Abstract
Background
Dove tree (Davidia involucrata Baill.) is a rare and endangered species. Natural reproduction of dove tree is extremely difficult due to its low fecundity. Serious seed abortion is one of the key factors restraining its sexual reproduction. Understanding the inducements of seed abortion is critical for addressing the issue of offspring production and the survivability of such an endangered species. However, studies on the molecular mechanism of seed abortion in woody plants are lacking, and the dearth of genomic resources for dove tree restricts further research.
Results
In this study, using the Illumina platform, we performed de novo transcriptome sequencing of the fruit and seed in dove tree. A total of 149,099 transcripts were isolated and then assembled into 72,885 unigenes. Subsequently, differentially expressed genes (DEGs) between normal and abortive seeds were screened. Genes involved in response to stress, hormone signal transduction, programmed cell death, lignin biosynthesis, and secondary cell wall biogenesis showed significant different expression levels between normal and abortive seeds.
Conclusion
Combined results indicated that the abortive seeds were under the adversity stress, which should be controlled by the maternal plant. Maternally controlled development of integument is assumed to be a critical process for abortion regulation. MYB and WRKY transcription factors, receptor kinase and laccase are considered to be important regulators in seed abortion. Moreover, mass sequence data facilitated further molecular research on this unique species.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0772-x) contains supplementary material, which is available to authorized users.
Keywords: Transcriptome, Adversity stress, Phytohormone, Seed abortion, Integument, Dove tree (Davidia involucrata Baill.)
Background
Davidia involucrata Baill., also known as dove tree or handkerchief tree, is a relic species of the Tertiary [1]. Davidia was a dominant part of the flora at many sites in the Paleocene of North America. However, it is demic to China today [1, 2]. It is considered by most researchers to be the sole member of the genus Davidia of the family Davidiaceae [3]. The most special characteristics of dove tree are its head inflorescences and intriguing pair of white bracts. Davidia is also an endangered species that has been listed as a first-grade nationally protected plant of China [2]. Currently, the distribution of natural Davidia population is rare and scattered, mainly due to its rigorous ecotope demand and low fecundity. In China, distribution areas of natural dove tree population are continuously decreasing, and most natural populations present the “Inverted Pyramid” structure, which indicates population depression [4]. For dove tree resources conservation, introduction and artificial breeding techniques of Davidia have been studied in China since 1979 [5]. However, studies did not progress smoothly as Davidia sexual reproduction was seriously restricted by the extremely long dormancy periods and high abortion ratio of its seeds [5]. Generally, only 1–3 well-developed seeds could be found in a Davidia fruit. Our observation found the manner of seed abortion in Davidia was independent with temperature, precipitation, biennial cycle and genotype. Moreover, seed abortion occurred in other endangered tree species such as Caryocar brasiliense [6], Magnolia denudate [7] and Liriodendron chinense [8], implying conserved mechanisms of seed abortion existed within these rare species.
Flower, fruit and seed abortion is pervasive in the plant kingdom. Many plant species, especially perennials, produce far more flowers than fruits and more ovules than seeds [9]. The low seed to ovule and fruit to flower ratios cause poor fecundity in some long-living tree species [10]. Evolutionary hypotheses propose that this “surplus of flowers or ovules” is a bet-hedging strategy that accounts for variable and unpredictable environments [11]. Diverse explanations have been proposed to interpret the mechanism underlying this phenomenon, including resource limitation [12, 13], pollen deficiency [14, 15], sibling rivalry [16] and genetic load [10, 17, 18]. Seed abortion could occur at different developmental stages of the embryo due to genotype, low vigor, inferior position or pathogen infection [19]. Abortion is considered to be a potentially beneficial mechanism that increases progeny quality [11]. Recent reports suggest that seed abortion is a complex plant behavior triggered by internal and external conditional cues [20]. However, for endangered species, such abortion mechanisms seriously limit proliferation, cultivation and conservation.
Despite numerous studies on seed abortion, most are focused on the physiological and morphological rather than molecular level. This is partly due to the fact that species with serious seed abortion are usually non-model plants, leading to a lack of genomic data. Recently research has focused on the genes and proteins involved in seed abortion in longan [21], peanut [22], chrysanthemum [23] and hazelnut [24] using transcriptome and proteome analysis.
To reveal the molecular events occurring in abortive seeds of Davidia, we used the Illumina platform and de novo sequenced the transcriptome to establish the first unigene library of fruit and seed of Davidia. Moreover, we identified the differentially expressed genes (DEGs) between normal and abortive seeds. Genes involved in cell proliferation, DNA replication, nutrient reservoir activity, and starch and sucrose metabolism were found to have significantly higher expression in normal seeds. In contrast, genes involved in response to stress, oxidoreductase activity, secondary metabolites biosynthesis and programmed cell death were found to be uniformly up-regulated in abortive seeds. DEGs encoding transcription factors, receptor kinase, proteinase and laccase were presumed to be critical regulators in seed abortion. These findings will bring valuable insight to the molecular regulatory mechanism of seed abortion in woody perennials.
Results
Seed abortion in Davidia
In order to investigate seed abortion situation in Davidia, we collected approximately 400 fruits from more than ten individual trees and recorded the numbers of normal and abortive seeds in them. The fruit of Davidia has an 8-carpel structure (sometimes 1–2 carpels degenerated from pistil development). In most fruits, the number of normal seeds was 1–3. As such, more than half of the seeds were aborted (Fig. 1a). The correlation between fruit weight and abortion ratio, and length-width ratio of fruit and abortion ratio were statistically analyzed, respectively. The results showed no significant correlation between either of them, indicating that seed abortion occurred at the early developmental stage and was independent to fruit development (Fig. 1b and c). We found fruits with normal seed numbers ranging from 1–8, indicating that all ovules had potency to develop well (Fig. 2c-i). Unlike some legume plants, the distances between the stigma and each ovule were approximately equivalent in Davidia so all ovules had equal opportunities for nutrient uptake. Position effect, a key cause of seed abortion in some legume plants, could be eliminated in Davidia. Moreover, the abortion was observed to occur in either consecutive or interval carpels, implying no obvious competition among siblings (Fig. 2d-h). For appearance, the well-developed seeds were spindle-shaped, white in color and rich in fat while the abortive seeds were noticeably smaller and more shriveled, with seed coats that were tan in color (Fig. 2j-m).
Fig. 1.
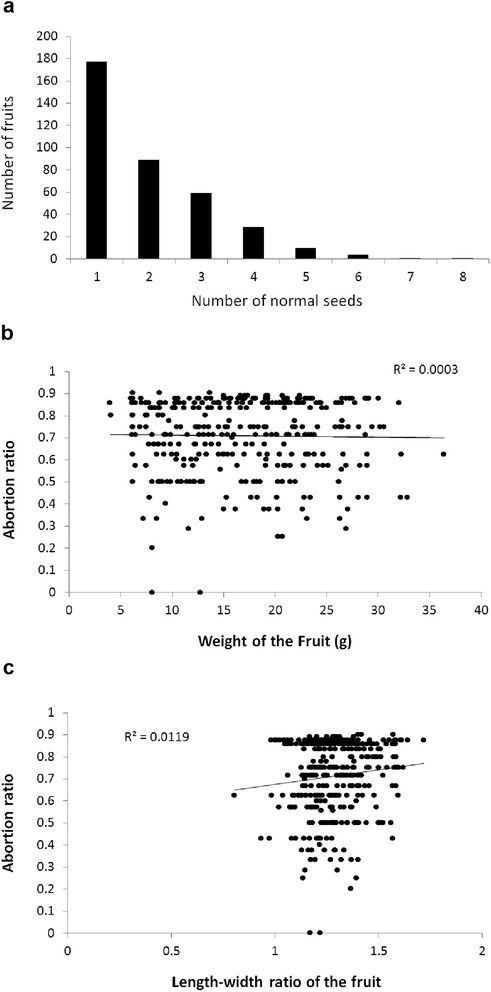
Seed abortion in Davidia. a Distribution of the numbers of normal seed in Davidia fruits; b Correlation between fruit weight and abortion ratio of seed; c Correlation between length-width ratio of fruit and abortion ratio of seed
Fig. 2.
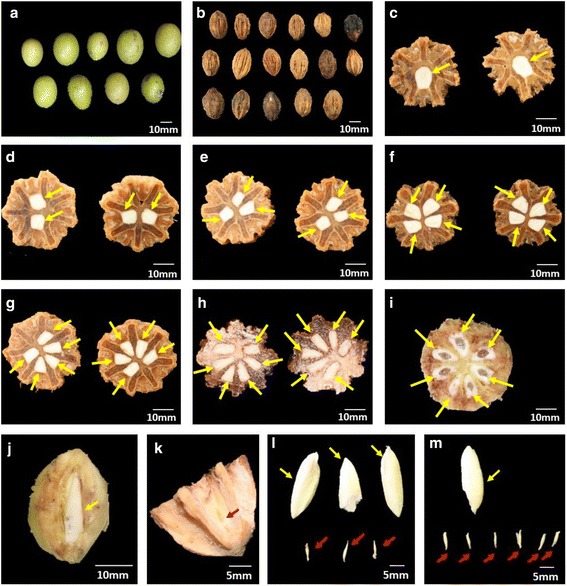
Fruits, normal seeds and abortive seeds of Davidia. a The intact fruits; b Fruits with sarcocarp removed; c-i Crosscutting sections of the kernels. The fruits contain 1 to 7 normal seeds are displayed in turn; j A normal seed in fruit; k An abortive seed in fruit; l Normal and abortive seeds collected from identical fruit. The fruit has 3 normal seeds and 3 abortive seeds; m Normal and abortive seeds collected from identical fruit. The fruit has 1 normal seeds and 6 abortive seeds. Normal and abortive seeds are represented by yellow and red arrows, respectively
We investigated the microstructure differences between normal and abortive seeds by microscopic observation. Paraffin sections demonstrated that the embryo sac in a normal seed was stacked well and the embryo was intact. On the contrary, in an abortive seed from the same fruit, the embryo sac was empty and flat, and the egg apparatus had been totally degenerated, indicating that the seed abortion occurred at the early stage of embryo development (Fig. 3).
