Dickson et al. find that the ER membrane lipid phosphatase Sac1 localizes to ER–plasma membrane (PM) contact sites and acts as a cellular sensor and controller of PM phosphoinositide homeostasis.
Abstract
Endoplasmic reticulum–plasma membrane (ER–PM) contact sites play an integral role in cellular processes such as excitation–contraction coupling and store-operated calcium entry (SOCE). Another ER–PM assembly is one tethered by the extended synaptotagmins (E-Syt). We have discovered that at steady state, E-Syt2 positions the ER and Sac1, an integral ER membrane lipid phosphatase, in discrete ER–PM junctions. Here, Sac1 participates in phosphoinositide homeostasis by limiting PM phosphatidylinositol 4-phosphate (PI(4)P), the precursor of PI(4,5)P2. Activation of G protein–coupled receptors that deplete PM PI(4,5)P2 disrupts E-Syt2–mediated ER–PM junctions, reducing Sac1’s access to the PM and permitting PM PI(4)P and PI(4,5)P2 to recover. Conversely, depletion of ER luminal calcium and subsequent activation of SOCE increases the amount of Sac1 in contact with the PM, depleting PM PI(4)P. Thus, the dynamic presence of Sac1 at ER–PM contact sites allows it to act as a cellular sensor and controller of PM phosphoinositides, thereby influencing many PM processes.
Introduction
This study investigates junctions between the ER and plasma membrane (PM) and their effect on PM phosphoinositides. Membranes of the ER are functionally connected to all membranes of the secretory and endocytic pathways via vesicular transport. There is growing evidence that intimate physical contacts between the ER membrane and membranes of other organelles play major roles in interorganelle communication and cell physiology (Prakriya et al., 2006; Friedman et al., 2011; Toulmay and Prinz, 2011; Dickson et al., 2012; Hoppins and Nunnari, 2012; Stefan et al., 2013). These membrane contact sites have been implicated in calcium homeostasis (Prakriya et al., 2006; Zhang et al., 2006; Lewis, 2007), the regulation of lipid exchange (Prinz, 2010; Schauder et al., 2014; Chung et al., 2015), and phosphoinositide metabolism (Manford et al., 2010; Stefan et al., 2011, 2013). In mammalian cells, the extended synaptotagmins (E-Syt1/2/3), orthologs of yeast tricalbins, mediate the steady-state formation of ER–PM contact sites via C2 domain-dependent interactions with the PM that require phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2; Creutz et al., 2004; Manford et al., 2012; Giordano et al., 2013).
PI(4,5)P2 is the signature phosphoinositide of the PM. There it regulates and orchestrates a plethora of activities, including cell organization, cell motility, membrane trafficking, ion channel function, and signal transduction (Hilgemann et al., 2001; Di Paolo and De Camilli, 2006; Balla, 2013; Hille et al., 2015). The immediate precursor of PM PI(4,5)P2 is PI(4)P. Two major pools of PI(4)P exist within the cell: one at the PM and one at the Golgi (Szentpetery et al., 2010; Dickson et al., 2014).
Mammalian Sac1 is an essential type II integral-membrane lipid phosphatase that dephosphorylates all phosphatidylinositol (PI) monophosphates, as well as PI(3,5)P2 (but not PI(4,5)P2) to PI (Guo et al., 1999; Nemoto et al., 2000; Liu et al., 2009; Zhong et al., 2012). It is localized to the ER in steady state but may traffic to the Golgi in nonproliferating cells (Blagoveshchenskaya et al., 2008; Mesmin et al., 2013). Overexpression of Sac1 in mammalian cells reduces the intensity of fluorescent reporters for PI(4)P at both the PM and the Golgi (Hammond et al., 2014). In yeast, Sac1 has been proposed to act at ER–PM contact sites to control PM PI(4)P in trans (Stefan et al., 2011; Manford et al., 2012), whereas reconstituted liposome experiments suggest it acts in cis (Mesmin et al., 2013). Knockout leads to significant increases in total cellular PI(4)P levels (Wenk et al., 2003). Thus, Sac1 is an important regulator of phosphoinositide metabolism.
In this study, we show that a portion of the Sac1 phosphatase resides in dynamic ER–PM contact sites in mammalian cells. There, ER Sac1 sits closely apposed (<10 nm) to the PM and dephosphorylates PM PI(4)P, reducing PM pools of PI(4)P and secondarily PI(4,5)P2. Our findings reveal that E-Syt2 increases the proportion of the ER surface in contact with the PM and the abundance of ER–PM Sac1 in a PI(4,5)P2-dependent manner. Activation of store-operated calcium entry (SOCE) also increases the amount of Sac1 in contact with the PM and initiates a decline in PM PI(4)P and PI(4,5)P2 levels. Finally, depletion of PM PI(4,5)P2 in response to G protein–coupled receptor activation disrupts ER–PM contacts and reduces the amount of Sac1 in contact with the PM, a novel feedback mechanism that results in PM phosphoinositide homeostasis.
Results
Some of the ER lipid phosphatase Sac1 localizes to ER–PM junctions
We begin by presenting evidence that a portion of the total cellular pool of Sac1 resides in regions of the ER membrane closely apposed to the PM (ER–PM contact sites). To visualize the ER, we overexpressed the ER-resident protein mCherry-Sec61β. Imaging of this protein in fixed or living tsA201 cells revealed the ER’s characteristic reticular appearance (Figs. 1 A and S1 B). Next, we assessed the subcellular distribution of the ER lipid phosphatase Sac1. Transfecting tsA201 cells with GFP-Sac1 (Rohde et al., 2003) revealed ramifying structures (Fig. 1 B and Video 1) that were significantly colocalized with mCherry-Sec61β (Fig. 1 C). These results confirm that Sac1 is distributed on ER membranes throughout the cell (Guo et al., 1999; Rohde et al., 2003; Blagoveshchenskaya et al., 2008). We looked for Sac1 proximal to the PM using total internal reflection fluorescence (TIRF) microscopy on cells expressing GFP-Sac1 (Fig. 1 D) or stained for anti-Sac1 (Fig. 1 E) and discovered discrete accumulations close to the PM. SuperresolutionTIRF (subscript denotes the mode of acquisition; see Materials and methods) localization maps of GFP-Sac1 (Fig. 1 F) or anti-Sac1–labeled cells revealed an abundance of nonrandomly distributed Sac1 clusters that have similar distributions throughout the cellular footprint (Fig. S1 D).
Figure 1.
The ER lipid phosphatase Sac1 is closely apposed to the PM. (A) Inverted (negative contrast), maximum-intensity projection of a pair of tsA201 cells expressing mCherry-Sec61β ER marker. Inset is 3× magnification. (B) Inverted (negative contrast), maximum-intensity projection of a tsA201 cell expressing GFP-Sac1. Inset is 2× magnification. (C) SuperresolutionEPI localization map of a region of interest from a fixed tsA201 cell stained for anti-mCherry against mCherry-Sec61β (left), anti-Sac1 (middle), and merge (right). (D) Inverted TIRF footprint of a tsA201 cell expressing GFP-Sac1. (E) Inverted TIRF footprint from a tsA201 cell fixed and stained for anti-Sac1. (F) SuperresolutionTIRF localization map from a fixed tsA201 cell expressing GFP-Sac1. (G, top) TIRF footprint from a single tsA201 cell expressing MAPPER (left), Sac1 (middle), and the region of the TIRF footprint with overlapping signals (Sac1–PM; right). Inset is 1.5× magnification. (bottom) Representative intensity profile of a region of the TIRF footprint. Note that the signals are coincident with one another. (H, top) Confocal micrograph of a single tsA201 cell expressing GFP-Sac1 (donor); middle: same cell, expressing RFP-LDR (acceptor); right: FRET signal. (middle) Representative profile plot taken between the arrows of the FRET image in the top panel. (H, bottom) Quantification of NFRET (means ± SEM, n = 8–11; t test; *, P < 0.05; **, P < 0.01).
To identify potential sites of contact between the ER and PM, we simultaneously overexpressed the ER–PM junction marker MAPPER (Chang et al., 2013; Tsai et al., 2014) and found significant colocalization with Sac1 (Fig. 1 G). We assessed the distance between Sac1 puncta and the PM using Förster resonance energy transfer (FRET; see Materials and methods) between GFP-Sac1 (donor) and an RFP protein that is palmitoylated and myristoylated to reside in the PM (RFP-LDR; acceptor; Fig. 1 H). We observed a punctate FRET signal around the periphery of cells (corresponding to the size of Sac1 puncta), indicating a distance of <10 nm between donor and acceptor (Fig. 1 H). Quantification of this FRET signal (normalized FRET [NFRET]; Hoppe et al., 2002; Ji et al., 2008; see Materials and methods) revealed that GFP-Sac1 had a significantly greater NFRET signal with LDR-RFP than did another ER-resident protein, GFP-STIM1. As a positive control we stimulated PM translocation of GFP-STIM1 by depleting ER calcium (thapsigargin; 500 nM). The NFRET signal between GFP-STIM1 and LDR-RFP increased in response to thapsigargin (Fig. 1 H), consistent with the store depletion–induced association of STIM1 with PM-resident Orai1 subunits.
Collectively, these observations show that the ER lipid phosphatase Sac1 is present at sites closely apposed to the PM. We refer to these contact sites, containing Sac1, as ER–PM Sac1 puncta.
Formation of ER–PM junctions reduces PM PI(4)P and PI(4,5)P2
To test whether Sac1 could dephosphorylate PM PI(4)P when present at these junctions, we increased the area of ER–PM contact by recruiting the ER acutely toward the PM using a rapamycin-mediated heterodimerization strategy (Inoue et al., 2005). Overexpression of the FK506 binding protein (FKBP) coupled to an ER localization signal (CFP-FKBP-Cb5) and the FKBP12 rapamycin-binding domain (FRB) coupled to a PM localizing signal (lyn11-FRB) allowed rapid recruitment of ER to the PM after the addition of rapamycin (Fig. 2 A). Simultaneously, we monitored PM PI(4)P or PI(4,5)P2 with the P4M domain from secreted bacterial effector protein SidM (P4M; Hammond et al., 2014) or the pleckstrin homology domain from phospholipase Cδ1 (PHPLCδ1), respectively. After the addition of rapamycin, the ER marker was recruited to the PM anchor, whereas the lipid indicators translocated away from the PM (Fig. 2, A [bottom] and D [bottom]). Thus, there was a decline in both PM PI(4)P and PM PI(4,5)P2 when the number of ER–PM junctions was increased.
Figure 2.
