Abstract
IL-1 antagonism has been hypothesized to preserve β-cell function in new onset Type 1 diabetes (T1D). However, the Anti-Interleukin-1 in Diabetes Action (AIDA) and TrialNet Canakinumab (TN-14) trials failed to show efficacy of IL-1 receptor antagonist (IL-1Ra) or canakinumab, as measured by stimulated C-peptide response. Additional measures are needed to define immune state changes associated with therapeutic responses. Here, we studied these trial participants with transcriptional analysis of plasma-induced PBMCs. In blinded analyses, 70.2% of AIDA and 68.9% of TN-14 participants were correctly called to their treatment arm. While the PBMC transcriptional signatures from the two groups were distinct, both therapies achieved varying immunomodulation consistent with IL-1 inhibition. On average, IL-1 antagonism resulted in modest normalization relative to healthy controls. At endpoint, signatures were quantified using a gene ontology-based inflammatory index, and an inverse relationship was observed between measured inflammation and stimulated C-peptide response in IL-1Ra- and canakinumab-treated patients. Cytokine neutralization studies showed that IL-1α and IL-1β additively contribute to the T1D inflammatory state. Finally, analyses of baseline signatures were indicative of later therapeutic response. Despite the absence of clinical efficacy by IL-1 antagonist therapy, transcriptional analysis detected immunomodulation and may yield new insight when applied to other clinical trials.
Keywords: type 1 diabetes, interleukin-1, anakinra, canakinumab, Transcriptional signatures, regulatory T cells, microarray
Introduction
Type 1 diabetes (T1D) is a T-cell dependent autoimmune disease that targets pancreatic β-cells, resulting in lifelong dependency on exogenous insulin [1, 2]. To date, clinical trials of immunomodulatory therapies in new onset T1D have been disappointing in that none have resulted in sustained disruption of the underlying disease process. However, there have been modest successes in slowing the decline in β-cell function in trials that impair adaptive immunity through targeting of T-cells and B-cells [3–5].
Growing evidence supports a role for innate immunity in T1D pathogenesis, and efforts have focused on the pro-inflammatory cytokine IL-1 as a therapeutic target [1]. IL-1 acts directly on β-cells, impairing insulin biosynthesis and release [6–8], and inducing cytokine- and hyperglycemia-induced β-cell death [8–10], effects that are prevented by IL-1 antagonism [11, 12]. IL-1 amplifies adaptive immunity by enhancing the expansion and survival of naive and memory T-cells and promoting T-helper-1 and Th17 effector T cell differentiation and proliferation [13]. In rodent models, IL-1 blockade slows progression [14–16] and impairs triggering [17] of T1D. In Type 2 diabetes, recombinant human interleukin-1 receptor antagonist (IL-1Ra, anakinra) and anti-IL-1β monoclonal antibody (gevokizumab) therapies reduced systemic inflammation, but only IL-1Ra resulted in improved β-cell function and glycemic control [18, 19].
IL-1 has been therapeutically targeted in newly diagnosed T1D patients [20]. In the Anti-Interleukin-1 in Diabetes Action (AIDA) trial, adult patients received IL-1Ra (anakinra). In the TrialNet Canakinumab (TN-14) trial, primarily pediatric T1D patients received human monoclonal anti-IL-1β antibody (canakinumab). While safe and well-tolerated, neither IL-1Ra nor canakinumab were overall effective in maintaining β-cell function as measured by stimulated C-peptide response [20].
New approaches are needed to better understand the immunomodulation achieved in T1D clinical trials. Therefore, we examined the immune state of subjects receiving IL-1Ra, canakinumab, and placebo using an array-based bioassay, whereby subject plasma is used to induce transcriptional responses in a well-controlled peripheral blood mononuclear cell (PBMC) reporter population drawn from a healthy blood donor. With this sensitive and comprehensive approach, we previously determined that pre- and recent onset T1D (ROT1D) plasma induces a disease-specific, partially IL-1 dependent signature relative to unrelated healthy controls (uHC) [21–23]. With it, we have also identified an elevated innate inflammatory state among healthy T1D family members that is temporally supplanted by an IL-10/TGF-β mediated regulatory state amongst sibling non-progressors possessing high-risk HLA haplotypes [23]. Emergence of this regulated state parallels peripheral increases in activated CD4+/CD45RA−/FoxP3high regulatory T-cell (Treg) frequencies, suggesting that failures in endogenous regulatory mechanisms that normally manage inherited T1D risk may underlie disease progression [23]. Here, we report that while the responses were distinct, both IL-1Ra and canakinumab resulted in modest normalization of the inflammatory state associated with ROT1D, and a relationship was observed between reduced inflammation and preserved β-cell function.
Results
Analysis of AIDA trial participants
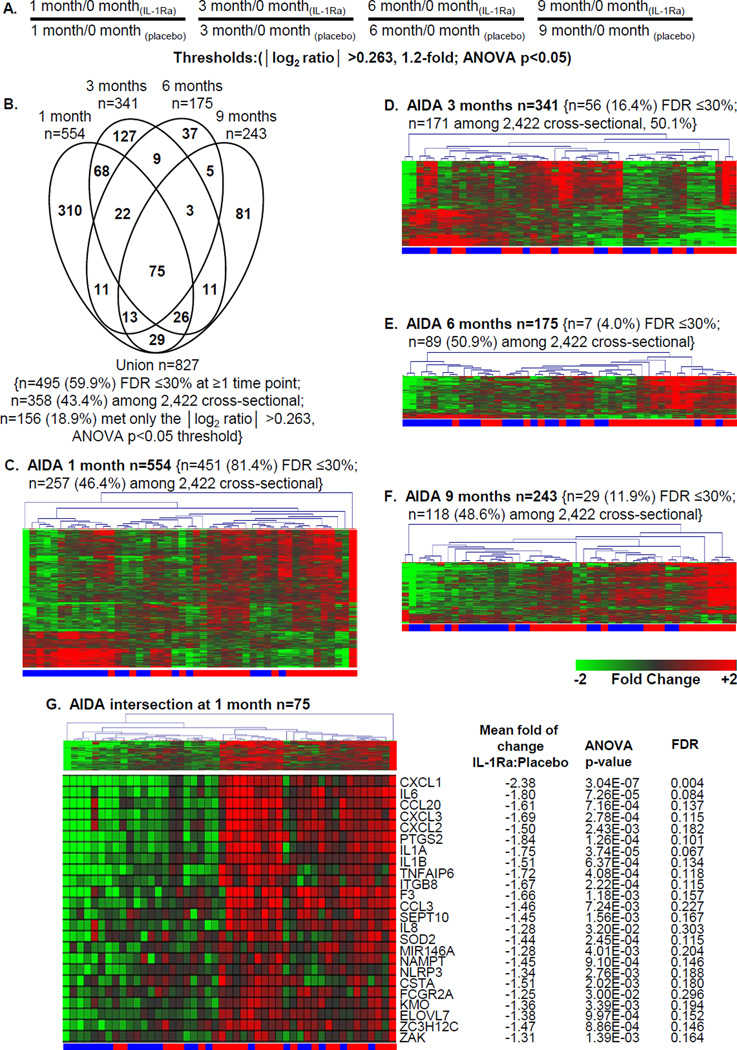
Signature analysis of plasma-induced PBMCs was conducted on AIDA trial participants without knowledge of treatment arm. To identify transcripts temporally regulated within each subject, data were baseline normalized. We identified, a priori, a set of IL-1 regulated genes that included IL1A, IL1B, IL1R1, and PTGS2. Based upon reduced expression of these genes at 1, 3, 6, and 9 months, 70.2% of the subjects were correctly identified to their treatment arm (33 of 47 subjects (p=0.006); 15/22 (68.2%) of the IL-1Ra-treated and 18/25 (72.0%) of the placebo-treated subjects). After unblinding, we identified 827 probe sets that were differentially induced (|log2 ratio|>0.263, 1.2-fold; ANOVA p<0.05) between the treatment and placebo arms at ≥1 time point (Figure 1A–1B; Supporting Information Table 2). Among these, 59.9% (495/827, 59.9%) exhibited an FDR of ≤30% at one or more time points (Supplemental Table 3), and as discussed below, 43% (358/827, X2 p<10E-06) were previously identified in cross-sectional analyses of ROT1D cases and healthy controls [23]. Among the 827 probe sets, 156 (18.9%) met only the |log2 ratio| >0.263, ANOVA p<0.05 threshold. There was heterogeneity in both study arms, as evidenced the imperfect hierarchical clustering observed at each time point (Figure 1C–1F). An intersection of 75 probe sets, regulated between the two arms at all time points was identified that consisted of transcripts down-regulated by plasma of IL-1Ra-treated participants (Figure 1G). These were primarily transcripts encoding inflammatory mediators regulated by IL-1, suggesting that plasma induced signature analysis could differentiate the two trial arms and despite a lack of overall efficacy, IL-1Ra therapy may have altered the inflammatory state.
Figure 1.
Plasma-induced signature analysis of AIDA trial participants. Among the 69 AIDA subjects, our analyses included 22 IL-1Ra treated subjects (age 26.0±2.9 years; range 18.1 – 33.6) and 25 placebo-treated subjects (age 25.7±2.0 years; range 19.8 – 34.1). These subjects were those with available sample at 0, 1, 3, 6, and 9 months and did not significantly differ from the total cohort. (A) Analysis strategy for identifying transcripts regulated to thresholds between the two treatment arms at 1, 3, 6, and 9 months. The selected thresholds (|log2 ratio| >0.263, 1.2-fold; ANOVA p<0.05) are based upon previous analyses [22, 23]. (B) Venn diagram illustrating the relationship of the union of 827 probe sets regulated to thresholds at 1, 3, 6, and 9 months between the IL-1Ra and placebo arms. Indicated are the number (and percentage) of probe sets that exhibited an FDR ≤30% as well as number (and percentage) of probe sets previously identified when cross-sectionally comparing ROT1D, related and unrelated healthy controls [23]. (C–F) Two-way hierarchical clustering (probe sets and subjects) for the regulated probe sets respectively identified between IL-1Ra-treated (blue bar) and placebo-treated (red bar) subjects at (C) 1 month (n=554 probe sets), (D) 3 months (n=341 probe sets), (E) 6 months (n=175 probe sets), and (F) 9 months (n=243 probe sets). (G) Two-way hierarchical clustering (probe sets and subjects) for the intersection of 75 commonly regulated probe sets regulated between IL-1Ra and placebo subjects at 1, 3, 6, and 9 months with tabulation of the mean fold of change, p-value, and FDR at the 1 month time point. All expression levels are baseline normalized.
