Abstract
Background
Vanillin is an industrially valuable molecule that can be produced from simple carbon sources in engineered microorganisms such as Saccharomyces cerevisiae and Escherichia coli. In E. coli, de novo production of vanillin was demonstrated previously as a proof of concept. In this study, a series of data-driven experiments were performed in order to better understand limitations associated with biosynthesis of vanillate, which is the immediate precursor to vanillin.
Results
Time-course experiments monitoring production of heterologous metabolites in the E. coli de novo vanillin pathway revealed a bottleneck in conversion of protocatechuate to vanillate. Perturbations in central metabolism intended to increase flux into the heterologous pathway increased average vanillate titers from 132 to 205 mg/L, but protocatechuate remained the dominant heterologous product on a molar basis. SDS-PAGE, in vitro activity measurements, and l-methionine supplementation experiments suggested that the decline in conversion rate was influenced more by limited availability of the co-substrate S-adenosyl-l-methionine (AdoMet or SAM) than by loss of activity of the heterologous O-methyltransferase. The combination of metJ deletion and overexpression of feedback-resistant variants of metA and cysE, which encode enzymes involved in SAM biosynthesis, increased average de novo vanillate titers by an additional 33 % (from 205 to 272 mg/L). An orthogonal strategy intended to improve SAM regeneration through overexpression of native mtn and luxS genes resulted in a 25 % increase in average de novo vanillate titers (from 205 to 256 mg/L). Vanillate production improved further upon supplementation with methionine (as high as 419 ± 58 mg/L), suggesting potential for additional enhancement by increasing SAM availability.
Conclusions
Results from this study demonstrate context dependency of engineered pathways and highlight the limited methylation capacity of E. coli. Unlike in previous efforts to improve SAM or methionine biosynthesis, we pursued two orthogonal strategies that are each aimed at deregulating multiple reactions. Our results increase the working knowledge of SAM biosynthesis engineering and provide a framework for improving titers of metabolic products dependent upon methylation reactions.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-016-0459-x) contains supplementary material, which is available to authorized users.
Keywords: E. coli, Vanillin, Methylation, S-adenosylmethionine, SAM, AdoMet, Methionine, Deregulation, Metabolic engineering
Background
Few biological molecules are as widely savored by humans as vanillin. Vanillin is the dominant flavor constituent in natural vanilla extract and the most heavily used food additive for flavoring annually by volume [1, 2]. Vanillin also has a variety of industrial uses, including as an intermediate in the chemical and pharmaceutical industries for production of herbicides, antifoaming agents, and drugs [3]. Biosynthesis of vanillin occurs naturally in the tropical vanilla orchid (Vanilla planifolia) and in trace amounts in other plants [3]. In V. planifolia, vanillin forms inside inedible pods or beans from which it must be extracted. Vanilla beans are harvested roughly 6–8 months after pollination and are subject to a curing process that can take more than 6 months to complete [3]. Inefficiencies stemming from V. planifolia cultivation are manifested in the high price of natural vanilla extract, which is estimated to be between $1000 and $4000 per kilogram [3]. Given that, researchers have sought to develop biosynthetic routes to vanillin in other host organisms. Ideally, these heterologous hosts could produce vanillin from glucose as a sole carbon source (de novo biosynthesis) given the abundance and affordability of glucose relative to other potential substrates [4].
De novo vanillin biosynthesis has been demonstrated in two of the industrial biotechnology workhorse organisms: Saccharomyces cerevisiae (baker’s yeast) [4] and Escherichia coli [5]. De novo production of vanillin in a heterologous host organism was first reported in both S. cerevisiae and Schizosaccharomyces pombe in 2009 [4]. The engineered metabolic pathway in yeast featured three heterologous enzymes: a dehydroshikimate dehydratase, an O-methyltransferase, and a carboxylic acid reductase. This pathway is similar to the E. coli pathway engineered by Frost and co-workers in 1998 [6]. The E. coli pathway was designed to produce vanillate in vivo using a heterologous dehydroshikimate dehydratase and O-methyltransferase. Subsequent reduction of purified vanillate into vanillin was catalyzed by a carboxylic acid reductase in vitro. The presumed rationale for the original two-stage vanillin pathway was that E. coli exhibits a high degree of aldehyde reductase activity catalyzed by numerous unknown endogenous enzymes, which would result in formation of vanillyl alcohol in vivo instead of vanillin. The problem of endogenous aromatic aldehyde reduction was recently addressed [5, 7], leading to a strain capable of accumulating aldehydes including vanillin (E. coli RARE, Addgene Catalog #61440). At that time, the de novo vanillin pathway was constructed in E. coli as one of two demonstrated and industrially relevant applications for the RARE strain.