Increasing Sac1 at the PM decreases PM PI(4)P and PI(4,5)P2. (A) Representative control cell. (top) Inverted TIRF micrographs of a tsA201 cells expressing CFP-FKBP-Cb5 ER anchor (top row) and mCherry-P4M (bottom row), before (left) and after (right) the addition of rapamycin (5 µM). Cells also express LDR PM anchor. Bar, 5 µm. (bottom) Normalized intensity kinetics of overexpressed proteins from TIRF footprint. (B and C) Same experiment as A, except Sac1 is overexpressed (B) or cells are treated with siRNA against Sac1 (C). Representative cells displayed. (D, left) Inverted TIRF footprint from a single tsA201 cell expressing CFP-FKBP-Cb5 and RFP-PHPLCδ1 (bottom row), before (left) and after (right) the addition of rapamycin (5 µM). (righti) Analysis of changes in PHPLCδ1 signal after the addition of rapamycin (means ± SEM, n = 12). (E) Same experiment as D, except GFP-Sac1 (top row) is overexpressed. Note that with GFP-Sac1 overexpression there is a faster decrease in PHPLCδ1 signal (means ± SEM, n = 11). (F, left) Time course of KCNQ2/3 inhibition after the recruitment of endogenous Sac1 (eSac1) to the PM. (right) Summary of the percent decrease in KCNQ2/3 current (n = 7). (G) Time course of KCNQ2/3 inhibition after the recruitment of overexpressed GFP-Sac1 (oSac1) to the PM. (right) Summary of the percent decrease in KCNQ2/3 current (n = 6; *, P < 0.05). (H) Mass spectrometry analysis of total cellular PS (left), PIP (center), and PIP2 (right) before and after the addition of rapamycin (5 µM) from tsA201 cells expressing CFP-FKBP-Cb5, LDR, and GFP-Sac1. Means ± SEM, n = 4, t test; N.S., P > 0.05; *, P < 0.05). (I, left) steady-state distribution of GFP-Evt2-x2PH in control (top) or Sac1-overexpressing (bottom) tsA201 cells. PM/cytoplasmic (Cyto.) intensity of GFP-Evt2-x2PH (means ± SEM, n = 10, t test, N.S., not significant). (middle) Same experiment as A, except GFP-Evt2-x2PH is expressed (n = 10). (right) Same experiment as B, except GFP-Evt2-x2PH is expressed (n = 11). Bars, 5 µm.
To test the role of Sac1 in these phosphoinositide changes, we monitored the lipid indicators under TIRF while inducing ER–PM contacts in cells with overexpressed Sac1 (oSac1) or reduced Sac1 (siRNA Sac1). With Sac1 overexpression, the amount and overall speed of P4M liberation from the PM was approximately doubled (compare Fig. 2, A and B, bottom) and the decline of RFP-PHPLCδ1 intensity was faster (compare Fig. 2, D and E, right). Knocking down endogenous Sac1 significantly reduced the decline in mCherry-P4M (Fig. 2 C).
To confirm that recruitment of the ER reduced PM phosphoinositide levels and did not merely sterically exclude fluorescent probes from ER–PM junctions, we recorded current from KCNQ2/3 channels (a sensitive proxy for PM PI(4,5)P2) and measured total cellular lipids using ultrahigh-performance liquid chromatography coupled with mass spectrometry (UHPLC/MS; for details, see Materials and methods). Recruiting the ER to the PM immediately initiated a slow decay in KCNQ2/3 current amplitude (Fig. 2 F) and total cellular PIP and PIP2 (Fig. 2 H). Overexpression of Sac1 significantly enhanced the reduction in current (Fig. 2 G).
Recently, oxysterol binding protein–related proteins (ORPs) have been reported to mediate PI(4)P/phosphatidylserine (PS) countertransport at ER–PM junctions (Chung et al., 2015). To test whether PI(4)P transfer could explain the observed decline in PI(4)P and PM PI(4,5)P2, we induced ER–PM junctions while simultaneously monitoring the fluorescence intensity of a high-affinity PS biosensor (GFP-Evt2-x2PH; Uchida et al., 2011). Similar to previous studies, the PS biosensor was distributed along the PM and cytoplasmic structures under steady-state conditions (Fig. 2 I, left; Uchida et al., 2011). With overexpression of Sac1 or induction of ER–PM junctions, there was no significant change in total cellular PS (Fig. 2 H) or GFP-Evt2-x2PH distribution (Fig. 2 I).
To further test ORP’s effect on steady-state PM PI(4)P and PI(4,5)P2, we analyzed the distribution of P4M and PHPLCδ1 with and without ORP5 overexpression. ORP5 overexpression reduced the intensity of PM P4M, as expected for PI(4)P countertransport, whereas PM PHPLCδ1 was unaltered (Fig. S1 G, right). Therefore PI(4)P transfer alone is not sufficient to explain the phosphoinositide changes after the induction of ER–PM junctions. Rather, our results are consistent with Sac1 directly dephosphorylating PM PI(4)P and leading to a secondary decline in PM PI(4,5)P2.
Sac1 at ER–PM junctions depletes PM PI(4)P and PI(4,5)P2
To quantify the effect of Sac1 on each phosphoinositide species, we used UHPLC/MS. As already shown in Fig. 2 H, this technique (Clark et al., 2011) allows for the simultaneous quantification of PS and each phosphoinositide and their different fatty-acyl isoforms (Fig. 3 and Fig. S2, A–D). We detected 7 PS species, 10 PI species, 9 PIP species, and 15 PIP2 species, the most abundant being fatty-acyl species 36:1 (Fig. S2, A–D). To validate the technique and confirm the accessibility of these species to phospholipase C (PLC), we activated transiently overexpressed Gq-coupled type-1 muscarinic receptors (M1R) and monitored the abundance of each phosphoinositide species. Application of the muscarinic agonist oxotremorine M (Oxo-M; 60 s; 10 µM) reduced total cellular PIP and PIP2 levels by ∼70% (Fig. S2, B and C). The PIP and PIP2 fatty-acyl species that were most strongly depleted were 36:1 and 38:4 (Fig. S2, B and C), suggesting that both are present on the PM. Overexpression of Sac1 reduced both total cellular PIP (Fig. 3 A) and PIP2 (Fig. 3 B) by 30%. Again, the 36:1 and 38:4 species of PIP and PIP2 were the most strongly depleted. This UHPLC/MS approach does not distinguish which inositol positions are phosphorylated, but because the most abundant PIP and PIP2 isoforms in mammalian cells are PI(4)P (∼80% of total PIP) and PI(4,5)P2 (∼92% of total PIP2; Pettitt et al., 2006), these results provide chemical evidence that Sac1 regulates PI(4)P and PI(4,5)P2 on the PM.
Figure 3.
Sac1 decreases cellular PIP and PIP2 and makes it easier for PLC to hydrolyze PI(4,5)P2. (A, left) Mass spectrometry analysis for PIP. Histogram of the most abundant PIP species in control (black bars) and Sac1-overexpressing (green bars) tsA201 cells (means ± SEM). (right) Summary of the total PIP signal in control and Sac1 overexpressing tsA201 cells (means ± SEM), n = 6; t test; *, P < 0.05). (B) Mass spectrometry analysis for PIP2. Same experiment as (A) but summarizing PIP2 species. (means ± SEM; n = 6; t test; *, P < 0.05). (C, left) Normalized change in RFP-PHPLCδ1 intensity from the TIRF footprint of a tsA201 cell after activation of the M1R. (right) Summary (means ± SEM, n = 7; t test; *, P < 0.05). (D) Same experiment as C, but with GFP-Sac1 overexpressed. (E and F) Same experiment as A and B, except with siRNA against endogenous Sac1 (means ± SEM, n = 6; *, P < 0.05). (G) Normalized, averaged time course of RFP-PHPLCδ1 within a TIRF footprint, after the activation of the M1R, in control (black line) or Sac1 siRNA-treated (red line) cells. (right) Summary histograms (n = 9; *, P < 0.05.). (H, left) Normalized, mean time courses of PHPLCδ1 FRET photometry signals in control, Sac1-overexpressing, and catalytically inactive (C389S) Sac1-expressing tsA201 cells. (right) Summary (means ± SEM, n = 6; t test; *, P < 0.05.). (I, left) Normalized, mean time courses of LibravIII FRET photometry signals in control or Sac1-overexpressing tsA201 cells. (right) Summary (means ± SEM, n = 6; t test; *, P < 0.05.). (J, left) two tsA201 cells expressing mCherry-P4M. (middle) Same experiment as left panel, except GFP-Sac1 is overexpressed. (right) Same experiments as left panel except cells have had hSac1tr recruited to their trans-Golgi network to deplete PI(4)P. (K) Ratio of the Golgi/cellular intensity in each experimental condition (scatter plot, means ± SEM n = 8–11; *, P < 0.05). Bars, 5 µm.
We next tested the effect of Sac1 in the context of physiological phosphoinositide changes. Using the muscarinic agonist Oxo-M (500 nM) for submaximal receptor activation, we asked whether overexpression of Sac1 synergizes the ability of muscarinic receptors to stimulate depletion of PM PI(4,5)P2. Without Sac1 overexpression, brief (20 s) application of 500 nM Oxo-M resulted in a small decrease (11 ± 8%; n = 7) in RFP-PHPLCδ1 intensity within a TIRF footprint (Fig. 3 C), whereas with Sac1 overexpression (oSac1), it led to a significantly larger decrease (25 ± 4%; n = 7; Fig. 3 D). Thus, overexpression of Sac1 made it significantly easier for submaximally activated Gq-coupled receptors to deplete PM PI(4,5)P2. Two indicators for resting PM PI(4,5)P2 (KCNQ2/3 current densities and normalized intensities of PHPLCδ1) supported results from mass spectrometry that Sac1 overexpression significantly reduced steady-state PM PI(4,5)P2 compared with control cells (Fig. S1, E and F). This reduction in the substrate pool would explain why fewer Gq-coupled receptors need to be activated to deplete PM PI(4,5)P2.
If Sac1 reduces the PM PI(4,5)P2 pool, then we would predict that knocking down Sac1 should allow PM phosphoinositide levels to rise. SuperresolutionTIRF localization maps from cells treated with siRNA against Sac1 revealed a significant reduction in the number and mean size of anti-Sac1 puncta in close proximity to the PM (Fig. S2 E). As anticipated, knocking down Sac1 resulted in a twofold increase in total cellular PIP and a 30% increase in PIP2 by mass spectrometry (Fig. 3, E and F) and a 30% increase in steady-state RFP-PHPLCδ1 intensity and KCNQ2/3 current density (Fig. S1 F). The increase of PM phosphoinositides with siRNA against Sac1 was also reflected in slower PI(4,5)P2 depletion after Gq-coupled receptor activation. In TIRF time-series experiments from cells expressing M1R and RFP-PHPLCδ1, knocking down Sac1 expression reduced the amount of PHPLCδ1 translocation somewhat (% decrease: control, 57 ± 6%; siRNA Sac1, 44 ± 4%; n = 9) and shortened the recovery time (time constant of recovery [τ]: control, 77 ± 16 s; siRNA Sac1, 41 ± 5 s; n = 9) after receptor activation (Fig. 3 G). Similarly, in photometry experiments measuring FRET between CFP-PHPLCδ1 and YFP-PHPLCδ1 as an assay for PM PI(4,5)P2, we found that overexpression of a catalytically inactive Sac1 mutant (Sac1 C389S; Hammond et al., 2014) reduced the ability of PLC to deplete PM PI(4,5)P2 (Fig. 3 H). Finally we asked if IP3 generation was impaired after Sac1 overexpression. Using the LibravIII, a PM-localized IP3 FRET reporter, we found overexpression of Sac1 decreased the amount of IP3 generated after Gq receptor activation (Fig. 3 I). Thus, Sac1 reduces PM phosphoinositides and thereby regulates the generation of PI(4,5)P2-derived second messengers.