The AIDA signature is independent of plasma IL-1Ra protein levels
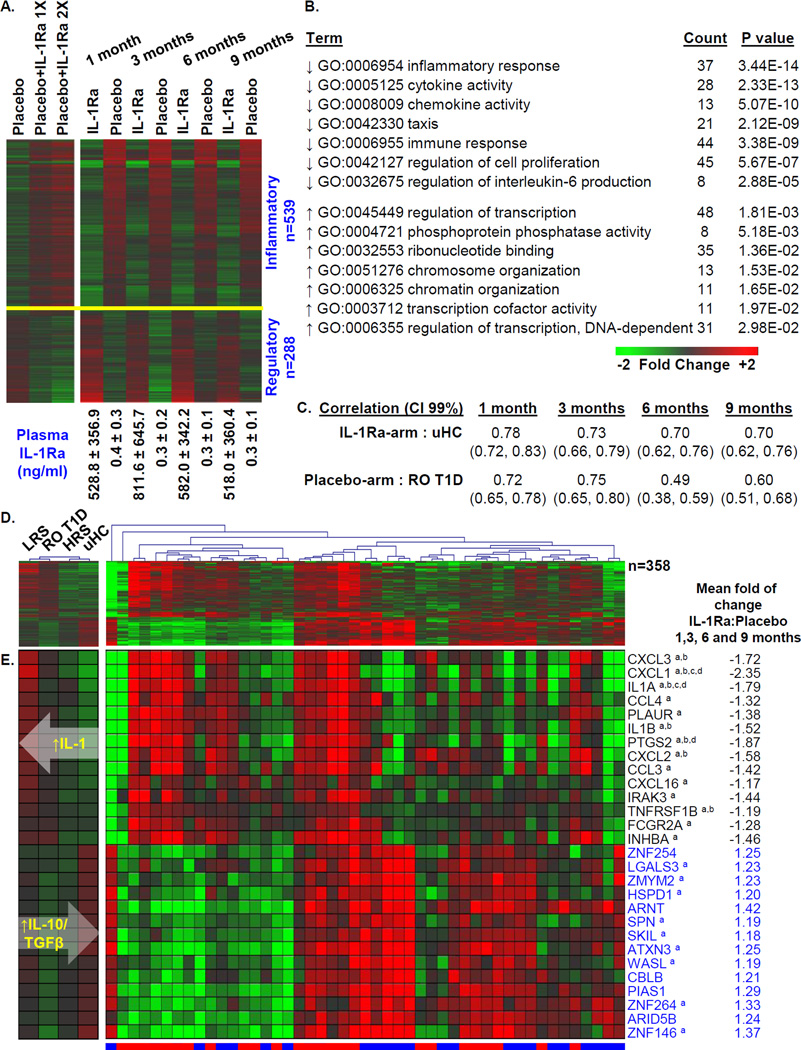
The half-life of IL-1Ra is <6 hours. Expectedly, IL-1Ra-treated subjects possessed higher plasma IL-1Ra concentrations compared to placebo-treated subjects (>500 ng/ml versus <0.5 ng/ml; Figure 2A). We reasoned that if the treatment arm signature was a direct consequence of carry-over IL-1Ra present in the plasma, it should be possible to largely recapitulate that signature in placebo samples by spiking the cultures with IL-1Ra. Therefore, plasma collected from AIDA placebo-treated participants were reanalyzed after supplementation with 0, 500, or 1000 ng/ml IL-1Ra. Even at the higher level, among the 827 probe sets differentially induced between the IL-1Ra and placebo groups at 1, 3, 6, and 9 months, 117 (14.1%) were directionally concordant, not considering a statistical threshold, and only 33 (4.0%) were directionally concordant and possessed false discovery rates (FDR) <20% (Supplemental Table 2). Expectedly, induction of IL-1 dependent transcripts was reduced upon introduction of IL-1Ra to cultures. However, most genes were oppositely induced when comparing in vitro to ex vivo conditions (Figure 2A), supporting that signatures of treated subjects were largely independent of carry-over IL-1Ra protein levels. These data suggest that differences between the signatures of the IL-1Ra-arm and placebo-arm were reflective of treatment-mediated alterations in the inflammatory state.
Figure 2.
Analysis of the union of 827 probe sets regulated between IL-1Ra and placebo treated patients at ≥1 time point. (A) One way clustering (probe sets only) and mean expression levels of probe sets regulated to thresholds: |log2 ratio| >0.263, 1.2-fold; ANOVA p<0.05 at the 1, 3, 6, and 9 month time points (right). The mean IL-1Ra plasma protein levels, as measured in [20], for the two study arms at each time point are indicated. The mean induced expression levels of the 827 probe sets by plasma of the 9 AIDA placebo-treated subjects used in the IL-1Ra add-back experiment are also shown (left). These samples were reanalyzed under 3 conditions: 1) 1 month samples + 0 ng/ml IL-1Ra; 2) 1 month samples + 500ng/ml IL-1Ra (~1× mean levels measured in the treated arm); and 3) 1 month samples + 1000 ng/ml IL-1Ra (~2× mean levels measured in the treated arm). Expression levels illustrated in heat maps are baseline normalized and the two panels are independently centered in terms of scale. (B) Probe sets down-regulated (n=539) and up-regulated (n=288) by plasma of the IL-1Ra arm were independently evaluated by DAVID to identify regulated GO terms. (C) Pearson Correlation Coefficients between 358 probe sets commonly identified through plasma induced signature analysis of AIDA participants and the previously described [23] cross-sectional analyses of ROT1D (n=47) and uHC (n=44) patients. Indicated in parentheses are, respectively, the lower and upper 0.99 confidence intervals (CI). (D) Two-way hierarchical clustering (probe sets and subjects) of IL-1Ra and placebo subjects using mean expression of 358/827 probe sets commonly identified through plasma induced signature analysis of AIDA trial participants at 1, 3, 6, and 9 months and the previously described [23] cross-sectional analyses of LRS (n=42), ROT1D, HRS (n=30) and uHC. IL-1Ra treated subjects are indicated by blue bar, placebo-treated subjects are indicated by red bar (right). The mean expression levels of these probe sets within the cross-sectional cohorts are also shown (left). (E) Mean expression levels of a subset of well annotated transcripts identified among 827 probe sets regulated between the two arms of the AIDA trial at 1, 3, 6, and 9 months. Black font indicates transcript annotated as being “inflammatory”; blue font indicates transcript annotated as being “regulatory”. The mean fold of change of IL-1Ra to placebo is tabulated for each probe set, expression differences that exhibited an FDR ≤30% at 1, 3, 6, or 9 months are indicated by a, b, c, or d, respectively. As indicated by the white overlaid arrows, an overall increasing IL10/TGFβ bias and decreasing IL-1 bias was identified across the LRS→ROT1D→HRS→uHC continuum.
IL-1Ra treatment alters the immune state
Pathway analysis was conducted independently on the 539 probe sets down-regulated and the 288 probe sets up-regulated by plasma of IL-1Ra-treated subjects (Figures 2A–2B). Representative down-regulated Gene Ontology Biological Processes and Molecular Functions (GO terms) were related to inflammation and immune activation, chemokine/cytokine activity and chemotaxis. Transcripts annotated under these terms included IL-1 family members, chemokines, and transcripts related to inflammatory mediator synthesis. Representative up-regulated GO terms were related to transcriptional regulation of immune activity and protein phosphatase activity. Transcripts annotated under these terms included transcriptional repressors and regulators of cytokine production and regulators of immune responses through ubiquitin mediated proteolysis. Overall, plasma of IL-1Ra-treated subjects induced greater regulatory transcription and decreased inflammatory transcription. In contrast, plasma of placebo-treated subjects more highly induced IL-1 dependent response genes and numerous chemokines and cytokines.
IL-1Ra treatment partially normalizes the signature associated with ROT1D
Recently, we reported cross-sectional plasma induced signature analyses of ROT1D, uHC, diabetes autoantibody-negative (AA−) high HLA risk (DR3 and/or DR4) siblings (“HRS”) and AA− low HLA risk (non-DR3/non-DR4) siblings (“LRS”) [23]. We identified 2,422 probe sets significantly regulated among these four cohorts that revealed a continuum of immune states. Relative to uHC, T1D family members exhibited an elevated, partially-IL-1 dependent, inflammatory state [23]. This familial inflammatory state was more regulated in HRS than LRS. ROT1D possessed signatures intermediate to those of LRS and HRS, such that inflammatory bias decreased and regulatory bias increased across the LRS→ROT1D→HRS→uHC continuum [23].
We investigated the degree to which IL-1Ra treatment modulated the immune state relative to the LRS→ROT1D→HRS→uHC continuum. Notably, 358/827 probe sets (43.3%, X2 p<10E-06) differentially induced between the IL-1Ra- and placebo-treated arms overlapped with those identified in cross-sectional analysis (Figure 1). Consistent with IL-1Ra treatment moderating the familial inflammatory state, for this intersection of 358 probe sets, the signatures of IL-1Ra subjects were more correlative to uHC, while signatures of the placebo patients were more correlative to those of ROT1D (Figure 2C). This relationship is further illustrated by the mean expression levels of the 358 probe set intersection by the cross-sectional cohorts and the AIDA participants at 1, 3, 6, and 9 months (Figures 2D–2E).
A relationship between reduced inflammation and preserved β-cell function in the AIDA trial
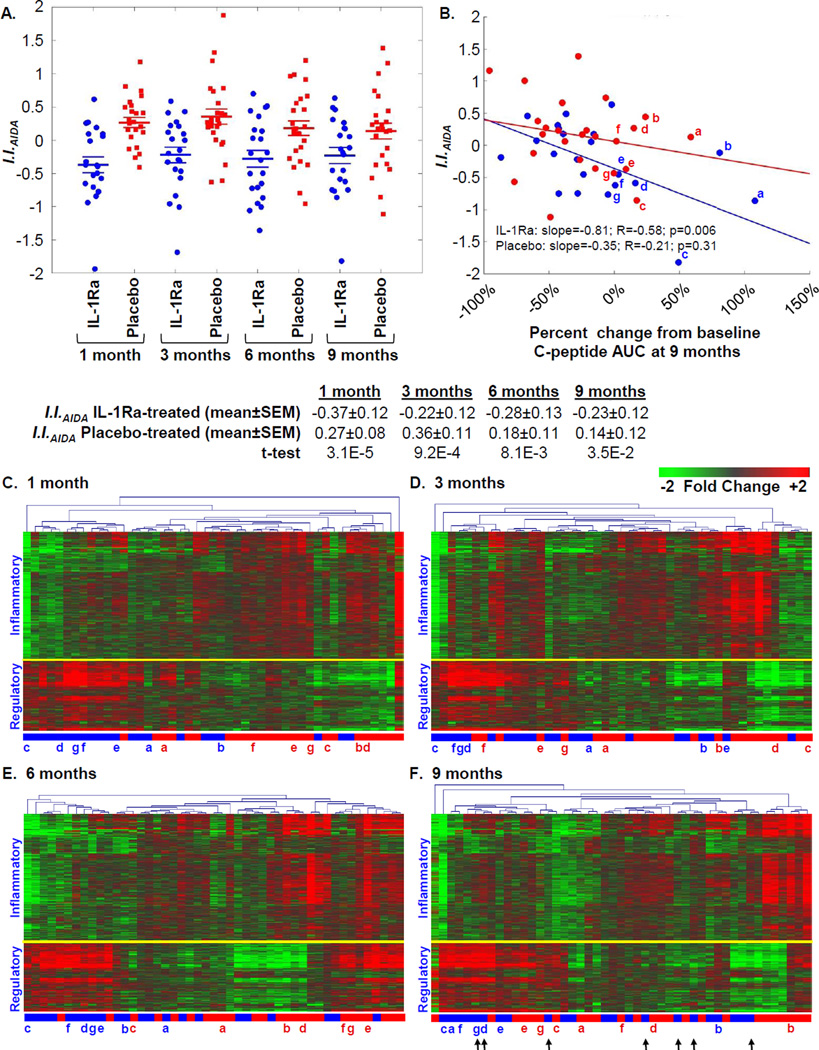
As defined by the ontological analyses (Figure 2B), the probe sets differentially induced between the two treatment arms can be broadly considered as “inflammatory” or “regulatory”. Based upon our previously described scoring strategy [23], the signatures of AIDA participants were quantitatively evaluated using an inflammatory index (I.I.AIDA). I.I.AIDA was calculated by determining the average ratio between the mean baseline normalized intensity of the 539 probe sets annotated as being “inflammatory” versus the mean baseline normalized intensity of the 288 probe sets annotated as being “regulatory”. The mean I.I.AIDA of IL-1Ra-treated patients was significantly lower than that of placebo-treated patients at each time-point (Figure 3A). Consistent with that reported for all 69 AIDA study subjects [20], the subset of 22 IL-1Ra- and 25 placebo- treated subjects studied here did not significantly differ in terms of mean baseline-adjusted 2-hour area under the curve (AUC) stimulated C-peptide response at the 9 month endpoint (−0.15 nmol/L, −0.22 nmol/L; p=0.39). However, at endpoint a significant inverse relationship was observed between I.I.AIDA and percent change from baseline C-peptide AUC among IL-1Ra-treated subjects (p=0.006), but not placebo-treated subjects, associating reductions in inflammation with preserved β-cell function (Figure 3B). While heterogeneity is evident, this subset of IL-1Ra treated subjects possessed signatures at 1, 3, and 6, and 9 months that exhibited greater regulatory bias; while placebo-treated patients retaining the highest percentages of baseline C-peptide at endpoint did not (Figures 3C–3F).
