Abstract
Although poorly understood, early-life infection is predicted to affect brain microglial cells, making them hypersensitive to subsequent stimuli. To investigate this, we assessed gene expression in hippocampal tissue obtained from a previously published study reporting increased microglial cell activity and reduced hippocampal-dependent learning in neonatal piglets infected with porcine reproductive and respiratory syndrome virus (PRRSV), a virus that induces interstitial pneumonia. Infection altered expression of 455 genes, of which 334 were up-regulated and 121 were down-regulated. Functional annotation revealed that immune function genes were enriched among the up-regulated differentially expressed genes (DEGs), whereas calcium binding and synaptic vesicle genes were enriched among the down-regulated DEGs. Twenty-six genes encoding part of the microglia sensory apparatus (i.e., the sensome) were up-regulated (e.g. IL1R1, TLR2, and TLR4), whereas 15 genes associated with the synaptosome and synaptic receptors (e.g. NPTX2, GABRA2, and SLC5A7) were down-regulated. As the sensome may foretell microglia reactivity, we next inoculated piglets with culture medium or PRRSV at PD 7 and assessed hippocampal microglia morphology and function at PD 28 when signs of infection were waning. Consistent with amplification of the sensome, microglia from PRRSV piglets had enhanced responsiveness to chemoattractants, increased phagocytic activity, and secreted more TNFα in response to lipopolysaccharide and Poly I:C. Immunohistochemical staining indicated PRRSV infection increased microglia soma length and length-to-width ratio. Bipolar rod-like microglia not evident in hippocampus of control piglets, were present in infected piglets. Collectively, this study suggests early-life infection alters the microglia sensome as well as microglial cell morphology and function.
Keywords: perinatal, infection, hippocampus, microglia, sensome, synapse
Introduction
Neuroimmune factors are thought to play important roles in the etiology of psychiatric disorders such as autism spectrum disorders and schizophrenia. The supposition is predicated on the prenatal and early postnatal brain being vulnerable to infectious insults that, if present, affect development and increase risk for behavioral disorders later (Andersen, 2003; Rees and Inder, 2005). Pro-inflammatory cytokines produced by immune cells, rather than the pathogen per se are strongly implicated (Dantzer et al., 2008). Therefore, an important role for microglia seems likely as they are resident immune cells in the brain, produce pro-inflammatory cytokines in response to infection, and play a key role in development by phagocytizing neural progenitors and engaging in synaptic pruning. Indeed, untimely activation of microglia has been shown to reduce prenatal cortical neurogenesis, resulting in fewer cortical neurons in adulthood (Cunningham et al., 2013). Furthermore, rat pups infected with E. coli to induce acute neuroinflammation, showed less cognitive resilience as adults when exposed to another immune stressor (Bilbo et al., 2006; Bilbo and Schwarz, 2009). A working hypothesis is that early-life infection alters microglial cells in a manner that renders them hypersensitive to subsequent stimuli (Meyer et al., 2011). However, there is a gap of knowledge regarding the effects of early postnatal viral infection on microglia activity and their sensitivity to subsequent insults.
Recently, we have conducted studies using a neonatal domestic piglet model. The piglet immune system is similar to human as is growth and development of its gyrencephalic brain (Conrad and Johnson, 2015). In our previous studies with neonatal piglets, infection with porcine reproductive and respiratory syndrome virus (PRRSV) resulted in reduced neurogenesis and altered pyramidal neuron structure in hippocampus (Conrad et al., 2015), reduced gray and white matter in several brain regions (Conrad et al., 2014), and compromised performance in a hippocampal-dependent spatial T-maze task (Elmore et al., 2014). In addition, peripheral infection was associated with robust activation of microglia in the hippocampus (Elmore et al., 2014). These studies indicated PRRSV infection induced a neuroinflammatory response and affected brain development.
To significantly extend these findings and to address the gap of knowledge regarding infection and microglia activity and subsequent sensitivity, the present study first assessed gene expression patterns in hippocampal tissue obtained from our previously published study reporting increased microglial cell activation and reduced hippocampal-dependent learning in piglets infected with PRRSV (Elmore et al., 2014). Results of this analysis suggested infection up-regulated a number of genes that are part of the newly defined microglia sensome (Hickman et al., 2013). As this sensory apparatus may foretell microglia reactivity, we subsequently assessed the effects of peripheral infection on hippocampal microglia morphology and function. The important findings suggest early-life infection alters the sensome, rendering microglia more responsive to subsequent immune stimuli.
Materials and Methods
Animals, Housing, Virus Inoculation and Detection
All animal care and experimental procedures were in accordance with the NRC Guide for the Care and Use of Laboratory Animals and approved by the University of Illinois at Urbana-Champaign Institutional Animal Care and Use Committee. A detailed description of animals and housing can be found in Elmore et al. (Elmore et al., 2014) and Conrad et al. (Conrad et al., 2015). Briefly, naturally farrowed crossbred piglets obtained at PD 2 were assigned to either control (control, sham-inoculated) or PRRSV treatment balancing for sex, body weight and litter of origin. Piglets were housed individually in cages (0.87 m × 0.87 m × 0.49 m; L × W × H) in biocontainment chambers appropriate for BSL-2 studies and fed a reconstituted nutritionally balanced commercial milk replacer (Advance Liqui-Wean, Milk Specialties). Milk was supplied at 285 mL/kg of body weight by an automated pump feeding system as previously described (Elmore et al., 2014). At PD7, pigs were inoculated intranasal with either 1 mL of sterile saline or 1 mL of 1 × 105 50% tissue culture infected dose (TCID50) of live PRRSV (strain P129-BV), obtained from the School of Veterinary Medicine at Purdue University (West Lafayette, Indiana). PRRSV, a single-stranded RNA virus, infects mononuclear myeloid cells in the lungs inducing an interstitial pneumonia (Chen et al., 2014; Duan et al., 1997). Rectal temperature and feeding score (0 = no attempt to consume the milk; 1 = attempted to consume the milk, but did not finish within 1 min; 2 = consumed all of the milk within 1 min) were determined daily beginning PD2. Piglets were euthanized at PD28 [21 d post inoculation (dpi)] and brains were dissected for collection of hippocampal tissue.
To detect the presence of PRRSV, serum samples collected at 0, 7, and 21 dpi were analyzed for PRRSV by quantitative PCR (qPCR) at the Veterinary Diagnostic Laboratory (College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Urbana, IL) to verify control piglets were PRRSV-free and that piglets inoculated with PRRSV became infected. All RNA-sequenced fragments from hippocampal tissue (described in the following section) were blasted against the PRRSV genome (PRRSV isolate 16244B, accession #AF046869, http://www.ncbi.nlm.nih.gov/nuccore/AF046869.1). The number of aligned transcripts was recorded for each sample.
