Abstract
Seed number per pod (SNPP) is one of the major yield components and breeding targets in rapeseed that shows great variation and is invaluable for genetic improvement. To elucidate the genetic architecture and uncover the mechanism of SNPP, we identified five quantitative trait loci (QTLs) using the BnaZNRIL population, which were integrated with those of previous studies by physical map to demonstrate a complex and relatively complete genetic architecture of SNPP. A major QTL, qSN.A6, was successfully fine-mapped from 1910 to 267 kb using near-isogenic line (NIL). In addition, qSN.A6 exhibited an antagonistic pleiotropy on seed weight (SW), which is caused by a physiological interaction in which SNPP acts “upstream” of SW. Because the negative effect of qSN.A6 on SW cannot fully counteract its positive effect on SNPP, it also enhanced the final yield (17.4%), indicating its great potential for utilization in breeding. The following genetic and cytological experiments further confirmed that the different rate of ovule abortion was responsible for the ~5 seed difference between Zhongshuang11 and NIL-qSN.A6. This systematic approach to dissecting the comprehensive genetic architecture of SNPP and characterizing the underlying mechanism has advanced the understanding of SNPP and will facilitate the development of high-yield cultivars.
Seed number per pod (SNPP) is one of three components (including pod number and seed weight) of yield and an important target trait of breeding in rapeseed (Brassica napus L.). The SNPP of rapeseed shows great variation in its germplasm resources (from 5 to 35 seeds per pod)1, which is invaluable for genetic improvement. As with other crops, SNPP is usually negatively correlated with the other two yield components2. These trade-offs are commonly explained as competition among sinks3,4. Genetically, the correlation between traits is generally considered to be either genetic linkage or pleiotropy5. Genetic linkage means the loci for different traits are physically near one another. Pleiotropy refers to the effect of a locus on two or more traits (Fig. 1). In addition, pleiotropy may be due to physiological interactions among traits in which one trait acts “upstream” of another6.
Figure 1. Diagram of the genetic causation of correlation among traits.
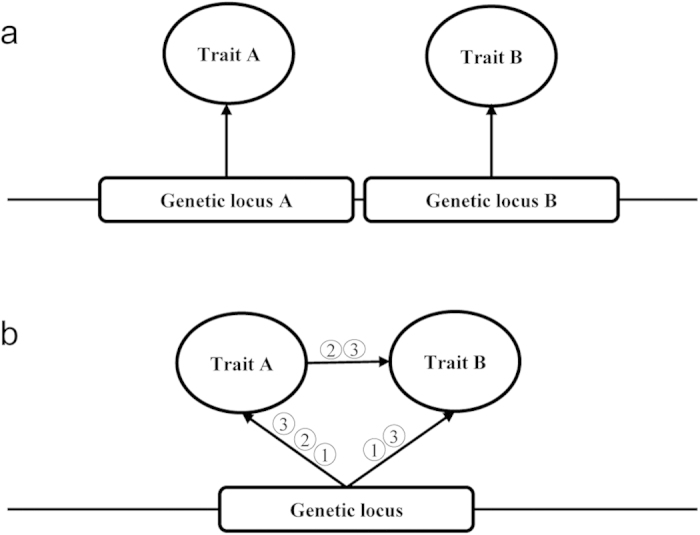
(a) Genetic linkage; (b) pleiotropy ① including physiological interaction ② and the combination of pleiotropy and physiological interaction ③.
SNPP in rapeseed is a highly complex trait determined by the number of ovules per ovary, the proportion of fertile ovules (ovule fertility), the proportion of ovules fertilized, and the proportion of fertilized ovules that develop into seeds. In rapeseed, the number of ovules per ovary and ovule fertility are determined by the process of ovule differentiation and development7,8. The proportion of ovules fertilized is determined by the fertilization process, which is dependent on many factors such as pollen sterility, the amounts of pollen deposited on the stigma, pollen grain germination9, pollination conditions10, and pollen tube growth11. The proportion of fertilized ovules that develop into seeds is determined by the process of zygote/seed development. Although the main developmental and biological processes that determine SNPP are largely clear, which mechanism(s) might be responsible for the natural variation of SNPP among the different germplasms of rapeseed remains unknown. The limited knowledge of the mechanism of SNPP variation in rapeseed has hindered its improvement.
Understanding the genetic architecture of SNPP is the first step towards its improvement. During the past decade, nearly ten linkage mapping studies2,12,13,14,15,16,17,18,19,20 and one association analysis study21 involving the SNPP of rapeseed have been reported. However, the genetic architecture of SNPP has remained fragmented and the underlying candidate gene(s) have remained unknown because these detected QTLs had not been integrated and the physical map was unavailable. In addition, only one major QTL of SNPP has been fine-mapped with a relatively large physical region of more than 1 Mb18. More importantly, the mechanisms of these QTLs have not been investigated and thus remain unclear. Therefore, it is difficult to narrow down and identify the candidate gene(s) for these SNPP QTLs. NIL is a basic strategy commonly used for fine-mapping. The traditional NIL strategy can fine-map any mendelizing variation in theory.
The main objectives of the current study were as follows: (1) to dissect the genetic architecture of SNPP variation in rapeseed by linkage mapping as well as physical map-based integration with previous studies; (2) fine-map the major QTL using NILs; (3) to dissect the mechanism of SNPP variation at a major QTL through an exclusion strategy using a series of ingenious genetic and cytological experiments based on the NILs; (4) to dissect the genetic causation of the opposite effect of the major QTLs on SNPP and SW; and (5) to evaluate the application potential of the major QTL by field testing of its effect on yield as well as on yield components and related traits.
Results
Mapping of QTLs for SNPP using the BnaZNRIL population
The two parents, Zhongshuang11 and No. 73290, showed extremely significant differences in SNPP in all four investigated environments. The SNPP of Zhongshuang11 was 21.7 ± 1.6, which was approximately twice the SNPP of No. 73290 (11.7 ± 1.4) (Table 1). The SNPP of the RIL population showed more or less normal distributions in all four investigated environments (Figure S1), indicating a quantitative inheritance suitable for QTL identification. In addition, the SNPP of the RIL population exhibited transgressive segregation but to a small degree, indicating that the favourable alleles were mainly distributed in one of the two parents. The broad-sense heritability (h2 = 81%) of SNPP in the RIL population was relatively high.
