Abstract
Although herpes simplex virus type-1 (HSV-1), and type-2 (HSV-2), Staphylococcus aureus and Candida albicans co-habit the oral and genital mucosa, their interaction is poorly understood. We determined the effect HSV has on bacterial and/or fungal adherence, the initial step in biofilm formation. HeLa229 cells were infected with HSV-1 (KOS) gL86 or HSV-2 (KOS) 333gJ− at a multiplicity of infection (MOI) of 50 and 10. S. aureus (ATCC 25923) and/or C. albicans (yeast forms or germ tube forms) were co-incubated for 30 min (37 °C; 5 % CO2; 5:1 organism: HeLa cell ratio; n = 16) with virus-infected HeLa cells or uninfected HeLa cell controls. Post-incubation, the monolayers were washed (3x; PBS), lysed (RIPA), and the lysate plated onto Fungisel and/or mannitol salts agar for standard colony count. The level of HeLa-associated S. aureus was significantly decreased (P < 0.05) for both HSV-1- and HSV-2-infected cells, as compared to virus-free HeLa cell controls (38 and 59 % of control, respectively). In contrast, HSV-1 and HSV-2 significantly (P < 0.05) enhanced HeLa cell association of C. albicans yeast forms and germ tube approximately two-fold, respectively. The effect of S. aureus on germ tube and yeast form adherence to HSV-1- and HSV-2-infected cells was specific for the Candida phenotype tested. Our study suggests that HSV, while antagonist towards S. aureus adherence enhances Candida adherence. Furthermore, the combination of the three pathogens results in S. aureus adherence that is either unaffected, or partially restored depending on both the herpes viral species and the fungal phenotype present.
Introduction
Adherence to cell surfaces is an essential initial stage in microbial colonization and subsequent biofilm formation [58, 77]. Shared sites of persistent colonization and chronic infection for Staphylococcus aureus,Candida albicans, and HSV are the oronasopharynx and genital tract. S. aureus and C. albicans, both commensals, are also the 2nd and 4th most common cause of bloodstream infections, respectively [54, 79]. Of the various sites of S. aureus and C. albicans co-colonization, the oronasopharynx serves as the reservoir for systemic infections [44]. Within the oronasopharynx, S. aureus,C. albicans, and HSV occupy two distinct geographic niches. In hosts with dentition, the oral mucosa is shared by HSV and C. albicans, while the anterior nasal nares are occupied by S. aureus [21]. Clinically, S. aureus is only rarely isolated from oral-pharyngeal specimens when normal tissue is present, despite in vitro findings that S. aureus adheres to buccal epithelial cells [22, 51]. Interestingly, in the presence of dentures, an abiotic surface, S. aureus forms a robust biofilm on the denture surface along with C. albicans [29, 63]. Little is known concerning genital tract co-colonization niches beyond the clinical findings that S. aureus infection is associated with genital inflammation, discharge, and dyspareunia, while C. albicans and HSV produce mucosal lesions similar to those observed in the oral cavity [25, 39, 46, 55, 59]. Whether present in the oronasopharynx or genital tract, it is a near certainty that S. aureus and C. albicans would interact at some point with HSV, a permanent resident of infection sites [5].
HSV, a major cause of morbidity and mortality, is a life-long pathogen present in >90 % of the world population [11, 71]. In immune-competent individuals, HSV-1 is a major cause of gingivostomatitis, as well as genital herpes, due to changes in sexual behaviors [5, 48]. Similar to HSV-1, HSV-2 causes oral lesions, although it has a higher association with genital lesions [7, 72]. A characteristic of herpes infections is chronic persistent viral shedding in the absence of symptoms [65, 67]. This permanent, albeit intermittent, presence of HSV virions may play a role in regulating the host microbiome. This could be accomplished via an alteration in available cell surface receptors for adherence by other members of the microbiome. [6, 8–10, 12, 16, 19, 21, 53]. Using a HeLa cell model of virus infection, the focus of this study was to determine whether HSV-1 or HSV-2 affect S. aureus and/or C. albicans germ tube and yeast form adherence, the initial step in biofilm formation.
Methods
Microbial Strains and Handling
Recombinant spread-deficient, entry proficient strains of HSV-1(KOS) gL86 and HSV-2 (KOS) 333gJ− encoding a beta-galactosidase reporter activity were used [3, 37, 68, 74]. Both virus strains enter and replicate, thus have the potential to induce cell signaling, but lack the genes essential for viral cell-to-cell spread. All virus used in this study were taken from a single lot. Virus stocks were maintained at −80 °C until use. HSV entry into HeLa cells was confirmed by o-nitrophenyl-β-d-galactopyranoside (ONPG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) assays, as previously described [52]. HeLa 229 cells were maintained and cultivated at 37 °C, 5 % CO2 in 1× Dulbecco’s modified Eagle’s medium (DMEM with 4.5 g/L glucose and l-glutamine, without sodium pyruvate; Mediatech) supplemented with 10 % heat-inactivated fetal bovine serum (FBS) and gentamicin (50 µg/ml).
Candida albicans, maintained at −80 °C until use, was initially subcultured onto Sabourad Dextrose agar. For use, C. albicans was cultured onto Fungisel medium (37 °C; 48 h;Troy Biologics). Yeast suspensions (YF) were prepared in Hanks Balanced Salts Solution (HBSS; 105 CFU/ml final concentration; 37 °C) immediately prior to use. Germ tube forms (GT) were generated by incubation in fetal bovine serum (FBS; 3 h; 37 °C; Abs600 0.3), followed by washing in HBSS (2×; 4000×g) and re-suspension in HBSS to 105 CFU/ml final concentration.
Staphylococcus aureus ATCC 25923, maintained at −80 °C until use, was subcultured onto mannitol salts medium (37 °C; 18 h) for use. S. aureus suspensions were prepared immediately prior to use in HBSS (105 CFU/ml final concentration; 37 °C).