Mammalian cells have two major pools of PI(4)P, the Golgi and PM, both of which make contributions to the maintenance of PM PI(4,5)P2 (Dickson et al., 2014). On which pool(s) does Sac1 act? To find out, we depleted trans-Golgi PI(4)P by recruiting a truncated version of Sac1 (mRFP-FKBP-Sac1tr) that is cytoplasmic. As we have previously demonstrated, recruitment of Sac1tr to the trans-Golgi network substantially reduces Golgi PI(4)P (Fig. 3 J, right) and reduces steady-state PM KCNQ2/3 currents (a sensitive proxy for PM PI(4,5)P2) by ∼25% (Dickson et al., 2014). In comparison, overexpression of the full-length ER-localized GFP-hSac1 protein reduced Golgi PI(4)P to a lesser extent (Fig. 3, J [middle] and K) but inhibited KCNQ2/3 currents twice as much as recruitment of Sac1tr to the trans-Golgi (50% reduction in KCNQ2/3 from Fig. S1 F). These results suggest that although Sac1 can reduce the trans-Golgi pool of PI(4)P, its impact on PM PI(4,5)P2 is primarily mediated by its action on PM PI(4)P.
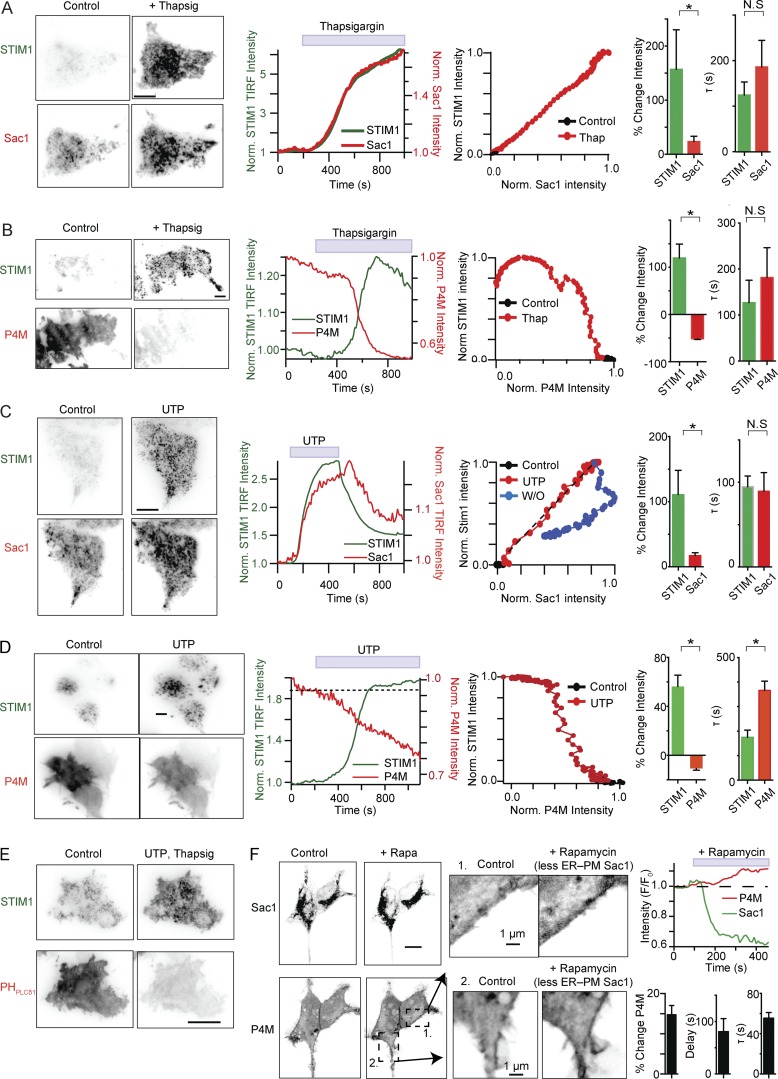
To approximate a native cell more closely, we tested the effect of endogenous Sac1 while increasing ER–PM contact sites via stimulated SOCE. Given that depletion of ER calcium stimulates STIM1-Orai interaction and increases ER–PM contacts, might it also increase ER–PM Sac1 abundance and activity? Depletion of ER calcium through blockade of the sarco-ER Ca2+ ATPase (SERCA) with thapsigargin (500 nM) resulted in an increase in the mean intensity of GFP-STIM1 in TIRF footprints (Fig. 4, A and B). Coupled to this increase was a concomitant increase in the intensity of Sac1-mCherry (Figs. 4 A and S3 A); temporal coupling was observed between STIM1 and Sac1 (Fig. 4 A, middle two panels). Thapsigargin led to the formation of STIM1-Orai ER–PM junctions, concurrent increase of Sac1 in proximity to the PM, and a subsequent decrease of P4M (Fig. 4 B, left two panels) and PHPLCδ1 (Fig. 4 E) intensities. To control for a direct effect of thapsigargin on phosphoinositides, we measured PHPLCδ1 intensity in the presence of only endogenous STIM1, Orai, and Sac1, and found the effect on PM PI(4,5)P2 to be minimal (Fig. S5, D and E). Similar reductions in PI(4)P and PI(4,5)P2 were observed when ER calcium stores were depleted by activating endogenous, low density Gq-coupled purinergic receptors (Fig. 4, C and D; and Fig. S3 B; Dickson et al., 2013; Falkenburger et al., 2013).
Figure 4.
Sac1 at ER–PM junctions reduces PM PI(4)P. (A, left) Inverted TIRF footprint from a single tsA201 cell expressing GFP-STIM1 (top row) and Sac1-mCherry (bottom row), before (left) and after (right) the addition of thapsigargin (500 nM). (second panel) Normalized kinetics of GFP-STIM1 and Sac1-mCherry changes after the addition of thapsigargin. (third panel) Temporal relationship between STIM1 and Sac1 recruitment to the PM (same cell as left panel). (right) Summary of the change in intensity and kinetics of recruitment to the PM of STIM1 and Sac1 (means ± SEM, n = 11). (B) Similar experiments as A, except P4M-mCherry is expressed and monitored (means ± SEM, n = 9). (C) and D) Same experimental design as before, except 100 µM UTP is used to initiate STIM1–Orai1 interactions (means ± SEM, n = 8–13). (E) Same experiment as D, except RFP-PHPLCδ1 is expressed. (F, left) Confocal micrographs of two tsA201 cells expressing Sac1 (top) and P4M-mCherry (bottom), before (left) and after (right) the addition of rapamycin (5 µM) to recruit the ER to the mitochondria. (middle) Higher magnification confocal images from the regions labeled in A, before (left) and after (right) the addition of rapamycin. (right) (top) Normalized kinetics. (bottom) Summary histograms (means ± SEM, n = 4). All changes in P4M intensity are significantly different compared with no application of rapamycin. Bars, 5 µm. N.S., P > 0.05; *, P < 0.05.
To determine the effect of endogenous Sac1 on phosphoinositide metabolism, we used the rapamycin-inducible dimerization system to disrupt steady-state ER–PM junctions. In this case, the ER is attracted away from ER–PM junctions to form junctions with mitochondria (rapamycin pair: FKBP-Cb5 and FRB-MOA), where ER-Sac1 would be expected to have little effect on PM phosphoinositide levels (Ueno et al., 2011). Recruiting the ER to mitochondria significantly reduced Sac1 near the PM (Fig. 4 F). It should be noted that not all ER–PM Sac1 was effectively redistributed to mitochondria, perhaps because mitochondrial FRB-MOA anchors became saturated with FKBP-Cb5 distributed across the substantial surface area of the ER. In cells where a significant proportion of Sac1 was redistributed to mitochondria, there was a delayed (90 ± 22 s), slow (τ = 55 ± 6 s) increase (15 ± 2%) in the intensity of mCherry-P4M, the PM PI(4)P label (Fig. 4 F, right). Collectively, these results suggest that ER–PM Sac1 is a direct negative regulator of PM PI(4)P and an indirect negative regulator of PM PI(4,5)P2. It controls the size of each phosphoinositide pool and tunes their sensitivities to Gq-coupled receptor activation.
PM PI(4,5)P2 is necessary for ER–PM Sac1 puncta
We have seen that ER–PM Sac1 can regulate PM phosphoinositide pools. Is there also regulatory feedback from phosphoinositides to control the amount of ER–PM Sac1? To answer this question, we monitored the intensity of ER–PM GFP-Sac1 using TIRF microscopy and activated the overexpressed M1R to deplete PM PI(4,5)P2 and PI(4)P. After M1R activation, GFP-Sac1 intensity was significantly reduced in the TIRF footprint (mean intensity decrease, 20 ± 2%; n = 12) and returned to prestimulation levels within a few hundred seconds (Fig. 5 A, left and middle; and Video 2). The kinetics of GFP-Sac1 were similar to those of a PM PI(4,5)P2 biosensor (PHPLCδ1; Fig. 5 B). To preclude the possibility that the PM moved out of the evanescent TIRF field, we simultaneously monitored a PM protein (LDR-CFP). Its intensity was unchanged (Fig. S3 C).
Figure 5.
PM PI(4,5)P2 is necessary for ER–PM Sac1 localization. (A, left) Inverted TIRF footprints from living tsA201 cells before, during, and after the addition of Oxo-M (10 µM). (middle, top) Kymograph of GFP-Sac1 taken from the red line in A. Bar, 5 µm. (middle, bottom) Normalized time course of GFP-Sac1 after the addition of Oxo-M. (right) Summary histograms (means ± SEM, n = 15, t test; *, P < 0.05). (B, left) TIRF footprint from a single tsA201 cell expressing GFP-Sac1 (left column), RFP-PHPLCδ1 (middle column), and merge, before (top row), during (middle row), and after (bottom row) M1R activation. (middle, top) Kymographs from the cell in left panel. Bars, 3 µm. (middle, bottom) Normalized kinetics of GFP-Sac1 and RFP-PHPLCδ1 from cell in left panel. (right) Summary (means ± SEM, n = 12, t test; *, P < 0.05) of the time constants of GFP-Sac1 (green bars) and RFP-PHPLCδ1 (red bars). (C, left) Experimental design: recruited PI(4)P 5-kinase phosphorylates PM PI(4)P into PI(4,5)P2. (right) Representative TIRF footprint kinetics of GFP-Sac1 and RFP-PHPLCδ1 after the addition of rapamycin to recruit PI(4)P 5-kinase to the PM. (D, left) Experimental design: VSP rapidly dephosphorylates PM PI(4,5)P2 into PI(4)P. (middle) Representative TIRF images from a patch-clamped tsA201 cell expressing GFP-Sac1 and VSP and held at −60 mV (top) and +100 mV to activate VSP (bottom). (right) Normalized kinetics of GFP-Sac1 from the same cell after two sequential steps to +100 mV for 4 s to activate VSP. Bar, 5 µm.
