Abstract
Advances in next-generation sequencing and mass spectrometry have revealed widespread messenger RNA modifications and RNA editing, with dramatic effects on mammalian transcriptomes. Factors introducing, deleting, or interpreting specific modifications have been identified, and analogous with epigenetic terminology, have been designated “writers,” “erasers,” and “readers.” Such modifications in the transcriptome are referred to as epitranscriptomic changes and represent a fascinating new layer of gene expression regulation that has only recently been appreciated. Here, we outline how RNA editing and RNA modification can rapidly affect gene expression, making both processes as well suited to respond to cellular stress and to regulate the transcriptome during development or circadian periods.
Introduction
Eukaryotic mRNA expression and regulation involves transcription, mRNA processing, and degradation of transcripts. All of these processes are regulated and coordinated to maintain transcript homeostasis and allow responses to intra- and extracellular stimuli (Maniatis and Reed, 2002; Keene, 2007; Bentley, 2014; Braun and Young, 2014). In addition to these classic mechanisms affecting transcriptome homeostasis, an ever-increasing list of RNA editing and modification events has been described in recent years. Tailored mRNA-sequencing projects have revealed that RNA modifications and RNA editing sites are present in almost every transcript and can be dynamically regulated (Li et al., 2009; Dominissini et al., 2012; Meyer et al., 2012; Peng et al., 2012; Carlile et al., 2014; Schwartz et al., 2014). These processes provide a direct and fast way to manipulate the existing transcriptome, bypassing conventional gene expression mechanisms mediated, for instance, by the activation of transcription factors. For example, RNA modifications can increase translation efficiency, thereby boosting expression of particular transcripts and allowing immediate responses to rapidly changing conditions (Wang et al., 2015; Zhou et al., 2015).
Here we discuss how RNA editing and modification mechanisms control the transcriptome (Fig. 1), focusing on m6A methylation and adenosine to inosine editing (A-to-I editing). For other epitranscriptomic mechanisms such as cytosine to uracil deamination (C-to-U editing) or pseudouridylation, we will mostly highlight their functions during environmental or cellular stress.
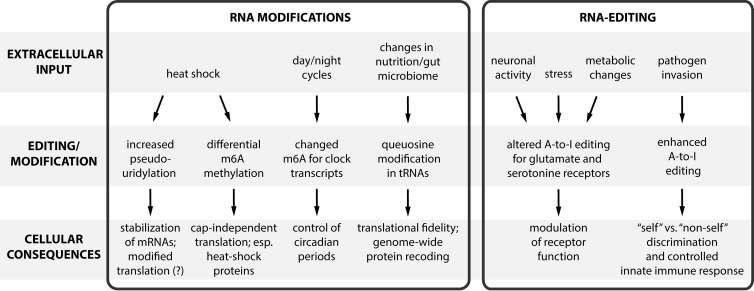
Figure 1.
Rapid modifications of the epitranscriptome in response to extracellular inputs. RNA modifications and editing can regulate the transcriptome. Both types of epitranscriptomic regulation are particularly suited to modulate the transcriptome in situations of stress because they allow a more rapid response compared with classic regulation mechanisms of gene expression.
m6A RNA methylation dynamically regulates the epitranscriptome
m6A RNA methylation (N6-methyladenosine) was discovered ∼40 years ago (Desrosiers et al., 1974; Perry and Kelley, 1974) and was shown to occur on ∼1–2% of all mRNA adenosines (Perry et al., 1975). This RNA modification more recently caught the attention of many researchers when m6A demethylating activity was demonstrated for the fat mass and obesity protein (FTO; Jia et al., 2011). In search for substrates, distinct m6A RNA methylation sites in mammalian transcriptomes were identified and characterized in two pioneering studies (Dominissini et al., 2012; Meyer et al., 2012). Both studies identified modification sites in transcripts originating from ∼7,000 human genes, using an m6A-specific immunoprecipitation assay combined with RNA sequencing. Excitingly, m6A RNA methylation was found to be dynamically associated with particular transcripts. Moreover, factors for writing, erasing, and reading m6A methylations have been identified (Fig. 2). Here, we will illustrate the role of m6A methylation during mammalian development and circadian regulation.
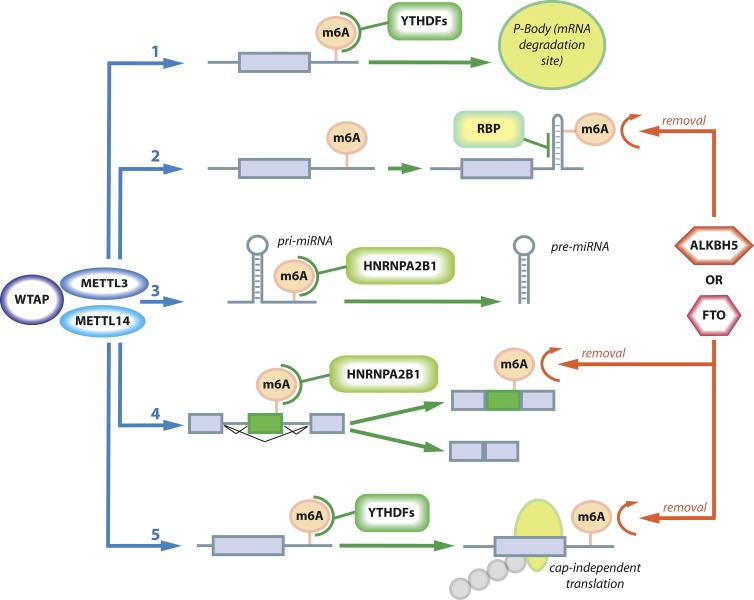
Figure 2.