Polymicrobic Adherence Assay
The number of HeLa cell-associated S. aureus and C. albicans was determined as an indicator of biofilm initiation (adherence). HeLa 229 were grown overnight in 96-well (4 × 104 cells/well) at 37 °C, 5 % CO2 to reach 85 % final confluence. Before infection with virus, the cells were washed with 1× Opti-MEM with HEPES, sodium bicarbonate and l-glutamine (Gibco). Virus (HSV-1 (KOS) gL86 or HSV-2 (KOS) 333gJ−) was added to HeLa 299 cells at a multiplicity of infection (MOI) of 50 and 10 for 3 h at 37 °C, 5 % CO2. After viral infection, the cells were washed once each with PBS then HBSS before incubation with C. albicans YF or GT with and without S. aureus (5:1 target to cell ratio; n = 16). After incubation (30 min; 37 °C; 5 % CO2), HeLa cell monolayers were washed to remove unbound microbes (PBSx3) and lysed (RIPA, Life Technologies, 1:50 dilution; filter sterilized). The cell lysate (50 µl) was spread plated onto mannitol salts and/or Fungisel agar to select for S. aureus and C. albicans, respectively. Controls consisted of HSV-uninfected HeLa cells handled as described for virus-infected HeLa cells. For each experiment, a separate control plate to confirm the viral MOI was performed.
Imaging studies of HSV-1- and HSV-2-infected (MOI 50; 3 h; 37 °C) HeLa cell monolayers (5 × 104 cells/coverslip) were performed by both fluorescent microscopy and bright field microscopy. Cell monolayers were incubated (30 min; 37 °C; 5 % CO2) with S. aureus and C. albicans (5:1 target to cell). HeLa cells were then washed free of non-adherent microbes (PBSx3) and fixed (methanol). For fluorescent microscopy, monolayers were stained with FITC-conjugated Herpes Simplex Virus Type 1+2 gD antibody (Abcam), and 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies) then examined by epi-fluorescent microscopy. Cells that were signal positive for HSV or Candida or S. aureus (100 individual cells per microbe signal per coverslip) were secondarily scanned for the presence of additional microbe co-localization signals (1000× initial magnification; Nikon). For bright field microscopy, cell monolayers were stained with Gram’s crystal violet (Troy Biologics), then examined by light microscopy. Cells that were positive for Candida or S. aureus (100 individual cells per microbe signal per coverslip) were secondarily scanned for the presence of additional microbe co-localization signals (1000× initial magnification).
Statistical Analysis
Each adherence experiment was conducted twice in octuplicate. Each imaging study was conducted twice in triplicate. Data were evaluated by analysis of variance (ANOVA; GraphPad InStat 3.10 for Windows, GraphPad Software Inc.). Mean values were considered significantly different at P < 0.05.
Results
Candida, HSV-1 and HSV-2 Modulation of S. aureus Adherence to HeLa Cells
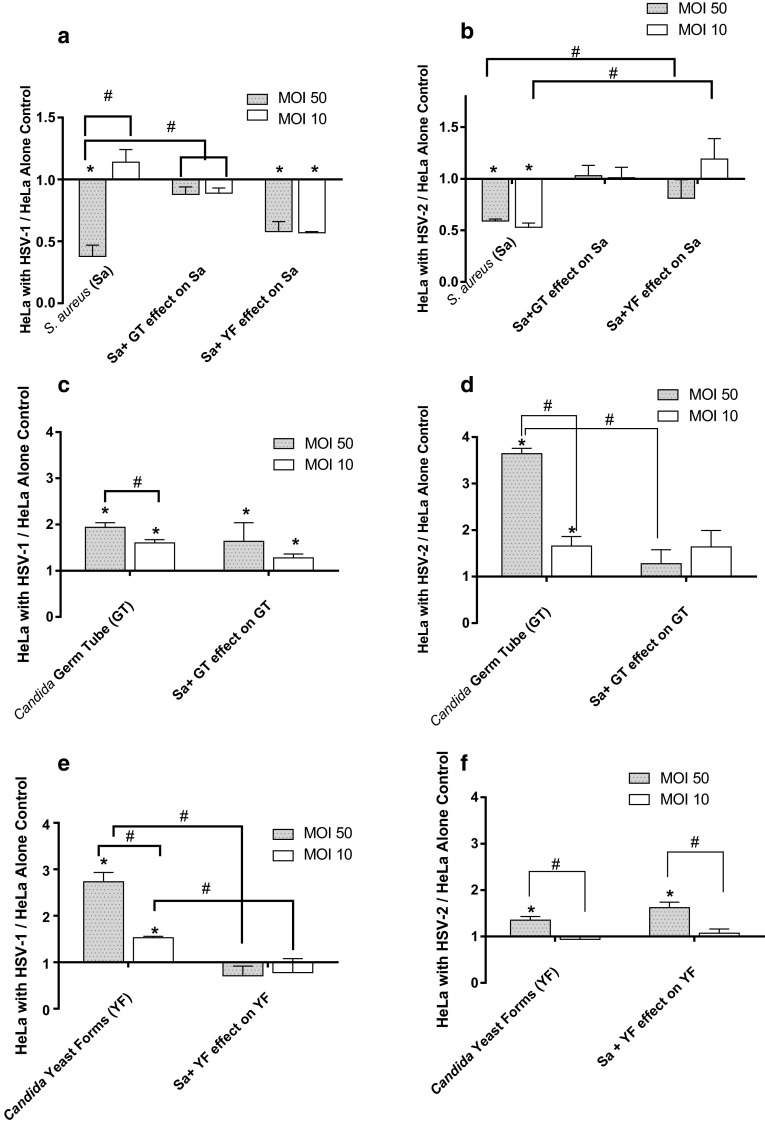
Since HSV-1 and HSV-2 co-localize in the oropharynx with C. albicans, we first determined their effect alone, and in concert, on S. aureus adherence to virus-infected HeLa cells as compared to S. aureus adherence to virus-free HeLa cells. Both HSV-1- and HSV-2-affected S. aureus adherence to HeLa cells (Fig. 1a, b). At the highest virus concentration (MOI 50) the level of S. aureus adhered to uninfected HeLa cells (24.2 CFU/well ± 3.19; n = 16) was significantly higher (2.7-fold; P < 0.05) than that measured for S. aureus adherence to HSV-1-infected HeLa cells (9.1 ± 1.1 CFU/well). Staphylococcal adherence to uninfected HeLa cells was also significantly (1.7-fold; P < 0.05) higher than that measured for adherence to HSV-2 (MOI 50)-infected HeLa cells. The level of S. aureus adherence to HSV-2-infected cells at a fivefold lower MOI (MOI 10) was similar to that measured for the HSV-2 MOI 50 (1.9-fold below virus-free control).