Figure 3.
Ontology-based scoring of AIDA participant plasma induced transcriptional signatures. (A) Mean Inflammatory Index (I.I.AIDA) of participants in the IL-1Ra (blue) and placebo (red) arms at 1, 3, 6, and 9 months. The mean I.I.AIDA was significantly different between the two arms at each time point. (B) Relationship between percentage change from baseline to 9 month C-peptide AUC and scored signatures using I.I.AIDA at 9 months. The plot is similar if data are considered as C-peptide AUC from the 9 month visit normalized by baseline: IL-1Ra: slope= −1.39; R=−0.55; p=0.009; Placebo: slope=−0.03; R=−0.13; p=0.52. The tertile of IL-1Ra and placebo –treated subjects that showed the highest percentage of baseline C-peptide at 9 months are indicated in lower case letters, these individuals are indicated in panels (C–F). (C–F) Two-way hierarchical clustering (probe sets and subjects) for 827 regulated probe sets respectively identified between IL-1Ra-treated and placebo-treated subjects at (C) 1 month, (D) 3 months, (E) 6 months, and (F) 9 months. As indicated, a yellow line separate transcripts generally annotated as being inflammatory or regulatory. A subset of IL-1Ra treated patients with intermediary residual β-cell function at baseline showed an increase in C-peptide relative to placebo (indicated with arrows).
In the AIDA trial, treatments were self-administered and plasma IL-1Ra levels exhibited a large range from 0.1 ng/ml to 2526.7 ng/ml (mean±S.D.: 465.9±415.3 ng/ml) [20]. However, among IL-1Ra-treated subjects, significant relationships between plasma IL-1Ra levels and I.I.AIDA were not observed, further supporting a negligible role for carryover IL-1Ra levels in the plasma influencing the measured signatures. Relationships between previously measured IL-6 or glycated hemoglobin levels [20] and I.I.AIDA were not identified.
Analysis of TN-14 trial participants
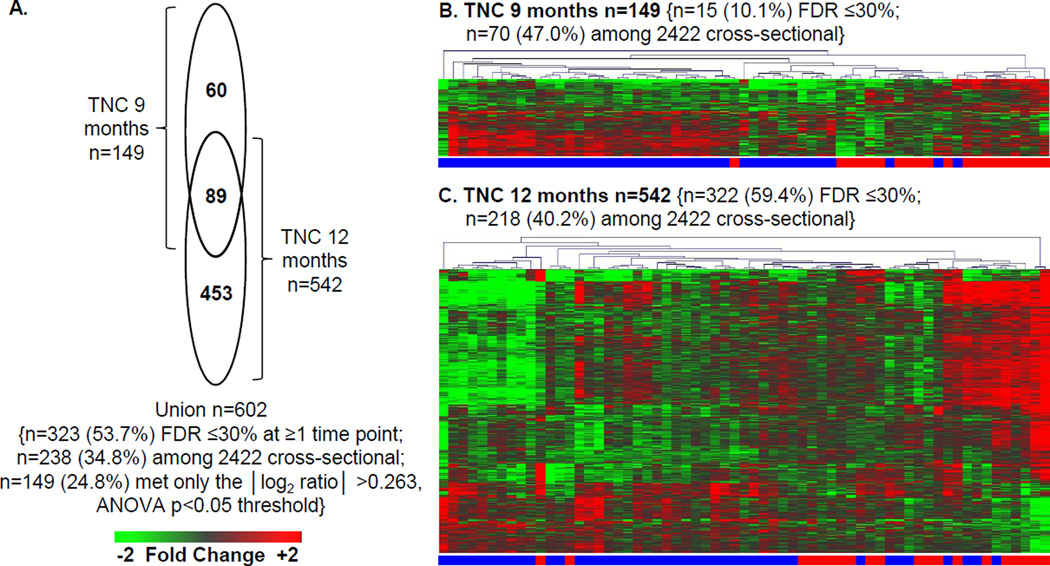
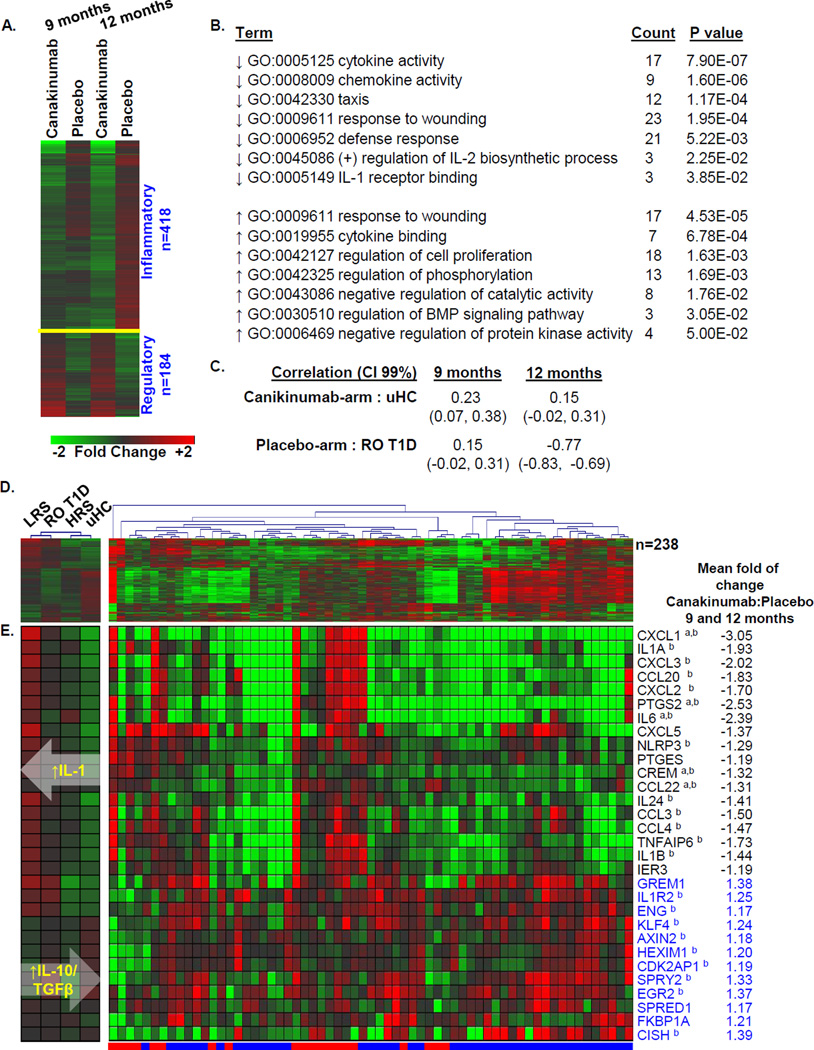
The primary endpoint for the TN-14 study was 12 months. Since the AIDA trial endpoint was 9 months, analyses of TN-14 data considered both time points. Samples of 43 canakinumab- and 20 placebo- treated subjects, collected at 0 (baseline), 9, and 12 months, were assayed without knowledge of treatment arm. Based upon reduced expression of known IL-1 regulated genes at the 9 and 12 month time points, 69.8% of the subjects were correctly identified to their treatment arm (44/63 (χ2 p=0.02) total; 34/43 (79.1%) of canakinumab-treated and 10/20 (50%) of placebo subjects). After unblinding, we compared the baseline normalized data at 9 and 12 months between the treatment arms and identified 602 significantly regulated probe sets (|log2 ratio|>0.263, 1.2-fold; ANOVA p<0.05; Figures 4A–C; Supplemental Table 2). Among these, 53.7% (323/602, 59.9%) exhibited an FDR of ≤30% at one or more time points (Supplemental Table 4), and as discussed below, 43% (238/602, X2 p<10E-06) were previously identified in cross-sectional analyses of ROT1D cases and healthy controls [23]. Among the 602 probe sets, 149 (24.8%) met only the |log2 ratio| >0.263, ANOVA p<0.05 threshold. There was heterogeneity in both study arms, as evidenced the imperfect hierarchical clustering observed at each time point (Figure 1B–C).
Figure 4.
Plasma induced signature analysis of TN-14 trial participants. Among the 69 TN-14 study participants, our analyses included samples of 43 canakinumab-treated subjects (age 12.3 ± 4.0 years; range 6.5 – 25.6 years) and 20 placebo-treated subjects (age 12.2 ± 6.0 years; range 6.1 – 32.0 years). These subjects represented those with available sample at 0, 9 and 12 months, and did not significantly differ from the total cohort. Data collected at 9 and 12 month time points was baseline normalized. After unblinding, differentially induced transcripts (|log2 ratio| >0.263, 1.2-fold; ANOVA p<0.05) between the treatment arms were identified. (A) Venn diagram illustrating the relationship of the union of 602 probe sets regulated to thresholds at 9 and 12 months between the canakinumab and placebo arms. Indicated are the number (and percentage) of probe sets that exhibited an FDR ≤30% as well as number (and percentage) of probe sets previously identified when cross-sectionally comparing ROT1D, related and unrelated healthy controls [23]. (B and C) Two-way hierarchical clustering (probe sets and subjects) for the regulated probe sets respectively identified between canakinumab (denoted by blue bar) and placebo (denoted by red bar) subjects at (B) 9 months (n=149 probe sets) and (C) 12 months (n=542 probe sets). All expression levels are baseline normalized.
Pathway analysis was conducted independently on the 418 probe sets down-regulated and the 184 probe sets up-regulated by plasma of canakinumab-treated subjects (Figures 5A–5B). As reflected by the enriched GO terms, representative down-regulated activities were related to inflammation and immune activation, chemokine/cytokine activity and chemotaxis. Transcripts related to these terms included IL-1 family members, cytokines and chemokines, and transcripts related to inflammatory mediator synthesis. Many of the 184 up-regulated probe sets are known to function in tempering immune responses: these include KLF4, a zinc-finger-containing transcription factor that regulates IL-10 expression [24] and ENG, a co-receptor of the TGF-β superfamily necessary for TGF-β mediated resolution of inflammation [25]; and the immunophilin protein family member FKBP1A. Additional transcripts annotated under these GO terms included negative regulators of protein kinase activity and negative regulators of signal transduction and growth. Overall, plasma of canakinumab-treated subjects induced greater regulatory transcription and decreased inflammatory transcription. In contrast, plasma of placebo-treated subjects more highly induced IL-1 dependent response genes and numerous chemokines and cytokines (Figure 5E).
Figure 5.
Pathway analysis of the union of 602 probe sets regulated between canakinumab- and placebo-treated patients. (A) One way clustering (probe sets only) and mean expression levels of probe sets regulated to thresholds (|log2 ratio| >0.263, 1.2-fold; ANOVA p<0.05) at the 9 and 12 month time points. (B) Probe sets down-regulated (n=418) and up-regulated (n=184) by plasma of the canakinumab arm were independently evaluated for biological pathway enrichment using DAVID to identify regulated GO terms. Representative pathway terms, the number of identified genes and significance of enrichment are tabulated. (C) Pearson Correlation Coefficients between the 238 probe sets commonly identified through plasma induced signature analysis of TN-14 trial participants and cross-sectional analyses of ROT1D patients and uHC. Indicated in parentheses are, respectively, the lower and upper 0.99 confidence intervals (CI). (D) Two-way hierarchical clustering (probe sets and subjects) of canakinumab-treated (blue bar) and placebo-treated (red bar) subjects using the 238 probe sets commonly identified through plasma induced signature analysis of TN-14 trial participants and cross-sectional analyses of LRS (n=42), ROT1D (n=47), HRS (n=30) and uHC (n=44) (right). The mean expression levels of these probe sets within the cross-sectional cohorts are shown (left). (E) Mean expression levels of a subset of well annotated transcripts identified among 602 probe sets regulated between the arms of the TN-14 trial. Black font indicates transcript annotated as being “inflammatory”; blue font indicates transcript annotated as being “regulatory”. The mean fold of change of IL-1Ra to placebo is tabulated for each probe set, expression differences that exhibited an FDR ≤30% at 9 or 12 months are indicated by a, or b, respectively. As indicated by the white overlaid arrows, an overall increasing IL10/TGFβ bias and decreasing IL-1 bias was identified across the LRS→ROT1D→HRS→uHC continuum. All expression levels are baseline normalized.