The dynamic interplay of factors writing, reading, and erasing m6A modifications. The m6A mark is written by METTL3 or METTL14 (blue) in complex with WTAP (purple), whereas the proteins ALKBH5 or FTO (red) are erasers of m6A modifications. Several reading mechanisms have been characterized: (1) YTHDF proteins (green) can detect the m6A mark and shift localization of the transcripts to P-bodies and thereby promote degradation; (2) m6A can cause structural changes and thereby lead to differential binding of RNA-binding proteins (RBP, green); (3 and 4) HNRNPA2B1 (green) can bind to m6A sites in pri-miRNAs or pre-mRNAs and either promote maturation of miRNAs (3) or cause alternative splicing (4); and finally, (5) under stress conditions, m6A can also cause cap-independent translation. m6A writing mechanisms are in blue, reading mechanisms are in green, and erasing mechanisms are in red.
Writers and erasers of m6A RNA methylation
To date, four major factors have been characterized that mediate m6A homeostasis in RNA. Methylation is achieved by two methyltransferase-like proteins, METTL3 and METTL14, that work in a complex. Demethylation, on the other hand, is achieved by FTO or the α-ketoglutarate–dependent dioxygenase homolog, ALKBH5 (Jia et al., 2011; Zheng et al., 2013).
METTL3 (also known as MT-A70) was identified as binding S-adenosyl-l-methionine and is the core component of a multiprotein complex catalyzing m6A methylation (Bokar et al., 1997). METTL14, another crucial subunit of the complex, forms a stable heterodimer with METTL3 (Liu et al., 2014; Ping et al., 2014; Wang et al., 2014b). A third constituent of the methylation complex, Wilms tumor 1–associated protein (WTAP), likely also regulates m6A methylation (Liu et al., 2014; Ping et al., 2014). Both METTL3 and METTL14 have essential functions, with knockdown of either protein eliminating m6A methylation and self-renewal capacity in embryonic stem cells (Wang et al., 2014b). Interestingly, METTL3 binding can be modulated by miRNAs that target transcripts containing m6A binding sites (Chen et al., 2015).
FTO was the first m6A demethylase to be identified (Jia et al., 2011; Fu et al., 2013). Consistently, overexpression of FTO in HEK293T cells leads to a reduction of m6A modifications in cellular RNA (Meyer et al., 2012). FTO belongs to the AlkB family of demethylases (Kurowski et al., 2003), and another member of the AlkB family, ALKBH5, also removes m6A methylation in RNA by oxidation (Zheng et al., 2013). Alkbh5-knockout mice are viable until adulthood, but fertility of male knockout mice is affected, suggesting that the dynamic removal of m6A modifications is fundamental for mammalian spermatogenesis (Zheng et al., 2013).
Readers of m6A RNA methylation
The discovery of proteins that read m6A RNA methylations was a milestone in elucidating the impact of m6A modifications in RNA. With m6A-modified RNA as bait for affinity purification and subsequent mass spectrometry, the proteins YTHDF1-3 and HuR were identified as m6A binders (Dominissini et al., 2012; Wang et al., 2014a). YTHDF2 binds more than 3,000 cellular RNAs—mostly mRNAs but also noncoding RNAs—and shifts them to sites of mRNA degradation, such as P-bodies (Wang et al., 2014a). In line with this finding, m6A methylation can impair binding of the mRNA stabilizing protein HuR and indirectly promote degradation (Wang et al., 2014b). On the other hand, YTHDF1 promotes translation and, in conjunction with YTHDF2, allows rapid regulation of the transcriptome (Wang et al., 2015).
Another nuclear m6A reader, HNRNPA2B1, binds to m6A modifications, influences alternative splicing, and promotes miRNA biogenesis (Alarcón et al., 2015a,b). Consistently, METTL3 knockdown also modulates alternative splicing (Alarcón et al., 2015a). m6A modifications can also affect many other RNA-binding proteins, such as HNRNPC, by changing local RNA structures (Liu et al., 2015). Approximately 3,000 so-called m6A switches have been identified in which m6A modification affects HNRNPC binding, leading to alternative splicing of mRNAs. However, many consequences of m6A-induced structural changes are not yet characterized and will provide an interesting basis for future studies.
miRNAs show the way
One of the first transcriptome-wide studies revealed a correlation of m6A patterns in 3′ UTRs with miRNA-binding sites; UTRs that contain miRNA-binding sites showed a significant enrichment for m6A sites (Meyer et al., 2012). Similarly, it was shown that targets of the 25 most highly expressed miRNAs in the brain are significantly enriched in m6A sites (Meyer et al., 2012). Overexpression of miRNAs leads to elevated m6A levels at corresponding target sites, whereas miRNA repression results in m6A down-regulation (Chen et al., 2015). Subsequent mutational analysis showed that these effects are sequence specific. The same study also showed that miRNAs regulate m6A levels by affecting METTL3 activity. As discussed earlier, m6A methylation also marks primary miRNAs for processing and promotes miRNA biogenesis (Alarcón et al., 2015a,b). Thus, there is a dynamic regulatory interplay between miRNAs and m6A modifications; m6A promotes miRNA biogenesis, whereas mature miRNAs, in turn, modulate METTL3 activity and increase m6A methylation.
The importance of m6A RNA methylation in development
m6A methylation is dynamically regulated and may play a role during development. For example, m6A modifications are relatively low during embryonic stages but display a dramatic increase during brain development, as well as tissue-specific regulation (Meyer et al., 2012). Recent studies have also linked m6A RNA methylation to the developmental switch from pluripotency to differentiation (Batista et al., 2014; Wang et al., 2014b; Geula et al., 2015). Knockout of Mettl3 in mouse embryonic stem cells leads to reduced m6A RNA methylation and promotes their self-renewal (Batista et al., 2014). Similarly, knockout of Mettl3 in human embryonic stem cells prevents differentiation (Batista et al., 2014). Moreover, METTL3 selectively targets mRNAs that regulate pluripotency, such as those of NANOG or SOX2 (Batista et al., 2014; Chen et al., 2015). Geula et al. (2015) further confirmed that METTL3 is required for differentiation in vivo, using mouse models. In agreement with earlier findings, increased half-lives of several METTL3 targets and elevated levels of transcripts affecting pluripotency were found in Mettl3-knockout cells and embryoid bodies derived from those cells (Geula et al., 2015). Consistently, combined knockdown of the pluripotency factors KLF4, NANOG, SOX2, and ZFP42 in Mettl3-knockout embryonic stem cells leads to an up-regulation of lineage commitment markers (Geula et al., 2015). These data suggest that ablation of METTL3 leads to increased stability of transcripts encoding pluripotency factors that inhibit differentiation. Therefore, m6A methylation facilitates resolution of murine naive pluripotency toward differentiation by mediating timely down-regulation of pluripotency factors at the onset of differentiation (Geula et al., 2015).