Fig. 1.
Effect of HSV-1 (panels a, c, e) and HSV-2 (panels b, d, f) at multiplicities of infection (MOI) of 50 and 10 on adherence of S. aureus and/or C. albicans to HeLa cells. a S. aureus (Sa) binding to HSV-1-infected cells in the presence of C. albicans germ tubes (GT) or yeast forms (YF); b S. aureus (Sa) binding to HSV-2-infected cells in the presence of C. albicans germ tubes (GT) or yeast forms (YF); c C. albicans germ tubes (GT) binding to HSV-1-infected cells in the presence of S. aureus (Sa); d C. albicans germ tubes (GT) binding to HSV-2-infected cells in the presence of S. aureus (Sa); e C. albicans yeast forms (YF) binding to HSV-1-infected cells in the presence of S. aureus (Sa); f C. albicans yeast forms (YF) binding to HSV-2-infected cells in the presence of S. aureus (Sa). All data points are mean ± SEM, n = 16 normalized to virus-free control. *Significantly different (P < 0.05) from uninfected HeLa cell control. #Significantly different (P < 0.05) from paired point indicated by bracket
Co-incubation of S. aureus with C. albicans germ tube (GT) forms restored the level of S. aureus adherence to HSV-1 and HSV-2-infected HeLa cells to that measured for uninfected HeLa cell controls. Similar to the effect of GT forms, S. aureus adherence to HSV-2-infected cells in the presence of C. albicans yeast forms (YF) resulted in staphylococcal adherence levels similar to that measured uninfected HeLa cells. In contrast, the presence of YF did not affect the HSV-1 mediated depression in S. aureus adherence. These results show that HSV viral entry is antagonist to S. aureus adherence, an antagonism that can be, in part, reversed by C. albicans GT and YF.
C. albicans Adherence to HSV-1- and HSV-2-Infected HeLa Cells in the Presence and Absence of S. aureus
In these assays, we determined whether C. albicans GT and YF adherence to HeLa cells was affected by HSV and S. aureus (Fig. 1c–f). Our findings showed that HSV-1 and HSV-2 significantly (P < 0.05) enhanced C. albicans GT and YF adherence in a virus dose-dependent manner; maximal adherence levels of GT and YF occurred at an MOI of 50. For both candidal phenotypes, adherence was significantly higher (P < 0.05) than that measured for adherence to virus-uninfected HeLa cell controls. Candida GT adherence to HSV-2-infected HeLa cells (normalized to virus-uninfected HeLa cell controls) was significantly (P < 0.05) higher than its adherence to HSV-1-infected HeLa cells (3.6-fold vs. 1.9-fold higher, respectively). The reverse pattern was observed for YF adherence. YF significantly (P < 0.05) and preferentially adhered to HSV-1-infected cells (2.7-fold over uninfected control) as compared to YF adherence to HSV-2-infected cells (1.9-fold over uninfected control).
The HSV-mediated enhancement of YF and GT adherence was significantly (P < 0.05) and preferentially affected by the presence of S. aureus. S. aureus decreased GT adherence to HSV-2-infected cells to levels measured for uninfected HeLa cell control. In contrast, S. aureus significantly (P < 0.05) decreased YF adherence to HSV-1-infected HeLa cells. S. aureus had no significant effect on YF adherence to HSV-2-infected cells or GT adherence to HSV-1-infected HeLa cells. Taken together, these results indicate that HSV promotes the association of C. albicans in a phenotypic-specific manner. Furthermore, these findings indicate that S. aureus’ effect on C. albicans adherence was HSV-type specific with regard to fungal phenotype.
Microscopic Evaluation of C. albicans and S. aureus Adherence Pattern to HSV-1- and HSV-2-Infected HeLa Cells
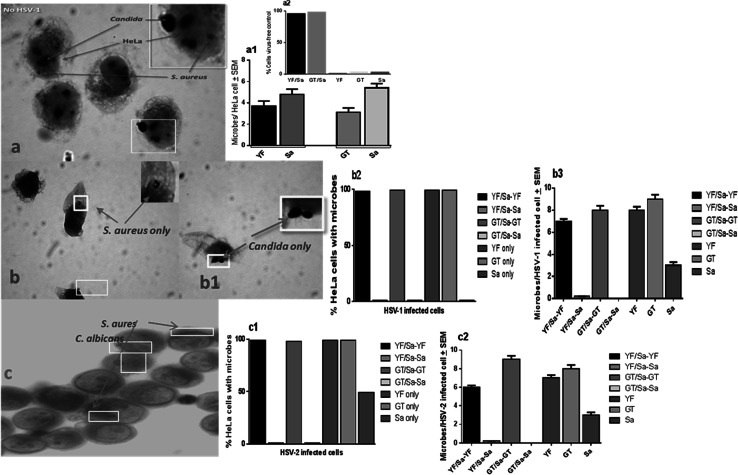
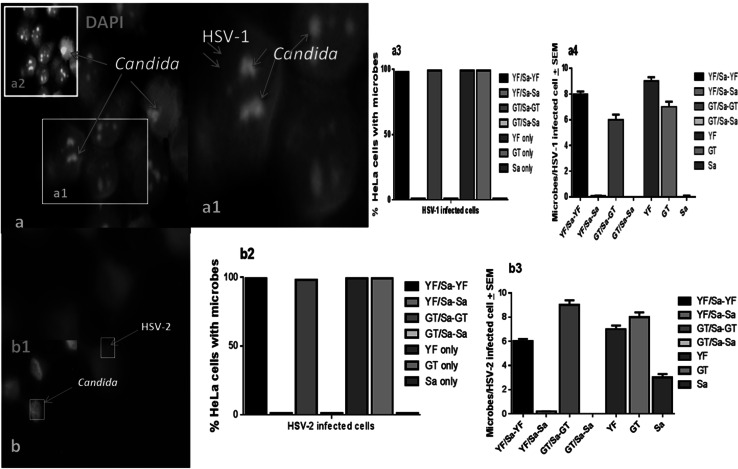
We visually examined the association of S. aureus and C. albicans with HSV-1- and HSV-2-infected HeLa cells using the size, morphology, and arrangement differences between S. aureus and C. albicans to distinguish between the organisms. In the absence of HSV-1 or HSV-2, S. aureus and C. albicans (GT and YF) co-localized onto uninfected HeLa control cells (Fig. 2a). In contrast, no co-localization of staphylococci with C. albicans was observed on HSV-1- or HSV-2-infected HeLa cells (Fig. 2b–d). Using fluorescent microscopy with FITC-conjugated anti-HSV-gD monoclonal antibody, we further confirmed that S. aureus (Fig. 3b, c) did not co-localize with C. albicans, HSV-1 or HSV-2 (Fig. 3a, a1, 3d). However, Candida co-localized with HSV-1 and HSV-2 (Fig. 3a1, c). S. aureus and C. albicans were not observed microscopically to interact with one another (data not shown). Furthermore, the co-localization adherence pattern of S. aureus and Candida, in the absence of HSV, indicates a lack of physical interference with each other’s adherence. These findings further suggest a depletion of available S. aureus adherence receptors subsequent to HSV cell entry.