Hippocampal Gene Expression
RNA Extraction, cDNA Synthesis, and RNA-Sequencing Library Preparation
To assess the neuroimmune effects of early life peripheral infection, gene expression in hippocampal tissue collected at PD 28 (21 dpi) from our previously published study wherein piglets were experimentally infected with PRRSV was assessed (Elmore et al., 2014). The experimental design, animal housing, feeding, virus inoculation and tissue collection protocol in the study by Elmore et al. (2014) was the same as in the current study. RNA was extracted using an AllPrep DNA/RNA Mini Kit (Qiagen) following the manufacturer’s protocol. RNA concentrations were determined using a NanoDrop Spectrophotometer (NanoDrop Technologies, Inc.).Samples were analyzed by the Carver High-Throughput DNA Sequencing and Genotyping Unit (HTS lab, University of Illinois, Urbana, IL, USA) using an Agilent bioanalyzer DNA7500 DNA chip (Agilent Technologies) to determine RNA integrity as well as the presence/absence of genomic DNA (gDNA). Only RNA samples with an RNA integrity number (RIN) greater than 7 were used for sequencing. This high-quality RNA was sent to the HTS lab (University of Illinois, Urbana, IL, USA) for the generation of TruSeq Stranded RNA-seq libraries using the TruSeq Stranded RNA Sample Preparation Kit (Illumina Inc.) following standard protocols. Briefly, messenger RNA was selected from 1ug of high quality, DNase treated, total RNA and first-strand synthesis performed with a random hexamer and SuperScript II (Life Technologies). Double stranded DNA was blunt-ended, 3’-end A-tailed, and ligated to indexed adaptors. The adaptor –ligated double-stranded cDNA was amplified by PCR for 10 cycles with the Kapa HiFi polymerase (Kapa Biosystems) to reduce the likeliness of multiple identical reads due to preferential amplification. The final libraries were quantified using a Qubit (Life Technologies) and the average size was determined on the bioanalyzer and diluted to 10nM. The 10nM dilution was further quantified by qPCR on an ABI 7900 (Applied Biosystems), which resulted in a high accuracy quantification for consistent pooling of barcoded libraries and maximized the number of clusters in the Illumina flowcell.
Illumina RNA-Sequencing
RNA-seq Illumina sequencing was performed by the HTS lab (University of Illinois, Urbana, IL, USA). Briefly, the libraries were multiplexed and loaded onto 8-lane flowcells for cluster formation and sequenced on an Illumina HiSeq2000 (Illumina, Inc.). One of the lanes was loaded with a PhiX Control library that provides a balanced genome for the calculation of the matrix, phasing and prephasing, which are essential for accurate base calling. The libraries were sequenced from both ends (paired-end sequencing) of the molecules to a total read length of 100 bp from each end. The run generated .bcl files which were converted into demultiplexed, compressed fastq files using Casava 1.8.2 (Illumina Inc.).
RNA-Sequencing Data Analysis
An average of 35.7 million raw paired-end reads were produced for each sample, ranging from 29.7 to 43 million. Raw reads were trimmed three times sequentially for adapter contamination: A-tails, minimum quality score (20), and minimum length (20 bp) using Trim Galore v.0.3.3 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Unpaired reads were retained with a minimum length of 35 bp. Trimmed paired and unpaired reads were initially aligned to the swine reference genome (Sscrofa10.2) to filter out reads extending the maximum number of alignments (-M option) followed by alignment to the Ensemble swine reference transcriptome (-G) and alignment to the genome using Tophat v.2.2.10 (Kim et al., 2013). The number of allowed alignment hits (-g option) was 20. Furthermore the –read-realign-edit-dist option was set to 0, the –mate-inner-dist option to 120, the –mate-std-dev option to 260 and included the fr-firststrand option. Aligned bam files were assessed for differential gene expression using Cufflinks v.2.2.1 (Trapnell et al., 2010). First, Cufflinks was used to assemble transcripts for each sample using the fr-firststrand option, followed by Cuffmerge to merge the assembled transcripts from all samples with the reference transcripts. Cuffquant was used to pre-compute gene expression levels for each sample using the –u option, which more accurately weights reads mapping to multiple locations, and the fr-firststrand option. Finally Cuffdiff was used for differential gene expression analysis using the –u and fr-firststrand options. The RNA-seq data sets supporting the results of this article are available in the European Nucleotide Archive and are available under accession number PRJEB11625 (www.ebi.ac.uk/ena/data/view/PRJEB11625).
Visualization and Functional Enrichment Analysis of Differentially Expressed Genes (DEGs)
For visualization of DEGs, a heatmap was created based on the DEG dataset using the heatmap.2 function of the R package for visualization. Each column represents the DEGs of an individual pig (control or PRRSV), and each row represents the relative expression level (row z-score) of one gene across all pigs. The columns were clustered based on similarity of expression patterns of each individual pig. Functional enrichment analysis was performed using DAVID v6.7 (Huang et al., 2009). Official gene symbols of up- and down-regulated DEG were uploaded and analyzed separately using the human genome as the background. The human genome was selected as the reference set because only 122 of the 455 DEGs were mapped in the swine genome, whereas 384 were mapped in the human genome. Default settings of annotation categories and sources for DAVID were accepted. The defaulted stringency settings were used for Gene Ontology enrichment analysis. Enriched GO terms (cellular component, molecular function, and biological process) and KEGG pathways were reported.
Microglia Activity and Morphology after Infection
Immunostaining of Microglia and Assessment of Soma Morphology
Free floating coronal hippocampal sections (40 μm) were prepared as described previously (Conrad et al., 2015). Three sections of each hippocampus were used for immunostaining of microglia with a primary antibody, rabbit anti-Iba1 (1:1200, Wako Chemicals) and a biotinylated goat anti-rabbit secondary antibody (1:250, Jackson ImmunoResearcd Laboratories). The ABC system (Vector Laboratories) and diaminobenzidine kit (DAB; Sigma) were used for the chromogen. Digital images were captured using a Nanozoomer Digital Pathology System at 40× magnification (Hamamatsu Photonics). Consecutive images of the CA1 region of each section were exported at 40× digital zoom. Microglial soma morphology (length, width, and total area) was measured using ImageJ. Soma length was defined as the direct distance connecting the two farthest points of the soma, while soma width was the widest distance of the soma orthogonal to the soma length. Approximately 170 cells from hippocampal CA1 region of each piglet were measured for soma morphology in both treatment group (n=6 piglets/treatment).
