Abstract
Genotype List (GL) Strings use a set of hierarchical character delimiters to represent allele and genotype ambiguity in HLA and KIR genotypes in a complete and accurate fashion. A RESTful web service called Genotype List Service was created to allow users to register a GL String and receive a unique identifier for that string in the form of a URI. By exchanging URIs and dereferencing them through the GL Service, users can easily transmit HLA genotypes in a variety of useful formats. The GL Service was developed to be secure, scalable, and persistent. An instance of the GL Service is configured with a nomenclature and can be run in strict or non-strict modes. Strict mode requires alleles used in the GL String to be present in the allele database using the fully qualified nomenclature. Non-strict mode allows any GL String to be registered as long as it is syntactically correct. The GL Service source code is free and open source software, distributed under the GNU Lesser General Public License (LGPL) version 3 or later.
Keywords: Genotype, GL String, RESTful Service, HLA, KIR
1. Introduction
Ambiguity in recording and reporting of HLA genotyping results continues to impact clinical decision support for donor/patient matching as well as understanding post-transplant outcomes research. Recommendations addressing this have been proposed [1]. These guidelines include listing all allele and genotype ambiguities at the highest resolution detected, documenting the typing method, and identifying the pertinent reference allele sequence database version used to perform the typing. To address the first part of these guidelines, a string format capable of fully representing HLA allele and genotype ambiguity was developed. This format, named Genotype List String (GL String), was developed by extending a proposed standard for reporting KIR genotyping data for use with HLA [2]. The GL String is parsed based on character delimiters that organize the alleles in terms of loci, alleles, lists of possible alleles, phased lists of alleles, genotypes, lists of possible genotypes, and multilocus unphased genotypes (MUGs). By applying character delimiters with defined precedence, GL Strings can be used to record allele and genotype ambiguity in a standard manner that does not increase ambiguity or lose information. The GL String format can also be used for other genetic systems with defined nomenclatures (e.g., KIR) as long as those nomenclatures do not employ the GL String character delimiters. These delimiters, parsed in order of precedence from first to last, are for loci (“^”), genotype ambiguity (“|”), copies of genes (“+”), phased genes (“~”), and allele ambiguity (“/”). For example, an allele ambiguity of HLA-A*01:01:01:01 and HLA-A*01:02 would be represented as HLA-A*01:01:01:01/HLA-A*01:02. A genotype consisting of this ambiguity together with another allele, HLA-A*01:03, would be reported as HLA-A*01:01:01:01/HLAA*01:02+HLA-A*01:03.
An important goal of the GL String format is to separate the encoding of genotype data from the management and presentation of those data. GL Strings are potentially quite long and difficult to read, and are expected to proliferate rapidly in number. However, they are easily machine generated and parsed, and the work of creating and displaying them should be left to machines. The remaining challenge is one of exchanging the strings easily.
Toward this end, we have developed the GL Service, a web service that allows users to register GL Strings they have generated, and that returns a unique Uniform Resource Identifier (URI) [3] for each registered GL String. When provided with a URI, the GL Service will return a GL in a variety of formats. In this way, use of the GL Service improves the portability of genotyping results; URIs can be shared between parties, and identical GL Strings can be obtained from the GL Service. As described in greater detail in sections 2 and 3, the use of the GL Service as a hub for sharing genotyping results allows the identification of the pertinent reference allele database version for a given GL String, and establishes the equivalence of GL Strings across difference reference allele database versions.
The GL Service is currently available at https://gl.nmdp.org. A GL Service Explorer page for IMGT/HLA Database release version 3.20.0 is available at https://gl.nmdp.org/imgt-hla/3.20.0/explorer/. While an individual can use the Explorer to register GL Strings and receive URIs, and to dereference URIs and receive GLs, the GL Service is primarily intended to interact with other services and programs in an automated fashion. The GL Service code is open-source and available, and anyone may run a public or private version of their own GL Service. Here, we describe the current implementation of the GL Service, and the available libraries, tools and approaches for interacting with the service.
2. Methods
2.1. Genotype List Service
The Genotype List Service is implemented in Java in a modular fashion. It was designed to be secure, scalable, and persistent. Web services based on REpresentational State Transfer (REST) architectural style [4, 5] with HTTP content negotiation were developed employing a Java library that manages genotyping results. Resources are identified with a structured URI.
The URI includes the namespace of the service and depends on its local implementation. For example, the URI of https://gl.nmdp.org/imgt-hla/3.20.0/allele/123 describes a strict service (described below) that uses a base name which includes the type of service (https), the domain name (gl.nmdp.org), the allele nomenclature (imgt-hla), nomenclature version (3.20.0), the resource type (allele), and an endpoint for the resource (123). It should be emphasized that the ID is the entire URI, not just the last field.
The service makes reference to the IMGT/HLA Database [6], and allows aggregation from alleles to lists of alleles, haplotypes, genotypes, lists of genotypes and multi-locus unphased genotypes. RESTful web service APIs are provided for programmatic storage and retrieval from other software. The service supports content negotiation, and can be configured with a nomenclature and allele reference database version in either strict or non-strict modes.
Public services include creating and retrieving these objects. While REST allows four CRUD operations (create, retrieve, update and delete), the GL Service only provides create (POST) and retrieve (GET) operations. No updating or deleting of registered resources is permitted.
2.2. Implementation
The core model of the Genotype List Service follows directly from the Genotype List String (GL String) grammar described previously [2]. An abstract superclass Genotype List resource provides URI and GL String representation data properties as defined in Table 1.