To test the importance of PI(4,5)P2 in ER–PM Sac1 puncta further, we manipulated PI(4,5)P2 levels in two additional ways: (1) by recruiting a PI(4)P 5-kinase to the PM to increase PM PI(4,5)P2 (Suh et al., 2006), and (2) by activating a voltage-sensing 5-phosphatase (VSP) to decrease PM PI(4,5)P2 (Murata et al., 2005). Recruiting PI(4)P 5-kinase to a PM anchor led to a simultaneous increase in GFP-Sac1 and RFP-PHPLCδ1 intensities within a TIRF footprint (Fig. 5 C). In contrast, activation of the voltage-sensing phosphatase resulted in a rapid and quickly reversible decrease in ER–PM GFP-Sac1 intensity (27 ± 4% decrease; τ recovery = 21 ± 3 s; n = 5; Fig. 5 D). The rate of recovery of ER–PM GFP-Sac1 intensity is indistinguishable from the known rapid rate of PI(4,5)P2 resynthesis after VSP activation (Falkenburger et al., 2010). These results indicate that PM PI(4,5)P2, but not PI(4)P, is required for ER–PM Sac1 puncta and that the ER–lipid interaction equilibrates in a few seconds.
E-Syt2 regulates the number of ER–PM Sac1 puncta
Recently, the family of extended synaptotagmin proteins was proposed to serve as tethers between the ER and the PM in mammalian cells (Giordano et al., 2013). These cortical ER proteins couple to the PM via C2-domain–dependent interactions that require PM PI(4,5)P2. We tested the hypothesis that E-Syt2 or E-Syt3 regulates the number of ER–PM Sac1 puncta at steady state. We rule out a role for E-Syt1 because its regulation requires both PM PI(4,5)P2 and calcium influx across the PM (Giordano et al., 2013; Idevall-Hagren et al., 2015). SuperresolutionEPI and superresolutionTIRF localization maps revealed that E-Syt2-mCherry was preferentially distributed to the cell periphery in distinct puncta or plaques (Figs. 6 A and S4 A), consistent with participation in ER–PM contact sites. Coexpressing E-Syt2-mCherry with GFP-Sac1 increased the accumulation of Sac1 in ER–PM puncta (Fig. 6 A).
Figure 6.
Extended synaptotagmin 2 helps regulate the number of ER–PM Sac1 puncta to control PM phosphoinositide metabolism. (A, left) SuperresolutionEPI localization maps of E-Syt2-mCherry, GFP-Sac1, and merge. Bar, 5 µm. (right) Intensity profile of GFP-Sac1 (green line) and E-Syt2-mCherry (red line) in the region between the white arrows. (B) SuperresolutionTIRF localization maps from a tsA201 cell expressing GFP-Sac1 (left) and E-Syt2-mCherry (right), and merge. Insets: enlarged images from the region within the white box. Bar, 5 µm. (C, left) Summary of the overlap coefficent between Sac1 and E-Syt2 (mean ± SEM, n = 6) and (right) histogram of relative distance of GFP-Sac1 to E-Syt2-mCherry (means ± SEM, n = 6). (D, left) SuperresolutionTIRF localization map of GFP-Sac1 from a single, fixed tsA201 cell after treatment with siRNA against E-Syt2. Bar, 5 µm. Inset is magnified 4×. (middle) SuperresolutionTIRF localization map of a tsA201 cell stained for anti-Sac1. Inset is magnified 3×. (right) SuperresolutionTIRF localization map of a tsA201 cell treated with siRNA against E-Syt2 and stained for anti-Sac1. Inset is magnified 3× (E) Histograms summarizing the effect of siRNA treatment against E-Syt2 on total PIP (left) and PIP2 (right; means ± SEM, n = 6, t test, *, P < 0.05). (F, left) Representative Inverted TIRF images from a tsA201 cell expressing GFP-Sac1 (left) and E-Syt2-mCherry (right) before (top row) and after (bottom row) the activation of the M1R. (right, top) Kymographs taken from the red dashed line in left panel. Bar, 5 µm. (right, bottom) Normalized time series kinetics of GFP-Sac1 (green line) and E-Syt2-mCherry (red line) after the addition of Oxo-M (10 µM). (G) Representative kymograph (top) and normalized time series kinetics (bottom) of GFP-Sac1 (green line) and E-Syn2 C2CΔ-mCherry (red line). Bars, 4 µm.
SuperresolutionTIRF localization maps for GFP-Sac1 and E-Syt2-mCherry suggested close proximity between E-Syt2 and Sac1. Quantitative analysis of superresolutionTIRF localization maps revealed that even at this enhanced resolution, there was strong overlap between Sac1- and E-Syt2–positive pixels compared with Sac1 and a random distribution (Fig. 6 B and Video 3; overlap coefficient, R = 0.26 ± 0.02; random distribution, R = 0.014 ± 0.04; n = 8), with a mean distance between Sac1- and E-Syt2–positive pixels of ∼25 nm (Fig. 6 C). These results demonstrate close proximity between E-Syt2 and Sac1. To test the concept that E-Syt2 helps regulate the number of steady-state ER–PM Sac1 puncta, we knocked down E-Syt2 using siRNA and assessed the distribution of Sac1 using superresolutionTIRF microscopy. This significantly reduced the number and size of ER–PM GFP-Sac1 and ER–PM anti-Sac1 puncta (Fig. 6 D and Fig. S4 C, top) and significantly increased PIP and PIP2 levels (Fig. 6 E), similar to Sac1 knockdown (Fig. 3, compare E and F). These results suggest that Sac1 is regulating PM PIP and PIP2 levels directly.
In agreement with Giordano et al. (2013), brief activation of M1R (10 µM Oxo-M, 20 s) to hydrolyze PM PI(4,5)P2 resulted in reversible decreases in the intensities of PHPLCδ1 and E-Syt2 in TIRF time-series experiments (Fig. S4 D). In experiments comparing the kinetics of E-Syt2-mCherry and GFP-Sac1, the intensity in the footprint fell in parallel for the two proteins, whereas the recovery was on average slower for GFP-Sac1 than for E-Syt2 (Fig. 6 F, middle). Overexpressing an E-Syt2 mutant (E-Syt2 C2CΔ-mCherry) that lacks the ability to bind PM PI(4,5)P2 and that accumulates in the noncortical ER (Giordano et al., 2013) reduced the number of ER–PM Sac1 puncta and diminished the response of the remaining GFP-Sac1 intensity to M1R activation (Fig. 6 G). We also compared the actions of another isoform, E-Syt3, with those of E-Syt2. E-Syt3 did show robust cortical ER expression (Fig. S4 B), and it increased the amount of ER–PM Sac1, but unlike GFP-Sac1, its distribution did not change with M1R-induced PI(4,5)P2 depletion (compare Fig. 5 A and Fig. S4 E). Although E-Syt3 may regulate ER–PM Sac1 puncta, our results provide stronger evidence for the involvement of E-Syt2 (see Discussion).
Localization and phosphoinositide regulatory functions of Sac1 are conserved in neurons
Does Sac1 behave similarly in native cells? We approached this question using quantitative confocal, TIRF, and superresolution microscopy on rat superior cervical ganglion (SCG) and hippocampal neurons. As in tsA201 cells, ER–PM Sac1 puncta were clearly visible in SCG and hippocampal cell somata (Fig. 7, A and E; and Fig. S5 A) and SCG neurites (Fig. 7 D). These ER–PM Sac1 puncta were closely apposed to the PM, because FRET was detected in SCG neurons coexpressing GFP-Sac1 and the PM marker LDR-RFP (Fig. 7 B). The size, number, and distribution of ER–PM Sac1 puncta in SCG neurons were similar to those in tsA201 cells (Fig. S5 B).
Figure 7.
Sac1 is found in ER–PM contact sites in superior cervical ganglia and hippocampal neurons. (A, left) Confocal image of a living SCG neuron expressing GFP-Sac1. (right) TIRF footprint of a SCG neuron, fixed and stained for anti-Sac1. (B) Confocal images of a single, living SCG neuron overexpressing GFP-Sac1 (left) and RFP-LDR (middle), and the resulting FRET image (right). (C) Summary (mean ± SEM, n = 5; t test; *, P < 0.05) of the KCNQ2/3 current densities from control neurons and SCG neurons overexpressing GFP-Sac1. (D, left) Representative inverted TIRF footprint from an SCG neuron overexpressing GFP-Sac1. (middle) Representative normalized time course of GFP-Sac1 from the footprint of a SCG neuron during the application of 10 µM Oxo-M (purple bar). (right) Same SCG neuron. Normalized time course of GFP-Sac1 from the distal end of an SCG neurite after addition of 10 µM Oxo-M. (E, left) Inverted, maximum-intensity projection of a hippocampal neuron. Inset is 2.5× magnification. (middle) TIRF footprint from a single, living hippocampal neuron expressing GFP-Sac1. (right) Normalized, mean time courses of GFP-Sac1 within the TIRF footprint of hippocampal neuron soma (n = 9). (F) Model illustrating the role of Sac1 in regulating “steady-state” PM phosphoinositide homeostasis and its change in distribution after hydrolysis of PM PI(4,5)P2. 1 and 2 represent Sac1 dephosphorylating PM PI(4)P in trans and cis, respectively.
To monitor changes in Sac1 distribution in live neurons, we overexpressed GFP-Sac1. Those neurons had significantly reduced endogenous KCNQ2/3 current densities, suggesting a loss of PI(4,5)P2 compared with control neurons (Fig. 7 C; control, 13 ± 2 pA/pF; GFP-Sac1, 6 ± 1 pA/pF). Finally, activation of endogenous Gq-coupled muscarinic receptors to deplete PM PI(4,5)P2 resulted in the translocation of Sac1 out of the near-PM evanescent field and into the cytosol (Fig. 7, D and E). The loss of ER–PM Sac1 occurred in nearly all regions of cells, including SCG neuron somata and the tips of neurites located ∼100 µm from the neuron soma (Fig. 7 D). These results suggest that the distribution of Sac1 and the mechanism through which it regulates PM phosphoinositide homeostasis are conserved in neurons.