Fig. 3.
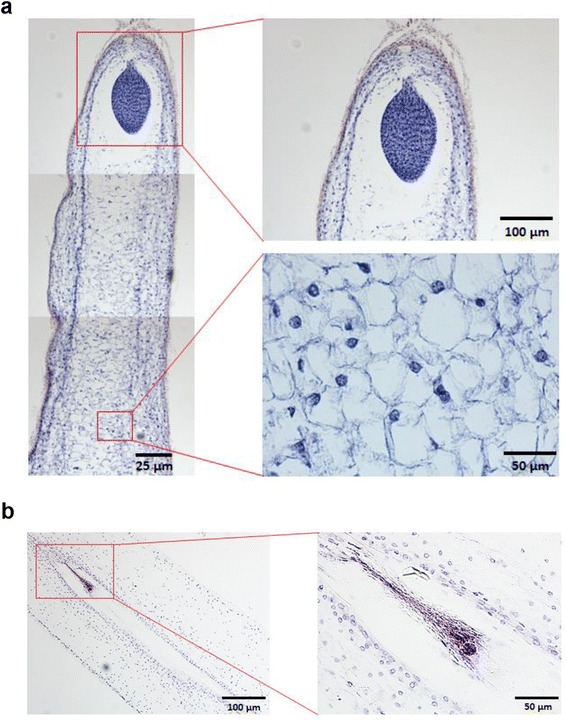
Microstructure of normal and abortive seeds. a The well-developed embryo and endosperm in normal seed. b The empty embryo sac and degenerative embryo in abortive seed
Overview of the Davidia transcriptome
One fruit sample, three normal seed samples and three abortive seed samples of Davidia were used to build a mixed library for high-throughput sequencing. RNA quality of each sample, represented by RNA integrity number (RIN), was 9.6 (Di-1 N), 9.1 (Di-1A), 9.5 (Di-2 N), 9.0 (Di-2A), 9.1 (Di-3 N), 9.1 (Di-3A) and 10.0 (Di-F), respectively. RNAs of different samples were mixed in equal quantities to construct the cDNA library. In total, the library produced 6,472,538,761 (6.47G) raw data with 89.2 % Q30 bases (percentage of sequences with sequencing error rates <0.1 %) by Illumina HiSeq 2500.
Using the Trinity de novo assembly program, short-read sequences were assembled into 149,099 transcripts with a mean length of 1,056.57 bp. The sequencing raw data was deposited to the NCBI Short Reads Archive (SRA) with the accession number SRP058736. The transcripts were then subjected to cluster and assembly analysis. Finally, we harvested a total of 72,885 unigenes with N50 length of 1150 and an average length of 656.61 bp (Additional file 1). An overview of the assembly contigs and unigenes is shown in Table 1.
Table 1.
Summary of Illumina transcriptome assembly for Davidia
| Length range | Contig | Transcript | Unigene |
|---|---|---|---|
| 200–300 | 3,948,088(98.63 %) | 35,712(23.95 %) | 29,975(41.13 %) |
| 300–500 | 25,956(0.65 %) | 26,996(18.11 %) | 18,692(25.65 %) |
| 500–1000 | 14,748(0.37 %) | 28,175(18.90 %) | 11,244(15.43 %) |
| 1000–2000 | 9,357(0.23 %) | 35,663(23.92 %) | 8,289(11.37 %) |
| 2000+ | 4,603(0.11 %) | 22,553(15.13 %) | 4,685(6.43 %) |
| Total Number | 4,002,752 | 149,099 | 72,885 |
| Total Length | 218,539,083 | 157,533,456 | 47,856,850 |
| N50 Length | 49 | 1,766 | 1,150 |
| Mean Length | 54.60 | 1,056.57 | 656.61 |
All the 73,885 assembled unigenes were searched against the Nr, Swiss-Prot, GO, COG and KEGG databases using the BLAST algorithm (E-value < 1E−5) (Table 2). Totally, 33,725 (45.6 %) unigenes were annotated (Additional file 2). Nr database queries revealed that a high percentage of Davidia sequences closely matched the sequences of Vitis vinifera (46.5 %), Theobroma cacao (11.2 %), Prunus persica (6.9 %), Populus trichocarpa (6.1 %), Solanum lycopersicum (5.7 %) and Ricinus communis (5.1 %) (Fig. 4). To ensure the accuracy of the annotation, the assembled unigenes were searched against the genomic database of Arabidopsis thaliana, Vitis vinifera, Theobroma cacao, Populus trichocarpa, Eucalyptus grandis and another relic species, Amborella trichopoda. 34.2 % - 39.9 % of total Davidia unigenes were annotated to the genomic data of these species (Additional file 3).
Table 2.
Summary for the annotation of unigenes of Davidia
| Annotated databases | Unigene | ≥300 nt | ≥1000 nt |
|---|---|---|---|
| COG | 8,375 | 7,605 | 5,268 |
| GO | 24,834 | 19,635 | 10,157 |
| KEGG | 6,257 | 5,162 | 2,980 |
| Swiss-Prot | 21,010 | 17,024 | 8,925 |
| Nr | 33,562 | 25,908 | 12,207 |
| All | 33,725 | 25,983 | 12,210 |
Fig. 4.
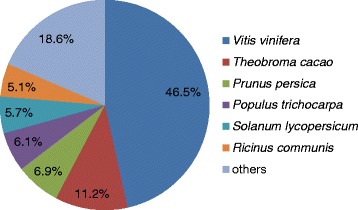
Species distribution of the BLASTX results of Davidia transcriptome. The numbers in the pies indicate the percentage of unique reads in each category
Among annotated unigenes, 24,834 unigenes were matched in the GO database and classified into 3 functional categories: molecular function (13,386, 53.9 %), biological process (4918, 19.8 %) and cellular component (6530, 26.3 %). “Binding”, “catalytic activity” and “transporter activity” were the largest GO terms of molecular function. “Metabolic process”, “cellular process” and “response to stimulus” were the largest GO terms of biological process. And “cell part”, “cell” and “organelle” were the largest GO terms of cellular component. KEGG pathway analysis showed 6257 unigenes were matched in the database and assigned to 116 KEGG pathways. The pathways containing the largest number of unigenes include “ribosome”, “plant hormone signal transduction”, “spliceosome”, “protein processing in endoplasmic reticulum”, “RNA transport”, “oxidative phosphorylation”, “purine metabolism”, “glycolysis/gluconeogenesis”, “starch and sucrose metabolism” and “plant-pathogen interaction”.
DEGs between normal and abortive seeds
A total of 61.27 M reads, including 3.12 G of raw data, were produced by RNA-seq. The high quality reads were aligned to the established Davidia unigene library and the proportions of mapped reads ranged from 74.3 % to 77.9 % (Table 3). The RPKM values of all genes were used to analyze the correlation between each of the two samples. The correlation coefficients between normal seed samples were higher than 0.82 (an exception is Di-2 N vs. Di-3 N, 0.78), and the correlation coefficients between abortive seed samples were higher than 0.94, indicating the slight variability among the biological replicates. The correlation coefficient between each normal seed sample and abortive seed sample was less than 0.10, indicating a significant expression difference (Additional file 4).
Table 3.
Summary for the alignment of reads to unigene library
| Sample | Total reads | Mapped reads | Unique mapped reads | Multiple mapped reads |
|---|---|---|---|---|
| Di-1N | 9,617,978 | 7,491,843(77.89 %) | 6,835,644(71.07 %) | 656,199(6.82 %) |
| Di-1A | 9,958,796 | 7,533,463(75.65 %) | 6,759,052(67.87 %) | 774,411(7.78 %) |
| Di-2N | 10,659,112 | 8,156,133(76.52 %) | 7,443,600(69.83 %) | 712,533(6.68 %) |
| Di-2A | 11,102,640 | 8,534,699(76.87 %) | 7,595,828(68.41 %) | 938,871(8.46) |
| Di-3N | 9,762,626 | 7,249,252(74.26 %) | 6,627,147(67.88) | 622,105(6.37 %) |
| Di-3A | 10,171,988 | 7,702,621(75.72 %) | 6,919,954(68.03 %) | 782,667(7.69 %) |
In total, 2770 DEGs were discovered between normal and abortive seeds. Among them, 978 genes were up-regulated and 1792 genes were down-regulated (Fig. 5, Additional file 5). 2631 DEGs were annotated by Nr, Swiss-Prot or genomic data of other species. Top 30 down-regulated and up-regulated genes are shown in Tables 4 and 5, respectively. A total of 1630 genes were annotated by GO. Compared to the unigene library, significantly enriched GO terms were found, such as “protein kinase binding”, “indole-3-acetic acid amido synthetase activity” and “peroxidase activity” in the “molecular function” category. “Cytokinesis by cell plate formation”, “regulation of DNA replication” and “cell proliferation” were found in the “biological process” category. And “nucleosome”, “chromocenter” and “microtubule associated complex” were found in the “cellular component” category. (Fig. 6). The top 50 GO terms for DEGs were shown in Fig. 7.
Fig. 5.
