Abstract
Extracellular Cu/Zn superoxide dismutases (SODs) are critical for balancing the level of reactive oxygen species in the extracellular matrix of eukaryotes. In the present study we have detected constitutive SOD activity in the haemolymph and defensive secretions of different leaf beetle species. Exemplarily, we have chosen the mustard leaf beetle, Phaedon cochleariae, as representative model organism to investigate the role of extracellular SODs in antimicrobial defence. Qualitative and quantitative proteome analyses resulted in the identification of two extracellular Cu/Zn SODs in the haemolymph and one in the defensive secretions of juvenile P. cochleariae. Furthermore, quantitative expression studies indicated fat body tissue and defensive glands as the main synthesis sites of these SODs. Silencing of the two SODs revealed one of them, PcSOD3.1, as the only relevant enzyme facilitating SOD activity in haemolymph and defensive secretions in vivo. Upon challenge with the entomopathogenic fungus, Metarhizium anisopliae, PcSOD3.1-deficient larvae exhibited a significantly higher mortality compared to other SOD-silenced groups. Hence, our results serve as a basis for further research on SOD regulated host-pathogen interactions. In defensive secretions PcSOD3.1-silencing affected neither deterrent production nor activity against fungal growth. Instead, we propose another antifungal mechanism based on MRJP/yellow proteins in the defensive exudates.
Using oxygen in aerobic metabolism provides more energy per glucose than glycolysis does and hence constitutes an evolutionary advantage. Anyhow, molecular oxygen can be reduced to highly reactive by-products, termed reactive oxygen species (ROS), that can inflict damage on almost all molecules1. Oxygen is therefore a stringent selection criterion acting on organisms to sustain its reactive variants2,3,4. O2•− is considered the “primary” ROS, which can further interact with other molecules to generate “secondary” ROS including hydroxyl radicals (•OH), hydroperoxyl radicals (HOO•) or hydrogen peroxide (H2O2)5,6. While organisms evolved antioxidant activity to counteract the deleterious effects (termed oxidative stress) of these oxygen variants, they have also developed mechanisms to use ROS as vital components in cellular signalling pathways or immune response5,7,8.
Superoxide dismutases (SOD, E.C. 1.15.1.1), common to all kingdoms of life, serve as first-line antioxidant enzymes6,9. They dismutate O2•− into O2 and H2O2 with diffusion-limited rate constants. SODs are classified according to their metal co-factors and localization10. In eukaryotes two intracellular forms are known. The dimeric Cu/Zn-binding SODs (SOD1; 32 kDa) are localized in the cytoplasm, outer mitochondrial space and nucleus11, whereas the tetrameric MnSODs (SOD2; 89 kDa) is exclusively present in the inner mitochondrial space12. The most recently discovered members of the SOD family are the tetrameric Cu/Zn SODs (SOD3; EC-SOD; 135 kDa)13,14,15,16 functioning in the extracellular matrix.
Although SOD3 has been conserved among metazoans, the first functional evidence for a SOD3 in insects has been reported from the ant Lasius niger not before the 21st century17. And still, the understanding of the function of extracellular SOD isoforms in insects is incomplete and only a few examples have been published to date. For Drosophila melanogaster a SOD3 was reported to exhibit a protective effect against oxidative stress caused by ROS generating agents such as paraquat18, UV-radiation, and has an impact on life-span regulation19. Further, it has been assumed that extracellular SODs play a role in insect-parasitoid interactions20. Parasitoid wasps, for example, produce extracellular SODs and secrete them into their venom21. These exogenous SODs are suggested to increase the survival of the parasitoid eggs upon injection of the venom into an insect host during oviposition.
However, extracellular variants of SODs do not seem to play only a role in wasp venoms but can be found also in other insect defensive exudates. In the present study, we have identified a SOD3 isoenzyme in the proteome of the larval defensive secretions collected from the mustard leaf beetle Phaedon cochleariae (family Chrysomelidae, subtribe Chrysomelina). Within the Chrysomelina, the larvae of all species defend themselves by semiochemicals produced and released from specialized exocrine glands upon disturbance22,23,24,25. The defensive glands function as “bioreactors” where pre-toxins are enzymatically converted into biological active compounds. In P. cochleariae, the defensive iridoid chrysomelidial is converted from imported 8-hydroxygeraniol glucoside26,27,28,29,30,31. This transformation involves the hydrolysis of the glucoside and oxidation of the two primary hydroxy groups to produce the dialdehyde 8-oxogeranial, followed by a cyclisation step29,32,33,34,35. SODs could function as superoxide scavengers during the FAD dependent oxidation of 8-hydroxygeraniol or as a general protector of secretions’ components from oxidative stress during storage. Besides in P. cochleariae, oxidation reactions in the defensive secretions of larvae can also be found in related Chrysomelina species29,36,37,38.
By means of P. cochleariae larvae, we report the identification of a glandular extracellular SOD together with an additional extracellular isoform in the haemolymph. Given the open circulation in insects, the SODs are in contact with all tissues and, therefore, it is hypothesized that these enzymes have an overall function in the insects, for example, in the innate immunity. Their importance in ROS balancing mechanisms prompted us to investigate extracellular SODs and their role in opposing microbial invaders in the haemolymph as well as their possible function in the defensive larval exudates. As we have found constitutive extracellular SOD activity in secretions and haemolymph in related leaf beetle species we propose a widespread function of SOD3 in extracellular fluids of insects.
Results
Distribution of extracellular SOD activity among juvenile leaf beetles
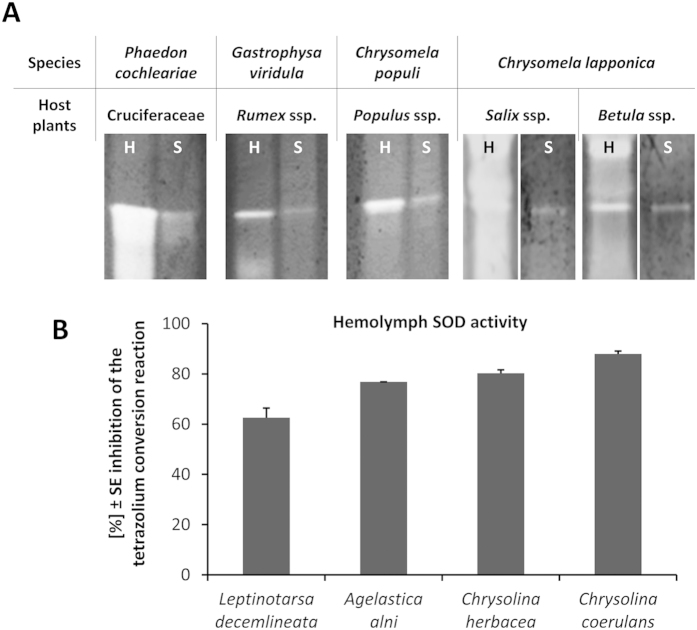
In order to examine SOD activity in extracellular fluids of leaf beetles, we have carried out in gel activity assays. We detected activity in the defensive secretions as well as in the haemolymph of all analysed larvae which were collected from different related species of the subtribe Chrysomelina (Fig. 1A). Interestingly, the extracellular SOD activity in the larval secretions is independent from their pathway of deterrent production. The biosynthesis of deterrents usually involves an oxidation reaction, as in the case of P. cochleariae, Gastrophysa viridula, Chrysomela populi, willow-feeding Chrysomela lapponica. However, birch-feeding C. lapponica larvae are devoid of an oxidation step in the deterrent synthesis and could therefore surrender SOD activity in the secretions38,39,40. Further analyses of other larvae of leaf beetles including members of the genera Chrysolina (Chrysomelini, Chrysolinina) Leptinotarsa (Chrysomelini, Doryphorina) and Agelastica (Galerucinae, Sermylini) showed activity in their haemolymph (Fig. 1B). It is, therefore, reasonable to assume that extracellular SOD activity is a widespread phenomenon in leaf beetles, which did not emerge in just a single subtribe. For our further experiments, we have chosen the mustard leaf beetle, P. cochleariae, as case study to investigate the function of extracellular SODs in insects.
Figure 1. Detection of SOD activity in defensive exudates and haemolymph collected from larvae of different leaf beetle species.
(A) From each species 1 μl haemolymph (H, left) and 0.3 μl secretion (S, right) was separated on native PAGE and functionally stained for SOD activity by using a qualitative zymography method based on the conversion of the tetrazolium salt, MTT. SOD activity inhibits the formation of a dark blue formazane dye and corresponds to white bands on a dark background. Here we show cropped gel sections. Full-length gels are presented in Supplementary Figs S14 and S15. (B) From pooled individuals of each species 0.8 μl of haemolymph was used in a 20 μl reaction mixture for the SOD assays using a quantitative microtiter plate method. SOD activity was determined colorimetrically at 440 nm as the inhibition of the reduction of the tetrazolium salt, WST-1 (n = 3).
Identification of extracellular Cu/Zn SODs in juvenile Phaedon cochleariae
Larval secretions as well as cell-free haemolymph were at first separated by 1D-SDS-PAGE. After recovering, the protein bands were then analysed by LC-MSE. For each of the two samples, the peptides derived from proteins of a molecular weight of 15 to 25 kDa were matched to data base proteins with similarity to Cu/Zn SODs (Sod_Cu (PF00080) EC 1.15.1.1). The full-length amplification and sequencing of the corresponding transcripts led to coding sequences of 516 bp for PcSOD3.1 and 534 bp for PcSOD3.2 for P. cochleariae. While PcSOD3.1 has been identified by LC-MSE analyses in the defensive secretions and haemolymph, PcSOD3.2 has been determined only in the haemolymph (Supplementary Table S1).
