with either saline (control) or 1mg/kg MK-801 diluted in 0.9% NaCl 30 min prior to being brought into the light. V1 visual cortex and S1 somatosensory cortex were isolated based on anatomical landmarks and immediately flash frozen. RNA and protein were harvested and measured as described above.
Quantification of inhibitory synapse density
Embryonic mouse cortical neurons were cultured on PDL/laminin coated glass coverslips (Bellco). Neurons were transfected with Ctrl1 or Carf shRNA1 by lipofectamine 2000 on DIV 3 and fixed in 4% paraformaldehyde at room temperature for 10mins on either DIV14 or DIV19. Neurons were blocked in 16% normal goat serum and permeabilized in 0.2% Triton X-100 prior to antibody incubation. Coverslips were incubated in primary antibodies overnight at 4°C. Secondary antibodies were incubated at room temperature for 1 hr. Hoechst dye (0.1μg/ml, Sigma) was used to label nuclei. Primary antibodies used in this study were mouse anti-Gephyrin, 1:500 (Synaptic Systems; 147021), rabbit anti-GAD65, 1:500 (Millpore; AB5082), chicken anti-GFP, 1:2000 (Millpore; AB16901). Images were acquired on a Leica SP8 confocal microscope with a 40X objective at 1024X1024 pixel resolution. Images were collected as a z-stack of 10-12 sections at 0.5μm step size, and maximum intensity projections generated from the z-stack were used for analysis. Synapse density was quantified with ImageJ using NeuronJ and SynapCountJ plugin. For each experiment, approximately 5-10 neurons from at least two different coverslips were analyzed, and between 2-3 experiments were conducted per condition.
Statistical Analyses
Unless otherwise indicated, all data presented are the average of at least three biological replicates from each of at least two independent experiments. Also unless otherwise indicated, data were analyzed by a Student’s unpaired t-test, and p<0.05 was considered significant. Bar and line graphs show mean values and all error bars show S.E.M.
Results
Loss of CaRF results in potentiation of NMDAR-dependent transcription
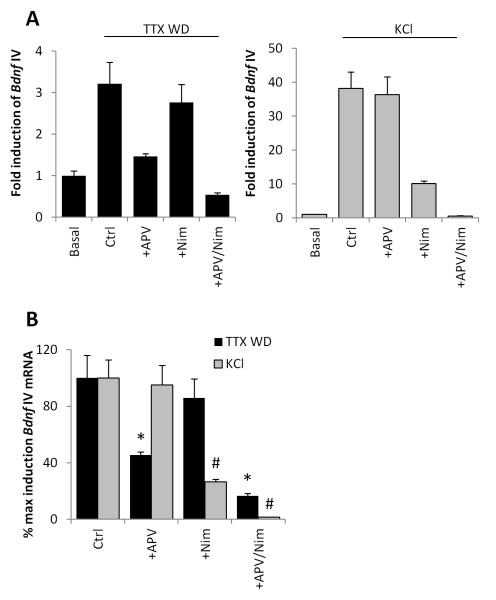
To assay NMDAR-dependent activation of Bdnf transcription, we stimulated dissociated mouse cortical neuron cultures with tetrodoxin withdrawal (TTX WD). This stimulus has previously been characterized for its ability to induce IEG expression in neurons in an NMDAR-dependent manner (Rao et al. 2006; Saha et al. 2011; Ghiretti et al. 2014), including exon IV-containing forms of Bdnf (Fig.1A). We confirmed that pretreatment with the NMDAR antagonist APV significantly attenuated TTX WD-induced Bdnf IV expression (p=0.0285). Conversely, pre-treatment with the L-VGCC antagonist Nimodipine alone had no significant effect on TTX WD-induced Bdnf IV expression (p=0.5372), though it blocked Bdnf IV transcription induced by membrane depolarization with elevated extracellular KCl (p= 0.0047) (Fig. 1A,B). Thus using these two stimulation paradigms (TTX WD and elevation of extracellular KCl) we can selectively activate gene transcription in an NMDAR- or L-VGCC-dependent manner.
Figure 1. Stimulus-selective induction of Bdnf exon IV transcription.
(A) Levels of Bdnf exon IV-containing mRNA in mouse cortical neurons either without stimulation (Basal) or 6 hrs following TTX withdrawal (TTX WD) or 55mM elevated extracellular potassium (KCl) prior to RNA harvesting. Stimulation was done in the presence of the following pharmacological blockers of NMDARs and L-VSCCs: none (Ctrl), +APV, nimodipine (+Nim), or APV + Nim (+APV/Nim). mRNA levels are reported as fold induction relative to Basal. n=3. (B) Relative induction Bdnf exon IV-containing mRNA by TTX WD or KCl replotted from (A) for direct comparison of the relative efficacy of NMDAR and L-VSCC inhibitors in the two different stimulation protocols. We set the fold induction reached by either TTX WD or KCl in the Ctrl group to 100%. *p<0.05 compared with Ctrl in TTX WD. #p<0.05 compared with Ctrl in KCl.