Isolation of Hippocampal Microglia and Flow Cytometry
Hippocampal CD11b+ cells were isolated using the Neural Tissue Dissociation Kit P (Miltenyi Biotec) based on the protocol described by Nikodemova et al. (Nikodemova and Watters, 2012). Briefly, both hippocampi of a piglet were dissected, homogenized/dissociated using a gentleMACS Dissociator (Miltenyi Biotec) and enzymatically digested at 37°C for 35 min. Aggregates were removed by passing the cell suspension through a 40 μm cell strainer to yield a single cell suspension. Myelin was removed by passing cells through 25 mL of a 30% percoll-plus (GE Healthcare) solution. Cells were incubated in buffer (1× PBS with 2 mM EDTA and 0.5% BSA) containing anti-CD11b magnetic MicroBeads at 4°C for 15 min. CD11b+ cells were separated in a magnetic field using MS columns (Miltenyi Biotec). Isolated cells (~6 × 106) from both hippocampi of the same pig were resuspended in DMEM and pooled for subsequent in vitro assays. The initial plan was to isolate a pure microglia fraction for in vitro assays by staining with CD45 antibody, which is a surface marker that is widely used to differentiate microglia (CD11b+/CD45low to intermediate) from macrophages (CD11b+/CD45high) in rodents (Hickman et al., 2013; Nikodemova and Watters, 2012; Sedgwick et al., 1991). However, microglial yield and viability was low. Therefore, CD11b+ cells were used for in vitro assays. Microglia (CD45low to intermediate) made up 80-90% of the CD11b+ cells for both control and PRRSV piglets (Fig. S2.B).
Chemotaxis Assay
Migration of CD11b+ cells towards chemoattractants (recombinant porcine CCL-2; Innovative Research Inc.; and recombinant porcine IL-8; R&D systems) was measured using the Neuro Probe ChemoTx system (101-8; Neuro Probe, Inc.) following the manufacture’s protocol. In brief, DMEM (control), CCL-2 (100 ng/mL DMEM), or IL-8 (100 ng/mL DMEM) were added to the bottom chamber of a 96-well plate of a chemotaxis system and 30 μl of DMEM with CD11b+ cells (1 × 106 cell/mL) were added to the upper chamber. After 3 h at 37°C with 5% CO2, plates were centrifuged at 300 ×g for 10 min to separate cells that had migrated through the filter paper. Adherent cells were stained with Hoechst 33342 solution (5 μg/mL; Life technologies) and relative fluorescence was determined.
Phagocytosis Assay
Phagocytic activity of CD11b+ cells was analyzed using the Vybrant Phagocytosis Assay Kit (Life technologies) following the manufacture’s protocol with minor modifications. Cells were diluted (~6×105 cells/mL) in DMEM containing 10% heat-inactivated FBS, 1% penicillin-streptomycin, and 10 ng/mL recombinant porcine GM-CSF (R&D Systems) and plated with 5 intended replicates in a 96-well cell culture plate (100 μL/well). Cells were incubated for 24 h at 37°C 5% CO2 and then given fresh DMEM, LPS (1 ng/mL DMEM; E.coli 0127:B8, Sigma), or Poly I:C (1 μg/mL DMEM; GE Healthcare/Amersham Biosciences). After 3 h incubation, 100 μL of fluorescein-labeled BioParticles were added to each well to assess phagocytosis. After a 90 min incubation, the BioParticle suspension was removed by aspiration and 100 μL of a 1× trypan blue suspension was added for 15 min at room temperature to quench the fluorescence of extracellular (non-engulfed) BioParticles. The plate was read at an excitation of 485 nm and an emission of 528 nm.
Cytokine Production and Microglial Gene Expression
CD11b+ cells were diluted (~5×105 cells/mL) in DMEM containing 10% heat-inactivated FBS, 1% penicillin-streptomycin, and 10 ng/mL recombinant porcine GM-CSF, and plated in duplicate in 24-well plates (~5×105 cells/well). After 24 h at 37°C 5% CO2, DMEM, LPS (1 ng/mL DMEM), or Poly I:C (1 μg/mL DMEM) was added to wells. Following 8 h incubation, medium was collected and assayed for TNFα using a commercially available porcine-specific ELISA kit (R&D systems). Conditioned supernatants were assayed for lactate dehydrogenase using a commercially available detection kit (Roche Diagnostics Corporation) following the manufacturer’s instructions to verify treatments were not cytotoxic. Total RNA from microglia was isolated using TRI Reagent protocol (Sigma), and cDNA was synthesized using a QuantiTect Reverse Transcription Kit (QIAGEN). The real-time PCR was performed using the Applied Biosystems Taqman Gene Expression Assay Protocol. Genes of interest (Table S1) were compared with and expressed as fold change relative to the reference gene (GAPDH, Ss03374854_g1, Applied Biosystem).
Statistical Analysis
Data were analyzed using the mixed procedure of SAS 9.4. Body temperature, feeding score, body and brain weight, serum cytokines, MHCII expression, microglia morphology were analyzed as a complete randomized design with PRRSV infection as a fixed effect and pig within treatment as a random term. For variables measured over time (e.g. body temperature), a repeated statement was used in the model. Data from the in vitro studies were analyzed as a split-plot design with PRRSV infection as a fixed effect of the main plot and in vitro treatment as a fixed effect of the subplot. Significant difference was declared at P < 0.05, tendency at P < 0.10.
Results
Respiratory viral infection affects the hippocampal transcriptome in neonatal piglets
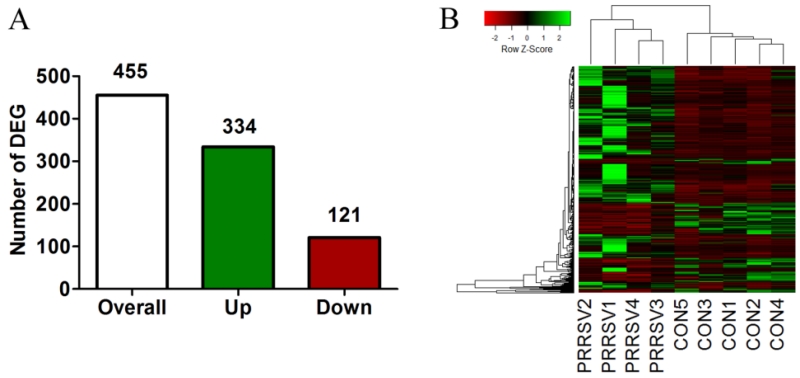
We first assessed gene expression patterns in hippocampal tissue obtained from our previously published study (Elmore et al., 2014). A total of 455 differentially expressed genes (DEGs) were revealed in the hippocampus in response to PRRSV (P < 0.05, Benjamini-Hochberg adjusted for multiplicity; Fig. 1A), of which 334 were up-regulated and 121 were down-regulated. The hierarchical clustering heatmap of the overall DEG pattern suggested a consistent effect of PRRSV infection on hippocampal transcriptomic profiles (Fig 1B). The Database for Annotation, Visualization and Integrated Discovery (DAVID v6.7) tool was used for functional enrichment analysis. Using DAVID, only 122 of the 455 DEGs were mapped with the swine genome, whereas 384 were mapped with the human genome. Due to limited gene annotation of the swine genome in the DAVID database, the human genome was selected as background for functional annotation.