Discussion
Our results in tsA201 cells and neurons demonstrate that PM phosphoinositide metabolism is actively regulated by the ER-resident lipid phosphatase Sac1. Localization of Sac1 close to the PM is critically dependent on the formation of E-Syt2–mediated ER–PM contact sites, the amount of ER calcium, and the presence of PM PI(4,5)P2. Depletion of PM PI(4,5)P2 leads to reversible loss of ER–PM Sac1 puncta. Dynamic retraction of the ER–PM Sac1 phosphatase allows PM PI(4,5)P2 to recover faster after Gq-coupled receptor activation as in the scheme of Fig. 7 F. The ER–PM activity of Sac1 initially revealed in yeast (Stefan et al., 2011) is thus also conserved in mammals. Here, we have extended this concept to a dynamic picture and determined the kinetic properties.
E-Syt2 regulates the number of ER–PM Sac1 puncta
The crystal structure of the cytoplasmic domain of yeast Sac1 reveals an electrostatically bipolar protein. One surface is negatively charged, and the opposite surface (including the catalytic residues) is enriched with positive charges (Manford et al., 2010). Such polarity likely influences the orientation of the phosphatase domain toward its negatively charged phospholipid substrates on lipid bilayers. Nevertheless, as we demonstrate, PI(4)P alone is not sufficient to localize Sac1 to ER–PM contact sites. Additional proteins mediate formation of these spatially extended compartments of the cortical ER that make close and stable contacts with the PM. ER–PM junctions were first observed in muscle cells (Porter and Palade, 1957), and subsequently, many functional roles and molecular determinants have been revealed. The best-characterized examples are the triads and diads that underlie excitation–contraction coupling in striated muscle. There, the intimate juxtaposition of PM voltage-gated calcium channels and sarcoplasmic reticulum ryanodine receptor channels generates an increase in local cytosolic calcium, the signal for contraction. A similar mechanism underlies SOCE in many cell types. Depletion of ER calcium initiates the oligomerization of ER-localized stromal interaction molecule proteins (STIMs) and their subsequent translocation into puncta near the PM, where they engage PM Orai channels to facilitate refilling of ER calcium (Prakriya et al., 2006; Zhang et al., 2006; Park et al., 2009).
In yeast, six ER-resident proteins mediate extensive interactions between the ER and PM needed for phosphoinositide regulation (Stefan et al., 2011, 2013; Manford et al., 2012). Three of these proteins (tricalbins 1–3) are homologs of the extended synaptotagmin family. The E-Syts have recently been implicated in the formation of PI(4,5)P2-dependent ER–PM contacts in mammalian cells (Giordano et al., 2013). We find much evidence for a role of E-Syt2 in regulating the number of ER–PM Sac1 puncta: (1) E-Syt2 and Sac1 proteins are closely localized to one another, sometimes <20 nm apart in ER–PM Sac1 puncta; (2) knockdown of E-Syt2 reduces ER–PM Sac1 and increases total PIP and PIP2; (3) increasing E-Syt2 increases the amount of ER–PM Sac1; (4), decreasing PI(4,5)P2 by Gq-coupled receptor activation decreases E-Syt2 in TIRF footprints; and (5) mutations in E-Syt2 reduce the number of ER–PM Sac1 puncta and reduce the amount of ER–PM Sac1 that translocates from the PM after PLC activation. Our results show that E-Syt2 promotes the presence of Sac1 at ER–PM junctions; additional proteins may be involved including E-Syt3, which also acts in a PI(4,5)P2-dependent manner and forms homo- and heterodimers with other E-Syt isoforms (Giordano et al., 2013; Schauder et al., 2014).
Outstanding questions include whether E-Syt2 physically associates with Sac1 in ER–PM junctions or simply brings more of the ER, with freely diffusing Sac1, into closer proximity of the PM. We show that forming E-Syt2–induced ER–PM contacts did not immediately localize Sac1 to ER–PM junctions. A requirement for coincident ER–PM contacts and PM PI(4)P may be reflected in the delay observed after the reformation of ER–PM contact sites and the accumulation of Sac1 into such sites after PLC activation. We know from previous observations that PM PI(4,5)P2 recovers more quickly after PLC activity than PM PI(4)P because the PI(4)P 5-kinase is 20-fold faster than the apparent rate of PI 4-kinase (Willars et al., 1998; Falkenburger et al., 2010). Thus, PI(4,5)P2-dependent ER–PM contacts may form first, with secondary recruitment of Sac1 as PI(4)P accumulates more slowly. Alternatively, the difference in recovery kinetics between E-Syt2 and Sac1 may reflect a slow oligomerization of E-Syt heteromers that are subsequently recognized by Sac1 or the aggregation of additional PI(4)P-dependent proteins that recruit Sac1 to ER–PM junctions, exemplified in yeast by Osh3 and VAP proteins at ER–PM junctions, Vps74 at ER–Golgi junctions, or some of the oxysterol-binding protein (OSBP)–related proteins (ORPs) at ER–PM/ER–Golgi junctions (Stefan et al., 2011; Cai et al., 2014; Chung et al., 2015). Together, our results provide a framework for future experiments designed to test such hypotheses.
Sac1: regulator of phosphoinositide metabolism at membrane contact sites
Sac1 has complicated substrate specificity, being able to dephosphorylate PI(3)P, PI(4)P, and PI(3,5)P2 (but not PI(4,5)P2) into PI (Guo et al., 1999; Nemoto et al., 2000; Rohde et al., 2003). This broad substrate preference and a predominant ER localization under normal growth conditions (Faulhammer et al., 2007; Blagoveshchenskaya et al., 2008) means that Sac1 could be a key homeostatic regulator of phosphoinositide metabolism at the interfaces of the ER with other organelle membranes. Indeed, Sac1 localized to ER–Golgi membrane junctions has been proposed to hydrolyze ER PI(4)P in cis (i.e., in the same membrane), thereby maintaining a PI(4)P gradient that drives OSBP-mediated sterol exchange across the ER–Golgi interface (Mesmin et al., 2013).
Here, in a mammalian cell, we invoke a regulatory role for Sac1 in PM phosphoinositide metabolism. We find that increasing ER–PM Sac1 puncta significantly reduces resting PM PI(4)P and PM PI(4,5)P2 levels, making it easier for active PLC to hydrolyze each pool. How do we know that Sac1 is acting directly at the PM? We provide several arguments supporting such a concept: (1) we see endogenous Sac1 at ER–PM junctions at high resolution; (2) overexpressing Sac1 decreases cellular PIP and PIP2, and the latter is almost exclusively at the PM; (3) similar chemical species of PIP2 are decreased after PM Gq-coupled receptor activation as after Sac1 overexpression; (4) knockdown of Sac1 results in a twofold increase in cellular PIP and a 30% increase in cellular PIP2; (5) depleting ER Ca2+ facilitates STIM1–Orai1 interactions to recruit Sac1 to the PM and reduce PM phosphoinositides; (6) dynamic removal of steady-state ER–PM Sac1 by rapamycin has immediate effects on both PM PI(4)P and PI(4,5)P2; and (7) knockdown of ER–PM E-Syt2 reduces the abundance of ER–PM Sac1 contacts and increases PM PI(4)P and PI(4,5)P2. We also determined that ER Sac1 dephosphorylates Golgi PI(4)P, as is expected given its parallel localization to ER–Golgi junctions (Mesmin et al., 2013), but we find this action to be less important in the maintenance of PM PI(4,5)P2.
Our experimental results do not distinguish whether ER–PM Sac1 is acting in cis on ER lipid that is imported from the PM by a lipid transfer protein (Chung et al., 2015) or in trans on PM lipid (Stefan et al., 2011). Measurements of ER–PM distances from the edges of mammalian cells revealed mean distances of 15–20 nm (Fernández-Busnadiego et al., 2015)—large enough to cast into question a direct action of Sac1 on its PM substrate. However, the long flexible linker between the catalytic domain of Sac1 and its ER membrane anchor (∼70 amino acids, >15 nm; Manford et al., 2010) could potentially allow it to bridge such a gap and dephosphosphorylate its PM substrates in a trans mode. This hypothesis has been challenged by Cai et al. (2014). From mutagenesis and phosphatase assays, they find that this flexible linker region of Sac1 is required for catalytic activity and suggest that it may not provide sufficient freedom for Sac1 to function in trans at organelle membrane contact sites. Further, there is growing evidence that OBPs can transport PI(4)P at plasma and Golgi membranes to ER membranes where Sac1 acting in cis can dephosphorylate it into PI (Chung et al., 2015). We find that overexpression of Sac1 did not alter cellular PS levels or the distribution of a PS biosensor, and although ORP5 did reduce PM PI(4)P, it had little effect on PM PI(4,5)P2. Taken together, these results do not support an obvious role for ORP5 in facilitating the Sac1-mediated decreases in PM PI(4)P and PI(4,5)P2. Future experiments are required to fully address Sac1’s preferred mode of action at ER–membrane junctions. In our simplified diagrammatic representation of phosphoinositide metabolism at ER–PM contact sites (Fig.7 F), we have ER Sac1 acting both in trans on the PM (Fig. 7 F, 1), similar to what has been proposed in yeast (Manford et al., 2010, 2012; Stefan et al., 2011), and in cis (Fig. 7 F, 2) as has been proposed at Golgi and PMs (Mesmin et al., 2013; Cai et al., 2014; Chung et al., 2015). If Sac1 acts on phosphoinositides in cis, PM PI(4)P would have to be transferred quickly to the ER membrane. Indeed, examples of lipid transfer exist at several interorganelle contact sites including ER–PM (Prinz, 2014; Schauder et al., 2014; Chung et al., 2015), ER–Golgi (Mesmin et al., 2013), and ER–mitochondria (Toulmay and Prinz, 2012; Voss et al., 2012).
SOCE increases PM Sac1 and reduces PM phosphoinositides
We find that SOCE presents Sac1 to the PM leading to a concurrent decrease in PM PI(4)P and PI(4,5)P2 (Fig. S5 C for model). These STIM1-Orai1–containing ER–PM assemblies seem independent of E-Syt2–mediated ER–PM Sac1-containing junctions as they are maintained during periods of net PM PI(4,5)P2 depletion. Previous studies have implicated PI(4)P in supporting store-operated Orai1-mediated entry of calcium, showing that this Ca2+ influx is sensitive to PI 4-kinase inhibition and PLC activation, but not to PI(4,5)P2 depletion (Rosado and Sage, 2000; Broad et al., 2001; Korzeniowski et al., 2009). Such observations suggest a role for PM PI(4)P in stabilizing Orai1 channel activity, perhaps similar to the role of PI(4,5)P2 for many other PM ion channels (Hille et al., 2015). In that event, recruitment of Sac1 to ER–PM junctions during SOCE could reduce calcium release–activated current (ICRAC) by depleting PM PI(4)P. Another possible result of Sac1 delivery to the PM during SOCE is gradual depletion of PI(4)P and PI(4,5)P2 at the PM, with consequent reduction in the production of calcium-mobilizing inositol trisphosphate (IP3). Both possible results of Sac1 translocation would function as self-limiting “brakes” on SOCE.
