Summary
HLA-DM is essential for editing peptides bound to MHC class II, thus influencing the repertoire of peptides mediating selection and activation of CD4+ T cells. Individuals expressing HLA-DQ2 or DQ8, and DQ2/8 trans-dimers, have elevated risk for type 1 diabetes (T1D). Cells co-expressing DM with these DQ molecules were observed to express elevated levels of CLIP (Class II associated invariant chain peptide). Relative resistance to DM-mediated editing of CLIP was further confirmed by HPLC-MS/MS analysis of eluted peptides, which also demonstrated peptides from known T1D-associated autoantigens, including a shared epitope from ZnT8 that is presented by all four major T1D-susceptible DQ molecules. Assays with purified recombinant soluble proteins confirmed that DQ2-CLIP complexes are highly resistant to DM editing, whereas DQ8-CLIP is partially sensitive to DM, but with an apparent reduction in catalytic potency. DM sensitivity was enhanced in mutant DQ8 molecules with disruption of hydrogen bonds that stabilize DQ8 near the DM-binding region. Our findings show that T1D-susceptible DQ2 and DQ8 share significant resistance to DM editing, compared with control DQ molecules. The relative resistance of the T1D-susceptible DQ molecules to DM editing and preferential presentation of T1D-associated autoantigenic peptides may contribute to the pathogenesis of T1D.
Keywords: MHC class II, antigen presentation, Type 1 diabetes, HLA-DM, HLA-DQ, invariant chain-derived CLIP peptides
Introduction
Type 1 diabetes (T1D) is an autoimmune disease characterized by selective destruction of pancreatic β cells. Multiple genetic and environmental factors have been implicated in the etiology of T1D. Among these factors, certain MHC haplotypes are associated with a high risk for development of T1D, with a potential role of MHC molecules in the selection or activation of autoreactive T cells involved in T1D [1]. Genetic studies have implicated DQ2 (DQA1*0501/DQB*0201) and DQ8 (DQA1*0301/DQB1*0302) as important risk factors. DQ2/8 heterozygotes, expressing two potential DQ trans-dimers as well as DQ2 and DQ8, have a further elevation in risk. By contrast, DQ6 (DQA1*0102/DQB*0602) has a dominant protective effect, while DQ1 (DQA1*0101/DQB1*0501) is neutral to T1D [2-8]. Beyond speculation, the mechanisms responsible for the disease risk associated with expression of specific MHC class II (MHCII) alleles remain to be established.
MHCII molecules present endogenous or exogenous peptides on the surface of APC to select or activate CD4+ T cells in the thymus or periphery [9]. Classical MHCII are initially assembled with the class II invariant chain (Ii) chaperone protein. Ii is partially released through a series of successive proteolytic events in endosomal compartments, leaving a fragment, CLIP, occupying the peptide-binding groove. Antigen presentation is critically dependent on the release of CLIP and exchange with antigenic peptides sampled in endosomal compartments, which is catalyzed by a non-classical MHC protein HLA-DM. DM functions optimally at acidic pH to catalyze CLIP dissociation and peptide exchange. It can catalyze multiple rounds of peptide exchange, and preferentially edit unstable peptide complexes, favoring the presentation of highly stable peptide complexes under physiological conditions. Thus, DM plays a critical role in editing the repertoire of peptides available for selection and recognition by CD4+ T cells [10, 11]. It has been noted that CD4+ T cells with specificity for self-peptides with low affinity for MHC can escape negative selection in the thymus [12-16]. The identification of autoreactive CD4+ T cells recognizing low-affinity peptides in both NOD mouse [17-19] and human [20] autoimmune diabetes suggests the possibility that incomplete DM editing may contribute to presentation of a unique repertoire of peptides important to the pathogenesis of T1D.
The recently published co-crystal structures of DM bound to the MHCII molecule DR1 demonstrates a major structural rearrangement in the region of the DR1 peptide-binding groove that accommodates the N-terminal segment of bound peptide proximal to the DM contact surface [21]. This conformation change precludes full occupancy of the peptide-binding groove. Polymorphisms in DM contact residues have the potential to impact DM binding affinity and catalytic potency [22]. In addition, polymorphisms in MHCII that might impact the conformational stability in the region that undergoes a major structural transition might impact DM editing function [23]. The potential effects of MHCII polymorphism on susceptibility to DM editing have been explored to a limited extent. Notably, it has been reported that DQ2 is a poor substrate for DM, due to a natural deletion of arginine in α chain of DQ2 [22, 24]. This allele is associated with celiac disease [25] as well as T1D. However, that natural deleted residue of DQ2 is intact in DQ8, and the susceptibility of DQ8 to DM editing is unknown. Different mechanism(s) might be responsible for the association of DQ8 with increased risk for T1D. In addition to the obvious impact on peptide binding specificity, MHCII polymorphisms might contribute to the pathogenesis of CD4+ T cell autoimmunity by influencing a variety of steps in the peptide loading, editing, and presentation process, possibly enabling alternative mechanisms for presentation of key autoantigens or selection of autoreactive T cells [22, 26, 27].
Given the potential importance of DM in modulating the repertoire of peptides that mediate thymic selection and/or peripheral activation of autoreactive T cells in T1D [17, 18, 20], we investigated the sensitivity of the major T1D-susceptible DQ molecules to DM editing. We co-expressed DQ, Ii and DM molecules in 293T cells to compare DM mediated release of Ii derived CLIP peptides. We found that DQ2, DQ8 or DQ2/8 trans-dimers were more resistant to DM editing than T1D-protective DQ6 or the T1D-neutral DQ1 molecules, as demonstrated by retention of CLIP. HPLC-MS/MS analysis confirmed the presence of DM-resistant CLIP, as well as known T1D-associated autoantigenic peptides specifically binding to T1D- susceptible DQ molecules. Fluorescence polarization (FP) assays with purified recombinant soluble DQ proteins demonstrated that DQ8-CLIP is partially sensitive to DM, but with an apparent reduction in catalytic potency. DM sensitivity was enhanced in mutant DQ8 molecules with disruption of hydrogen bonds that stabilize the DQ8 in the DM-binding region. We hypothesize that a relative DM editing resistance of T1D associated DQ2, DQ8 and DQ2/8 trans-dimer molecules may contribute to the pathogenesis of T1D, by affecting the selection and activation of autoreactive CD4+ T cells.