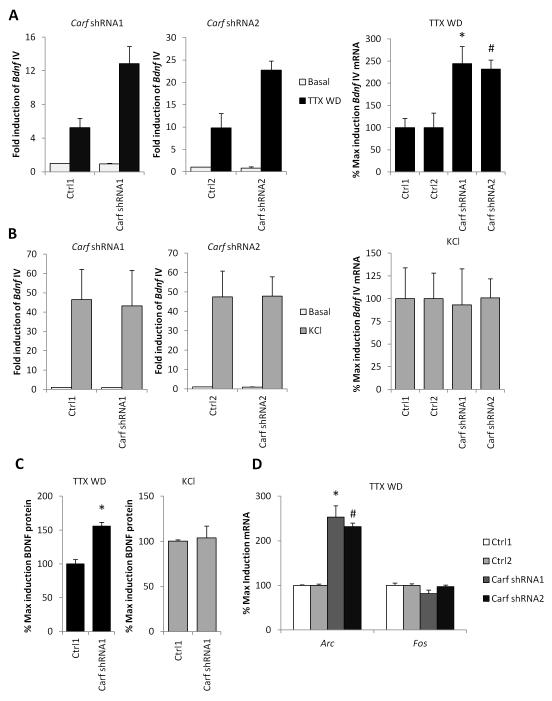
To test the requirement for CaRF in NMDAR-dependent activation of Bdnf IV, we characterized two independent shRNAs in lentiviral vectors that knockdown (KD) expression of CaRF in cultured neurons (Fig. S1). We then measured Bdnf IV mRNA following TTX WD. To our surprise, given that we have previously characterized CaRF as an activator of Bdnf transcription, we found that Bdnf IV induction was significantly potentiated upon TTX WD in CaRF KD neurons compared with neurons that were infected with paired control viruses (Carf shRNA1: p= 0.0171; Carf shRNA2: p= 0.0269; Fig. 2A). This effect was selective for the activation of Bdnf IV transcription by NMDARs because we did not observe any significant difference in Bdnf IV induction between CaRF control and KD neurons when we stimulated L-VGCC-dependent transcription with the elevation of extracellular KCl (Carf shRNA1: p= 0.8989; Carf shRNA2: p= 0.9814; Fig. 2B). The transcriptional activation of Bdnf IV by both TTX WD and KCl leads to increased expression of BDNF protein as measured by ELISA (Fold induction BDNF after KCl: 4.54 ± 0.17, n=6; after TTX WD: 3.06 ± 0.49, n=6). Importantly, the potentiation of NMDAR-dependent Bdnf IV transcription we observed in CaRF KD neurons resulted in enhanced expression of BDNF protein in CaRF KD compared with control neurons (p< 0.0001; Fig. 2C). By contrast, KCl-mediated membrane depolarization induced BDNF protein expression to similar levels in both CaRF control and KD neurons (p= 0.7889; Fig. 2C).
Figure 2. Knockdown of Carf selectively enhances NMDAR-dependent transcription.
(A) Levels of Bdnf exon IV mRNA in cortical neurons infected with the indicated lentiviruses then stimulated with 6hrs TTX WD. Left, relative basal and stimulus fold induction of Bdnf IV mRNA in WT and CaRF KD neurons. Right, induced mRNA levels for neurons infected with each shRNA targeting Carf (shRNA1, shRNA2) are reported as percentages of induction relative to their respective control vectors (Ctrl1 or Ctrl2). n=3-4, *p<0.05 compared with Ctrl1, #p<0.05 compared with Ctrl2. (B) Levels of Bdnf exon IV mRNA in cortical neurons infected with the indicated lentiviruses then stimulated with 55mM extracellular KCl for 6hrs. Left, relative basal and stimulus fold induction of Bdnf IV mRNA in Ctrl and CaRF KD neurons. Right, induced mRNA levels for neurons infected with each shRNA targeting Carf (shRNA1, shRNA2) are reported as percentages of induction relative to their respective control vectors (Ctrl1 or Ctrl2). n=3-4 (C) Levels of BDNF protein as measured by ELISA from neurons treated for 6 hrs as in (A) and (B). Protein levels are reported as percentages of induction by TTX WD or KCl relative to control shRNA (Ctrl1) n=6, *p<0.05 compared with Ctrl1. (D) Levels of Arc and Fos mRNA in cortical neurons infected with the indicated lentiviruses then stimulated with 90min TTX WD. n=3-4 *p<0.05 compared with Ctrl1, #p<0.05 compared with Ctrl2.
In addition to Bdnf, other IEGs including Fos and Arc are strongly induced by TTX WD stimulation and the activation of NMDARs (Saha et al. 2011). We found that KD of CaRF did not lead to a nonspecific increase in all NMDAR-activated transcription (Fig. 2D), because the TTX WD-induced expression of Fos was not different between CaRF control and KD neurons (Carf shRNA1: p=0.1926; Carf shRNA2: p=0.6159). However KD of CaRF did lead to a potentiation of TTX WD-induced Arc expression compared with that seen in control-infected neurons (Carf shRNA1: p= 0.0264; Carf shRNA2: p< 0.0001), demonstrating that the effects of CaRF on NMDAR-dependent gene expression extend beyond the regulation of Bdnf. These data reveal for the first time that CaRF regulates NMDAR-dependent neuronal plasticity by selectively inhibiting NMDAR-dependent activation of a subset of IEGs, which include not only Bdnf IV but also Arc.
The NMDAR subunit gene Grin3a is a regulatory target of CaRF
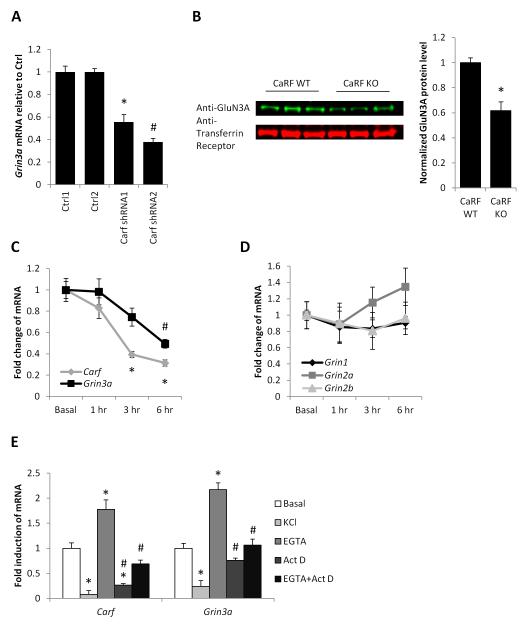
CaRF is a DNA binding transcription factor that in neurons is bound to a large number of regulatory elements dispersed across the genome (Pfenning et al. 2010). Because the effects of CaRF KD were selective for NMDAR-dependent transcription, we reasoned that CaRF might confer this specificity via regulation of components of the NMDAR itself. In addition to the obligatory GluN1 subunit encoded by the Grin1 gene, forebrain neurons most highly express the GluN2A and B subunits encoded by Grin2a and Grin2b as well as the GluN3A subunit encoded by Grin3a (Fig. S2). Of the genes encoding these subunits, only Grin3a was among those we found significantly changed in microarray analysis of gene expression in mouse cortical neurons lacking Carf (Whitney et al. 2014). Q-PCR confirmed that CaRF knockdown had consistent and significant effects expression on the Grin3a gene, which was reduced by more than 50% in CaRF KD neurons relative to control (Carf shRNA1: p= 0.0001; Carf shRNA2: p< 0.0001; Fig. 3A). Furthermore, levels of GluN3A were significantly reduced in membrane fractions taken from the cortex of postnatal day 21 (P21) CaRF knockout (KO) mice as compared with their wildtype (WT) littermates (p=0.0013; Fig. 3B).