Table 1. Phenotypic variation of SNPP for Zhongshuang11 and No. 73290 as well as the derived RIL population in four investigated environments.
| Experiment code | Parents |
RIL population |
||||
|---|---|---|---|---|---|---|
| Zhongshuang11 | No. 73290 | pt-test | Min | Max | Mean | |
| W11 | 22.2 ± 1.6 | 12.7 ± 2.2 | 3.7E–10 | 6.3 | 25.1 | 15.6 ± 3.0 |
| Z11 | 21.4 ± 1.6 | 11.1 ± 1.9 | 1.2E–17 | 7.3 | 27.3 | 17.7 ± 3.7 |
| W12 | 21.7 ± 1.4 | 11.6 ± 1.1 | 5.6E–21 | 7.3 | 19.2 | 11.1 ± 3.8 |
| Z12 | 21.3 ± 1.7 | 12.2 ± 1.4 | 2.1E–08 | 7.3 | 22.9 | 14.7 ± 3.6 |
| Mean | 21.7 ± 1.6 | 11.7 ± 1.4 | 2.0E–55 | 7.0 | 23.6 | 14.8 ± 2.8 |
The RIL genetic map consisted of 19 linkage groups and 2264 unique loci/bins (Table S1), which covered a total length of 2107 cM, with an average distance of 0.93 cM. Eight significant QTLs and three suggestive QTLs were detected for the SNPP. After deleting two non-reproducible suggestive QTLs, nine QTLs were identified (Fig. 2), which were distributed on the A02, A06, C01, C04, and C06 linkage groups (Table S2A). After the interaction of overlapping identified QTLs in different environments (Table S2B), five consensus QTLs were obtained, which explained 5.3–24.8% of phenotypic variance (Table 2). Of the two repeatable consensus QTLs, qSN.A6 (defined as an 18.9 cM region between the markers Bn-A06-p22608192 and Bn-A06-p24274213) was consistently detected in all four environments and showed a relatively large effect (mean R2 = 20.1%; mean additive effect = 1.58), and thus it could be treated as a major QTL for further dissection.
Figure 2. SNPP QTL scanning curves for the 19 linkage groups in four environments.
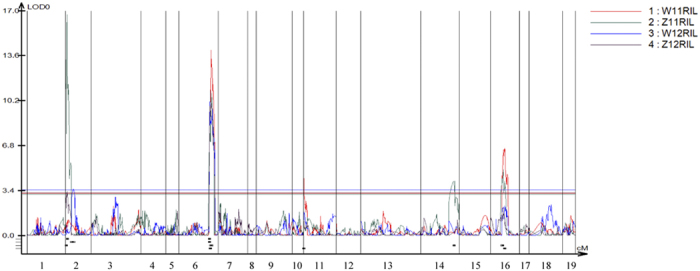
The horizontal and vertical axes represent the genetic distance and LOD value, respectively. The lines and curves indicate the threshold and true LOD values, respectively. The different environments are represented using different colours, as indicated in the legend. W11, W12, Z11, and Z12 are the codes for the environments detailed in Materials and Methods.
Table 2. Consensus QTLs for SNPP.
| Consensus QTL | Linkage group | Peak position | Confidence Interval | Genomic regions (Mb) | Additive effect | LOD value | R2(%) | Experiment code |
|---|---|---|---|---|---|---|---|---|
| qSN.A02 | A02 | 4.2 | 3.0–5.0 | 32.8–39.4 | 2.16 | 16.7 | 24.8 | Z11RIL |
| qSN.A06 | A06 | 122.6 | 113.5–133.7 | 21.6–23.3 | 1.58 | 11.6 | 20.1 | W11RIL/Z11RIL/W12RIL/Z12RIL |
| qSN.C01 | C01 | 0.0 | 0.0–3.4 | 1.2–1.6 | −0.77 | 4.3 | 6.2 | W11RIL |
| qSN.C04 | C04 | 129.6 | 129.2–129.6 | 45.5–45.7 | −0.87 | 4.1 | 5.3 | Z11RIL |
| qSN.C06 | C06 | 54.5 | 50.7–58.2 | 18.9–20.4 | 0.96 | 5.74 | 8.18 | W11RIL/Z11RIL |
In silico integration of QTLs for SNPP in Brassica napus
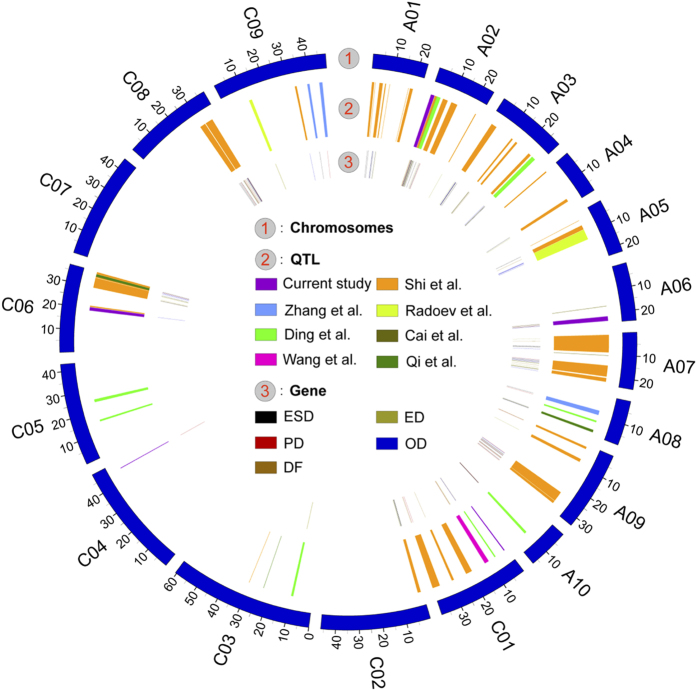
Of the total of 100 QTLs and seven association signals identified in the current and reported QTL mapping and association mapping studies (Table S3), 81 QTLs and four association signals were successfully anchored to the physical map, which were then subjected to meta-analysis, resulting in 70 consensus QTLs (57 non-overlapping QTLs and 13 overlapping QTLs) (Fig. 3). These consensus QTLs were distributed on almost all of the 19 chromosomes (except for C07). The R2 of these consensus QTLs ranged from 2.3 to 22.5. Most (70%) showed only small effects (R2 < 10%), and only five should be considered as major QTLs.
Figure 3. Integration of SNPP QTLs and candidate genes for on the reference genome of Brassica napus.