The deduced amino acid sequences PcSOD3.1 and PcSOD3.2 possess about 50% identity with each other. For PcSOD3.1 and PcSOD3.2, N-terminal signal peptides were specified by cleavage site predictions suggesting those to be extracellular proteins. Further analyses of their structure by homology modelling using Phyre241 revealed for both proteins active sites and binding motifs for the metal cofactors which are summarized in Table 1 and visualized in Supplementary Figs S1 and S2. PcSOD3.2 possesses amino acid exchanges in position H85 and H102 into proline and asparagine, respectively.
Table 1. Main characteristics of Cu/Zn SOD protein sequences identified in P. cochleariae.
| PcSOD3.1 | PcSOD3.2 | PcSOD1 | |
|---|---|---|---|
| Protein (aa) | 171 | 177 | 153 |
| Signal peptide | Yes (aa 1–16) | Yes (aa 1–20) | no |
| Molecular mass (kDa) | 17.8 | 18.8 | 15.9 |
| N-Glycosylation | N in position 36 | N in position 43 | N in position 23 |
| Cysteine residues forming disulphide bonds | Cys74-Cys163 | Cys79-Cys169 | Cys56-Cys144 |
| Zn2+ binding site | H80, H88, H97, D100 | P85, H93, N102, D105 | H62, H70, H79, D82 |
| Cu2+ binding site | H63, H65, H80, H137 | H68, H70, P85, H143 | H45, H47, H62, H119 |
| Active site | H63, H65, H80, H97, D100, H137 | H68, H70, P85, N102, D105, H143 | H45, H47, H62, H79, D82, H119 |
In order to demonstrate the in vitro activity of PcSOD3.1 and PcSOD3.2, both enzymes have been expressed heterologously in E. coli and purified subsequently. In the enzyme assays, recombinant PcSOD3.1 displayed an activity of 36% ± 8% whereas PcSOD3.2 showed an activity of only 1% ± 0.6% (see Supplementary Fig. S3). Hence, PcSOD3.2 seems to have a strongly reduced activity compared to PcSOD3.1, most likely due to the amino acid exchanges in its active centre.
Phylogenetic analyses including intra- and extracellular Cu/Zn SODs encoding sequences inferred from our transcriptome reference libraries, other insect and mammalian homologs showed a clustering of the identified PcCu/Zn SODs into the group of extracellular SODs (Supplementary Fig. S4A). While one group of intracellular SODs cluster together, the other group is formed by the extracellular variants. Human SOD3 is grouped to the extracellular enzymes. Sequences similar to PcSOD3.1 and PcSOD3.2 were found in related Chrysomelina species with bootstrap values of 43% and 100%, respectively. Chrysomeline PcSOD3.2 homologs also possess amino acid changes in the positions as in PcSOD3.2 (Supplementary Fig. S4B). Remarkably, within the extracellular proteins conserved sequences were found with a high bootstrap value of 70%. A physiological function, however, has not been described as yet for any of these proteins.
Localization of Cu/Zn SODs in juvenile P. cochleariae
We performed qPCR to analyse the transcript abundance of the extracellular SODs in P. cochleariae larvae. The defensive glands predominantly expressed PcSOD3.1. In the fat-body tissue, we detected a high expression of both PcSOD3.1 and PcSOD3.2 (Supplementary Fig. S5). Due to the relatively low mRNA levels in all the other tested tissues, including gut, Malpighian tubules, haemolymph containing haemocytes, and head we consider the defensive glands and fat body tissue as the main synthesis sites of PcSOD3.1 and PcSOD3.2.
In human SOD3, the C-terminus is highly rich in positively charged lysine and arginine residues that are involved in anchoring to heparan sulfate proteoglycans on cell surfaces and connective tissue matrix42,43,44. This C-terminal domain can be proteolytically cleaved to facilitate entrance to extracellular fluids. The two extracellular insect PcSODs, however, lack such a heparin binding peptide (Supplementary Fig. S1). In order to test the membrane association of extracellular SODs, the tissues of Malpighian tubules and fat body from juvenile P. cochleariae have been separated into membrane and cytosolic fractions. All fractions exhibited SOD activity (Supplementary Fig. S6). By using LC-MSE proteome analyses, we were, however, not able to detect extracellular SODs in any of the samples. Instead, we identified an additional Cu/Zn SOD (Supplementary Table S1). The corresponding full-length sequence does not have a predicted peptide signal suggesting that it is a cytoplasmic variant. It has been therefore named PcSOD1 (Table 1). This PcSOD1 is expressed in all tested larval tissues (Supplementary Fig. S5). The activity of recombinant PcSOD1 has also been assayed in vitro and was found to be active with 59% ± 6% (Supplementary Fig. S3). As SOD1 has been reported to play an important role for ROS detoxification and immune response in other insects we have included PcSOD1 into our further analyses as a positive control of RNAi effects45,46,47,48,49.
Effects of PcSOD3.1 silencing on the defensive secretions of P. cochleariae
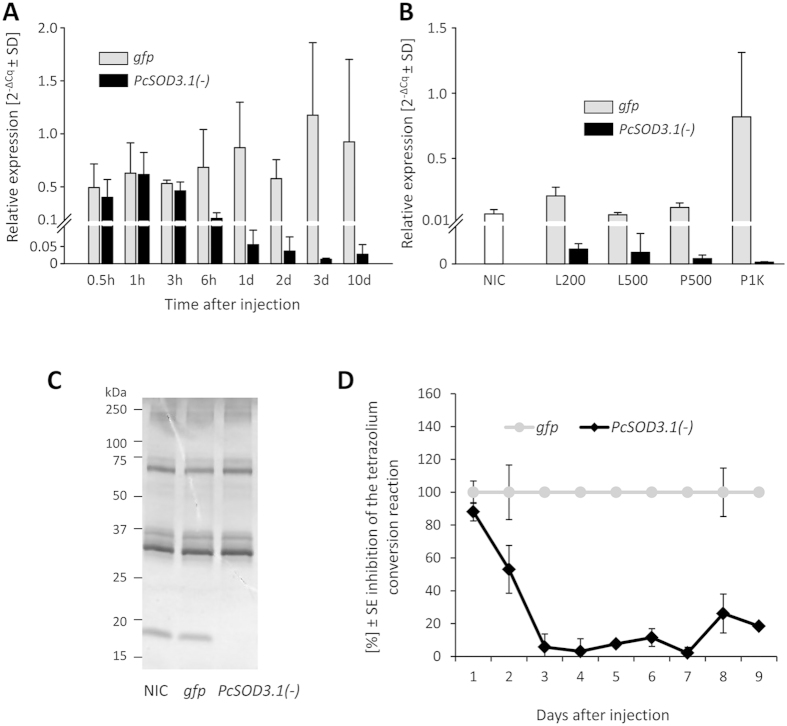
To study the function of extracellular SODs in leaf beetles in vivo we have tested RNAi by targeting the first candidate PcSOD3.1. We injected dsRNA-PcSOD3.1 into each of early second-instar larvae. A significant reduction of the transcript in the whole larvae compared to the control group has been measured already 6 h after injection (29.5% ± 7.9), which continued dropping down to 1% ± 1 within 24 h and revealed to be stable until pupation 10 days later (0.6% ± 0.6) (Fig. 2A). Though the reduction of the transcript was stable even in adult beetles until their death (Fig. 2B), eight weeks after maturation (4.5% ± 7.4), the silencing effects were not transmitted into the F1-generation (Supplementary Fig. S7) (cf. Bucher et al.50).
Figure 2. RNAi effects on transcript and protein abundance, and SOD activity in P. cochleariae.
(A) Transcript abundance of PcSOD3.1 after injection with 200 ng PcSOD3.1-dsRNA or gfp as a control, respectively. RNA from whole larvae has been used as template. The efficiency corrected Cq values of PcSOD3.1-silenced samples have been normalized to the reference genes PcRPL6 and PcRPS3 (n = 3). (B) Resulting transcript abundance of whole female adult beetles eight weeks after maturation, which were injected with PcSOD3.1-dsRNA or gfp-dsRNA during different developmental stages (n = 5). L200, larvae injected with 200 ng dsRNA; L500, larvae injected with 500 ng dsRNA; P500, pupae injected with 500 ng dsRNA; P1K, pupae injected with 1 μg dsRNA. (C) Coomassie stained SDS-PAGE with 0.3 μl of larval defence secretions from each experimental group, nine days after RNAi induction as described in (A). Here we show a cropped gel section. The full-length gel is presented in Supplementary Fig. S16. NIC, Non-injected control. (D) Time series of SOD activity of defence secretions, quantitatively determined by using a colorimetric microtiter plate assay. From pooled individuals from each experimental group 0.3 μl of defensive secretions was used in a 20 μl reaction mixture for the SOD assays. Values of the PcSOD3.1(−)-group were normalized to gfp-control (n = 3).
Consistently with the reduction of the transcript level, both, the protein and the enzymatic activity, decreased rapidly in P. cochleariae larvae (Fig. 2C,D). PcSOD3.1 was not detectable anymore in the defence secretome nine days after dsRNA-PcSOD3.1 injection whereas the protein pattern of the remaining proteins compared to the controls remained unchanged. SOD activity of the secretion was reduced to 5.7% ± 7.1 compared to gfp-control and stayed reduced by more than 80% until prepupal stage.
In addition, we checked the amount of chrysomelidial in the defence secretion, as an indicator of an RNAi-related alteration of the de novo synthesis of this defence allomone, but observed no significant differences among the populations (Supplementary Fig. S8). Hence, the silencing of PcSOD3.1 in the defensive secretions did not affect the production of the defensive chemistry.