Table 1.
| Resource Type | Definition | Syntax comments |
|---|---|---|
| Locus | genomic location at which alleles may be defined. | The Locus is found in the prefix of the allele, separated from the rest of the allele by a ‘*’. The locus representation in GL String format must not contain any of the following characters: '*', '/', '~', '+', '|', '^' |
| Allele | variation at a particular locus. It provides an accession number data property. | The allele representation in GL String format must not contain any of the following characters: '/', '~', '+', '|', '^' |
| AlleleList | unordered collection of one or more alleles representing allelic ambiguity. | If more than one allele is present, they are separated in GL String format by the character '/' |
| Haplotype | unordered collection of one or more allele lists in phase. | If more than one allele list is present, they are separated in GL String format by the character '~' |
| Genotype | unordered collection of one or more haplotypes. | If more than one haplotype is present, they are separated in GL String format by the character '+' |
| GenotypeList | unordered collection of one or more genotypes representing genotypic ambiguity. | If more than one genotype is present, they are separated in GL String format by the character '|' |
| MultilocusUnphasedGenotype | unordered collection of one or more genotype lists. | If more than one genotype list is present, they are separated in GL String format by the character '^' |
The core model is implemented as a Java library, defined as an ontology in OWL/RDF format [7, 8], and described in XML Schema [9, 10], in both object graph form and linked data form using XLink [11].
The Genotype List service builds on this core model and provides Java APIs for registering GL resources given a GL String and for retrieving GL resources given a URI identifier. An instance of the service is composed of various implementation modules using dependency injection. This allows the same code base to be deployed in various configurations according to operational environment characteristics and/or performance considerations.
The Genotype List service can be configured to use several persistence back ends through Java Database Connectivity (JDBC) libraries. A relational database persistence implementation module supports both MySQL [12] and Oracle [13]. Additional No-SQL implementation modules offer potential read performance and horizontal scalability beyond that of a RDBMS. Finally, a RAM-only cache persistence implementation module is provided for functional testing, which runs as part of a continuous integration build process. The current implementation residing at https://gl.nmdp.org is using a MySQL persistence back end.
While public use of the service is strongly encouraged, authentication and authorization of services have been designed into the system using OpenID for user identification and OAuth2 (http://oauth.net/) for authorization. This allows third-party applications to obtain limited access to an HTTP service. The initial purpose of this is to assign and enforce quotas for ensure performance, and gathering usage statistics. This can be turned on or off depending on the needs of the organization providing the service. Use cases include anonymous and authorized access, and administrative processes including account management, quota configuration and scoping authorization. Other API gateways are available that include authorization, quotas, and monitoring of back-end services. Choosing from existing components allows anyone to pick and choose based on features, cost, licensing, and support.
The GL Service is available as free open source software, with the source code distributed under the GNU Lesser General Public License (LGPL) version 3 or later.
Multiple versions of the GL Service, the API Explorer, and the liftover service configured with the IMGT/HLA Database reference alleles in strict mode are available at https://gl.nmdp.org.
The GL resource ontology in OWL/RDF format and in XML Schema, both in object graph form and in linked data form, are also available.
Source code and documentation can be found on https://github.com/nmdp-bioinformatics/genotype-list.
3. Results
3.1. Genotype List Service
3.1.1. RESTful web service APIs
The GL Service advertises its functionality via web service APIs in the REST architectural style [4, 5]. An endpoint URL is provided for each GL resource type. HTTP requests to these endpoints perform operations based on HTTP verbs (i.e., HTTP GET and HTTP POST).
To retrieve information about a GL resource identified by a URI, a client uses HTTP GET (e.g. HTTP GET https://gl.nmdp.org/imgt-hla/3.20.0/allele-list/fz). If the service does not know anything about a particular URI, an HTTP 404 Not Found error is returned.
To register a new GL resource described by a string in GL String representation, a client uses HTTP POST to the appropriate endpoint (e.g. for allele lists, HTTP POST https://gl.nmdp.org/imgt-hla/3.20.0/allele-list) with the GL String representation in the body of the request. On success, the service will return a response with a HTTP 201 Created status code, the new URI in the Location header field, and the GL String representation of the newly created GL resource in the response body (as text/plain media type). On failure, an HTTP 400 Bad Request error is returned.
HTTP requests using other HTTP verbs (PUT, PATCH, DELETE) are not supported by the RESTful web service APIs.
3.1.2. Content negotiation
For convenience, each GL resource is available in several different representations that are useful for different applications. GL String is used for plain text representation, HTML can be used for internet browsers, XML and JSON for structured data exchange, OWL/RDF and N3 for semantic web applications, and QR Code for scanning printed forms to avoid manual entry. The GL Service uses content negotiation to return the representation that best meets the needs of the client (or user agent). The user agent expresses its preferences by providing an Accept HTTP header that lists acceptable media types and associated quality factors.
The GL service will return text/plain media type by default for command line tools such as curl or wget, text/html media type for web browsers such as Firefox or Chrome, and RDF/XML media type for semantic web browsers and reasoners (e.g. Protégé [14] or Jena [15]).
In the case that content negotiation may not work as intended, the GL service also supports the use of file extensions in the query URL (e.g., .json, .rdf, .n3, .xml, .xml-xlink). For example, to force text/n3 media type in a web browser user agent, use the .n3 file extension in the query URL. (e.g., HTTP GET https://gl.nmdp.org/imgt-hla/3.20.0/allele-list/fz.n3). In this case, depending on your browser, you will prompted to download a file containing allele list in this format which would be useful for semantic web applications.