Comparison of the canakinumab treatment signature with that of a cross-sectional data set
Among the 602 probe sets regulated between the canakinumab- and placebo- treated arms, 238 (34.8%, X2 p<10E-6) overlapped with those previously identified in cross-sectional studies of uHC, LRS, HRS, and ROT1D [23]. For this intersection, the average signature of the canakinumab-treated arm showed a low direct correlation with that of the uHC cohort, and the average signature of the placebo-treated arm did not directly correlate with that of the ROT1D cohort (Figure 5C). The induced expression levels of the 238 probe set intersection by TN-14 participants (Figure 5D) show that the lack of overall correlation is a reflection of variability observed in both trial arms. However, the data show that the regulated immune state observed in uHC subjects was recapitulated in a subset of canakinumab-treated subjects, particularly through reduced induction of transcripts for inflammatory mediators (Figure 5D–5E, rightmost cluster).
The relationship between I.I.TN-14 and β-cell function
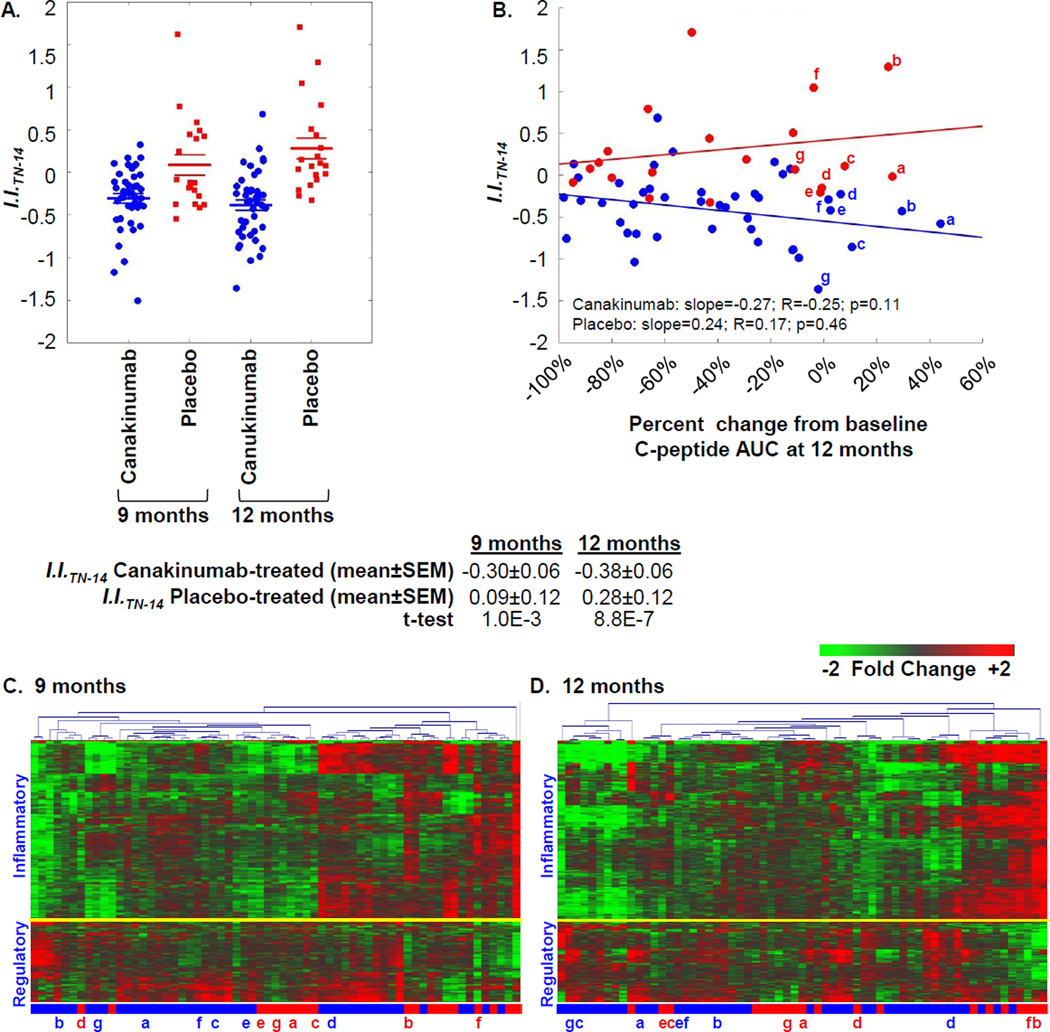
Paralleling the AIDA study, signatures of TN-14 participants were quantitatively evaluated using an inflammatory index (I.I.TN-14). I.I.TN-14 was calculated by determining the average ratio between the mean baseline-normalized intensity of 418 probe sets annotated as “inflammatory” versus the mean baseline-normalized intensity of the 184 probe sets annotated as “regulatory”. The mean I.I.TN-14 of canakinumab-treated patients was significantly lower than that of the placebo-treated patients at 9 months and 12 months (Figure 6A). Consistent with that reported for all 69 TN-14 study subjects [20], the subset of 43 canakinumab- and 20 placebo-treated subjects studied here did not significantly differ at 12 months in terms of mean baseline-adjusted 2-hour AUC stimulated C-peptide response (−0.12 nmol/L, −0.12 nmol/L; p=0.91). The inverse relationship observed between I.I.TN-14 and the percent change from baseline to 12 month C-peptide AUC (Figure 6B) did not reach statistical significance (p=0.11). However, canakinumab-treated subjects possessing the lowest I.I.TN-14 tended to exhibit more regulated signatures compared to placebo-treated patients that retained the highest percentages of C-peptide at endpoint (Figures 6C–6D).
Figure 6.
Ontology-based scoring of TN-14 participant signatures. (A) Mean Inflammatory Index (I.I.TN-14) of participants in the canakinumab (blue) and placebo (red) arms at 9 and 12 months. As tabulated, the mean I.I.TN-14 was significantly different between the two arms at each time point. (B) Relationship between percentage change in C-peptide AUC from baseline to 12 months for each subject and scored signatures using I.I.TN-14 at 12 months. The plot is similar and the relationship is not significant if data are considered as C-peptide AUC from the 12 month visit normalized by baseline: Canakinumab: slope= −0.21; R=−0.07; p=0.66; Placebo: slope=0.55; R=0.16; p=0.49. The tertile of canakinumab and placebo-treated subjects showing the highest percentage of baseline C-peptide at 12 months are indicated in lower case letters, these individuals are indicated in panels (C–D). (C–D) Two-way hierarchical clustering (probe sets and subjects) for the 602 regulated probe sets respectively identified between canakinumab and placebo subjects at (C) 9 months and (D) 12 months. As indicated, a yellow line separate transcripts generally annotated as being inflammatory or regulatory.
Comparison of responses observed in the AIDA and TN-14 trials
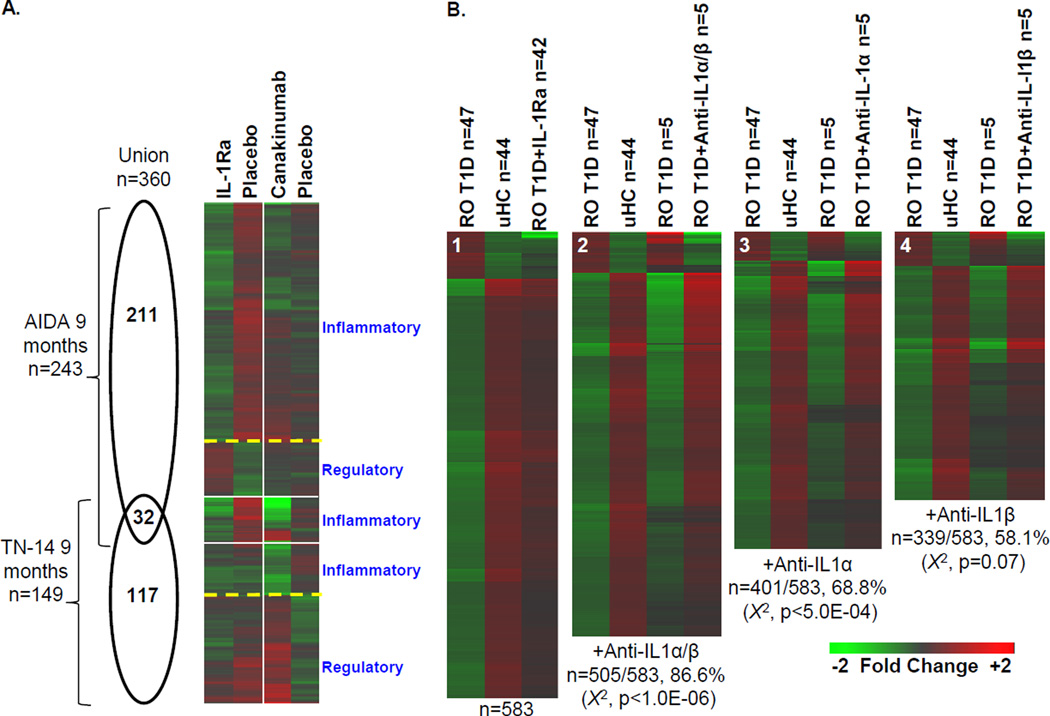
Plasma induced signatures were measured at 9 months in the AIDA and TN-14 trials where, respectively, 243 and 149 regulated probe sets were identified when comparing the treatment and placebo study arms (Figure 7A). The Pearson correlation coefficient between the 360 probe set union of these two data sets was 0.29, while the Pearson correlation coefficient for the 32 probe set intersection was 0.85. The transcripts that exhibited reduced induction by plasma collected from the treatment arms of the two trials were annotated as being “inflammatory” (AIDA n=204/243; TN-14 n=62/149); each of these data subsets shared identity when compared to the other trial (Pearson correlation coefficients of 0.70 and 0.84, respectively). The up-regulated transcripts (AIDA n=39/243; TN-14 n=87/149), annotated as being “regulatory”, did not show identity when compared to the other trial (Pearson correlation coefficients of 0.05 and −0.13, respectively).
Figure 7.
The distinctiveness of plasma induced signatures of AIDA and TN-14 participants at 9 months. (A) Venn diagram illustrating the relationship of the union of 360 probe sets regulated to thresholds (|log2 ratio| >0.263, 1.2-fold; ANOVA p<0.05) between treatment and placebo-treated arms of the AIDA and TN-14 trials at 9 months (left). One way clustering (probe sets only) and mean expression levels of probe sets regulated to thresholds at 9 months. For each data set and intersection, an indication of the general function of the cluster is indicated based on ontological analyses (right). (B) Additive effects of IL-1α and IL-1β on the ROT1D signature. Previously [23], adding 4 µg/mL IL-1Ra to ROT1D cultures (n = 47) modulated IL-1–dependent components of the ROT1D:uHC signature, directionally altering expression of 583/762 genes (76.5%; χ2 p < 10E-6; panel B1). Here, 5 pediatric ROT1D subjects with mean plasma IL-1α levels of 89.4 +/− 56.4 pg/ml and IL-1β levels of 4.6 +/− 5.0 pg/ml were studied. Addition of both IL-1α –neutralizing antibodies (15 µg/mL) and IL-1β –neutralizing antibodies (0.3 µg/mL) to the 5 ROT1D cultures directionally altered 505/583 of the previously identified IL-1Ra dependent transcripts (86.6%; χ2 p < 1.0E-6; panel B2). Addition of IL-1α–neutralizing antibodies (15 µg/mL) to the 5 ROT1D cultures directionally altered 401/583 of the previously identified IL-1Ra-dependent transcripts (68.8%; χ2 p < 5.0E-4; panel B3). Addition of IL-1β–neutralizing antibodies (0.3 µg/mL) to the 5 ROT1D cultures directionally altered 339/583 of the previously identified IL-1Ra dependent transcripts (58.1%; χ2 p=0.07; panel B4). Nonspecific isotypic control antibodies did not significantly modulate expression. Regulated transcripts are provided in Supporting Information Table 2.
