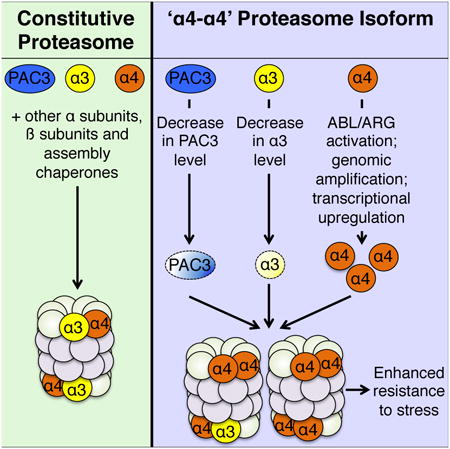
Summary
Targeted intracellular protein degradation in eukaryotes is largely mediated by the proteasome. Here we report formation of an alternative proteasome isoform in human cells, previously found only in budding yeast, which bears an altered subunit arrangement in the outer ring of the proteasome core particle. These proteasomes result from incorporation of an additional α4 (PSMA7) subunit in the position normally occupied by α3 (PSMA4). Assembly of ‘α4-α4’ proteasomes depends on the relative cellular levels of α4 and α3, and on the proteasome assembly chaperone PAC3. The oncogenic tyrosine kinases ABL and ARG and the tumor suppressor BRCA1 regulate cellular α4 levels and formation of α4-α4 proteasomes. Cells primed to assemble α4-α4 proteasomes exhibit enhanced resistance to toxic metal ions. Taken together, our results establish the existence of a novel mammalian proteasome isoform and suggest a potential role in enabling cells to adapt to environmental stresses.
Graphical abstract
Introduction
Efficient and selective degradation of cellular proteins is essential for protein quality control and maintenance of cellular homeostasis (Lecker et al., 2006). Impaired protein quality control and degradation is associated with many human diseases including cancer, cardiovascular diseases, and aging-related pathophysiological conditions such as Alzheimer's disease and Parkinson's disease (Paul, 2008). The ubiquitin-proteasome pathway mediates the majority of targeted protein degradation in eukaryotic cells under normal conditions (Goldberg, 2003; Tomko and Hochstrasser, 2013). In most cases, proteins destined for degradation are first post-translationally tagged with a polyubiquitin chain(s), which then targets them to the 26S proteasome, a 2.6 MDa proteolytic complex (Tomko and Hochstrasser, 2013).
The eukaryotic 26S proteasome consists of a 20S core particle (CP) harboring the proteolytic active sites and a 19S regulatory particle (RP), which binds to one or both ends of the CP cylinder and regulates substrate binding and target protein entry into the CP (Tomko and Hochstrasser, 2013; Tanaka, 2013). The CP is composed of four heptameric rings stacked along a central axis in the order α1-7ß1-7ß1-7α1-7 with a central catalytic chamber bearing the protease active sites (Groll et al., 1997). In the canonical CP, the α rings have seven different but related α subunits and the ß rings each have seven different ß subunits. The catalytic residues of the proteasome are contributed by the ß1, ß2 and ß5 subunits.
CP assembly is an ordered process requiring several dedicated proteasome ‘assembly chaperones' (Le Tallec et al., 2007; Murata et al., 2009; Ramos et al., 1998; Tomko and Hochstrasser, 2013). Individual α subunits are thought to first assemble into an α ring, which then serves as a template for ß-ring assembly followed by the joining of two half-proteasomes; however, alternative intermediates involving α-ß subunit subcomplexes may also form (Panfair et al., 2015). The proteasome assembly chaperone POMP (Ump1 in yeast) is important for ß-ring assembly, whereas the heterodimeric chaperones PAC1-PAC2 (Pba1-Pba2 in yeast) and PAC3-PAC4 (Pba3-Pba4 in yeast) are involved in the efficient assembly of the α ring. Although it forms a heterodimer with PAC4, PAC3 has also been suggested to function as a homodimer (Murata et al., 2009).
In addition to the canonical CP, alterations in CP assembly in mammalian cells yield specialized proteasomes containing distinct paralogs of the active-site ß subunits (Murata et al., 2009) (Tanaka, 2013). One such specialized proteasome is the interferon γ (IFN-γ)-inducible ‘immunoproteasome,’ and another is the thymus-specific ‘thymoproteasome’. Immunoproteasomes are formed by preferential incorporation of the IFN-γ inducible ß1i, ß2i and ß5i subunits in place of the canonical ß1, ß2 and ß5 subunits. These proteasomes are important for generating peptides for antigen presentation by MHC class I molecules. Thymoproteasomes are specifically expressed in thymic cortical epithelial cells. They are formed by incorporating the thymus-specific ß5t subunit in the place of the ß5i subunit in the immunoproteasome and are important in producing peptides involved in the positive selection of CD8+ T cells (Murata et al., 2009).
Thus, subtle variations in proteasome CP composition yield non-canonical proteasomes with altered substrate selectivity and specialized functions. While specialized proteasomes formed by alterations in ß-ring assembly are now well documented in mammalian cells, there is no evidence of somatic mammalian cells with alternative α rings. Mouse male germ cells have been shown to utilize another version of α4, called α4s; these paralogous subunits are 85% identical (94% similar) in sequence, and the functional differences, if any, between α4s proteasomes and canonical ones remain unknown (Uechi et al., 2014).
In the baker's yeast Saccharomyces cerevisiae, the proteasome is required for viability, but one subunit, α3 (Pre9), is unique in being the only non-essential CP component (Emori et al., 1991). Yeast lacking α3 assemble an alternative ‘α4-α4’ proteasome by substituting an additional α4 subunit in the position normally occupied by α3 (Velichutina et al., 2004). Furthermore, the proteasome assembly chaperone Pba3-Pba4 was identified as a factor that normally prevents assembly of alternative α4-α4 proteasomes (Kusmierczyk et al., 2008). Interestingly, yeast cells that form such alternative proteasomes exhibit a growth advantage when exposed to the heavy metal cadmium (Kusmierczyk et al., 2008). This suggests that α4-α4 proteasomes might enable yeast cells to adapt to certain cellular stresses, although such proteasomes have not yet been observed in wild-type (WT) yeast.
Here, we provide the first evidence for formation of the alternative α4-α4 proteasome isoform in mammalian cells, establishing its conservation from yeast to humans. The relative levels of α3 (PSMA4) and α4 (PSMA7) subunits and the level of the PAC3 assembly chaperone are important determinants for the assembly of such proteasomes in human cells. Notably, assembly of α4-α4 proteasomes is enhanced under conditions found in certain cancers such as those with high levels of the ABL tyrosine kinase. Further, mammalian cells primed to assemble these alternative proteasomes exhibit enhanced resistance to cellular stress induced by metal ions. Taken together, our work establishes that the capacity to form alternative α4-α4 proteasomes is evolutionarily conserved and implicates this proteasomal isoform in human disease.
Results
Human α4 can rescue growth of yeast lacking both α3 and α4
In our earlier study, we had found that the Arabidopsis thaliana proteasome subunit α4 (PAD1) can take the place not only of S. cerevisiae α4 (Pre6) but also α3 (Pre9) (Velichutina et al., 2004). This is reflected in the ability of the plant PAD1 gene to rescue the lethal phenotype of both α4Δ and α3Δ α4Δ mutants. To test the ability of the human α4 (PSMA7) subunit to replace α4 and α3 in the yeast proteasome, we performed plasmid-shuffle experiments. A plasmid carrying human α4 was transformed into α4Δ and α3Δ α4Δ strains containing a URA3-marked plasmid with the WT yeast α4 gene. Eviction of the original α4 plasmid on plates containing 5-fluoroorotic acid (FOA), which is lethal to URA3-expressing cells, resulted in viable colonies with both mutants (Fig. 1A). This result suggests that, similar to plant PAD1, human α4 can incorporate into the yeast proteasome and is also capable of occupying two positions in the CP, supporting the idea that this ability is conserved among higher eukaryotes.