Results
Relative resistance to DM editing of T1D-susceptible cis- and trans-DQ molecules
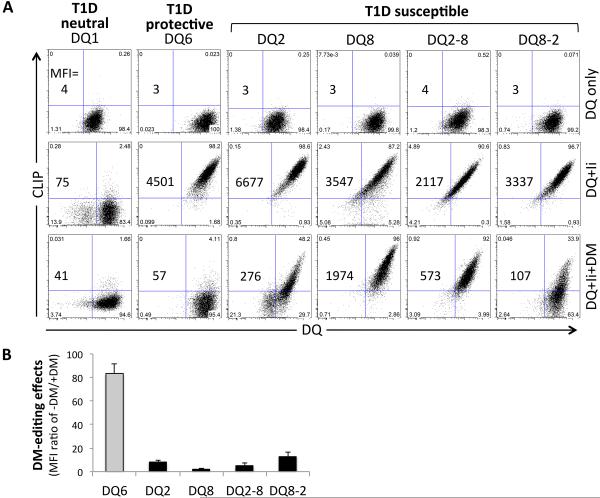
To investigate the DM editing sensitivity of the T1D-susceptible DQ2, DQ8 and DQ2/8 molecules, we expressed each of the four DQ molecules or the controls, T1D-protective DQ6 or T1D-neutral DQ1 molecules, in the 293T cells, which do not express endogenous Ii or MHCII. All six DQ molecules were efficiently expressed at the cell surface in the absence or presence of Ii, confirming that the two trans-dimers potentially expressed in DQ2/8 heterozygotes, DQ2-8 and DQ8-2, are efficiently assembled and transported to the cell surface [28, 29] (Fig.1). Cell surface CLIP levels, measured by flow cytometry with CerCLIP.1, a mAb specifically recognizing CLIP1 peptides with N-terminal extensions (Supporting Information Fig.1), were high for each of the T1D-susceptible DQ molecules and the DQ6 control on cells co-expressing Ii in the absence of DM, demonstrating functional association with Ii, proteolytic processing to generate CLIP, and inefficient release of CLIP in the absence of DM (Fig.1). Co-expression of DM resulted in a complete loss of cell surface CLIP on cells expressing the control molecule DQ6. By contrast, DQ2-CLIP complexes were relatively resistant to DM editing, confirming previous reports demonstrating that DQ2 is a poor substrate for DM [22, 24]. Strikingly, the three other T1D-susceptible DQ molecules (DQ8, DQ2-8 and DQ8-2) were also observed to retain high levels of cell surface CLIP in the presence of DM (Fig.1, Supporting Information Fig.2E). The cell lines used in these experiments expressed similar levels of DQ, Ii, and DM (Fig.1, Supporting Information Fig.2D). Interestingly, there is no detectable CLIP peptide presented by DQ1 even in the absence of DM (Fig.1A). A hierarchy in DM sensitivity was observed for these molecules, with DQ8 being the least sensitive, followed by DQ2-8, DQ2, and DQ8-2. However, all of the four T1D-susceptible DQ molecules were relatively resistant to DM editing of CLIP as compared with DQ6. Although 293T cells have been used in MHC class II antigen presentation studies [30], they are not professional APC. To explore the generality of the results with 293T cells, we performed additional experiments with T2 lymphoblastoid cells. The T2 cell line, a hybrid human B cell line with endogenous Ii expression but with the deletion of entire MHC class II region in the genome [31, 32], has been used extensively as a model professional APC. We expressed the six different DQ alleles in T2 cells in the absence or presence of HLA-DM and determined the effectiveness of DM in editing CLIP by flow cytometry. The results were consistent with those obtained with 293T cells (Supporting Figure 3A and 3B).
FIGURE 1.
DM editing sensitivity of Ii-derived CLIP peptides in T1D neutral DQ1, protective DQ6 or susceptible DQ2, DQ8 and DQ2/8 trans-dimers. (A) CLIP and DQ molecules on 293T cells transduced with DQ only (top), DQ+Ii (middle) or DQ+Ii+DM (bottom) were cell surface stained with fluorescence conjugated Cer.CLIP1 and SPVL3 antibodies as described in Materials and Methods. The mean fluorescence intensity (MFI) is stated in the panels, and indicates cell surface expression level of CLIP bound to DQ. (B) Relative DM editing effects indicated by comparison of the MFI ratio of cell surface CLIP and DQ levels in the absence or presence of DM expression. Cells were surface stained for CLIP and DQ and gated on the same level of GFP and/or mCherry expression. Data are representative of 3 independent experiments and shown as mean + SD in panel B.
Direct Analysis of DQ-bound CLIP and T1D-associated autoantigen-derived peptides
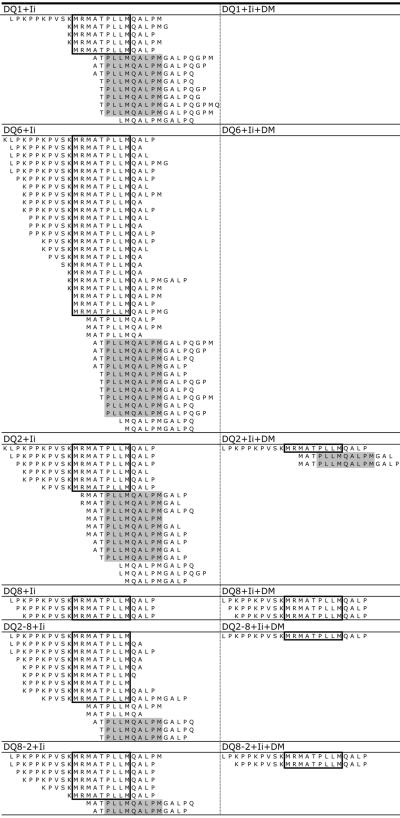
We next sought to confirm DM-resistant retention of CLIP by the four T1D-associated DQ molecules by direct identification of peptides eluted from DQ molecules. Peptides were eluted from affinity-purified DQ molecules isolated from 293T cells co-expressing Ii with or without DM, and samples were analyzed by HPLC-MS/MS. Consistent with the findings from flow cytometry, CLIP peptides were prominently presented by all DQ alleles in cells expressing Ii but not DM (Fig.2, left). A large number of nested peptides ranging from 9-24 amino acids in length were identified that contained the classical CLIP1 core sequence (boxed) [33] as well as the alternative CLIP2 register (gray highlight). Consistent with the flow cytometry results, CLIP was not detected in peptide eluates from cells expressing DQ6+Ii+DM, indicating efficient DM editing on DQ6-CLIP complexes (Fig.2, right). By contrast, CLIP1 was detected in peptide samples from cells co-expressing DM and each of the T1D- susceptible DQ molecules, supporting the conclusion that CLIP1 bound to these alleles is resistant to DM editing. Strikingly, Ii-derived CLIP peptides, including both CLIP1 and CLIP2 registers, were also detected in peptide samples co-expressing DQ1 and Ii, but not in samples co-expressing DM, indicating that DQ1 molecules do present CLIP peptides and DM efficiently mediates the editing of DQ1-CLIP complexes (Fig.2). However, most of the CLIP1 peptides presented by DQ1 molecules lack the N-terminal extensions, the epitope recognized by the CerCLIP.1 mAb [34], which is consistent with the flow cytometry observations and CerCLIP1 mAb specificity (Fig.1A, Supporting Information Fig.1). CLIP2 peptides were highly represented in peptide samples eluted from DQ2 [35-37]. It is not known if DQ2-CLIP2 is generated by an alternative association of Ii with DQ2 in the ER, or from rebinding of Ii fragments in this register in endosomal compartments [24]. Our results demonstrate that CLIP2 associates with DQ molecules other than DQ2, including the T1D-protective DQ6 and T1D-neutral DQ1 molecules. It is also interesting to note that CLIP2 peptides were also present in samples from cells expressing DQ2+Ii+DM, consistent with prior findings indicating the CLIP2 register has a relatively high affinity for DQ2 [35-37].
FIGURE 2.
Pattern of eluted unique CLIP peptides. 293T cells expressing different DQ molecules in the absence (left, DQ+Ii) or presence (right, DQ+Ii+DM) of DM were used to elute peptides bound to each DQ molecules captured by anti-DQ mAb (SPVL3) immobilized with protein A beads. Bound peptides were eluted and further separated by HPLC and analyzed by HPLC-MS/MS. The two well-defined registers of Ii-derived peptides are highlighted as CLIP1 (boxed) and CLIP2 (gray). Data shown are representative of 2 independent experiments.