Comparing the silencing effects of extracellular and cytoplasmic Cu/Zn SODs in the secretions and haemolymph of P. cochleariae
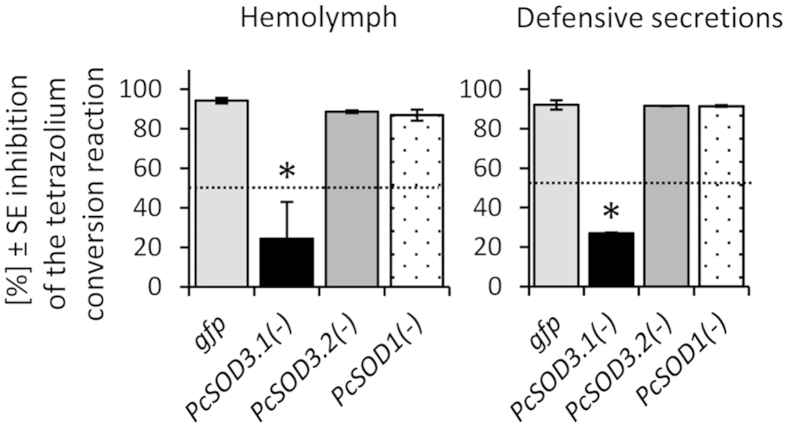
Besides PcSOD3.1 we have knocked-down PcSOD3.2 and PcSOD1 individually. By comparing the transcript levels of the Cu/ZnPcSODs we observed no significant co-silencing or up-regulation in whole larvae 10 days after dsRNA injection (Supplementary Fig. S9). A quantitative SOD assay revealed an activity significantly reduced only in the PcSOD3.1(−)-group; PcSOD3.1 seems to be the responsible enzyme for the constitutive SOD activity found in haemolymph and secretions (Fig. 3). Hence, no other SOD putatively existing in the genome seems to take over the function to the extent of PcSOD3.1 in haemolymph and secretions after its silencing.
Figure 3. RNAi effects after individual silencing of Cu/Zn PcSODs in extracellular fluids of P. cochleariae larvae.
SOD activity was measured ten days after injection with 200 ng of PcSOD3.1-, PcSOD3.2-, PcSOD1- or gfp-dsRNA. The used colorimetric assay was based on the inhibition of the reduction of the tetrazolium salt, WST-1. From pooled individuals of each experimental group 0.5 μl of haemolymph (left graph) and 0.3 μl of defensive secretions (right graph) was assayed regarding SOD activity in a 20 μl reaction mixture (n = 3). Level of significance: *p < 0.05 (ANOVA on ranks, Turkey test).
Effect of Cu/Zn SOD silencing on beetle development and life-span
We induced RNAi in second-instar larvae and monitored long-term effects due to decreased SOD activity in P. cochleariae. The period of time in which individuals of one group remained in one developmental stage, appeared not to differ significantly in the transition from larval to pupal and to adult stage (Supplementary Fig. S10). Monitoring the longevity revealed that the silencing of PcSOD1 resulted in an earlier death of individuals after 115 days compared to controls or the extracellular SOD-deficient groups (Supplementary Fig. S11). This observation is in accordance with papers showing an impact of SOD1 in life-span of e.g. D. melanogaster knockout mutants e.g.49. Statistic tests applied to our data, however, did not demonstrate significance for the observed phenomenon. Concluding, the probability of survival of P. cochleariae is not impaired by the silencing of the single PcSOD genes.
Consequences of SOD-knock-down after microbial challenge
As O2•− and hydrogen peroxide play important roles on innate immune response against potentially invasive organisms45,51, we tested whether the deficiency in one Cu/Zn PcSOD alters survival of P. cochleariae larvae exposed to microbes after inoculation with the entomopathogenic fungus M. anisopliae, the Gram(−)bacterium E. coli K12 and the Gram(+)bacterium M. luteus.
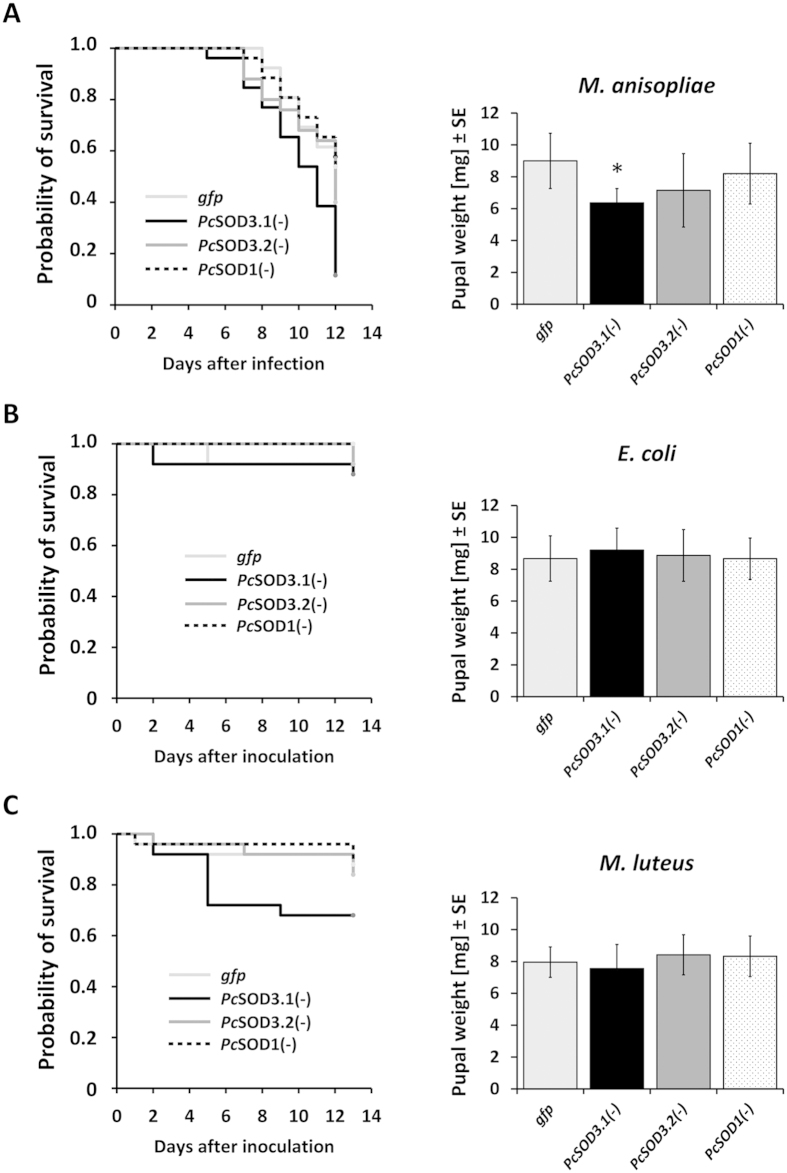
Testing pathogenicity of M. anisopliae, we could observe a fast increase in mortality of all groups, starting 6 days after inoculation (Fig. 4A). The probability of the survival of the PcSOD3.1(−) group was significantly reduced compared to that of the other groups. Also the pupal weight was reduced to average 6 mg which was significantly less than the 9 mg of gfp control. Hence, the PcSOD3.1(−) larvae suffering during the whole larval phase from fungal infection were possibly compromised in optimal nutrition compared to each of the other groups with intact PcSOD3.1 enzyme.
Figure 4. Immunologic consequences of Cu/Zn PcSOD silencing in P. cochleariae.
Thirty larvae were injected each with 200 ng of PcSOD3.1-, PcSOD3.2-, PcSOD1-dsRNA or gfp-dsRNA. Five days after this initial injection, larvae were challenged with microbes. The probability of survival is depicted as Kaplan-Meier curves by applying Gehan-Breslow statistic test. The weight of freshly emerged pupa was determined after RNAi silencing and infection of larvae. (A) Survival upon fungal challenge. After emptying of the defence reservoirs, the larvae of each group were submerged at the same time in the same solution of M. anisopliae 1*106 conidia/ml. Overall level of significance: p = 0.014 (Gehan-Breslow test); p = 0.04 (Log Rank (Mantel-Cox)). Pairwise multiple comparison procedures for gfp vs. PcSOD3.1(−): p = 0.04 (Holm-Sidak method). Pupal weight, level of significance: p = 0.023* (two-tailed t-test). (B) Survival after injection of 1*106 E. coli-cells/200 nl. (C) Survival after injection of 1*106 M. luteus-cells/200 nl.
LC-MSE analyses of haemolymph revealed the induction of antimicrobial peptides as part of the humoral defence upon microbial challenge (Supplementary Table S2)52,53,54,55. For example, a protein sequence with a similarity of 77% to defensin from Sitophilus zeamais56 was detectable only in the infected haemolymph samples and not in the control. A protein, 61% similar to S. zeamais attacin C56, was about 20-fold increased after M. anisopliae infection. And also an acaloleptin A similar protein (83% similarity to the protein from Acalolepta luxuriosa57) was about 7-fold induced compared to the non-infected control. However, the antimicrobial peptides could apparently not prevent a lethal phenotype upon fungal infection.
Further time-dependent expression analyses comparing fungal-infected and non-infected larvae deficient in Cu/Zn SOD activity showed significant induction of PcSOD3.2 and PcSOD1 expression two days after inoculation (Supplementary Fig. S12). PcSOD3.1 was not significantly induced over a period of six days. LC-MSE analyses of the haemolymph of PcSOD3.1(−) larvae demonstrated that there is no induction of any other extracellular SOD upon infection compared to non-infected PcSOD3.1(−) larvae (Supplementary Table S3).