Figure 3. Carf and Grin3a are co-regulated.
(A) Grin3a mRNA levels in mouse cortical neurons infected with the indicated lentiviruses. n=8, *p<0.05 compared with Ctrl1. #p<0.05 compared with Ctrl2. (B) Representative image (left) and quantification (right) of Western blot showing GluN3A level in membrane fractions from the cortex of p21 CaRF WT or CaRF KO mice. Transferrin receptor is shown as a loading control. Band density in each lane was quantified using ImageJ. n= 6WT, 3KO. *p<0.05 compared with CaRF WT. (C) Carf and Grin3a mRNA levels in the hippocampus of mice injected with either saline (Basal) or pilocarpine for the indicated amounts of time. n=5-6, *p < 0.01 compared with Carf Basal. #p<0.05 compared with Grin3a Basal. (D) Indicated mRNA levels in the hippocampus of mice injected with either saline (Basal) or pilocarpine for the indicated amounts of time. n=5-6. (E) Carf and Grin3a mRNA levels in mouse cortical neurons treated with KCl, EGTA, Actinomycin D or EGTA + Actinomycin D for 6hrs. n=5-6, *p < 0.05 compared with Basal. #p<0.05 compared with EGTA.
If Grin3a is a regulatory target of CaRF, these gene products should be co-regulated. Grin3a is most highly expressed during a transient period in early postnatal forebrain development (Ciabarra et al., 1995; Sucher et al., 1995), and notably, CaRF expression follows a similar pattern of temporal regulation (McDowell et al. 2010). Furthermore our data show that, in addition to this slow developmental regulation, expression of CaRF is under the more acute control of activity-regulated signaling pathways. Following pilocarpine-induced seizure, which rapidly upregulated expression of Bdnf IV and Fos in hippocampus (Fold induction Bdnf IV 1hr after pilocarpine injection: 9.24 ± 1.53, n=5-6; Fold induction Fos 1hr after pilocarpine injection: 105.73 ± 23.5, n=5-6), Carf expression was significantly reduced (F1,21=19.15, p<0.0001; Fig. 3C). Grin1, Grin2a, and Grin2b showed no significant change in their expression over this timecourse (Grin1: F1,21=0.42, p=0.7342; Grin2a: F1,21=0.96, p=0.4296; Grin2b: F1,21=0.17, p=0.9148; Fig. 3D), however like Carf, Grin3a expression was significantly reduced following pilocarpine treatment (F1,21=7.26, p=0.002; Fig. 3C). The downregulation of both Carf and Grin3a was recapitulated by elevation of extracellular KCl in cultured hippocampal neurons, suggesting a role for calcium signaling pathways in this regulation (Carf: p< 0.0001; Grin3a: p= 0.0001; Fig. 3E). Consistent with this hypothesis, treating cultures with 2.5mM EGTA, which chelates free extracellular calcium, drove a significant increase in both Carf and Grin3a mRNA expression (Carf: p=0.0051; Grin3a: p=0.0001; Fig. 3E). This induction was transcription-dependent, because the EGTA-dependent increase of Carf and Grin3a mRNA expression was blocked by pre-treatment with the transcriptional inhibitor Actinomycin D (Carf: p= 0.0002; Grin3a: p< 0.0001; Fig. 3E).
Because CaRF can function as a DNA sequence-specific transcriptional activator (Tao et al. 2002), it could regulate the expression of Grin3a directly by binding to the Grin3a promoter. Although a search of the proximal Grin3a promoter revealed two elements with partial homology to the consensus CaRF binding site, electrophoretic mobility shift assays revealed no evidence for CaRF binding at these sites (Fig. S3) and furthermore CaRF binding was not detected at the proximal Grin3a promoter by ChIP-seq (Pfenning et al. 2010). Thus these data suggest that CaRF does not directly bind the proximal Grin3a promoter.
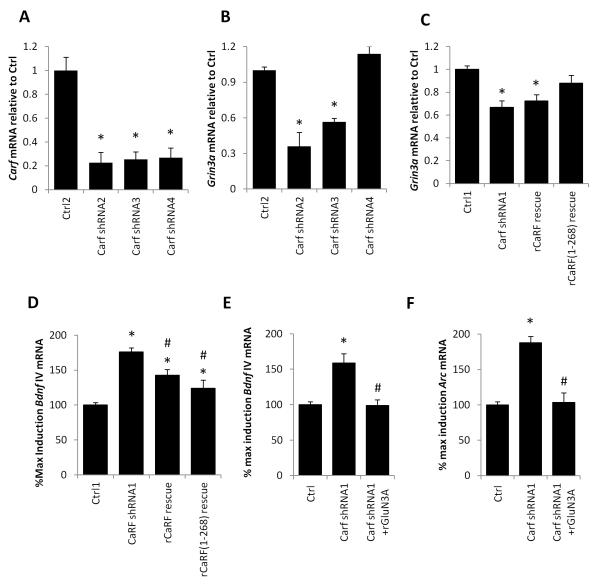
The Carf gene encodes several splice variants, some of which lack the C-terminal activation domain and are predicted to encode truncated CaRF isoforms that function as transcriptional repressors (Fig. S4, S5) (Tao et al. 2002; McDowell et al. 2010; Pfenning et al. 2010). To determine which forms of CaRF are required for regulation of Grin3a we tested the consequences of knocking down CaRF using shRNAs that are either specific to the longer forms or common to sequences contained within all variants (Fig. S4). Whereas all shRNAs reduced the expression of CaRF mRNA (Fig. 4A) only the shRNAs that targeted all of the truncated and the full-length CaRF variants resulted in reduced expression of Grin3a (Carf shRNA2: p= 0.0018; Carf shRNA3: p<0.0001; Carf shRNA4: p=0.0882; Fig. 4B). Because these data suggested that expression of the shorter truncated version of CaRF might be sufficient to promote expression of Grin3a, we selectively re-expressed rat isoforms of either truncated or full-length CaRF in CaRF KD neurons, which due to sequence variation in the region targeted by Carf shRNA1 are resistant to knockdown. Though both rescue constructs were overexpressed relative to endogenous levels of CaRF (Fig. S5), only the truncated form of rat CaRF was able to restore Grin3a expression in CaRF KD neurons (Carf shRNA1: p<0.0001; rCaRF rescue: p=0.009; rCaRF(1-268): p=0.1018; Fig. 4C). Since this variant of CaRF lacks the transcriptional activation domain (Fig. S4, S5), taken together with the lack of CaRF binding to the Grin3a promoter (Fig. S3), these data suggest that CaRF functions in an indirect manner to drive Grin3a expression.