The GL service also provides the generation of a QR Code PNG image encoded with the URI identifier of the requested resource by use of the .png file extension. This could provide for example a small 2D barcode representing the full allelic and genotypic ambiguity of a typing result across several loci (i.e., a multilocus unphased genotype) in a small space on a typing report. For example https://gl.nmdp.org/imgt-hla/3.20.0/allele-list/fz.png will return an image of a QR Code that redirects the scanning tool to the URI representing HLA-A*01:01:01:01/HLA-A*01:02.
See Supplementary Material for more examples of different formatted views of the same genotype.
3.1.3. IMGT/HLA Database release versions
To support our primary use case, where the IMGT/HLA Database [6] provides loci and alleles used to compose higher-level GL String representations, an instance of the GL Service can be configured with a specific release version of an allele reference database.
All of the loci and alleles provided by this database are registered as the service initializes. For the most recent version of the IMGT/HLA Database (Release 3.21.0, 2015-07), this consists of 36 loci and 13,412 alleles.
3.1.4. Strict/non-strict mode
Complementary to the nomenclature configuration parameter, the GL Service can be configured in either strict or non-strict mode. Strict mode requires alleles used in the GL String to be present in a particular version of a reference allele database (e.g., IMGT/HLA version 3.20.0). Non-strict mode allows any GL String to be registered as long as it is syntactically correct.
In the default non-strict mode, the GL Service will create new loci and alleles as necessary to register higher-level GL resources. For example, if an allele list represented in GL String format by “HLA-A*01:01:01:01/HLA-Z*99:99:99:99” was registered against a non-strict-mode instance of the GL Service, a new HLA-Z locus and HLA-Z*99:99:99:99 allele would be created in order to correctly represent the allele list.
In strict mode instances of the GL Service, pre-populated with specific reference allele lists, this default behavior is overridden, so that GL resources that do not adhere to the allele database release version in question cannot be registered. Any attempts to register an invalid GL resource will result in an error.
In strict mode, the namespace includes the name (e.g. “imgt-hla”, “ipd-kir”) and version number of the reference database. In non-strict mode, the name and version of the reference database are not included in namespace of the service. For example, the service found at https://gl.nmdp.org/imgt-hla/3.20.0 only accepts alleles found in version 3.20.0 of the IMGT/HLA Database. Using strict mode, a new instance of the service must be created for each reference database release. In strict mode, nomenclatures may not be mixed (e.g., HLA and KIR) within a GL String unless the nomenclature databases have been preloaded into the instance.
In strict mode, each allele must be represented as a complete object, including the full locus name with prefix. For example, HLA-A*01:01:01:01 is acceptable, but A*01:01:01:01 is not. The latter would be acceptable in non-strict mode, but this mode loses the ability to validate the allele. In this case, validation would be a responsibility of the creator of the GL-String and/or the consumer of the GL String referenced by the URI.
In strict mode, IMGT/HLA G groups are recognized as accepted HLA nomenclature (e.g., HLA-A*01:01:01G). They are accepted in strict mode as a courtesy to those who wish to exchange validated GL Strings containing G groups. G groups are registered as alleles, although in reality they are in fact lists of alleles, and so represents an exception to the definition of allele used in the service. In the future, a separate service may be developed to translate GL Strings containing G groups into IMGT/HLA Database version specific lists of alleles. P groups were not initially included because P groups reflect the alleles that are actually expressed and so do not include null alleles. Since null alleles are important in many of our use cases and because we did not wish to further conflate the definition of allele in the service, P groups were not added to the GL Service. P groups may be added in the future if users require them.
At the time of this writing, 65 strict GL Services are available representing IMGT/HLA Database versions from 1.5.0 to 3.20.0.
3.2. Client libraries and tools
Since RESTful web service APIs are based on HTTP, the service endpoints are accessible from web browsers, browser-based RESTful web service clients (e.g. RESTClient [16], REST Easy [17]), and from command-line HTTP clients.
For example, to register a GL String via HTTP POST, the curl command may be used:
$ curl -v --header "content-type: text/plain" \ --data "HLA-A*01:01:01:01/HLA-A*01:02+HLA-A*01:03" \ -X POST https://gl.nmdp.org/imgt-hla/3.20.0/genotype
This above command will register the GL String and return the URI in the Location field of the response. In this case the returned URI is https://gl.nmdp.org/imgt-hla/3.20.0/genotype/jn. The last field of the URI (e.g., “jn”) returned cannot be predicted. The service will attempt to reuse a URI if it finds an existing one that matches the GL String being registered, but this is not guaranteed (or necessary). URI’s should never be directly compared with each. Rather, comparisons should only be made with the GL String that is being returned by the URI.
To retrieve the contents of this URI, the wget command may be used. For example,
$ wget -q -O - https://gl.nmdp.org/imgt-hla/3.20.0/genotype/jn
returns the GL String HLA-A*01:01:01:01/HLA-A*01:02+HLA-A*01:03
The GL Service project provides functionality beyond these generic REST clients, such as an interactive API Explorer, client libraries, and command line tools.
3.2.1. API Explorer
An interactive web tool demonstrating the GL Service API is available at https://gl.nmdp.org/imgt-hla/3.20.0/explorer/.