Biological consequences of m6A dynamics: Tuning the circadian clock
The circadian clock is a fascinating, self-regulated circuit that is fine-tuned by external cues (Asher and Sassone-Corsi, 2015). Fustin et al. (2013) provided compelling evidence that m6A RNA methylation affects the speed of the circadian clock, supported by the fact that many “clock” transcripts contain methylation sites. Knockdown of METTL3 leads to elongation of the circadian cycle, whereas METTL3 overexpression shortens it. In addition, depletion of METTL3 increases the duration of nuclear retention of the clock transcripts Arntl and Per2, which may explain the effect on circadian rhythm (Fustin et al., 2013).
In line with these findings, earlier results showed that ALKBH5-deficient cells exhibit accelerated nuclear RNA export (Zheng et al., 2013), as might be expected for an m6A RNA-demethylase acting in opposition to METTL3. Furthermore, Zheng et al. (2013) demonstrated that depletion of ALKBH5 strongly affects protein factors involved in pre-mRNA processing. The splicing factor ASF/SF2 was found to colocalize with ALKBH5, and upon ALKBH5 depletion, nuclear staining patterns for ASF/SF2 were diminished. Moreover, the kinase SRPK1 relocalized from the nucleus to the cytoplasm upon ALKBH5 knockdown. Notably, SRPK1 is one of the main kinases that phosphorylate ASF/SF2. This may explain the effect of accelerated RNA export in ALKBH5-deficient cells because ASF/SF2 promotes mRNA export when dephosphorylated. Zheng et al. (2013) speculated that the methylation status of transcripts targeted by pre-mRNA/mRNA processing factors may feed back on the activity of these factors. Very recently, it was demonstrated that pre-mRNA processing factors such as HNRNPA2B1 act as m6A readers, supporting this idea (Alarcón et al., 2015a).
Thus, m6A RNA methylation affects the rates of mRNA export from the nucleus to the cytoplasm and, in this way, further influences protein translation. This can affect circadian rhythms and may also have an impact on other cellular functions.
Pseudouridylation and m6A: An anti-stress therapy for the transcriptome?
Pseudouridylation was the first RNA modification to be discovered (Cohn, 1951). Pseudouridine (Ψ) is an isomer of uridine and leads to more stable base pairing with adenosine compared with U-A base pairs (Karijolich et al., 2015). Pseudouridylation is well documented in the context of spliceosomal small nuclear RNAs (snRNAs) and ribosomal RNAs. Pseudouridine was believed to be a constitutive and stable RNA modification but was recently reported to be induced in the yeast spliceosomal RNA U2 upon stress, such as nutrient deprivation or heat shock (Wu et al., 2011). Subsequently, more than 200 stress-induced Ψ sites were found in yeast and human cells (Carlile et al., 2014; Schwartz et al., 2014; Li et al., 2015). Thus, it seems that pseudouridylation actively adjusts the transcriptome in response to stress. It is known that pseudouridylation stabilizes RNA structures and may therefore be well suited to stabilize transcripts under heat shock conditions (Hall and McLaughlin, 1991; Arnez and Steitz, 1994; Davis et al., 1998). Moreover, pseudouridylation allows alternative decoding of particular transcripts and increases translational efficiency, thereby offering different possibilities for how specific pseudouridylation sites may improve stress tolerance (Anderson et al., 2010; Karijolich and Yu, 2011; Fernández et al., 2013).
Like Ψ, m6A modifications can also be modulated after heat shock or other sorts of stress (Meyer et al., 2015; Zhou et al., 2015). Upon heat shock, localization of the m6A reader YTHFD2 is shifted to the nucleus, where it blocks FTO-mediated demethylation. Consequently, RNAs produced during heat shock carry more m6A marks in their 5′ end, leading to increased cap-independent translation (Zhou et al., 2015). Collectively, these findings suggest that different RNA modifications may mediate a fast response to stressful environmental conditions.
The gut microbiome and tRNA modification
Queuosine is a rare nucleoside in tRNAs of bacteria and eukaryotes that is structurally similar to guanine. Most interestingly, animals cannot synthesize queuosine and rely on the gut microbiome to provide the precursor queuine (Fergus et al., 2015). The gut microbiome thus determines the bioavailability of queuine and thereby influences the synthesis of queuosine and the ratio of this modified nucleotide in tRNAs (Müller et al., 2015). Queuosine has been found only in the tRNA anticodon, suggesting a role in translation. Indeed, it was shown that queuosine in the wobble position affects codon choice (Meier et al., 1985). Moreover, data from Drosophila suggest that the presence of queuosine in tRNA alters translational fidelity (Zaborske et al., 2014). Thus, the nutritional environment and the gut microbiome can directly control a host’s tRNA composition and its translational fidelity. Ultimately, this may provide an elegant and direct link to how the microbiome affects translation and therefore the proteome of a host.