We hypothesized that differences in the signatures of IL-1Ra and canakinumab-treated subjects could be due to each agent’s unique mechanism of action. IL-1Ra blocks action of both IL-1α and IL-1β, while canakinumab neutralizes only IL-1β. In our recent report [23], the partial IL-1 dependence of the ROT1D:uHC signature was confirmed by introducing IL-1Ra into cultures possessing ROT1D plasma, which directionally altered expression of 583/762 probe sets (76.5%, X2 p<10E-6). To investigate whether the distinct signatures observed in these trials may be in part due to the unopposed action of IL-1α in TN-14, we conducted plasma induced transcription whereby replicate cultures of ROT1D subjects were supplemented with both IL-1α and IL-1β –neutralizing antibodies, only IL-1α–neutralizing antibodies, or only IL-1β–neutralizing antibodies. On average, cultures possessing neutralizing antibodies for both ligands reversed a significant proportion of the previously identified signature (n=505/583; χ2 p< 10E-6; Figure 7B). This contrasted with cultures independently supplemented with antibodies towards either IL-1α or IL-1β, where expression of fewer transcripts was reversed (401/583 and 339/583, respectively). These data suggest that both IL-1 isoforms contribute to the inflammatory state associated with ROT1D and that the unmitigated action of IL-1α in TN-14 may contribute to the distinct signatures measured in the two trials.
A relationship between the baseline plasma induced signature and therapeutic response
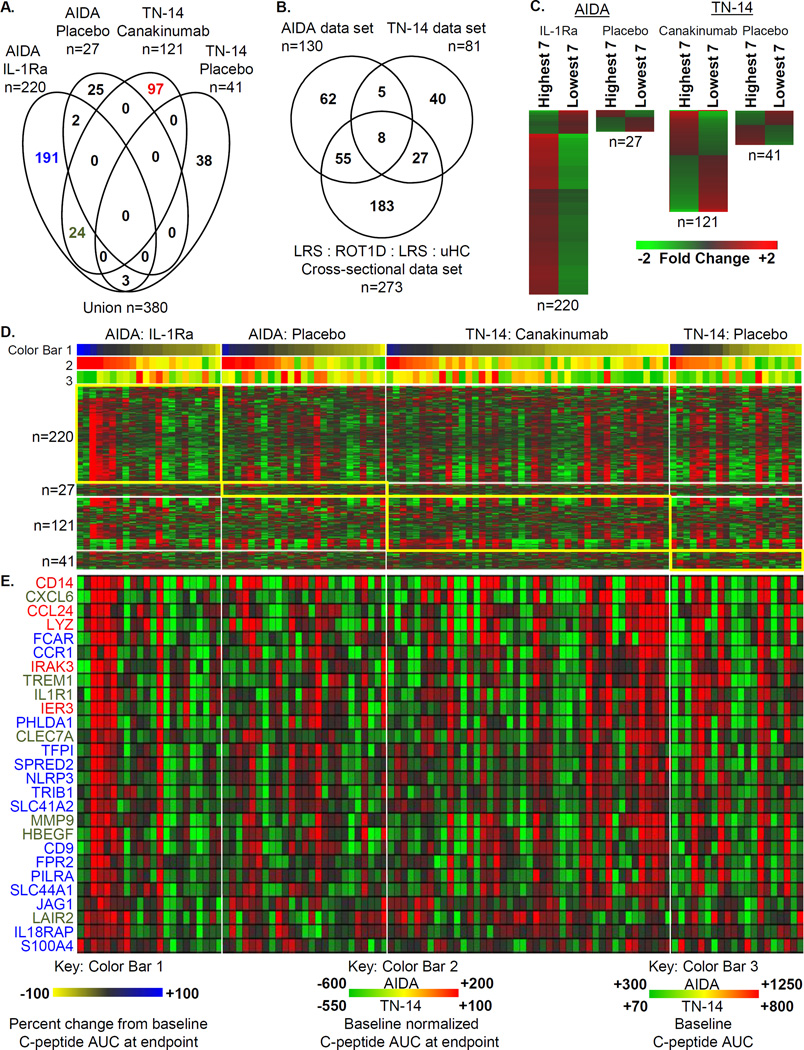
We investigated the relationship between the baseline plasma induced signature and outcome in the AIDA and TN-14 trials. Since three of the four trial arms possessed ~20 subjects, for each trial arm we identified differentially expressed genes from the seven subjects (tertile) with the highest and lowest percentage change in C-peptide AUC from baseline to study endpoint (Figure 8A–8C). Given the limited number of subjects compared, this analysis was restricted to the union of 3,206 significantly regulated probe sets identified when comparing the treatment and placebo arms of the AIDA (n=827) and TN-14 (n=602) trials, as well those previously identified in the cross-sectional studies of uHC, LRS, HRS, and ROT1D patients (n=2,422) [23]. A total of 380 differentially induced transcripts were identified. Consistent with immunomodulation, significantly more regulated probe sets were identified when comparing subjects within the treatment arms (AIDA: n=220, p<7.6E-15, odds ratio=4.26; TN-14: n=121, p<4.9E-9, odds ratio=2.84) versus when comparing subjects within the placebo arms (AIDA: n=27; TN-14: n=41). Significantly identified GO terms associated with this data set were again related to inflammation, innate immune activation, and chemotaxis. Transcripts annotated under these terms (Figure 8D) were more highly induced by baseline plasma of IL-1Ra-treated subjects that had the highest percentages of baseline C-peptide AUC at endpoint, suggesting that those AIDA subjects with the highest levels of innate inflammation may have benefited the most from IL-1Ra treatment. Notably, these subjects generally had the lowest C-peptide AUC levels at baseline suggesting that baseline signatures in this trial may be informative of therapeutic response. This relationship was not seen in the canakinumab-treated subjects.
Figure 8.
The relationship between the plasma induced signature at the baseline sampling and the percentage change in C-peptide AUC from baseline to study endpoint (9 months for AIDA and 12 months for TN-14). (A) Venn diagram illustrating the relationship of the union of 380 probe sets regulated to thresholds (|log2 ratio| >0.263, 1.2-fold; ANOVA p<0.05) when comparing the tertile (n=7) of subjects possessing the highest and the lowest percent change from baseline C-peptide AUC at study endpoint within each of the 4 study arms. (B) Venn diagram showing the distribution of the 380 regulated probe sets among the 3 source data sets: AIDA (n=827 total probe sets), TN-14 (n=602 total probe sets) and the uHC/LRS/HRS/ROT1D cross-sectional (n=2,422 total probe sets). (C) Mean expression levels of regulated probe sets identified when comparing, within each arm, 7 subjects with the highest and lowest percent change from baseline C-peptide AUC at study endpoint. (D) One-way hierarchical clustering (probe sets) of the 4 data sets illustrated in (A). As reflected by Color Bar 1, subjects within each study arm (D and E) are sorted by percent change from baseline C-peptide AUC at study endpoint (highest to lowest, blue to yellow). Color Bar 2 provides a measure of baseline normalized C-peptide AUC (highest to lowest, red to yellow to green) and shows the general rank order is conserved. Color Bar 3 provides a measure of the baseline C-peptide AUC. (E) Mean expression levels of a subset of well annotated transcripts identified within the IL-1Ra and canakinumab treatment arms. The font colors used for gene symbols are coded to the Venn diagram in (A). Conducting the analysis by comparing subjects with the highest and lowest baseline normalized C-peptide AUC yielded similar results and did not alter the overall biological interpretation.
Discussion
Overall, the treatment and placebo arms of the AIDA and TN-14 trials did not differ in terms of stimulated C-peptide AUC at endpoint. This brings into question whether therapies directed at innate immune pathways should continue to be pursued as a monotherapy in new onset T1D. Our application of plasma induced transcription to these trials began with blinded analyses, where we correctly identified the majority of subjects to their treatment arm by examining induction of genes central to IL-1 action. Though the transcripts differentially regulated between the treatment and placebo arms of the AIDA and TN-14 trials were different, ontological analyses suggested that inflammation was reduced and Treg activity was enhanced in both trials, as reflected by the induction of IL-10 and TGF-β dependent genes that were not modulated in the IL-1Ra add-back studies.
We compared the signatures of AIDA and TN-14 participants to those previously identified by cross-sectional analysis in ROT1D and uHC [23]. Both IL-1Ra and canakinumab modulated the immune state, respectively normalizing 14.8% (358/2,422) and 9.8% (238/2,422) of the probe sets identified in historical cross-sectional analyses. This modest normalization suggests that the immune state associated with T1D arises from a complex milieu of mediators extending beyond IL-1. Because of the recognized complexity of diabetes pathogenesis, combinatorial approaches that therapeutically target multiple immune pathways are now being considered. Preclinical studies suggest that IL-1 blockade in combination with other therapies may preserve β-cell function more effectively than using it alone [26, 27]. Relevant to this shifting paradigm, the unmodified portions of the ROT1D signature may represent the combination of pathways that must be targeted in new onset patients to more fully recapitulate the immune state observed in healthy controls.
Plasma induced signatures were scored with an ontology-based inflammatory index. This enabled quantitative assessment of the immunomodulation achieved in each subject across the duration of the intervention and alignment of the signature with other clinical measures. The significant correlation between I.I.AIDA and stimulated C-peptide AUC in IL-1Ra treated subjects at 9 months suggested that in a subset of patients exhibiting the greatest reduction in inflammation, there was preserved β-cell function.
One possible limitation of the plasma induced transcription assay is the necessity to identify a representative responder cell population, as heterogeneity may exist between fresh cells collected from different healthy individuals or even between successive draws of the same person. Vendors now offer highly viable, cryopreserved PBMC that have been collected by aphaeresis. Quantities of cells, sufficient for thousands of assays, can be prepared from a single draw of a healthy well-characterized donor. This greatly simplifies the process of testing a panel of candidates and identifying a representative responder cell donor. We have previously reported the characteristics of plasma induced transcription assays when using cryopreserved PBMCs of a single donor [22], where in the analysis of the same plasma samples in 5 independent assays, we observed a low mean inter-assay coefficient of variation (762 probe set ROT1D:uHC signature=0.095±0.062). It is therefore unlikely that inter-assay variation was the basis for the distinctive signatures associated with IL-1Ra and canakinumab treatment.
IL-1Ra binds IL-1R1, competitively inhibiting the binding and activities of IL-1β and IL-1α. In contrast, canakinumab neutralizes IL-1β, while not acting on other IL-1 family members. We observed an additive effect when cultures possessing ROT1D plasma were supplemented with IL-1α and IL-1β neutralizing antibodies. These findings are consistent with our recent report [23], where we measured elevated IL-1α levels in ROT1D patients and their healthy AA− siblings relative to uHC, and other studies that support a role for both IL-1 isoforms in the inflammatory state associated with T1D susceptibility [28, 29]. Interestingly, these data parallel results found in adults with Type 2 diabetes, in that IL-1β specific blockade did not offer the same benefit to β-cell function as IL-1Ra [19].