Fig. 2.
Lack of S. aureus and C. albicans interactions on HSV-1- and HSV-2-infected HeLa cells. HSV-1 and HSV-2-infected (MOI 50) HeLa cell monolayers with S. aureus and C. albicans (5:1 target to cell). For bright field microscopy, cell monolayers were stained with Gram’s crystal violet, then examined by light microscopy. Cells that were positive for Candida or S. aureus (100 individual cells per microbe signal/per coverslip) for were secondarily scanned for the presence of additional microbe co-localization signals (×1000 initial magnification). a—a1 S. aureus (Sa) and C. albicans yeast forms (YF) or germ tube forms (GT) co-localize on uninfected HeLa cells; a2 (insert) Percent of HeLa cells with co-localized or individual microbes; b—b3 Lack of S. aureus and C. albicans co-localization in the presence of HSV-1; c—c2 Lack of S. aureus and C. albicans co-localization in the presence of HSV-2; mean ± SEM
Fig. 3.
Lack of co-localization of S. aureus with C. albicans on HSV-1- or HSV-2-infected HeLa cells. HSV-infected HeLa cell monolayers challenge with S. aureus and C. albicans (5:1 target to cell) stained (FITC-conjugated anti-HSV gD antibody, and DAPI). Pictures are representative of findings from screening of cells that were signal positive for HSV then scanned for Candida and S. aureus (100 individual cells per microbe signal per coverslip) that were then secondarily scanned for the presence of additional microbe co-localization signals (×1000 initial magnification; Nikon). a—a4 C. albicans (a2 insert; DAPI staining) co-localize with HSV-1; b-b3 C. albicans co-localized with HSV-2 (b1 insert, C. albicans DAPI staining); mean ± SEM
Discussion
In this report, we describe an in vitro model system to study the polymicrobial interactions which occur at the initiating step in biofilm formation, i.e., adherence. The use of HeLa cells in this model system presents a unique advantage in the study of microbial interactions regarding cell surface receptors as they lack surface expression of fibronectin [4, 13, 27]. This absence of surface fibronectin more closely mimics that observed in colonization and natural infection, since the apical surface of mucosal epithelia normally lacks fibronectin [18, 33, 38, 49, 66]. This absence of fibronectin also allows for the study of alternative mechanisms by which S. aureus and C. albicans adhere to HSV-infected cells since both microbes adhere to fibronectin [2, 4, 13, 40, 69]. This model system for the study of virus effects on bacterial and fungal adherence was further enhanced through the use of entry and replication proficient, but non-spreading strains of HSV-1 and HSV-2 [3, 74].
HSV enters cells via endocytosis [56, 57]. Endocytosis of the virus-receptor complex results in a dynamic display of cell surface receptors available to S. aureus and C. albicans for their adherence. Interestingly there is an opposing interaction of S. aureus and C. albicans with heparan sulfates on the mammalian cell surface. S. aureus adherence to various sulfonated heparans, particularly a heparan component syndecan-1, is important in its interaction with epithelial cells [31, 36, 42, 47, 61, 73]. In contrast, the presence of cell surface heparan sulfates block C. albicans adherence to extracellular matrix proteins, e.g., laminin, and types I and IV collagen [43]. HSV cell entry causes a species specific (HSV-1 vs. HSV-2) differential depletion of heparan sulfates, and induction of heparanase secretion, which further depletes cell surface sulfonated heparans [1, 14, 20, 28, 30, 35, 41, 45, 56, 57, 70, 75]. Both these events seem critical in our observations. For instance, loss of S. aureus heparan sulfate receptors could explain the lack of its adherence to HeLa cells infected with either HSV-1 or HSV-2 (Fig. 4). Conversely, Candida preferred receptors would be unmasked by heparanase production and HSV entry depletion of surface heparan sulfates. The HSV-1 vs. HSV-2 differential depletion of sulfonated heparan receptors could explain our findings of enhanced adherence to virus-infected cells that was Candida phenotype (YF-GT) dependent (Fig. 4) [45].
Fig. 4.
Proposed model of the key steps involved in polymicrobic interaction (see text for details). Sa S. aureus, YF yeast form, GT germ tube form, HS heparan sulfate, HSV herpes simplex virus, ECM extracellular matrix proteins other than glycosaminoglycans, e.g., laminin and collagen
Although this model begins to address the patterns measured for S. aureus and C. albicans adherence to HSV-infected cells, a secondary mechanism is likely responsible for the effects of S. aureus and C. albicans on each other’s adherence in the presence of both uninfected and HSV-infected (50 % HeLa cells infected/well, MOI50) cells. HSV-infected cells release interferons alpha and gamma, which in turn cause perturbation of lipid rafts altering membrane fluidity (hydrophobicity) and the cell cytoskeletal complex in non-virally infected cells [24, 34, 60, 64, 72]. Both S. aureus and C. albicans interact with hydrophobic surfaces [2, 21, 32]. The secondary extrinsic HSV-mediated effects on uninfected cells, i.e., surface hydrophobicity changes, could result in increased S. aureus and C. albicans adherence and the apparent restoration of cell binding. The lack of cell-to-cell spread of the virus in this model system will allow for determination of which virally mediated cell alteration, i.e., intrinsic, extrinsic, or a combination of changes, is responsible for the differential adherence by S. aureus and C. albicans [2, 21].