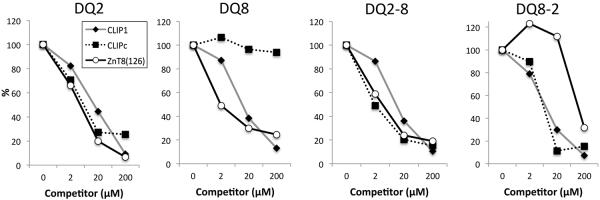
Previous reports (http://www.ebi.ac.uk/arrayexpress/experiments/E-GEOD-24733/) indicate that 293T cells express variable levels of some T1D-associated autoantigens, including heat shock protein 60 (HSP60), zinc transporter 8 (ZnT8), and insulinoma antigen-2 (IA-2) [38]. Interestingly, we identified peptides from these autoantigens in samples eluted from T1D-susceptible DQ molecules but not from T1D-neutral DQ1 or T1D-protective DQ6 (Supporting Information Table I). Strikingly, a peptide from ZnT8 was identified in samples from all four T1D-susceptible DQ molecules. Peptides with overlapping sequence from HSP60, were detected in DQ8, DQ2-8, and DQ8-2 samples. These peptide sequences, as well as an additional HSP60 peptide eluted from DQ2-8 and three IA-2 peptide sequences eluted from DQ8, DQ2-8, and DQ8-2 samples have been previously reported as positive stimulators in autoreactive CD4+ T cell activation assays (Table I) [39-52]. We also confirmed that the ZnT8 peptide binds T1D-susceptible DQ molecules with relatively higher binding affinity than the pan-DQ binding peptide, CLIP1 (Fig.3). These results confirm that functionally relevant autoantigenic peptides can be identified by HPLC-MS/MS analysis of peptide isolated from cells expressing autoantigen proteins, although the physiologic processing mechanisms for presentation of these epitopes in professional APC might be different, and they raise the possibility that key self-peptide epitopes can be presented by multiple different T1D-susceptible DQ molecules.
TABLE I.
Eluted T1D-associated autoantigen-derived peptides with previously reported positive function in autoreactive CD4+ T cell activation assays.
| AutoAg | Methods (ref.) | Start AA | Sequence |
|---|---|---|---|
| ZnT8 | DQ2, 8, 8-2 LC-MS | 126 a | K P P S K R L T F G W H R A E I L G A L |
| (Q8IWU4) | DQ8,2-8, 8-2 LC-MS | 126 | K P P S K R L T F G W H R A E I L G A L L S |
| ELISPOTc [41] | 120-146 | S L W L S S K P P S K R L T F G W H R A E I L G A L L | |
| ELISPOT [41] | 134-174b | F G W H R A E I L G A L L S I L C I W V V T G V L V Y L A C E R L L Y P D Y Q I Q | |
| Cytokine [41] | 124-138 | S S K P P S K R L T F G W H R | |
| ELISPOT [40] | 106-132 | H L L I D L T S F L L S L F S L W L S S K P P S K R L | |
|
| |||
| IA-2 | DQ8 LC-MS | 876 | P A E G T P A S T R P L L D F R R K V N K C Y R |
| (Q16849) | 3H thymidine [43] | 889-904 | D F R R K V N K C Y R G R S C P |
| DQ2-8 LC-MS | 622 | G D T T F E Y Q D L C R Q | |
| 3H thymidine [46] | 616-633 | G P E G A H G D T T F E Y Q D L C R | |
| DQ8-2 LC-MS | 956 | K D Q F E F A L T A V A E E V N A I L K A L P | |
| 3H thymidine [46] | 961-979 | F A L T A V A E E V N A I L K A L P Q | |
| 3H thymidine [49] | 957-969 | D Q F E F A L T A V A E E | |
| 3H thymidine [43] | 959-974 | F E F A L T A V A E E V N A I L | |
| 3H thymidine [47] | 955-975 | S K D Q F E F A L T A V A E E V N A I L K | |
| ELISPOT [48] | 955-976 | S K D Q F E F A L T A V A E E V N A I L K A | |
|
| |||
| HSP60 | DQ8, DQ2-8 LC-MS | 448 | I P A L D S L T P A N E D Q K |
| (P10809) | 3H thymidine [39, 42, 44, 50] |
437-460 | V L G G G V A L L R V I P A L D S L T P A N E D |
| ELISPOT [51] | 437-460 | V L G G G V A L L R V I P A L D S L T P A N E D | |
| Cytokine [45] | 437-460 | V L G G G V A L L R V I P A L D S L T P A N E D | |
| DQ2-8 LC-MS | 523 | K V V R T A L L D | |
| 3H thymidine [39] | 511-530 | V N M V E K G I I D P T K V V R T A L L | |
The sequence identified in different samples of this study by LC-MS is bold.
The bold and italic sequence shows the overlapped portion of peptide compared with the peptide identified in this study.
The listed method that was used to identify the function of each peptide in each of the references. The Immune Epitope Database (www.iedb.org) was applied for the search of T cell response to specific epitope as described [52].
FIGURE 3.
Binding analysis of the ZnT8-derived ZnT8(126) peptide to purified fDQ molecules. Peptide competition assays were performed with purified fDQ proteins and bCLIP, using unlabeled peptides CLIP1, CLIPc or ZnT8(126) peptides as competitor. CLIPc is a control peptide derived from the C-terminus of Ii that was detected by HPLC-MS/MS from all of the samples (DQ2, DQ2-8 and DQ8-2), except DQ8. Data shown are representative of 3 independent experiments.
DM catalytic potency in CLIP dissociation from soluble DQ6, DQ2 and DQ8
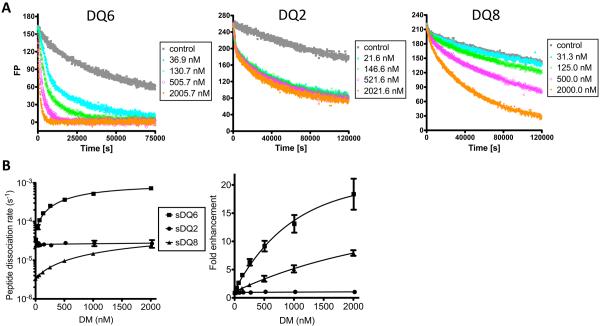
To further characterize the biochemical properties of these DQ molecules, soluble DQ (sDQ) proteins, sDQ6, sDQ2 and sDQ8 were generated. The sDQ-peptide complexes were pre-loaded with labeled CLIP peptide (488CLIP), and peptide dissociation rates were measured as a function of DM concentration (Fig.4). DM efficiently catalyzed the dissociation of CLIP from the control DQ6 protein. Peptide dissociation rates increased with increasing DM concentration. Note that rates appeared to saturate with increasing DM concentration. This might reflect a relatively high affinity in DM binding to DQ6-CLIP complexes. By contrast, DQ2-CLIP complexes were resistant to DM activity, showing little enhancement in dissociation with high DM concentrations. An intermediate level of DM potency was observed with DQ8-CLIP complexes. A clear catalytic effect is evident with DQ8, but the rate enhancement was less than that observed with DQ6 as judged by the initial slope of rate versus DM concentration. Furthermore, less rate saturation was observed, consistent with the idea that DM may have a lower affinity for DQ8-CLIP as compared with DQ6-CLIP. This is illustrated in normalized plots of fold-enhancement in rates versus DM concentration (Fig.4B, right). Thus, experiments with purified recombinant proteins demonstrate a striking resistance of DQ2 to DM-mediated CLIP editing and an intermediate DM-resistance phenotype for DQ8.