The ER–PM Sac1 E-Syt2 system acts as a cellular “thermostat” controlling PM PI(4)P levels
Because of Sac1’s dynamic relocalization, ER–PM phosphoinositide metabolism is self-regulating. As demonstrated, PM PI(4,5)P2 is required for the maintenance of ER–PM contact sites. Net depletion of PM PI(4,5)P2 reduces the number of ER–PM contacts and reduces the population of Sac1 proximate to the PM. Loss of this lipid phosphatase relieves an inhibitory brake on the PM PI(4)P pool, allowing PM PI(4)P and PI(4,5)P2 levels to rise. Rising PM PI(4,5)P2 helps reestablish ER–PM contacts, and, after PI(4)P accumulation, brings Sac1 back into close proximity of the PM, reapplying the brake on PI(4)P and PI(4,5)P2 accumulation. In this scenario, the ER Sac1–E-Syt2 system could be considered a thermostat controlling PM PI(4)P levels. Physiologically, this negative feedback on lipid dephosphorylation would be beneficial for the recovery of PI(4,5)P2 after activation of Gq-coupled receptors. Although ER–PM assemblies containing VAMP-associated protein (VAP) and phosphoinositide transfer protein Nir2 are enhanced in response to PLC signaling (Kim et al., 2015), we show that E-Syt2–containing junctions are disrupted by PI(4,5)P2 depletion. Thus, multiple ER–PM junctions appear to work in concert to replenish PI(4,5)P2 after PLC-mediated hydrolysis.
In conclusion, we provide evidence in mammalian cells that the ER-resident lipid phosphatase, Sac1, is present at distinct sites intimately apposed to the PM via a PI(4,5)P2–E-Syt2 interaction. The phosphoinositide-dependent localization of Sac1 makes it a dynamic regulator of PM phosphoinositide metabolism.
Materials and methods
Cell culture, transfection, and RNAi
tsA201 cells and rat SCG neurons were cultured in DMEM containing 10% FBS and 0.2% penicillin/streptomycin. Cells were incubated at 37°C and 5% CO2. TsA201 cells were transfected at ∼75% confluency with cDNA: CFP-FKBP-Cb5 (Komatsu et al., 2010); DR-VSP (Murata et al., 2005); LDR (Suh et al., 2006); E-Syt2, E-Syt3, and E-Syt2 C2C (Giordano et al., 2013); GFP-Sac1 (Blagoveshchenskaya et al., 2008); mCherry-P4M (Hammond et al., 2014); KCNQ2 and KCNQ3 (Selyanko et al., 2000); MOA-FRB (Komatsu et al., 2010); myc-Orai1 (Park et al., 2009); Sac1 C389S-mCherry (Hammond et al., 2014); Sec61β-mCherry (49155; Addgene); STIM1-eGFP (Ji et al., 2008); recruitable PI(4)P 5-kinase (Suh et al., 2006), using Lipofectamine 3000 (Invitrogen). Double-stranded siRNAs were transfected using similar protocols with sequences were derived from the following sources: human Sac1 siRNA, sc-78323 (Santa Cruz Biotechnology, Inc.); human ESYT2 (HSC.RNAI.N020728.12.5; Integrated DNA Technologies [IDT]); and control (NC1 negative control duplex from IDT). Cells treated with RNAi plasmids were cultured for 48 h posttransfection before experimentation. For confocal, TIRF, and superresolution imaging and for photometry experiments, transfected cells were subcultured onto poly-d-lysine–coated coverglass chips (#0; Thomas Scientific) and cultured overnight (24 h). Patch-clamp experiments required KCNQ2/3 channel–expressing cells to be cultured for a further 24 h to allow for channel subunits to express at the PM.
Animals were handled according to guidelines approved by the University of Washington Institutional Animal Care and Use Committee. Neurons were isolated from SCG of 7- to 12-wk-old male Sprague-Dawley rats by enzymatic digestion (Vivas et al., 2014). Isolated neurons were plated on poly-L-lysine–coated (Sigma-Aldrich) glass chips and incubated in 5% CO2 at 37°C in medium supplemented with 10% FBS. SCG neurons were transfected, via intranuclear injection (Vivas et al., 2014), 18 h after isolation, using an Eppendorf 5242 pressure microinjector and 5171 micromanipulator system (Eppendorf). cDNA was dissolved in 1 mg/ml dextran-fluorescein solution (10,000 kD; Molecular Probes) to yield a final concentration of 50–150 ng/µl. Subsequently, neurons were cultured at 37°C for a further 12 h before experimentation.
Fluorescence microscopy
Confocal microscopy
One day posttransfection, tsA201 cells and SCG neurons were transferred from culture medium to a recording chamber containing modified Krebs–Ringer solution (see Materials and solutions). Fluorophores were excited with an argon-ion (CFP, GFP, and YFP) and a helium-neon (mCherry or RFP) laser and monitored using an inverted microscope under a Plan-Apochromat 63×/1.40 oil DIC M27 objective (confocal, LSM 710; ZEISS). Images were acquired every 2–10 s at room temperature (21–23°C) using Zen software.
FRET
To measure FRET signals between GFP and RFP proteins, tsA201 cells were transfected with expression constructs for GFP-Sac1 and LDR-RFP. FRET from GFP-Sac1 (donor) to LDR-RFP (acceptor) was measured on a confocal laser-scanning microscope (LSM 710) equipped with a plan-apochromat 63×/1.40 oil-immersion objective. An argon-ion laser was used to excite cells at 488 nm. Emitted light was collected between 492–557 and 591–675 nm. The resultant raw donor and acceptor images were corrected for background and bleed-through of GFP emission into the RFP channel. Intensity measurements from the corrected images, referred to as GFPG and RFPG, respectively (where subscript G refers to excitation of GFP with 488-nm light), were extracted using ImageJ software. FRET was expressed as normalized FRET (NFRET; Hoppe et al., 2002; Ji et al., 2008).
TIRF microscopy
Culture conditions and solutions were similar to those for confocal microscopy experiments. TIRF footprints were acquired using a Nikon TiE microscope equipped with a TIRF 60×/1.25 oil differential interference contrast H objective and Photometrics QuantEM electron-multiplying charge-coupled device camera (Nikon). Time series images were acquired every 2–20 s at room temperature (21–23°C) using NIS elements (Nikon).
Superresolution microscopy
For superresolution image analysis, tsA201 cells and SCG neurons were (1) fixed (3.2–4% paraformaldehyde and 0.1% glutaraldehyde in PBS) for 30 min at room temperature, (2) washed (with PBS, five times for 5 min), (3) cleared (50 mM glycine or freshly prepared sodium borohydride) at 4°C for 10 min, (4) blocked and permeabilized for 1 h at room temperature (50% Seablock [Thermo Fisher Scientific] and 0.5% Triton X-100) in PBS, (5) incubated with primary antibody (20% Seablock and 0.5% Triton X-100) for 1 h at room temperature or at 4°C overnight, (6) rinsed (five times for 5 min with PBS) and washed (five times for 5 min with 20% Seablock in PBS), (7) incubated with secondary antibody (20% Seablock, 0.5% Triton X-100) for 1 h at room temperature, and (8) washed (five times for 5 min with PBS). Cells were exposed to primary antibodies targeting anti-GFP (rabbit; 1:500; Thermo Fisher Scientific), anti-mCherry (rat; 1:1,000; Thermo Fisher Scientific), anti–α tubulin (rat; 1:1,000; Abcam), or anti-Sac1 (rabbit; 1:500; Rohde et al., 2003; a gift from P. Mayinger, Oregon Health and Science University, Portland, OR). We used secondary antibodies conjugated to Alexa Fluor 568 (goat; 1:1,000; Thermo Fisher Scientific) or Alexa Fluor 647 (donkey or chicken; 1:1,000; Thermo Fisher Scientific). During imaging at room temperature (21–23°C), cells were kept in PBS, pH 7.4, containing 30 mM mercaptoethylamine. Superresolution TIRF (superresolutionTIRF) or superresolution epifluorescent (superresolutionEPI) localization maps were acquired using a Leica ground-state depletion system (Leica Biosystems). The system is coupled to an inverted microscope (DMI6000B; Leica Biosystems) equipped with a 160× HCX Plan-Apochromat (NA 1.47) oil-immersion lens and an electron-multiplying charge-coupled device camera (iXon3 897; Andor Technology). Fluorophores were excited with 300-mW 568- and 647-nm lasers. The localization of fluorescent particles was determined by fitting single-molecule fluorescence signals with 2D Gaussian or histogram functions using LASAF software (Leica Biosystems; Dixon et al., 2015). High-resolution localization images were reconstructed using the coordinates of centroids obtained from fluorescent particles in 40,000–100,000 images. To confirm that the fixation and permeabilization processes did not disrupt ER integrity, ER tubule length was measured from cells expressing mCherry-Sec61β (Fig. S1, B and C), In accordance with previous measurements from old and new tubules, we found a mean ER tubule size of ∼90 nm (n = 100; Shim et al., 2012). Finally, to estimate the resolution we achieve using this specific type of superresolution microscopy, we fixed and stained tsA201 cells for α-tubulin (antitubulin; ab6160; Abcam). Imaging of microtubules revealed a full-width half-maximum of ∼33 nm (Fig. S1 A), in accordance with published measurements (Wade and Chrétien, 1993; Dempsey et al., 2011; Olivier et al., 2013). Therefore, we estimate our resolution to be <30 nm.
Patch-clamp recordings and photometric FRET measurements
KCNQ2/3 currents were recorded in whole-cell, gigaseal voltage-clamp configuration at room temperature (21–23°C). Recordings were made using an EPC9 amplifier with Patchmaster 2.35 software (HEKA). Cells were held at a holding potential of −60 mV, and 500-ms test pulses to −20 mV were given every 4 s. Currents were quantified by measuring KCNQ2/3 tail currents 400 ms after repolarization to −60 mV using a custom-written algorithm (IGOR Pro software 6.0; WaveMetrics). For calculation of current densities, KCNQ2/3 currents were measured simultaneously with membrane electrical capacitance using the lock-in extension of the Patchmaster online program.
UHPLC/MS
The extraction, derivatization, and detection of phosphoinositides were modified from Clark et al. (2011).