Figure 4. CaRF regulates Grin3a expression indirectly.
(A) Levels of Carf mRNA in mouse cortical neurons infected with lentiviruses encoding shRNAs targeting three independent sequences in Carf. Carf mRNA levels are shown normalized to expression in cells infected with the paired control virus (Ctrl2). n=3. *p<0.05 compared with Ctrl2. (B) Grin3a mRNA levels in mouse cortical neurons infected with the indicated lentiviruses. n=3, *p<0.05 compared with Ctrl2. (C) Grin3a mRNA levels in mouse cortical neurons infected with the indicated lentiviruses. n=4-10, *p<0.05 compared with Ctrl1. (D) Bdnf exon IV mRNA level in cortical neurons infected with the indicated lentiviruses following stimulation with 6hrs TTX WD. n=6-10, *p<0.05 compared with Ctrl1. #p<0.05 compared with Carf shRNA1. (E) Bdnf exon IV and (F) Arc mRNA level in cortical neurons infected with the indicated lentiviruses following stimulation with 6hrs TTX WD. n=5-6, *p<0.05 compared with Ctrl1. #p<0.05 compared with Carf shRNA1.
Regardless of the mechanism by which CaRF regulates expression of Grin3a, if the potentiation of NMDAR-dependent transcription in CaRF KD neurons is due to the reduced GluN3A expression we observed, then rescuing the expression of GluN3A in these neurons should be sufficient to restore the regulation of TTX WD-induced Bdnf IV expression to control levels. To test this hypothesis, we first asked whether selective re-expression of the truncated form of rat CaRF would restore NMDAR-regulated Bdnf IV induction to control levels in CaRF KD neurons. Consistent with this hypothesis we found that expression of an shRNA-resistant form of CaRF significantly reduced NMDAR-dependent Bdnf IV induction in shRNA-expressing neurons toward control levels (rCaRF(1-268): p=0.0005; Fig. 4D). Notably, re-expressing full-length rat CaRF also partially rescued Bdnf IV inducibility despite the fact that it did not rescue Grin3a expression (rCaRF: p=0.0031; Fig. 4C,D). Because our previous microarray and chromatin immunoprecipitation experiments have identified a large number of putative CaRF target genes (Pfenning et al. 2010; Whitney et al. 2014), we interpret these latter data to suggest that full length CaRF may regulate gene products other than Grin3a that can also impact Bdnf inducibility. Thus to more directly determine whether restoration of GluN3A can rescue Bdnf inducibility in neurons lacking CaRF, we overexpressed rat GluN3A or GFP as a control in both CaRF KD and control neurons and measured the induction of Bdnf IV mRNA upon TTX WD. Whereas CaRF KD neurons expressing GFP showed potentiated induction of Bdnf IV and Arc relative to control neurons, when we expressed rat GluN3A together with the Carf shRNA, the induction of Bdnf IV and Arc was not significantly different from control (Bdnf IV: p= 0.8960; Arc: p= 0.8243; Fig. 4E and 4F). Together these data place CaRF as an indirect upstream regulator of Grin3a, and Grin3a upstream of Bdnf IV and Arc induction, establishing a mechanism by which CaRF can selectively modulate NMDAR-dependent processes in the developing brain.
Enhanced sensory experience induced gene transcription in the developing cortex of CaRF knockout mice
NMDARs and activity-regulated genes like Bdnf play an important role in the sensory experience-dependent refinement of cortical organization in the postnatal brain. Given that both CaRF and GluN3A show peak expression during of the first two postnatal weeks of brain development (McDowell et al. 2010; Ciabarra et al. 1995; Sucher et al. 1995), we hypothesized that the CaRF-GluN3A pathway we have elucidated here may be important for regulating NMDAR-dependent gene expression by sensory stimuli in the developing postnatal brain. To test this hypothesis, we assayed light-dependent activation of transcription in the visual cortex of mice that were dark-adapted from postnatal day 14 (P14) to P21.
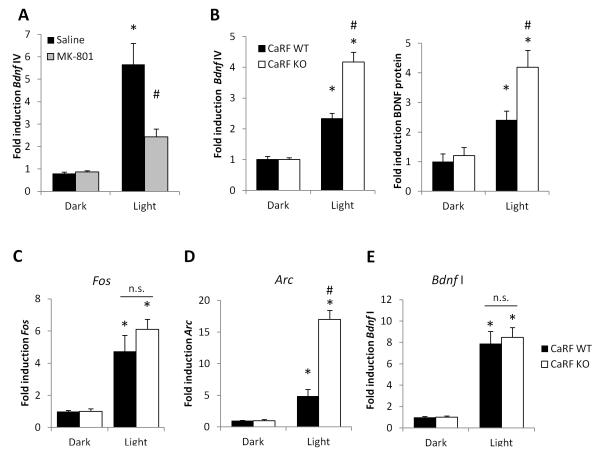
Light exposure drives rapid upregulation of Bdnf expression in visual cortex (Castrén et al. 1992). This induction is NMDAR-dependent because when we pretreated dark-adapted mice with the NMDAR inhibitor MK-801 prior to light exposure we significantly inhibited the induction of Bdnf IV mRNA by light (p= 0.0309; Fig. 5A). Because we found that GluN3A expression is reduced in the developing cortex of CaRF KO mice relative to their WT littermates at P21 (Fig. 3B) we predicted that light-dependent activation of NMDAR-dependent gene transcription at this time point would be enhanced in the visual cortex of these mice. Indeed we found that light-induced expression of both Bdnf IV mRNA and BDNF protein were potentiated in visual cortex of P21 dark-adapted CaRF KO mice compared with their identically treated WT littermate controls (mRNA: p= 0.0003; protein: p=0.0156; Fig. 5B). The induction of gene expression was specific to brain regions activated by the light stimulus, because we saw no light-dependent regulation of activity-dependent genes in somatosensory cortex of the same mice (Fold induction Fos: 1.21 ± 0.56; Arc: 0.90 ± 0.32; Bdnf IV: 0.67 ± 0.12. Fig. S6). The potentiation of Bdnf IV transcription in CaRF KO mice did not reflect a general increase in all NMDAR-dependent transcription because we saw no difference between CaRF WT and KO mice in the light-dependent induction of Fos (p= 0.2446; Fig. 5C). However, similar to our results in cultured neurons, the potentiation of NMDAR-dependent transcription in CaRF KO mice did extend to other NMDAR-dependent genes including Arc (p= 0.0001; Fig. 5D). Interestingly, although transcriptional regulation of Bdnf promoter I is also regulated by NMDARs expression of Bdnf exon I does not show potentiation of light-induced expression in the visual cortex of CaRF KO mice (p= 0.7034; Fig. 5E). Taken together these data demonstrate that CaRF plays an important role in limiting sensory stimulus-regulated transcription of NMDAR-dependent genes in the developing postnatal brain.