This tool is not meant to be for production use, but rather provides an educational interface to the service, showing how GL Strings can be registered in the service, and how URIs can be dereferenced to retrieve GL Strings. The Explorer can also be used to show how different content types can be retrieved and display their output. In the following examples, a GL String representing a genotype list is being followed from being registered and then retrieved in different formats.
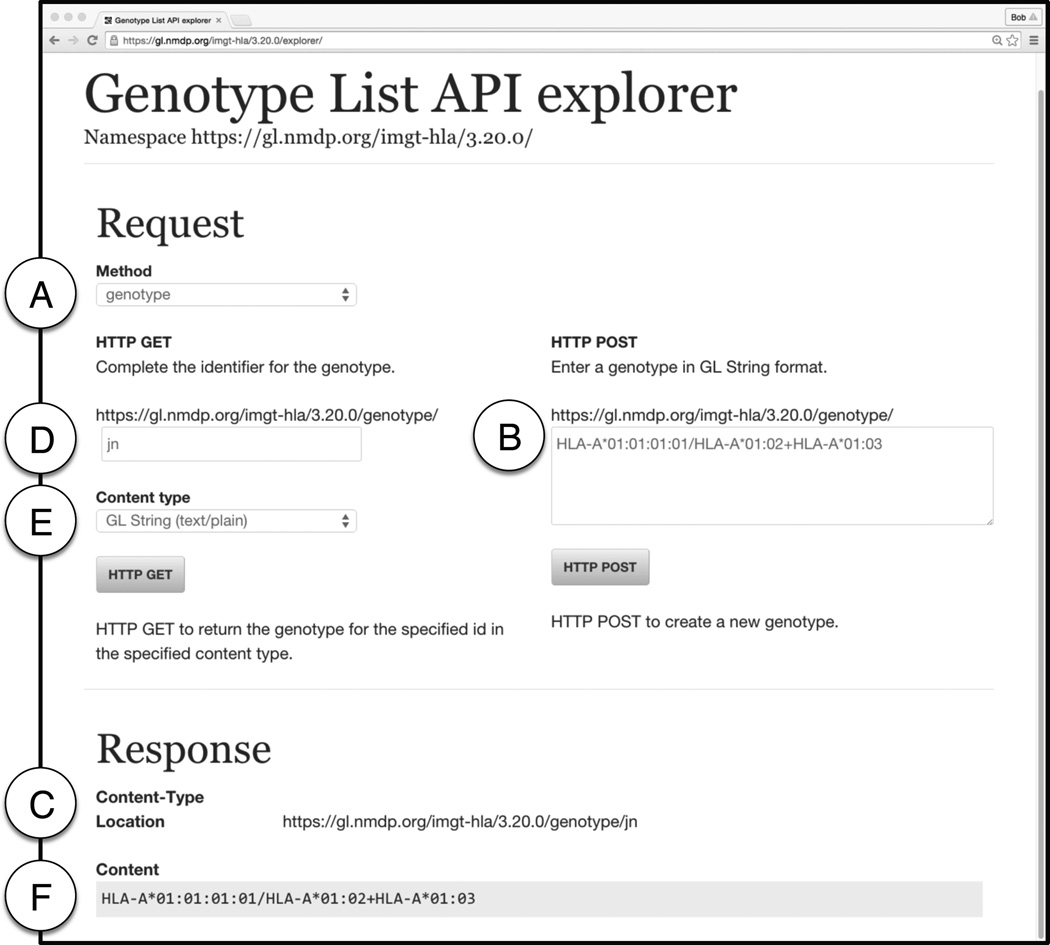
Figure 1 shows the interface to the API Explorer. The user selects the method that describes the object that is being registered (Figure 1A). In this example, the GL String represents a genotype consisting of a list of two alleles and a single allele: HLA-A*01:01:01:01/HLA-A*01:02+HLA-A*01:03. The string is entered into the text entry box on the right and the HTTP POST button is clicked (Figure 1B). The response from the service is displayed at the bottom of the page showing the Location of the resource in the form of a URI. In this example the URI is https://gl.nmdp.org/imgt-hla/3.20.0/genotype/jn (Figure 1C).
Figure 1.
Interface of GL Service API Explorer found at https://gl.nmdp.org/imgt-hla/3.20.0/explorer/
A – Selecting method in the API Explorer. Options from a dropdown menu include locus, allele, allele-list, haplotype, genotype, genotype-list, and multilocus-unphased genotype. Here the Method selected is genotype.
B – Using HTTP POST to register a GL String. Here the GL String representing the genotype HLA-A*01:01:01:01/HLA-A*01:02+HLA-A*01:03 is entered.
C – After clicking the HTTP POST button, the URI returned is displayed in the Location field of the page. Here the URI listed it https://gl.nmdp.org/imgt-hla/3.20.0/genotype/fn.
D – Using HTTP GET to retrieve a GL String. The URI for the genotype is completed using the data entry box. Here fn is entered to complete the URI https://gl.nmdp.org/imgt-hla/3.20.0/genotype/fn.
E – Selecting content type. This dropdown menu is used to select the format when retrieving the dereference URI. Options include GL String (text/plain), HTML (text/html), JSON (application/json), RDF (application/rdf+xml), N3 (text/n3), QR Code (image/png), XML object graph style (txt/xml), and XML refs style (text/xml). In this case, GL String (text/plain) is selected.