Figure 1. Human α4 can replace yeast α4 (Pre6) in yeast proteasomes and can form alternative ‘α4-α4’ proteasomes in mammalian cells.
(A) Human α4 (α4-Hs) expression in yeast allows growth in cells lacking Pre6 (α4-Sc) or both Pre6 and Pre9 (α3-Sc). Eviction of YCplac33-α4-Sc in α4Δ and α3Δ α4Δ yeast strains carrying either p424-GPD- α4-Hs or pRS424-α4-Sc results in viable colonies. The empty vector control showed no growth.
(B) Plate growth assay using serially diluted α4Δ (pre6Δ) and α3Δ α4Δ (pre9Δ pre96Δ) yeast cells transformed with either pRS424-PRE6 (α4-Sc) or p424GPD-PSMA7 (α4-Hs), which express yeast and human α4, respectively, and spotted on medium lacking tryptophan in 10-fold serial dilutions.
(C) Cartoon illustrating disulfide bond formation between engineered cysteines in α4 subunits if two α4 subunits are adjacent to each other in the α-ring.
(D) α4-Hs can form α4-α4 proteasomes in yeast. α4-Hs-CC (α4 T77C:T152C) subunits were disulfide crosslinked under oxidizing conditions (CuCl2) in lysates, resolved by nonreducing SDS-PAGE, and immunoblotted with anti-α4. α4 monomer and dimer bands are indicated by black arrowheads. Additional uncharacterized oxidation products accumulated in both samples. Samples marked 4* and 8* were the same as those in lanes 4 and 8, respectively, but were treated with reducing agent (DTT). Also see Fig. S1A.
(E) Crosslinking of neighboring human α4 subunits can be detected in functional yeast 26S proteasomes purified (using native gel separation) from α3Δ α4Δ cells expressing α4-Hs-CC. No α4-α4 dimers are seen in the α4Δ single mutant, which still has endogenous α3 (lane 1), or with WT α4 that lacks the engineered cysteine residues (lane 2). Also see Fig. S1B.
(F) Ectopic expression of α4-Hs-CC results in formation of α4-α4 proteasomes in HEK293T cells. Whole cell extracts bearing disulfide-crosslinked α4 were analyzed as in panel D with yeast extracts except no CuCl2 was added.
The growth of the α4Δ and α3Δ α4Δ mutants expressing human α4 was then compared to the equivalent strains expressing yeast α4. We performed serial dilution growth assays at 30°C and 36.5°C. The matched strains grew similarly at 30°C, the optimal growth temperature, regardless of the species source for the α4 subunit, and the same was true when complementation was tested in the α4Δ single mutant at the higher temperature (Fig. 1B). In contrast, the α3Δ α4Δ yeast mutant expressing either the yeast or human α4 gene barely grew at 36.5°C. The double mutant transformant still lacked α3, so this result is consistent with previously published data showing temperature-sensitive growth of α3Δ cells (Velichutina et al., 2004).
To verify that human α4 forms alternative ‘α4-α4’ proteasomes in α3Δ α4Δ yeast, we adapted a previously developed disulfide-engineering approach (Velichutina et al., 2004). In our earlier study, we mutated residues N79 and I155 of yeast α4 to cysteines, which enabled detection of disulfide crosslinks between neighboring α4 subunits in the alternative proteasome isoform (Fig. 1C). The structures of 20S proteasomes from yeast and mammals are highly similar (Unno et al., 2002), and alignment of their α4 sequences suggested that human α4 residues T77 and T152 correspond to the residues in yeast α4 that were efficiently crosslinked when changed to cysteine. These human α4 residues were therefore also mutated to cysteines by site-directed mutagenesis.
We first tested whether the engineered double-cysteine mutation in human α4 (α4-Hs-CC) could be used to detect α4-α4 proteasome assembly in yeast. Cell lysates from α3Δ α4Δ yeast expressing either wild type human α4 (α4-Hs-WT) or the cysteine-substitution mutants were treated with oxidant (CuCl2) and analyzed by non-reducing SDS-PAGE and immunoblotting. A ∼60 kDa band was specifically detected in cells expressing α4-Hs-CC but not the α4-Hs-WT or single cysteine-substitution mutants (Figs. 1D and S1A). The intensity of the crosslinked species increased in a time-dependent fashion upon treating the cell lysate with oxidant. Loss of the ∼60 kDa species after treating the sample with the reducing agent dithiothreitol (DTT) was consistent with this species representing a divalent α4-α4 dimer.
To determine whether the α4 interaction detected by crosslinking of whole cell lysates reflected the α-ring configuration in fully assembled functional 26S proteasomes rather than dead-end intermediates, we isolated active 26S proteasomes from human α4-expressing α3Δ α4Δ yeast cells by nondenaturing polyacrylamide gel electrophoresis (native PAGE). The active complexes were identified by a substrate overlay assay. The excised complexes were then subjected to non-reducing SDS-PAGE and immunoblotting (as in Fig. 1D) to detect the crosslinked α4-α4 species. Indeed, we saw the ∼60 kDa crosslinked α4-α4 band specifically in 26S proteasomes from cells expressing α4-CC but not α4-WT or the single cysteine substitution mutants (Figs. 1E and S1B). No crosslinked α4-α4 dimers were seen in proteasomes isolated from the α4Δ single mutant, consistent with the presence of endogenous α3, which limits α4 incorporation at the α3 position (Velichutina et al., 2004). In addition to establishing the shared positional plasticity of yeast and human α4 subunits, these results confirmed the utility of the specific engineered cysteine mutations in human α4 as a means to examine alternative α4-α4 proteasomes.
Human cells can assemble α4-α4 proteasomes
To determine whether the ability to assemble alternative α4-α4 proteasomes is unique to budding yeast or conserved in other eukaryotes, we transfected HEK293T cells with either the α4-Hs-WT or cysteine substitution mutants. Expression of α4-CC mutant, but not α4-WT or the single cysteine mutants, yielded the ∼60 kDa species corresponding to the disulfide crosslinked α4-α4 dimer (Fig. 1F). Unlike in yeast, the formation of α4 crosslinks in these cells did not require addition of external oxidizing agents to the cell lysate, suggesting that the lysis (or cellular) conditions were already sufficiently oxidizing to induce disulfide bond formation. Additional oxidation products were also observed, as was the case in yeast, but these species were not characterized further. We also tested the ability of multiple human cell types (including the IMR-90 primary cell line derived from human fetal lung) to assemble alternative proteasomes (Fig. S1C). Our results demonstrate that the ability to assemble α4-α4 proteasomes is conserved across transformed and primary human cell lines. Finally, we found that α4 dimers were also seen in fully assembled 26S proteasomes isolated from human cells, and not just assembly intermediates or dead-end products (Fig. S1D; also see Fig. 2D).