Concentric circles show the different features that were drawn using the Circos program. 1, 19 chromosomes of rapeseed. 2, Consensus QTLs of SNPP, different colours represent different studies. 3, Genes referred to QTL mapping results for SNPP, ESD (embryo sac development), ED (embryo development), PD (pollen development), OD (ovule development), and DF (double fertilization), which are represented by different colours. QTL mapping range and genes are represented by squares and lines, respectively.
A total of 306 genes (Table S4) (for the main biological processes affecting SNPP) collected from Arabidopsis were aligned to 855 loci on the genome of Darmor by BLAST. Of these, 117 genes were found to underlie the corresponding 155 loci genomic regions of 41 consensus QTLs, which therefore should be considered as candidates. These candidates involve embryo sac development (ESD), ovule development (OD), pollen development (PD), double fertilization process (DF) and embryo development (ED).
Fine-mapping of qSN.A6 using BC4F2 population and BC4F3 progeny
The QTL-NILs of BC4F1 were obtained by the successive backcrossing of F1 with Zhongshuang11. During the backcrossing process, the progenies of each backcross were selected using two flanking SSR/InDel markers (BrSF47–389 and ni108) of the target region, which were very close to Bn-A06-p22608192 and Bn-A06-p24274213, respectively. For the BC4F1 lines, 111 plants were surveyed using not only two flanking markers for the selected foreground but also a total of 80 SSR/SNP markers that were evenly distributed on the 19 linkage groups to screen the genetic background (Table S5). The background proportions of these plants ranged from 87.3% to 97.6%, and several individuals with > 95% were self-crossed to produce BC4F2.
As expected, the frequency distribution of SNPP of the BC4F2 population deviated from the normal distribution (D = 0.16, p < 0.01) and appeared to be a bimodal distribution (Fig. 4). The ratio of the two phenotypic types was a good fit to the expected ratio of 1:3 (χ2 = 3.51, p = 5.6E–1), which indicated a single-locus Mendelian segregation. In the BC4F2 population, the numbers of three types of QTL genotypes (494:886:457), i.e., homozygous for the Zhongshuang11 allele (ZZ), heterozygous (ZN) and homozygous for the No. 73290 allele (NN), showed an expected ratio of 1:2:1 (χ2 = 3.79, p = 1.5E–1), indicating an absence of distorted segregation. Clearly, the SNPP of the BC4F2 plants with the ZZ genotype (21.6 ± 1.4) was slightly higher (p = 3.2E–2) than the SNPP of the heterozygotes (20.6 ± 1.6), and both were much higher (p = 1.1E–18 and 1.0E–15) than for the NN genotype (16.0 ± 0.4). These results suggested that the favourable allele of qSN.A6 was from Zhongshuang11 and the mode-of-inheritance of qSN.A6 was partially dominant. As expected, in the BC4F2 population, SNPP was significantly positively correlated with seed yield per plant (SY) (r = 0.51; p < 0.0001). Interestingly, SNPP was also significantly negatively correlated with seed weight (SW) (r = −0.30; p < 0.0001) but not significantly correlated with pod number (PN) (r = 0.10; p = 0.1587).
Figure 4. Frequency distribution of SNPP in the BC4F2 population.
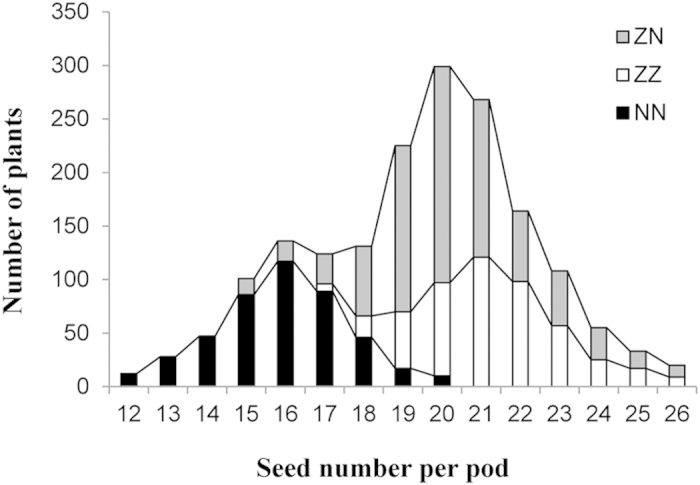
The horizontal axis represents the trait value of SNPP. The vertical axis represents the number of individuals. The three types of genotype are represented by the three column colours, as shown in the legend.
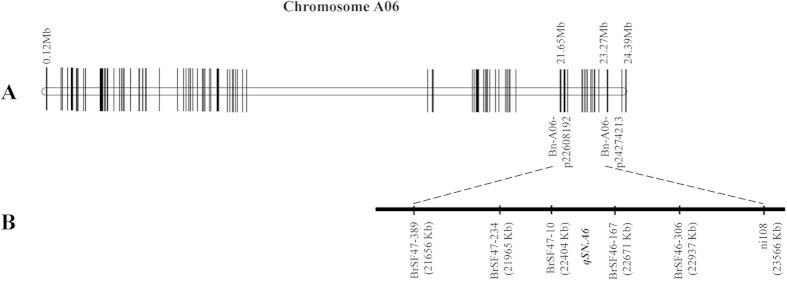
Six evenly distributed co-dominant SSR/InDel markers within the 18.9 cM interval of qSN.A6 (between SNP markers Bn-A06-p22608192 and Bn-A06-p24274213) were used to genotype the 2586 plants of the BC4F2 population (Fig. 5). A total of 20 individuals belonging to ten possible types of recombination were identified. Based on progeny testing of these lines, the ten types of recombination were classified into two groups based on the comparison of their SNPP with the recurrent parent Zhongshuang11 (21.2 ± 1.3) (Table 3). The first group included four types (type 1, 2, 9, and 10) whose SNPP (21.2 ± 1.7 to 21.8 ± 1.0) were not significantly different from Zhongshuang11. The SNPP (15.9 ± 0.8 to 16.7 ± 2.2) of the other group (including type 3, 4, 5, 6, 7, and 8) was significantly lower than for Zhongshuang11. All six types of recombination have a common introgression fragment between markers BrSF47–10 and BrSF46–167, which delimited qSN.A6 to this interval.
Figure 5. Fine-mapping of qSN.A6.
(A) Position of qSN.A6 on the A6 linkage group. The hollow vertical column shows the linkage group in which the position of each SNP marker is indicated by horizontal short lines. (B) The location of fine-mapped qSN.A6. The vertical solid columns show the physical map of qSN.A6, on which the physical position of each SSR/InDel marker is indicated with short lines.