F – The content returned after a HTTP GET request is made is displayed in the Content field of the page. In this case, the content that the URI https://gl.nmdp.org/imgt-hla/3.20.0/genotype/fn is the GL String HLA-A*01:01:01:01/HLA-A*01:02+HLA-A*01:03
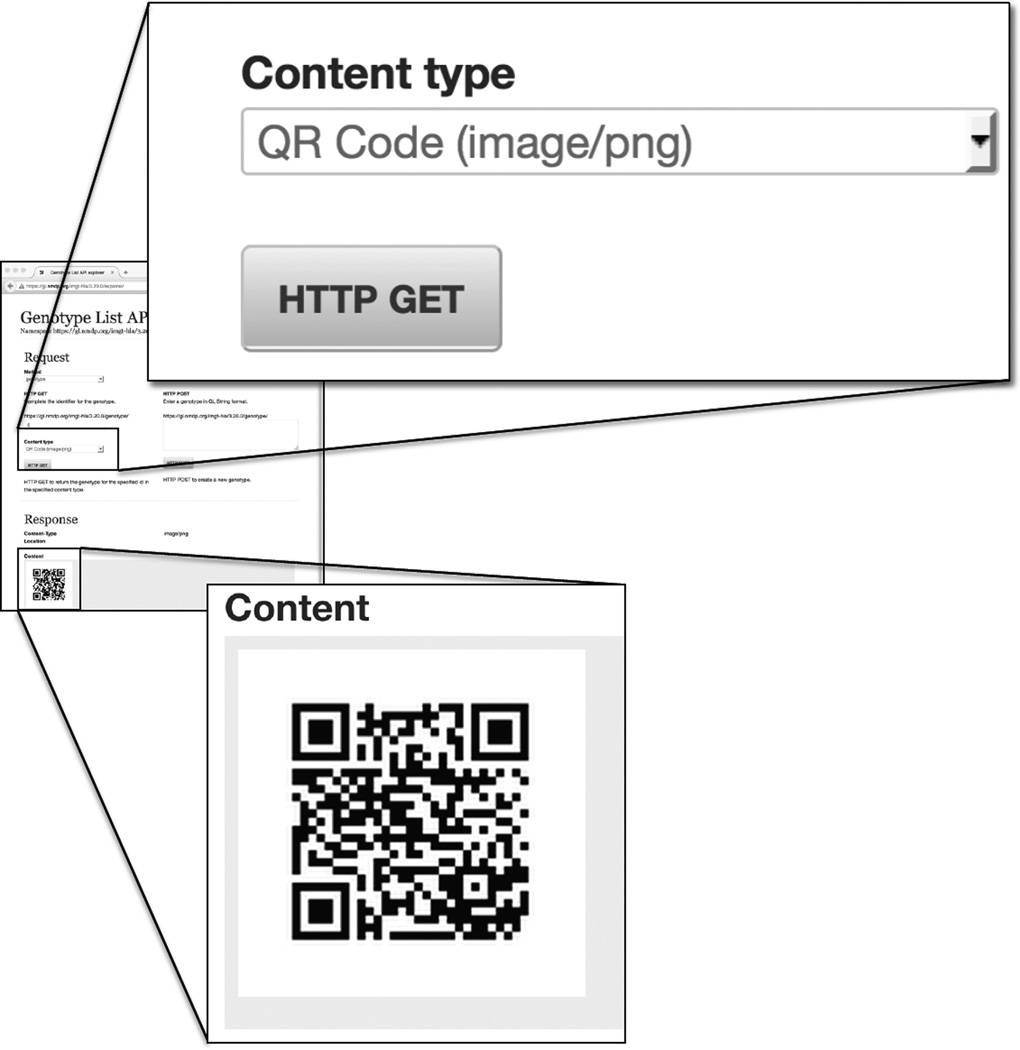
The URI can then be used to retrieve the genotype in GL String format, or any one of other formats including HTML, XML, JSON, OWL/RDF, N3 and QR Code. The URI is completed by entering the last field in the text box in (Figure 1D) and the output selected through a dropdown menu (Figure 1E). Clicking the HTTP GET button retrieves the object and displays it in the selected content type. Figure 1F shows the response of the service in GL String format. Figure 2 shows the result of a request for the object in QR Code format. Scanning this QR Code with a scanning application found on smart phones will result in the URI opening in a web browser, showing the object in HTML format, which can be drilled deeper into the structure by following links.
Figure 2.
Using HTTP to GET a QR Code. The Response when QR Code is selected from Content Type. When scanned, the URI representing the GL String is returned. If the scanner is configured to open a web page, it will return the HTML representation of the URI. In this case, the QR Code returns the URI https://gl.nmdp.org/imgt-hla/3.20.0/genotype/fn which will open a page displaying the HTML format of the GL String containing HLA-A*01:01:01:01/HLA-A*01:02+HLA-A*01:03.
3.2.2. Client libraries
For integration into larger software systems, a Java client library is provided that wraps HTTP interaction with the RESTful web service API behind simple method calls.
Code listing (excerpt) GlClient.java:
public interface GlClient {
String registerAlleleList(String glstring) throws GlClientException;
AlleleList getAlleleList(String uri) throws GlClientException;
AlleleList createAlleleList(String glstring) throws GlClientException;
...
}
The register methods accept a GL String representation and return the URI of the newly registered GL resource. The get methods accept a URI identifier and return the e.g., AlleleList object representation of the allele list identified by that URI, if such exists. The create methods perform a call to the register method followed by a call to the get method, so for an e.g., allele list in GL String representation the caller receives an AlleleList object representation of the newly registered allele list resource.