A significant relationship between the inflammatory index and β-cell function was not observed in the TN-14 trial. While this may be related to the action of IL-1α, the studied AIDA and TN-14 participants also differed in age (25.9+/−4.9 versus 12.3+/−4.7, respectively, p<6.8E-6). Pediatric onset T1D is characterized by a more aggressive disease process compared to adult onset T1D. Indeed, in agreement with previous reports [30, 31], lower baseline stimulated C-peptide levels among subjects diagnosed at earlier ages was observed. This relationship showed age dependency in subjects between 5 and 19 years (p<1.0E-4, Supplemental Table 1). These differences in T1D pathogenesis may have contributed to the weaker relationship between I.I.TN-14 and β-cell function in TN-14. They may also have contributed to the contrasting relationship between innate inflammation at baseline and percentage baseline C-peptide AUC at endpoint observed between the two trials. These and other results [5, 32], suggest that interventions in one population may not have therapeutic equivalence when applied to the other.
These analyses represent a step towards meeting the need for measures of immunological efficacy in T1D clinical trials. Plasma induced signature analyses of patients undergoing IL-1 antagonism showed reduced induction of IL-1 -dependent genes and increased induction of IL-10/TGF-β -dependent genes, suggesting that both trials achieved varying levels of the anticipated immunomodulation. Through the analyses subjects after the initiation of therapy, these data show it is possible to differentiate between a complete absence of immune efficacy from insufficient immunomodulation to provide a significant clinical benefit. Through analyses of baseline samples, these data also suggest that it may be possible to identify subjects more likely to experience therapeutic response. This study provides a rationale for additional investigations of therapies targeting innate immunity in T1D, perhaps in selected subjects, earlier in the disease process. It also provides a rationale for future studies aimed at broadening the understanding of diabetes pathogenesis by defining the common and unique effects other completed clinical trials had on the immune state associated with T1D.
Materials and Methods
Study subjects
Local IRB approval was previously granted for both the AIDA and TN-14 trials and informed consent was obtained for all subjects. In the AIDA trial, patients were randomly assigned to subcutaneous dosing of IL-1Ra 100 mg daily for 9 months [20]. In the TN-14 trial, subjects were randomized in a 2:1 ratio to receive canakinumab 2 mg/kg monthly for 12 months [20]. We procured samples collected at 0 (baseline), 1, 3, 6, and 9 months from 47/69 AIDA participants (22 IL-1Ra-treated; 25 placebo-treated); and samples collected at 0 (baseline), 9 and 12 months from 63/69 TN-14 participants (43 canakinumab-treated; 20 placebo-treated). Participants were <100 days post-diagnosis and possessed mixed meal tolerance test stimulated C-peptide ≥0.2 nmol/L. Subject characteristics are described in [20] and Supplemental Table 1.
Plasma induced transcription
Cryopreserved PBMCs of a healthy 34 year old blood donor (HLA-A2, Caucasian male designated UPN727, purchased from Cellular Technology Ltd., Shaker Heights, OH) were cultured with 40% subject plasma in RPMI 1640 medium and induced transcription was measured using Affymetrix GeneChip Human Genome U133 plus 2.0 arrays (Affymetrix, Santa Clara, CA) as described [22, 23]. Global median normalization was accomplished with the Robust Multi-array Analysis (RMA) algorithm derived by the Bioconductor group (http://www.bioconductor.org) [33]. To identify temporally regulated transcripts, data collected at 1, 3, 6, 9, and 12 months for each subject was normalized with that of the baseline visit. Baseline normalized probe sets that revealed a 1.2-fold difference (|log2 ratio| > 0.263) between the treatment arms were identified and used in subsequent analyses. The significance of differentially induced transcription was assessed through ANalysis of VAriation (ANOVA) and the rate of type I errors in multiple testing was assessed through the determination of FDRs using Partek Genomics Suite 6.5 (Partek, Saint Louis, MO) as described [22, 23]. Ontological analysis utilized the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [34]. Hierarchical clustering was conducted with Genesis [35]. While investigated, the data were not adjusted for variables such as age, disease duration, or glycated hemoglobin. Data files are available through the National Center for Biotechnology Information Gene Expression Omnibus (accession numbers GSE37025 and GSE68049, http://www.ncbi.nlm.nih.gov/geo/) [36].
Comparison of the ex vivo versus in vitro effect of IL-1Ra
PBMCs were pre-treated with human recombinant IL-1Ra (R&D Systems, Minneapolis, MN) at 0 ng/mL, 500 ng/mL, and 1000 ng/mL for 45 minutes prior to the addition of 40% AIDA placebo subject plasma in RPMI 1640 medium. Transcriptional analysis was performed as detailed above.
Individual contributions of IL-1α and IL-1β to the T1D-associated inflammatory state
Plasma was collected from ROT1D subjects exhibiting good glycemic control 2–7 months post-diagnosis. These subjects were recruited at Children’s Hospital of Wisconsin through a protocol approved by its institutional review board. Written informed consent was obtained from subjects or their parents/legal guardians. Cytokine levels were assayed as described [22, 23]. Plasma was pre-treated with IL-1 neutralizing or isotype control antibodies for 45 minutes prior to adding PBMCs. Antibodies were introduced into cultures at levels reported to neutralize activity [23, 37, 38]: anti-IL-1α at 15 µg/m L, anti-IL-1β at 0.3 µg/mL, mouse IgG2α isotype control at 15 µg/mL, and mouse IgG1 isotype control at 0.3 µg/mL (R&D Systems, clones 4414, 8516, 20102, and 11711, respectively). Transcriptional analysis was performed as described above.
Supplementary Material
Acknowledgments
The authors thank the patients that participated in the studies.
This ancillary study was supported by The Juvenile Diabetes Research Foundation International (grants 1-2008-1026, 5-2012-220, 17-2012-621, 2-SRA-2015-109-Q-R to MJH); American Diabetes Association (grant 7-12-BS-075 to MJH); National Institutes of Health (grants R01AI078713 to MJH, DP3DK098161 to MJH/CJG, R01DK080100 to XW, and the National Center for Advancing Translational Sciences, National Institutes of Health grant 8UL1TR000055); and The Children’s Hospital of Wisconsin Foundation.
The canakinumab trial was sponsored by the Type 1 Diabetes TrialNet Study Group. The Type 1 Diabetes TrialNet Study Group is a clinical trials network funded by the National Institutes of Health through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and The Eunice Kennedy Shriver National Institute of Child Health and Human Development, through the cooperative agreements U01 DK061010, U01 DK061016, U01 DK061034, U01 DK061036, U01 DK061040, U01 DK061041, U01 DK061042, U01 DK061055, U01 DK061058, U01 DK084565, U01 DK085453, U01 DK085461, U01 DK085463, U01 DK085466, U01 DK085499, U01 DK085505, U01 DK085509, and contract HHSN267200800019C; the National Center for Research Resources, through Clinical Translational Science Awards UL1 RR024131, UL1 RR024139, UL1 RR024153, UL1 RR024975, UL1 RR024982, UL1 RR025744, UL1 RR025761, UL1 RR025780, UL1 RR029890, UL1 RR031986, and General Clinical Research Center Award M01 RR00400; the JDRF; and the American Diabetes Association. Novartis (Basel, Switzerland) provided canakinumab (Ilaris), input regarding dosage, and other suggestions but had no direct involvement with study design, conduct, or management; data collection, analysis or interpretation; or manuscript preparation. Roche Diagnostics provided blood glucose monitoring meters and strips to research participants in the canakinumab trial.
Abbreviations
- AIDA
Anti-Interleukin-1 in Diabetes Action
- AA−
diabetes autoantibody negative
- AUC
area under the curve
- β-cell
beta cell
- CCL
chemokine (C-C motif) ligand
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- FDR
false discovery rate
- GO
Gene Ontology
- HRS
high risk sibling to someone with type 1 diabetes, possessing DR3 and/or DR4 HLA haplotype
- I.I.AIDA
inflammatory index of AIDA participants
- I.I.TN-14
inflammatory index of TN-14 participants
- IL-1Ra
interleukin-1 receptor antagonist
- LRS
low risk sibling to someone with type 1 diabetes, with non-DR3/non-DR4 HLA haplotype
- PBMC
peripheral blood mononuclear cell
- ROT1D
subjects with recent onset type 1 diabetes mellitus
- T1D
type 1 diabetes mellitus
- TN-14
TrialNet Canakinumab Trial
- uHC
unrelated healthy control
AIDA Trial Investigators
Steering and Writing Committee: Thomas Mandrup-Poulsen, PI and Trial Sponsor (University of Copenhagen), Marc Y. Donath (University Hospital of Basel), Flemming Pociot (University Hospital of Glostrup) and Charles Dinarello (University of Colorado Health Science Center).
Data Management Unit (DMU): Jeffrey Krischer, PI of DMU, Linda Shanker, DMU Coordinator, Brian Bundy, Biostatistician, Franz Badias, Senior Applications Developer, David Cuthbertson, Biostatistician, and Sureka Bollepalli, Endocrinologist (Pediatrics Epidemiology Center, University of South Florida).
Data and Safety Monitoring Board: Edwin AM Gale, Chair (University of Bristol), Gisela Dahlquist, Bioethicist (University of Umeaa), Mikael Knip (University of Helsinki) and Mark Peakman (King’s College London).
Central Laboratories - Steno Diabetes Center Central Laboratory: Merete Frandsen, Head, Helle Niebling, Coordinating Technician and Charlotte Leth, Technician. University Hospital of Zurich Central Laboratory: Arnold von Eckardstein, Head of Institute of Clinical Chemistry, Stephan Regenass, Laboratory Director Clinical Immunology, Thorsten Hornemann, Lab IT Coordinator, and Isabelle Peereboom and Monika Seiler, Coordinating Technicians.
Investigators (numbers in parentheses are screened/randomised subjects, respectively, after which local funding sources are listed if any) -
Denmark: Hans-Henrik Lervang, Site PI, Mette Pilegaard Rasmussen, Site coordinator and Lene Holm Pedersen, Technician (Aalborg Hospital, 1/1). Joan Bach Nielsen and Jens Friis Bak, Site PIs, Jørgen Rungby, Investigator (Aarhus University Hospital, 7/4). Hans Perrild, Site PI and Anne Frederiksen, Study Nurse (Bispebjerg University Hospital, 1/1). Thomas Mandrup-Poulsen, Site PI, Linda Pickersgill, Central Site Coordinator, Lise Tarnow, Head, Clinical Research Unit, Henrik Ullits Andersen and Ulla Bjerre-Christensen, Investigators, Birgitte Hemmingsen, Study Nurse, Hanne Foght, Central Site Technician, Annette Thyde, Central Site Administrator (Steno Diabetes Center, Gentofte, 4/3, Øresund Diabetes Academy, Novo Nordisk). Germany: Nanette Schloot, Site PI, Bettina Nowotny, Christian Herder, Dorothea Krog and Sabine Kahl, Investigators, and Petra Heidkamp, Study Nurse (German Diabetes Center Duesseldorf, 7/6). Klaus Badenhoop, Site PI, Gesine Meyer, Investigator, Maria Sandler, Site Coordinator, and Ellen Althaus, Study Nurse (University of Frankfurt-am-Main, 8/6). Anette-G. Ziegler, Site PI, David Wiesenäcker, Maike Wallner and Eleni Giannopoulou, Site Coordinators, Markus Walter, Investigator, and Melanie Bunk and Melanie Herbst, Study Nurses (Munich University of Technology, Forschergruppe Diabetes e.V., and Helmholtz Center Munich, 16/13, Charlotte Fievet Foundation, Forschergruppe Diabetes e.V., and The German Center for Diabetes Research, Helmholtz Center Munich). Bernhard Böhm, Country Coordinator and Site PI, Silke Rosinger, Site Coordinator, and Julia Aufschild, Sigrun Merger and Roza Blagieva, Investigators (Ulm University, 14/10). Holland: Eelco de Koning, Site PI and Coordinator, Bart Roep and Fleur Kleijwegt, Investigators, and Marja Dijk, Study Nurse (Leiden University Medical Center, 16/14). Italy: Paolo Pozzilli, Site PI, Chiara Guglielmi, Site Coordinator, Nicoletta Onori, Giovanni Sironi and Andrea Soare, Investigators, and Luciana Valente, Technician (University Campus Biomedico, Rome, 5/4). Spain: Luis Castaño, Site PI, Federico Vázquez, Site Coordinator, Sonia Gaztambide and Javier González Mielgo, Investigators, and Rosa María Axpe Pascual, Study Nurse (Hospital Universitario Cruces, UPV/EHU, Ciberdem, 3/1). Ana M Wägner, Country Coordinator and Site PI, Maria del Pino Alberiche, María A García Núñez, Dunia Marrero, Javier Nóvoa, and Alicia Diez, Investigators, and Lucía Martin and Jose Aide, Study Nurses (Hospital Universitario Insular de Gran Canaria, Las Palmas de Gran Canaria, 1/1). Didac Mauricio, Site PI, Marta Hernández Garcia, Site Coordinator, and Lidia Carrasco and Sala Núria Garcia, Study Nurses (Hospital Arnau de Vilanova, Lleida, 3/3). Switzerland: Marc Donath, Site PI, Patrizia Zala, Site Coordinator, and Eleonora Seelig and Katarina Timper, Investigators (University Hospital Basel, 3/2).