FIGURE 4.
CLIP dissociation rate and DM catalytic potency measured with purified DQ molecules. (A) The FP measurement of CLIP dissociation and DM editing effects. Thrombin cleaved sDQ proteins were pre-loaded with 488CLIP peptides. Peptide dissociation rates were measured in the presence of different concentration of sDM and 100 μM of unlabeled CLIP1 as competitor peptides. The control represents the sample without adding extra sDM or competitor peptides. (B) Peptide dissociation rates of sDQ6, sDQ2 and sDQ8 measured by FP assay in the presence of different concentration of DM (left) and the DM-catalyzed rate enhancements normalized relative to dissociation rates measured in the absence of DM (right). Data are representative of 3 independent experiments and shown as mean ± SD in panel B.
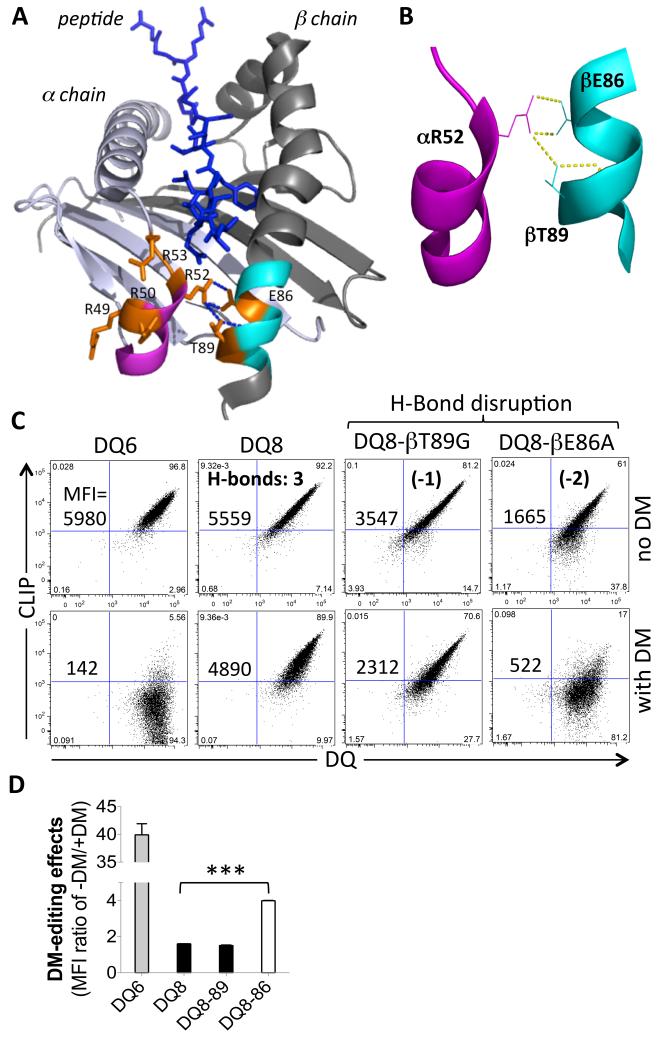
Contribution of unique hydrogen bonds in DQ8 to its DM-editing resistance
It has previously been demonstrated that a natural deletion in the DQ2 α chain contributes to the resistance of this protein to DM catalytic activity. Introduction of Gly at position 53 in DQ2 restored DM sensitivity [22]. The natural deletion in DQ2α is positioned in the DM contact interface [21] and thus it may reduce the affinity of the DM interaction with DQ2 or alter the structure of the interface such that catalytic potency is reduced. This would also provide a structural explanation for the DM resistant phenotype of the DQ2-8 trans-dimer, which shares the α chain with DQ2. However, the DQ8 α chain does not have this natural deletion and the structural basis of DQ8 and DQ8-2 trans-dimer for a relative resistance to DM editing remained to be explored.
Comparison of the crystal structures of different DQ molecules, as well as DR1 bound to different peptides and the DM-DR1 co-crystal structure, demonstrates conformational differences in the helix-β-sheet region near the P1 pocket of the peptide binding groove, the DM interaction site [23]. Compared with the other DQ molecules, there are multiple positively charged arginine residues in the 310-helix region of DQ8 α chain (Fig. 5A). Notably, three extra hydrogen bonds (H-bonds) are present between the two helixes near the P1 pocket of DQ8 peptide binding groove (Fig. 5A, 5B), as well as one H-bond in DQ8-2, but none in DQ1, DQ6, DQ2 or DQ2-8 [23]. To test whether these H-bonds contribute to the relative resistance of DQ8 to DM editing, we replaced the residues in the DQ8 β chain that participate in these H-bonds with the amino acids present in DQ6 with the goal of disrupting the extra H-bond(s) formed with the 310-helix of DQ8α. The mutant DQ8 proteins were expressed with Ii in the absence or presence of DM in 293T cells (Fig.5C). The βT89G mutation, intended to disrupt one H-bond between αR52 and βT89, resulted in little change in DM sensitivity, such that CLIP levels remained high in the presence of DM. However, disruption of two H-bonds through the βE86A substitution resulted in restoration of a substantial increase in sensitivity to DM editing, as determined by comparing CLIP levels with and without co-expression of DM (Fig.5C, D). These findings support the idea that the additional H-bonds between the α and β chain helices in DQ8 may contribute to a phenotype of relative resistance to DM editing, possibly by stabilizing the region of MHCII that undergoes a marked conformation transition when bound to DM.
FIGURE 5.
Impact of hydrogen bonds between the two helical regions of α and β chains near the P1 pocket of DQ8 molecule. (A) Top view of the two helical regions of DQ8 α and β chains near the P1 pocket of the peptide binding groove with bound peptide (PDB: 1JK8). The R52 residue in the 310-helix (purple) of the α chain, which is in rich of arginine in DQ8, forms three H-bonds with E86 and T89 in the helix (cyan) near the P1 pocket of the β chain. DQ8 specific residues are labeled orange. (B) A close-up view of the three H-bonds formed between the two helixes in DQ8. (C) Comparison of CLIP level on 293T cells transduced with Ii and DQ6, DQ8, or DQ8 mutants, in the absence or presence of DM expression. The DQ8 mutants were designed to disrupt one (βT89G) or two (βE86A) H-bonds, respectively. The MFI of cell surface CLIP is shown. (D) The relative DM editing effects indicated by comparison of the ratio of CLIP/DQ in the absence or presence of DM expression. Cells were surface stained for CLIP and DQ and gated on the same level of GFP and/or mCherry expression. Data are representative of 3 independent experiments and shown as mean + SD in panel D. ***, P<0.001; two tail t-test.
Discussion
Collectively, our findings indicate that the four T1D-susceptible DQ molecules, DQ2, DQ8, and the trans-dimers expressed in DQ2/8 heterozygotes, share a distinct property of being relatively resistant to DM-mediated editing of CLIP, compared with the T1D-protective DQ6 or T1D-neutral DQ1 molecules that have the classical DM-sensitive phenotype. The most direct evidence for this conclusion was obtained by analyzing surface CLIP levels on cells with or without co-expression of DM. We observed little or no reduction in CLIP levels on 293T cells co-expressing DM with DQ8 or DQ2-8, indicating a significant DM editing resistance of CLIP peptides. The phenotype was less striking for the DQ2 and DQ8-2 trans-dimer (Fig.1). Similar results were obtained with T2 lymphoblastoid cells, however DQ2 and DQ2-8 were less sensitive to DM than DQ8 in these cells.