Table 3. QTL Genotypes and SNPP of ten types of recombinant NILs and the two parents.
| Lines | Number in BC4F3 | Genotype of the six markers within QTL interval |
SNPP in the BC4F3 lines |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BrSF47-389 | BrSF47-234 | BrSF47-10 | BrSF46-167 | BrSF46-306 | ni108 | Mean ± SD | P | |||
| Parents | Zhongshuang11 | 30 | A | A | A | A | A | A | 21.2 ± 1.3 | – |
| No. 73290 | 30 | B | B | B | B | B | B | 12.2 ± 1.5 | < 0.0001 | |
| NILs | Type−1 | 35 | B | A | A | A | A | A | 21.2 ± 1.7 | 0.3620 |
| Type−2 | 34 | B | B | A | A | A | A | 21.4 ± 2.2 | 0.7411 | |
| Type−3 | 40 | B | B | B | A | A | A | 16.4 ± 1.9 | < 0.0001 | |
| Type−4 | 32 | B | B | B | B | A | A | 16.4 ± 1.7 | < 0.0001 | |
| Type−5 | 33 | B | B | B | B | B | A | 16.7 ± 2.2 | < 0.0001 | |
| Type−6 | 36 | A | B | B | B | B | B | 15.9 ± 0.8 | < 0.0001 | |
| Type−7 | 58 | A | A | B | B | B | B | 16.1 ± 2.2 | < 0.0001 | |
| Type−8 | 30 | A | A | A | B | B | B | 16.5 ± 0.3 | < 0.0001 | |
| Type−9 | 37 | A | A | A | A | B | B | 21.5 ± 1.6 | 0.8382 | |
| Type−10 | 33 | A | A | A | A | A | B | 21.8 ± 1.0 | 0.6545 | |
Genotype A and B represented homologous alleles from Zhongshuang11 and No. 73290, respectively.
Dissection of the cytological mechanism of qSN.A6 by the exclusion method
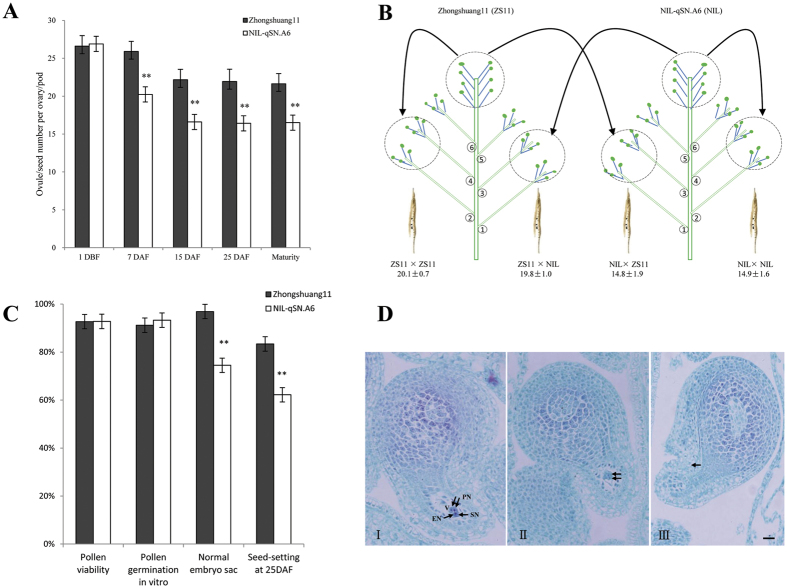
To determine the stage in which the SNPP difference formed between Zhongshuang11 and NIL-qSN.A6, continuous observations of ovule/seed number per ovary/pod from bud to maturity were conducted. At 1 DBF (day before flowering), the ovule number per ovary of Zhongshuang11 (26.6 ± 1.4) and NIL-qSN.A6 (26.9 ± 1.4) did not differ significantly (p = 4.8E–1) (Fig. 6A). From 1 DBF to 7 DAF (day after flowering), the SNPP of both Zhongshang11 and NIL-qSN.A6 decreased quickly, but the decrease speed of NIL-qSN.A6 was faster than for Zhongshang11, and thus the SNPP of NIL-qSN.A6 (20.2 ± 1.3) was significantly (p = 2.1E–13) lower than for Zhongshuang11 (25.9 ± 1.3). From 7 to 15 DAF, the SNPP of both Zhongshuang11 and NIL-qSN.A6 decreased faster, with almost the same speed (Zhongshuang11: 22.2 ± 1.3; NIL-qSN.A6: 16.6 ± 2.3, p = 8.7E–15), and thus the difference between the two lines did not change from the previous stage (p = 8.8E–1). From 15 DAF to 25 DAF and to the maturity stage, the SNPP of both Zhongshuang11 and NIL-qSN.A6 reached a stable period. These results implied that the SNPP difference between Zhongshang11 and NIL-qSN.A6 forms before 7 DAF. From 1 DBF to 7 DAF, many factors affect the seed setting rate, such as pollen fertility, ovule fertility, and the fertilization process (pollen tube elongation). To identify the key factor responsible for the seed setting rate difference between Zhongshang11 and NIL-qSN.A6, the following genetic and cytological experiments were performed.
Figure 6. The cytological mechanism of qSN.A6.
(A) Continuous observations of ovule/seed number per ovary/pod in Zhongshuang11 and NIL-qSN.A6 from budding to maturity. The horizontal and vertical axes show the stages and number of ovules/seeds, respectively. The two lines, Zhongshuang11 and NIL-qSN.A6, are represented by black and white, respectively. The bars show the standard deviation of each value. DBF and DAF stand for days before flowering and days after flowering, respectively. **represents a significance level of P < 0.01. (B) The SNPP of self- and cross-pollinations between Zhongshuang11 and NIL-qSN.A6. (C) The fertility-related traits of Zhongshuang11 and NIL-qSN.A6. (D) Normal embryo sac (I) and two types of abnormal embryo sac (II and III). A normal embryo sac consists of seven cells including eight nuclei: a single egg nucleus (EN), two polar nuclei (PN), two synergid nuclei (SN), and a central vacuole (V). The antipodal cells are degenerated or not clearly observed. The abnormal embryo sac exhibited two types of incomplete cellularization: (II) only two large cells were present in the embryo sac; (III) the embryo sac had not differentiated and lacked any visible cells. Bar equals 10 μm.