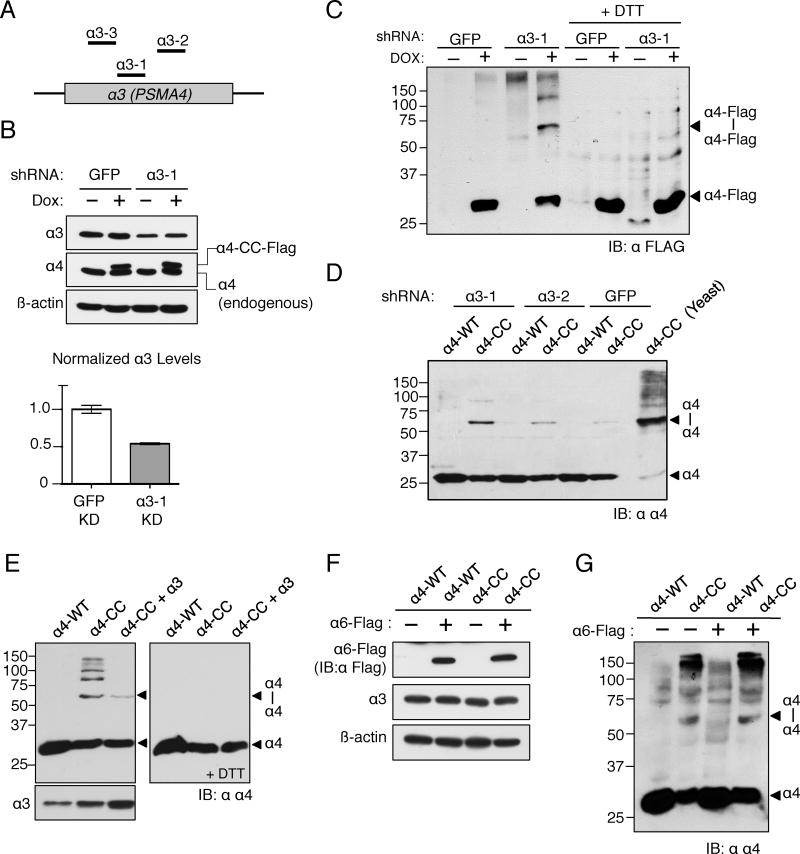
Figure 2. α3 regulates α4-α4 proteasome assembly in human cells.
(A) Cartoon illustrating regions of the α3 gene targeted by different shRNAs.
(B) shRNA-mediated knockdown of α3 in stable α4-CC-Flag (HeLa) cells. The α3-1 shRNA cause a ∼50% decrease in α3 levels. α4-CC-Flag expression was induced by the addition of doxycycline (Dox).
(C) α3 knockdown in α4-CC-Flag stable HeLa cells results in increased assembly of α4-α4 proteasomes. Also see Fig. S2.
(D) Confirmation of the increase in α4-α4 proteasomes upon α3 knockdown using gel-isolated HeLa cell 26S proteasomes.
(E) Ectopic overexpression of α3 in α4-overexpressing HeLa cells causes a decrease in α4-α4 proteasome formation based on α4- α4 crosslinking.
(F) α6-Flag expression was induced in stable α6-Flag HeLa cells by addition of Dox; α4-WT or α4-CC was transiently expressed in these cells.
(G) α6 expression level does not affect α4-α4 proteasome assembly. α6-Flag expression was induced in α6-Flag stable HeLa cells by addition of Dox (see Fig. 2F).
Changes in α3 levels modulate α4-α4 proteasome assembly in human cells
Inasmuch as deletion of the α3 gene induces assembly of the alternative α4-α4 proteasome isoform in yeast (Velichutina et al., 2004), we tested the effect of reducing α3 levels on α4-α4 proteasome assembly in human cells. We first created an isogenic Flp-In HeLa cell line stably expressing α4-CC-Flag under a doxycycline (Dox)-responsive promoter and then knocked down α3 levels by ∼50% with a lentivirus-delivered shRNA (Fig. 2A, B). α3 knockdown enhanced assembly of α4-α4 proteasomes as indicated by the increase in the ∼60 kDa α4-α4 crosslinked species (Fig. 2C). Multiple shRNAs targeting different regions of α3 also resulted in an increase in alternative proteasome assembly in HeLa cells transiently transfected with α4-CC (Figs. 2A, S2A and S2B).
Similar to the results obtained with HeLa cells, knockdown of α3 in U2OS cells also resulted in increased α4-α4 levels, indicating a general effect in different cell types (Fig. S2C). The increase in α4-α4 crosslinking observed upon α3 knockdown using whole cell extracts was also observed in gel-isolated active 26S proteasomes (Fig. 2D). This confirmed that the difference in abundance of proteasome isoforms detected by disulfide crosslinking using cell lysates mirrors changes in fully assembled proteasomes. Conversely, overexpression of ectopic α3 resulted in decreased alternative proteasome assembly in α4-CC overexpressing HeLa cells (Fig. 2E), consistent with an inverse relationship between α3 levels and α4-α4 proteasome formation. No effect on α4-α4 proteasome assembly was observed upon overexpression of another CP α subunit, α6 (PSMA1), confirming that the effect of α3 on α4-α4 proteasome assembly was specific (Figs. 2F and 2G). Taken together, these results establish cellular α3 levels as an important determinant in regulating the assembly of this alternative proteasome isoform in human cells.
PAC3 levels and cadmium regulate α4-α4 proteasome assembly
The heterodimeric CP assembly chaperone Pba3-Pba4 was identified as an important factor that limits α4-α4 proteasome formation in yeast (Kusmierczyk et al., 2008). To determine whether PAC3, the mammalian ortholog of Pba3, influences assembly of this isoform in mammalian cells in a similar way, we used shRNA to knock down PAC3 in isogenic Flp-In stable HeLa cells stably expressing α4-CC-Flag (Figs. 3A and 3B). Knockdown of PAC3 enhanced α4-α4 proteasome assembly (Fig. 3C). This result was further verified using two different shRNAs against PAC3, both of which caused an increase in α4-α4 proteasome assembly in HeLa cells transiently transfected with α4-CC (Figs. 3A, S3A and S3B). By contrast, overexpression of ectopic PAC3 in cells stably expressing α4-CC-Flag strongly decreased α4-α4 proteasome assembly (Figs. 3D and 3E). Thus, as in yeast, PAC3 (presumably as part of the PAC3-PAC4 dimer) is an important regulator of α4-α4 proteasome assembly in human cells.
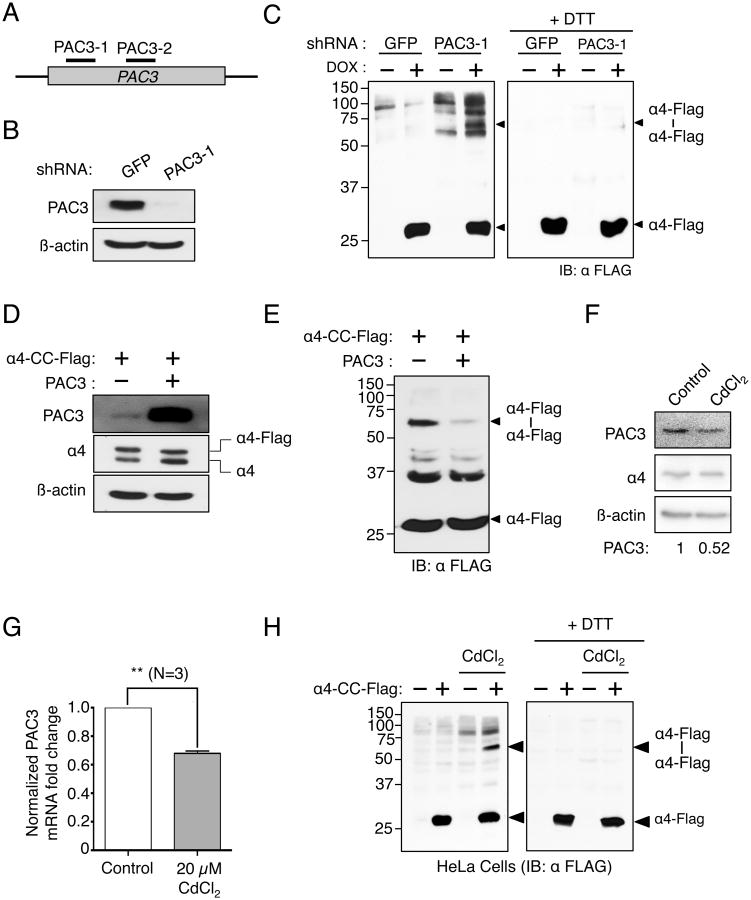
Figure 3. Proteasome assembly chaperone PAC3 regulates α4-α4 proteasome assembly in human cells.
(A) Cartoon illustrating different PAC3 gene regions targeted by PAC3 shRNAs.