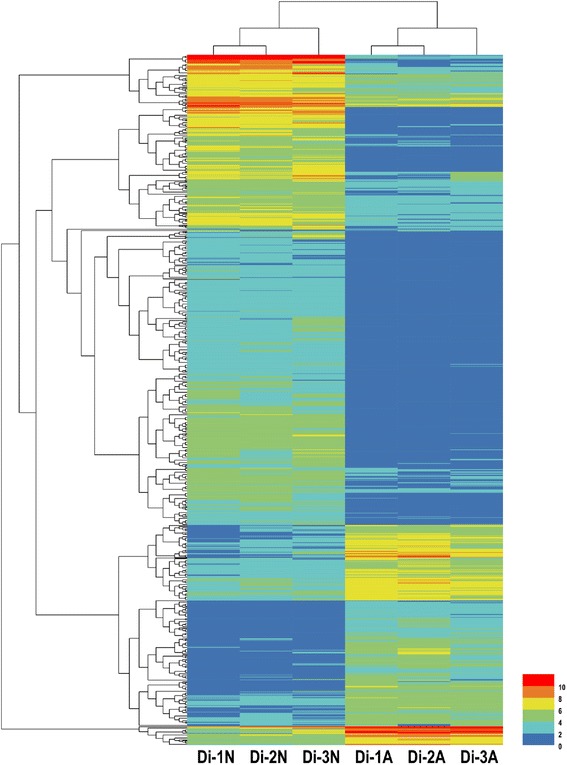
Expression pattern of DEGs in Davidia seeds. Hierarchical clustering analysis of DEGs between normal and abortive seeds based on expression data
Table 4.
Top 30 down-regulated DEGsR
| ID | FDR | log2FC | Species | Annotation |
|---|---|---|---|---|
| c21628.graph_c1 | 2.87E-08 | -11.11 | Capsella rubella | ECAGL3 - ECA1 gametogenesis related family protein precursor |
| c38878.graph_c0 | 1.56E-41 | -10.57 | Theobroma cacao | ABI3, putative isoform 1 |
| c34912.graph_c0 | 9.52E-41 | -10.48 | Populus trichocarpa | GDSL-like Lipase/Acylhydrolase superfamily protein |
| c20515.graph_c0 | 5.74E-7 | -10.47 | Populus trichocarpa | bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein |
| c32703.graph_c0 | 2.42E-15 | -10.35 | Vitis vinifera | Lipid transfer protein |
| c20459.graph_c0 | 1.33E-41 | -10.34 | Ricinus communis | GDSL-like Lipase/Acylhydrolase superfamily protein |
| c16559.graph_c0 | 9.77E-16 | -10.31 | Theobroma cacao | Nuclear factor Y, subunit C2 |
| c42905.graph_c0 | 8.30E-09 | -10.26 | Vitis vinifera | Late embryogenesis abundant protein-related/LEA protein-related |
| c17720.graph_c0 | 1.47E-31 | -10.23 | Ricinus communis | Uncharacterized protein |
| c28950.graph_c1 | 1.02E-15 | -10.13 | Glycine max | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein |
| c32557.graph_c0 | 3.54E-17 | -10.13 | Ricinus communis | pfkB-like carbohydrate kinase family protein |
| c17814.graph_c0 | 2.49E-08 | -10.10 | Vitis vinifera | PEBP (phosphatidylethanolamine-binding protein) family protein |
| c37825.graph_c0 | 2.46E-34 | -10.10 | Vitis vinifera | PA-domain containing subtilase family protein |
| c37890.graph_c0 | 3.95E-09 | -10.05 | Glycine max | Early nodulin-like protein 9 |
| c40930.graph_c0 | 4.57E-28 | -10.04 | Vitis vinifera | Core-2/I-branching |
| Beta-1,6-N-acetylglucosaminyltransferase family protein | ||||
| c40314.graph_c0 | 5.25E-27 | -9.99 | Vitis vinifera | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| c26337.graph_c0 | 1.44E-20 | -9.95 | Arabidopsis thaliana | 2S sulfur-rich seed storage protein |
| c17716.graph_c0 | 2.29E-28 | -9.94 | Vitis vinifera | Uncharacterized protein |
| c39633.graph_c1 | 2.40E-11 | -9.92 | Sesamum indicum | RmlC-like cupins superfamily protein |
| c46499.graph_c1 | 2.81E-42 | -9.87 | Petunia integrifolia subsp. inflata | RmlC-like cupins superfamily protein |
| c17661.graph_c0 | 3.02E-10 | -9.83 | Solanum tuberosum | Aluminium induced protein with YGL and LRDR motifs |
| c17693.graph_c0 | 3.89E-23 | -9.83 | Magnolia salicifolia | RmlC-like cupins superfamily protein |
| c37540.graph_c1 | 1.61E-10 | -9.81 | Setaria italica | PREDICTED: ZF-HD homeobox protein |
| c46966.graph_c0 | 7.42E-45 | -9.80 | Prunus persica | Aquaporin-like superfamily protein |
| c47330.graph_c0 | 1.15E-10 | -9.80 | Vitis vinifera | uncharacterized protein |
| c42176.graph_c0 | 1.20E-34 | -9.77 | Vitis vinifera | PREDICTED: hydroxycinnamoyl-Coenzyme A shikimate/quinate hydroxycinnamoyltransferase |
| c40562.graph_c0 | 6.40E-09 | -9.75 | Fragaria vesca subsp. vesca | Uncharacterized protein |
| c18277.graph_c0 | 1.16E-11 | -9.69 | Solanum lycopersicum | Homolog of Medicago truncatula MTN3 |
| c32587.graph_c0 | 2.25E-37 | -9.65 | Populus trichocarpa | Uncharacterized protein |
| c17717.graph_c0 | 1.14E-17 | -9.64 | Arabidopsis thaliana | Seed storage albumin 5 |
Table 5.
Top 30 up-regulated DEGs
| ID | FDR | log2FC | Species | Annotation |
|---|---|---|---|---|
| c29359.graph_c0 | 0.003579 | 8.84 | Vitis vinifera | FAD-binding Berberine family protein |
| c40611.graph_c1 | 0.009777 | 8.36 | Fragaria vesca subsp. vesca | PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like |
| c15946.graph_c0 | 0.000386 | 7.82 | Fagus crenata | Transcription factor MYB251 |
| c40565.graph_c0 | 8.72E-13 | 7.81 | Solanum lycopersicum | Soybean gene regulated by cold-2 |
| c31002.graph_c0 | 0.002082 | 7.78 | Vitis vinifera | Heavy metal transport/detoxification superfamily protein |
| c37038.graph_c1 | 3.80E-20 | 7.77 | Ricinus communis | Auxin-responsive GH3 family protein |
| c28235.graph_c0 | 0.001514 | 7.00 | Diospyros kaki | Putative MYB transcription factor |
| c37059.graph_c0 | 0.001769 | 6.80 | Populus trichocarpa | PLANT CADMIUM RESISTANCE 2 |
| c36067.graph_c0 | 3.16E-20 | 6.74 | Guillardia theta CCMP2712 | Hypothetical protein GUITHDRAFT_76875, partial |
| c15251.graph_c0 | 1.51E-08 | 6.69 | Vitis vinifera | Basic helix-loop-helix (bHLH) DNA-binding family protein |
| c49458.graph_c0 | 0.001387 | 6.65 | Theobroma cacao | Uncharacterized protein |
| c17425.graph_c0 | 1.54E-09 | 6.59 | Solanum lycopersicum | PREDICTED: CASP-like protein |
| c39858.graph_c0 | 2.63E-15 | 6.52 | Vitis vinifera | myb domain protein |
| c35722.graph_c0 | 0.004717 | 6.46 | Vitis vinifera | Laccase-14 |
| c21576.graph_c0 | 0.00219 | 6.45 | Vitis vinifera | Hexose transporter |
| c48162.graph_c0 | 1.82E-05 | 6.42 | Vitis vinifera | Respiratory burst oxidase protein F |
| c35307.graph_c0 | 0.009666 | 6.40 | Vitis cinerea var. helleri x Vitis riparia | Tumor-related protein |
| c15785.graph_c0 | 0.000308 | 6.35 | Vitis vinifera | myb domain protein |
| c33981.graph_c0 | 1.23E-05 | 6.26 | Vitis vinifera | Nitrate transmembrane transporters |
| c21046.graph_c0 | 6.81E-06 | 6.24 | Vitis vinifera | Allergen-related protein |
| c39360.graph_c1 | 5.14E-14 | 6.17 | Vitis vinifera | Squamosa promoter-binding-like protein 8 |
| c17796.graph_c0 | 3.31E-08 | 6.16 | Populus trichocarpa | Uncharacterized protein |
| c8841.graph_c0 | 3.04E-07 | 6.12 | Vitis vinifera | PREDICTED: protein MKS1-like |
| c36602.graph_c0 | 8.46E-09 | 6.11 | Vitis vinifera | Lateral organ boundaries (LOB) domain family protein |
| c45306.graph_c0 | 2.67E-13 | 6.09 | Vitis vinifera | Seven transmembrane MLO family protein |
| c32231.graph_c0 | 0.000386 | 6.01 | Prunus persica | Uncharacterized protein |
| c38398.graph_c1 | 7.24E-20 | 5.99 | Populus trichocarpa | Putative membrane lipoprotein |
| c19374.graph_c0 | 3.32E-07 | 5.99 | Theobroma cacao | myb domain protein |
| c46658.graph_c0 | 2.34E-26 | 5.95 | Theobroma cacao | Cytokinin oxidase 5 |
| c52887.graph_c0 | 6.95E-07 | 5.93 | Vitis vinifera | FAD-dependent oxidoreductase family protein |
Fig. 6.