Because RNAi is systemic in beetles, a loss of PcSOD3.1 in the defensive secretions could have an effect on the germination of the conidia on the surface of the larvae. Agar diffusion tests showed, however, that silencing of PcSOD3.1 did not alter antifungal activity of the defensive secretions (Supplementary Fig. S13A). It is therefore reasonable to assume, that constitutive PcSOD3.1 plays a pivotal role for ROS homeostasis in the haemolymph in immune response to fungal challenge. By using LC-MSE analyses we have identified in the larval defensive exudates a protein similar to members of the yellow-like protein family characterized by the presence of a major royal jelly domain58,59 (Supplementary Table S4). In pilot experiments its silencing caused the loss of the antifungal activity of larval secretions against M. anisopliae (Supplementary Fig. S13B). Hence, the mode of action of antifungal activity in defensive secretions differs from that found in the haemolymph and is currently under investigation.
In case of inoculation by 1*106 E. coli cells, we could not detect any difference on the survival or the pupal weight between the groups (Fig. 4B). In a similar experiment, using 1*106 of M. luteus cells, we could detect a reduced, but statistically not significant, survival in the PcSOD3.1(−) group compared to gfp and PcSOD1(−) and PcSOD3.2(−) groups (Fig. 4C).
LC-MSE analyses of haemolymph revealed also the induction of antimicrobial peptides after E. coli and after M. luteus infection, including defensin, attacin C (50-fold induced compared to the non-infected control), and also an acaloleptin A similar protein (4–10-fold induced compared to the non-infected control) (Supplementary Table S2). Apparently, in our experiments the lack of SOD activity did not interfere significantly with the antibacterial activity of the immune response in the insects most likely due to the action of the antimicrobial peptides.
Discussion
The insect’s innate immune system is comprised of both cellular and humoral elements60,61,62,63,64. Invasive organisms that enter the host’s hemocoel encounter reactive haemocytes and an array of specific, e.g. antimicrobial peptides52,54,65,66, and non-specific, e.g. ROS, cytotoxic molecules45,51. Particularly, SODs have a paramount role in balancing ROS in an organism. We have identified two extracellular SODs, PcSOD3.1 and PcSOD3.2, and one cytosolic variant, PcSOD1, in juvenile P. cochleariae. Unlike the human extracellular SODs, the extracellular isoforms from the beetles are most likely not attached to the extracellular matrix, but secreted as soon as they are synthesized. This phenomenon has already been described for the extracellular proteins of Drosophila19. Fat body and defensive glands of juvenile P. cochleariae are suggested to be the main production sites of the two extracellular SODs. On the basis of in vitro activity assays, PcSOD3.1 displayed a higher activity compared to the activity of PcSOD3.2. As PcSOD3.2 is present in the haemolymph in much higher quantity compared to PcSOD3.1, we hypothesize a function for PcSOD3.2 in regulating ROS equilibrium in the haemolymph by e.g. trapping H2O2.
Processes causing ROS production in insects include the immune responses against microbes67,68. Our data show the vital role of PcSOD3.1 in the haemolymph particularly against entomopathogenic fungi compared to treatments with bacteria. Immune response against fungal infection includes melanisation leading to the physical encapsulation of intruders in a dense melanin coat which is accompanied by the production of ROS together with the redoxactive melanogenic intermediates63,69,70,71. In our opinion, a direct participation of PcSOD3.1 in the phenoloxidase cascade leading to melanin capsules could be excluded by the fact, that O2•− is needed to drive melanisation. It has been shown, that the enzymatic oxidation of DOPA as intermediate of the melanisation cascade is SOD sensitive in vitro72, and a SOD in the process of encapsulation is contra-productive since it slows down melanisation73. Instead, we hypothesize two possible functions for PcSOD3.1 identified in the beetle’s haemolymph. First, it could be intended to produce H2O2 which will fuel Fenton reactions. Second, H2O2 serves as signal intermediate, necessary for the transcriptional regulation of an adequate immune response74. H2O2 signalling, which is involved in immune response, proceeds via Nox-enzymes. Superoxide anions have to be converted in the extracellular space to H2O2 to be active in the cytosol after reimport74. This path is often described to rely on spontaneous dismutation of O2•− but SODs work an order of magnitude faster yielding H2O2 and prohibit the depolarization of the membrane due to too many negative charges75,76. To conclude, if PcSOD3.1 is missing in the haemolymph upon fungal infection, the melanisation cascade may be activated anyway, but the production of antimicrobial radicals or H2O2 for the regulation of downstream reactions leading to an effective immune response might be unbalanced.
Due to their sugar content, the larval defensive secretions of P. cochleariae could be a culture medium for microbes34. Therefore, it is not surprising that the secretions contain components of antimicrobial activity. For example, it has been demonstrated that chrysomelidial possesses an effect against Gram(−)bacteria and yeast77. In agar diffusion tests using defensive secretions from PcYellow-like(−) larvae against the entomopathogen M. anisopliae we found antifungal activity mediated presumably by a yellow-like protein. To date, the function of these proteins in insects is, however, largely not understood59. In D. melanogaster, for example, yellow members seem to be involved in the process of melanization78. In Apis mellifera these proteins are found as a major component in royal jelly, a substance which is fed to larvae for triggering their development into queens58. Yellow proteins have also been identified in the bee venoms, but a function has not been described as yet79. To what extent these proteins in bee venoms also have antifungal effects analogous to the defensive secretions, needs to be tested in the future.
In the chrysomelidial pathway, most likely PcSOD3.1 could have assisted the GMC oxidoreductase, Pc8HGO, being active in the defensive secretions32. But on the one hand, it has been shown, that the GMC oxidoreductases are producing directly H2O2 as a by-product of oxidation, without superoxide intermediates80. On the other hand, as the silencing of PcSOD3.1 did not interfere with the amount of chrysomelidial produced, we are not able to infer a direct function of PcSOD3.1 for deterrent production from our experiments. Regardless, a possible function of PcSOD3.1 could be the general protection of other secretions’ components, not yet identified, from oxidative stress during storage. This may explain why SOD activity is present in the defensive exudates of different chrysomeline species independent from the pathway of deterrent synthesis.
In summary, our results demonstrate that insects secrete extracellular SODs constitutively into extracellular fluids and that these proteins contribute to fungal defence in their innate immunity. As extracellular SOD activity seems to be a widespread phenomenon in the open circulation of insects, we created a suitable source on which to base current and future concepts regarding SOD regulated antimicrobial response in ecologically relevant non-model organisms.
Methods
Information on biological replicates and the individuals pooled within is given in the figure legends. Statistical calculations are indicated in the figure legends; Excel (Microsoft, Redmond, WA, USA), SPSS (IBM, Armonk, NJ, USA) and SigmaPlot (Systat Software, Inc., CA, USA) have been used for the calculations. All primer used in this study are listed in Supplementary Table S5. The accession numbers of all sequences identified from P. cochleariae and all sequences used in this study are listed in Supplementary Table S6. All chemicals are purchased from Serva (Heidelberg, Germany), Sigma-Aldrich (St. Louis, MO, USA) or Carl Roth (Karlsruhe, Germany), if not stated other.
Beetle rearing and sample collection of extracellular fluids
P. cochleariae was reared on Brassica rapa subsp. pekinensis “Cantonner Witkrop” (Quedlinburger Saatgut, Quedlinburg, Germany) in a Snijder chamber (Snijders scientific, Tilburg, Netherlands) in a light/dark cycle of 16 h light and 8 h darkness (LD 16/8) and 13 °C/11 °C ± 1 °C, 70% humidity. After microbial infection and during developmental experiments the insects were incubated at LD 14/10, 21 ± 2 °C, 70% humidity.
Larval secretions were collected in glass capillaries (i.d.: 0.28 mm, o.d.: 0.78 mm, length 100 mm; Hirschmann, Eberstadt, Germany). Secretions were weighed in the sealed capillaries on an ultra-microbalance (Mettler-Toledo, Greifensee, Switzerland) three times; the weight of the capillaries was subtracted and the final weight was averaged. Sealed capillaries were stored at −20 °C until needed. Haemolymph from P. cochleariae as well as from juveniles from related leaf beetle species was collected in graduated 5 μl glass capillaries (Hirschmann, Eberstadt, Germany) after scissoring of one meta-thoracal legs at the coxa and also stored at −20 °C after sealing.
Identification of SOD encoding sequences and their phylogenetic relationships
For the identification of putative Cu/Zn SODs we analysed computationally the transcriptome libraries of the three Chrysomelina species P. cochleariae81, C. populi82, and C. lapponica32. Using tool “transeq” from package EMBOSS (V6.3.1), the cDNAs of these libraries were translated into all six possible open reading frames. We annotated these protein sequences by applying pfamscan against the protein families’ database (Pfam, updated in Jan. 2013) with default parameters83. Those sequences that were annotated to possess a Sod_Cu domain (PF00080) and to be longer than 130 amino acids were selected for further analyses. Sequences were also searched via BLAST approach against the NCBI non-redundant database to validate the prediction84. In addition, SignalP (SignalP 4.1: http://www.cbs.dtu.dk/services/SignalP/)85 has been used to identify signal peptides. The tool GlycoEP has been used to predict N-Glycosylation sites (http://www.imtech.res.in/raghava/glycoep/)86. Selected sequences have been further analysed by using Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index)41.
To analyse the phylogenetic relationships of the identified Cu/Zn SOD sequences from the three Chrysomelina leaf beetle species and other insects, a phylogenetic tree has been constructed. In addition to the Chrysomelina sequences mentioned above, we included predicted Cu/Zn SODs of G. viridula and Leptinotarsa decemlineata identified in corresponding transcriptome libraries87, and of other Coleoptera, Hemiptera, Diptera, Hymenoptera, Lepidoptera, and Mammalia. The sequence of a bacterium was used as outgroup. The accession numbers of these sequences are listed in Supplementary Table S6. The identified protein sequences were aligned by using MAFFT (v7.023b) with default parameters and L-INS-i method. Afterwards, the randomized accelerated maximum likelihood tool (RAxML, V7.2.8) was applied with the parameter of ARTREVF (bootstrap value = 1000) to generate the phylogenetic tree.