Usage:
GlClient client = ...;
Allele a0 = client.getAllele("https://gl.nmdp.org/imgt-hla/3.18.0/allele/0");
Allele a1 = client.getAllele("https://gl.nmdp.org/imgt-hla/3.18.0/allele/1");
AlleleList alleleList0 = client.createAlleleList(a0, a1);
String uri = client.registerAlleleList("HLA-A*01:01:01:01/HLA-A*01:01:01:02N");
AlleleList alleleList1 = client.getAlleleList(uri);
Note the client library caches frequently used resources in order to speed up consequent queries. The client library (and by extension, the command line tools described below) can be configured to use either the application/json media type or the application/xml media type when accessing the Genotype List service RESTful web service APIs.
The JSON representation uses a linked data style format, where associations between parent and child resources are described using URI identifiers. Thus a single request for a high-level GL resource type, such as multilocus unphased genotype, may result in several separate queries against the service to populate the full hierarchy of resources. The response to each individual request will be relatively small in content size.
The XML representation uses a fully specified nested object representation, where associations between parent and child resources are contained as nested XML elements in a single file. This results in only a single request to the service for a high-level resource type. The response to the request will be much larger in content size, however.
This configuration choice allows the user to optimize for response content size or number of requests as necessary.
3.2.3. Command-line tools
For straightforward integration into scripting applications such as typing report submission or sequence analysis pipelines, a suite of command line tools is provided. These command line tools wrap the client library and allow for batch processing of large numbers of GL String resources at one time.
One command line tool for each GL resource type is available. Resources in GL String representation are read via the standard input stream (stdin) or an input file. The URI identifiers are written to the standard output stream (stdout) or an output file.
For example,
$ gl-register-allele-lists \
--namespace https://gl.nmdp.org/imgt-hla/3.20.0/ \
--glstrings glstrings.txt \
--identifiers identifiers.txt
reads allele lists in GL String representation from the file glstrings.txt, registers them against the instance of Genotype List Service at the namespace https://gl.nmdp.org/imgt-hla/3.20.0/, and writes the URI identifiers of the newly created allele list resources to the file identifiers.txt.
3.3. Accessory services
As a low-level utility service, the GL Service provides a robust building block on which to create accessory services. Two such services are already available, a service that provides the means to lift over a GL resource defined in one version of the IMGT/HLA allele reference database to an equivalent GL resource defined in another version, and a service that maps allelic ambiguity represented by an allele list GL resource to an arbitrary ambiguity encoding.
3.3.1. Liftover service
Given two or more instances of the GL Service configured with different versions the IMGT/HLA Database nomenclature in strict mode, it may be more straightforward to compare GL resources if they are both registered in the same instance of the GL service.
A liftover service has been implemented that given a source namespace, a URI identifying a GL resource in the source namespace, and a target namespace will attempt to lift over the source GL resource to the GL Service at the target namespace. The liftover service is nominal at this point and uses very simple rules. It uses the Allelelist_history.txt file from IMGT/HLA as a reference table of allele nomenclature versions mapped to an accession number. For each allele in the input GL String in the source nomenclature, the service will find its accession number and look up the new allele in the target nomenclature. If it does not exist, it is dropped. If it does exist, the service will create a new output GL String in the target nomenclature from the new alleles.
For example, an allele list resource identified by the URI https://gl.nmdp.org/imgt-hla/3.17.0/allele-list/0 with GL String representation “HLA-A*01:01:01:01/HLA-A*01:01:01:02N” may lift over to the target namespace https://gl.nmdp.org/imgt-hla/3.18.0/ via the liftover service, which if successful will return the URI identifying the newly created allele list in the target namespace, e.g. https://gl.nmdp.org/imgthla/3.18.0/allele-list/0. Otherwise an error will result. While in this example, the last field of the URI happens to be the same as the last field of the original URI (0), this is not guaranteed and should not be expected.
Like the GL Service, the liftover service provides RESTful web service APIs . Documentation about the liftover service is found on the source repository at github.com (See the availability section above).
3.3.2. Ambiguity service
Many legacy or arbitrary encodings of allelic ambiguity are in use in the HLA and KIR communities (e.g., NMDP Allele Codes [18]). To adapt these encodings to allele list GL resources registered in instances of the GL Service configured with the IMGT/HLA Database nomenclature in strict mode, an ambiguity service has been developed. This is a RESTful service for assigning a short name (e.g., NMDP Allele Code) to an allele list representing allelic ambiguity. This service could also be used for serology mappings, epitope group mappings, search determinant mappings, protein level allele name to strict-mode IMGT/HLA nomenclature mappings, or P group mappings, etc.
4. Discussion
While standards are critical for HLA data interoperability, they are not meaningful until useful tools are developed and made available for community use. Here we have described the development of web services to create and retrieve complete HLA typing data in standardized formats. GL Strings containing KIR loci may be registered in a non-strict service since this type of GL Service does not have any restriction on the nomenclature of loci/alleles. Services that are strict for IPD-KIR nomenclature may also be created using a similar way as was done for HLA, i.e., preloading the service with KIR loci and alleles from the IPD-KIR database.