First, to identify the male or female origin of the SNPP difference between Zhongshang11 and NIL-qSN.A6, self- and cross-pollinations were performed by hand using the two lines (Fig. 6B). The results showed that, whether Zhongshang11 or NIL-qSN.A6 was used as the mother plant, the SNPP of the self and cross-pollinated pods was not significantly different. These results strongly suggested that the seed setting rate difference of Zhongshang11 and NIL-qSN.A6 was not related to pollen. To confirm this result, the main characteristics (including pollen viability, germination efficiency, and adherence to stigma) (Fig. 6C) reflecting the quality and quantity of pollen for both lines were checked, and no significant differences were found (Figure S2A–F). Second, to determine whether the seed setting rate difference between Zhongshuang11 and NIL-qSN.A6 was related to the fertilization, the process of pollen tube elongation (Figure S2G,H) was observed, and no significant difference was found.
The results of the above experiments strongly indicated that the SNPP difference between Zhongshuang11 and NIL-qSN.A6 should be related to the quality of ovules. Therefore, we conducted serial sectioning to investigate embryo sac fertility. As expected, the embryo sac fertility of NIL-qSN.A6 was only 74.5 ± 4.5%, which was significantly (p = 9.0E–15) lower than for Zhongshang11 (96.9 ± 3.1%), which accurately explained the ~5 seeds per pod difference between Zhongshuang11 and NIL-qSN.A6 (Fig. 6C). Generally, the fertile embryo sac contains seven cells including eight nuclei (Fig. 6D(I)). However, the cellularization of the embryo sac was incomplete, with one of the following two scenarios: (Fig. 6D(II)) only two large cells were present in the embryo sac or (Fig. 6D(III)) the embryo sac had not differentiated and lacked any visible cells. These results showed that the cytological mechanism of qSN.A6 was due to ovule abortion caused by incomplete cellularization of the embryo sac.
Effect of qSN.A6 on yield and other important traits
In the large-scale field test at Wuhan in 2014–2015, the SNPP of Zhongshuang11 (21.2 ± 0.5) was significantly (p = 6.3E–10) higher (percentage = 25.1%) than in NIL-qSN.A6 (15.9 ± 1.2) (Table 4). Regarding the other two yield component traits, the SW of Zhongshuang11 (4.29 ± 0.06) was significantly (p = 8.4E–07) lower (percentage = 9.1%) than in NIL-qSN.A6 (4.68 ± 0.24), but the PN of the two lines did not differ. Strikingly, the SY of Zhongshuang11 was significantly (p = 4.7E–02) higher (percentage = 17.4%) than in NIL-qSN.A6. The other traits related to yield did not significantly differ between the two lines. In conclusion, qSN.A6 exerts significant opposite effects on two of the three yield components but not on PN; however, the decrease in SW cannot counteract the increase in SNPP (which is also reflected by the moderately negative correlation between the two traits in the BC4F2 population), and the final seed yield is increased.
Table 4. Yield traits at harvest for Zhongshuang11 and NIL-qSN.A6.
| Trait | Zhongshuang11 | NIL-qSN.A6 | P-Value | Rate | |
|---|---|---|---|---|---|
| Yield | SY (g) | 19.4 ± 0.9 | 16.0 ± 1.0 | 4.7E–02 | 17.4% |
| Yield component | SNPP | 21.2 ± 0.5 | 15.9 ± 1.2 | 6.3E–10 | 25.1% |
| SW (g) | 4.29 ± 0.06 | 4.68 ± 0.24 | 8.4E–07 | −9.09% | |
| PN | 261 ± 5 | 273 ± 8 | 8.8E–01 | −4.6% | |
| Yield related | PL (mm) | 84.9 ± 2.1 | 81.9 ± 3.8 | 9.3E–02 | 3.5% |
| PH (cm) | 185 ± 3 | 182 ± 3 | 6.2E–02 | 1.6% | |
| BN | 6.20 ± 0.81 | 6.63 ± 1.10 | 3.9E–01 | −6.5% | |
| MIL (cm) | 46.2 ± 4.4 | 47.4 ± 3.1 | 7.7E–01 | −2.2% | |
| Flowering time (days) | 191 ± 3 | 191 ± 5 | 7.3E–01 | 0.0% | |
| Maturity time (days) | 214 ± 6 | 216 ± 4 | 4.5E–01 | −0.9% |
Trait data are expressed as the means ± SD measured from 30 different plants. SY, SW, PL, PN, PH, BH and MIL represented seed yield per plant, seed weight, pod length, pod number, plant height, branch number and main inflorescence, respectively.
To further investigate the genetic causation of the effects of qSN.A6 on both SNPP and SW, conditional QTL analysis was performed on the RIL population. Interestingly, when SNPP was conditioned by SW, the LOD value and R2 of qSN.A6 showed small variation and remained significant (Table 5). However, the effect of qSN.A6 became non-significant when SW was conditioned by SNPP (SW|SNPP). These results supported that the causation of the genetic effect of qSN.A6 on both SNPP and SW was pleiotropy (rather than tight linkage) through a physiological interaction in which SNPP acts “upstream” of SW.
Table 5. Conditional analysis of qSN.A6 on SNPP and SW in the RIL population.
| Experiment code | LOD/R2(%) |
|||
|---|---|---|---|---|
| SNPP | SNPP|SW | SW | SW|SNPP | |
| W11 | 14.5/25.7 | 14.0/24.1 | 5.4/5.4 | NA |
| Z11 | 10.3/24.5 | 7.8/16.9 | 3.5/9.0 | NA |
| W12 | 3.1/4.6 | 8.4/12.9 | 2.9/4.5 | NA |
| Z12 | 5.2/12.0 | 4.6/5.3 | NA | NA |
NA represented non-significant.
Discussion
The complexity of the genetic basis of the SNPP natural variation in rapeseed
By integrating our detected QTLs with the previous studies based on the recently completed physical map of rapeseed, we obtain a complex genetic architecture of SNPP that includes several major QTLs and numerous small-effect QTLs with underlying candidate genes. This result is understandable because SNPP is the final consequence of many developmental/biological processes, each of which is controlled by many genes. To our knowledge, this is the first comprehensive genetic framework of SNPP, which will deepen our understanding of this trait and provide a blueprint for the genetic improvement of SNPP.
To obtain a high-resolution genetic basis of SNPP variation in rapeseed, fine-mapping of the identified major QTL qSN.A6 was performed using the NIL strategy from a preliminary interval of 1910 to 267 kb (Fig. 5). The mendelizing of qSN.A6 in the BC4F2 population enabled an accurate estimate of its mode-of-inheritance. Similarly to the dominant inheritance of qSS.C918, qSN.A6 showed a partial dominant inheritance. These results supported the hypothesis of dominance rather than over-dominance as the genetic basis of the SNPP heterosis in rapeseed, which suggested the potential of qSN.A6 and qSS.C9 in the utilization of heterosis.