The finding herein that HSV-1 and HSV-2 renders cells refractory to S. aureus adherence, while enhancing phenotype specific C. albicans adherence, as well as predilection for differential fungal phenotype adherence mediated by HSV-1 (yeast form) vs. HSV-2 (germ tube form) begin to explain host colonization sites and may play a role directing maintenance of the candidal commensal (YF) versus pathogenic state (GT) in vivo, e.g., HSV-2 may impact the incidence of vaginal candidiasis. These positive interactions between HSV and C. albicans may also provide an environment more conducive to HSV survival. Recently it has been shown that HSV-1 becomes embedded in the fungal biofilm where it remains viable and protected from antiviral agents, as well host factors, e.g., antibodies [50]. This protection of HSV from host factors is augmented by Candida’s ability to downregulate host proinflammatory cytokine secretion, which would be detrimental to HSV replication [17, 23, 26, 76]. HSV-1 in turn has been shown to protect Candida by down-regulating antifungal immune response [15]. Both HSV and Candida would benefit from the exclusion of S. aureus from their neighborhood. S. aureus is a proinflammatory pathogen [80]. In addition to the induction of various cytokines, including interferon gamma, the chronic presence of S. aureus elicits a leukocytic infiltrate comprised neutrophils, T cells and natural killer cells [62, 78]. Such a proinflammatory response would be detrimental to both HSV and candidal survival.
To our knowledge this is the first report demonstrating the ability of HSV to regulate the adherence of multiple opportunistic pathogens that are part of the inter-kingdom microbiome. This study demonstrates that HSV-1 and HSV-2 modulate both fungal and bacterial adherence to cells, likely, in part, through HSV alteration of heparan sulfate cell surface display. These findings represent a paradigm shift from the current view that host factors solely control microbiome population members, to one where a life-long latent viral pathogen could co-opt the host by a specific molecular mechanism that alters biofilm initiation, thus, usurping regulatory control of microbiome membership. Further studies are needed to define the specific role the various HSV viral entry receptors play in modulation of staphylococcal and candidal adherence. By understanding the initial interactions that occur between HSV-1 and HSV-2 as permanent members of the host microbiome, and chronic colonizers, e.g., S. aureus and C. albicans, another avenue opens with regard to biofilm inhibition and eradication.
Acknowledgments
This Project was supported by research funds from Office of Research and Sponsored Programs (ORSP), Midwestern University, IL and College of Dental Medicine-Illinois (CDMI) at Midwestern University.
Compliance with Ethical Standards
Conflict of interests
The authors have no conflict of interests.
Footnotes
Balbina J. Plotkin and Ira M. Sigar have contributed equally to this work.
Vaibhav Tiwari and Scott Halkyard have contributed substantively to this work.
References
- 1.Adamiak B, Trybala E, Mardberg K, Johansson M, Liljeqvist JA, Olofsson S, Grabowska A, Bienkowska-Szewczyk K, Szewczyk B, Bergstrom T. Human antibodies to herpes simplex virus type 1 glycoprotein C are neutralizing and target the heparan sulfate-binding domain. Virology. 2010;400(2):197–206. doi: 10.1016/j.virol.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama H, Yamasaki O, Kanzaki H, Tada J, Arata J. Adherence characteristics of Staphylococcus aureus and coagulase-negative staphylococci isolated from various skin lesions. J Dermatol Sci. 1998;18(2):132–136. doi: 10.1016/S0923-1811(98)00036-X. [DOI] [PubMed] [Google Scholar]
- 3.Ali M, Karasneh G, Jarding MJ, Tiwari V, Shukla D. 3-O-sulfated heparan sulfate binding peptide preferentially targets herpes simplex virus 2-infected cells. J Virol. 2012;86(12):6434–6443. doi: 10.1128/JVI.00433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alva-Murillo N, Lopez-Meza JE, Ochoa-Zarzosa A. Nonprofessional phagocytic cell receptors involved in Staphylococcus aureus internalization. Biomed Res Int. 2014 doi: 10.1155/2014/538546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arduino PG, Porter SR. Herpes simplex virus type I infection: overview on relevant clinico-pathological features. J Oral Pathol Med. 2008;37(2):107–121. doi: 10.1111/j.1600-0714.2007.00586.x. [DOI] [PubMed] [Google Scholar]
- 6.Arendrup MC, Fuursted K, Gahrn-Hansen B, Schonheyder HC, Knudsen JD, Jensen IM, Bruun B, Christensen JJ, Johansen HK. Semi-national surveillance of fungaemia in Denmark 2004–2006: increasing incidence of fungaemia and numbers of isolates with reduced azole susceptibility. Clin Microbiol Infect. 2008;14(5):487–494. doi: 10.1111/j.1469-0691.2008.01954.x. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein DI, Bellamy AR, Hook EW, Levin MJ, Wald A, Ewell MG, Wolff PA, Deal CD, Heineman TC, Dubin G, Belshe RB. Epidemiology, clinical presentation, and antibody response to primary infection with herpes simplex virus type 1 and type 2 in young women. Clin Infect Dis. 2013;56(3):344–351. doi: 10.1093/cid/cis891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas K, Hoggard M, Jain R, Taylor MW, Douglas R. The nasal microbiota in health and disease: variation within and between subjects. Front Microbiol. 2015 doi: 10.3389/fmicb.2018.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjarnsholt T. The role of bacterial biofilms in chronic infections. Apmis. 2013;121:1–58. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 10.Blumberg HM, Jarvis WR, Soucie JM, Edwards JE, Patterson JE, Pfaller MA, Rangel-Frausto MS, Rinaldi MG, Saiman L, Wiblin RT, Wenzel RP, Grp NS. Risk factors for candidal bloodstream infections in surgical intensive care unit patients: The NEMIS Prospective Multicenter Study. Clin Infect Dis. 2001;33(2):177–186. doi: 10.1086/321811. [DOI] [PubMed] [Google Scholar]