The α chain of DQ2 (DQA1*0501) contains a deletion of residue 53. Insertion of Gly at this position was shown to restore sensitivity to DM as judged by a marked increase in DM-mediated rate enhancement in the dissociation of a DQ2 binding peptide [22]. Given the location of α53 in the DM contact interface [21], it is tempting to consider the possibility that the natural α53 deletion in DQ2 (and the DQ2-8 trans-dimer) confers a general resistance to DM-mediated peptide exchange, reducing the impact of DM editing on the peptide repertoire presented by these molecules. However, additional structural elements may also contribute to the DM editing sensitivity. The DQA1*0501/DQB1*0301 DQ7 allele shares DQA1*0501 with DQ2, yet it is not associated with T1D. Other DQA alleles represented in human populations have a natural deletion at α53, including DQA1*0201 and DQA1*04. Further work will be needed to determine the DM sensitivity of the proteins generated from these alleles and the structural features that affect DM catalytic efficiency. There is an inverse relationship between the intrinsic stability of peptide complexes and sensitivity to DM-catalyzed peptide dissociation, such that less stable complexes are selectively edited, and more stable complexes survive for recognition by CD4+ T cells [10, 53, 54]. Reduction in DM editing might broaden the array of self-peptide complexes available to activate autoreactive T cells, and unstable complexes may be more susceptible to alternative peptide loading pathways for exogenous autoantigenic peptides [55].
In experiments with sDQ molecules, high concentrations of DM were observed to have little effect on the rate of dissociation of CLIP from sDQ2, whereas DM was clearly able to enhance CLIP dissociation from DQ8 in a dose-dependent manner. DM catalytic potency was intermediate between DQ2 and the DM-sensitive DQ6 control. The stability of sDQ8-CLIP complexes was observed to be considerably greater than that of sDQ6-CLIP, with intrinsic half-lives of 58 h and 7 h, respectively. The increased stability of sDQ8-CLIP might explain the relative resistance to DM. While DQ8 does not have the α53 deletion found in DQ2, it does have three H-bonds, not present in DQ2, DQ2-8, DQ1 or DQ6 molecules. The H-bonds connect the helices of the α and β chains near the DM contact site [23]. Disruption of two H-bonds through a βE86A substitution was observed to partially restore sensitivity to DM in cells expressing the substituted DQ protein. It is possible that the mutation reduces the intrinsic stability of DQ8 molecule as well as the complex of DQ8-CLIP, as a potential mechanism for enhancing DM sensitivity. Alternatively, the additional H-bonds in DQ8 might serve to stabilize the DM-binding region of DQ8, making it more difficult for the molecule to undergo the conformational rearrangement required for interaction with DM [21]. It is interesting to note that the DQ8-2 trans-dimer has one additional H-bond [23] that could potentially stabilize the conformation of the peptide-binding groove and contribute to the partial DM-resistant phenotype observed in cells expressing this mixed haplotype DQ molecule.
HPLC-MS/MS analysis confirmed that Ii-derived peptides lacking the core CLIP1 sequence and containing the alternative CLIP2 binding register were highly represented in samples from cells expressing DQ2+Ii. The CLIP2 loading mechanism is unknown, reflecting either an alternative mode of assembly of Ii with DQ2 or an exchange mechanism in which CLIP1 is replaced by CLIP2 [24]. CLIP2 peptides were also prominently represented in samples from cells expressing DQ1, DQ6, DQ8 or the DQ8/2 trans-dimers in the absence of DM expression. While the biological significance of CLIP2 is unknown, it is clear that CLIP2 is not uniquely associated with DQ2 or T1D-associated DQ molecules. The detection of CLIP peptides in DQ1 peptide samples by HPLC-MS/MS, but not by CerCLIP.1 mAb surface staining, reflects the specificity of the CerCLIP.1 mAb, for only long CLIP1 peptides with N-terminal sequence extensions. The peptide elution data confirm that DQ1-CLIP is sensitive to DM editing. However, the mechanism responsible for selective loading of DQ1 with short CLIP peptides remains to be investigated, possibly reflecting an increased sensitivity to proteolytic trimming.
A number of peptides from well known T1D-associated autoantigens were identified in the HPLC-MS/MS experiments with samples from T1D-susceptible DQ molecules. The 293T human embryonic kidney cell line fortuitously expresses some of the proteins relevant in T1D pathogenesis. Further work will be needed to determine whether specific peptides are dependent on DM for presentation, or sensitive to DM editing such that DM-independent presentation mechanisms would be required for T cell recognition. It was interesting to observe that several peptides were identified with multiple different T1D-susceptible DQ molecules, but not with DQ1 or DQ6. Strikingly, a ZnT8 peptide was eluted from cells expressing each of the T1D-susceptible DQ alleles. This raises the question of whether presentation of selected peptides by the multiple different DQ molecules expressed on the cells in DQ8/2 heterozygotes might play a role in inducing cross-reacting autoreactive T cell responses in these individuals [56]. It was also interesting that most of the peptide sequences identified from T1D autoantigens had previously been defined as epitopes for autoreactive CD4+ T cell responses in T1D. This provides some level of validation that mass spectrometric analysis of MHCII-associated peptides may be useful in identifying candidate autoantigenic epitopes.
Overall, we conclude that the four T1D-susceptible DQ alleles have in common a shared resistance to DM-mediated editing of CLIP. Different structural elements appear to be responsible for this phenotype in DQ2, DQ8, and the DQ2/8 trans-dimers. Cell surface CLIP levels have been reported to be elevated in the peripheral blood of people with T1D [57]. It is possible that high levels of CLIP in the thymus might result in less efficient negative selection of a population of self-reactive T cells with the potential to participate in initiating or propagating immune responses to islet antigens. Alternatively, reduced susceptibility to DM editing function could potentially facilitate peripheral presentation of self-peptides loaded under inflammatory conditions through DM-independent mechanisms. Unanue and colleagues have demonstrated that DM-independent loading of free peptides can result in the formation of peptide-MHCII complexes with alterative binding registers [58]. These alternatively-generated peptide complexes can activate a population of unconventional “type B” T cells that are not eliminated through conventional mechanisms that maintain self tolerance [55]. While there is little doubt that the impact of MHCII polymorphism on CD4+ T cell repertoire selection and epitope specificity must play a major role in determining genetic susceptibility to specific autoimmune diseases, allelic differences that impact the peptide loading pathway may also be important.
Materials and methods
Expression of DQ, Ii and DM molecules and purification of full length and soluble DQ in 293T cells
Constructs encoding the full length α and β chains of DQ1, DQ2, DQ6 or DQ8 were fused with the T2A sequence [59] and cloned into pWPI vector (Addgene). Human Ii cDNA was cloned into another pWPI vector. The DM construct was cloned from MigR1-DM [60] and inserted behind the IRES site within lentiviral HIV\CS\Ub-ChIT (ChIT) vector provided by Dr. Vicente Planelles (University of Utah), which contains an mCherry gene driven by a human ubiquitin promoter (Supporting Information Fig.2A, 2B). High-titer lentiviral supernatants were generated by transfection of 293T cells (ATCC), a human embryonic kidney cell line having no endogenous expression of Ii, DM or DQ, with pWPI-DQ, pWPI-Ii or ChIT-DM construct, as previously described [61]. The 293T transductants stably expressing DQ (DQ only) or DQ and Ii (DQ+Ii) were sorted for GFP and DQ expression with a FACSAria™ cell sorter (BD Biosciences). DQ+Ii cells were further transduced with ChIT-DM lentivirus and sorted for GFP, DQ and mCherry expression to obtain DQ+Ii+DM cells (Supporting Information Fig.2C). Full length DQ (fDQ) was purified from DQ+Ii 293T cells by SPVL3 affinity column [53]. Full length DM (fDM) was purified as previously described [53]. Soluble DQ proteins were generated from pWPI-sDQ lentiviral transduced 293T cells using the construct as previously described [62]. The culture media were pre-cleared and incubated with His-Tag Purification Resin (Roche), and the eluates were further purified by SPVL3 affinity column. Purified sDQ were overnight cleaved with immobilized thrombin (EMD Millipore) in PBS at room temperature to facilitate the release of endogenous CLIP peptide. Soluble DM was purified as previously described [60].