Quantitative real time PCR
For generation of cDNA insects were dissected in 0.9% NaCl solution under an stereomicroscope (Zeiss, Jena, Germany), where fat body, gut and Malpighian tubules were frozen directly on the wall of liquid nitrogen cooled reaction tubes. Defensive glands were collected with a pipette in 200 μl RNAlater (Qiagen, Hilden, Germany). Whole larvae and adults were placed alive in 2 ml safe-lock reaction tubes (Eppendorf) containing 6–10 silica beads (Ø2mm) and poured with 200 μl lysis-buffer (kit see below). These samples were homogenized using a genogrinder2010 (Spex samplePrep, Metuchen, NJ, USA). All samples were stored until further processing at −80 °C. RNA extraction and cDNA synthesis were performed according to88 using the RNAqueous kit (Ambion, Thermo scientific), superscript III enzyme and oligo dT(12–18) primer (both Invitrogen, Thermo scientific). Quality was assessed by TA gel electrophoresis, and the A260/A280 value.
cDNA from haemolymph, head, gut, Malpighian tubules, fat body and accessory defence glands, whole larvae or adults of P. cochleariae was used as a template for expression analysis of Cu/Zn PcSODs using a CFX96 qPCR system (Biorad, Hercules, CA, USA) and Brilliant III SYBR green Mastermix (Agilent, Santa Clara, CA, USA). Cq values of PcSODs from three biological and two technical replicates were normalized to PcRPL6 and PcRPS3 (Supplementary Tables S5 and S6). Resulting data were analysed using qBASE PLUS software89. Assays were performed following the MIQE guidelines90.
Cloning of Cu/Zn PcSODs
Coding sequences of PcSOD3.1, PcSOD3.2, PcSOD1, and PcYellow-like were amplified from a cDNA pool of all tissues using Pfx-Polymerase (Invitrogen, Thermo scientific). The fragments were cloned into pIB/V5-HIS TOPO vectors (Invitrogen, Thermo scientific), which is lacking T7-promotor sites, interfering with dsRNA synthesis. For that, this part of each coding region has been chosen, which resulted to be unique after off-target-prediction88. These fragments were amplified using PCR with primers containing both a 5′-T7-promotor-sequence. Also, the unique part of the coding sequence of the green fluorescent protein from pcDNA3.1/CT-GFP-TOPO (Invitrogen, Thermo scientific) was subjected to the described protocol and cloned into a pIB-vector. The cloning products were primarily amplified using E. coli TOP10F’ cells (Invitrogen, Thermo scientific) and sequenced, to confirm unaltered dsRNA templates.
dsRNA synthesis
Sequenced pIB-plasmids were used to re-amplify DNA templates via PCR. The amplicons were subjected to T7-polymerase based in vitro transcription reactions (MEGAscript RNAi kit, Ambion, Thermo scientific) according to manufacturer’s instructions. The resulting dsRNA was eluted after nuclease digestion three times with 50 μl hot injection buffer (3.5 mM Tris-HCl, 1 mM NaCl, 50 nM Na2HPO4, 20 nM KH2PO4, 3 mM KCl, 0.3 mM EDTA, pH 7.0). For control of the influence of the activation of the RNAi-machinery, we by default raised a control group injected with corresponding amounts of dsRNA according to the gfp nucleotide sequence, which has no counterpart in P. cochleariae transcriptome. The concentration of dsRNA was measured spectrophotometrically, calculated with A260 = 1 = 45 mg/ml and adjusted to 2 μg/μl. The quality of dsRNA was checked by TBE-agarose-electrophoresis.
RNAi induction
Second instars of P. cochleariae with three mm body length and >1.0 mg body weight were injected individually with 0.2 μg of dsRNA about five days after hatching. Injections were accomplished with ice-chilled larvae using a Nano2010 injector (WPI, Sarasota, FL, USA) driven by a three-axis micromanipulator mounted to a stereomicroscope (Zeiss). The larvae were injected dorso-medial between the pro- and mesothorax.
Heterologous expression of Cu/Zn PcSODs
For the in vitro functional validation of Cu/Zn PcSODs, we used heterologously expressed 6xHIS-N-terminal tagged proteins. The coding sequences of PcSOD3.1, PcSOD3.2, and PcSOD1 omitting the signal peptide-sequence in cases of the extracellular variants were cloned directionally into pET100/D TOPO vectors (Invitrogen, Thermo scientific). Using the Calbiochem Autoinduction system 1 (Merck, Darmstadt, Germany), recombinant (r)PcSOD1, rPcSOD3.1, and rPcSOD3.2 were produced in E. coli strain BL21(DE3)star (Invitrogen, Thermo scientific) and purified according to Frick et al.27. A negative control has been included with a recycled pET100-TOPO vector with no insert.
Microbial infection
L2-larvae of P. cochleariae were treated with Cu/Zn PcSOD-dsRNA or gfp. Five days later, developed third-instar larvae from were injected with bacteria or dipped into fungal conidia solution. Hereafter they were kept at room temperature, to allow the pathogens to develop.
Metarhizium anisopliae (DSM-1490, DSMZ, Braunschweig, Germany) was maintained on potato-dextrose-agar (Sigma, 2% glucose), and mycelia were used for subsequent propagation. Conidia were harvested from liquid culture (potato-dextrose, 200 ml in 500 ml Erlenmeyer flask), which was incubated at room temperature for three weeks without shaking, after overnight agitation of cultures with a 10 cm magnetic stirring bar at 4 °C. Mycelium was separated by the filtration of the suspension through fine gauze. The supernatant, containing the conidia was centrifuged (10.000 × g, 4 °C, 10 min.) and washed twice with 0.1% aqueous tween20 solution. The concentration of conidia was adjusted to 1*106/ml (hemocytometer) and infection was performed according to Rostas et al.91. All larvae of one RNAi-induced group were submerged for 30 sec. in 1 ml of conidia suspension after swabbing the larvae on lab tissue, to get rid of defence secretions. The suspension was poured on filter paper and the larvae were air dried for 1 min. Only those larvae were assigned as killed by the fungus, which showed mycelia growth out of the carcass after incubating the corps on humid sterile filter paper in sealed petri dishes.
For bacterial infections, RNAi has been induced and 1*106 cells of E. coli K12 (NEB, Ipswich, MA, USA) or Micrococcus luteus (DSM 20030, DSMZ, Germany) in 100 nl were injected into third-instar larvae. Both bacterial species were maintained on LB-agar-plates and fresh bacteria were purified from inoculated liquid LB cultures, agitating for four days at 37 °C, 200 rpm. The cells were pelleted (5 min, 8.000 × g, 4 °C) in 50 ml Falcon tubes and washed twice in Ringer solution92 and adjusted to 1*1010 cells/ml. The experiment ended again with the death or the successful pupation of the species.
Probabilities of survival after the treatments have been statistically analysed by using Kaplan-Meier estimation with Gehan-Breslow statistic test and pairwise multiple comparison procedures (Holm-Sidak method) by applying SigmaPlot software (version 11.0). Log Rank (Mantel Cox) tests have been carried out by using SPSS software (version 17.0).
Haemolymph samples for analysing antimicrobial peptides by LC-MSE were collected from third-instar larvae four days after challenging with bacteria or M. anisopliae as described above.
Antifungal activity of secretions was testes in agar diffusion assays using conidia of M. anisopliae. Secretions of PcSOD3.1 or yellow-like silenced larvae were tested eight days after the triggering of RNAi. Potato-dextrose-agar (Sigma, 2% glucose) was inoculated with 1*106 conidia/100 ml agar.
Sample preparation for protein analyses
Samples of snap-frozen tissues of 8–10 larvae were placed into 50 μl Ringer’s solution with Protease-Inhibitor Mix HP (Serva, Heidelberg, Germany). Tissues were grinded with sea sand. After the centrifugation of the samples (10 min., 2000 × g, 4 °C), supernatant was stored at −20 °C in a fresh reaction tube.
Haemolymph samples were also centrifuged and the supernatant checked for being cell-less with a transmitted light microscope before activity assays and proteome analyses. Protein concentrations were determined spectrophotometrically or by using bicinchoninic acid assay (Pierce, Thermo scientific). For membrane protein isolation the Native Membrane Protein Extraction Kit (Calbiochem, Proteo Extract) was used. Frozen tissue samples were processed in accordance with the manufacturer’s instructions.
Protein digestion and nanoUPLC-MS/MS analysis
Protein samples were divided in two groups: for shotgun LC-MS/MS analysis and for Gel-LC-MS/MS approach. For gel-based LC-MS/MS analysis, protein samples were separated by any-kD gradient gels (Bio-Rad Laboratories, Munich, Germany) in SDS-PAGE93. Protein bands of Coomassie Brilliant blue R250 stained gels were cut from the gel matrix and tryptic digestion was carried out as described94. For LC-MS analysis the extracted tryptic peptides were reconstructed in 10 μl aqueous 1% formic acid. For semi-quantitative analyses of antimicrobial peptides, Waters MassPREP BSA digestion standard (Waters, Manchester, UK) as internal standard was added to the samples in the concentration of 50 fmol/μl.