- 11.Bradley H, Markowitz LE, Gibson T, McQuillan GM. Seroprevalence of herpes simplex virus types 1 and 2-United States, 1999–2010. J Infect Dis. 2014;209(3):325–333. doi: 10.1093/infdis/jit458. [DOI] [PubMed] [Google Scholar]
- 12.Brogden KA, Guthmiller JM, Taylor CE. Human polymicrobial infections. Lancet. 2005;365(9455):253–255. doi: 10.1016/S0140-6736(05)70155-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderone RA, Scheld WM. Role of fibronectin in the pathogenesis of candidal infections. Rev Infect Dis. 1987;9(Suppl 4):S400–S403. doi: 10.1093/clinids/9.Supplement_4.S400. [DOI] [PubMed] [Google Scholar]
- 14.Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev Med Virol. 2000;10(5):305–319. doi: 10.1002/1099-1654(200009/10)10:5<305::AID-RMV286>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 15.Cermelli C, Orsi CF, Ardizzoni A, Lugli E, Cenacchi V, Cossarizza A, Blasi E. Herpes simplex virus type 1 dysregulates anti-fungal defenses preventing monocyte activation and downregulating toll-like receptor-2. Microbiol Immunol. 2008;52(12):575–584. doi: 10.1111/j.1348-0421.2008.00074.x. [DOI] [PubMed] [Google Scholar]
- 16.Champion AE, Goodwin TA, Brolinson PG, Werre SR, Prater MR, Inzana TJ. Prevalence and characterization of methicillin-resistant Staphylococcus aureus isolates from healthy university student athletes. Ann Clin Microbiol Antimicrob. 2014 doi: 10.1186/s12941-014-0033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandra JMT, Imamura Y, Mukherjee PK, Ghannoum MA. Interaction of Candida albicans with adherent human peripheral blood mononuclear cells increases C. albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect Immun. 2007;75:2612–2620. doi: 10.1128/IAI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christopher RA, Kowalczyk AP, McKeown-Longo PJ. Localization of fibronectin matrix assembly sites on fibroblasts and endothelial cells. J Cell Sci. 1997;110(Pt 5):569–581. doi: 10.1242/jcs.110.5.569. [DOI] [PubMed] [Google Scholar]
- 19.Clark DW, Wenaas A, Luong A, Citardi MJ, Fakhri S. Staphylococcus aureus prevalence in allergic fungal rhinosinusitis vs other subsets of chronic rhinosinusitis with nasal polyps. Int Forum Allergy Rhinol. 2013;3(2):89–93. doi: 10.1002/alr.21090. [DOI] [PubMed] [Google Scholar]
- 20.Clement C, Tiwari V, Scanlan PM, Valyi-Nagy T, Yue B, Shukla D. A novel role for phagocytosis-like uptake in herpes simplex virus entry. J Cell Biol. 2006;174(7):1009–1021. doi: 10.1083/jcb.200509155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colacite J, Scoaris DD, Yamada-Ogatta SF, Ueda-Nakamura T, Nakamura CV, Dias BP. Pathogenic potential of Staphylococcus aureus strains isolated from various origins. Ann Microbiol. 2011;61(3):639–647. doi: 10.1007/s13213-010-0184-3. [DOI] [Google Scholar]
- 22.Colombo AV, Barbosa GM, Higashi D, di Micheli G, Rodrigues PH, Simionato MRL. Quantitative detection of Staphylococcus aureus, Enterococcus faecalis and Pseudomonas aeruginosa in human oral epithelial cells from subjects with periodontitis and periodontal health. J Med Microbiol. 2013;62:1592–1600. doi: 10.1099/jmm.0.055830-0. [DOI] [PubMed] [Google Scholar]
- 23.da Silva BR, de Freitas VAA, Nascimento-Neto LG, Carneiro VA, Arruda FVS, de Aguiarc ASW, Cavada BS, Teixeira EH. Antimicrobial peptide control of pathogenic microorganisms of the oral cavity: a review of the literature. Peptides. 2012;36(2):315–321. doi: 10.1016/j.peptides.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Delboy MG, Patterson JL, Hollander AM, Nicola AV. Nectin-2-mediated entry of a syncytial strain of herpes simplex virus via pH-independent fusion with the plasma membrane of Chinese hamster ovary cells. Virol J. 2006;3:105. doi: 10.1186/1743-422X-3-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donders GGG, Vereecken A, Bosmans E, Dekeersmaecker A, Salembier G, Spitz B. Definition of a type of abnormal vaginal flora that is distinct from bacterial vaginosis: aerobic vaginitis. BJOG. 2002;109(1):34–43. doi: 10.1111/j.1471-0528.2002.00432.x. [DOI] [PubMed] [Google Scholar]
- 26.Feller L, Khammissa RAG, Chandran R, Altini M, Lemmer J. Oral candidosis in relation to oral immunity. J Oral Pathol Med. 2014;43(8):563–569. doi: 10.1111/jop.12120. [DOI] [PubMed] [Google Scholar]
- 27.Fowler T, Wann ER, Joh D, Johansson S, Foster TJ, Hook M. Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell beta1 integrins. Eur J Cell Biol. 2000;79(10):672–679. doi: 10.1078/0171-9335-00104. [DOI] [PubMed] [Google Scholar]
- 28.García B, Fernández-Vega I, García-Suárez O, Castañón S, Quirós L. The role of heparan sulfate proteoglycans in bacterial infections. J Med Microb Diagn. 2014;3:157–164. [Google Scholar]
- 29.Griffiths EC, Pedersen AB, Fenton A, Petchey OL. The nature and consequences of coinfection in humans. J Infect. 2011;63(3):200–206. doi: 10.1016/j.jinf.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadigal SR, Agelidis AM, Karasneh GA, Antoine TE, Yakoub AM, Ramani VC, Djalilian AR, Sanderson RD, Shukla D. Heparanase is a host enzyme required for herpes simplex virus-1 release from cells. Nat Commun. 2015;6:6985. doi: 10.1038/ncomms7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashida A, Amano S, Gallo RL, Linhardt RJ, Liu J, Park PW. 2-O-sulfated domains in syndecan-1 heparan sulfate inhibit neutrophil cathelicidin and promote Staphylococcus aureus corneal infection. J Biol Chem. 2015;290(26):16157–16167. doi: 10.1074/jbc.M115.660852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazen KC, Plotkin BJ, Klimas DM. Influence of growth-conditions on cell-surface hydrophobicity of Candida albicans and Candida glabrata. Infect Immun. 1986;54(1):269–271. doi: 10.1128/iai.54.1.269-271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heino J, Kapyla J. Cellular receptors of extracellular matrix molecules. Current Pharm Des. 2009;15(12):1309–1317. doi: 10.2174/138161209787846720. [DOI] [PubMed] [Google Scholar]