Abs and flow cytometry
The following mAbs, DM-PE (clone Map.DM1; BD Pharmingen), CLIP- PerCP-Cy5.5 (clone Cer.CLIP1; Santa Cruz Biotechnology) and isotype-matched negative controls, were used in cell staining. To staining DQ, purified mAb to DQ (clone SPVL3, ATCC) was labeled with Biotinamidohexanoic acid N-hydroxysuccinimide ester (Sigma) then accordingly stained by streptavidin-APC or -PE-Cy7 (Invitrogen). Purified mAb to Ii (clone PIN-1), provided by Dr. Peter Cresswell (Yale), was conjugated with Alexa Fluor® 647 (Invitrogen). Intracellular staining was performed with the Cytofix/Cytoperm kit (BD Biosciences). Stained cells were analyzed by LSRFortessa™ flow cytometer (BD Biosciences). The data were analyzed using FlowJo software (Tree Star). Based on GFP, mCherry and surface DQ expression, cells were sorted with FACSAria™ cell sorter (BD Biosciences) at the core facility of University of Utah. The sDQ 293T transductants were sorted for high GFP expression, and sDQ secretion in culture media was measured by immunoassay [63], using SPVL3 and biotin-IVA12 mAbs.
Peptide elution and HPLC-MS/MS sequence analysis
To elute peptides bound to each DQ molecules, ~1×109 of each DQ+Ii or DQ+Ii+DM transductant 293T cells were lysed and the DQ molecules were captured by anti-DQ mAb (SPVL3) immobilized with protein A beads. Bound peptides were eluted and further separated by HPLC for HPLC-MS/MS analysis as previously described [64].
Peptide competition and fluorescence polarization assay
N-acetylated CLIP (80-103, KLPKPPKPVSKMRMATPLLMQALC), in which the last residue was substituted by cysteine for labeling, was commercially synthesized (CelTek Pepitdes) and labeled with BMCC-biotin (Pierce), designated as bCLIP. The N-acetylated peptide CLIP (VSKMRMATPLLMQ) was labeled on lysine (underlined) with Alexa Fluor 488 (Molecular Probes) as previously described, designated as 488CLIP [60]. The peptides, CLIP1 (LPKPPKPVSKMRMATPLLMQA), CLIPc (PSSGLGVTKQDLGPVPM) and ZnT8(126) (KPPSKRLTFGWHRAEILGAL), were used as competitor peptides in competition assays. Briefly, 100 nM fDQ, 1 μM bCLIP, 200 nM fDM and the competitor peptide (0, 2, 20 or 200 μM) were incubated at 37°C for 3 h in 100 mM citrate-phosphate buffer (pH 5.0) with 0.2% NP-40. Sample was loaded into 96-well plate coated with SPVL3 mAb, and detected by immunoassay [63]. The relative loading of bCLIP to specific DQ molecules was normalized to the reaction without competitor peptides. Thrombin-cleaved sDQ was pre-loaded with extra 488CLIP (1:5 molar ratio) in FP buffer (10 mM citrate-phosphate buffer, pH 5.0, 150 mM NaCl, 0.05% Tween 20), at 37°C for 3 h (for sDQ6) or overnight (for sDQ2) with 100 nM sDM, or at 37°C overnight without sDM (for sDQ8), due to the different DM sensitivity. Unbound 488CLIP peptide was removed by buffer exchange with 30-kD cut-off spin column (Millipore). In FP assay, 50 nM sDQ-488CLIP complex, sDM (0 to 2000 nM) and 100 μM competitor peptide were incubated in FP buffer and measured at 37°C with a Tecan Infinite F200 plate reader. FP value was normalized by the background FP signal of free 488CLIP peptides, and the data were processed with Prism 5.0 (GraphPad Software), as previously described [60].
Supplementary Material
Acknowledgements
This research was supported by NIH Grants AI30554 and AI33614 awarded to PEJ. We thank Dr. Matthew A. Williams for critical reading of the manuscript, Dr. Adam P. Barker for technical assistance in protein purification and peptide labeling, and the members of our laboratories for technical comments.
Abbreviations
- bCLIP
biotin-labeled CLIP peptide
- fDQ
purified full-length HLA-DQ
- Ii
invariant chain
- MHCII
MHC class II
- sDQ
soluble recombinant HLA-DQ
- T1D
type 1 diabetes
- FP
Fluorescence polarization
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Pociot F, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G, Nierras CR, Todd JA, Rich SS, Nerup J. Genetics of type 1 diabetes: what's next? Diabetes. 2010;59:1561–1571. doi: 10.2337/db10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorodezky C, Alaez C, Murguia A, Rodriguez A, Balladares S, Vazquez M, Flores H, Robles C. HLA and autoimmune diseases: Type 1 diabetes (T1D) as an example. Autoimmun Rev. 2006;5:187–194. doi: 10.1016/j.autrev.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Cifuentes RA, Rojas-Villarraga A, Anaya JM. Human leukocyte antigen class II and type 1 diabetes in Latin America: a combined meta-analysis of association and family-based studies. Hum Immunol. 2011;72:581–586. doi: 10.1016/j.humimm.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Rojas-Villarraga A, Botello-Corzo D, Anaya JM. HLA-Class II in Latin American patients with type 1 diabetes. Autoimmun Rev. 2010;9:666–673. doi: 10.1016/j.autrev.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 5.van Autreve JE, Weets I, Gulbis B, Vertongen F, Gorus FK, van der Auwera BJ, Belgian Diabetes R. The rare HLA-DQA1*03-DQB1*02 haplotype confers susceptibility to type 1 diabetes in whites and is preferentially associated with early clinical disease onset in male subjects. Hum Immunol. 2004;65:729–736. doi: 10.1016/j.humimm.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Aly TA, Ide A, Jahromi MM, Barker JM, Fernando MS, Babu SR, Yu L, Miao D, Erlich HA, Fain PR, Barriga KJ, Norris JM, Rewers MJ, Eisenbarth GS. Extreme genetic risk for type 1A diabetes. Proc Natl Acad Sci U S A. 2006;103:14074–14079. doi: 10.1073/pnas.0606349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, Mychaleckyj JC, Todd JA, Bonella P, Fear AL, Lavant E, Louey A, Moonsamy P, Type 1 Diabetes Genetics, C. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pociot F, McDermott MF. Genetics of type 1 diabetes mellitus. Genes Immun. 2002;3:235–249. doi: 10.1038/sj.gene.6363875. [DOI] [PubMed] [Google Scholar]
- 9.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 10.Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- 11.Sant AJ, Chaves FA, Leddon SA, Tung J. The control of the specificity of CD4 T cell responses: thresholds, breakpoints, and ceilings. Front Immunol. 2013;4:340. doi: 10.3389/fimmu.2013.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairchild PJ, Wildgoose R, Atherton E, Webb S, Wraith DC. An autoantigenic T cell epitope forms unstable complexes with class II MHC: a novel route for escape from tolerance induction. Int Immunol. 1993;5:1151–1158. doi: 10.1093/intimm/5.9.1151. [DOI] [PubMed] [Google Scholar]