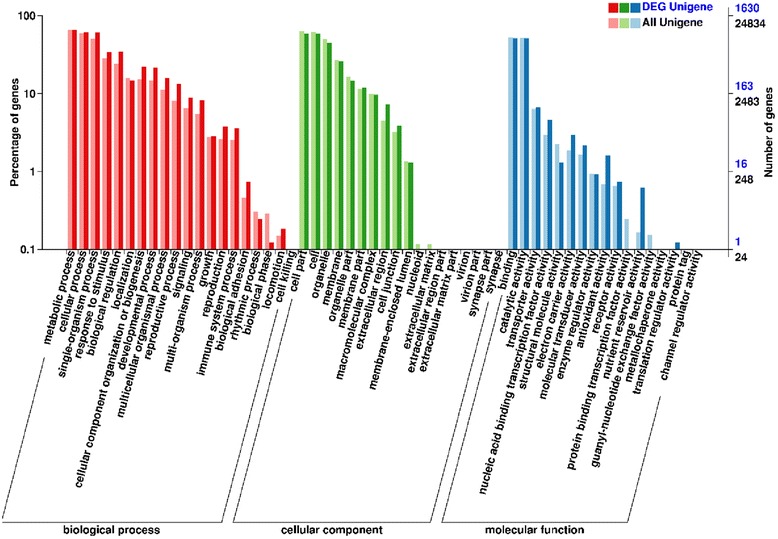
Gene Ontology classification of DEGs between normal and aborted seeds. Unigenes were annotated in three categories: cellular components, molecular functions, and biological processes. Numbers in black represent the numbers of all unigenes in GO terms, and numbers in blue represent the numers of DEGs in GO terms. DEGs were significantly enriched in the terms such as “biological adhesion”, “extracellular region”, “antioxidant activity” and “nutrient reservoir activity”
Fig. 7.
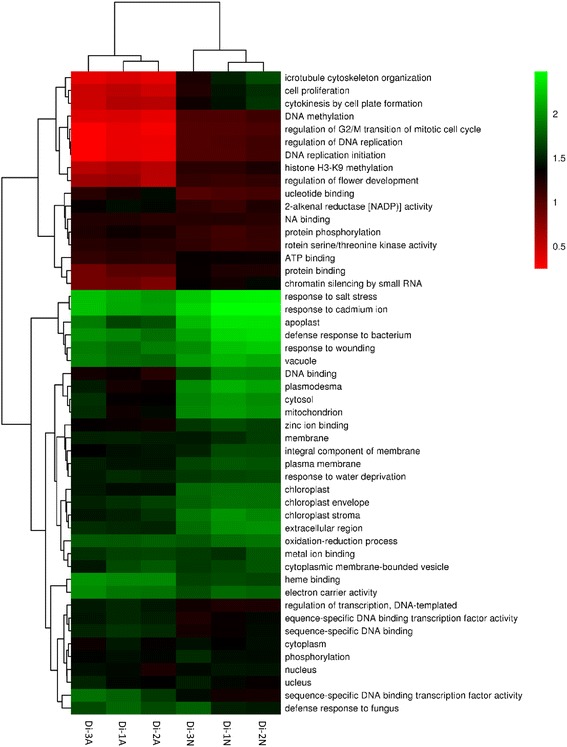
Top 50 different GO terms enriched in normal and abortive seeds. The heat map shows the fold change based on RPKM values of the DEGs in these GO terms in normal and aborted seeds. A q value cutoff of 0.05 calculated by the RPKM of all genes in corresponding GO terms was used to select enriched GO terms
Then DEGs were aligned to the KEGG database and assigned to 79 pathways. Among them, a large number of genes were involved in the pathways related to metabolism, such as “starch and sucrose metabolism”, “cysteine and methionine metabolism”, “pyruvate metabolism”, “pyrimidine metabolism”, “purine metabolism” and “phenylalanine metabolism”. A number of DEGs enriched in the pathways of genetic information processing, such as “ribosome”, “DNA replication” and “spliceosome” were involved. Another large group of DEGs were enriched in pathways of biosynthesis, including “phenylpropanoid biosynthesis”, “fatty acid biosynthesis”, “steroid biosynthesis”, “terpenoid backbone biosynthesis” and “zeatin biosynthesis”. These results were consistent with the status of normal and abortive seeds, which are quite different in nutrient accumulation, cell proliferation, tissue development and secondary metabolism. Remarkably, a number of genes were enriched in the pathways of “plant hormone signal transduction”, “plant-pathogen interaction”, “endocytosis” and “phagosome”, which are presumed to play critical roles in seed abortion regulation (Fig. 8).
Fig. 8.
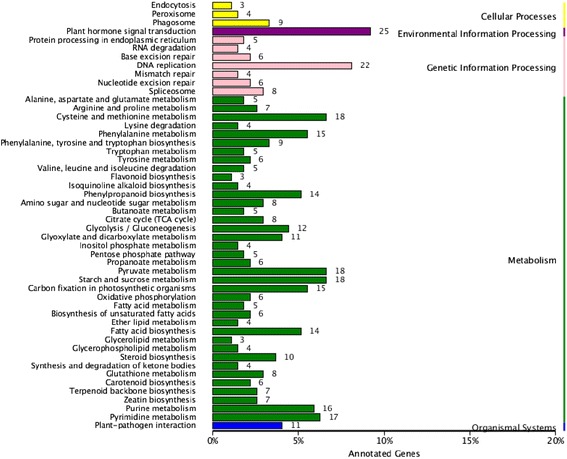
Top 50 enriched KEGG pathways between normal and abortive seeds. Number at the right of each column indicates the number of unigenes included in corresponding pathway. The pathways were divided into five categories: “cellular processes”, “environmental information processing”, “genetic information processing”, “metabolism” and “organismal systems”. A q value cutoff of 0.05 calculated by the RPKM of all genes in corresponding KEGG pathways was used to select enriched KEGG pathways
One thousand seventy-four DEGs were matched in the COG database. Similar to the results of GO and KEGG analysis, COG analysis showed that several DEGs were enriched in the biological processes such as “transcription”, “replication, recombination and repair”, “signal transduction mechanisms” and “carbohydrate transport and metabolism”. On the contrary, the fewest DEGs were enriched in “cell motility”, “intracellular trafficking, secretion, and vesicular transport” and “nucleotide transport and metabolism”. Compared to COG analysis of all unigenes, DEGs were significantly enriched in the terms such as “cell cycle control, cell division, chromosome partitioning”, “lipid transport and metabolism” and “secondary metabolites biosynthesis, transport and catabolism”, while less enriched in the terms of “translation, ribosomal structure and biogenesis”, “posttranslational modification, protein turnover, chaperones” and “intracellular trafficking, secretion and vesicular transport” (Fig. 9).
Fig. 9.
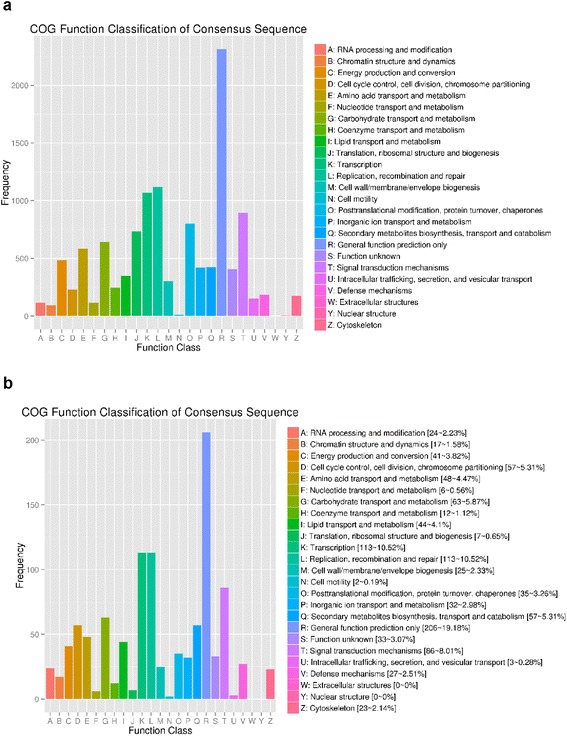
Cluster of orthologous groups (COG) classification. a In total, 8,375 of 72,885 unigenes were grouped into 25 COG classifications. b 1,074 of the 2,770 DEGs were grouped into 25 COG classifications. DEGs were enriched in the biological processes such as “transcription”, “replication, recombination and repair”, “signal transduction mechanisms” and “carbohydrate transport and metabolism”. Compared to COG analysis of all unigenes, DEGs were significantly enriched in the terms such as “cell cycle control, cell division, chromosome partitioning”, “lipid transport and metabolism” and “secondary metabolites biosynthesis, transport and catabolism”, while less enriched in the terms of “translation, ribosomal structure and biogenesis”, “posttranslational modification, protein turnover, chaperones” and “intracellular trafficking, secretion and vesicular transport”
Functional analysis of DEGs
DNA replication and cell proliferation are seriously impaired in abortive seeds
Among the DEGs, all genes encoding DNA polymerase alpha catalytic subunit, DNA replication licensing factor, ATP-dependent DNA helicase, DNA topoisomerase, DNA mismatch repair protein and condensin complex subunit showed dramatically decreased transcript abundance in abortive seeds. Consistently, genes encoding histone, chromatin assembly factor, structural maintenance of chromosomes protein and mini-chromosome maintenance complex-binding protein showed uniformly decreased expression in abortive seeds.
Genes involved in cytokinesis and microtubule cytoskeleton organization, including kinesin-like protein, 125 kDa kinesin-related protein, early nodulin-like protein, 65-kDa microtubule-associated protein, microtubule-associated protein RP/EB family, DNA (cytosine-5)-methyltransferase, high mobility group B protein, MAR-binding filament-like protein, callose synthase, thaumatin-like protein and tubulin showed significantly decreased expression in abortive seeds.
Cell cycle was observed to be disturbed in abortive seeds for nine genes encoding cyclin, four genes encoding G2/mitotic-specific cyclin, two genes encoding cyclin-dependent kinase and eight genes encoding formin-like protein, which were globally down-regulated to a large extent in abortive seeds.