Lipid extraction
Each measurement used a 3.5-mm dish of 75% confluent cells. tsA201 cells were washed with a modified Ringer’s solution (160 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 8 mM glucose, pH 7.4) and spun down at 300–500 g for 3 min at 4°C. Cells were resuspended in 40 µl ice-cold molecular biology–grade water and 100 µl n-butanol (Sigma-Aldrich) added. The resulting solution was vortexed and allowed to sit on ice for 10 min. Next, 12 µl 6N HCl was added, vortexed, and again incubated on ice for a further 10 min. The mixture was centrifuged at 18,000–20,000 g for 3 min at 4°C and 80 µl of the n-butanol phase removed and stored in a fresh tube. An additional 100 µl n-butanol was then added to the cell mixture and again vortexed and spun down at 18,000–20,000 g for 3 min at 4°C. After centrifugation, the n-butanol phase was collected and added to the tube containing the 80 µl of the n-butanol phase. 100 μl chloroform was added to the tube containing the remaining cell suspension and vortexed and centrifuged for 2.5 min at 18,000 g and 4°C. The chloroform phase was collected and combined with the n-butanol extracts. The chloroform wash was repeated twice more and collected. After lipid extractions, the combined organic solution was dried with a Nitrogen evaporator (TurboVap LV; Biotage).
Derivatization
90 µl methanol/CH2Cl2 at a ratio of 4:5 was added to samples and vortexed before adding 20 µl 2 M TMS-diazomethane (Sigma-Aldrich). The mixture was then incubated at room temperature for 1 h. Samples were dried under N2 and resuspended in 50–100 µl methanol for injection into the UHPLC/MS.
UHPLC/MS
Samples were resuspended in 100% methanol (LC-MS Optima grade; Thermo Fisher Scientific) before chromatographic separation at room temperature on a C4 column (Waters Acquity UPLC Protein BEH C4, 300A, 1.1 × 100; 1.7 µm). The mobile phase consisted of a gradient started at 10 mM formic acid in water (A) and 10 mM formic acid in acetonitrile (B) (50:50 vol/vol) delivered at a flow rate of 0.1 ml/min by an ACQUITY UHPLC (Waters). For quantitative analysis, the effluent was monitored by an XEVO TQ-S MS/MS (Waters) in multiple reaction monitoring in positive-ion mode. Phosphoinositide species were quantified by integrating the area under the reconstructed chromatographs. The resulting ionic intensities were normalized to (1) quantitative nonendogenous lipid standards (PI(4)P standard: C17:0-20:4; PI(4,5)P2 standard: C17:0-20:4; Avanti Polar Lipids, Inc.) and (2) protein levels (Bradford assay; Thermo Fisher Scientific) to account for different extraction efficiencies, cell numbers, and effect of protein overexpression on cell integrity. It should be noted when making quantitative interpretations of mass spectrometry results, the cells in a dish will have a heterogeneous range of expression of transiently overexpressed M1R and Sac1.
Statistical analysis
Superresolution image analysis
All image data were analyzed using ImageJ (National Institutes of Health [NIH]).
Calculation GFP-Sac1–PM contact sites
The MAPPER channel of confocal or TIRF micrographs was background corrected and converted into a binary mask. The resulting mask is a binary representation of ER–PM contacts. Subsequently, the mask was applied to the Sac1-mCherry channel to reveal Sac1-mCherry–PM contacts. The area of Sac1-mCherry–PM contacts was calculated using ImageJ and the data transferred to GraphPad Prism for analysis. For presentation purposes, the Sac1-mCherry–PM contacts were displayed using a 16-color lookup table (Fig. 1 G).
Estimation of GFP-Sac1, anti-Sac1, E-Syt2, and E-Syt3 densities
SuperresolutionTIRF localization maps were exported with a pixel size of 10 nm and converted into binary masks. The binary masks were then subjected to dilation followed by erosion operations designed to fill in and smooth objects. Care was taken to ensure that the resulting images were similar to the original mask. Area of cluster size was then calculated and a frequency histogram generated using GraphPad Prism.
Nearest neighbor distance
Distances between single-channel puncta was calculated using the BioVoxxel Toolbox plugin for FIJI (NIH).
Calculation of distance between E-Syt2 and Sac1 puncta
SuperresolutionTIRF localization maps were exported with a pixel size of 10 nm. E-Syt2 and Sac1 channels were split and dilation was followed by erosion operations performed on both chanels. Next, the E-Syt2 channel was used to generate a mask, which was subsequently applied to the Sac1 channel. Application of this mask allowed us to determine the number of Sac1-positive pixels within E-Syt2–positive puncta. Such a procedure is analogous to a traditional correlation coefficient calculation. Next, E-Syt2 masks of increasing sizes were created by sequentially swelling each E-Syt2–positive pixel by 20–200 nm. The resulting masks were then applied to the Sac1 channel, allowing the generation of a distribution histogram detailing the proportion of Sac1-positive pixels within a given distance of E-Syt2–positive pixels.
All data were analyzed using IGOR Pro software (WaveMetrics) or FIJI. Means are shown ± SEM, and significance is assessed by Student’s t test or ANOVA using GraphPad Prism software. Differences were considered significant when P < 0.05.
Materials and solutions
All experiments used Ringer’s solution containing 160 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, and 8 mM glucose, pH 7.4 (NaOH). Intracellular pipette solution for KCNQ2/3 recordings contained 175 mM KCl, 5 mM MgCl2, 5 mM Hepes, 0.1 mM K4BAPTA, 3 mM Na2ATP, and 0.1 mM Na3GTP, adjusted with NaOH to pH 7.2. Rapamycin (LC Laboratories), uridine-5′-triphosphate, and oxotremorine-M (both Sigma-Aldrich) were applied in the superfusate.
Online supplemental material
Fig. S1 provides a quantitative analysis of ER morphology and ER–PM contact sites. Fig. S2 is an UHPLC/MS analysis of cellular PIP and PIP2 signals in tsA201 cells. Fig. S3 visualizes STIM1–Sac1 colocalized pixels after depletion of ER luminal calcium. Fig. S4 provides a quantitative characterization of the distribution, kinetics, and consequences of extended synaptotagmin overexpression. Fig. S5 describes the distribution of Sac1 in SCG neurons. Video 1 shows a 3D superresolutionEPI localization map of GFP-Sac1. Video 2 is a TIRF movie of two tsA201 cells expressing RFP-PHPLCδ1 (left) and GFP-Sac1 (right) before, during, and after the activation of the muscarinic type-I receptor to deplete PM PI(4,5)P2. Video 3 shows a 3D superresolution localization map of GFP-Sac1 (green) and E-Syt2-mCherry (red). Note that this is a movie of the magnified area of Fig. 7 B. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201508106/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Tamas Balla (NIH), Pietro De Camilli (Yale University), Gerald Hammond (NIH), Takanari Inoue (The Johns Hopkins University), and Peter Mayinger (Oregon Health and Science University) for generously providing constructs and antibodies; Lea M. Miller for technical help; Drs. Fernando Santana, Rose Ellen Dixon, and Claudia Moreno for use, help, and advice with superresolution microscopy; Dale Whittington and the other members of the UW School of Pharmacy Mass Spectrometry Center for help and advice; and Drs. Michael Ailion, Gucan Dai, Seung Ryoung Jung, Martin Kruse, Alex J. Merz, Jong Bae Seo, John D. Scott, Nephi Stella, Haijie Yu, and William N. Zagotta for comments and discussions on the manuscript.
This work was supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke grant R37NS008174 (to B. Hille), and National Institutes of Health grants HL085870 and HL085686 (both to L.F. Santana).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- FRET
- Förster resonance energy transfer
- Oxo-M
- oxotremorine M
- PI
- phosphatidylinositol
- PIP
- phosphatidylinositol phosphate
- PM
- plasma membrane
- PS
- phosphatidylserine
- SCG
- superior cervical ganglion
- SOCE
- store-operated calcium entry
- TIRF
- total internal reflection fluorescence
- UHPLC/MS
- ultrahigh-performance liquid chromatography coupled with mass spectrometry
- VSP
- voltage-sensing 5-phosphatase
References
- Balla T. 2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya A., Cheong F.Y., Rohde H.M., Glover G., Knödler A., Nicolson T., Boehmelt G., and Mayinger P.. 2008. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J. Cell Biol. 180:803–812. 10.1083/jcb.200708109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad L.M., Braun F.J., Lievremont J.P., Bird G.S., Kurosaki T., and Putney J.W. Jr. 2001. Role of the phospholipase C-inositol 1,4,5-trisphosphate pathway in calcium release-activated calcium current and capacitative calcium entry. J. Biol. Chem. 276:15945–15952. 10.1074/jbc.M011571200 [DOI] [PubMed] [Google Scholar]
- Cai Y., Deng Y., Horenkamp F., Reinisch K.M., and Burd C.G.. 2014. Sac1-Vps74 structure reveals a mechanism to terminate phosphoinositide signaling in the Golgi apparatus. J. Cell Biol. 206:485–491. 10.1083/jcb.201404041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.L., Hsieh T.S., Yang T.T., Rothberg K.G., Azizoglu D.B., Volk E., Liao J.C., and Liou J.. 2013. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Reports. 5:813–825. 10.1016/j.celrep.2013.09.038 [DOI] [PubMed] [Google Scholar]
- Chung J., Torta F., Masai K., Lucast L., Czapla H., Tanner L.B., Narayanaswamy P., Wenk M.R., Nakatsu F., and De Camilli P.. 2015. INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 349:428–432. 10.1126/science.aab1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J., Anderson K.E., Juvin V., Smith T.S., Karpe F., Wakelam M.J., Stephens L.R., and Hawkins P.T.. 2011. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods. 8:267–272. 10.1038/nmeth.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutz C.E., Snyder S.L., and Schulz T.A.. 2004. Characterization of the yeast tricalbins: membrane-bound multi-C2-domain proteins that form complexes involved in membrane trafficking. Cell. Mol. Life Sci. 61:1208–1220. 10.1007/s00018-004-4029-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey G.T., Vaughan J.C., Chen K.H., Bates M., and Zhuang X.. 2011. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 8:1027–1036. 10.1038/nmeth.1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson E.J., Duman J.G., Moody M.W., Chen L., and Hille B.. 2012. Orai-STIM-mediated Ca2+ release from secretory granules revealed by a targeted Ca2+ and pH probe. Proc. Natl. Acad. Sci. USA. 109:E3539–E3548. 10.1073/pnas.1218247109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson E.J., Falkenburger B.H., and Hille B.. 2013. Quantitative properties and receptor reserve of the IP3 and calcium branch of Gq-coupled receptor signaling. J. Gen. Physiol. 141:521–535. 10.1085/jgp.201210886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson E.J., Jensen J.B., and Hille B.. 2014. Golgi and plasma membrane pools of PI(4)P contribute to plasma membrane PI(4,5)P2 and maintenance of KCNQ2/3 ion channel current. Proc. Natl. Acad. Sci. USA. 111:E2281–E2290. 10.1073/pnas.1407133111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., and De Camilli P.. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Dixon R.E., Moreno C.M., Yuan C., Opitz-Araya X., Binder M.D., Navedo M.F., and Santana L.F.. 2015. Graded Ca2+/calmodulin-dependent coupling of voltage-gated CaV1.2 channels. eLife. 4:4 10.7554/eLife.05608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburger B.H., Jensen J.B., and Hille B.. 2010. Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol. 135:99–114. 10.1085/jgp.200910345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburger B.H., Dickson E.J., and Hille B.. 2013. Quantitative properties and receptor reserve of the DAG and PKC branch of G(q)-coupled receptor signaling. J. Gen. Physiol. 141:537–555. 10.1085/jgp.201210887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulhammer F., Kanjilal-Kolar S., Knödler A., Lo J., Lee Y., Konrad G., and Mayinger P.. 2007. Growth control of Golgi phosphoinositides by reciprocal localization of sac1 lipid phosphatase and pik1 4-kinase. Traffic. 8:1554–1567. 10.1111/j.1600-0854.2007.00632.x [DOI] [PubMed] [Google Scholar]