- 13.Geluk A, van Meijgaarden KE, Schloot NC, Drijfhout JW, Ottenhoff TH, Roep BO. HLA-DR binding analysis of peptides from islet antigens in IDDM. Diabetes. 1998;47:1594–1601. doi: 10.2337/diabetes.47.10.1594. [DOI] [PubMed] [Google Scholar]
- 14.Gross DA, Graff-Dubois S, Opolon P, Cornet S, Alves P, Bennaceur-Griscelli A, Faure O, Guillaume P, Firat H, Chouaib S, Lemonnier FA, Davoust J, Miconnet I, Vonderheide RH, Kosmatopoulos K. High vaccination efficiency of low-affinity epitopes in antitumor immunotherapy. J Clin Invest. 2004;113:425–433. doi: 10.1172/JCI19418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu GY, Fairchild PJ, Smith RM, Prowle JR, Kioussis D, Wraith DC. Low avidity recognition of self-antigen by T cells permits escape from central tolerance. Immunity. 1995;3:407–415. doi: 10.1016/1074-7613(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 16.McNeil LK, Evavold BD. Dissociation of peripheral T cell responses from thymocyte negative selection by weak agonists supports a spare receptor model of T cell activation. Proc Natl Acad Sci U S A. 2002;99:4520–4525. doi: 10.1073/pnas.072673899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc Natl Acad Sci U S A. 2010;107:10978–10983. doi: 10.1073/pnas.1006545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford F, Stadinski B, Jin N, Michels A, Nakayama M, Pratt P, Marrack P, Eisenbarth G, Kappler JW. Specificity and detection of insulin-reactive CD4+ T cells in type 1 diabetes in the nonobese diabetic (NOD) mouse. Proc Natl Acad Sci U S A. 2011;108:16729–16734. doi: 10.1073/pnas.1113954108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levisetti MG, Lewis DM, Suri A, Unanue ER. Weak proinsulin peptide-major histocompatibility complexes are targeted in autoimmune diabetes in mice. Diabetes. 2008;57:1852–1860. doi: 10.2337/db08-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Chow IT, Sosinowski T, Torres-Chinn N, Greenbaum CJ, James EA, Kappler JW, Davidson HW, Kwok WW. Autoreactive T cells specific for insulin B:11-23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc Natl Acad Sci U S A. 2014;111:14840–14845. doi: 10.1073/pnas.1416864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pos W, Sethi DK, Call MJ, Schulze MS, Anders AK, Pyrdol J, Wucherpfennig KW. Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell. 2012;151:1557–1568. doi: 10.1016/j.cell.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou T, Macmillan H, Chen Z, Keech CL, Jin X, Sidney J, Strohman M, Yoon T, Mellins ED. An insertion mutant in DQA1*0501 restores susceptibility to HLA-DM: implications for disease associations. J Immunol. 2011;187:2442–2452. doi: 10.4049/jimmunol.1100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z, Jensen PE. Structural Characteristics of HLA-DQ that May Impact DM Editing and Susceptibility to Type-1 Diabetes. Front Immunol. 2013;4:262. doi: 10.3389/fimmu.2013.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fallang LE, Roh S, Holm A, Bergseng E, Yoon T, Fleckenstein B, Bandyopadhyay A, Mellins ED, Sollid LM. Complexes of two cohorts of CLIP peptides and HLA-DQ2 of the autoimmune DR3-DQ2 haplotype are poor substrates for HLA-DM. J Immunol. 2008;181:5451–5461. doi: 10.4049/jimmunol.181.8.5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundin KE, Gjertsen HA, Scott H, Sollid LM, Thorsby E. Function of DQ2 and DQ8 as HLA susceptibility molecules in celiac disease. Hum Immunol. 1994;41:24–27. doi: 10.1016/0198-8859(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 26.Busch R, De Riva A, Hadjinicolaou AV, Jiang W, Hou T, Mellins ED. On the perils of poor editing: regulation of peptide loading by HLA-DQ and H2-A molecules associated with celiac disease and type 1 diabetes. Expert Rev Mol Med. 2012;14:e15. doi: 10.1017/erm.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan MA, Muller PS, Mould A, Newland SA, Nichols J, Robertson EJ, Cooke A, Bikoff EK. The nonconventional MHC class II molecule DM governs diabetes susceptibility in NOD mice. PLoS One. 2013;8:e56738. doi: 10.1371/journal.pone.0056738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Lummel M, van Veelen PA, Zaldumbide A, de Ru A, Janssen GM, Moustakas AK, Papadopoulos GK, Drijfhout JW, Roep BO, Koning F. Type 1 diabetes-associated HLA-DQ8 transdimer accommodates a unique peptide repertoire. J Biol Chem. 2012;287:9514–9524. doi: 10.1074/jbc.M111.313940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tollefsen S, Hotta K, Chen X, Simonsen B, Swaminathan K, Mathews II, Sollid LM, Kim CY. Structural and functional studies of trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) protein molecule. J Biol Chem. 2012;287:13611–13619. doi: 10.1074/jbc.M111.320374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Testa JS, Apcher GS, Comber JD, Eisenlohr LC. Exosome-driven antigen transfer for MHC class II presentation facilitated by the receptor binding activity of influenza hemagglutinin. J Immunol. 2010;185:6608–6616. doi: 10.4049/jimmunol.1001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovats S, Whiteley PE, Concannon P, Rudensky AY, Blum JS. Presentation of abundant endogenous class II DR-restricted antigens by DM-negative B cell lines. Eur J Immunol. 1997;27:1014–1021. doi: 10.1002/eji.1830270431. [DOI] [PubMed] [Google Scholar]
- 32.Hosken NA, Bevan MJ. Defective presentation of endogenous antigen by a cell line expressing class I molecules. Science. 1990;248:367–370. doi: 10.1126/science.2326647. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh P, Amaya M, Mellins E, Wiley DC. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature. 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 34.Denzin LK, Robbins NF, Carboy-Newcomb C, Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 35.Wiesner M, Stepniak D, de Ru AH, Moustakis AK, Drijfhout JW, Papadopoulos GK, van Veelen PA, Koning F. Dominance of an alternative CLIP sequence in the celiac disease associated HLA-DQ2 molecule. Immunogenetics. 2008;60:551–555. doi: 10.1007/s00251-008-0310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Wal Y, Kooy YM, Drijfhout JW, Amons R, Koning F. Peptide binding characteristics of the coeliac disease-associated DQ(alpha1*0501, beta1*0201) molecule. Immunogenetics. 1996;44:246–253. doi: 10.1007/BF02602553. [DOI] [PubMed] [Google Scholar]
- 37.Vartdal F, Johansen BH, Friede T, Thorpe CJ, Stevanovic S, Eriksen JE, Sletten K, Thorsby E, Rammensee HG, Sollid LM. The peptide binding motif of the disease associated HLA-DQ (alpha 1* 0501, beta 1* 0201) molecule. Eur J Immunol. 1996;26:2764–2772. doi: 10.1002/eji.1830261132. [DOI] [PubMed] [Google Scholar]