(B) shRNA-mediated knockdown of PAC3 in α4-CC-Flag stable HeLa cells using PAC3-1 shRNA.
(C) PAC3 knockdown results in increased α4-α4 proteasome assembly in stable α4-CC-Flag expressing cells. Also see Fig. S3.
(D) Ectopic overexpression of PAC3 does not affect α4 levels.
(E) Overexpression of PAC3 reduces α4-α4 proteasome assembly in Dox-induced α4-CC-Flag cells.
(F) Treatment of HeLa cells with 10 μM CdCl2 for 24 hours results in a ∼50% decrease in PAC3 levels.
(G) CdCl2 causes a decrease in PAC3 mRNA levels. Results represent three independent experiments in which mRNA levels were measure by RT-qPCR. ** p<0.05 (student's t-test).
(H) CdCl2 treatment (10 μM) leads to enhanced α4-α4 proteasome assembly in α4-CC-Flag stable HeLa cells. α4-CC-Flag expression was induced in α4-CC-Flag stable HeLa cells by addition of Dox.
During our efforts to identify conditions that might affect α4-α4 proteasome assembly, we found that exposure to the heavy metal cadmium caused a ∼50% decrease in cellular PAC3 levels (Fig. 3F). Most or all of the effect of CdCl2 on PAC3 protein levels appeared to be through reduction of PAC3 mRNA (Fig. 3G). Since PAC3 plays an important role in regulating the assembly of α4-α4 proteasomes, we tested the effect of CdCl2 on α4-α4 proteasome levels. As shown in Figs. 3H and S3C, CdCl2 treatment led to an increase in α4-α4 proteasome assembly in both HeLa and HEK293T cells. CdCl2 is known to induce an oxidizing cellular environment, which could potentially have caused an increase in in vivo disulfide crosslinking (Nemmiche et al., 2011). Hence, we wanted to establish whether the observed increase in α4-α4 crosslinking reflects an actual increase in α4-α4 proteasomes in response to reduced PAC3 levels or is an artifact of altered cellular oxidative conditions. Because short-term treatment of cells with CdCl2 is sufficient to alter cellular redox but not enough to decrease PAC3 levels, we analyzed α4-α4 crosslinking in cells subjected to CdCl2 treatment for 3 h or 24 h (Fig. S3D) (Nair et al., 2013). Unlike the 24 h treatment, short-term exposure to CdCl2 had no effect on α4-α4 dimer levels, suggesting that reduced PAC3, rather than enhanced disulfide-bond induction, is likely to mediate the CdCl2-induced assembly of the α4-α4 proteasome variant (Fig. S3E).
α4 overexpression rescues viability of α3 knockdown cells
Unlike in yeast where α3 deletion is well tolerated under normal growth conditions, potent α3 knockdown caused strong phenotypic abnormalities and a decrease in cell viability in human cells. Cultured human cells with reduced α3 levels formed long branched structures resembling neuron-like projections (Fig. 4A). This change in cell morphology was absent in the corresponding GFP knockdown control cells. The α3 knockdown-induced changes in morphology were consistently observed using multiple shRNAs targeting different regions of α3 (data not shown). Furthermore, prolonged α3 knockdown resulted in decreased cell viability, suggesting mammalian cells are more dependent on α3 for cell survival than are yeast. All protein analyses shown in this study were performed before any decrease in cell viability was observed.
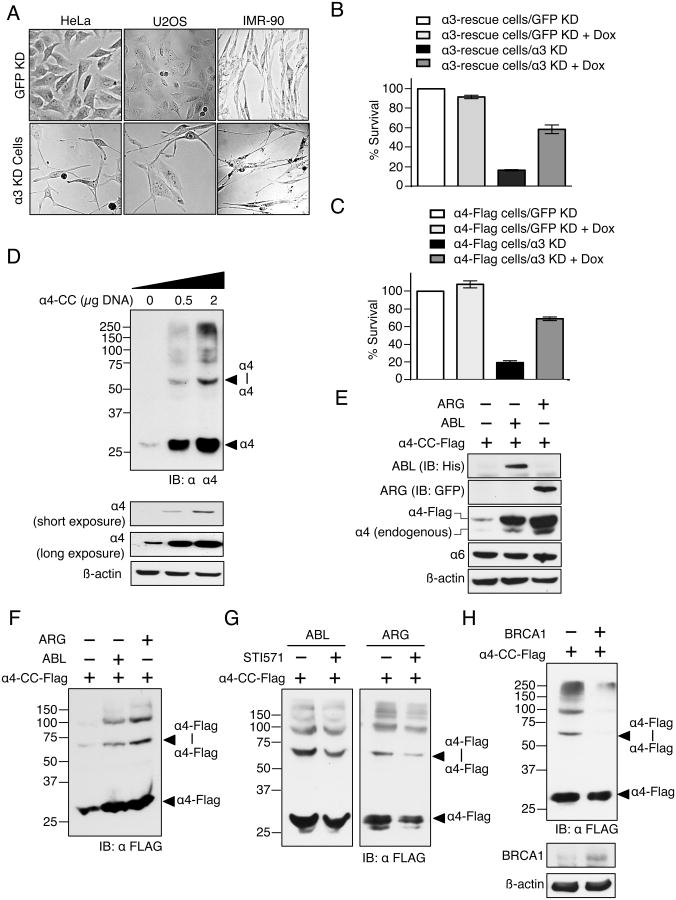
Figure 4. ABL, ARG, and BRCA1 regulate α4 levels and α4-α4 proteasome assembly and elevated α4 levels rescues α3 knockdown induced loss of cell viability.
(A) α3 knockdown results in changes in cell morphology. α3 shRNA induces formation of neuron-like cell protrusions in HeLa, U2OS and IMR-90 cells.
(B) α3 shRNA-induced loss of cell viability is rescued by ectopic expression of shRNA-resistant α3. Flp-In HeLa cells containing stably integrated α3-Flag with silent mutations that make it resistant to α3-3 shRNA (“α3-rescue cells”) were treated with either α3-3 shRNA or control GFP shRNA for 4 d. α3-Flag expression was induced in these cells by Dox at the same time as shRNA treatment. Cell viability was measured on day 4 using the CellTiter-Glo® luminescent cell viability assay. Data from three experiments were quantified; error bars represent standard deviation. Also see Fig. S4A.
(C) Ectopic expression of α4 also rescues α3 shRNA-induced loss of cell viability. Flp-In HeLa cells containing stably integrated α4-Flag was treated with either α3-2 shRNA or control GFP shRNA for 8 days. α4-Flag expression was induced in these cells by Dox at the same time as shRNA treatment. Cell viability was measured on day 8 as in Fig. 3B. Data from three experiments were quantified; error bars represent standard deviation. Also see Fig. S4D.
(D) The level of α4-α4 proteasomes in cells correlates with the level of α4 (top panel). Transfection of increasing amounts of α4-CC in HEK293T cells resulted in corresponding increases in α4-α4 proteasomes. The bottom panel shows total α4 protein levels in cell lysates used for the α4-α4 crosslinking assay (top panel).
(E) Transient overexpression of ABL-6xHis and ARG-EYFP in HEK293T cells results in a concomitant increase in both endogenous α4 and co-transfected α4-CC-Flag.
(F) Expression of ABL or ARG tyrosine kinase in HEK293T cells expressing α4-CC-Flag increases α4-α4 proteasome levels.
(G) Increased formation of α4-α4 proteasomes upon ABL and ARG overexpression is dependent on their tyrosine kinases activity. Treatment of HEK293T cells co-expressing α4-CC-Flag and either ABL or ARG with inhibitor STI571 (Immatinib) decreases α4-α4 levels.
(H) Overexpression of BRCA1 (bottom panel) causes a decrease in α4-α4 proteasomes in HEK293T cells (top panel).