The tools being developed here provide the HLA researcher, clinician and lab technician a common resource for managing HLA data in a standardized fashion. We envision these tools augmenting workflows through creation of new instances of HLA typing objects when needed, and retrieval of those objects and their associated metadata when called upon. By exchanging URIs and dereferencing them through the GL Service, users can easily transmit HLA genotypes in a variety of useful formats (GL String, HTML, XML, JSON, OWL/RDF, N3 and QR Code). The GL Service may be combined with other tools and services to create useful workflows for different use cases, such as sending genotyping data to registries, or research pipelines. They may be used as persistent identifiers for genotyping results in publications, and exchanged between research collaborators, and integrated with semantic web genotype-phenotype and disease association resources.
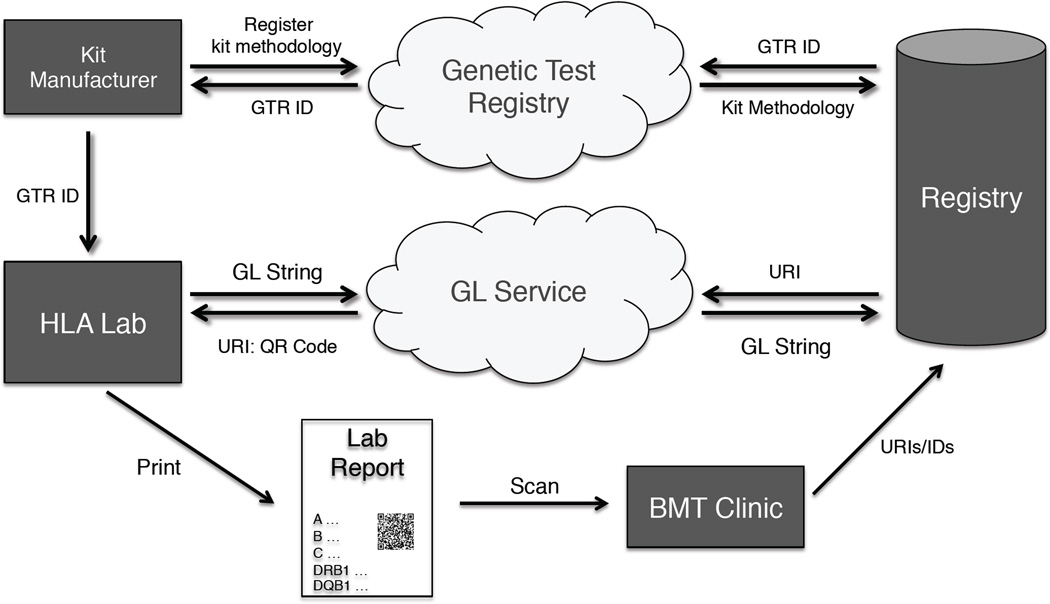
While full allele and genotype ambiguity can be recorded into GL Strings, these strings can be quite large and legacy systems with limited data formats in their transport mechanisms may not be able to accommodate them. Newer formats for HLA typing (e.g., HML 1.0) and clinical data exchange (e.g., HL7 CDA, HL7 FHIR) often provide mechanisms to include pointers to data rather than the data itself. The GL Service is well suited for this model of clinical reporting. Here, the GL String would be registered in the GL Service and the URI may be used in a genotyping report, perhaps with a QR code included in a printed document if needed. Using separate service to register the methodology of the lab test (e.g., NCBI Genetic Testing Registry), the IDs may be sent to the registry, which can then dereference them in their systems. This example is illustrated in Figure 3. Components of this example are being developed today. For example, HML 1.0 contains new data structures specifically to hold a GL String or a URI which points to one for reporting allele assignment from a HLA genotyping test as well as structures to hold pointers to registered test methodologies.
Figure 3.
A workflow describing how data generated by an HLA Lab may be sent to a registry by using a combination of the GL Service and the NCBI Genetic Test Registry (GTR). Here, test methodologies are registered with the GTR and later retrieved using a GTR Test ID while the allele assignments are recorded with GL Strings and registered with the GL Service which returns a URI (in this example in the form of a QR Code).
The QR Code format could be used to augment a current HLA laboratory typing report so make it machine readable. By encoding the data using the GL Service and obtaining a QR Code included with the report the recipient of the typing report might then use their smart phone or desktop computer to decode the URI identifier in the QR Code and then retrieve the full ambiguity in GL String representation using the Genotype List service.
Toward these ends, tools leveraging the GL Service to store and exchange genotype data are under development. For example, the Toolkit for Immunogenomic Data-Exchange and Storage (TIDES), accepts genotype data generated by a variety of platforms (e.g., HLA Fusion and Conexio Genomics Assign ATF), generates compact, standardized GL Strings for these data, registers them with the GL Service, disambiguates strings, provides reports in a variety of formats, and traffics formatted genotype data directly to data-analysis software. The use of TIDES and other applications that leverage the GL Service will foster the sharing, reuse, revaluation and meta-analysis of genotype data in ways that have not previously been possible.
The promise of the ‘cloud’ is that data will persist and that service is secure and reliable, not only for today, but for the future. Anyone can now set up a service to register and retrieve GL Strings. For those who create a service, expectations on security, performance, and reliability must be communicated and met.
Supplementary Material
Acknowledgements
This work was supported by Office of Naval Research (ONR) grant N00014-12-1-0142 (MM and RPM), National Institutes of Health (NIH) grants U01AI067068 (SJM and JAH), awarded by the National Institute of Allergy and Infectious Disease (NIAID), and R01GM109030 (SJM, JAH, MM, and RPM), awarded by the National Institute of General Medical Sciences (NIGMS). The content presented is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, NIAID, NIGMS, ONR, Department of Office of Naval Research, the Department of the Navy, the Department of Defense, or the US Government.