RNA editing
RNA editing is a posttranscriptional process in which a genomically templated sequence is altered at the RNA level. Two major types of RNA editing exist in mammals: adenosine to inosine (A-to-I) and cytidine to uracil (C-to-U). A-to-I editing is catalyzed by the ADAR (adenosine deaminase acting on RNA) class of enzymes that bind double-stranded RNA (dsRNA). During this deamination reaction, an adenosine is converted to inosine, which has the base pairing properties of guanosine and is thus interpreted as guanosine by cellular machines (Fig. 3 A). A-to-I editing is primarily a nuclear event and is believed to occur cotranscriptionally. Thus, A-to-I editing can affect several steps during gene expression and regulation, such as splicing, RNA stability, localization, miRNA function, and translation (Fig. 3, A and B; Nishikura, 2010; Daniel et al., 2015; Tajaddod et al., 2016). A-to-I editing sites mostly reside in noncoding parts of the human transcriptome, such as introns or 3′ UTRs, but can also be found in coding regions (Li et al., 2009; Peng et al., 2012; Ramaswami et al., 2013). Interestingly, levels of editing are very diverse and range from barely detectable to almost 100%, depending on the tissue, developmental stage, and substrate (Li et al., 2009; Wahlstedt et al., 2009; Stulić and Jantsch, 2013). This suggests that editing is dynamically regulated, possibly in response to cellular or extracellular stimuli. In fact, A-to-I editing levels are deregulated during cancer and can promote cancer progression, suggesting they could be an important parameter for cancer treatment strategies (Chen et al., 2013; Fumagalli et al., 2015; Han et al., 2015; Paz-Yaacov et al., 2015).
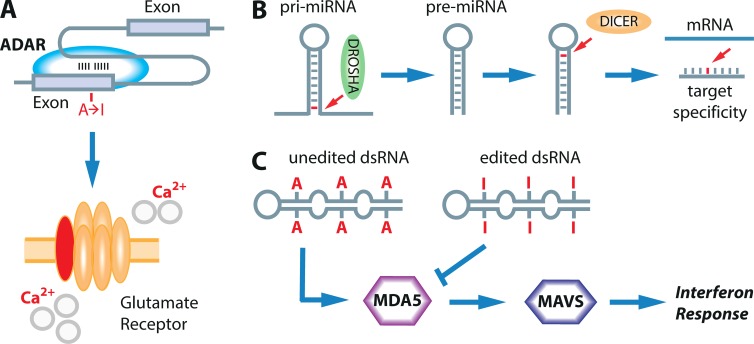
Figure 3.
The consequences of A-to-I-editing are diverse. (A) Editing sites in protein-coding transcripts are frequently defined by double-stranded structures between exons and introns. The extent of editing can have various consequences. In the case of the glutamate receptor subunit GRIA2, it influences the receptor permeability for Ca2+ ions. (B) A-to-I editing can affect miRNA biogenesis and interfere with DROSHA or DICER cleavage, as well as alter target specificity of miRNAs. Editing sites are marked in red; arrows point to the editing site. (C) Unedited foreign dsRNA is sensed by MDA5 (purple) and promotes the interferon response via MAVS (blue). Edited dsRNA does not activate the interferon response and is treated as self.
The second most abundant editing mediated by deamination occurs on cytidines that are converted to uracil (C-to-U editing). C-to-U editing is catalyzed by a diverse family of 10 different cytidine deaminases called APOBECs that can target RNA and DNA (Smith et al., 2012). The substrate specificity and the targets of the different family members are only sporadically identified, but C-to-U editing is involved in various cellular processes such as host cell defense, antiviral defense, immunity, or recoding of transcripts (Prohaska et al., 2014). For the APOBEC1 protein—the first protein of the family to be identified—more than 50 C-to-U editing events have been identified in the mouse transcriptome (Rosenberg et al., 2011; Blanc et al., 2014). For APOBEC3A—the second APOBEC protein shown to exhibit RNA editing activity—several hundred editing sites have been identified (Sharma et al., 2015).
In contrast to ADAR deaminases that act exclusively on RNA, many APOBEC members primarily act on DNA of both viral and endogenous origin. Misregulation of DNA deamination can cause mutations and has been associated with cancer (Alexandrov et al., 2013; Roberts et al., 2013).
A-to-I RNA editing enzymes
Two catalytically active ADAR deaminases are known in mammals, ADAR1 and ADAR2. Mice lacking ADAR1 deaminase activity die at early embryonic stages (Hartner et al., 2004, 2009; Mannion et al., 2014; Liddicoat et al., 2015), but ADAR1-deficient mice can be rescued until after birth if they also lack proteins sensing dsRNA (Mannion et al., 2014; Liddicoat et al., 2015). This suggests that ADAR1 editing “marks” endogenous dsRNAs to discriminate them from foreign or viral RNA and thereby plays an important role in the innate immune response (see following section and Fig. 3 C). Mice deficient of ADAR2 die within 3 wk after birth but can be rescued when a preedited allele encoding glutamate receptor subunit 2 (Gria2) is coexpressed, arguing for Gria2 as the major target of ADAR2 (Higuchi et al., 2000). For a third adenosine deaminase, ADAR3, no catalytic activity has yet been discovered (Chen et al., 2000; Schneider et al., 2014). However, ADAR3 may have regulatory functions by competing with ADAR1 and ADAR2 for substrate binding (Chen et al., 2000).