Figure 1. Differentially expressed genes (DEGs) in hippocampus of neonatal piglets 21 dpi with PRRSV.
(A) The number of DEGs (FDR < 0.05) in PRRSV piglets (n=4) compared to control piglets (n=5). (B) Visualization of relative expression of the 455 DEGs in each piglet (green: up-regulation, red: down-regulation). Each row represents a gene and its relative expression. Piglets were clustered based on resemblance of overall expression pattern.
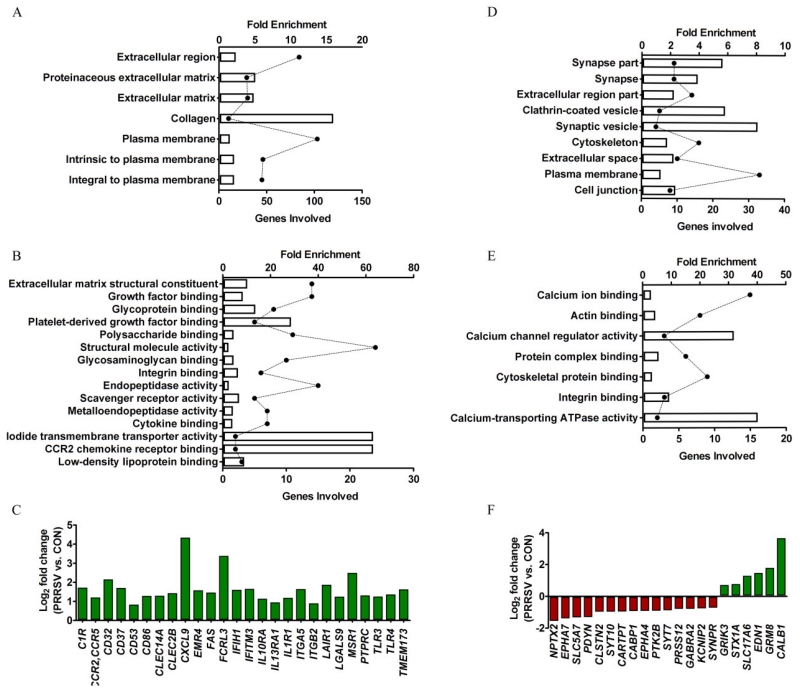
On the one hand, Gene Ontology (GO) Biological Process (BP) terms significantly enriched for up-regulated DEGs (Bonferroni-adjusted P < 0.05) were predominantly involved in immune response (e.g., adaptive and innate immune responses and classical complement activation pathway; Fig. 2A,B; Fig S1), indicating robust activation of neuroimmune defense mechanisms. Cytokine receptors (IL1R1, IL10RA, FAS and IL13RA1), chemokine receptors (CCR2/CCR5), complement (ITGB2), toll-like receptors (TLR3 and TLR4), integrins (ITGA5 and ITGB2), Fc receptor (CD32), scavenger receptor (MSR1), leukocyte antigens (CD37, CD53, CD86, and PTPRC), and transmembrane proteins associated with the immune response (IFITM3, and TMEM173) were coordinately up-regulated in the hippocampus of PRRSV piglets (Fig. 2C). The protein products of these genes were previously defined as being part of the microglial “sensome” (Hickman et al., 2013), which serves as a “sensory apparatus” to pathogen-associated molecular patterns and danger-associated molecular patterns. This was surprising because there was little evidence that the brain was infected by PRRSV: First, with the >33.5M sequenced cDNA fragments from hippocampal tissue of PRRSV piglets, a BLAST search of the PRRSV genome revealed on average only 8 hits, a number so small it is most likely explained by contaminating blood; second, virus was not detected in microglia isolated from PRRSV piglets even though PRRSV preferentially infects mononuclear myeloid cells (data not shown). In addition to immune-related genes, GO BP terms for extracellular matrix (ECM) organization, blood vessel development, and adhesion molecules were significantly enriched (Bonferroni-adjusted P < 0.05). Of note, adhesion molecules mediate migration and recruitment of immune cells across the endothelial barrier, although we have not seen evidence of brain infiltration of leukocytes in this model (Conrad et al., 2015).
Figure 2. Gene ontology (GO) functional enrichment analysis using DAVID bioinformatics 6.7 (A, B, D, E) and clusters of sensome (C) and synaptic genes (F).
(A, B) Top enriched GO biological process (A) and molecular function (B) associated with up-regulated DEGs (bars correspond to fold enrichment; solid circles correspond to number of gene involved; statistics for enrichment, Bonferroni-adjusted P < 0.05). (C) Manually identified DEGs that are part of the microglia sensome. (D, E) Top enriched GO cellular component (D) and molecular function (E) associated with down-regulated DEGs (statistics for enrichment, non-adjusted P < 0.05). (F) Manually identified DEGs encoding synaptic proteins.
On the other hand, GO Cellular Component (CC) and Molecular Function (MF) terms significantly enriched for down-regulated DEGs (unadjusted P < 0.05) were mainly associated with synapse and calcium channel, respectively (Fig. 2D, E). A closer look at the 21 DEGs encoding synaptic proteins that were manually identified revealed 15 were down-regulated and only 6 were up-regulated (Fig. 2F).