For shotgun analysis the samples were subjected to TCA precipitation according to the protocol from Gorg et al.95. Prior precipitation each sample was spiked with BSA (Pierce, Thermo scientific) used as an internal standard for protein quantification, for 1 μg of protein 1 pmol of BSA was spiked. The protein pellets subjected for shotgun-LC-MS/MS were suspended in 100 mM NH4HCO3 by vortexing, heated to 90 °C for 20 min, cooled on ice and diluted with occasional vortexing with methanol to produce a sample solution containing 60% methanol. The samples were not fully dissolved and therefore stepwise digested with trypsin at a trypsin/protein ratio of 1:30 at 37 °C for approximately 12 hours.
The samples were acquired on a nanoAcquity UPLC system on-line connected to a Q-ToF Synapt HDMS mass spectrometer (Waters, Manchester, UK). The peptides were concentrated on a Symmetry C18 trap-column (20 × 0.18 mm, 5 μm particle size) using a mobile phase of 0.1% aqueous formic acid at a flow rate of 15 μL min−1 and separated on a nanoAcquity C18 column (200 mm × 75 μm ID, C18 BEH 130 material, 1.7 μm particle size) by in-line gradient elution at a flow rate of 0.350 μl min−1 using an increasing acetonitrile gradient from 1% to 95% B over 90 min (Buffers: A, 0.1% formic acid in water; B, 100% acetonitrile in 0.1% formic acid).
The eluted peptides were transferred into a Synapt HDMS Q-Tof tandem mass spectrometer equipped with a nanolockspray ion source (Waters, Manchester, UK). LC-MS data were collected using data-independent LC-MSE analysis96. Full scan LC-MS data were collected using alternating mode of acquisition: low energy (MS) and elevated energy (MSE) mode over 1.5 sec intervals in the range m/z of 50–1700 with an interscan delay of 0.2 sec. A reference compound, human Glu-Fibrinopeptide B [650 fmol/mL in 0.1% formic acid/ACN (v/v, 1:1)], was infused through a reference sprayer at 30 s intervals for external calibration. The data acquisition was controlled by MassLynx v4.1 software (Waters).
Data processing, protein identification and quantification
The acquired continuum LC-MSE data were processed using ProteinLynx Global Server (PLGS) version 2.5.2 (Waters) to generate product ion spectra for database searching according to Ion Accounting algorithm described97. The thresholds for low/ high energy scan ions and peptide intensity were set at 150, 30 and 750 counts, respectively. The processed data were searched against the P. cochleariae protein subdatabases constructed from in-house transcriptome-database by their translation from all six reading frames combined with Swissprot database downloaded on 13 August, 2011 from http://www.uniprot.org/. The database searching was performed at a False Discovery Rate (FDR) of 4%, following searching parameters were applied for the minimum numbers of: product ion matches per peptide (3), product ion matches per protein (5), peptide matches (1), and maximum number of missed tryptic cleavage sites (1). Searches were restricted to tryptic peptides with a fixed carbamidomethyl modification for Cys residues. Proteins were quantified using the top 3 matched peptides (Hi3 method) from the spiked internal standard (BSA) as described by Silva et al.98
SOD activity assays
SOD activity was determined by a qualitative in gel activity assay (in gel zymography) and a quanitative SOD microtiter plate assay. For conducting this zymography method, native gels were loaded with 15 μl of protein sample mixed with nondenaturing, nonreducing loading buffer. After performing electrophoresis at 150 V in a Native-PAGE running buffer, gels were incubated in the assay solution (80 ml 0.5 M Tris-HCl, 10 mg 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), 3 mg 5-methylphenazinium methyl sulfate (PMS), pH 8.0) for 15 min at 4 °C in the dark. Gels were then exposed to sunlight. SOD activity inhibited the reduction of the tetrazolium salt into the formazan dye and got visible, thus, as yellow bands on a dark-blue background.
Quantitative SOD activity assays, based on the conversion of the tetrazolium salt, WST-1 (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) into a formazan dye upon reduction with a superoxide anion, were carried out by using the SOD Determination Kit (Sigma-Aldrich, MO, USA) following the instructions of the manufacturer. The SOD standard inhibition curve was determined according to the manual with bovine erythrocyte SOD (15000 U/ml) and an incubation time of 20 min. Data sets were statistically evaluated by applying ANOVA on ranks (Kruskal-Wallis One Way Analysis of Variance on Ranks) together with a pairwise multiple comparison procedure (Tukey Test) by using SigmaPlot software (version 11.0).
Additional Information
How to cite this article: Gretscher, R.R. et al. A common theme in extracellular fluids of beetles: extracellular superoxide dismutases crucial for balancing ROS in response to microbial challenge. Sci. Rep. 6, 24082; doi: 10.1038/srep24082 (2016).
Supplementary Material
Acknowledgments
We gratefully acknowledge Angelika Berg, Yvonne Hupfer, Dr. Maritta Kunert, Gerhard Pauls, and Kerstin Ploss for technical assistance. We wish to thank Dr. Peter Rahfeld for help with the silencing experiments of yellow-like. We also like to thank our student assistants Christin Meißner, Toni Krause, Tim Baumeister, Anthea Wirges and Franziska Eberl for their enthusiastic help on daily lab-work over the last three years. Further we would like to thank Andreas Weber from the greenhouse team for the supply of Chinese cabbage. This work was supported by the Deutsche Forschungsgemeinschaft (BU 1862/2-1; 1862/2-2) and the Max Planck Society.
Footnotes
Author Contributions R.R.G., W.B. and A.B. designed the study and developed the experiments. R.R.G., P.S. and A.S.S. performed the research and analysed output data together with A.B.; M.S. analysed transcriptome data and performed in silico off-target-prediction for all RNAi targets. D.W. performed the phylogenetic analyses. N.W. carried out all proteomic analyses and data processing. R.R.G. and A.B. wrote the manuscript. All authors reviewed the manuscript.
References
- Miller D. M., Buettner G. R. & Aust S. D. Transition-metals as catalysts of autoxidation reactions. Free Radic. Biol. Med. 8, 95–108, 10.1016/0891-5849(90)90148-c (1990). [DOI] [PubMed] [Google Scholar]
- Gerschman R., Gilbert D. L., Nye S. W., Dwyer P. & Fenn W. O. Oxygen poisoning and x-irradiation-a mechanism in common. Science 119, 623–626, 10.1126/science.119.3097.623 (1954). [DOI] [PubMed] [Google Scholar]
- Nohl H. Generation of superoxide radicals as by-product of cellular respiration. Ann. Biol. Clin. (Paris) 52, 199–204 (1994). [PubMed] [Google Scholar]
- Valko M., Morris H. & Cronin M. T. D. Metals, toxicity and oxidative stress. Curr. Med. Chem. 12, 1161–1208, 10.2174/0929867053764635 (2005). [DOI] [PubMed] [Google Scholar]
- Valko M. et al. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 39, 44–84, 10.1016/j.biocel.2006.07.001 (2007). [DOI] [PubMed] [Google Scholar]
- Miller A.-F. Superoxide dismutases: Ancient enzymes and new insights. FEBS Lett. 586, 585–595, 10.1016/j.febslet.2011.10.048 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F. & Giulivi C. Superoxide dismutases and their impact upon human health. Mol. Aspects Med. 26, 340–352, 10.1016/j.mam.2005.07.006 (2005). [DOI] [PubMed] [Google Scholar]
- Jomova K. & Valko M. Advances in metal-induced oxidative stress and human disease. Toxicology 283, 65–87, 10.1016/j.tox.2011.03.001 (2011). [DOI] [PubMed] [Google Scholar]
- Bannister W. H. From haemocuprein to copper-zinc superoxide dismutase: a history on the fiftieth anniversary of the discovery of haemocuprein and the twentieth anniversary of the discovery of superoxide dismutase. Free Radic. Res. Commun. 5, 35–42, 10.3109/10715768809068556 (1988). [DOI] [PubMed] [Google Scholar]
- Zelko I. N., Mariani T. J. & Folz R. J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 33, 337–349, 10.1016/s0891-5849(02)00905-x (2002). [DOI] [PubMed] [Google Scholar]
- McCord J. M. & Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244, 6049–6055 (1969). [PubMed] [Google Scholar]
- McCord J. M. Iron- and manganese-containing superoxide dismutases: structure, distribution, and evolutionary relationships. Adv. Exp. Med. Biol. 74, 540–550, 10.1007/978-1-4684-3270-1_45 (1976). [DOI] [PubMed] [Google Scholar]
- Marklund S. L. Human copper-containing superoxide dismutase of high molecular weight. Proc. Natl. Acad. Sci. USA 79, 7634–7638, 10.1073/pnas.79.24.7634 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund S. L. In Methods Enzymol. Vol. 349 (ed Packer L.) 74–80 (Academic Press, 2002). [DOI] [PubMed] [Google Scholar]
- Nozik-Grayck E., Suliman H. B. & Piantadosi C. A. Extracellular superoxide dismutase. Int. J. Biochem. Cell Biol. 37, 2466–2471, 10.1016/j.biocel.2005.06.012 (2005). [DOI] [PubMed] [Google Scholar]