The cytological mechanism of SNPP variation first uncovered via the systematic investigation of qSN.A6
Although the developmental processes (such as the fertility of ovule and pollen) that affect the SNPP of rapeseed were relatively clear8,22, which process is responsible for the natural variation of SNPP was unknown. The mendelizing of qSN.A6 provides a unique opportunity to investigate the mechanism of SNPP variation at a single-locus level. A series of well-designed experiments were performed to exclude the abovementioned processes one by one, and finally, the different rate of ovule abortion (due to incomplete cellularization of the embryo sac) was found to be responsible for the ~5 seed difference between Zhongshuang11 and NIL-qSN.A6. To our knowledge, this report is the first to address the mechanism responsible for SNPP variation in rapeseed. The demonstration of the detailed cellular mechanism of qSN.A6 also enhanced the accuracy of the identification of its candidate gene(s).
The incomplete cellularization of the embryo sac is commonly observed in other plants, such as Arabidopsis, rice, and maize23,24,25,26. Further research showed that abnormal meiosis of the megasporocyte or abnormal mitosis of the functional megasporocyte is the main reason27,28. Based on two detailed structures (Fig. 6D (II and III)), the incomplete cellularization of the embryo sac might be the product of a single or lacking division of the megaspore mother cell. Therefore, the incomplete cellularization of the embryo sac of qSN.A6 observed in our study is likely caused by abnormal meiosis of the megasporocyte.
The genetic causation of trade-off (antagonistic pleiotropy) among sink traits
Source limitation forces an organism to allocate energy to processes in a competitive manner29. In seed plants, major trade-offs among “sink” traits (such as the number and size of seeds) are commonly observed30,31, as reflected by typical negative correlations. For rapeseed, negative correlations among the main sink traits, i.e., the three yield components, PN, SNPP, and SW, are also commonly present17,18. However, the genetic causation of the trade-off among these sink traits has not been previously investigated and remains unknown.
The opposite effects of qSN.A6 on SNPP and SW provide an ideal example to investigate the genetic causation of the trade-off among sink traits. The results of conditional QTL analysis showed that it was caused by antagonistic pleiotropy rather than tight linkage. The detailed results of reciprocal conditional QTL analysis further revealed that this type of antagonistic pleiotropy was due to a physiological interaction in which SNPP acts “upstream” of SW, which is highly consistent with the commonly accepted physiological mechanism of negative feedback among sinks. To our knowledge, this study is the first that clearly demonstrates the genetic causation of trade-off among sink/yield-component traits in rapeseed as well as in other crops. Regarding the three main sink traits of rapeseed, in addition to SNPP, qSN.A6 has a pleiotropic effect on SW but not on PN. This finding is understandable because (1) the determination stage of PN is earlier than SN, and both are earlier than SW and (2) after flowering, the pod wall (rather than the leaf) is the main “source” organ responsible for the size of its “sink”, which is determined by the number and size of the seeds.
Utilization potential of qSN.A6 in rapeseed breeding
The negative/decreasing effect of qSN.A6 on seed weight cannot fully counteract its positive/increasing effect on SNPP, which is consistent with the coefficients of negative correlation between SNPP and SW in the BC4F2 population. In fact, the coefficients of negative correlation among the three yield component traits were also moderate in all previous studies. As expected, qSN.A6 increased the final seed yield by a proportion of 17.4%, which indicated the great potential of the breeding utilization of qSN.A6 in rapeseed.
Methods
Plant materials, field trial, and trait measurement
The F7 generation recombinant inbred line (BnaZNRIL) population of 184 families was derived by single-seed descent from a cross between two sequenced rapeseed cultivars, Zhongshuang11 (high seed number) and No. 73290 (low seed number). The near-isogenic lines (NILs) of the BC4F1 generation were developed by successive backcrossing of F1 with Zhongshuang11 as the recurrent parent four times (Figure S3). The BC4F3 progeny of the recombinant and reference lines were phenotyped to determine the qSN.A6 genotypes of these lines and to further fine-map the target region.
The BnaZNRIL population was arranged in a randomized complete block design with two replications. Each block contained 3 rows with a spacing of 33.3 cm between rows and 16.7 cm between individual plants. It was planted in Wuhan and Zhengzhou from Oct. 2010 to May 2011 and from Oct. 2011 to May 2012 (code W11RIL, W12RIL, Z11RIL and Z12RIL). All populations and the two parents were sown by hand with 15 plants per row, and the field management followed standard agriculture practice. At maturity, ten representative individuals in the middle of the second row of each block were harvested.
Harvested plants were air-dried and stored at room temperature for approximately two weeks before testing. SNPP was calculated as the average number of well-filled seeds from a whole plant’s well-developed pods. The other traits were tested according to previous studies2,32.
SSR/InDel and SNP genotyping
The SSR/InDel markers used to genotype the BC4F2 were previously developed by our laboratory32,33,34. The PCR procedure, electrophoresis, and silver staining were performed as previously described34. The Brassica 60 K Illumina® Infinium SNP array, which was recently developed by the international Brassica Illumina SNP consortium, was used to genotype the BnaZNRIL population. The array hybridization and data processing was conducted by Emei Tongde Co. (Beijing) according to the manufacturer’s protocol (http://www.illumina.com/technology/infinium_hd_assay.ilmn). Leaf tissue was collected from seedlings of the BnaZNRIL, BC1F1, BC2F1, BC3F1, BC4F2 and BC4F3 populations. Genomic DNA was extracted according to the CTAB method35.
Map construction and QTL mapping and integration
The genetic linkage map was constructed using the JoinMap 4.0 software (http://www.kyazma.nl/index.php/mc.JoinMap) with a threshold for goodness-of-fit of ≦ 5, a recombination frequency of < 0.4, and a minimum logarithm of odds (LOD) score of 2.0. All genetic distances were expressed in centimorgans (cM), as derived by the Kosambi function.