PRRSV infection causes sickness and affects microglia phenotype and morphology
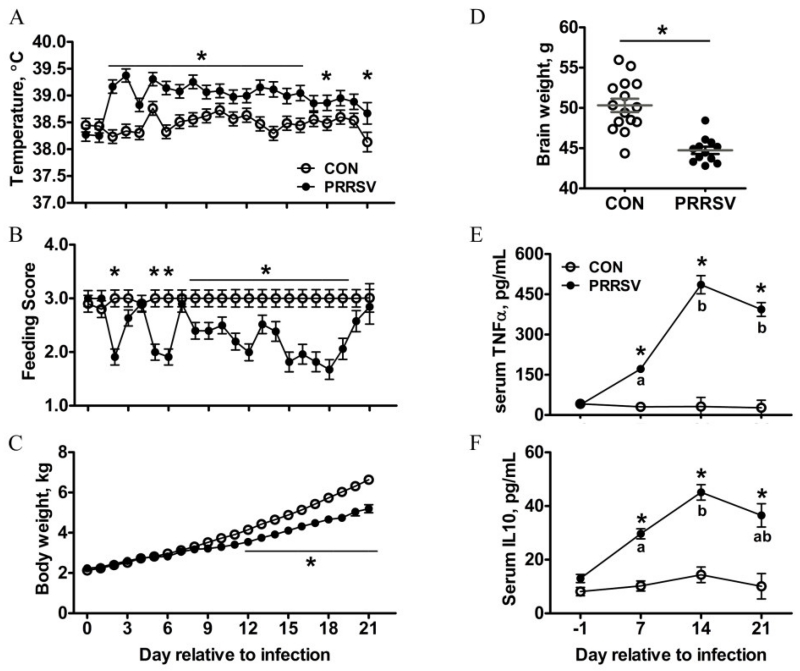
The RNA sequencing data suggested peripheral viral infection induced a robust neuroimmune response. Furthermore, the up-regulation of genes that are part of the microglia sensome suggested peripheral infection altered microglial cell activity, although this had not been previously established. Therefore, in a subsequent study piglets were inoculated (PD 7) intranasal with PRRSV or given sterile culture medium and euthanized 21d later (PD 28) to assess microglial cell phenotype, morphology, and function. All experimental procedures were identical to that described by Elmore et al.(Elmore et al., 2014), except spatial learning was not assessed. Repeated-measures ANOVA of body temperature, feeding activity, body weight, and serum TNFα and IL-10 revealed main effects of day and treatment, and a day × treatment interaction (at least P < 0.03 for all except the main effect of treatment for body weight was P < 0.06). In general, piglets showed clinical signs of infection beginning 3 d after inoculation and showed signs of recovery by the end of the study. At the end of the study, whole brain weight was less in PRRSV piglets (P < 0.001; Fig. 3B).
Figure 3. Assessment of PRRSV infection. (A-C) Daily rectal temperature (A), feeding score (B), and body weight (C) of control (n=15) and PRRSV (n=12) piglets from 0 to 21 dpi.
(D) Brain weight at 21 dpi. (E,F) Serum concentration of TNF-α (E) and IL-10 (F) at 0, 7, 14, and 21 dpi (n=6). Data are presented as mean ± SEM. Statistical analysis, one-way ANOVA with dpi as repeated measurement; * tukey-adjusted multiple comparisons between control and PRRSV piglets at specified time points, P < 0.05. a,b tukey-adjusted multiple comparison across different time points within the treatment, different small letter P < 0.05.
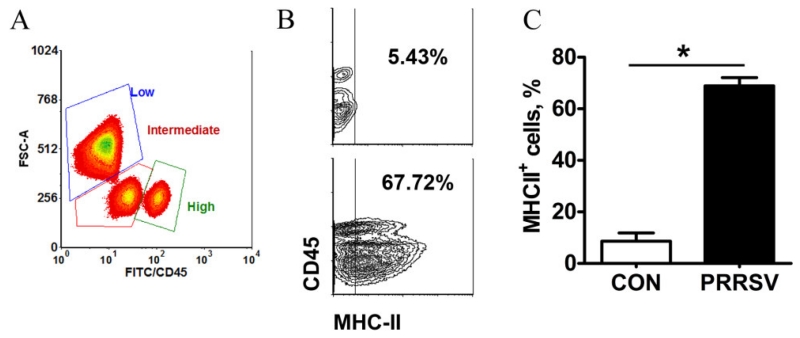
Microglia were isolated from hippocampal tissue at PD 28 following a procedure involving enzymatic digestion of tissue and separation of CD11b+ cells by antibody-coated magnetic beads as was used in rodents (Nikodemova and Watters, 2012). CD11b+ cells were subsequently stained for CD45, which differentiates microglia (CD11b+CD45low to intermediate) from infiltrating macrophage/monocytes (CD11b+CD45high) (Hickman et al., 2013; Nikodemova and Watters, 2012; Sedgwick et al., 1991). Three populations that differed in CD45 expression (low, intermediate, or high) were identified in CD11b+ cells by flow cytometry (Fig 4A). The three populations exhibited a similar side scatter, but the forward scatter of the CD45low fraction differed from that of CD45intermediate and CD45high, suggesting the CD45low cells were larger. This is consistent with a study in rats that found neonatal infection resulted in larger CD11b+ cells in brain (Bilbo et al., 2012). In PRRSV piglets the percentage of cells CD11b+CD45low decreased (P = 0.028) and CD11b+CD45intermediate increased (P = 0.051). Consistent with this, hippocampal expression of PTPRC, which encodes CD45, increased in response to infection. These results suggest respiratory viral infection shifted CD45low cells towards CD45intermediate and affected cell morphology (Fig S2B). No differences were observed for the CD45high fraction, suggesting infection did not result in infiltration of peripheral monocytes, although it should be noted that infiltrating monocytes can downregulate CD45 after entering the brain parenchyma (Wohleb et al., 2013). Consistent with an earlier study (Elmore et al., 2014), infection dramatically increased the proportion of isolated cells that stained for MHC-II, a marker of microglial cell activation (Fig 4B and C). The increase in MHC-II was evident in CD45+ cells (low, intermediate and high; Fig 4B).
Figure 4. PRRSV infection increased the percentage of MHC-II+ microglia.
(A) A representative density plot showing subpopulations of CD11b+ cells based on expression level of CD45 and FCS-A pattern. (B) A representative contour plot showing the gating threshold used to define MHC-II+ microglia for control (top) and PRRSV (bottom) piglets. (C) The percentage of MHC-II+ microglia at 21 dpi (n=6). Statistical analysis: one-way ANOVA; * P < 0.001. MHC-II was increased similarly in CD11b+ cells irrespective of level of CD45 expression (Fig. S2).