- 38.Roep BO, Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb Perspect Med. 2012;2:a007781. doi: 10.1101/cshperspect.a007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abulafia-Lapid R, Elias D, Raz I, Keren-Zur Y, Atlan H, Cohen IR. T cell proliferative responses of type 1 diabetes patients and healthy individuals to human hsp60 and its peptides. J Autoimmun. 1999;12:121–129. doi: 10.1006/jaut.1998.0262. [DOI] [PubMed] [Google Scholar]
- 40.Chujo D, Foucat E, Nguyen TS, Chaussabel D, Banchereau J, Ueno H. ZnT8-Specific CD4+ T cells display distinct cytokine expression profiles between type 1 diabetes patients and healthy adults. PLoS One. 2013;8:e55595. doi: 10.1371/journal.pone.0055595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dang M, Rockell J, Wagner R, Wenzlau JM, Yu L, Hutton JC, Gottlieb PA, Davidson HW. Human type 1 diabetes is associated with T cell autoimmunity to zinc transporter 8. J Immunol. 2011;186:6056–6063. doi: 10.4049/jimmunol.1003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dosch H, Cheung RK, Karges W, Pietropaolo M, Becker DJ. Persistent T cell anergy in human type 1 diabetes. J Immunol. 1999;163:6933–6940. [PubMed] [Google Scholar]
- 43.Honeyman MC, Stone NL, Harrison LC. T-cell epitopes in type 1 diabetes autoantigen tyrosine phosphatase IA-2: potential for mimicry with rotavirus and other environmental agents. Mol Med. 1998;4:231–239. [PMC free article] [PubMed] [Google Scholar]
- 44.Huurman VA, van der Meide PE, Duinkerken G, Willemen S, Cohen IR, Elias D, Roep BO. Immunological efficacy of heat shock protein 60 peptide DiaPep277 therapy in clinical type I diabetes. Clin Exp Immunol. 2008;152:488–497. doi: 10.1111/j.1365-2249.2008.03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jonson CO, Lernmark A, Ludvigsson J, Rutledge EA, Hinkkanen A, Faresjo M. The importance of CTLA-4 polymorphism and human leukocyte antigen genotype for the induction of diabetes-associated cytokine response in healthy school children. Pediatr Diabetes. 2007;8:185–192. doi: 10.1111/j.1399-5448.2007.00245.x. [DOI] [PubMed] [Google Scholar]
- 46.Kudva YC, Deng YJ, Govindarajan R, Abraham RS, Marietta EV, Notkins AL, David CS. HLA-DQ8 transgenic and NOD mice recognize different epitopes within the cytoplasmic region of the tyrosine phosphatase-like molecule, IA-2. Hum Immunol. 2001;62:1099–1105. doi: 10.1016/s0198-8859(01)00308-1. [DOI] [PubMed] [Google Scholar]
- 47.Peakman M, Stevens EJ, Lohmann T, Narendran P, Dromey J, Alexander A, Tomlinson AJ, Trucco M, Gorga JC, Chicz RM. Naturally processed and presented epitopes of the islet cell autoantigen IA-2 eluted from HLA-DR4. J Clin Invest. 1999;104:1449–1457. doi: 10.1172/JCI7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrich de Marquesini LG, Fu J, Connor KJ, Bishop AJ, McLintock NE, Pope C, Wong FS, Dayan CM. IFN-gamma and IL-10 islet-antigen-specific T cell responses in autoantibody-negative first-degree relatives of patients with type 1 diabetes. Diabetologia. 2010;53:1451–1460. doi: 10.1007/s00125-010-1739-3. [DOI] [PubMed] [Google Scholar]
- 49.Schulz RM, Hawa M, Leslie RD, Sinigaglia F, Passini N, Rogge L, Picard JK, Londei M. Proliferative responses to selected peptides of IA-2 in identical twins discordant for Type 1 diabetes. Diabetes Metab Res Rev. 2000;16:150–156. doi: 10.1002/1520-7560(0000)9999:9999<::aid-dmrr101>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 50.Shpigel E, Elias D, Cohen IR, Shoseyov O. Production and purification of a recombinant human hsp60 epitope using the cellulose-binding domain in Escherichia coli. Protein Expr Purif. 1998;14:185–191. doi: 10.1006/prep.1998.0929. [DOI] [PubMed] [Google Scholar]
- 51.Szebeni A, Schloot N, Kecskemeti V, Hosszufalusi N, Panczel P, Prohaszka Z, Fust G, Uray K, Hudecz F, Meierhoff G. Th1 and Th2 cell responses of type 1 diabetes patients and healthy controls to human heat-shock protein 60 peptides AA437-460 and AA394-408. Inflamm Res. 2005;54:415–419. doi: 10.1007/s00011-005-1362-9. [DOI] [PubMed] [Google Scholar]
- 52.Vaughan K, Peters B, Mallone R, von Herrath M, Roep BO, Sette A. Navigating diabetes-related immune epitope data: resources and tools provided by the Immune Epitope Database (IEDB) Immunome Res. 2013;9 doi: 10.4172/1745-7580.1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 54.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 55.Mohan JF, Unanue ER. Unconventional recognition of peptides by T cells and the implications for autoimmunity. Nat Rev Immunol. 2012;12:721–728. doi: 10.1038/nri3294. [DOI] [PubMed] [Google Scholar]
- 56.Kooy-Winkelaar Y, van Lummel M, Moustakas AK, Schweizer J, Mearin ML, Mulder CJ, Roep BO, Drijfhout JW, Papadopoulos GK, van Bergen J, Koning F. Gluten-specific T cells cross-react between HLA-DQ8 and the HLA-DQ2alpha/DQ8beta transdimer. J Immunol. 2011;187:5123–5129. doi: 10.4049/jimmunol.1101179. [DOI] [PubMed] [Google Scholar]
- 57.Silva DG, Socha L, Correcha M, Petrovsky N. Elevated lymphocyte expression of CLIP is associated with type 1 diabetes and may be a useful marker of autoimmune susceptibility. Ann N Y Acad Sci. 2004;1037:65–68. doi: 10.1196/annals.1337.009. [DOI] [PubMed] [Google Scholar]
- 58.Pu Z, Lovitch SB, Bikoff EK, Unanue ER. T cells distinguish MHC-peptide complexes formed in separate vesicles and edited by H2-DM. Immunity. 2004;20:467–476. doi: 10.1016/s1074-7613(04)00073-1. [DOI] [PubMed] [Google Scholar]
- 59.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 60.Zhou Z, Callaway KA, Weber DA, Jensen PE. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J Immunol. 2009;183:4187–4191. doi: 10.4049/jimmunol.0901663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gauthier L, Smith KJ, Pyrdol J, Kalandadze A, Strominger JL, Wiley DC, Wucherpfennig KW. Expression and crystallization of the complex of HLA-DR2 (DRA, DRB1*1501) and an immunodominant peptide of human myelin basic protein. Proc Natl Acad Sci U S A. 1998;95:11828–11833. doi: 10.1073/pnas.95.20.11828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tompkins SM, Rota PA, Moore JC, Jensen PE. A europium fluoroimmunoassay for measuring binding of antigen to class II MHC glycoproteins. J Immunol Methods. 1993;163:209–216. doi: 10.1016/0022-1759(93)90124-p. [DOI] [PubMed] [Google Scholar]
- 64.Escobar H, Reyes-Vargas E, Jensen PE, Delgado JC, Crockett DK. Utility of characteristic QTOF MS/MS fragmentation for MHC class I peptides. J Proteome Res. 2011;10:2494–2507. doi: 10.1021/pr101272k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.