The linkage mapping of QTL was performed by composite interval mapping36 using the WinQTL Cartographer 2.5 software (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). The experiment-wise LOD threshold was determined by permutation analysis37 with 1,000 repetitions. LOD scores corresponding to p = 0.05 were used to identify significant QTLs, and p = 0.5 was used to identify suggestive QTLs. The overlapping suggestive QTLs and all significant QTLs were admitted and termed identified QTLs2. QTL meta-analysis was used to estimate the number and positions of the meta-QTLs, which were repeatedly detected in different environments and populations located on the same chromosomal region38. The computation was conducted according to the methods of a previous study2,32. The availability of the pseudochromosomes of B. napus enabled the physical map-based comparison of the detected QTLs. The corresponding genomic intervals of these QTLs were determined by BLAST/e-PCR analysis using the associated markers (within confidence intervals) with available probe/primer sequences against the physical map of B. napus. A total of 306 genes of Arabidopsis with known functions relating to the number of fertile ovules per ovary, the number of ovules fertilized, and the number of fertilized ovules developing into seeds in this study were collected from the TAIR website (http://www.arabidopsis.org/).
To dissect the genetic basis (pleiotropy or tight linkage) of the co-localization of the SNPP and SW QTLs, conditional analysis was performed as described previously32. The conditional phenotypic values y (T1|T2) were obtained by the mixed model approach for the conditional analysis of quantitative traits using QGAStation 1.0 (http://ibi.zju.edu.cn/software/qga/index.htm), where T1|T2 indicates that trait 1 is conditioned by trait 2. Then, conditional QTL mapping was conducted using the conditional phenotypic values as in the unconditional QTL mapping.
Observation of ovule/seed number per ovary/pod in Zhongshuang11 and NIL-qSN.A6
The observation was conducted in five stages, i.e., one day before flowering (DBF), seven, 15, and 25 days after flowering (DAF), and at maturity. The ovule/seed number per ovary/pod was measured as the average number of normal ovules/seeds of three ovaries/pods sampled from ten individuals. To maintain the developmental identity of the samples, flowers from the same day and in the middle of the main inflorescence from individuals with the same flowering time were marked (with red rope) for sampling. At every stage, samples of three ovaries/pods in every plant were taken from at least 30 plants.
Identification of the female and/or male origin of the qSN.A6 effect
To identify the female and/or male origin of the qSN.A6 effect, an ingenious genetic-mating experiment was designed. The alternate branches on the same mother plant of Zhongshuang11/NIL-qSN.A6 were hand-pollinated using its own pollen and that of NIL-qSN.A6/Zhongshuang11, respectively. Each self/out-cross was repeated three times, and pollinations were completed within one day. At maturity, the hand-pollinated pods were harvested and threshed to measure the SNPP. Then a multiple comparison of SNPP was conducted among the hand-pollinated pods of Zhongshuang11, NIL-qSN.A6, and the reciprocal cross.
Evaluation of pollen vitality, pollen germination efficiency, and pollen tube growth as well as embryo sac fertility
To test pollen vitality, pollen of Zhongshuang11 and NIL-qSN.A6 was collected from recently completely opened flowers and stained with 1% acetocarmine39. Then, the cytoplasm integrity of these stained pollen grains was checked individually under a light fluorescence microscope (IX-71; Olympus, Tokyo, Japan), according to their staining and shape.
To investigate in vitro pollen germination efficiency, pollen of Zhongshuang11 and NIL-qSN.A6 was collected and shaken off to spread evenly on a PGM (10% sucrose, 0.005% H3BO3, 10 mM CaCl2, 0.05 mM KH2PO4, 6% PEG 4000) glass slide with 0.3% agar and was then left at room temperature for 3 h, as described previously9. To observe in vivo pollen germination efficiencies and the path of pollen tubes inside the pistil, pistils at one and three DAF were cut (ovary wall was removed) and fixed in 50% FAA (50% ethanol, 5% glacial acetic acid, 3.7% formaldehyde, v/v) for 24 h, softened in 8 M NaOH at 65 °C for 3 h, washed with 50 mM K-phosphate buffer (pH 7.5), and stained in 0.1% aniline blue. The stained pistils and pollen tubes were observed using a light fluorescence microscope (IX-71; Olympus, Tokyo, Japan) under UV light11.
To investigate the fertility of the embryo sac before flowering, paraffin sections were prepared as described by Li et al.40. The buds of one DBF were sampled and fixed in 50% FAA solution, dehydrated through an ethanol series of 75%, 85%, 95% and 100% (v/v), embedded in paraffin wax according to the method of a previous study41, and observed using a light fluorescence microscope (IX-71; Olympus, Tokyo, Japan).
Statistical analysis
The broad-sense heritability was calculated as h2 = 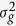 /
/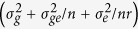 , as described previously, where
, as described previously, where 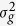 ,
, 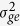 , and
, and 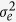 are the variance of genotype, genotype × environment, and error, respectively, and n and r are the number of environments and replications, respectively. The values of
are the variance of genotype, genotype × environment, and error, respectively, and n and r are the number of environments and replications, respectively. The values of 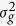 ,
, 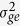 , and
, and 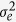 were estimated using the SAS ANOVA procedure. Pearson’s correlation coefficients, the Kolmogorov-Smirnov test, Duncan’s t-test, and multiple comparisons were performed using SAS software version 8.1.
were estimated using the SAS ANOVA procedure. Pearson’s correlation coefficients, the Kolmogorov-Smirnov test, Duncan’s t-test, and multiple comparisons were performed using SAS software version 8.1.
Additional Information
How to cite this article: Yang, Y. et al. Genetic architecture and mechanism of seed number per pod in rapeseed: elucidated through linkage and near-isogenic line analysis. Sci. Rep. 6, 24124; doi: 10.1038/srep24124 (2016).
Supplementary Material
Acknowledgments
This research was supported by the National Basic Research and Development Program (2015CB150202), the Natural Science Foundation (31101181), the Rapeseed Industry Technology System (CARS-13), the Agricultural Science and Technology Innovation Project (CAAS-ASTIP-2013-OCRI), the Core Research Budget of the Non-profit Governmental Research Institution (1610172014001), and the Hubei Agricultural Science and Technology Innovation Center of China.
Footnotes
Author Contributions J.S. and H.W. designed the study. J.S performed the construction, genotyping, and phenotyping of the RIL and the development of NILs. Y.Y. performed the NIL-based fine-mapping, cytological analysis, and yield testing. X.W. and G.L. performed the field experiments. Y.Y. and J.S. performed the data analysis. Y.Y. and J.S wrote the manuscript.