To confirm that the decrease in viability of cells treated with α3 shRNA was not an off-target effect of the shRNA, we tested the ability of ectopic shRNA-resistant α3 to rescue the decreased viability. For this, we generated a Dox-inducible stable HeLa cell line expressing α3 with silent mutations that protected it from the shRNA against α3 (Fig. S4A). The expression of shRNA-resistant α3 protected the cells from α3 knockdown-induced cell death; in contrast, cells overexpressing WT (shRNA-sensitive) α3 failed to rescue growth, indicating that it is the decrease in α3 levels that reduced cell viability (Figs. 4B, S4B and S4C).
The results in Fig. 4A and 4B suggest that endogenous α4 is either incapable of overcoming the loss of α3 through α4-α4 proteasome assembly or is expressed at insufficient levels to allow rescue. If the latter explanation is correct, then overexpression of α4 in α3 knockdown cells should enhance assembly of α4-α4 proteasomes and rescue the decreased viability of these cells. Indeed, α4 overexpression in cells resulted in increased tolerance of α3 knockdown (Figs. 4C, S4D and S4E). Overexpression of another proteasome alpha subunit, α6, had no effect on either α4-α4 proteasome formation or α3 knockdown-induced cell death (Figs. S4F and S4G).
Oncogenic ABL and ARG kinases and the tumor suppressor BRCA1 have reciprocal effects on assembly of α4-α4 proteasomes
Increasing cellular levels of α4 caused corresponding increases in α4-α4 proteasome assembly (Fig. 4D). Interestingly, the related cytoplasmic tyrosine kinases ABL and ARG were recently reported to reduce degradation of α4 protein and thereby promote its accumulation (Li et al., 2015). This was accompanied by an increase in total proteasome levels and proteolytic capacity of the cells (Li et al., 2015) (see Fig. S5B). We hypothesized that the enhanced proteasome levels and activity observed in cells overexpressing these oncogenic kinases results at least in part through increased assembly of α4-α4 proteasomes. To determine whether ABL and ARG overexpression causes such an increase, α4-CC-Flag was co-expressed along with either ABL or ARG in HEK293T cells. Concomitant expression of ABL or ARG kinase along with α4-CC-Flag resulted in increased levels of α4 and α4-α4 proteasomes (Figs. 4E and 4F). This increased assembly of α4-α4 proteasomes in the presence of ABL or ARG required their kinase activity as the effect was suppressed by addition of the ABL/ARG kinase inhibitor STI571 (Immatinib) (Fig. 4G). We note that overexpression of ABL and ARG had no effect on PAC3 levels in HEK293T cells (Fig. S4H).
The tumor suppressor protein BRCA1 was previously identified as a ubiquitin ligase that targets free α4 for proteasome-mediated degradation (Li et al., 2015). Correspondingly, we found that co-expression of BRCA1 with α4-CC-Flag in HEK293T cells resulted in decreased levels of α4-α4 proteasomes (Fig. 4H). These results suggest that the unique regulation of α4 levels by the ABL and ARG kinases and the BRCA1 ubiquitin ligase also leads to changes in levels of the α4-α4 proteasome isoform.
Increased ABL kinase activity resulting from chromosomal translocation of ABL to the BCR locus has a causal role in chronic myelogenous leukemia (CML) (Ren, 2005). Amplification of the genomic locus harboring α4 (PSMA7) (20q13) has been reported in several cases of cervical and colon cancer (Hu et al., 2008; Scotto et al., 2008). Consistently, higher α4 protein levels have been detected in 38.8% (24/62) of colorectal cancer primary sites, 52.9% (18/34) of lymph node metastases and 100% (13/13) of liver metastases (Hu et al., 2008). Further, genome-wide gene expression studies have shown that α4 mRNA levels are elevated in testicular and breast cancers (Fig. S4I) (Korkola et al., 2006; Richardson et al., 2006). Increased α4 expression is also observed in castration-recurrent prostate cancer (Romanuik et al., 2010). These data taken together suggest that α4 levels are selectively up-regulated in various cancers through multiple mechanisms. Our results show that this can lead to increased assembly of α4-α4 proteasomes, which may have a role in promoting these malignancies.
Assembly of α4-α4 proteasomes correlates with enhanced heavy metal resistance
Since elevated α4 levels are observed in several cancers, we sought to clarify the effect of increased α4 levels on the proteolytic capacity of cells (Hu et al., 2008; Romanuik et al., 2010). Overexpression of α4-Flag resulted in increased levels of cellular proteasome activity (Figs. 5A and 5B), as observed previously (Li et al., 2015). Overexpression of α4 also resulted in increased levels of 26S and 20S proteasomes (Figs. 5C and S5A). As expected, overexpression of ABL or ARG concomitantly with ectopic α4 appeared to further enhance proteasome activity as determined by in-gel assays (Fig. S5B). To evaluate if this increased proteolytic capacity translates into higher resistance to proteotoxic stresses, we challenged cells expressing high levels of α4 with increasing concentrations of the toxic metal ions cadmium or copper. These chemicals have the potential to alter cellular redox states and cause oxidative protein damage, which increases the load on cellular proteasomes (Pourahmad and O'Brien, 2000). Cells expressing elevated levels of α4 are primed to assemble α4-α4 proteasomes, and they exhibited significantly higher resistance to the toxicity induced by CdCl2 and CuCl2 (Figs. 5D, 5E and S5C). In contrast to α4 overexpression, ectopic expression of α6 had little or no effect on the ability of cells to resist metal-induced toxicity (Figs. 5F, 5G and S5D). Taken together, these results demonstrate that multiple mechanisms leading to increases in α4 levels cause elevated levels of active proteasomes. As our data also indicate that high α4 levels increase assembly of the α4-α4 proteasome isoform (Fig. 4D), it is possible that this variant helps cells overcome proteotoxic stresses such as those when cells are exposed to heavy metals (Fig. 5D).
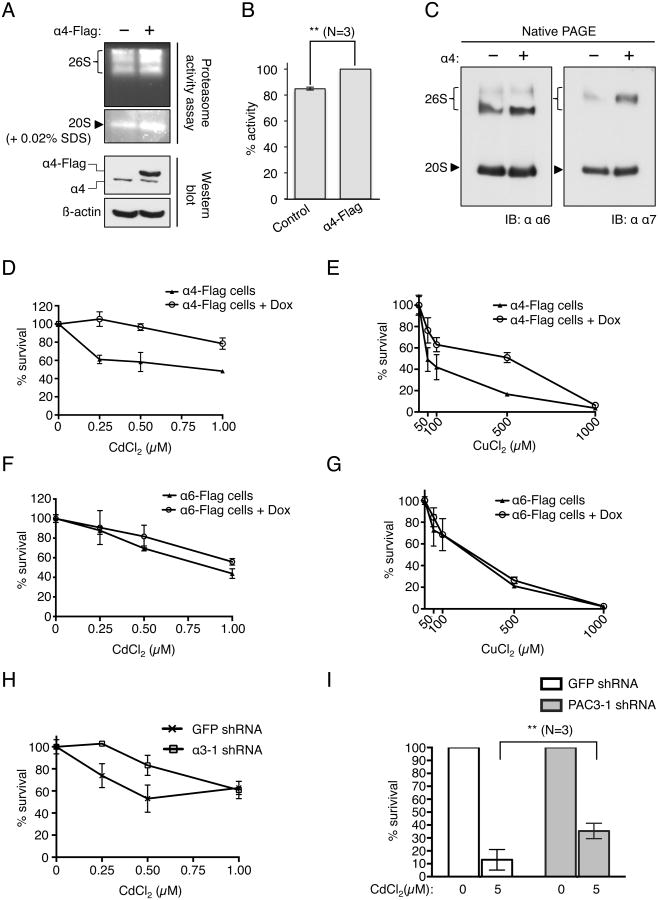
Figure 5. High α4 levels correlate with elevated proteasome activity and enhanced resistance to toxic metals.