Fatty acid, starch and sucrose metabolism are at low levels in abortive seeds
Fatty acid content is significantly different between normal and abortive seeds (unpublished data). The high content of fatty acid in normal Davidia seed might explain how it survived the Tertiary. Genes involved in fatty acid biosynthesis, including acetyl-coenzyme A carboxylase carboxyl transferase, hydroxyacyl-ACP dehydratase, lipoxygenase, acyl carrier protein, long chain acyl-CoA synthetase and protein ECERIFERUM (which are highly expressed in normal seeds) are uniformly down-regulated in abortive seeds. Furthermore, genes encoding products involved in unsaturated fatty acid biosynthetic process, such as cycloartenol synthase, dihydrolipoyllysine-residue acetyltransferase, peroxygenase, acyl-[acyl-carrier-protein] desaturase, omega-3 fatty acid desaturase and omega-6 fatty acid desaturase are consistently down-regulated in abortive seeds (Fig. 10).
Fig. 10.
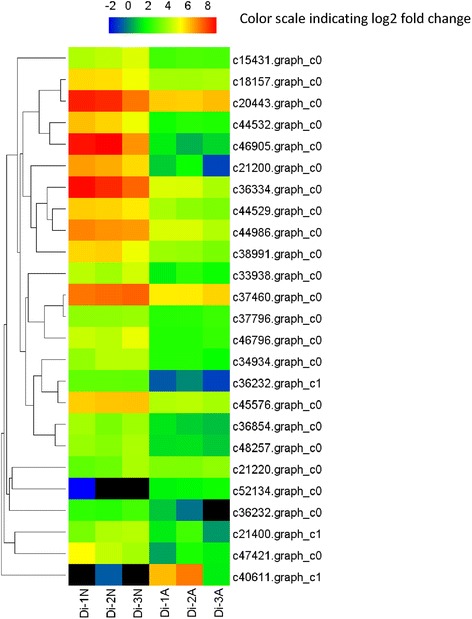
Expression patterns of genes involved in fatty metabolism. The codes of transcripts are indicated at the side of each step. The sample names are showed at the bottom. Black blocks represent that the expression is undetected
Genes involved in starch biosynthesis and catabolism, such as fructokinase, Alpha-xylosidase, granule-bound starch synthase, catalase and beta-amylase show uniformly decreased transcript abundance, indicating a low starch level in abortive seeds. For sucrose biosynthesis, 3 genes encoding sucrose synthase are dramatically down-regulated in abortive seeds.
Nutrient and ion transport are possibly restricted in abortive seeds. Three genes encoding the bidirectional sugar transporter, three genes encoding the cationic amino acid transporter and three genes encoding the nitrate transporter are significantly down-regulated. Moreover, various genes encoding copper, polyol, boron, zinc, sulfate and potassium transporter, respectively, are also down-regulated at different levels. Moreover, three genes encoding aquaporin are down-regulated to undetected levels in abortive seeds. These results demonstrate that the basic nutrition supply is greatly impaired in abortive seeds.
Nutrient reservoir and seed development are constrained in abortive seeds
Almost all of the genes encoding seed storage protein, such as globulin, albumin, sulfur-rich seed storage protein and legumin, were found to be down-regulated at the largest degree among DEGs. A number of key regulators of embryo development, such as genes encoding B3 domain-containing protein, ZF-HD homeobox protein, LOB domain-containing protein and zinc finger CCCH domain-containing protein were dramatically down-regulated in abortive seeds. The significantly low expressions of these genes confirmed the developmental defects in abortive seeds.
Notably, two genes encoding protein TRANSPARENT TESTA 12, which is essential for cell elongation in the integument, showed significantly decreased expression. On the other hand, six genes encoding receptor-like protein kinase HAIKU2 (with an exception), which control endosperm growth and modulate integument cell elongation, showed increased expression.
Difference of plant hormone signal transduction between normal and abortive seeds
Seven genes encoding indole-3-acetic acid-amido synthetase were found in DEGs, five of them showed decreased expression and two of them showed increased expression. Seven genes encoding auxin response factor were found; five of them were down-regulated and two of them were up-regulated. Three genes encoding auxin-responsive protein were found; two of them showed decreased expression. Two genes encoding auxin efflux carrier component were down-regulated. Two genes encoding auxin-induced in root cultures protein were up-regulated.
Three genes encoding gibberellin receptor were up-regulated. Three genes encoding gibberellin 2-beta-dioxygenase were up-regulated while a gene encoding gibberellin 3-beta-dioxygenase was down-regulated. Two genes encoding gibberellin 20 oxidase were undetected in abortive seeds. A number of genes response to gibberellin, such as two genes encoding monogalactosyldiacylglycerol synthase, a gene encoding transcription factor HB29, two gene encoding transcription factor RAX2 and two genes encoding transcription factor TCP15 showed increased expression in abortive seeds.
Five genes encoding cytokinin dehydrogenase/oxidase were found among DEGs; three of them showed increased expression and two of them showed decreased expression. Nine genes encoding ethylene-responsive transcription factor were found, and eight of them showed increased expression with an exception. A gene encoding abscisic acid receptor and two genes encoding abscisic stress-ripening protein showed increased expression.
Genes involved in response to stress and reactive oxygen species scavenging
Genes response to biotic and abiotic stress included seven genes encoding protein phosphatase 2C, sixteen genes encoding WRKY transcription factor, three genes encoding sugar transport protein, seven genes encoding MYB transcription factor (with an exception), seven genes encoding LRR receptor-like serine/threonine-protein kinase, six genes encoding zinc finger protein ZAT and four genes encoding heavy metal-associated isoprenylated plant protein, showing increased transcript abundance in abortive seeds. These results indicated that abortive seeds were under adversity stress. These up-regulated genes, including various transcription factors and protein kinases, might have initiated corresponding pathways to restrain the growth of the seeds.
Biotic and abiotic stress often induced a high content of reactive oxygen species (ROS) in plants. Genes encoding reactive oxygen species scavengers included a gene encoding cationic peroxidase, four genes encoding respiratory burst oxidase homolog protein and two genes encoding reticuline oxidase-like protein, showing increased expression in abortive seeds. Eleven genes encoding peroxidase were found among DEGs; eight of them showed increased expression and three showed decreased expression.
Calcium may be an important second messenger in abortion regulation
A number of calcium related genes were found in DEGs. Four genes encoding calcium-binding protein CML, two genes encoding calmodulin-like protein, three genes encoding cation/calcium exchanger and a gene encoding autoinhibited calcium ATPase showed increased expression. Two genes encoding CBL-interacting serine/threonine-protein kinase 14 showed increased expression and one gene encoding CBL-interacting serine/threonine-protein kinase 7 showed decreased expression. On the other hand, a gene encoding calcium-dependent protein kinase, a gene encoding calmodulin-binding transcription activator and three genes encoding calreticulin showed decreased expression. These results implied that calcium played an important role in abortion regulation.
Cell apoptosis and programmed cell death in abortive seeds
A number of genes involved in cell apoptosis, such as a gene encoding BAG family molecular chaperone regulator and nine genes encoding CASP-like protein (with an exception) showed increased expression in abortive seeds. Ten genes encoding F-box protein that might have participated into the apoptosis process were also found. Six showed increased expression and four showed decreased expression.
Genes involved in programmed cell death, including six genes encoding aspartic proteinase, four genes encoding cysteine-rich receptor-like protein kinase, a gene encoding leucine-rich repeat receptor-like protein kinase PXL2 and two genes encoding NAC transcription factor, were found to be significantly up-regulated in abortive seeds. Remarkably, a gene encoding cysteine proteinase showed decreased expression, while three genes encoding cysteine proteinase inhibitor showed dramatically decreased expression, indicating subtle mechanisms in cysteine proteinase activity regulation.
Lignin biosynthesis and secondary cell wall biogenesis
Almost all DEGs involved in lignin biosynthesis showed significantly increased expression, including seven genes encoding laccase, a gene encoding caffeoyl-CoA O-methyltransferase, a gene encoding cinnamyl alcohol dehydrogenase, a gene encoding shikimate O-hydroxycinnamoyltransferase and a gene encoding COBRA-like protein. On the other hand, various genes involved in secondary cell wall biogenesis showed uniformly increased expression, including five genes encoding cellulose synthase A catalytic subunit, a gene encoding secondary cell wall-related glycosyltransferase, a gene encoding protein IRX15-like, a gene encoding UDP-glucuronate:xylan alpha-glucuronosyltransferase and two genes encoding NAC domain-containing protein. Some previously described genes, such as genes encoding COBRA-like protein, MYB transcription factor and laccase, were also involved in secondary cell wall biogenesis. Laccase family is also involved in the oxidation-reduction process. We compared the expression of all identified Davidia laccase genes between normal and abortive seeds, and the results demonstrated that they were significantly up-regulated in abortive seeds (Fig. 11). Moreover, we found eight genes encoding wall-associated receptor kinase were uniformly up-regulated in abortive seeds. These results indicated that lignin accumulation and cell wall organization were changed in abortive seeds, critically affecting the regulation of seed development.
Fig. 11.