Figure 5. Potentiation of light-induced gene transcription in the visual cortex of CaRF knockout mice.
(A) Levels of Bdnf exon IV mRNA in the primary visual cortex of adult mice adapted to darkness for 7 days. Mice were injected with either saline or the NMDAR blocker MK-801 in the dark, then 30 min later they were either maintained in darkness (Dark) or exposed to light for 2hrs (Light) prior to tissue harvesting. n= 3, *p<0.05, Saline vs MK-801. (B) Levels of Bdnf exon IV mRNA and BDNF protein in the primary visual cortex of P21 CaRF WT and KO mice after 7 days of constant darkness (Dark) or following exposure to light for 6hrs (Light) prior to tissue harvesting. n=8-11 animals per condition, *p<0.05, Dark vs. Light. #p<0.05 WT vs. KO. (C-E) Fos (C), Arc (D), and Bdnf exon I (E) mRNA levels in primary visual cortex of mice from (B). n=8-11 animals per condition, *p<0.05, Dark vs. Light. #p<0.05 WT vs. KO.
Enhanced BDNF expression in CaRF-knockdown neurons accelerates inhibitory synapse development
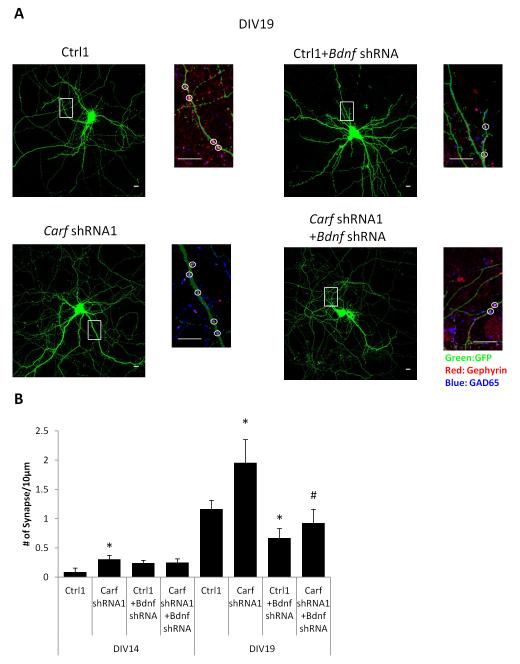
The best characterized function of neuronal activity-dependent BDNF transcription in the developing brain is its role in promoting the formation of GABAergic synapses (Hong et al. 2008). We previously reported that we found increased expression of GABAergic synaptic proteins in the striatum of adult Carf KO mice (McDowell et al. 2010). To determine whether the enhanced NMDAR-dependent BDNF expression we detected in the CaRF KO and KD neurons could contribute to increases in GABAergic synapse formation, we assayed the development of GABAergic synapses by co-localization of pre- and post-synaptic markers in cultured control and Carf KD neurons. The density of GABAergic synapses increased between Day 14 in culture (DIV14) and DIV19 (Ctrl1: p<0.0001; Fig. 6). This developmental increase in GABAergic synapses was attenuated by transfection of an shRNA that knocks down expression of Bdnf (Fig. S7), demonstrating the importance of BDNF as a modulator of GABAergic synapse number (DIV19, Ctrl1+Bdnf shRNA: p=0.0417). This increase was significantly greater in CaRF KD neurons compared with control (DIV19, Carf shRNA1: p=0.0425) indicating that CaRF functions to limit GABAergic synapse formation during development. The effect of losing CaRF on GABAergic synapse number was BDNF-dependent because knocking down BDNF in CaRF KD neurons blocked the increase of GABAergic synapses compared to control on DIV19 (DIV19, Carf shRNA1+ Bdnf shRNA: p=0.0473; Fig. 6). Together these data demonstrate that CaRF-dependent modulation of BDNF transcription is important for regulating GABAergic synapse formation during development.
Figure 6. Knockdown of Carf accelerates inhibitory synapse formation in a BDNF dependent manner.
(A) Representative low and high magnification images of shRNA transfected mouse cortical neurons immunostained with indicated antibodies. The smaller images are the enlargement of the boxed area in low magnification images shows details of dendrites. Scale bar, 10 μm. (B) Quantification of the average density of GAD65/Gephyrin coclusters along the dendrites of mouse cortical neurons transfected with indicated shRNA. 10-15 cells per condition, *p<0.05 compared with Ctrl. #p<0.05 compared with Carf shRNA1.
Discussion
NMDA-type glutamate receptors couple sensory experience with synapse maturation and neuronal survival in the developing brain (Scheetz and Constantine-Paton 1994; Cull-Candy and Leszkiewicz 2004). NMDARs exert long-lasting effects on brain development at least in part via their ability to activate calcium-regulated gene transcription (Bading 2013). Our studies provide a new angle on the molecular mechanisms of this experience-dependent brain development by showing for the first time that the transcription factor CaRF regulates NMDAR-dependent gene transcription. Specifically we find that neurons lacking CaRF have enhanced NMDAR-dependent activation of Bdnf and Arc, transcription of which mediates experience-dependent synaptic plasticity in the developing cortex (Leslie and Nedivi 2011). Interestingly our data further suggest that CaRF acts upstream of NMDARs to limit synaptic activity-dependent gene transcription in the developing brain by regulating NMDAR subunit composition.