- 34.Heldwein EE, Krummenacher C. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci. 2008;65(11):1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henry-Stanley MJ, Hess DJ, Erlandsen SL, Wells CL. Ability of the heparan sulfate proteoglycan syndecan-1 to participate in bacterial translocation across the intestinal epithelial barrier. Shock. 2005;24(6):571–576. doi: 10.1097/01.shk.0000184286.95493.78. [DOI] [PubMed] [Google Scholar]
- 36.Hess DJ, Henry-Stanley MJ, Erlandsen SL, Wells CL. Heparan sulfate proteoglycans mediate Staphylococcus aureus interactions with intestinal epithelium. Med Microbiol Immunol. 2006;195(3):133–141. doi: 10.1007/s00430-005-0007-5. [DOI] [PubMed] [Google Scholar]
- 37.Hubbard S, Choudhary S, Maus E, Shukla D, Swenson S, Markland FS, Tiwari V. Contortrostatin, a homodimeric disintegrin isolated from snake venom inhibits herpes simplex virus entry and cell fusion. Antiviral Therapy. 2012;17(7):1319–1326. doi: 10.3851/IMP2291. [DOI] [PubMed] [Google Scholar]
- 38.Hynes RO, Ali IU, Destree AT, Mautner V, Perkins ME, Senger DR, Wagner DD, Smith KK. A large glycoprotein lost from the surfaces of transformed cells. Ann N Y Acad Sci. 1978;312:317–342. doi: 10.1111/j.1749-6632.1978.tb16811.x. [DOI] [PubMed] [Google Scholar]
- 39.Jarvis WR. The epidemiology of colonization. Infect Cont Hosp Epidemiol. 1996;17(1):47–52. doi: 10.2307/30142366. [DOI] [PubMed] [Google Scholar]
- 40.Kanangat S, Postlethwaite A, Cholera S, Williams L, Schaberg D. Modulation of virulence gene expression in Staphylococcus aureus by interleukin-1beta: novel implications in bacterial pathogenesis. Microbes Infect. 2007;9(3):408–415. doi: 10.1016/j.micinf.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 41.Karoli T, Liu LG, Fairweather JK, Hammond E, Li CP, Cochran S, Bergefall M, Trybala E, Addison RS, Ferro V. Synthesis, biological activity, and preliminary pharmacokinetic evaluation of analogues of a phosphosulfomannan angiogenesis inhibitor (PI-88) J Med Chem. 2005;48(26):8229–8236. doi: 10.1021/jm050618p. [DOI] [PubMed] [Google Scholar]
- 42.Kiefer F, Herrmann M, Peschel A, Xia G. Identification and characterization of heparan sulphate-binding proteins in Staphylococcus aureus. Int J Med Microbiol. 2012;302:72-72. [Google Scholar]
- 43.Klotz SA, Smith RL. Glycosaminoglycans inhibit Candida albicans adherence to extracellular-matrix proteins. FEMS Microbiol Lett. 1992;99(2–3):205–208. doi: 10.1111/j.1574-6968.1992.tb05567.x. [DOI] [PubMed] [Google Scholar]
- 44.Lam OLT, McMillan AS, Samaranayake LP, Li LSW, McGrath C. Effect of oral hygiene interventions on opportunistic pathogens in patients after stroke. Amer J Infect Cont. 2013;41(2):149–154. doi: 10.1016/j.ajic.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 45.Lembo D, Donalisio M, Lame C, Cagno V, Civra A, Bianchini EP, Zeghbib N, Bouchemal K. Auto-associative heparin nanoassemblies: a biomimetic platform against the heparan sulfate-dependent viruses HSV-1, HSV-2, HPV-16 and RSV. Euro J Pharm Biopharm. 2014;88(1):275–282. doi: 10.1016/j.ejpb.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Li JR, McCormick J, Bocking A, Reid G. Importance of vaginal microbes in reproductive health. Repro Sci. 2012;19(3):235–242. doi: 10.1177/1933719111418379. [DOI] [PubMed] [Google Scholar]
- 47.Liang OD, Ascencio F, Fransson LA, Wadstrom T. Binding of heparan-sulfate to Staphylococcus aureus. Infect Immun. 1992;60(3):899–906. doi: 10.1128/iai.60.3.899-906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Looker KJ, Garnett GP. A systematic review of the epidemiology and interaction of herpes simplex virus types 1 and 2. Sex Transmit Infect. 2005;81(2):103–107. doi: 10.1136/sti.2004.012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mao Y, Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24(6):389–399. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 50.Mazaheritehrani E, Sala A, Orsi CF, Neglia RG, Morace G, Blasi E, Cermelli C. Human pathogenic viruses are retained in and released by Candida albicans biofilm in vitro. Virus Res. 2014;179:153–160. doi: 10.1016/j.virusres.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 51.Merghni A, Ben Nejma M, Hentati H, Mahjoub A, Mastouri M. Adhesive properties and extracellular enzymatic activity of Staphylococcus aureus strains isolated from oral cavity. Microb Pathogen. 2014;73:7–12. doi: 10.1016/j.micpath.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87(3):427–436. doi: 10.1016/S0092-8674(00)81363-X. [DOI] [PubMed] [Google Scholar]
- 53.National Nosocomial Infections Surveillance (NNIS) System Report data summary from January 1992 to June 2002, issued August 2002. (2002) Am J Infect Control, 30(8), 458–475 [DOI] [PubMed]
- 54.National Nosocomial Infections Surveillance (NNIS) System Report data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control, 32(8), 470-485 [DOI] [PubMed]
- 55.Nenoff P, Kruger C, Schaller J, Ginter-Hanselmayer G, Schulte-Beerbuhl R, Tietz HJ. Mycology - an update Part 2: dermatomycoses: Clinical picture and diagnostics. J Der Deutschen Dermatol Gesellschaft. 2014;12(9):749–779. doi: 10.1111/ddg.12420. [DOI] [PubMed] [Google Scholar]