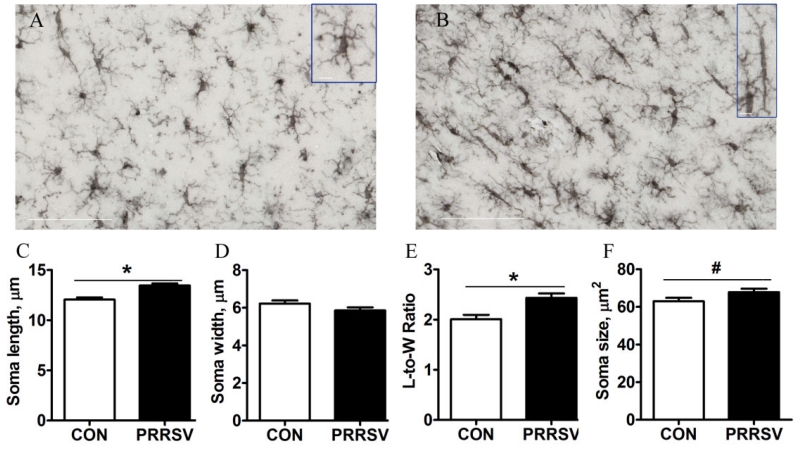
Coronal sections of the hippocampus from a subset of control and PRRSV piglets were stained with Iba-1to assess microglial cell morphology. Representative photomicrographs of microglia in the CA1 region of the hippocampus of a control and PRRSV piglet are shown in Fig 5, as are summary graphs showing effects of treatment on soma length, width, length-to-width ratio, and overall size. PRRSV infection increased soma length and the length-to-width ratio (P < 0.05), and tended to increase overall soma size (P < 0.10). An increased in microglial cell volume was also observed in adult rats that was suffered from bacterial infection at neonatal age (Bland et al., 2010). In healthy brain, microglia can show heterogeneous shapes, ranging from amoeboid to spindle or rod-shaped (Taylor et al., 2014). Herein we observed more bipolar rod-like microglia in PRRSV piglets.
Figure 5. PRRSV infection increased the number of microglia in CA1 region displaying an elongated soma at 21 dpi.
(A, B) Representative photomicrographs of Iba1-stained microglia in a control (A) and PRRSV (B) piglet (40×, scale bar = 100 μm). Inserts highlight a typical microglial cell seen in controls and a rod-like microglial cell seen in PRRSV piglets (63×, scale bar = 10 μm). (C, D, E, F) Soma length (C), width (D), length-to-width ratio (E), and soma size (F). Statistical analysis: one-way ANOVA; * P < 0.05, # P < 0.10; n=6)
PRRSV infection affects microglia function
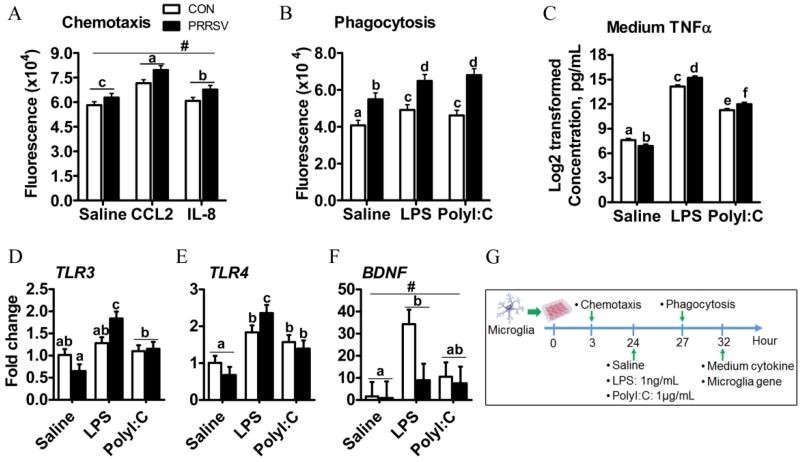
The effects of PRRSV infection on the transcriptional signature of hippocampal tissue as well as the phenotype and morphology of microglia suggested microglial cell function was altered. Therefore, we next sought to assess in microglia isolated from control and PRRSV piglets chemotaxis, phagocytosis, and cytokine production in response to LPS or Poly I:C. The yield of viable CD11b+CD45low-intermediate cells was low. Therefore, the in vitro functional assays were preformed using CD11b+ cells, which were estimated to comprise 80-90% microglia (i.e., CD11b+CD45low-intermediate) in both control and PRRSV piglets (Fig. 4). Microglia from PRRSV piglets tended to exhibit higher migratory activity than that of control piglets without addition of a chemoattractant (P = 0.056). Both CCL2 and IL-8 increased migration of microglia from control and PRRSV piglets (P < 0.001; Fig. 6A), with the response to CCL2 being more robust (P < 0.05). In another study phagocytic activity of microglia was assessed. Microglia from PRRSV piglets were more phagocytic than those from control piglets in both the absence and presence of in vitro stimulation with LPS and Poly I:C. A significant interaction between in vivo treatment (PRRSV vs control) and in vitro treatment (saline vs LPS or Poly I:C) indicated microglia from PRRSV piglets were hypersensitive to phagocytic stimuli (P = 0.012; Fig. 6B). This finding was supported by a similar interaction for TNFα secretion (Fig. 6C): An exaggerated TNFα response to both LPS (P < 0.001) and Poly I:C (P < 0.001) was seen in microglia from PRRSV pigs. Interestingly, basal TNFα secretion was lower in PRRSV microglia (P < 0.05).
Figure 6. Responses of microglia (CD11b+ cells) isolated from control (n=8) and PRRSV (n=6) piglets to in vitro stimulation.
(A) Microglia chemotaxis in response to CCL-2 or IL-8 (PRRSV, P = 0.056; in vitro treatment, P < 0.001; and PRRSV × in vitro treatment P = 0.27). (B) Microglia phagocytic activity in response to TLR agonists (PRRSV, P < 0.001; in vitro treatment P < 0.001; and PRRSV × in vitro treatment, P = 0.012). (C) Microglia secretion of TNFα in response to TLR agonists (PRRSV, P = 0.14; in vitro treatment P < 0.001; and PRRSV × in vitro treatment, P < 0.001). (D, E, F) Microglia mRNA expression of TLR3 (PRRSV, P = 0.596; in vitro treatment P < 0.001; and PRRSV × in vitro treatment, P = 0.003), TLR4 (PRRSV, P = 0.98; in vitro treatment P < 0.001; and PRRSV × in vitro treatment P = 0.023) and BDNF (PRRSV, P = 0.099; in vitro treatment, P = 0.021; and PRRSV × in vitro treatment, P = 0.166) in response to TLR agonists. (G) Timeline for in vitro experiments. Statistical analysis: two-way ANOVA, significance was declared at P < 0.05, and tendency at P < 0.10. Mean separation was performed and denoted for significance with different letters (a,b,c,d,e,f) when significant interaction is detected. # denotes tendency of significance (P < 0.10) due to main effect of infection.