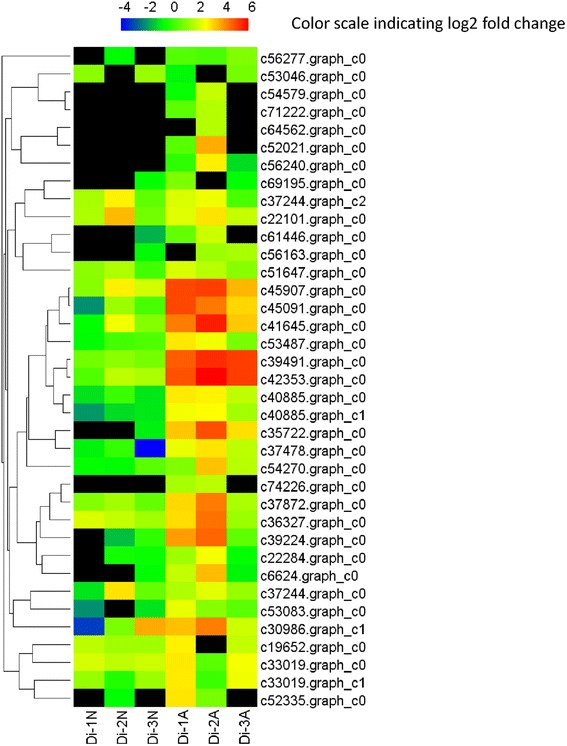
Expression pattern of laccase genes in normal and abortive seeds. The codes of transcripts are indicated at the side of each step. The sample names are showed at the bottom. Black blocks represent that the expression is undetected
Validation of differentially expressed genes by qPCR
To confirm the accuracy and reproducibility of the RNA-Seq results, 19 genes (detailed information is shown in Additional file 3: Table S1) with distinct expression profiles in normal and abortive seeds were chosen for qPCR validation. The expression levels of selected genes in normal and aborted seeds were detected by qPCR (Additional file 6). Correlation between RNA-Seq results (RPKM) and qPCR results (2-ΔΔCT) was calculated using the log2 fold change measurements to generate the scatterplots. The results showed that the qPCR results had significant similarity (r2 = 0.55) with the RNA-Seq data (Fig. 12).
Fig. 12.
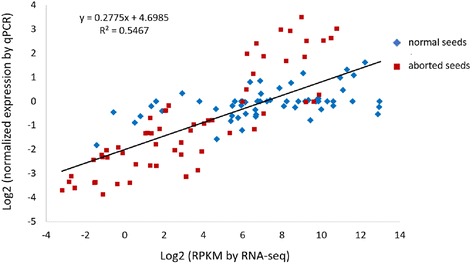
Correlation of gene expression results. The x-axis represents the value of Log2 RPKM and the y-axis represents the value of Log2 normalized expression level. Violet diamonds represent normal seeds. Red squares represent aborted seeds. R2 value represent the correlation between RNA-seq and qPCR results
Discussion
Davidia transcriptome
As an ancient relic species, Davidia has unique features in its fruits, seeds and bracts. The endosperm of normal Davdia seeds has abundant fatty acid and storage protein, which makes it nutritious and cold-resistant. During fruit development, lignin rapidly accumulates in the endocarp after approximately 20 days. This process makes the endocarp extremely hard and the space between the endocarp and seeds nearly contiguous (Fig. 2). These properties were presumed to be important reasons why Davida was able to survive the Quaternary Period [1]. Davidia transcriptome analysis showed that corresponding genes involved in fatty acid metabolism and lignin biosynthesis was highly expressed. High activity of secondary metabolites biosynthesis such as phenylpropanoid, flavonoid, terpene and steroid were indicated from the expression levels of related genes, implying that diverse secondary metabolites exist in Davidia. Compared to the genomic data of Arabidopsis thaliana, Vitis vinifera, Theobroma cacao, Populus trichocarpa, Eucalyptus grandis and Amborella trichopoda, Davidia sequences showed relatively low similarity to them (Additional file 4: Table S2), indicating its unique position in the evolution of angiosperm. Only 45.6 % transcripts were annotated, which was relatively low, suggesting that plenty of novel genes (which should include genes closely related to the unique phynotype of the fruit and seed of Davidia) were discovered from our data. In addition, approximately 0.2 % sequences were homologous to algae, bacteria, fungi and yeast, implying genetic integration events occurred, or multiple endogenous microorganism existed in Davidia.
Possible inducements of seeds abortion in Davidia
The serious seed abortion in Davida has been reconfirmed through our investigations. The breeding system of dove tree is an outcrossing type and partially self-compatible. It produces excessive flowers and pollens in the reproduction period, and the pollination rate is very high [25]. No abnormal development has been observed in gametogenesis or female and male gamete development [26]. Therefore, inbreeding depression is considered to be an important reason for abortion. The identified down-regulated genes, which are involved in energy and metabolic pathways, confirm the low vigor and rivalry power of abortive seeds, which might due to the genetic load caused by inbreeding.
The alteration of phytohormone levels was a significant sign in abortive seeds. From our results, major indole-3-acetic acid-amido synthetase genes and auxin response factor genes were significantly down-regulated. On the contrary, ethylene-responsive transcription factor genes and gibberellin receptor genes were globally up-regulated. These results implied that auxin levels were decreased, while gibberellin, ethylene and ABA levels were increased in abortive seeds. Gibberellins are essential regulators in plant development. Ectopic expression of a pea GA 2-oxidase2 cDNA induced gibberellin-deficiency and caused seed abortion in Arabidopsis [27]. It was also reported that high levels of gibberellin could cause seed abortion in grape [28]. Similarly, high levels of ethylene could induce kernel abortion and suppression of grain maturation in wheat [29]. However, the role of gibberellin and ethylene in seed formation and germination has remained controversial due to different results observed in different plant species [30]. ABA level was closely related to the response to stress in plants, and the increased ABA levels had been described in aborted seeds of maize and chrysanthemum [23, 31].
Signal transduction messagers involved in seed abortion
It is critical for maternal plants to recognize “bad offspring” and selectively restrain their growth. How the messages are exchanged between seeds and maternal plants remains unclear. According to our data, we infer that calcium ion is an important messenger in the abortion pathway. The increased expression of cation/calcium exchanger should change the Ca2+ levels in the seeds and regulate the activity of Ca2+-dependent protein kinases, subsequently controlling the seeds’ development [32].
Sucrose, which plays a critical role in seed development, is presumed to be another molecular messenger in abortion regulation. Suppression of sucrose synthase gene expression in cotton inhibits endosperm and embryo development and blocks the formation of adjacent seed integument transfer cells [33]. Overexpression of a potato sucrose synthase gene in cotton improved early seed development and reduced seed abortion [34]. We identified three genes encoding sucrose synthase that showed more than a 100-fold decreased expression in abortive seeds, demonstrating that the deficiency of sucrose synthase activity was critical for seed abortion in Davidia.
Programmed cell death in abortive seeds
Programmed cell death (PCD) is closely related to the vegetative and reproductive development of a plant [35]. Cysteine and aspartic proteinases are essential protelytic enzymes involved in PCD [36]. We identified five genes encoding aspartic proteinase that were uniformly up-regulated in abortive seeds, demonstrating the regulatory roles of this gene family in abortive seeds. Similar results were reported in comparative proteomic analysis of longan seed abortion, in which three cysteine protease protein were highly accumulated in abortive seeds at 50 d after pollution, suggesting that PCD was a common mechanism of seed abortion in different species [21]. Interestingly, we found one cysteine proteinase gene that was down-regulated in abortive seeds while three cysteine proteinase inhibitor genes were drastically down-regulated in abortive seeds. This finding indicated that cysteine proteinase activity was subtly regulated by its inhibitor in Davidia.
Lignin biosynthesis and seed integument development
We identified a series of genes related to secondary cell wall biogenesis and lignin biosynthesis that were significantly up-regulated in abortive seeds. Laccase genes were the most significant DEGs found to have uniformly improved expression levels in abortive seeds (Fig. 11). Laccase is a multiple function enzyme that can induce the flavonoid oxidation, which is also a resistance mechanism against biotic and abiotic stress [37]. The function of laccase was nonredundant with peroxidase for lignin polymerization [38], and most peroxidase genes were also significantly up-regulated in abortive seeds. A gene family encoding cellulose synthase A catalytic subunit, also involved in the lignin biosynthetic pathway, was found uniformly up-regulated in abortive seeds. Notably, some laccase and cellulose synthase were specially expressed in seed integument [37, 39]. Maternal control of integument cell elongation was validated to determine seed size in Arabidopsis [40]. Significantly decreased expression of protein TRANSPARENT TESTA indicated the development of integument was restricted in abortive seeds. Altogether, we assumed the growth of abortion seeds were controlled by maternal plants through the seed integument. The rapid accumulation of lignin or cellulose might have occurred in the seed integument, thus forming a compact and hard structure, which would restrain endosperm development.
Candidate regulators in seed abortion
For transcription factor, most stress-responsive transcription factors, such as AP2, MYB and WRKY transcription factor showed uniformly increased expression. Most development-related transcription factors, such as B3 domain-containing transcription factor, showed decreased expression in abortive seeds.
MYB domain protein was reported to act as a key determinant for proanthocyanidin accumulation [41]. Related genes, including three genes encoding anthocyanin regulatory C1 protein also showed increased expression. Proanthocyanidin accumulation was involved in seed integument development of Arabidopsis [42]. Some MYB transcription factors were also involved in lignification and secondary cell wall formation [43, 44].
Two Arabidopsis genes, MINISEED3 (MINI3) and HAIKU2 (IKU2), are proven regulators of seed size [45]. MINISEED 3 encodes Arabidopsis AtWRKY10, and a wrky10 mutant produces significantly smaller seeds. HAIKU2 encodes a protein kinase, and the haiku mutant produces seeds of reduced size, which results from impaired communication between the endosperm and maternal seed integument. Interestingly, among almost all WRKY genes and the six HAIKU2 genes in our DEG data, most genes showed increased expression. Only one HAIKU2 gene showed decreased expression. These findings implied different regulatory mechanisms in Davidia.
It is notable that most wall-associated receptor kinases, which are required for cell expansion and disease resistance [46], show uniformly increased expression in abortive seeds. Whether this gene family is involved in the signal transduction of abortion needs further investigation.