- 56.Nicola AV, McEvoy AM, Straus SE. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and chinese hamster ovary cells. J Virol. 2003;77(9):5324–5332. doi: 10.1128/JVI.77.9.5324-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicola AV, Straus SE. Cellular and viral requirements for rapid endocytic entry of herpes simplex virus. J Virol. 2004;78:7508–7517. doi: 10.1128/JVI.78.14.7508-7517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohlsen K, Oelschlaeger TA, Hacker J, Khan AS. Carbohydrate receptors of bacterial adhesins: implications and reflections. Glycosci Microb Adhes. 2009;288:109–120. doi: 10.1007/128_2008_10. [DOI] [PubMed] [Google Scholar]
- 59.Okonofua FE, Akonai KA, Dighitoghi MD. Lower genital-tract infections in infertile nigerian women compared with controls. Genitourin Med. 1995;71(3):163–168. doi: 10.1136/sti.71.3.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palu G, Biasolo MA, Sartor G, Masotti L, Papini E, Floreani M, Palatini P. Effects of herpes-simplex virus type-1 infection on the plasma-membrane and related functions of HeLa S3 cells. J Gen Virol. 1994;75:3337–3344. doi: 10.1099/0022-1317-75-12-3337. [DOI] [PubMed] [Google Scholar]
- 61.Park PW, Hayashida A. Syndecan-1 in microbial pathogenesis, host defense, and inflammation. Trends Glycosci Glycotechnol. 2005;17(98):271–284. doi: 10.4052/tigg.17.271. [DOI] [Google Scholar]
- 62.Parker D, Planet PJ, Soong G, Narechania A, Prince A. Induction of type I interferon signaling determines the relative pathogenicity of Staphylococcus aureus strains. Plos Pathog. 2014;10(2):e1003951. doi: 10.1371/journal.ppat.1003951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pereira CA, Toledo BC, Santos CT, Pereira Costa ACB, Back-Brito GN, Kaminagakura E, Jorge AOC. Opportunistic microorganisms in individuals with lesions of denture stomatitis. Diag Microbiol Infect Dis. 2013;76(4):419–424. doi: 10.1016/j.diagmicrobio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Rahn E, Petermann P, Hsu MJ, Rixon FJ, Knebel-Morsdorf D. Entry pathways of herpes simplex virus type 1 into human keratinocytes are dynamin- and cholesterol-dependent. Plos One. 2011;6(10):e25464. doi: 10.1371/journal.pone.0025464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sacks SL, Griffiths PD, Corey L, Cohen C, Cunningham A, Dusheiko GM, Self S, Spruance S, Stanberry LR, Wald A, Whitley RJ. HSV shedding. Antiviral Res. 2004;63:S19–S26. doi: 10.1016/j.antiviral.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 66.Schwarzbauer JE, DeSimone DW. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb Perspect Biol. 2011;3(7):a005041. doi: 10.1101/cshperspect.a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott DA, Coulter WA, Lamey PJ. Oral shedding of herpes simplex virus type 1: a review. J Oral Pathol Med. 1997;26(10):441–447. doi: 10.1111/j.1600-0714.1997.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 68.Shukla SY, Singh YK, Shukla D. Role of Nectin-1, HVEM, and PILR-α in HSV-2 entry into human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2009;50(6):2878–2887. doi: 10.1167/iovs.08-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skerl KG, Calderone RA, Segal E, Sreevalsan T, Scheld WM. In vitro binding of Candida albicans yeast cells to human fibronectin. Can J Microbiol. 1984;30(2):221–227. doi: 10.1139/m84-033. [DOI] [PubMed] [Google Scholar]
- 70.Stump JD, Sticht H. Mutations in herpes simplex virus gD protein affect receptor binding by different molecular mechanisms. J Molecu Model. 2014;20(4):1. doi: 10.1007/s00894-014-2192-x. [DOI] [PubMed] [Google Scholar]
- 71.Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. Evolution and diversity in human herpes simplex virus genomes. J Virol. 2014;88(2):1209–1227. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taylor TJ, Brockman MA, McNamee EE, Knipe DM. Herpes simplex virus. Front Biosci. 2002;7:D752–D764. doi: 10.2741/taylor. [DOI] [PubMed] [Google Scholar]
- 73.Teng YHF, Aquino RS, Park PW. Molecular functions of syndecan-1 in disease. Matrix Biol. 2012;31(1):3–16. doi: 10.1016/j.matbio.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tiwari V, Liu J, Valyi-Nagy T, Shukla D. Anti-heparan sulfate peptides that block herpes simplex virus infection in vivo. J Biol Chem. 2011;286:25406–25415. doi: 10.1074/jbc.M110.201103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tiwari V, Clement C, Xu D, Valyi-Nagy T, Yue B, Liu J, Shukla D. Role for 3-O-sulfated heparan sulfate as the receptor for herpes simplex virus type 1 entry into primary human corneal fibroblasts. J Virol. 2006;80(18):8970–8980. doi: 10.1128/JVI.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tlaskalová-Hogenová H, Stepánková R, Hudcovic T, Tucková L, Cukrowska B, Lodinová-Zádníková R, Kozáková H, Rossmann P, Bártová J, Sokol D, Funda D, Borovská D, Reháková Z, Sinkora J, Hofman J, Drastich P, Kokesová A. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 2004;93:97–108. doi: 10.1016/j.imlet.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 77.Veerachamy S, Yarlagadda T, Manivasagam G, Yarlagadda P. Bacterial adherence and biofilm formation on medical implants: a review. Proceed Institut Mech Engin Part H-J Engin Med. 2014;228(10):1083–1099. doi: 10.1177/0954411914556137. [DOI] [PubMed] [Google Scholar]
- 78.Wagner C, Iking-Konert C, Hug F, Stegmaier S, Heppert V, Wentzensen A, Hänsch G. Cellular inflammatory response to persistent localized Staphylococcus aureus infection: phenotypical and functional characterization of polymorphonuclear neutrophils (PMN) Clin Exper Immunol. 2006;143(1):70–77. doi: 10.1111/j.1365-2249.2005.02963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wisplinghoff H, Bischoff T, Tallent M, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(7):309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 80.Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, Monticelli S, Lanzavecchia A, Sallusto F. Pathogen-induced human T(H)17 cells produce IFN-gamma or IL-10 and are regulated by IL-1 beta. Nature. 2012;484(7395):514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]