(A) Ectopic expression of α4 (bottom panel) results in higher 26S and 20S proteolytic activity (top panel). Cell lysates were resolved by native PAGE, and proteasome activity was measured by an in-gel substrate overlay assay using the fluorogenic peptide Suc-LLVY-AMC.
(B) α4 overexpression results in approx. 15% increase in proteolytic capacity of cells. In vitro proteasome activity assay was performed on the whole cell extract using the fluorogenic peptide substrate Suc-LLVY-AMC. Fluorescence was measured at Ex380nm/Em460nm. The relative fluorescence intensity was calculated from the mean ± SE of three independent experiments. ** p<0.05 (student's t-test).
(C) Native PAGE-western blot analysis of HEK293T cell lysate reveals increase in 26S and 20S proteasome levels upon ectopic α4 expression. Also see Fig. S5A.
(D) Dox-induced expression of stably integrated α4-Flag in Flp-In HeLa cells enables the cells to resist exposure to the toxic metal cadmium (CdCl2). Cell viability was measured using the CellTiter-Glo® luminescent cell viability assay. N=3, and error bars represent standard deviations. Also see Fig. S5C.
(E) Dox-induced expression of stably integrated α4-Flag in Flp-In HeLa cells enhances copper (CuCl2) resistance. Cell viability was measured as in Fig. 4C. N=3, and error bars represent standard deviations. Also see Fig. S5C.
(F and G) Dox-induced α6 overexpression in α6-Flag stable HeLa cells had no effect on the ability of the cells to resist toxicity due to CdCl2 (F) or CuCl2 (G). Cell viability was measured as in Fig. 5E. N=3 and error bars represent standard deviations. Also see Fig. S5D.
(H) α3 knockdown in HeLa cells using α3-1 shRNA enhances cadmium (CdCl2) resistance. Cell viability was measured as in Fig. 5E. N=3, and error bars represent standard deviations. Also see Fig. S5E.
(I) PAC3 knockdown in HeLa cells using PAC3-1 shRNA results in slightly enhanced ability of HeLa cells to resist cadmium (CdCl2) toxicity. shRNA-mediated knockdown of PAC3 in HeLa cells was carried out for 8 days. Cells were treated with 5 μM CdCl2 for 24 hours prior to measuring cell viability using the CellTiter-Glo® luminescent cell viability assay. N=3, and error bars represent standard deviations. ** p<0.05 (Student's t-test). Also see Fig. S5F.
The enhanced resistance to heavy metals observed in α4-overexpressing cells could in principle be due to the resulting higher overall proteasome levels (Fig. 5) or to the increased assembly of the α4-α4 proteasome isoform (Fig. 4D), or a combination of these effects. It is noteworthy that yeast cells expressing α4-α4 proteasomes also display greater cadmium resistance even though they assemble fewer proteasomes overall (Kusmierczyk et al., 2008). Knockdown of either α3 or PAC3 in human cells also lowered both proteasome activity and levels (Figs. S5E, S5F and S5G). At the same time, as shown above, reduction of cellular α3 and PAC3 promoted assembly of α4-α4 proteasomes (Figs. 2 and 3). Interestingly, we observed that moderate reduction of α3 (using α3-1 shRNA) resulted in enhanced resistance to cadmium (Figs. 5H and S5E), whereas more drastic reduction of α3 levels (α3-2 shRNA) failed to provide a survival advantage to the cells when challenged with CdCl2 (Figs. S5H and S5E). This suggests that while enhanced α4-α4 proteasome assembly might enable the cells to resist Cd-induced proteotoxic stress, reduction in total proteasome levels below a certain threshold reverses this advantage. PAC3 knockdown also resulted in modestly enhanced resistance to Cd toxicity (Figs. 5I and S5F). It is possible that knockdown of α3 and PAC3 also leads to up-regulation of oxidative stress-response genes, which can potentially help cells resist Cd-induced stress. Interestingly, α6 overexpression also caused an increase in proteasome levels and activity (S5I-K), but unlike α4 overexpression, had no effect on cellular resistance to Cd or Cu, again suggesting that the ability of cells to assemble an alternative form of the proteasome, rather than just changing its levels, is important in cellular resistance to heavy metal-induced proteotoxic stress (Figs. 5F-G).
Discussion
Capacity to assemble α4-α4 proteasomes is conserved from yeast to humans
In this work, we have presented the first evidence that human cells can alter proteasome α-ring assembly to generate a novel proteasome isoform, which our data predict will accumulate in a variety of tumors. The α4-α4 proteasome is formed by incorporation of an additional α4 subunit in the position usually occupied by α3 in either one or both α rings of the CP. Such α4-α4 proteasomes were previously identified only in certain yeast mutants (Velichutina et al., 2004; Kusmierczyk et al., 2008). Our results establish that the capacity to form this alternative isoform is evolutionarily conserved and can occur in non-mutant primary human cell lines.
The human α4 (PSMA7) gene can successfully complement not only yeast lacking endogenous α4 (Pre6) but also those that lack both α3 (Pre9) and α4. The human α4 protein thus allows formation of yeast-human hybrid α4-α4 proteasomes with four of 28 positions occupied by the human subunit. Human α4 is 59% identical in sequence to yeast α4 and only 37% identical to yeast α3. It is therefore remarkable that these cross-species subunit substitutions are so well tolerated.
In our disulfide crosslinking experiments with mammalian cells expressing ectopic α4-CC, the sensitivity of the assay is expected to be reduced due to competition between the ectopically introduced α4-CC protein and endogenous α4, which lacks the engineered cysteines necessary for α4-α4 crosslinking. Since disulfide crosslinking requires two α4-CC subunits to be present adjacent to each other in the α-ring, the level of α4-α4 proteasomes we observed is likely to be an underestimate of the total amount of this proteasomal isoform in these cells. Nevertheless, the assay is sufficiently robust to see changes in the assembly of alternative α4-α4 proteasomes in cells with different relative levels of the α3 and α4 subunits or altered amounts of the CP assembly chaperone PAC3 (Figs. 2, 3, 4 and 6). These observations are very similar to what had been seen in yeast cells (Velichutina et al., 2004), suggesting a similar mechanistic basis for their assembly in yeast and humans.
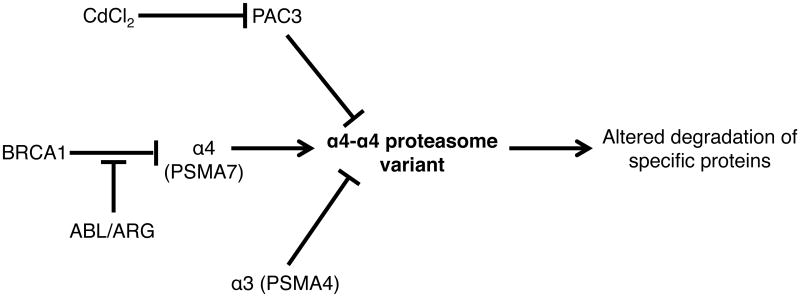
Figure 6. Model summarizing the factors regulating assembly of the alternative α4-α4 proteasome isoform in human cells, and their potential physiological relevance.
See main text for details.
α3 knockdown induces morphologic changes and cell death in human cells
Although several factors important in α4-α4 proteasome assembly are conserved between yeast and humans, some key differences exist. Most obviously, loss of the CP α3 subunit is not lethal to S. cerevisiae but is in all the human cell lines we tested, including primary cells. Despite reduced proteasome assembly efficiency, α3Δ yeast cells can still form sufficient levels of the alternative α4-α4 proteasome to maintain viability (Velichutina et al., 2004). In contrast, knockdown of α3 in human cells results in striking morphological changes and ultimately a loss of cell viability (Fig. 4). Unlike in yeast, the endogenous levels of α4 in mammalian cells fail to overcome α3 loss in the cell lines we tested. Ectopic expression of α4, however, substantially suppresses the defects caused by reduced α3 levels (Fig. 4C). This result supports the idea that under physiological conditions which increase cellular α4 levels, mammalian cells can also accommodate a reduction in α3 by assembling α4-α4 proteasomes. This might promote cell survival under specific conditions or in certain cancers.