The Type 1 Diabetes TrialNet Canakinumab Study Group investigators
Steering committee present members: Australia P Colman, J Wentworth (Walter and Eliza Hall Institute of Medical Research, Parkville, VIC); Canada D Wherrett (University of Toronto Toronto, ON); Finland O Simell (Hospital District of Southwest Finland, Turku); Italy E Bosi (San Raffaele Hospital, Milan), M G Roncarolo (San Raffaele Scientific Institute, Milan); UK P Bingley (University of Bristol, Bristol), M Peakman (Guy’s, King’s, and St Thomas’ School of Medicine, London); USA J S Skyler (Chairman; University of Miami Diabetes Research Institute, Miami, FL); M Anderson, S Gitelman (University of California San Francisco, San Francisco, CA), M Atkinson, M Clare-Salzler, D Schatz, W Winter (University of Florida, Gainesville, FL), K Bourcier (National Institute of Allergy and Infectious Diseases, Bethesda, MD), D Becker, M Trucco (University of Pittsburgh, Pittsburgh, PA), J Blum, L DiMeglio (Indiana University, Indiana, PA), J Buckner (Benaroya Research Institute, Seattle, WA), H P Chase, G S Eisenbarth*, P Gottlieb (University of Colorado Barbara Davis Center for Childhood Diabetes, Aurora, CO), C G Fathman, D M Wilson (Stanford University, Stanford, CA), R Goland (Columbia University, New York, NY), G Grave (Eunice Kennedy Shriver National Institute of Child Health and Human Development, Rockville, MD), C Greenbaum (Benaroya Research Institute, Seattle, WA), B Hering, A Moran (University of Minnesota, Minneapolis, MN), K Herold, R Sherwin (Yale University, New Haven, CT), R Insel (Juvenile Diabetes Research Foundation, New York, NY), J P Krischer (University of South Florida, Tampa, FL), J Marks, A Pugliese (University of Miami Diabetes Research Institute, Miami, FL), J P Palmer (University of Washington, Seattle, WA), P Raskin, M Siegelman (University of Texas Southwestern Medical School, Dallas, TX), W Russell, J Thomas (Vanderbilt University, Nashville, TN), J Fradkin (ex-officio), E Leschek (ex-officio), L Spain (ex-officio), P Savage (NIDDK, Bethesda, MD). Steering committee past members: Australia L Harrison (Walter and Eliza Hall Institute of Medical Research, Parkville, VIC); Canada J Mahon (University of Western Ontario, London, ON); Finland K Nanto-Salonen (Hospital District of Southwest Finland, Turku); Germany A Ziegler (Helmholtz Center Munich Diabetes Research Group at the Technical University of Munich, Munich); USA C Benoist, T Orban (Joslin Diabetes Center, Boston, MA), J Bluestone (University of California San Francisco, San Francisco, CA), D Brown, J Wagner (University of Minnesota, Minneapolis, MN), C Cowie (NIDDK, Bethesda, MD), S Jordan (Cedars-Sinai Medical Center, Los Angeles, CA), F R Kaufman, R Parkman (Childrens Hospital Los Angeles, Los Angeles, CA), J M Lachin (George Washington University, Washington, DC), G Nepom (Benaroya Research Institute, Seattle, WA), M Pescovitz†, H Rodriguez (Indiana University, Indiana, PA), J Peyman, J Ridge (National Institute of Allergy and Infectious Diseases, Bethesda, MD). Executive committee present members: USA J S Skyler (University of Miami Diabetes Research Institute, Miami, FL), K Bourcier (National Institute of Allergy and Infectious Diseases, Bethesda, MD), C J Greenbaum (Benaroya Research Institute, Seattle, WA), J P Krischer (University of South Florida, Tampa, FL), E Leschek, P Savage, L Spain (NIDDK, Bethesda, MD), L Rafkin (University of Miami Diabetes Research Institute, Miami, FL). Executive committee past members: USA C Cowie, S Malozowski (NIDDK, Bethesda, MD), M Foulkes, H Krause-Steinrauf, J M Lachin, S J Zafonte (George Washington University, Washington, DC), J Peyman, J Ridge (National Institute of Allergy and Infectious Diseases, Bethesda, MD). Chairman’s office (University of Miami Diabetes Research Institute, Miami, FL): J S Skyler, C J Greenbaum, N S Kenyon, L Rafkin, I Santiago, J M Sosenko. TrialNet coordinating center (University of South Florida, Tampa, FL, USA) present members: J P Krischer, B Bundy, A L Ritzie, D M Amado, M Abbondondolo, T Adams, P Alies, F Badias, C Beam, M Boonstra, D Boulware, D Cuthbertson, C Eberhard, J Ford, J Ginem, H Guillette, B Hays, M Henry, P Law, C Linton, S Liu, J Lloyd, S Muller, R O’Donnell, Y Parrimon, K Paulus, J Pilger, J Ramiro, A Roberts, K Sadler, A Terry, M Wootten, P Xu, K Young. TrialNet coordinating center (University of South Florida, Tampa, FL, USA) past members: M Bassi, D Freeman, M Granger, M Kieffer, L Nallamshetty, A Shor. Data safety and monitoring board present members: USA E Blumberg (Chair; University of Pennsylvania, Philadelphia, PA); G Beck (Cleveland Clinic, Cleveland, OH), J Braun (University of California Los Angeles, Los Angeles, CA), D Brillon (Cornell University, Ithaca, NY), R Gubitosi-Klug (Case Western Reserve University, Cleveland, OH), L Laffel (Joslin Diabetes Center, Boston, MA), R Veatch (Georgetown University, Washington, DC), D Wallace (Research Triangle Institute, Durham, NC). Data safety and monitoring board past members: Canada B Zinman (University of Toronto, Toronto, ON); Denmark J Nerup (University of Copenhagen, Copenhagen); Sweden A Lernmark (Lund University, Lund); USA B Lo (University of California San Francisco, San Francisco, CA), H Mitchell (Rho, Chapel Hill, NC), A Naji (University of Pennsylvania, Philadelphia, PA), T Orchard (University of Pittsburgh, Pittsburgh, PA), M Steffes (University of Minnesota, Minneapolis, MN), A Tsiatis (North Carolina State University, Raleigh, NC). Infectious disease safety committee: USA B Loechelt (Medical Monitor; Children’s National Medical Center, Washington, DC), L Baden (Harvard University, Cambridge, MA), M Green (University of Pittsburgh, Pittsburgh, PA), A Weinberg (University of Colorado, Boulder, CO). Laboratory directors: USA S Babu, L Yu, G S Eisenbarth (University of Colorado Barbara Davis Center for Childhood Diabetes, Aurora, CO), S Marcovina, J P Palmer (University of Washington, Seattle, WA), A Weinberg (University of Colorado, Boulder, CO), W Winter (University of Florida, Gainesville, FL). Protocol committee: UK M Peakman (King’s College London, London); USA A Moran (Chair; University of Minnesota, Minneapolis, MN), C Greenbaum, S Sanda (Benaroya Research Institute, Seattle, WA), E Leschek, L Spain (NIDDK, Bethesda, MD), D Matheson (University of Miami Diabetes Research Institute, Miami, FL), M Pescovitz† (Indiana University, Indiana, PA), A Pugliese, J S Skyler, L Rafkin (University of Miami Diabetes Research Institute, Miami, FL). Clinical Center Staff involved in this protocol: Benaroya Research Institute (Seattle, WA, USA) C Greenbaum, J Bollyky, S Sanda, D Tridgell, M McCulloch-Olson, H Vendettuoli, D Hefty, M Ramey, C Webber, K Kuhns, N Hilderman, A Dove, M Hammond, J Klein, E Batts; Columbia University (New York, NY, USA) R Goland, E Greenberg, M P Gallagher, J Trast, M Chan, K Smith; Indiana University (Indianapolis, IN, USA) M Pescovitz†, L DiMeglio, C Evans-Molina, M Spall, J Terrell, R Hufferd, L Ford; Stanford University (Stanford, CA, USA) D M Wilson, B A Buckingham, T Aye, T Esrey, A Soto, B Baker, B Berry; University of California San Francisco (San Francisco, CA, USA) S E Gitelman, S M Rosenthal, M Anderson, S Adi, K Breen, C Hamilton; University of Florida (Gainesville, FL, USA) D Schatz, M Haller, M Clare-Salzler, M Atkinson, R Cook, D Mancini, A Abraham, J Ferguson, G Cole; University of Miami Diabetes Research Institute (Miami, FL, USA) J B Marks, A Pugliese, D Matheson, C Blaschke, L Arazo, R Arce, M Cisneros, B Acosta; University of Minnesota (Minneapolis, MN, USA) A Moran, B Nathan, J Wagner, B Hering, J Smith, A Street, J Leschyshyn, C Gibson, C Kwong; University of Pittsburgh (Pittsburgh, PA, USA) D Becker, I Libman, K Riley, K Delallo, N Gurtunca, M Trucco, B Copemen, B Elnyczky; University of Texas Southwestern Medical School (Dallas, TX, USA) P Raskin, J Ricahrd, E Roe, S Mirfakhraee, P White, M Alford, S Fernandez, J Arthur, M Hutchins, R Davis; University of Toronto (Toronto, ON, Canada) D K Wherrett, R Kovalakovska, L A Eisel, B Ahenkorah, M Sriskandarajah, M-J Ricci, J Cevallos, R Steger; Yale University (New Haven, CT, USA) K C Herold, L Feldman, R Sherwin. *Dr Eisenbarth died in November, 2012. †Dr Pescovitz died in December, 2010.
For the Type 1 Diabetes TrialNet see http://www.diabetestrialnet.org/
For AIDA see http://www.aidastudy.org
Footnotes
Author Contributions: SMC researched data and wrote, reviewed, and edited the manuscript. MLK designed and conducted experiments and interpreted data. YGC designed and interpreted experiments. SJ and XW developed/tested algorithms, conducted data analysis and reviewed manuscript. CJG and TMP discussed data and reviewed and edited the manuscript. The AIDA and TrialNet Study groups provided subject specimens and clinical data as well as reviewed the manuscript. MJH designed experiments, analyzed data, wrote, reviewed, and edited the manuscript. MJH is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Conflict of Interest Disclosure: The authors declare no commercial or financial conflict of interest.