Permissive and instructive actions of CaRF on activity-regulated genes
It is well established that the promoters and enhancers of activity-regulated genes including Bdnf and Arc are bound by multiple activity-responsive transcription factors (e.g. CREB, MEF2, SRF) (Lyons and West 2011). Under basal conditions these activity-dependent transcription factors are either inactive or they actively repress their target genes. In response to synaptic activity, calcium signaling pathways rapidly convert these factors into potent transcriptional activators. CaRF is also bound to the promoters of many activity-regulated genes including Bdnf (Tao et al. 2002; Pfenning et al. 2010). However unlike CREB and MEF2, full-length CaRF is competent to mediate transcription even in the absence of synaptic activity and it undergoes a comparatively small increase in its ability to promote transcription following membrane depolarization (Tao et al. 2002; West 2011). CaRF can likely also act as a basal repressor, because among the set of genes that have CaRF bound at their promoters, equal numbers are up- versus down-regulated in CaRF knockout neurons compared with control (Pfenning et al. 2010). Although the mechanisms that allow CaRF to both activate and repress genes are not fully understood, the Carf gene does encode short variants that contains the DNA binding domain but lack the transcriptional activation domain (Tao et al. 2002) and our evidence that selective re-expression of the truncated form of CaRF in CaRF KD neurons is sufficient to rescue both Grin3a expression and NMDAR-dependent Bdnf IV induction (Fig. 4C,D) suggests that the repressor forms contribute to neuronal gene regulation. Taken together, we interpret these data to suggest that CaRF primarily acts at activity-regulated gene promoters not as a calcium-inducible activator, but rather as a basal activator or repressor.
Stimuli that increase synaptic activity decrease CaRF expression (Fig. 3C). Thus we propose that it is the loss of the basal activator or repressor activity at CaRF-regulated promoters following synaptic activity that permits transcription of these genes to rise or fall. This model helps to explain our previous microarray analysis of L-VGCC regulated transcription in CaRF knockout and knockdown neurons (Whitney et al. 2014). In those experiments we discovered a set of genes that increase or decrease their expression following membrane depolarization in CaRF wildtype neurons, but which show either basally increased or decreased expression, respectively, in neurons that lack CaRF. Furthermore, these genes show no change in their expression following membrane depolarization in neurons lacking CaRF. The occlusion of L-VGCC-dependent regulation in CaRF-lacking neurons suggests that the activity-induced loss of CaRF expression mediates the transcriptional regulation of these target genes. Conversely, since CaRF expression is elevated by pharmacological inhibition of intracellular calcium (Fig. 3E) our model also predicts that CaRF may play an instructive role in activating or repressing gene transcription following the inhibition of synaptic activity. This is interesting because although increases in transcription are required to mediate homeostatic synaptic scaling following inhibition of neuronal firing (Ibata et al. 2008), very few transcription factors have been described that show increases in their transcriptional activity under conditions of reduced neuronal activity. Other than CaRF the only known transcription factor that may show inactivity-induced increases in its transcriptional activity is CCAT, which is derived from an alternative promoter in an intron of the gene encoding the L-VGCC subunit Cacna1c and which translocates to the nucleus of neurons under conditions of low intracellular calcium (Gomez-Ospina et al. 2006; Gomez-Ospina et al. 2013). Whether CaRF or CCAT is required for inactivity-dependent glutamatergic synaptic scaling remains to be determined.
Modulation of NMDAR-dependent transcription
In addition to regulating a subset of activity-responsive genes directly, our data suggest that CaRF also impacts NMDAR-dependent transcription by promoting the expression of the Grin3a gene, which encodes the NMDAR subunit GluN3A. NMDARs are comprised of an obligatory GluN1 subunit together with variable GluN2(A-D) and GluN3(A-B) subunits (Köhr 2006). Expression of GluN3A is strongly developmentally regulated in the mouse forebrain, with highest levels of expression occurring during the first two postnatal weeks of life (Al-Hallaq et al. 2002; Sasaki et al. 2002; Wong et al. 2002). Transient expression of GluN3A in this time period is required for the proper temporal regulation of excitatory synapse maturation (Roberts et al. 2009; Kehoe et al. 2014; Henson et al. 2012). Our data are the first to suggest that GluN3A may contribute to activity-dependent synapse development by modulating NMDAR-dependent changes in gene transcription. Specifically our observation that rescuing the expression of GluN3A in CaRF knockdown neurons is sufficient to restore the induction of Bdnf and Arc to control levels (Fig. 4D,E), places the actions of GluN3A downstream of CaRF and upstream of NMDAR-dependent Bdnf IV and Arc inducibility.
The presence of GluN3A could impact NMDAR-dependent transcription either by altering the biophysical or the biochemical signaling properties of NMDARs. NMDARs containing GluN3A have reduced calcium permeability (Kehoe et al. 2013), and because calcium is a key mediator of NMDAR-induced gene transcription (Lyons and West 2011) it is possible that GluN3A inhibits the induction of activity-dependent genes by limiting the synaptic activity-induced elevation of postsynaptic calcium. However this model fails to explain why only a subset of activity regulated genes (e.g. Bdnf IV and Arc, but not Fos and Bdnf I) appear to be sensitive to GluN3A expression. An alternative possibility, in analogy to the differential ability of GluN2A versus GluN2B containing NMDARs to activate CREB (Martel et al. 2012), is that GluN3A-containing and GluN3A-lacking NMDARs differ in their abilities to activate specific downstream transcription factors. This could explain the specificity of gene regulation because different activity-regulated genes are induced by distinct complements of transcription factors (Lyons and West 2011). The long intracellular C-terminal tail of GluN3A, which is distinct from that of other NMDAR subunits, provides docking sites for an assortment of intracellular signaling molecules (Henson et al. 2010). If GluN3A-mediated recruitment of one or more of these signaling molecules to NMDARs were to change the ability of NMDARs to signal to specific transcription factors in the nucleus, this could impact the transcriptional outcome of NMDAR activation. Identifying whether specific stimulus-regulated transcription factors are differentially activated in the presence or absence of GluN3A will help advance understanding of the mechanisms by which subunit composition can affect the specificity of NMDAR-dependent transcriptional regulation.