Contrary to the up-regulation of microglia sensome genes in homogenized hippocampal tissue from PRRSV piglets (Fig. 2C), isolated microglia from PRRSV and control piglets expressed similar levels of TLR3 and TLR4 mRNA when unstimulated (Figure 6D, E). This may have been due to microglia being cultured for more than 24 h prior to isolating RNA, but we cannot exclude that the expression profile of hippocampus is also impacted by other cell types (i.e. astrocyte). Nonetheless, a significant interaction between in vivo PRRSV treatment and in vitro treatment affected microglia expression of TLR3 (P = 0.003) and TLR4 (P = 0.023) mRNA. Compared to microglia from control piglets, microglia from PRRSV piglets expressed more TLR3 and TLR4 mRNA after LPS (Fig. 6D,E). Poly I:C increased TLR3 and TLR4 mRNA similarly in microglia from control and PRRSV piglets (Fig. 6D,E). Finally, microglia from PRRSV piglets tended to have lower expression of the neurotrophic factor BDNF (P = 0.099) regardless of in vitro treatment. Collectively, these findings suggest respiratory viral infection heightens the activity of hippocampal microglial cells.
Discussion
The hippocampal transcriptome induced by PRRSV revealed a gene expression pattern indicative of (a) neuroinflammation as the up-regulated DEGs were predominantly involved in the immune response; and (b) reduced plasticity as synaptic genes were mostly down-regulated. This was evident 21 d after inoculation at a time piglets showed signs of recovery and in the absence of compelling evidence that virus entered the brain. By isolating microglia from adult mice and direct RNA sequencing, Hickman et al. (Hickman et al., 2013) reported microglia express a cluster of transcripts encoding proteins for sensing changes in the brain’s milieu, including endogenous ligands and microbes. In the present study, although the transcriptome was not assessed for isolated microglia, the hippocampus transcriptome showed up-regulation of 26 genes that are consistent with the microglia sensome, suggesting peripheral viral infection up-regulates this sensory apparatus. It warrants future research to characterize potential long-lasting consequences associated with up-regulated sensome genes. Nonetheless, these data are interesting in light of the proposed neuroimmune mechanism of psychiatric disorders and a recent transcriptome analysis of brain samples acquired through the Autism Tissue Program (Gupta et al., 2014). The analysis implicated dysregulated microglial responses in concert with altered neuronal activity-related genes in autism brains. The analysis further suggested activation of the type I interferon signaling pathway in autism brain, which is notable because type I interferons are part of the innate immune response to viruses and type I interferon genes were up-regulated in the present study. These findings are largely consistent with another study that used gene co-expression network analysis and identified two network modules highly correlated with autism: (a) a down-regulated neuronal module including genes involved in synaptic function; and (b) an up-regulated module that included genes related to astrocyte markers, markers of activated microglia, and immune and inflammatory responses (Voineagu et al., 2011). Increased microglia density and activation as well as elevated pro-inflammatory cytokines have been observed in autism spectrum disorders and schizophrenia patients of various ages (Doorduin et al., 2009; Li et al., 2009; Morgan et al., 2010). The present data showing increased expression of immune response genes and decreased expression of synaptic genes in hippocampus are also interesting in light of previous studies with this model showing decreased hippocampal neurogenesis, altered granule cell morphology, and impaired hippocampal-dependent spatial learning and memory (Conrad et al., 2015; Elmore et al., 2014).
In the current study, the percentage of MHC-II+ microglia was markedly elevated 21 d after inoculation with PRRSV, suggesting sustained microglial activation. Early-life infection has been suggested to “prime” or sensitize microglia to subsequent immune stimuli, however, the molecular mechanism responsible for this is unknown. As microglia constantly survey the brain parenchyma (Nimmerjahn et al., 2005), the microglia sensome is implicated herein as many genes related to this sensory apparatus were up-regulated by PRRSV infection. Indirect support for this is provided by the enhanced ex vivo responses of microglia from PRRSV piglets. Compared to microglia isolated from control piglets, microglia from PRRSV piglets (a) showed greater mobility in response to two chemokines; (b) exhibited a higher rate of phagocytosis; (c) secreted more TNFα in response to two TLR agonists; and (d) exhibited enhanced expression of TLR3 and TLR4 mRNA after LPS challenge in vitro. Thus, in the young developing brain of animals with an infection, an up-regulated microglia sensome seems to foretell an exaggerated neuroinflammatory response.
The morphology of hippocampal microglia was also altered by PRRSV, resulting in more microglia throughout the CA1 region with a rod-shape/elongated soma. The so-called “rod microglia” have been reported in a number of pathological conditions and are distinct from ramified and amoeboid microglia (Taylor et al., 2014). Upon brain injury, rod microglia align end-to-end in close proximity to damaged neurons (Ziebell et al., 2012). In vitro they are highly proliferative, transform to an amoeboid morphology, and produce pro-inflammatory cytokines in response to LPS (Tam and Ma, 2014). In the present study, we did not see evidence of end-to-end alignment of rod microglia, which is not surprising since the change in morphology was a response to peripheral infection as opposed to focal brain injury. The role of rod microglia in the developing hippocampus in the context of peripheral infection is not known. Whether or not they are neuroprotective or neurotoxic, how they respond to subsequent immune insults, and if they have a role in the etiology of behavioral disorders are yet to be determined.
Collectively, the results presented herein suggest early-life infection alters the sensome, rendering microglia more responsive to subsequent stimuli. Several limitations should be noted, however. First, enhanced activity was determined ex vivo with microglia isolated from control and infected piglets. If microglia of infected and recovered piglets behave similarly to a secondary in vivo challenge is not currently known. Second, all assessments related to microglia activity were done at a single time point after inoculation. A more complete picture of the induction, maintenance, and cessation of microglia activity would be useful, as would knowing if changes to microglia persisted into adulthood. Clearly more work is needed to understand how this might impact brain development and associated behavioral outcomes. Nonetheless, the results highlight a new potential role for the microglial sensome in response to peripheral infection.
Supplementary Material
Highlights.