References
- 1.Mandrup-Poulsen T, Pickersgill L, Donath MY. Blockade of interleukin 1 in type 1 diabetes mellitus. Nat Rev Endocrinol. 2010;6:158–166. doi: 10.1038/nrendo.2009.271. [DOI] [PubMed] [Google Scholar]
- 2.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Stiller CR, Dupre J, Gent M, Jenner MR, Keown PA, Laupacis A, Martell R, Rodger NW, von Graffenried B, Wolfe BM. Effects of cyclosporine immunosuppression in insulin-dependent diabetes mellitus of recent onset. Science. 1984;223:1362–1367. doi: 10.1126/science.6367043. [DOI] [PubMed] [Google Scholar]
- 4.Keymeulen B, Vandemeulebroucke E, Ziegler AG, Mathieu C, Kaufman L, Hale G, Gorus F, Goldman M, Walter M, Candon S, Schandene L, Crenier L, De Block C, Seigneurin JM, De Pauw P, Pierard D, Weets I, Rebello P, Bird P, Berrie E, Frewin M, Waldmann H, Bach JF, Pipeleers D, Chatenoud L. Insulin needs after CD3-antibody therapy in new-onset type 1 diabetes. N Engl J Med. 2005;352:2598–2608. doi: 10.1056/NEJMoa043980. [DOI] [PubMed] [Google Scholar]
- 5.Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, Becker DJ, Gitelman SE, Goland R, Gottlieb PA, Marks JB, McGee PF, Moran AM, Raskin P, Rodriguez H, Schatz DA, Wherrett D, Wilson DM, Lachin JM, Skyler JS. Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N Engl J Med. 2009;361:2143–2152. doi: 10.1056/NEJMoa0904452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandler S, Andersson A, Hellerstrom C. Inhibitory effects of interleukin 1 on insulin secretion, insulin biosynthesis, and oxidative metabolism of isolated rat pancreatic islets. Endocrinology. 1987;121:1424–1431. doi: 10.1210/endo-121-4-1424. [DOI] [PubMed] [Google Scholar]
- 7.Spinas GA, Hansen BS, Linde S, Kastern W, Molvig J, Mandrup-Poulsen T, Dinarello CA, Nielsen JH, Nerup J. Interleukin 1 dose-dependently affects the biosynthesis of (pro)insulin in isolated rat islets of Langerhans. Diabetologia. 1987;30:474–480. doi: 10.1007/BF00279615. [DOI] [PubMed] [Google Scholar]
- 8.Mandrup-Poulsen T, Bendtzen K, Nerup J, Dinarello CA, Svenson M, Nielsen JH. Affinity-purified human interleukin I is cytotoxic to isolated islets of Langerhans. Diabetologia. 1986;29:63–67. doi: 10.1007/BF02427283. [DOI] [PubMed] [Google Scholar]
- 9.Maedler K, Sergeev P, Ris F, Oberholzer J, Joller-Jemelka HI, Spinas GA, Kaiser N, Halban PA, Donath MY. Glucose-induced beta cell production of IL-1beta contributes to glucotoxicity in human pancreatic islets. J Clin Invest. 2002;110:851–860. doi: 10.1172/JCI15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada K, Takane-Gyotoku N, Yuan X, Ichikawa F, Inada C, Nonaka K. Mouse islet cell lysis mediated by interleukin-1-induced Fas. Diabetologia. 1996;39:1306–1312. doi: 10.1007/s001250050574. [DOI] [PubMed] [Google Scholar]
- 11.Dayer-Metroz MD, Wollheim CB, Seckinger P, Dayer JM. A natural interleukin 1 (IL-1) inhibitor counteracts the inhibitory effect of IL-1 on insulin production in cultured rat pancreatic islets. J Autoimmun. 1989;2:163–171. doi: 10.1016/0896-8411(89)90152-2. [DOI] [PubMed] [Google Scholar]
- 12.Zumsteg U, Reimers JI, Pociot F, Morch L, Helqvist S, Brendel M, Alejandro R, Mandrup-Poulsen T, Dinarello CA, Nerup J. Differential interleukin-1 receptor antagonism on pancreatic beta and alpha cells. Studies in rodent and human islets and in normal rats. Diabetologia. 1993;36:759–766. doi: 10.1007/BF00401148. [DOI] [PubMed] [Google Scholar]
- 13.Dinarello CA, Simon A, van der Meer JW. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11:633–652. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaldunski M, Jia S, Geoffrey R, Basken J, Prosser S, Kansra S, Mordes JP, Lernmark A, Wang X, Hessner MJ. Identification of a serum-induced transcriptional signature associated with type 1 diabetes in the BioBreeding rat. Diabetes. 2010;59:2375–2385. doi: 10.2337/db10-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dayer-Metroz M-D, Duhamel D, Rufer N, Izui S, Carmichaels D, Wollheim CB, Thompson RC, Dayer J-M. IL-1 receptor antagonist delays spontaneous autoimmune diabetes in BB rats. European Journal of Clinical Investigation. 1992;22 Abstract 285. [Google Scholar]
- 16.Nicoletti F, Di Marco R, Barcellini W, Magro G, Schorlemmer HU, Kurrle R, Lunetta M, Grasso S, Zaccone P, Meroni P. Protection from experimental autoimmune diabetes in the non-obese diabetic mouse with soluble interleukin-1 receptor. Eur J Immunol. 1994;24:1843–1847. doi: 10.1002/eji.1830240818. [DOI] [PubMed] [Google Scholar]
- 17.Hara N, Alkanani AK, Dinarello CA, Zipris D. Modulation of virus-induced innate immunity and type 1 diabetes by IL-1 blockade. Innate Immun. 2013;20:574–584. doi: 10.1177/1753425913502242. [DOI] [PubMed] [Google Scholar]
- 18.Larsen CM, Faulenbach M, Vaag A, Volund A, Ehses JA, Seifert B, Mandrup-Poulsen T, Donath MY. Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N Engl J Med. 2007;356:1517–1526. doi: 10.1056/NEJMoa065213. [DOI] [PubMed] [Google Scholar]
- 19.Cavelti-Weder C, Babians-Brunner A, Keller C, Stahel MA, Kurz-Levin M, Zayed H, Solinger AM, Mandrup-Poulsen T, Dinarello CA, Donath MY. Effects of gevokizumab on glycemia and inflammatory markers in type 2 diabetes. Diabetes Care. 2012;35:1654–1662. doi: 10.2337/dc11-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moran A, Bundy B, Becker DJ, DiMeglio LA, Gitelman SE, Goland R, Greenbaum CJ, Herold KC, Marks JB, Raskin P, Sanda S, Schatz D, Wherrett DK, Wilson DM, Krischer JP, Skyler JS, Pickersgill L, de Koning E, Ziegler AG, Boehm B, Badenhoop K, Schloot N, Bak JF, Pozzilli P, Mauricio D, Donath MY, Castano L, Wagner A, Lervang HH, Perrild H, Mandrup-Poulsen T. Interleukin-1 antagonism in type 1 diabetes of recent onset: two multicentre, randomised, double-blind, placebo-controlled trials. Lancet. 2013;381:1905–1915. doi: 10.1016/S0140-6736(13)60023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Jia S, Geoffrey R, Alemzadeh R, Ghosh S, Hessner MJ. Identification of a molecular signature in human type 1 diabetes mellitus using serum and functional genomics. J Immunol. 2008;180:1929–1937. doi: 10.4049/jimmunol.180.3.1929. [DOI] [PubMed] [Google Scholar]
- 22.Levy H, Wang X, Kaldunski M, Jia S, Kramer J, Pavletich SJ, Reske M, Gessel T, Yassai M, Quasney MW, Dahmer MK, Gorski J, Hessner MJ. Transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes. Genes Immun. 2012;13:593–604. doi: 10.1038/gene.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YG, Cabrera SM, Jia S, Kaldunski ML, Kramer J, Cheong S, Geoffrey R, Roethle MF, Woodliff JE, Greenbaum CJ, Wang X, Hessner MJ. Molecular signatures differentiate immune States in type 1 diabetic families. Diabetes. 2014;63:3960–3973. doi: 10.2337/db14-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Zhang H, Liu Y, Wang K, Feng Y, Liu M, Xiao X. KLF4 regulates the expression of interleukin-10 in RAW264.7 macrophages. Biochem Biophys Res Commun. 2007;362:575–581. doi: 10.1016/j.bbrc.2007.07.157. [DOI] [PubMed] [Google Scholar]
- 25.Peter MR, Jerkic M, Sotov V, Douda DN, Ardelean DS, Ghamami N, Lakschevitz F, Khan MA, Robertson SJ, Glogauer M, Philpott DJ, Palaniyar N, Letarte M. Impaired resolution of inflammation in the Endoglin heterozygous mouse model of chronic colitis. Mediators Inflamm. 2014;2014:767185. doi: 10.1155/2014/767185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ablamunits V, Henegariu O, Hansen JB, Opare-Addo L, Preston-Hurlburt P, Santamaria P, Mandrup-Poulsen T, Herold KC. Synergistic reversal of type 1 diabetes in NOD mice with anti-CD3 and interleukin-1 blockade: evidence of improved immune regulation. Diabetes. 2012;61:145–154. doi: 10.2337/db11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagni PP, Bresson D, Rodriguez-Calvo T, Bel Hani A, Manenkova Y, Amirian N, Blaszczak A, Faton S, Sachithanantham S, von Herrath MG. Combination therapy with an anti-IL-1beta antibody and GAD65 DNA vaccine can reverse recent-onset diabetes in the RIP-GP mouse model. Diabetes. 2014;63:2015–2025. doi: 10.2337/db13-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain MJ, Peakman M, Gallati H, Lo SS, Hawa M, Viberti GC, Watkins PJ, Leslie RD, Vergani D. Elevated serum levels of macrophage-derived cytokines precede and accompany the onset of IDDM. Diabetologia. 1996;39:60–69. doi: 10.1007/BF00400414. [DOI] [PubMed] [Google Scholar]
- 29.Dogan Y, Akarsu S, Ustundag B, Yilmaz E, Gurgoze MK. Serum IL-1beta, IL-2, and IL-6 in insulin-dependent diabetic children. Mediators Inflamm. 2006;2006:59206. doi: 10.1155/MI/2006/59206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crino A, Schiaffini R, Manfrini S, Mesturino C, Visalli N, Beretta Anguissola G, Suraci C, Pitocco D, Spera S, Corbi S, Matteoli MC, Patera IP, Manca Bitti ML, Bizzarri C, Pozzilli P. A randomized trial of nicotinamide and vitamin E in children with recent onset type 1 diabetes (IMDIAB IX) Eur J Endocrinol. 2004;150:719–724. doi: 10.1530/eje.0.1500719. [DOI] [PubMed] [Google Scholar]
- 31.Greenbaum CJ, Beam CA, Boulware D, Gitelman SE, Gottlieb PA, Herold KC, Lachin JM, McGee P, Palmer JP, Pescovitz MD, Krause-Steinrauf H, Skyler JS, Sosenko JM. Fall in C-peptide during first 2 years from diagnosis: evidence of at least two distinct phases from composite Type 1 Diabetes TrialNet data. Diabetes. 2012;61:2066–2073. doi: 10.2337/db11-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherry N, Hagopian W, Ludvigsson J, Jain SM, Wahlen J, Ferry RJ, Jr, Bode B, Aronoff S, Holland C, Carlin D, King KL, Wilder RL, Pillemer S, Bonvini E, Johnson S, Stein KE, Koenig S, Herold KC, Daifotis AG. Teplizumab for treatment of type 1 diabetes (Protege study): 1-year results from a randomised, placebo-controlled trial. Lancet. 2011;378:487–497. doi: 10.1016/S0140-6736(11)60931-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 36.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc Natl Acad Sci U S A. 2009;106:17031–17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olaru F, Jensen LE. Staphylococcus aureus stimulates neutrophil targeting chemokine expression in keratinocytes through an autocrine IL-1alpha signaling loop. J Invest Dermatol. 2010;130:1866–1876. doi: 10.1038/jid.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.