References
- Chen F., Zhang J., Qi C., Pu H. & Chen S. The analys on diversity of germplasm resource in Brassica napus L. Jiangsu Agricultural Sciences 40, 98–99 (2013). [Google Scholar]
- Shi J. et al. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182, 851–861 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K. M. & Latta R. G. Shared quantitative trait loci underlying the genetic correlation between continuous traits. Molecular ecology 16, 4195–4209 (2007). [DOI] [PubMed] [Google Scholar]
- Geritz S. A., van der Meijden E. & Metz J. A. Evolutionary dynamics of seed size and seedling competitive ability. Theor Popul Biol 55, 324–343 (1999). [DOI] [PubMed] [Google Scholar]
- Wagner G. P. & Zhang J. The pleiotropic structure of the genotype–phenotype map: the evolvability of complex organisms. Nat Rev Genet 12, 204–213 (2011). [DOI] [PubMed] [Google Scholar]
- Fletcher R. S., Mullen J. L., Heiliger A. & McKay J. K. QTL analysis of root morphology, flowering time, and yield reveals trade-offs in response to drought in Brassica napus. J Exp Bot 66, 245–256, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Why do plants produce so many more ovules than seeds ? Nature 338, 21–22 (1989). [Google Scholar]
- Bouttier C. & Morgan D. Ovule development and determination of seed number per pod in oilseed rape (Brassica napus L.). J Exp Bot 43, 709–714 (1992). [Google Scholar]
- Schreiber D. N. & Dresselhaus T. In vitro pollen germination and transient transformation of Zea mays and other plant species. Plant Mol Biol Rep 21, 31–41 (2003). [Google Scholar]
- Brown A. P., Brown J. & Dyer A. Optimal pollination conditions for seed set after a self-pollination, an intraspecific cross and an interspecific cross of marrow-stem kale (Brassica oleracea var. acephala). Euphytica 51, 207–214 (1990). [Google Scholar]
- Lu Y. et al. Pollen tubes lacking a pair of K + transporters fail to target ovules in Arabidopsis. Plant Cell 23, 81–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijada P. A., Udall J. A., Lambert B. & Osborn T. C. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor Appl Genet 113, 549–561 (2006). [DOI] [PubMed] [Google Scholar]
- Udall J. A., Quijada P. A., Lambert B. & Osborn T. C. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor Appl Genet 113, 597–609 (2006). [DOI] [PubMed] [Google Scholar]
- Chen W. et al. Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor Appl Genet 115, 849–858 (2007). [DOI] [PubMed] [Google Scholar]
- Radoev M., Becker H. C. & Ecke W. Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179, 1547–1558 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basunanda P. et al. Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.). Theor Appl Genet 120, 271–281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. PNAS 108, 12539–12544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Li S., Chen L. & Yang G. Identification and mapping of a major dominant quantitative trait locus controlling seeds per silique as a single Mendelian factor in Brassica napus L. Theor Appl Genet 125, 695–705 (2012). [DOI] [PubMed] [Google Scholar]
- Ding G. et al. Identification and multiple comparisons of QTL and epistatic interaction conferring high yield under boron and phosphorus deprivation in Brassica napus. Euphytica 198, 337–351 (2014). [Google Scholar]
- Shi J. et al. Linkage and regional association analysis reveal two new tightly-linked major-QTLs for pod number and seed number per pod in rapeseed (Brassica napus L.). Sci Rep 5, 14481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D. et al. Association mapping of six yield-related traits in rapeseed (Brassica napus L.). Theor Appl Genet 127, 85–96 (2014). [DOI] [PubMed] [Google Scholar]
- Pechan P. M. Ovule fertilization and seed number per pod determination in oil seed rape (Brassica napus). Ann Bot-London 61, 201–207 (1988). [Google Scholar]
- Siddiqi I., Ganesh G., Grossniklaus U. & Subbiah V. The dyad gene is required for progression through female meiosis in Arabidopsis. Development 127, 197–207 (2000). [DOI] [PubMed] [Google Scholar]
- Zhao Z. et al. Fine mapping of S31, a gene responsible for hybrid embryo-sac abortion in rice (Oryza sativa L.). Planta 226, 1087–1096 (2007). [DOI] [PubMed] [Google Scholar]
- Sheridan W. F., Avalkina N. A., Shamrov I. I., Batygina T. B. & Golubovskaya I. N. The mac1 gene: controlling the commitment to the meiotic pathway in maize. Genetics 142, 1009 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C. & Doyle G. Double meiotic mutants of maize: implications for the genetic regulation of meiosis. J Hered 82, 156–163 (1991). [Google Scholar]
- Ouyang X. Ultrastructure and ACPase ultracytochemical localization of abortive functional megaspores in intersubspecific F1 hybrids rice (Oryza sativa L.). Chinese Rice Research Newsletter 4, 2–3 (1996). [Google Scholar]
- Rim Y., Beuselinck P., McGraw R. & Somers D. Megagametophyte development in Lotus corniculatus, L. conimbricensis, and their protoplast fusion hybrid. Am J Bot 1990, 1084–1094 (1990). [Google Scholar]
- Obeso & Ramón J. The costs of reproduction in plants. New Phytol 155, 321–348 (2002). [DOI] [PubMed] [Google Scholar]
- Eriksson O. Seed size variation and its effect on germination and seedling performance in the clonal herb Convallaria majalis. Acta Oecologica 20, 61–66 (1999). [Google Scholar]
- Jakobsson A. & Eriksson O. A comparative study of seed number, seed size, seedling size and recruitment in grassland plants. Oikos 88, 494–502 (2000). [Google Scholar]
- Li N., Shi J., Wang X., Liu G. & Wang H. A combined linkage and regional association mapping validation and fine mapping of two major pleiotropic QTLs for seed weight and silique length in rapeseed (Brassica napus L.). BMC Plant Biol 14, 114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. et al. Identification of genome-wide single nucleotide polymorphisms in allopolyploid crop Brassica napus. BMC Genomics 14, 717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. et al. Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res 21, 53–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem bull 19, 11–15 (1987). [Google Scholar]
- Zeng Z.-B. Precision mapping of quantitative trait loci. Genetics 136, 1457–1468 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A. & Doerge R. W. Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffinet B. & Gerber S. Quantitative trait loci: a meta-analysis. Genetics 155, 463–473 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks G. An aceto-carmine glycerol jelly for use in pollen-fertility counts. Biotech Histochem 29, 277–277 (1954). [DOI] [PubMed] [Google Scholar]
- Li N. et al. The natural variation of seed weight is mainly controlled by maternal genotype in rapeseed (Brassica napus L.). PLoS One 10, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. et al. Unusually large oilbodies are highly correlated with lower oil content in Brassica napus. Plant Cell Rep 28, 541–549 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.