- Fernández-Busnadiego R., Saheki Y., and De Camilli P.. 2015. Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc. Natl. Acad. Sci. USA. 112:E2004–E2013. 10.1073/pnas.1503191112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., and Voeltz G.K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F., Saheki Y., Idevall-Hagren O., Colombo S.F., Pirruccello M., Milosevic I., Gracheva E.O., Bagriantsev S.N., Borgese N., and De Camilli P.. 2013. PI(4,5)P2-dependent and Ca2+-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 153:1494–1509. 10.1016/j.cell.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Stolz L.E., Lemrow S.M., and York J.D.. 1999. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274:12990–12995. 10.1074/jbc.274.19.12990 [DOI] [PubMed] [Google Scholar]
- Hammond G.R., Machner M.P., and Balla T.. 2014. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205:113–126. 10.1083/jcb.201312072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgemann D.W., Feng S., and Nasuhoglu C.. 2001. The complex and intriguing lives of PIP2 with ion channels and transporters. Sci. STKE. 2001:re19. [DOI] [PubMed] [Google Scholar]
- Hille B., Dickson E.J., Kruse M., Vivas O., and Suh B.C.. 2015. Phosphoinositides regulate ion channels. Biochim. Biophys. Acta. 1851:844–856. 10.1016/j.bbalip.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe A., Christensen K., and Swanson J.A.. 2002. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys. J. 83:3652–3664. 10.1016/S0006-3495(02)75365-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., and Nunnari J.. 2012. Cell Biology. Mitochondrial dynamics and apoptosis--the ER connection. Science. 337:1052–1054. 10.1126/science.1224709 [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O., Lü A., Xie B., and De Camilli P.. 2015. Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. EMBO J. 34:2291–2305. 10.15252/embj.201591565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Heo W.D., Grimley J.S., Wandless T.J., and Meyer T.. 2005. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods. 2:415–418. 10.1038/nmeth763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Xu P., Li Z., Lu J., Liu L., Zhan Y., Chen Y., Hille B., Xu T., and Chen L.. 2008. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc. Natl. Acad. Sci. USA. 105:13668–13673. 10.1073/pnas.0806499105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Guzman-Hernandez M.L., Wisniewski E., and Balla T.. 2015. Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev. Cell. 33:549–561. 10.1016/j.devcel.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T., Kukelyansky I., McCaffery J.M., Ueno T., Varela L.C., and Inoue T.. 2010. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat. Methods. 7:206–208. 10.1038/nmeth.1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski M.K., Popovic M.A., Szentpetery Z., Varnai P., Stojilkovic S.S., and Balla T.. 2009. Dependence of STIM1/Orai1-mediated calcium entry on plasma membrane phosphoinositides. J. Biol. Chem. 284:21027–21035. 10.1074/jbc.M109.012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R.S. 2007. The molecular choreography of a store-operated calcium channel. Nature. 446:284–287. 10.1038/nature05637 [DOI] [PubMed] [Google Scholar]
- Liu Y., Boukhelifa M., Tribble E., and Bankaitis V.A.. 2009. Functional studies of the mammalian Sac1 phosphoinositide phosphatase. Adv. Enzyme Regul. 49:75–86. 10.1016/j.advenzreg.2009.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford A., Xia T., Saxena A.K., Stefan C., Hu F., Emr S.D., and Mao Y.. 2010. Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J. 29:1489–1498. 10.1038/emboj.2010.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford A.G., Stefan C.J., Yuan H.L., Macgurn J.A., and Emr S.D.. 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 23:1129–1140. 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., and Antonny B.. 2013. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 155:830–843. 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Murata Y., Iwasaki H., Sasaki M., Inaba K., and Okamura Y.. 2005. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 435:1239–1243. 10.1038/nature03650 [DOI] [PubMed] [Google Scholar]
- Nemoto Y., Kearns B.G., Wenk M.R., Chen H., Mori K., Alb J.G. Jr., De Camilli P., and Bankaitis V.A.. 2000. Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J. Biol. Chem. 275:34293–34305. 10.1074/jbc.M003923200 [DOI] [PubMed] [Google Scholar]
- Olivier N., Keller D., Gönczy P., and Manley S.. 2013. Resolution doubling in 3D-STORM imaging through improved buffers. PLoS One. 8:e69004 10.1371/journal.pone.0069004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., Hoover P.J., Mullins F.M., Bachhawat P., Covington E.D., Raunser S., Walz T., Garcia K.C., Dolmetsch R.E., and Lewis R.S.. 2009. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 136:876–890. 10.1016/j.cell.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt T.R., Dove S.K., Lubben A., Calaminus S.D., and Wakelam M.J.. 2006. Analysis of intact phosphoinositides in biological samples. J. Lipid Res. 47:1588–1596. 10.1194/jlr.D600004-JLR200 [DOI] [PubMed] [Google Scholar]
- Porter K.R., and Palade G.E.. 1957. Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. J. Biophys. Biochem. Cytol. 3:269–300. 10.1083/jcb.3.2.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M., Feske S., Gwack Y., Srikanth S., Rao A., and Hogan P.G.. 2006. Orai1 is an essential pore subunit of the CRAC channel. Nature. 443:230–233. 10.1038/nature05122 [DOI] [PubMed] [Google Scholar]
- Prinz W.A. 2010. Lipid trafficking sans vesicles: where, why, how? Cell. 143:870–874. 10.1016/j.cell.2010.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz W.A. 2014. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205:759–769. 10.1083/jcb.201401126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H.M., Cheong F.Y., Konrad G., Paiha K., Mayinger P., and Boehmelt G.. 2003. The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J. Biol. Chem. 278:52689–52699. 10.1074/jbc.M307983200 [DOI] [PubMed] [Google Scholar]
- Rosado J.A., and Sage S.O.. 2000. Phosphoinositides are required for store-mediated calcium entry in human platelets. J. Biol. Chem. 275:9110–9113. 10.1074/jbc.275.13.9110 [DOI] [PubMed] [Google Scholar]
- Schauder C.M., Wu X., Saheki Y., Narayanaswamy P., Torta F., Wenk M.R., De Camilli P., and Reinisch K.M.. 2014. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 510:552–555. 10.1038/nature13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selyanko A.A., Hadley J.K., Wood I.C., Abogadie F.C., Jentsch T.J., and Brown D.A.. 2000. Inhibition of KCNQ1-4 potassium channels expressed in mammalian cells via M1 muscarinic acetylcholine receptors. J. Physiol. 522:349–355. 10.1111/j.1469-7793.2000.t01-2-00349.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S.H., Xia C., Zhong G., Babcock H.P., Vaughan J.C., Huang B., Wang X., Xu C., Bi G.Q., and Zhuang X.. 2012. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. USA. 109:13978–13983. 10.1073/pnas.1201882109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan C.J., Manford A.G., Baird D., Yamada-Hanff J., Mao Y., and Emr S.D.. 2011. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 144:389–401. 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Manford A.G., and Emr S.D.. 2013. ER-PM connections: sites of information transfer and inter-organelle communication. Curr. Opin. Cell Biol. 25:434–442. 10.1016/j.ceb.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh B.C., Inoue T., Meyer T., and Hille B.. 2006. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 314:1454–1457. 10.1126/science.1131163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpetery Z., Várnai P., and Balla T.. 2010. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc. Natl. Acad. Sci. USA. 107:8225–8230. 10.1073/pnas.1000157107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A., and Prinz W.A.. 2011. Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr. Opin. Cell Biol. 23:458–463. 10.1016/j.ceb.2011.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A., and Prinz W.A.. 2012. A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 125:49–58. 10.1242/jcs.085118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F.C., Seki A., Yang H.W., Hayer A., Carrasco S., Malmersjö S., and Meyer T.. 2014. A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nat. Cell Biol. 16:133–144. 10.1038/ncb2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y., Hasegawa J., Chinnapen D., Inoue T., Okazaki S., Kato R., Wakatsuki S., Misaki R., Koike M., Uchiyama Y., et al. 2011. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc. Natl. Acad. Sci. USA. 108:15846–15851. 10.1073/pnas.1109101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T., Falkenburger B.H., Pohlmeyer C., and Inoue T.. 2011. Triggering actin comets versus membrane ruffles: distinctive effects of phosphoinositides on actin reorganization. Sci. Signal. 4:ra87 10.1126/scisignal.2002033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas O., Kruse M., and Hille B.. 2014. Nerve growth factor sensitizes adult sympathetic neurons to the proinflammatory peptide bradykinin. J. Neurosci. 34:11959–11971. 10.1523/JNEUROSCI.1536-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss C., Lahiri S., Young B.P., Loewen C.J., and Prinz W.A.. 2012. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J. Cell Sci. 125:4791–4799. 10.1242/jcs.105635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R.H., and Chrétien D.. 1993. Cryoelectron microscopy of microtubules. J. Struct. Biol. 110:1–27. 10.1006/jsbi.1993.1001 [DOI] [PubMed] [Google Scholar]
- Wenk M.R., Lucast L., Di Paolo G., Romanelli A.J., Suchy S.F., Nussbaum R.L., Cline G.W., Shulman G.I., McMurray W., and De Camilli P.. 2003. Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nat. Biotechnol. 21:813–817. 10.1038/nbt837 [DOI] [PubMed] [Google Scholar]
- Willars G.B., Nahorski S.R., and Challiss R.A.. 1998. Differential regulation of muscarinic acetylcholine receptor-sensitive polyphosphoinositide pools and consequences for signaling in human neuroblastoma cells. J. Biol. Chem. 273:5037–5046. 10.1074/jbc.273.9.5037 [DOI] [PubMed] [Google Scholar]
- Zhang S.L., Yeromin A.V., Zhang X.H., Yu Y., Safrina O., Penna A., Roos J., Stauderman K.A., and Cahalan M.D.. 2006. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc. Natl. Acad. Sci. USA. 103:9357–9362. 10.1073/pnas.0603161103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Hsu F., Stefan C.J., Wu X., Patel A., Cosgrove M.S., and Mao Y.. 2012. Allosteric activation of the phosphoinositide phosphatase Sac1 by anionic phospholipids. Biochemistry. 51:3170–3177. 10.1021/bi300086c [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.