α3 knockdown results in cells becoming less rounded and developing long, thin protrusions. Since this morphology resembles that of senescent cells, we stained α3-knockdown and control cells for ß-galactosidase activity, a known marker for senescence. ß-galactosidase staining was similar in the two conditions, suggesting that the α3-deficient cells are not senescent (data not shown). The cause of the altered morphology remains to be determined.
Potential role of alternative α4-α4 proteasomes in human disease
Amplification of the genomic locus 20q13, which harbors α4 (PSMA7), has been reported in several cases of colon and cervical cancer and α4 protein levels have also been shown to be elevated at both primary colon cancer and metastatic sites (Hu et al., 2008; Scotto et al., 2008). Moreover, analysis of published gene expression data reveals selective upregulation of α4 mRNA in multiple cases of testicular and breast cancer (Fig. S4I) (Korkola et al., 2006; Richardson et al., 2006). Significant reduction in α3 expression is simultaneously observed in two of the testicular cancer datasets (mixed germ cell tumor and embryonal carcinoma), which should further enhance α4-α4 proteasome formation. On the other hand, both α3 and α6 levels remained unchanged in the breast cancer dataset (Fig S4I and data not shown) (Korkola et al., 2006; Richardson et al., 2006). α4 expression was also found to be elevated in castration-recurrent prostate cancer (Romanuik et al., 2010). These data highlight potential physiological scenarios during carcinogenesis where increased endogenous α4 levels, particularly when α3 levels are simultaneously reduced, would likely result in enhanced α4-α4 proteasome assembly, which might contribute to the deregulated growth of cancer cells (Fig. 6).
The α4-α4 proteasome has properties that are distinct from those of the canonical CP (Emori et al., 1991; Velichutina et al., 2004). The absence of α3 in the α ring eliminates the closed-gate conformation, which normally prevents substrate entry through the central pore (Osmulski et al., 2009). This leads to higher basal peptidase activity of the α4-α4 CP variant. The change in the α ring alters the interface with the RP and other CP regulators such as PA28 and PA200, potentially changing the regulation of CP activity (Stadtmueller and Hill, 2011). When α4-α4 proteasomes form in yeast, changes in RP assembly are also observed (Kusmierczyk et al., 2008). Finally, yeast cells that only contain the α4-α4 proteasome isoform have selective defects in the degradation of ubiquitin-proteasome system substrates (Velichutina et al., 2004). This may be due to reduced assembly efficiency of the alternative isoform relative to the canonical CP or to one or more of the aforementioned changes.
The oncogenic tyrosine kinases ABL and ARG phosphorylate α4, which stabilizes the free subunit and increases its levels in cells (Li et al., 2015). We show that ABL or ARG overexpression also increases α4-α4 proteasome assembly (Figs. 4F) and cellular proteolytic capacity (Fig. S5B; Li et al., 2015). ABL/ARG overexpression causes an increase in total cellular proteasome levels (Li et al., 2015). We hypothesize that the increased proteasome levels upon ABL and ARG overexpression is at least partly accounted for by the additional α4-α4 proteasomes induced by elevated cellular α4 (Fig. 6). ABL activity is up-regulated in certain cancers, especially CML, where the translocation of ABL to the BCR locus causes high expression of a constitutively active BCR-ABL fusion protein (Ren, 2005). Elevated proteasome activity has also been reported in these cancer cells; this is consistent with data from Li et al. (2015) and in this report showing a direct correlation between the increased ABL activity and cellular proteasomal proteolytic capacity (Crawford et al., 2009).
Phosphorylation of α4 at Tyr106 by ABL/ARG protects α4 from BRCA1-mediated ubiquitylation at Lys116 (Li et al., 2015). Ubiquitylation of α4 by BRCA1 targets it to the proteasome for degradation (Li et al., 2015). BRCA1 is a known tumor suppressor and inactivating mutations in BRCA1 are observed in breast and ovarian cancers (Welcsh and King, 2001). We find that ectopic BRCA1 expression decreases alternative proteasome formation (Fig. 4H). Taken together, these results suggest a potential role for α4-α4 proteasomes in the progression of certain cancers. Given the importance of ABL and ARG kinases in neuronal development, it will also be interesting to explore potential roles of α4-α4 proteasomes in developing neurons and in the pathology of neurodegenerative diseases (Koleske et al., 1998; Schlatterer et al., 2011).
The ability to assemble α4-α4 proteasomes provides mammalian cells a mechanism to alter their proteolytic capacity and specificity in response to cellular stress. Cells with elevated levels of α4 are primed to assemble α4-α4 proteasomes and exhibit greater tolerance for CdCl2 and CuCl2 toxicity (Fig. 5). CdCl2 and CuCl2 can cause oxidative protein damage, resulting in an increased load on cellular protein degradation pathways. Higher cellular proteolytic capacity or shifts in the specificity of proteasomes engendered by the α3-to-α4 subunit swap (or both) could help these cells overcome the increased load on the proteasome due to exposure to toxic heavy metals (Nemmiche et al., 2011; Pourahmad and O'Brien, 2000). Increased levels of reactive oxygen species are also observed in tumor cells, and tumor cells have a higher demand for protein turnover to support their rapid proliferation (Balch et al., 2008; Liou and Storz, 2010). Consistent with this, enhanced proteasome levels and activity are often observed in cancers, and several cancer types have been shown to be highly sensitive to proteasome inhibition (Buac et al., 2013).
The proteasome inhibitors bortezomib and carfilzomib are used in the treatment of hematological malignancies such as multiple myeloma and mantle cell lymphoma (Buac et al., 2013). However, a growing concern with these active site-targeted inhibitors is the emergence of drug resistance (Lu and Wang, 2013). The need to counter emerging resistance to such inhibitors makes it important to develop alternative strategies for reducing proteasome activity or assembly. The selective increase in α4 in certain cancers suggests a potential role for α4-α4 proteasomes in sustaining the rapid proliferation of cells in these malignancies. Thus, strategies that would enable us to interfere specifically with the assembly of α4-α4 proteasomes could have therapeutic potential in tumors where its function is relevant. This approach might also have fewer side effects and reduced toxicity compared to existing drugs that target total proteasome function across all cells. The development of such strategies will require confirmation of the role of α4-α4 proteasomes in cancer cell proliferation and an in-depth understanding of their assembly and function.
Experimental Procedures
Reagents and strains
E.coli strain TOP10 F′ was used for DNA cloning. Yeast strains used in this study are listed in Table S1. shRNA constructs used in this study as listed in Table S2. Plasmids used in this study are listed in Table S3. Antibodies used are listed in Table S4.
Mammalian Cell Culture
Mammalian cell lines (HEK293T, HeLa, U2OS and IMR-90) were grown using standard cell culture techniques in DMEM media supplemented with 10% fetal bovine serum. IMR-90 cells were purchased from ATCC. HEK293T, HeLa, and U2OS cells were obtained as a gift from Christian Schlieker (Yale University). Low passage HEK293T cells were used as the packaging cell line for lentivirus production. See supplementary experimental procedures for details on generation of isogenic Flp-In HeLa stable cell lines.
Disulfide crosslinking assay to detect α4-α4 proteasomes
Yeast cells grown to mid-log phase were harvested, spheroblasted and lysed by vortexing. Disulfide crosslinking assays in yeast were performed as described previously (Kusmierczyk et al., 2008). Human cells were lysed in in lysis buffer (50 mM Tris-HCl pH 7.5, 75 mM NaCl, 0.5% NP-40, 5 mM MgCl2) on ice for 1-1.5 h prior to crosslinking. See supplementary experimental procedures for details.