The genetic transformation system of Davidia is not available; therefore, further study on the function of the candidate gene, especially up-regulated transcription factors and gene families, should be performed in other species such as Arabidopsis. On the other hand, our data indicates limited nutrient and phytohormone regulation is essential for abortion. Therefore, exogenous nutrients and exogenous hormone imposing might be effective methods to alleviate abortion. If seed abortion in Davidia can be alleviated, it will bring great advantages for propagation and conservation of the tree.
Conclusion
De novo transcriptome sequencing of Davidia involucrata Baill. was performed in the present study using Illumina paired-end sequencing technology. In total, 72,885 unigenes from the fruits and seeds of Davidia were isolated. Focus on the regulatory mechanism of serious seed abortion in Davidia, the differentially expressed genes between normal and abortive seeds, were analyzed. We proposed that genetic load, resource limitation and phytohormone regulation were critical determinants for Davidia seed abortion. According to gene expression profiles, biological processes such as response to stress, starch and sucrose metabolism, PCD, secondary cell wall biogenesis and lignin biosynthesis were identified to be critical for abortion regulation. We assumed that maternally controlled development of integument was a critical process for abortion regulation. Calcium and sucrose were proposed to be important messengers in the abortion pathway. MYB and WRKY transcription factors, receptor kinase and laccase were identified as candidate regulators in seed abortion. The genomic data of Davidia will facilitate the further research on such endangered and low-fecundity species, and provide theoretical basis for protecting and utilizing these precious resources.
Methods
Plant materials
The fruits and seeds of Davidia were collected from three individual flowering trees of the naturally distributed Davidia population at Badagong Moutain Natural Reserve, Sangzhi County, Hunan Province (110°5’30”E, 28°46’60”N, 1383 m altitude). To eliminate genetic variance, we selected three trees that were grafted at the same time in 1983. The scions used for grafting were collected from the identical plant, known as the “King of Dove Trees”, the oldest dove tree in China (approximate age is 400 years). The fruits were collected on July 14, 2014, approximately 1 month after the bracts abscission, which represented the rough age of the seeds, or 60 to 90 days. The seeds were obtained by immediately dissecting the fruits after collection. Abortion ratio was calculated by NA/NT. (NA, number of abortive seeds in a fruit; NT, number of total seeds in a fruit). The normal seeds, abortive seeds and other parts of the fruit were separated and quickly frozen in liquid nitrogen and stored at −80 °C. Another group of the collected seeds were immediately fixed in formalin-aceto-alcohol (FAA) for microscopic observation. Three fruits of each tree were collected for seed sample preparation. Total normal seeds (3–5 grains) and total abortive seeds (15–20 grains) from the identical fruits were mixed and prepared as individual samples, respectively. The seed samples were named Di-1 N (mixed normal seeds from fruits of tree 1), Di-1A (mixed abortive seeds from the same fruits of tree 1), Di-2 N (mixed normal seeds from fruits of tree 2), Di-2A (mixed abortive seeds from the same fruits of tree 2), Di-3 N (mixed normal seeds from fruits of tree 3), and Di-3A (mixed abortive seeds from the same fruits of tree 3). All collected fruit samples (with seeds removed) were mixed and named as Di-F.
Microscopic observation
The normal and abortive seeds were fixed in FAA (50 % alcohol: acetic acid: formaldehyde solution = 89: 6: 5) immediately after dissection and stored at room temperature. Samples were washed in 50 % alcohol, dehydrated using an ethyl alcohol series, cleared in xylene and embedded in paraffin wax. The specimens were sectioned to a thickness of 8 μm. Sections were stained with hematoxylin, examined and photographed using an OLYMPUS BX-51 microscope.
RNA extraction,quantification and qualification
Total RNA was extracted using E.Z.N.A. Plant RNA Kit (Omega, R6827-01). RNA degradation and contamination was monitored on 1 % agarose gels. RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA concentration was measured using Qubit® RNA Assay Kit in Qubit®2.0 Flurometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Library construction and RNA-Seq
Construction of the library and RNA-Seq was performed by Biomarker Biotechnology Corporation (Beijing, China). RNA of the fruits, normal seeds and abortive seeds were combined in equal quantity to construct a large pool. Sequencing libraries were generated using NEBNext®Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. To select cDNA fragments at a preferential length of 150 ~ 200 bp, library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μl USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The cDNA library was sequenced on Illumina HiSeq™ 2500 using paired-end technology in a single run.
Sequence analysis and De novo assembly
Clean data was obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. The clean reads were assembled into contigs using the Trinity method, which recovers more full-length transcripts across a broad range of expression levels, with sensitivity similar to methods that rely on genome alignments [47]. We used the Trinity method with an optimized k-mer length of 25 for de novo assembly. Subsequently, the contigs were linked into transcripts according to the paired-end information of the sequences. Transcripts were then clustered based on nucleotide sequence identity. The longest transcripts in the cluster units were regarded as unigenes to eliminate redundant sequences, and then the unigenes were combined to produce the final assembly used for annotation.
Gene functional annotation
All the assembled unigenes were searched against the Nr (NCBI non-redundant protein sequences) database to identify the putative mRNA functions using the BLAST algorithm [48] with an E-value cut-off of 10−5. The BLAST algorithm was also used to align unique sequences to the Nt (NCBI non-redundant nucleotide sequences) and Swiss-Prot (a manually annotated and reviewed protein sequence database). Additionally, to improve the accuracy of the annotation, the assembled unigenes were aligned against the available genomic data of several species, including Arabidopsis thaliana (http://www.arabidopsis.org/), Populus trichocarpa, Vitis vinifera, Theobroma cacao, Eucalyptus grandis and Amborella trichopoda (http://phytozome.jgi.doe.gov/pz/portal.html).
GO (Gene Ontology) terms were extracted from the best hits obtained from the BLASTx against the Nr (non-redundant protein database) using the Blast2GO program [49]. COG (Clusters of Orthologous Groups of proteins) and KO (KEGG Ortholog database [50]) (with E-value cut-off of 10−5) analysis was conducted to predict possible functional classifications and molecular pathways.
Differential gene expression analysis
All reads from three normal seed samples and three abortive seed samples were mapped onto the nonredundant reference transcriptome by Tophat Bowtie software [51] to quantify the abundance of transcripts. Uni-transcript abundance differences between the samples were calculated based on the ratio of the RPKM values [52], and the false discovery rate (FDR). Differential expression analysis of normal and abortive seeds was performed using the DESeq R package (1.10.1). DESeq provided statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value <0.05 found by DESeq were assigned as differentially expressed. Uni-transcripts with an absolute value of log2 ratio ≥2 and an FDR significance score <0.001 was used for subsequent analysis. The identified DEGs were performed with GO, KEGG and COG analysis using the method described in the “Gene functional annotation” section. GO terms and KEGG pathways with a corrected P-value <0.05 (calculated by RPKM of genes involved in) were identified as differentially expressed.
qPCR analysis
The extracted RNA of seed samples were converted into cDNA using PrimeScriptTM One Step RT-PCR Kit Ver. 2 (Takara, Japan). Then the cDNA were 10 × diluted and used as templates for qPCR. qPCR reaction was performed using 2 × SYBR Green qPCR Master Mix (Biotool, USA) on ABI StepOneTM. Two independent biological replicates of each sample and three technical replicates of each biological replicate were used for qPCR analysis. A Davidia gene, DiActin, was used as the reference gene for data normalization. Primers used in qPCR are shown in Additional file 7. The relative expression fold of each sample was calculated by its CT value normalized to the CT value of reference gene using the 2-ΔΔCT method described by Livak and Schmittgen, 2001 [53]. The normalized values of relative expression and RPKM values were calculated by log2, respectively, and the values were used to analyze the correlation between qPCR and RNA-seq results.
Ethics
The authority responsible for the Davidia resources is the Badagong Mountain Nature Reserve Management Division, who provided permission to collect the samples for our scientific research.
Consent to publish
Not applicable.
Availability of data and materials
The sequencing raw data was deposited to the NCBI Short Reads Archive (SRA) with the accession number SRP058736. The BioSample accession is SAMN03733273 and the BioProject ID is PRJNA284915. The data was set to be released at 2018-5-24.
Acknowledgement
We thank Mr. Zhuang and Mr. Liao for sample collection. It is difficult and dangerous to climb trees more than 20 m tall for the fruits. We thank Axios Review for their valuable advices and professional service, which largely improved this paper. This work is supported by grants from the special funds of “The One-hundred-talents Scheme of Hunan Province” (112–0991) and Youth Foundation of Central South University of Forestry and Technology (QJ201512).
Abbreviations
- Davidia
Davidia involucrata Baill.
- DEG
differentially expressed gene
- PCD
programmed cell death
Additional files
Sequences of all assembled unigenes. (FA 48010 kb)
All annotated unigenes. (XLS 29890 kb)
Summary for the annotation of unigenes against database of other species. (PDF 9 kb)
Correlation coefficient between RPKM of unigenes of samples. (PDF 93 kb)
DEGs between normal and abortive seeds. (XLS 1148 kb)
Gene expression in normal and aborted seeds detected by qPCR. (PDF 313 kb)
Primer sequences used in qPCR. (PDF 34 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ML, FC and ZL conceived and designed the experiments. ML and XD performed the experiments. ML, XD, and JP analyzed the data. WX and XD provided the microstructure data. ML and RR provide the qPCR data. ML wrote the paper. ML, FC, ZL and JL revised and approved. All authors read and approved the final manuscript.
Contributor Information
Meng Li, Email: limeng0422@foxmail.com.
Xujie Dong, Email: 63930846@qq.com.
Jiqing Peng, Email: 125813393@qq.com.
Wen Xu, Email: 183659125@qq.com.
Rui Ren, Email: 1437154124@qq.com.
Jane Liu, Email: 3135118@qq.com.
Fuxiang Cao, Email: csfucao@163.com.