A-to-I editing dynamics during the innate immune response
One of the most recent and exciting findings in the A-to-I editing field is the demonstration that ADAR1 regulates the innate immune response. The innate immune response uses so-called pattern-recognition receptors (PRRs), which recognize specific molecular patterns that are inevitable for the survival of a pathogen (Wu and Chen, 2014). A particular class of PRRs can detect pathogen-derived nucleic acids in the cytosol. Proteins sensing these nucleic acids and transmitting the signal include retinoic acid inducible gene-I (RIG-I), melanoma differentiation associated gene 5 (MDA5), and the mitochondrial antiviral signaling protein (MAVS; Wu and Chen, 2014). However, one of the most intriguing questions is how PRRs discriminate between endogenous and pathogen-derived nucleic acids to distinguish “self” from “nonself.” Here, A-to-I editing plays a key role. Particular mutations in the human ADAR1 gene have been found to be associated with the autoimmune disorder Aicardi-Goutières syndrome, which results from up-regulation of interferon-stimulated genes (Rice et al., 2012). Consistently, deletion of ADAR1 in mouse models causes an elevated interferon response (Hartner et al., 2009). Interestingly, dsRNA containing multiple I-U base pairs has been shown to efficiently suppress the induction of interferon-stimulated genes (Vitali and Scadden, 2010). However, key evidence from rescue experiments in mice showed that ADAR1 and A-to-I editing represses nucleic acid–induced inflammation signaling and discriminates self from nonself dsRNA. Mice carrying homozygous deletions for either Mda-5 or Mavs can rescue ADAR−/− deficiency, albeit to different extents (Mannion et al., 2014; Liddicoat et al., 2015). Mannion et al. (2014) showed that mice lacking the entire ADAR1 protein can be rescued until birth when Mavs is also deleted. Liddicoat et al. (2015) showed that a knock-in mouse, in which only the catalytic activity of ADAR1 was inhibited, can be rescued till adulthood and is apparently fertile when Mda-5 is deleted. Together, these data are consistent with a model in which ADAR1 edits endogenous dsRNA to mark it as self. Upon virus infection, dsRNA is produced and triggers the antiviral interferon response. After ∼24 h, expression of the interferon-inducible cytoplasmic version of ADAR1 is increased. Cytoplasmic ADAR1 then edits all cytoplasmic RNA to dampen the interferon response to limit self-damage (Samuel, 2011; Mannion et al., 2014). Thus, editing can dynamically control the interferon response.
A-to-I editing levels are affected by external stimuli
The consequences of A-to-I editing for protein-coding transcripts are established for only a handful of targets. One of the first recoding events analyzed is the Q-to-R editing site in the Gria2 (GluR-B) transcript coding for the glutamate receptor subunit B. Editing leads to a change of a CAG codon to CIG (read as CGG) and thus an exchange from genomically encoded glutamine to arginine in the edited transcript (Higuchi et al., 1993; Fig. 3 A). Editing at this so-called Q/R site is of particular physiological interest, as ADAR2-knockout mice die with epileptic seizures caused by increased neuronal Ca2+ influx (Sommer et al., 1991). Most interestingly, lethality can be rescued by homozygous replacement of endogenous Gria2 with a preedited allele (Higuchi et al., 2000). Editing at the Q/R site decreases the Ca2+ permeability of the encoded receptor and affects the assembly of the glutamate receptor (Hume et al., 1991; Burnashev et al., 1992; Greger et al., 2003).
Another prominent editing target is the transcript coding for the serotonin receptor 2c (HTR2C), where editing at different sites increases the diversity of the encoded proteins and results in different protein isoforms (Burns et al., 1997). The serotonin receptor belongs to the family of G protein–coupled receptors. Editing also affects desensitization and trafficking of HTR2C isoforms (Marion et al., 2004). Mice with altered editing of the transcript coding for this serotonin receptor exhibit phenotypes similar to Prader-Willi syndrome, underlining the physiological importance of Htr2c editing in vivo (Kawahara et al., 2008; Mombereau et al., 2010; Morabito et al., 2010).
A growing body of evidence suggests that editing levels of the serotonin receptor 2c, the glutamate receptor, and other targets are dynamically adjusted. Editing levels of the Htr2c transcript, for instance, respond to serotonin levels and thereby influence the interaction between the receptor and the coupled G protein (Gurevich et al., 2002). This may be a mechanism by which the receptor adjusts its sensitivity and responds to very high or low levels of serotonin (Gurevich et al., 2002). Moreover, editing levels of the receptor also change in response to stress (Englander et al., 2005; Bhansali et al., 2007). Another article suggests that the level of editing of the glutamate receptor and the expression of ADAR2 are metabolically regulated, depending on energy and nutrient status (Gan et al., 2006). Moreover, in a subregion of the hippocampus, it was demonstrated that neuronal activity affects editing of the glutamate receptor (Balik et al., 2013). These examples show that editing levels respond to external stimuli but also demonstrate the advantage of having a mechanism such as A-to-I editing rather than hard-wiring a particular codon change in the genome itself. Editing gives the flexibility to quickly respond to specific inputs and adjust gene expression accordingly.
RNA editing is elevated during hypoxia and inflammation
C-to-U editing mediated by APOBEC3A is induced under hypoxic and inflammatory conditions in monocytes or macrophages (Baysal et al., 2013; Sharma et al., 2015). Because monocytes routinely enter a hypoxic environment as soon as they leave the oxygen-rich bloodstream and enter inflamed tissue, it seems likely that this environmental change is used to reprogram the transcriptome for these conditions (Strehl et al., 2014). Similar to C-to-U editing, A-to-I editing can also be induced under hypoxic conditions (Nevo-Caspi et al., 2011). When screening for Drosophila mutants that make flies more susceptible to oxygen deprivation, Ma et al. (2001) identified the Drosophila A-to-I editing deaminase dADAR as a susceptibility factor. Flies lacking the deaminase needed approximately twice as long to recover from anoxic conditions. Moreover, knockout flies were also more susceptible to heat shock. Thus, these data further suggest that A-to-I and C-to-U editing improve responsiveness to environmental stressors.
Concluding remarks
The role of the epitranscriptome in dynamic regulation of gene expression at the transcript level is a fairly novel concept (Saletore et al., 2012). However, the discovery of dynamic changes and readouts in RNA modifications sheds new light on well-known RNA modifications, such as pseudouridylation, that were primarily attributed to rRNAs, tRNAs, or spliceosomal snRNAs. New sequencing techniques tailored to the detection of modified nucleotides have been developed and applied to detect RNA modifications such as m6A or cytosine methylation (Li et al., 2015; Linder et al., 2015; Schaefer, 2015). Most importantly, the inputs and readouts of the dynamic epitranscriptomic marks are increasingly appreciated, and many laboratories aim to decipher the regulators and factors involved. Moreover, previously unknown links, such as the connection between innate immunity and A-to-I editing, are being established. Thus, the field of RNA editing and modification is interlinked with many cell biological, physiological, and organismic processes that lead to exciting and unexpected findings that connect to all areas of biology.