Activity-dependent transcription in the developing brain
Our evidence that young CaRF knockout mice show enhanced light-dependent induction of Bdnf and Arc in the visual cortex (Fig. 5B and 5D) suggests that CaRF-dependent modulation of NMDAR-dependent transcription may contribute to the fidelity of experience-dependent synapse maturation in the developing brain. Consistent with this model we find that CaRF knockdown neurons show enhanced GABAergic synapse development in culture that is BDNF-dependent (Fig. 6). The expression of both Bdnf and Arc is under tight spatial and temporal control in the developing cortex and even subtle disruption of this regulation can have a substantial impact on cortical development. Activity-dependent transcription of Bdnf mediated by CREB activation is required for proper development of inhibitory GABAergic synapses in the developing brain (Hong et al. 2008). Inhibition has been tightly linked to timing the critical period for plasticity in visual cortex (Hensch 2005), and indeed transgene expression of BDNF early in postnatal development both accelerates maturation of GABAergic synapses and leads to premature closure of the critical period for ocular dominance plasticity (Huang et al. 1999). Arc functions in excitatory synapse maturation and the loss of Arc in the postnatal brain is associated with impaired sensory-dependent synapse plasticity in the visual cortex (McCurry et al. 2010; Wang et al. 2006). Given that expression of CaRF is highly developmentally regulated, with peak expression in the early postnatal brain (McDowell et al. 2010), we propose that CaRF may act to refine the fidelity of sensory- and NMDAR-dependent gene transcription in the developing cortex in ways that optimize the timing of critical period plasticity. Adult CaRF knockout mice have enhanced expression and synaptic localization of GABAergic synapse proteins (McDowell et al. 2010) and display aberrant learning in Morris Water Maze, novel object, and interval timing tasks (Agostino et al. 2013; McDowell et al. 2010). Determining whether these defects arise from impaired postnatal expression of CaRF will enhance our understanding of the importance of tuning activity-dependent gene regulation in this critical period of brain development.
Supplementary Material
Acknowledgements
We thank Marty Yang, Jessi Cruger, and Marguerita Klein for technical assistance. This work was supported in part by the Ruth K. Broad Biomedical Research Foundation and NIH grants 1R01DA033610 and 1R21MH096310 (A.E.W.).
Footnotes
ARRIVE guidelines have been followed:
Yes
=> if No, skip complete sentence
=> if Yes, insert “All experiments were conducted in compliance with the ARRIVE guidelines.”
Conflicts of interest: None
=> if 'none', insert “The authors have no conflict of interest to declare.”
=> otherwise insert info unless it is already included
References
- Agostino PV, Cheng R-K, Williams CL, West AE, Meck WH. Acquisition of response thresholds for timed performance is regulated by a calcium-responsive transcription factor, CaRF. Genes. Brain. Behav. 2013;12:633–44. doi: 10.1111/gbb.12059. [DOI] [PubMed] [Google Scholar]
- Al-Hallaq RA, Jarabek BR, Fu Z, Vicini S, Wolfe BB, Yasuda RP. Association of NR3A with the N-methyl-D-aspartate receptor NR1 and NR2 subunits. Mol. Pharmacol. 2002;62:1119–27. doi: 10.1124/mol.62.5.1119. [DOI] [PubMed] [Google Scholar]
- Bading H. Nuclear calcium signalling in the regulation of brain function. Nat. Rev. Neurosci. 2013;14:593–608. doi: 10.1038/nrn3531. [DOI] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–14. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–5. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrén E, Zafra F, Thoenen H, Lindholm D. Light regulates expression of brain-derived neurotrophic factor mRNA in rat visual cortex. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9444–8. doi: 10.1073/pnas.89.20.9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciabarra AM, Sullivan JM, Gahn LG, Pecht G, Heinemann S, Sevarino KA. Cloning and characterization of chi-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J. Neurosci. 1995;15:6498–508. doi: 10.1523/JNEUROSCI.15-10-06498.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci. STKE. 2004;2004:re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science. 2001;294:333–9. doi: 10.1126/science.1063395. [DOI] [PubMed] [Google Scholar]
- Ghiretti AE, Moore AR, Brenner RG, Chen L-F, West AE, Lau NC, Hooser S. D. Van, Paradis S. Rem2 is an activity-dependent negative regulator of dendritic complexity in vivo. J. Neurosci. 2014;34:392–407. doi: 10.1523/JNEUROSCI.1328-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ospina N, Panagiotakos G, Portmann T, Pasca SP, Rabah D, Budzillo A, Kinet JP, Dolmetsch RE. A promoter in the coding region of the calcium channel gene CACNA1C generates the transcription factor CCAT. PLoS One. 2013;8:e60526. doi: 10.1371/journal.pone.0060526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Huang S, Chang MC, Worley P, Kirkwood A, Quinlan EM. Obligatory role for the immediate early gene NARP in critical period plasticity. Neuron. 2013;79:335–46. doi: 10.1016/j.neuron.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat. Neurosci. 2002;5:405–14. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- Hensch TK. Critical period mechanisms in developing visual cortex. Curr. Top. Dev. Biol. 2005;69:215–37. doi: 10.1016/S0070-2153(05)69008-4. [DOI] [PubMed] [Google Scholar]