Abbreviations
- API
Application Program Interface
- AWS
Amazon Web Services
- CDISC
Clinical Data Interchange Standards Consortium
- DaSH
Data Standard Hackathon
- EMBL
European Molecular Biology Laboratory
- EMDIS
European Marrow Donor Information System
- ENA
European Nucleotide Archive
- FDA
Food and Drug Administration
- FHIR
Fast Healthcare Interoperability Resources
- GL
Genotype List
- GNU
GNU’s Not Unix
- HL7
Health Level Seven, International
- HLA
Human Leucocyte Antigen
- HML
Histoimmunogenetics Markup Language
- HTML
Hypertext Markup Language
- HTTP
Hypertext Transfer Protocol
- IMGT
ImMunoGeneTics
- ISO
International Organization for Standardization
- JDBC
Java Database Connectivity
- JSON
Javascript Object Notation
- KIR
Killer-cell Immunoglobulin-like Receptor
- LGPL
Lesser General Public License
- LSDAM
Life Sciences Domain Analysis Model
- MHC
Major Histocompatibility Complex
- MIRING
Minimum Information for Reporting Immunogenomic NGS Genotyping
- MUG
Multilocus Unphased Genotype
- N3
Notation 3
- NCI
National Cancer Institute
- NGS
Next Generation Sequencing
- NMDP
National Marrow Donor Program
- OID
Object Identifier
- OWL
Web Ontology Language
- PNG
Portable Network Graphics
- QR Code
Quick Response Code
- RAM
Random Access Memory
- RDBMS
Relational Database Management System
- RDF
Resource Description Language
- REST
Representational State Transfer
- SBT
Sequence Based Typing
- SDK
Software Development Kit
- SQL
Structured Query Language
- SSO
Sequence Specific Oligonucleotide
- SSP
Sequence Specific Primer
- URI
Uniform Resource Identifier
- URL
Uniform Resource Locator
- XML
Extensible Markup Language
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest:
There are no conflicts of interest.
References
- 1.Hollenbach JA, Mack SJ, Gourraud P-A, Single RM, Maiers M, Middleton D, et al. A community standard for immunogenomic data reporting and analysis: proposal for a STrengthening the REporting of Immunogenomic Studies statement. Tissue Antigens. 2011;78:333–344. doi: 10.1111/j.1399-0039.2011.01777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milius RP, Mack SJ, Hollenbach Ja, Pollack J, Heuer ML, Gragert L, et al. Genotype List String: a grammar for describing HLA and KIR genotyping results in a text string. Tissue Antigens. 2013;82:106–112. doi: 10.1111/tan.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berners-Lee T, Fielding RT, Masinter L. Uniform Resource Identifier (URI): Generic Syntax. IETF RFP 3986 (standards Track), Internet Eng Task Force. 2005 [Google Scholar]
- 4.Fielding RT. Architectural styles and the design of network-based software architectures. Irvine: University of California; 2000. [Google Scholar]
- 5.Fielding RT, Taylor RN. Principled design of the modern Web architecture. ACM Trans Internet Technol. 2002;2:115–150. [Google Scholar]
- 6.Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SGE. The IMGT/HLA Database. Nucleic Acids Res. 2013;41:D1222–D1227. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.W3C OWL Working Group. OWL 2 Web Ontology Language: Document Overview - W3C Recommendation 11 December 2012. Second 2012. [Google Scholar]
- 8.Gandon F, Schreiber G. [last accessed 18 March 2015];RDF 1.1 XML Syntax - W3C Recommendation 25 February 2014. http://www.w3.org/TR/2012/REC-owl2-overview-20121211/,
- 9.Peterson D, Gao S, Malhotra A, Sperberg-McQueen CM, Thompson HS. [last accessed 18 March 2015];W3C XML Schema Definition Language (XSD) 1.1 Part 2: Datatypes - W3C Recommendation 5 April 2012. http://www.w3.org/TR/xmlschema11-2/,
- 10.Gao S, Sperberg-McQueen CM, Thompson HS. [last accessed 18 March 2015];W3C XML Schema Definition Language (XSD) 1.1 Part 1: Structures - W3C Recommendation 5 April 2012. http://www.w3.org/TR/xmlschema11-1/,
- 11.DeRose S, Maier E, Orchard D, Walsh N. [last accessed 18 March 2015];XML Linking Language (XLink) Version 1.1 - W3C Recommendation 06 May 2010. http://www.w3.org/TR/xlink11/,
- 12. [last accessed 18 March 2015];MySQL. http://www.mysql.com,
- 13. [last accessed 18 March 2015];Oracle. http://www.oracle.com,
- 14. [last accessed 18 March 2015];Protégé - A free open-source ontology editor and framework for building intellegent systems. http://protege.stanford.edu/,
- 15. [last accessed 18 March 2015];Apache Jena - A free and open source Java framework for building Semanic Web and Linked Data applications. https://jena.apache.org/,
- 16. [last accessed 18 March 2015];RESTClient, a debugger for RESTful web services. https://addons.mozilla.org/en-us/firefox/addon/restclient/,
- 17. [last accessed 18 March 2015];RESTEasy. http://resteasy.jboss.org/,
- 18.Maiers M, Hurley CK, Perlee L, Fernandez-Vina M, Baisch J, Cook D, et al. Maintaining updated DNA-based HLA assignments in the National Marrow Donor Program Bone Marrow Registry. Rev Immunogenet. 2000;2:449–460. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.