- Fattman C. L., Schaefer L. M. & Oury T. D. Extracellular superoxide dismutase in biology and medicine. Free Radic. Biol. Med. 35, 236–256, 10.1016/s0891-5849(03)00275-2 (2003). [DOI] [PubMed] [Google Scholar]
- Parker J. D., Parker K. M. & Keller L. Molecular phylogenetic evidence for an extracellular CuZn superoxide dismutase gene in insects. Insect Mol. Biol. 13, 587–594, 10.1111/j.0962-1075.2004.00515.x (2004). [DOI] [PubMed] [Google Scholar]
- Blackney M. J., Cox R., Shepherd D. & Parker J. D. Cloning and expression analysis of Drosophila extracellular Cu Zn superoxide dismutase. Biosci. Rep. 34, 851–863, 10.1042/bsr20140133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I., Kim T. Y. & Kim-Ha J. Identification of Drosophila SOD3 and its protective role against phototoxic damage to cells. FEBS Lett. 585, 1973–1978, 10.1016/j.febslet.2011.05.033 (2011). [DOI] [PubMed] [Google Scholar]
- Zhu J. Y., Ze S. Z., Stanley D. W. & Yang B. Parasitization by Scleroderma guani influences expression of superoxide dismutase genes in Tenebrio molitor. Arch. Insect Biochem. Physiol. 87, 40–52, 10.1002/arch.21179 (2014). [DOI] [PubMed] [Google Scholar]
- Colinet D., Cazes D., Belghazi M., Gatti J. L. & Poirie M. Extracellular superoxide dismutase in insects: characterization, function, and interspecific variation in parasitoid wasp venom. J. Biol. Chem. 286, 40110–40121, 10.1074/jbc.M111.288845 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteels J. M., Rowellrahier M., Braekman J. C. & Daloze D. Chemical defenses in leaf beetles and their larvae-the ecological, evolutionary and taxonomic significance. Biochem. Syst. Ecol. 12, 395–406, 10.1016/0305-1978(84)90071-1 (1984). [DOI] [Google Scholar]
- Pasteels J. M. & Rowell-Rahier M. Defensive glands and secretions as taxonomical tools in the Chrysomelidae. Entomography 6, 423–432 (1989). [Google Scholar]
- Renner K. Über die ausstülpbaren Hautblasen der Larven von Gastroidea viridula De Geer und ihre ökologische Bedeutung (Coleoptera: Chrysomelidae). Beitr. Entomol. 20, 527–533 (1970). [Google Scholar]
- Hinton H. E. On a little-known protective device of some chrysomelid pupae (Coleoptera). Proc. R. Ent. Soc. Lond. (A) 26, 67–73, 10.1111/j.1365-3032.1951.tb00123.x (1951). [DOI] [Google Scholar]
- Burse A. et al. Iridoid biosynthesis in Chrysomelina larvae: Fat body produces early terpenoid precursors. Insect Biochem. Mol. Biol. 37, 255–265, 10.1016/j.ibmb.2006.11.011 (2007). [DOI] [PubMed] [Google Scholar]
- Frick S. et al. Metal ions control product specificity of isoprenyl diphosphate synthases in the insect terpenoid pathway. Proc. Natl. Acad. Sci. USA 110, 4194–4199, 10.1073/pnas.1221489110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith M., Lorenz M., Boland W., Simon H. & Dettner K. Biosynthesis of iridoid monoterpenes in insects: Defensive secretions from larvae of leaf beetles (Coleoptera: Chrysomelidae). Tetrahedron 50, 6859–6874, 10.1016/S0040-4020(01)81338-7 (1994). [DOI] [Google Scholar]
- Lorenz M., Boland W. & Dettner K. Biosynthesis of iridodials in the defense glands of beetle larvae (Chrysomelinae). Angew. Chem. Int. Ed. Engl. 32, 912–914, 10.1002/anie.199309121 (1993). [DOI] [Google Scholar]
- Oldham N. J., Veith M., Boland W. & Dettner K. Iridoid monoterpene biosynthesis in insects-evidence for a de novo pathway occurring in the defensive glands of Phaedon armoraciae (Chrysomelidae) leaf beetle larvae. Naturwissenschaften 83, 470–473, 10.1007/s001140050318 (1996). [DOI] [Google Scholar]
- Pasteels J. M., Duffey S. & Rowell-Rahier M. Toxins in chrysomelid beetles, possible evolutionary sequence from de novo synthesis to derivation from food-plant chemicals. J. Chem. Ecol. 16, 211–222, 10.1007/BF01021280 (1990). [DOI] [PubMed] [Google Scholar]
- Rahfeld P. et al. Independently recruited oxidases from the glucose-methanol-choline oxidoreductase family enabled chemical defences in leaf beetle larvae (subtribe Chrysomelina) to evolve. Proc. R. Soc. B 281, 10.1098/rspb.2014.0842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert M. et al. Beetles do it differently: Two stereodivergent cyclisation modes in iridoid-producing leaf-beetle larvae. ChemBioChem 14, 353–360, 10.1002/cbic.201200689 (2013). [DOI] [PubMed] [Google Scholar]
- Rahfeld P. et al. Glandular β-glucosidases in juvenile Chrysomelina leaf beetles support the evolution of a host-plant-dependent chemical defense. Insect Biochem. Mol. Biol. 58, 28–38, 10.1016/j.ibmb.2015.01.003 (2015). [DOI] [PubMed] [Google Scholar]
- Veith M., Oldham N. J., Dettner K., Pasteels J. M. & Boland W. Biosynthesis of defensive allomones in leaf beetle larvae-stereochemistry of salicylalcohol oxidation in Phratora vitellinae and comparison of enzyme substrate and stereospecificity with alcohol oxidases from several iridoid producing leaf beetles. J. Chem. Ecol. 23, 429–443, 10.1023/B:JOEC.0000006369.26490.c6 (1997). [DOI] [Google Scholar]
- Brueckmann M., Termonia A., Pasteels J. M. & Hartmann T. Characterization of an extracellular salicyl alcohol oxidase from larval defensive secretions of Chrysomela populi and Phratora vitellinae (Chrysomelina). Insect Biochem. Mol. Biol. 32, 1517–1523, 10.1016/S0965-1748(02)00072-3 (2002). [DOI] [PubMed] [Google Scholar]
- Michalski C., Mohagheghi H., Nimtz M., Pasteels J. & Ober D. Salicyl alcohol oxidase of the chemical defense secretion of two chrysomelid leaf beetles-Molecular and functional characterization of two new members of the glucose-methanol-choline oxidoreductase gene family. J. Biol. Chem. 283, 19219–19228, 10.1074/jbc.M802236200 (2008). [DOI] [PubMed] [Google Scholar]
- Kirsch R. et al. Host plant shifts affect a major defense enzyme in Chrysomela lapponica. Proc. Natl. Acad. Sci. USA 108, 4897–4901, 10.1073/pnas.1013846108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilker M. & Schulz S. Composition of larval secretion of Chrysomela lapponica (Coleoptera, Chrysomelidae) and its dependence on host-plant. J. Chem. Ecol. 20, 1075–1093, 10.1007/bf02059744 (1994). [DOI] [PubMed] [Google Scholar]
- Geiselhardt S., Hilker M., Muller F., Kozlov M. V. & Zvereva E. L. Inter- and intrapopulation variability in the composition of larval defensive secretions of willow-feeding populations of the leaf beetle Chrysomela lapponica. J. Chem. Ecol. 41, 276–286, 10.1007/s10886-015-0558-x (2015). [DOI] [PubMed] [Google Scholar]
- Kelley L. A. & Sternberg M. J. E. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371, 10.1038/nprot.2009.2 (2009). [DOI] [PubMed] [Google Scholar]
- Sandstrom J., Carlsson L., Marklund S. L. & Edlund T. The heparin-binding domain of extracellular superoxide dismutase C and formation of variants with reduced heparin affinity. J. Biol. Chem. 267, 18205–18209 (1992). [PubMed] [Google Scholar]
- Petersen S. V. et al. Extracellular superoxide dismutase (EC-SOD) binds to type I collagen and protects against oxidative fragmentation. J. Biol. Chem. 279, 13705–13710, 10.1074/jbc.M310217200 (2004). [DOI] [PubMed] [Google Scholar]
- Nguyen A. D. et al. Fibulin-5 is a novel binding protein for extracellular superoxide dismutase. Circ. Res. 95, 1067–1074, 10.1161/01.res.0000149568.85071.fb (2004). [DOI] [PubMed] [Google Scholar]
- Nappi A. J., Vass E., Frey F. & Carton Y. Superoxide anion generation in Drosophila during melanotic encapsulation of parasites. Eur. J. Cell Biol. 68, 450–456 (1995). [PubMed] [Google Scholar]
- Sun X. P. et al. Nutrient-dependent requirement for SOD1 in lifespan extension by protein restriction in Drosophila melanogaster. Aging Cell 11, 783–793, 10.1111/j.1474-9726.2012.00842.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G. N. & Tower J. Superoxide dismutase evolution and life span regulation. Mech. Ageing Dev. 126, 365–379, 10.1016/j.mad.2004.08.012 (2005). [DOI] [PubMed] [Google Scholar]
- Martin I., Jones M. A. & Grotewiel M. Manipulation of Sod1 expression ubiquitously, but not in the nervous system or muscle, impacts age-related parameters in Drosophila. FEBS Lett. 583, 2308–2314, 10.1016/j.febslet.2009.06.023 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. P., Campbell S. D., Michaud D., Charbonneau M. & Hilliker A. J. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc. Natl. Acad. Sci. USA 86, 2761–2765, 10.1073/pnas.86.8.2761 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher G., Scholten J. & Klingler M. Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 12, R85–R86, 10.1016/S0960-9822(02)00666-8 (2002). [DOI] [PubMed] [Google Scholar]
- Munoz-Munoz J. L. et al. Generation of hydrogen peroxide in the melanin biosynthesis pathway. Biochim. Biophys. Acta 1794, 1017–1029, 10.1016/j.bbapap.2009.04.002 (2009). [DOI] [PubMed] [Google Scholar]
- Yi H. Y., Chowdhury M., Huang Y. D. & Yu X. Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 98, 5807–5822, 10.1007/s00253-014-5792-6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. M., Xiang Q., Zhang Q. H., Huang Y. D. & Su Z. J. Overview on the recent study of antimicrobial peptides: Origins, functions, relative mechanisms and application. Peptides 37, 207–215, 10.1016/j.peptides.2012.07.001 (2012). [DOI] [PubMed] [Google Scholar]