- Henson MA, Roberts AC, Pérez-Otaño I, Philpot BD. Influence of the NR3A subunit on NMDA receptor functions. Prog. Neurobiol. 2010;91:23–37. doi: 10.1016/j.pneurobio.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson MA, Larsen RS, Lawson SN, Pérez-Otaño I, Nakanishi N, Lipton SA, Philpot BD. Genetic deletion of NR3A accelerates glutamatergic synapse maturation. PLoS One. 2012;7:e42327. doi: 10.1371/journal.pone.0042327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–24. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J-H, Park JM, Park S, Xiao B, Dehoff MH, Kim S, Hayashi T, et al. Homeostatic scaling requires group I mGluR activation mediated by Homer1a. Neuron. 2010;68:1128–42. doi: 10.1016/j.neuron.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Kirkwood a, Pizzorusso T, Porciatti V, Morales B, Bear MF, Maffei L, Tonegawa S. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98:739–55. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- Ibata K, Sun Q, Turrigiano GG. Rapid Synaptic Scaling Induced by Changes in Postsynaptic Firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Karpova A, Mikhaylova M, Bera S, Bär J, Reddy PP, Behnisch T, Rankovic V, et al. Encoding and transducing the synaptic or extrasynaptic origin of NMDA receptor signals to the nucleus. Cell. 2013;152:1119–33. doi: 10.1016/j.cell.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Kehoe LA, Bernardinelli Y, Muller D. GluN3A: an NMDA receptor subunit with exquisite properties and functions. Neural Plast. 2013;2013:145387. doi: 10.1155/2013/145387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe LA, Bellone C, Roo M. De, Zandueta A, Dey PN, Pérez-Otaño I, Muller D. GluN3A promotes dendritic spine pruning and destabilization during postnatal development. J. Neurosci. 2014;34:9213–21. doi: 10.1523/JNEUROSCI.5183-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhr G. NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res. 2006;326:439–46. doi: 10.1007/s00441-006-0273-6. [DOI] [PubMed] [Google Scholar]
- Leslie JH, Nedivi E. Activity-regulated genes as mediators of neural circuit plasticity. Prog. Neurobiol. 2011;94:223–37. doi: 10.1016/j.pneurobio.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–72. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Lyons MR, Schwarz CM, West AE. Members of the myocyte enhancer factor 2 transcription factor family differentially regulate Bdnf transcription in response to neuronal depolarization. J. Neurosci. 2012;32:12780–5. doi: 10.1523/JNEUROSCI.0534-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog. Neurobiol. 2011;94:259–95. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel M-A, Ryan TJ, Bell KFS, Fowler JH, McMahon A, Al-Mubarak B, Komiyama NH, et al. The subtype of GluN2 C-terminal domain determines the response to excitotoxic insults. Neuron. 2012;74:543–56. doi: 10.1016/j.neuron.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurry CL, Shepherd JD, Tropea D, Wang KH, Bear MF, Sur M. Loss of Arc renders the visual cortex impervious to the effects of sensory experience or deprivation. Nat. Neurosci. 2010;13:450–7. doi: 10.1038/nn.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell KA, Hutchinson AN, Wong-Goodrich SJE, Presby MM, Su D, Rodriguiz RM, Law KC, Williams CL, Wetsel WC, West AE. Reduced cortical BDNF expression and aberrant memory in Carf knock-out mice. J. Neurosci. 2010;30:7453–65. doi: 10.1523/JNEUROSCI.3997-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenning AR, Kim T-K, Spotts JM, Hemberg M, Su D, West AE. Genome-wide identification of calcium-response factor (CaRF) binding sites predicts a role in regulation of neuronal signaling pathways. PLoS One. 2010;5:e10870. doi: 10.1371/journal.pone.0010870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VR, Pintchovski SA, Chin J, Peebles CL, Mitra S, Finkbeiner S. AMPA receptors regulate transcription of the plasticity-related immediate-early gene Arc. Nat. Neurosci. 2006;9:887–95. doi: 10.1038/nn1708. [DOI] [PubMed] [Google Scholar]
- Roberts AC, Díez-García J, Rodriguiz RM, López IP, Luján R, Martínez-Turrillas R, Picó E, et al. Downregulation of NR3A-containing NMDARs is required for synapse maturation and memory consolidation. Neuron. 2009;63:342–56. doi: 10.1016/j.neuron.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang J-Y, Daigle KR, Fenn JD, Adelman K, Dudek SM. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat. Neurosci. 2011;14:848–56. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YF, Rothe T, Premkumar LS, Das S, Cui J, Talantova MV, Wong H-K, et al. Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J. Neurophysiol. 2002;87:2052–63. doi: 10.1152/jn.00531.2001. [DOI] [PubMed] [Google Scholar]
- Scheetz AJ, Constantine-Paton M. Modulation of NMDA receptor function: implications for vertebrate neural development. FASEB J. 1994;8:745–52. doi: 10.1096/fasebj.8.10.8050674. [DOI] [PubMed] [Google Scholar]
- Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, Wu MK, Yuan JP, Jones EG, Lipton SA. Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J. Neurosci. 1995;15:6509–20. doi: 10.1523/JNEUROSCI.15-10-06509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, West AE, Chen WG, Corfas G, Greenberg ME. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 2002;33:383–95. doi: 10.1016/s0896-6273(01)00561-x. [DOI] [PubMed] [Google Scholar]
- Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- West AE. Biological functions and transcriptional targets of CaRF in neurons. Cell Calcium. 2011;49:290–5. doi: 10.1016/j.ceca.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb. Perspect. Biol. 2011;3:1–21. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Pruunsild P, Timmusk T. In: Neurotrophic Factors. Lewin GR, Carter BD, editors. Vol. 220: Handbook of Experimental Pharmacology. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. [Google Scholar]
- Whitney O, Pfenning AR, Howard JT, Blatti CA, Liu F, Ward JM, Wang R, et al. Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science. 2014;346:1256780-1–1256780-11. doi: 10.1126/science.1256780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayatunge R, Chen L-F, Cha YM, Zannas AS, Frank CL, West AE. The histone lysine demethylase Kdm6b is required for activity-dependent preconditioning of hippocampal neuronal survival. Mol. Cell. Neurosci. 2014;61:187–200. doi: 10.1016/j.mcn.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H-K, Liu X-B, Matos MF, Chan SF, Pérez-Otaño I, Boysen M, Cui J, et al. Temporal and regional expression of NMDA receptor subunit NR3A in the mammalian brain. J. Comp. Neurol. 2002;450:303–17. doi: 10.1002/cne.10314. [DOI] [PubMed] [Google Scholar]
- Zhou P, Porcionatto M, Pilapil M, Chen Y, Choi Y, Tolias KF, Bikoff JB, Hong EJ, Greenberg ME, Segal RA. Polarized signaling endosomes coordinate BDNF-induced chemotaxis of cerebellar precursors. Neuron. 2007;55:53–68. doi: 10.1016/j.neuron.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.