Zhiming Liu, Phone: +1 575 562 2495, Email: 13319516033@163.com.
References
- 1.Manchester SR. Leaves and fruits of Davidia (Cornales) from the Paleocene of North America. Syst Bot. 2002;27:368–82. [Google Scholar]
- 2.Fu L, Jin J. China plant red data book - rare and endangered plants (Volume 1) Beijing: Science Press; 1992. p. 44. [Google Scholar]
- 3.Li H. Davidia as the type of a new family Davidiaceae. Lloydia. 1954;17:329–31. [Google Scholar]
- 4.Zhang Q, Guo Q, Xu D, Yan H. Influence of climate changes on geographical distribution of Davidia involucrata, the precious and endangered species native to China. Scientia Silvae Sinicae. 2000;36(2):7–52. [Google Scholar]
- 5.Zhang J, Li J, Zhou B, Lian X. Natural distribution of Davidia involucrata and introduction analysis. J Beijing Forestry Univ. 1995;01:25–30. [Google Scholar]
- 6.Collevatti RG, Estolano R, Garcia SF, Hay JD. Seed abortion in the bat pollinated Neotropical tree species, Caryocar brasiliense (Caryocaraceae) Botany. 2009;87:1110–5. doi: 10.1139/B09-054. [DOI] [Google Scholar]
- 7.Wang R, Jia H, Wang J, Zhang Z. Flowering and pollination patterns of Magnolia denudata with emphasis on anatomical changes in ovule and seed development. Flora-Morphology, Distribution, Func Ecol Plants. 2010;205:259–65. doi: 10.1016/j.flora.2009.04.003. [DOI] [Google Scholar]
- 8.Huang SQ, Guo YH. Variation of pollination and resource limitation in a low seed-set tree, Liriodendron chinense (Magnoliaceae) Bot J Linn Soc. 2002;140:31–8. doi: 10.1046/j.1095-8339.2002.00080.x. [DOI] [Google Scholar]
- 9.Stephenson A. Flower and fruit abortion: proximate causes and ultimate functions. Annu Rev Ecol Syst. 1981;12:253–79. doi: 10.1146/annurev.es.12.110181.001345. [DOI] [Google Scholar]
- 10.Wiens D, Calvin CL, Wilson CA, Davern CI, Frank D, Seavey SR. Reproductive success, spontaneous embryo abortion, and genetic load in flowering plants. Oecologia. 1987;71(4):501–9. doi: 10.1007/BF00379288. [DOI] [PubMed] [Google Scholar]
- 11.Burd M. “Excess” flower production and selective fruit abortion: a model of potential benefits. Ecology. 1998;79(6):2123–32. [Google Scholar]
- 12.Lloyd DG. Sexual strategies in plants. New Phytol. 1980;86(1):69–79. doi: 10.1111/j.1469-8137.1980.tb00780.x. [DOI] [Google Scholar]
- 13.Nakamura RR. Seed abortion and seed size variation within fruits of Phaseolus vulgaris: pollen donor and resource limitation effects. Am J Bot. 1988;75:1003–10. doi: 10.2307/2443768. [DOI] [Google Scholar]
- 14.McDade LA. Pollination intensity and seed set in Trichanthera gigantea (Acanthaceae) Biotropica. 1983;15:122–4. doi: 10.2307/2387955. [DOI] [Google Scholar]
- 15.Burd M. Bateman’s principle and plant reproduction: the role of pollen limitation in fruit and seed set. Bot Rev. 1994;60(1):83–139. doi: 10.1007/BF02856594. [DOI] [Google Scholar]
- 16.Ganeshaiah KN, Shaanker RU. Seed abortion in wind-dispersed pods of Dalbergia sissoo: maternal regulation or sibling rivalry? Oecologia. 1988;77(1):135–9. doi: 10.1007/BF00380936. [DOI] [PubMed] [Google Scholar]
- 17.Krebs SL, Hancock JF. Embryonic genetic load in the high bush blueberry, Vaccinium corymbosum (Ericaceae) Am J Bot. 1991;78:1427–37. doi: 10.2307/2445281. [DOI] [Google Scholar]
- 18.Kärkkäinen K, Savolainen O, Koski V. Why do plants abort so many developing seeds: bad offspring or bad maternal genotypes? Evol Ecol. 1999;13(3):305–17. doi: 10.1023/A:1006746900736. [DOI] [Google Scholar]
- 19.Korbecka G, Klinkhamer PGL, Vrieling K. Selective embryo abortion hypothesis revisited - a molecular approach. Plant Biol. 2002;4(3):298–310. doi: 10.1055/s-2002-32331. [DOI] [Google Scholar]
- 20.Meyer KM, Soldaat LL, Auge H, Thulke HH. Adaptive and selective seed abortion reveals complex conditional decision making in plants. Am Nat. 2014;183(3):376–83. doi: 10.1086/675063. [DOI] [PubMed] [Google Scholar]
- 21.Liu H, Liu Y, Zheng S, Jiang J, Wang P, Chen W. Comparative proteomic analysis of longan (Dimocarpus longan Lour.) seed abortion. Planta. 2010;231:847–60. doi: 10.1007/s00425-009-1093-1. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Zhu W, Azam S, Li H, Zhu F, Li H, et al. Deep sequencing analysis of the transcriptomes of peanut aerial and subterranean young pods identifies candidate genes related to early embryo abortion. Plant Biotechnol J. 2013;11:115–27. doi: 10.1111/pbi.12018. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, Wang Z, Dong W, Sun C, Wang H, Song A, et al. Transcriptomic and proteomic analysis reveals mechanisms of embryo abortion during chrysanthemum cross breeding. Sci Rep. 2014;4:6536. doi: 10.1038/srep06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y, Liu J, Zhang H, Wang J, Zhao Y, Geng W. Transcriptome analysis and gene expression profiling of abortive and developing ovules during fruit development in hazelnut. PLoS One. 2015;10:e0122072. doi: 10.1371/journal.pone.0122072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng H, Su Z, Wang Y, Miao L, Shen Y. Study on flowering dynamics and breeding system of Davidia involucrata Baill. J Anhui Agr Sci. 2009;37(18):8445–8. [Google Scholar]
- 26.Li X, Chen F, Zhuang J, Li F. Cytological observation of mega-/micro-sporogenesis and female-/male-gametogenesis in Davidia involucrata Baill. J Zhejiang Agr Sci. 2008;5:546–50. [Google Scholar]
- 27.Singh DP, Jermakow AM, Swain SM. Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell. 2002;14(12):3133–47. doi: 10.1105/tpc.003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agüero C, Vigliocco A, Abdala G, Tizio R. Effect of gibberellic acid and uniconazol on embryo abortion in the stenospermocarpic grape cultivars Emperatriz and Perlon. Plant Growth Regul. 2000;30(1):9–16. doi: 10.1023/A:1006207614388. [DOI] [Google Scholar]
- 29.Hays DB, Do JH, Mason RE, Morgan G, Finlayson SA. Heat stress induced ethylene production in developing wheat grains induces kernel abortion and increased maturation in a susceptible cultivar. Plant Sci. 2007;172(6):1113–23. doi: 10.1016/j.plantsci.2007.03.004. [DOI] [Google Scholar]
- 30.Matilla AJ. Ethylene in seed formation and germination. Seed Sci Res. 2000;10(02):111–26. doi: 10.1017/S096025850000012X. [DOI] [Google Scholar]
- 31.Kakumanu A, Ambavaram MM, Klumas C, Krishnan A, Batlang U, Myers E, et al. Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 2012;160:846–67. doi: 10.1104/pp.112.200444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anil VS, Harmon AC, Rao KS. Spatio-temporal accumulation and activity of calcium-dependent protein kinases during embryogenesis, seed development, and germination in sandalwood. Plant Physiol. 2000;122:1035–44. doi: 10.1104/pp.122.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruan YL, Llewellyn DJ, Furbank RT. Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell. 2003;15(4):952–64. doi: 10.1105/tpc.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu SM, Brill E, Llewellyn DJ, Furbank RT, Ruan YL. Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Mol Plant. 2012;5(2):430–41. doi: 10.1093/mp/ssr090. [DOI] [PubMed] [Google Scholar]
- 35.Pennell RI, Lamb C. Programmed cell death in plants. Plant Cell. 1997;9(7):1157. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beers EP, Woffenden BJ, Zhao C. Plant proteolytic enzymes: possible roles during programmed cell death. 2000. pp. 155–71. [DOI] [PubMed] [Google Scholar]
- 37.Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I. Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci. 2007;12(1):29–36. doi: 10.1016/j.tplants.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, et al. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25(10):3976–87. doi: 10.1105/tpc.113.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harpaz‐Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, et al. Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J. 2011;68:941–53. doi: 10.1111/j.1365-313X.2011.04760.x. [DOI] [PubMed] [Google Scholar]
- 40.Garcia D, Gerald JNF, Berger F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell. 2005;17:52–60. doi: 10.1105/tpc.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell. 2003;15:2514–31. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C. Characterisation of a pine MYB that regulates lignification. Plant J. 2003;36:743–54. doi: 10.1046/j.1365-313X.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21(1):248–66. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:17531–6. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He ZH, He D, Kohorn BD. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 1998;14:55–63. doi: 10.1046/j.1365-313X.1998.00092.x. [DOI] [PubMed] [Google Scholar]
- 47.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korf I. Gene finding in novel genomes. BMC bioinformatics. 2004;5(1):1. [DOI] [PMC free article] [PubMed]
- 49.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–35. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(D1):D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C, Pachter L, Salzberg SL. Tophat: discovering splice junctions with RNA-seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing raw data was deposited to the NCBI Short Reads Archive (SRA) with the accession number SRP058736. The BioSample accession is SAMN03733273 and the BioProject ID is PRJNA284915. The data was set to be released at 2018-5-24.