Postnatal infection up-regulated microglial sensome genes in piglet hippocampus
Microglia isolated from hippocampus of infected piglets had increased activity
Bipolar rod-like microglia were evident in hippocampus of infected piglets
The microglial sensome may foretell an exaggerated neuroinflammatory response
Acknowledgements
This work was supported by grants from the NIH (R01 HD069899) to RWJ; Jeju National University of Rural Development of Administration of the Republic of Korea (538 JNU Korea 2012-06052) to LBS (for RNA-sequencing associated analysis). We thank Dr. William Van Alstine for provision of PRRS virus, and Jennifer Rytych for her help in microglial cell isolation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Dataset S1. List of differentially expressed genes in response to PRRSV infection.
Dataset S2. Results of functional annotation of differentially expressed genes using DAVID Bioinformatics Resources 6.7.
Reference
- Andersen SL. Trajectories of brain development: point of vulnerability or window of opportunity? Neuroscience and biobehavioral reviews. 2003;27:3–18. doi: 10.1016/s0149-7634(03)00005-8. [DOI] [PubMed] [Google Scholar]
- Bilbo SD, Rudy JW, Watkins LR, Maier SF. A behavioural characterization of neonatal infection-facilitated memory impairment in adult rats. Behavioural brain research. 2006;169:39–47. doi: 10.1016/j.bbr.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Bilbo SD, Schwarz JM. Early-life programming of later-life brain and behavior: a critical role for the immune system. Frontiers in Behavioral Neuroscience. 2009;3 doi: 10.3389/neuro.08.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbo SD, Smith SH, Schwarz JM. A lifespan approach to neuroinflammatory and cognitive disorders: a critical role for glia. Journal of neuroimmune pharmacology: the official journal of the Society on NeuroImmune Pharmacology. 2012;7:24–41. doi: 10.1007/s11481-011-9299-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland ST, Beckley JT, Young S, Tsang V, Watkins LR, Maier SF, Bilbo SD. Enduring consequences of early-life infection on glial and neural cell genesis within cognitive regions of the brain. Brain Behav Immun. 2010;24:329–338. doi: 10.1016/j.bbi.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NH, Chen XZ, Hu DM, Yu XL, Wang LL, Han W, Wu JJ, Cao Z, Wang CB, Zhang Q, Wang BY, Tian KG. Rapid differential detection of classical and highly pathogenic North American Porcine Reproductive and Respiratory Syndrome virus in China by a duplex real-time RT-PCR. Journal of virological methods. 2009;161:192–198. doi: 10.1016/j.jviromet.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Chen XX, Quan R, Guo XK, Gao L, Shi J, Feng WH. Up-regulation of pro-inflammatory factors by HP-PRRSV infection in microglia: implications for HP-PRRSV neuropathogenesis. Vet Microbiol. 2014;170:48–57. doi: 10.1016/j.vetmic.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Conrad MS, Harasim S, Rhodes JS, Van Alstine WG, Johnson RW. Early postnatal respiratory viral infection alters hippocampal neurogenesis, cell fate, and neuron morphology in the neonatal piglet. Brain Behav Immun. 2015;44:82–90. doi: 10.1016/j.bbi.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad MS, Johnson RW. The domestic piglet: an important model for investigating the neurodevelopmental consequences of early life insults. Annual review of animal biosciences. 2015;3:245–264. doi: 10.1146/annurev-animal-022114-111049. [DOI] [PubMed] [Google Scholar]
- Conrad MS, Sutton BP, Dilger RN, Johnson RW. An in vivo three-dimensional magnetic resonance imaging-based averaged brain collection of the neonatal piglet (Sus scrofa) PloS one. 2014;9:e107650. doi: 10.1371/journal.pone.0107650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Martinez-Cerdeno V, Noctor SC. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–57. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorduin J, de Vries EF, Willemsen AT, de Groot JC, Dierckx RA, Klein HC. Neuroinflammation in schizophrenia-related psychosis: a PET study. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 2009;50:1801–1807. doi: 10.2967/jnumed.109.066647. [DOI] [PubMed] [Google Scholar]
- Duan X, Nauwynck HJ, Pensaert MB. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV) Vet Microbiol. 1997;56:9–19. doi: 10.1016/S0378-1135(96)01347-8. [DOI] [PubMed] [Google Scholar]
- Elmore MR, Burton MD, Conrad MS, Rytych JL, Van Alstine WG, Johnson RW. Respiratory viral infection in neonatal piglets causes marked microglia activation in the hippocampus and deficits in spatial learning. J Neurosci. 2014;34:2120–2129. doi: 10.1523/JNEUROSCI.2180-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J, West AB, Arking DE. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nature communications. 2014;5:5748. doi: 10.1038/ncomms6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chauhan A, Sheikh AM, Patil S, Chauhan V, Li XM, Ji L, Brown T, Malik M. Elevated immune response in the brain of autistic patients. Journal of neuroimmunology. 2009;207:111–116. doi: 10.1016/j.jneuroim.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U, Feldon J, Dammann O. Schizophrenia and autism: both shared and disorder-specific pathogenesis via perinatal inflammation? Pediatric research. 2011;69:26R–33R. doi: 10.1203/PDR.0b013e318212c196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JT, Chana G, Pardo CA, Achim C, Semendeferi K, Buckwalter J, Courchesne E, Everall IP. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biological psychiatry. 2010;68:368–376. doi: 10.1016/j.biopsych.2010.05.024. [DOI] [PubMed] [Google Scholar]
- Nikodemova M, Watters JJ. Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. Journal of neuroinflammation. 2012;9:147. doi: 10.1186/1742-2094-9-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Rees S, Inder T. Fetal and neonatal origins of altered brain development. Early human development. 2005;81:753–761. doi: 10.1016/j.earlhumdev.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Sedgwick JD, Schwender S, Imrich H, Dorries R, Butcher GW, ter Meulen V. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:7438–7442. doi: 10.1073/pnas.88.16.7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam WY, Ma CH. Bipolar/rod-shaped microglia are proliferating microglia with distinct M1/M2 phenotypes. Scientific reports. 2014;4:7279. doi: 10.1038/srep07279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SE, Morganti-Kossmann C, Lifshitz J, Ziebell JM. Rod microglia: a morphological definition. PloS one. 2014;9:e97096. doi: 10.1371/journal.pone.0097096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohleb ES, Powell ND, Godbout JP, Sheridan JF. Stress-induced recruitment of bone marrow-derived monocytes to the brain promotes anxiety-like behavior. J Neurosci. 2013;33:13820–13833. doi: 10.1523/JNEUROSCI.1671-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell JM, Taylor SE, Cao T, Harrison JL, Lifshitz J. Rod microglia: elongation, alignment, and coupling to form trains across the somatosensory cortex after experimental diffuse brain injury. Journal of neuroinflammation. 2012;9:247. doi: 10.1186/1742-2094-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.