Acknowledgments
We thank Mamta Jain, Prajakta Bajad, and Laura Cimatti for critically reading the manuscript.
This work was supported by grants from the Austrian Science Foundation (F4313 and P26845) to M.F. Jantsch and a fellowship by the Deutsche Forschungsgemeinschaft (LI 2431 2-1) to K. Licht.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ADAR
- adenosine deaminase acting on RNA
- dsRNA
- double-stranded RNA
- PRR
- pattern-recognition receptor
- snRNA
- small nuclear RNA
References
- Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., and Tavazoie S.F.. 2015a HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 162:1299–1308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón C.R., Lee H., Goodarzi H., Halberg N., and Tavazoie S.F.. 2015b N6-methyladenosine marks primary microRNAs for processing. Nature. 519:482–485. 10.1038/nature14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., et al. ICGC PedBrain . 2013. Signatures of mutational processes in human cancer. Nature. 500:415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., and Karikó K.. 2010. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38:5884–5892. 10.1093/nar/gkq347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnez J.G., and Steitz T.A.. 1994. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 33:7560–7567. 10.1021/bi00190a008 [DOI] [PubMed] [Google Scholar]
- Asher G., and Sassone-Corsi P.. 2015. Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 161:84–92. 10.1016/j.cell.2015.03.015 [DOI] [PubMed] [Google Scholar]
- Balik A., Penn A.C., Nemoda Z., and Greger I.H.. 2013. Activity-regulated RNA editing in select neuronal subfields in hippocampus. Nucleic Acids Res. 41:1124–1134. 10.1093/nar/gks1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K., et al. 2014. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 15:707–719. 10.1016/j.stem.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal B.E., De Jong K., Liu B., Wang J., Patnaik S.K., Wallace P.K., and Taggart R.T.. 2013. Hypoxia-inducible C-to-U coding RNA editing downregulates SDHB in monocytes. PeerJ. 1:e152 10.7717/peerj.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.L. 2014. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15:163–175. 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhansali P., Dunning J., Singer S.E., David L., and Schmauss C.. 2007. Early life stress alters adult serotonin 2C receptor pre-mRNA editing and expression of the alpha subunit of the heterotrimeric G-protein G q. J. Neurosci. 27:1467–1473. 10.1523/JNEUROSCI.4632-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V., Park E., Schaefer S., Miller M., Lin Y., Kennedy S., Billing A.M., Ben Hamidane H., Graumann J., Mortazavi A., et al. 2014. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 15:R79 10.1186/gb-2014-15-6-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., and Rottman F.M.. 1997. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Braun K.A., and Young E.T.. 2014. Coupling mRNA synthesis and decay. Mol. Cell. Biol. 34:4078–4087. 10.1128/MCB.00535-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N., Monyer H., Seeburg P.H., and Sakmann B.. 1992. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 8:189–198. 10.1016/0896-6273(92)90120-3 [DOI] [PubMed] [Google Scholar]
- Burns C.M., Chu H., Rueter S.M., Hutchinson L.K., Canton H., Sanders-Bush E., and Emeson R.B.. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 387:303–308. 10.1038/387303a0 [DOI] [PubMed] [Google Scholar]
- Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., and Gilbert W.V.. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 515:143–146. 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.X., Cho D.S., Wang Q., Lai F., Carter K.C., and Nishikura K.. 2000. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 6:755–767. 10.1017/S1355838200000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li Y., Lin C.H., Chan T.H., Chow R.K., Song Y., Liu M., Yuan Y.F., Fu L., Kong K.L., et al. 2013. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 19:209–216. 10.1038/nm.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Hao Y.J., Zhang Y., Li M.M., Wang M., Han W., Wu Y., Lv Y., Hao J., Wang L., et al. 2015. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 16:289–301. 10.1016/j.stem.2015.01.016 [DOI] [PubMed] [Google Scholar]
- Cohn W.E. 1951. Some results of the applications of ion-exchange chromatography to nucleic acid chemistry. J. Cell. Physiol. Suppl. 38(S1, Suppl. 1):21–40. 10.1002/jcp.1030380405 [DOI] [PubMed] [Google Scholar]
- Daniel C., Lagergren J., and Öhman M.. 2015. RNA editing of non-coding RNA and its role in gene regulation. Biochimie. 117:22–27. 10.1016/j.biochi.2015.05.020 [DOI] [PubMed] [Google Scholar]
- Davis D.R., Veltri C.A., and Nielsen L.. 1998. An RNA model system for investigation of pseudouridine stabilization of the codon-anticodon interaction in tRNALys, tRNAHis and tRNATyr. J. Biomol. Struct. Dyn. 15:1121–1132. 10.1080/07391102.1998.10509006 [DOI] [PubMed] [Google Scholar]
- Desrosiers R., Friderici K., and Rottman F.. 1974. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA. 71:3971–3975. 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 485:201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Englander M.T., Dulawa S.C., Bhansali P., and Schmauss C.. 2005. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J. Neurosci. 25:648–651. 10.1523/JNEUROSCI.3895-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergus C., Barnes D., Alqasem M.A., and Kelly V.P.. 2015. The queuine micronutrient: Charting a course from microbe to man. Nutrients. 7:2897–2929. 10.3390/nu7042897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández I.S., Ng C.L., Kelley A.C., Wu G., Yu Y.T., and Ramakrishnan V.. 2013. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 500:107–110. 10.1038/nature12302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Jia G., Pang X., Wang R.N., Wang X., Li C.J., Smemo S., Dai Q., Bailey K.A., Nobrega M.A., et al. 2013. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 4:1798 10.1038/ncomms2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli D., Gacquer D., Rothé F., Lefort A., Libert F., Brown D., Kheddoumi N., Shlien A., Konopka T., Salgado R., et al. 2015. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Reports. 13:277–289. 10.1016/j.celrep.2015.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin J.M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., Isagawa T., Morioka M.S., Kakeya H., Manabe I., and Okamura H.. 2013. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 155:793–806. 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- Gan Z., Zhao L., Yang L., Huang P., Zhao F., Li W., and Liu Y.. 2006. RNA editing by ADAR2 is metabolically regulated in pancreatic islets and beta-cells. J. Biol. Chem. 281:33386–33394. 10.1074/jbc.M604484200 [DOI] [PubMed] [Google Scholar]
- Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S., et al. 2015. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 347:1002–1006. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Greger I.H., Khatri L., Kong X., and Ziff E.B.. 2003. AMPA receptor tetramerization is mediated by Q/R editing. Neuron. 40:763–774. 10.1016/S0896-6273(03)00668-8 [DOI] [PubMed] [Google Scholar]
- Gurevich I., Englander M.T., Adlersberg M., Siegal N.B., and Schmauss C.. 2002. Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. J. Neurosci. 22:10529–10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall K.B., and McLaughlin L.W.. 1991. Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry. 30:1795–1801. 10.1021/bi00221a010 [DOI] [PubMed] [Google Scholar]
- Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M., Eterovic A.K., Yuan Y., et al. 2015. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 28:515–528. 10.1016/j.ccell.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner J.C., Schmittwolf C., Kispert A., Müller A.M., Higuchi M., and Seeburg P.H.. 2004. Liver disintegration in the mouse embryo caused by deficiency in the RNA editing enzyme ADAR1. J. Biol. Chem. 279:4894–4902. 10.1074/jbc.M311347200 [DOI] [PubMed] [Google Scholar]
- Hartner J.C., Walkley C.R., Lu J., and Orkin S.H.. 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 10:109–115. 10.1038/ni.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., Single F.N., Köhler M., Sommer B., Sprengel R., and Seeburg P.H.. 1993. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron-exon structure determines position and efficiency. Cell. 75:1361–1370. 10.1016/0092-8674(93)90622-W [DOI] [PubMed] [Google Scholar]
- Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., and Seeburg P.H.. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA editing enzyme ADAR2. Nature. 406:78–81. 10.1038/35017558 [DOI] [PubMed] [Google Scholar]
- Hume R.I., Dingledine R., and Heinemann S.F.. 1991. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 253:1028–1031. 10.1126/science.1653450 [DOI] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., and He C.. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7:885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J., and Yu Y.T.. 2011. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 474:395–398. 10.1038/nature10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J., Yi C., and Yu Y.T.. 2015. Transcriptome-wide dynamics of RNA pseudouridylation. Nat. Rev. Mol. Cell Biol. 16:581–585. 10.1038/nrm4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y., Grimberg A., Teegarden S., Mombereau C., Liu S., Bale T.L., Blendy J.A., and Nishikura K.. 2008. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J. Neurosci. 28:12834–12844. 10.1523/JNEUROSCI.3896-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D. 2007. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 8:533–543. 10.1038/nrg2111 [DOI] [PubMed] [Google Scholar]
- Kurowski M.A., Bhagwat A.S., Papaj G., and Bujnicki J.M.. 2003. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 4:48 10.1186/1471-2164-4-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.B., Levanon E.Y., Yoon J.K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., and Church G.M.. 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 324:1210–1213. 10.1126/science.1170995 [DOI] [PubMed] [Google Scholar]
- Li X., Zhu P., Ma S., Song J., Bai J., Sun F., and Yi C.. 2015. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11:592–597. 10.1038/nchembio.1836 [DOI] [PubMed] [Google Scholar]
- Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., Li J.B., Seeburg P.H., and Walkley C.R.. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 349:1115–1120. 10.1126/science.aac7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., and Jaffrey S.R.. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 12:767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., et al. 2014. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10:93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Dai Q., Zheng G., He C., Parisien M., and Pan T.. 2015. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 518:560–564. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E., Gu X.Q., Wu X., Xu T., and Haddad G.G.. 2001. Mutation in pre-mRNA adenosine deaminase markedly attenuates neuronal tolerance to O2 deprivation in Drosophila melanogaster. J. Clin. Invest. 107:685–693. 10.1172/JCI11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., and Reed R.. 2002. An extensive network of coupling among gene expression machines. Nature. 416:499–506. 10.1038/416499a [DOI] [PubMed] [Google Scholar]
- Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D., Nellåker C., Vesely C., Ponting C.P., McLaughlin P.J., et al. 2014. The RNA editing enzyme ADAR1 controls innate immune responses to RNA. Cell Reports. 9:1482–1494. 10.1016/j.celrep.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion S., Weiner D.M., and Caron M.G.. 2004. RNA editing induces variation in desensitization and trafficking of 5-hydroxytryptamine 2c receptor isoforms. J. Biol. Chem. 279:2945–2954. 10.1074/jbc.M308742200 [DOI] [PubMed] [Google Scholar]
- Meier F., Suter B., Grosjean H., Keith G., and Kubli E.. 1985. Queuosine modification of the wobble base in tRNAHis influences ‘in vivo’ decoding properties. EMBO J. 4:823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., and Jaffrey S.R.. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 149:1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., and Jaffrey S.R.. 2015. 5′ UTR m(6)A promotes cap-independent translation. Cell. 163:999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombereau C., Kawahara Y., Gundersen B.B., Nishikura K., and Blendy J.A.. 2010. Functional relevance of serotonin 2C receptor mRNA editing in antidepressant- and anxiety-like behaviors. Neuropharmacology. 59:468–473. 10.1016/j.neuropharm.2010.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morabito M.V., Abbas A.I., Hood J.L., Kesterson R.A., Jacobs M.M., Kump D.S., Hachey D.L., Roth B.L., and Emeson R.B.. 2010. Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader-Willi syndrome. Neurobiol. Dis. 39:169–180. 10.1016/j.nbd.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Hartmann M., Schuster I., Bender S., Thüring K.L., Helm M., Katze J.R., Nellen W., Lyko F., and Ehrenhofer-Murray A.E.. 2015. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 43:10952–10962. 10.1093/nar/gkv980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo-Caspi Y., Amariglio N., Rechavi G., and Paret G.. 2011. A-to-I RNA editing is induced upon hypoxia. Shock. 35:585–589. 10.1097/SHK.0b013e31820fe4b7 [DOI] [PubMed] [Google Scholar]
- Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79:321–349. 10.1146/annurev-biochem-060208-105251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Yaacov N., Bazak L., Buchumenski I., Porath H.T., Danan-Gotthold M., Knisbacher B.A., Eisenberg E., and Levanon E.Y.. 2015. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Reports. 13:267–276. 10.1016/j.celrep.2015.08.080 [DOI] [PubMed] [Google Scholar]
- Peng Z., Cheng Y., Tan B.C., Kang L., Tian Z., Zhu Y., Zhang W., Liang Y., Hu X., Tan X., et al. 2012. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 30:253–260. 10.1038/nbt.2122 [DOI] [PubMed] [Google Scholar]
- Perry R.P., and Kelley D.E.. 1974. Existence of methylated messenger-RNA in mouse L cells. Cell. 1:37–42. 10.1016/0092-8674(74)90153-6 [DOI] [Google Scholar]
- Perry R.P., Kelley D.E., Friderici K., and Rottman F.. 1975. The methylated constituents of L cell messenger RNA: Evidence for an unusual cluster at the 5′ terminus. Cell. 4:387–394. 10.1016/0092-8674(75)90159-2 [DOI] [PubMed] [Google Scholar]
- Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S., et al. 2014. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24:177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohaska K.M., Bennett R.P., Salter J.D., and Smith H.C.. 2014. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdiscip. Rev. RNA. 5:493–508. 10.1002/wrna.1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G., Zhang R., Piskol R., Keegan L.P., Deng P., O’Connell M.A., and Li J.B.. 2013. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods. 10:128–132. 10.1038/nmeth.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Kasher P.R., Forte G.M., Mannion N.M., Greenwood S.M., Szynkiewicz M., Dickerson J.E., Bhaskar S.S., Zampini M., Briggs T.A., et al. 2012. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 44:1243–1248. 10.1038/ng.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G., et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45:970–976. 10.1038/ng.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg B.R., Hamilton C.E., Mwangi M.M., Dewell S., and Papavasiliou F.N.. 2011. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 18:230–236. 10.1038/nsmb.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletore Y., Meyer K., Korlach J., Vilfan I.D., Jaffrey S., and Mason C.E.. 2012. The birth of the epitranscriptome: Deciphering the function of RNA modifications. Genome Biol. 13:175 10.1186/gb-2012-13-10-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E. 2011. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 411:180–193. 10.1016/j.virol.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M. 2015. RNA 5-methylcytosine analysis by bisulfite sequencing. Methods Enzymol. 560:297–329. 10.1016/bs.mie.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Schneider M.F., Wettengel J., Hoffmann P.C., and Stafforst T.. 2014. Optimal guideRNAs for re-directing deaminase activity of hADAR1 and hADAR2 in trans. Nucleic Acids Res. 42:e87 10.1093/nar/gku272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S., et al. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 159:148–162. 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Patnaik S.K., Taggart R.T., Kannisto E.D., Enriquez S.M., Gollnick P., and Baysal B.E.. 2015. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 6:6881 10.1038/ncomms7881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.C., Bennett R.P., Kizilyer A., McDougall W.M., and Prohaska K.M.. 2012. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 23:258–268. 10.1016/j.semcdb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B., Köhler M., Sprengel R., and Seeburg P.H.. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 67:11–19. 10.1016/0092-8674(91)90568-J [DOI] [PubMed] [Google Scholar]
- Strehl C., Fangradt M., Fearon U., Gaber T., Buttgereit F., and Veale D.J.. 2014. Hypoxia: How does the monocyte-macrophage system respond to changes in oxygen availability? J. Leukoc. Biol. 95:233–241. 10.1189/jlb.1212627 [DOI] [PubMed] [Google Scholar]
- Stulić M., and Jantsch M.F.. 2013. Spatio-temporal profiling of Filamin A RNA editing reveals ADAR preferences and high editing levels outside neuronal tissues. RNA Biol. 10:1611–1617. 10.4161/rna.26216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajaddod M., Jantsch M.F., and Licht K.. 2016. The dynamic epitranscriptome: A to I editing modulates genetic information. Chromosoma. 125:51–63. 10.1007/s00412-015-0526-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P., and Scadden A.D.. 2010. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat. Struct. Mol. Biol. 17:1043–1050. 10.1038/nsmb.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstedt H., Daniel C., Ensterö M., and Ohman M.. 2009. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 19:978–986. 10.1101/gr.089409.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., et al. 2014a N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 505:117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., and He C.. 2015. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 161:1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., and Zhao J.C.. 2014b N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16:191–198. 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., and Chen Z.J.. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32:461–488. 10.1146/annurev-immunol-032713-120156 [DOI] [PubMed] [Google Scholar]
- Wu G., Xiao M., Yang C., and Yu Y.T.. 2011. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 30:79–89. 10.1038/emboj.2010.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborske J.M., DuMont V.L., Wallace E.W., Pan T., Aquadro C.F., and Drummond D.A.. 2014. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol. 12:e1002015 10.1371/journal.pbio.1002015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vågbø C.B., Shi Y., Wang W.L., Song S.H., et al. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 49:18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., and Qian S.B.. 2015. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 526:591–594. 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]