- Vilcinskas A. Evolutionary plasticity of insect immunity. J. Insect Physiol. 59, 123–129, 10.1016/j.jinsphys.2012.08.018 (2013). [DOI] [PubMed] [Google Scholar]
- Vogel H., Badapanda C., Knorr E. & Vilcinskas A. RNA-sequencing analysis reveals abundant developmental stage-specific and immunity-related genes in the pollen beetle Meligethes aeneus. Insect Mol. Biol. 23, 98–112, 10.1111/imb.12067 (2014). [DOI] [PubMed] [Google Scholar]
- Anselme C. et al. Identification of the Weevil immune genes and their expression in the bacteriome tissue. BMC Biol. 6, 43, 10.1186/1741-7007-6-43 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura M. et al. Multipeptide precursor structure of acaloleptin A isoforms, antibacterial peptides from the Udo longicorn beetle. Acalolepta luxuriosa. Dev. Comp. Immunol. 33, 1120–1127, 10.1016/j.dci.2009.06.004 (2009). [DOI] [PubMed] [Google Scholar]
- Buttstedt A., Moritz R. F. A. & Erler S. Origin and function of the major royal jelly proteins of the honeybee (Apis mellifera) as members of the yellow gene family. Biol. Rev. (Camb.) 89, 255–269, 10.1111/brv.12052 (2014). [DOI] [PubMed] [Google Scholar]
- Ferguson L. C., Green J., Surridge A. & Jiggins C. D. Evolution of the insect yellow gene family. Mol. Biol. Evol. 28, 257–272, 10.1093/molbev/msq192 (2011). [DOI] [PubMed] [Google Scholar]
- Buchon N., Silverman N. & Cherry S. Immunity in Drosophila melanogaster-from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810, 10.1038/nri3763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumer A., Van Loy T. & Schoofs L. The complexity of Drosophila innate immunity. ISJ-Invertebr. Surviv. J. 7, 32–44 (2010). [Google Scholar]
- Williams M. J. Drosophila hemopoiesis and cellular immunity. J. Immunol. 178, 4711–4716, 10.4049/jimmunol.178.8.4711 (2007). [DOI] [PubMed] [Google Scholar]
- Kim T. & Kim Y. J. Overview of innate immunity in Drosophila. J. Biochem. Mol. Biol. 38, 121–127, 10.5483/bmbrep.2005.38.2.121 (2005). [DOI] [PubMed] [Google Scholar]
- Hoffmann J. A. The immune response of Drosophila. Nature 426, 33–38, 10.1038/nature02021 (2003). [DOI] [PubMed] [Google Scholar]
- Charroux B. & Royet J. Drosophila immune response: From systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract. Fly 4, 40–47, 10.4161/fly.4.1.10810 (2010). [DOI] [PubMed] [Google Scholar]
- Lehrer R. I. & Ganz T. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 11, 23–27, 10.1016/s0952-7915(99)80005-3 (1999). [DOI] [PubMed] [Google Scholar]
- Nappi A. J. & Christensen B. M. Melanogenesis and associated cytotoxic reactions: Applications to insect innate immunity. Insect Biochem. Mol. Biol. 35, 443–459, 10.1016/j.ibmb.2005.01.014 (2005). [DOI] [PubMed] [Google Scholar]
- Nappi A. J. & Vass E. In Phylogenetic Perspectives on the Vertebrate Immune System Vol. 484 Advances in Experimental Medicine and Biology (eds Beck G., Sugumaran M. & Cooper E. L.) 329–348 (Kluwer Academic, 2001). [Google Scholar]
- Arvanitis M., Glavis-Bloom J. & Mylonakis E. Invertebrate models of fungal infection. Biochim. Biophys. Acta 1832, 1378–1383, 10.1016/j.bbadis.2013.03.008 (2013). [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Imler J. L., Hetru C. & Hoffmann J. A. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7, 862–874, 10.1038/nri2194 (2007). [DOI] [PubMed] [Google Scholar]
- Levitin A. & Whiteway M. Drosophila innate immunity and response to fungal infections. Cell. Microbiol. 10, 1021–1026, 10.1111/j.1462-5822.2008.01120.x (2008). [DOI] [PubMed] [Google Scholar]
- Komarov D. A., Slepneva I. A., Glupov V. V. & Khramtsov V. V. Superoxide and hydrogen peroxide formation during enzymatic oxidation of DOPA by phenoloxidase. Free Radical Res. 39, 853–858, 10.1080/10715760500166693 (2005). [DOI] [PubMed] [Google Scholar]
- Winterbourn C. C., French J. K. & Claridge R. F. C. Superoxide-dismutase as an inhibitor of reactions of semi-quinone radicals. FEBS Lett. 94, 269–272, 10.1016/0014-5793(78)80953-3 (1978). [DOI] [PubMed] [Google Scholar]
- Rhee S. G. H2O2 a necessary evil for cell signaling. Science 312, 1882–1883, 10.1126/science.1130481 (2006). [DOI] [PubMed] [Google Scholar]
- Fridovich I. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64, 97–112, 10.1146/annurev.bi.64.070195.000525 (1995). [DOI] [PubMed] [Google Scholar]
- Rada B. K., Geiszt M., Kaldi K., Timar C. & Ligeti E. Dual role of phagocytic NADPH oxidase in bacterial killing. Blood 104, 2947–2953, 10.1182/blood-2004-03-1005 (2004). [DOI] [PubMed] [Google Scholar]
- Gross J., Muller C., Vilcinskas A. & Hilker M. Antimicrobial activity of exocrine glandular secretions, hemolymph, and larval regurgitate of the mustard leaf beetle Phaedon cochleariae. J. Invertebr. Pathol. 72, 296–303, 10.1006/jipa.1998.4781 (1998). [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Vaccaro K. & Carroll S. B. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 12, 1547–1556, 10.1016/s0960-9822(02)01113-2 (2002). [DOI] [PubMed] [Google Scholar]
- Peiren N. et al. The protein composition of honeybee venom reconsidered by a proteomic approach. Biochim. Biophys. Acta 1752, 1–5, 10.1016/j.bbapap.2005.07.017 (2005). [DOI] [PubMed] [Google Scholar]
- Sampson N. S. Dissection of a flavoenzyme active site: The reaction catalyzed by cholesterol oxidase. Antioxid. Redox Signal. 3, 839–846, 10.1089/15230860152665019 (2001). [DOI] [PubMed] [Google Scholar]
- Stock M. et al. Putative sugar transporters of the mustard leaf beetle Phaedon cochleariae: their phylogeny and role for nutrient supply in larval defensive glands. PLoS ONE 8, e84461, 10.1371/journal.pone.0084461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss A. S. et al. Tissue-specific transcript profiling for ABC transporters in the sequestering larvae of the phytophagous leaf beetle Chrysomela populi. PLos ONE 9, e98637, 10.1371/journal.pone.0098637 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301, 10.1093/nar/gkr1065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402, 10.1093/nar/25.17.3389 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G. & Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786, 10.1038/nmeth.1701 (2011). [DOI] [PubMed] [Google Scholar]
- Chauhan J. S., Rao A. & Raghava G. P. S. In silico platform for prediction of N-, O- and C-glycosites in eukaryotic protein sequences. PLos ONE 8, e67008, 10.1371/journal.pone.0067008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauchet Y., Wilkinson P., Chauhan R. & ffrench-Constant R. H. Diversity of beetle genes encoding novel plant cell wall degrading enzymes. PLos ONE 5, e15635, 10.1371/journal.pone.0015635 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodemann R. R. et al. Precise RNAi-mediated silencing of metabolically active proteins in the defence secretions of juvenile leaf beetles. Proc. R. Soc. B 279, 4126–4134, 10.1098/rspb.2012.1342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F. & Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, 14, 10.1186/gb-2007-8-2-r19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 55, 611–622, 10.1373/clinchem.2008.112797 (2009). [DOI] [PubMed] [Google Scholar]
- Rostas M. & Hilker M. Indirect interactions between a phytopathogenic and an entomopathogenic fungus. Naturwissenschaften 90, 63–67, 10.1007/s00114-002-0395-y (2003) [DOI] [PubMed] [Google Scholar]
- Arbi M., Pouliliou S., Lampropoulou M., Marmaras V. J. & Tsakas S. Hydrogen peroxide is produced by E. coli challenged haemocytes and regulates phagocytosis, in the medfly Ceratitis capitata. The active role of superoxide dismutase. Dev. Comp. Immunol. 35, 865–871, 10.1016/j.dci.2011.03.020 (2011). [DOI] [PubMed] [Google Scholar]
- Sambrook J. & Russell D. W. Molecular cloning: A laboratory manual. (Cold Spring Harbor Laboratory Press, 2001). [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J. V. & Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860, 10.1038/nprot.2006.468 (2006). [DOI] [PubMed] [Google Scholar]
- Gorg A. et al. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21, 1037–1053, (2000). [DOI] [PubMed] [Google Scholar]
- Levin Y., Hradetzky E. & Bahn S. Quantification of proteins using data-independent analysis (MSE) in simple and complex samples: A systematic evaluation. Proteomics 11, 3273–3287, 10.1002/pmic.201000661 (2011). [DOI] [PubMed] [Google Scholar]
- Li G. Z. et al. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719, 10.1002/pmic.200800564 (2009). [DOI] [PubMed] [Google Scholar]
- Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P. C. & Geromanos S. J. Absolute quantification of proteins by LCMSE-A virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156, 10.1074/mcp.M500230-MCP200 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.