Cell viability assay
5000 cells were plated in each well of a 96-well plate. Cells were treated with different concentrations of CdCl2 and CuCl2 in triplicate for 24 h. The number of viable cells was quantified using CellTiter-Glo® Luminescent Cell Viability Assay (Promega) according to the manufacturer's instructions. The effect of α3 shRNA on cell viability was determined 48 h after plating the cells. The values reported are the mean ± SD of multiple independent experiments.
In-gel substrate overlay proteasome activity assay
Substrate overlay assay was performed as described previously (Hochstrasser and Funakoshi, 2012). To visualize the pepidase activity of intact 26S proteasomes, cell lysates were resolved by native PAGE. The gel was incubated in developing buffer (50 mM Tris-HCl pH 7.5, 1 mM ATP, 5 mM MgCl2, 10% glycerol) along with 0.1 mM of fluorogenic substrate N-succinyl-Leu-Leu-Val-Tyr-AMC (Suc-LLVY-AMC, Sigma) for 30 min at 30°C (yeast) or 37°C (mammalian cells). Fluorescence (proteasome activity) was visualized by exposing the gel to UV light on a UV transilluminator.
In vitro proteasome activity assay
To perform proteasome activity assays in crude lysates from mammalian cells, the cells were harvested and resuspended in lysis buffer (50 mM HEPES (pH 7.8), 10 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 250 mM sucrose, 5 mM EDTA). Cell lysis was achieved by sonicating twice for 10 sec (microtip setting, ∼2). Protein concentration of the supernatant was determined by Bradford assay (Bio-Rad Protein Assay Kit). Fifty μg protein were used per reaction in a 96-well plate. Reaction buffer (lysis buffer + 2 mM ATP) containing 0.1 mM Suc-LLVY-AMC was added to a final volume of 200 μL. Fluorescence was measured using a plate reader (SynergyMX, BioTek) at Ex380nm/Em460nm. The relative fluorescence intensity was calculated from the mean ± SE of three independent experiments.
Supplementary Material
Highlights.
Capacity to assembly an α4-α4 proteasome isoform is conserved from yeast to humans.
α3, α4, and PAC3 levels modulate α4-α4 proteasome assembly in humans.
Conditions favoring assembly of α4-α4 proteasomes are observed in certain cancers.
α4-α4 proteasome levels correlate with cellular resistance to toxic metal ions.
Acknowledgments
We thank Christian Schlieker for sharing reagents and laboratory resources, Anthony Koleske for pCDNA3.1-ABL-6xHis and pEYFP-ARG, and Narendra Wajapeyee for pBabe-puro-HA-BRCA1. We also thank Jason Berk and Yanjie Li for critically reading the manuscript and for helpful comments. This work was supported by NIH grants GM083050 and GM046904 to M.H.
Footnotes
Author Contributions: A.P., S.A.V., and M.H. designed and performed the experiments. A.P. and M.H. analyzed the results and wrote the manuscript.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Buac D, Shen M, Schmitt S, Kona FR, Deshmukh R, Zhang Z, Neslund-Dudas C, Mitra B, Dou QP. From bortezomib to other inhibitors of the proteasome and beyond. Current pharmaceutical design. 2013;19:4025–4038. doi: 10.2174/1381612811319220012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford LJ, Windrum P, Magill L, Melo JV, McCallum L, McMullin MF, Ovaa H, Walker B, Irvine AE. Proteasome proteolytic profile is linked to Bcr-Abl expression. Experimental hematology. 2009;37:357–366. doi: 10.1016/j.exphem.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Emori Y, Tsukahara T, Kawasaki H, Ishiura S, Sugita H, Suzuki K. Molecular cloning and functional analysis of three subunits of yeast proteasome. Molecular and cellular biology. 1991;11:344–353. doi: 10.1128/mcb.11.1.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Funakoshi M. Disulfide engineering to map subunit interactions in the proteasome and other macromolecular complexes. Methods in molecular biology. 2012;832:349–362. doi: 10.1007/978-1-61779-474-2_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XT, Chen W, Wang D, Shi QL, Zhang FB, Liao YQ, Jin M, He C. The proteasome subunit PSMA7 located on the 20q13 amplicon is overexpressed and associated with liver metastasis in colorectal cancer. Oncology reports. 2008;19:441–446. [PubMed] [Google Scholar]
- Koleske AJ, Gifford AM, Scott ML, Nee M, Bronson RT, Miczek KA, Baltimore D. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 1998;21:1259–1272. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- Korkola JE, Houldsworth J, Chadalavada RS, Olshen AB, Dobrzynski D, Reuter VE, Bosl GJ, Chaganti RS. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer research. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nature structural & molecular biology. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Molecular cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. Journal of the American Society of Nephrology. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- Li D, Dong Q, Tao Q, Gu J, Cui Y, Jiang X, Yuan J, Li W, Xu R, Jin Y, et al. c-Abl regulates proteasome abundance by controlling the ubiquitin-proteasomal degradation of PSMA7 subunit. Cell reports. 2015;10:484–496. doi: 10.1016/j.celrep.2014.12.044. [DOI] [PubMed] [Google Scholar]
- Liou GY, Storz P. Reactive oxygen species in cancer. Free radical research. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Wang J. The resistance mechanisms of proteasome inhibitor bortezomib. Biomarker research. 2013;1:13. doi: 10.1186/2050-7771-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nature reviews. Molecular cell biology. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- Nair AR, Degheselle O, Smeets K, Van Kerkhove E, Cuypers A. Cadmium-Induced Pathologies: Where Is the Oxidative Balance Lost (or Not)? International journal of molecular sciences. 2013;14:6116–6143. doi: 10.3390/ijms14036116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemmiche S, Chabane-Sari D, Kadri M, Guiraud P. Cadmium chloride-induced oxidative stress and DNA damage in the human Jurkat T cell line is not linked to intracellular trace elements depletion. Toxicology In Vitro. 2011;25:191–198. doi: 10.1016/j.tiv.2010.10.018. [DOI] [PubMed] [Google Scholar]
- Osmulski PA, Hochstrasser M, Gaczynska M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure. 2009;17:1137–1147. doi: 10.1016/j.str.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panfair D, Ramamurthy A, Kusmierczyk AR. Alpha-ring Independent Assembly of the 20S Proteasome. Scientific reports. 2015;5:13130. doi: 10.1038/srep13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. BioEssays. 2008;30:1172–1184. doi: 10.1002/bies.20852. [DOI] [PubMed] [Google Scholar]
- Pourahmad J, O'Brien PJ. A comparison of hepatocyte cytotoxic mechanisms for Cu2+ and Cd2+ Toxicology. 2000;143:263–273. doi: 10.1016/s0300-483x(99)00178-x. [DOI] [PubMed] [Google Scholar]
- Ramos PC, Hockendorff J, Johnson ES, Varshavsky A, Dohmen RJ. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nature reviews Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Romanuik TL, Wang G, Morozova O, Delaney A, Marra MA, Sadar MD. LNCaP Atlas: gene expression associated with in vivo progression to castration-recurrent prostate cancer. BMC medical genomics. 2010;3:43. doi: 10.1186/1755-8794-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatterer SD, Acker CM, Davies P. c-Abl in neurodegenerative disease. Journal of molecular neuroscience : MN. 2011;45:445–452. doi: 10.1007/s12031-011-9588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes, chromosomes & cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- Stadtmueller BM, Hill CP. Proteasome activators. Molecular cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. The proteasome: from basic mechanisms to emerging roles. Keio journal of medicine. 2013;62:1–12. doi: 10.2302/kjm.2012-0006-re. [DOI] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annual review of biochemistry. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uechi H, Hamazaki J, Murata S. Characterization of the testis-specific proteasome subunit alpha4s in mammals. Journal of biological chemistry. 2014;289:12365–12374. doi: 10.1074/jbc.M114.558866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- Velichutina I, Connerly PL, Arendt CS, Li X, Hochstrasser M. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. The EMBO journal. 2004;23:500–510. doi: 10.1038/sj.emboj.7600059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Human molecular genetics. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.