When de novo biosynthesis was first reported in yeast and in E. coli, vanillin titers achieved were similarly low under comparable flask-scale conditions (65 mg/L in S. pombe supplied with yeast extract-based media [4] versus 119 or 56 mg/L in E. coli supplied with glucose in LB or M9 minimal media [5], respectively). Production of vanillin or vanillin-β-d-glucoside in yeast has since been enhanced [8, 9] and even commercialized by the company Evolva [10]. Numerous differences between eukaryotic yeast and prokaryotic E. coli led us to suspect that similar de novo pathways might suffer from different host-dependent limitations. Metabolic engineering reviews that discuss host selection not only compare natural differences in metabolic fluxes and regulation across model organisms such as yeast and E. coli [11] but also highlight dissimilarities in these respects among different species of yeast [12] and even among different strains of E. coli [13].
In this report, we describe a series of metabolic engineering experiments used to improve the production of vanillate from glucose as a sole carbon source using E. coli. We initially focus on increasing flux into the engineered pathway by perturbing pool sizes of precursor molecules and observing the effect on heterologous metabolite titers as a function of time in order to identify pathway bottlenecks downstream. To simplify our analysis, we exclude the confounding presence of aldehyde products, which exert negative effects on titer by inhibiting cell growth [5, 14]. We achieve this by omitting the carboxylic acid reductase CarNi from the pathway, which results in vanillate as the intended final product rather than vanillin (Fig. 1a).
Fig. 1.
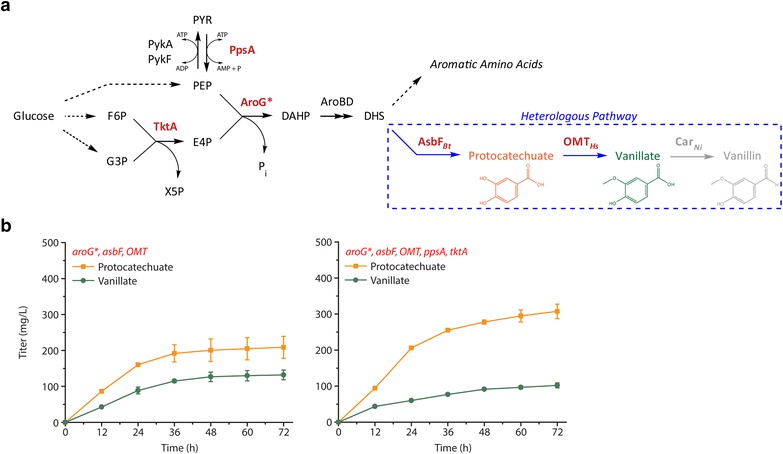
Conversion of protocatechuate to vanillate limits engineered de novo vanillin biosynthesis in E. coli. a Metabolic pathway diagram depicting engineered route from glucose to vanillin. Genes corresponding to enzymes labeled in red are overexpressed in this study. Enzymes written without subscripts are native to E. coli. Dashed blue lines indicate the heterologous portion of the pathway. The gene corresponding to CarNi (shown in gray) is not overexpressed throughout this study to avoid confounding presence of aldehydes during pathway troubleshooting. Thus, only solid blue arrows represent heterologous reaction steps pertaining to this study, where vanillate is the desired end product. Maximum theoretical yields of vanillin and vanillate using this pathway are 0.395 and 0.414 mol/mol_glucose, respectively. b Time-course experiment in which the PTS− glu+ RAREʹ host strain was transformed to express aroG*, asbF Bt, and OMT Hs or ppsA and tktA in addition to the other genes. Titers of heterologous metabolites protocatechuate and vanillate were measured every 12 h by HPLC and reveal bottleneck in conversion of protocatechuate to vanillate. Genes overexpressed in each experiment are labeled in red. Titers of protocatechuate increase by roughly 50 % when ppsA and tktA are overexpressed, suggesting flux into the heterologous pathway increased. However, vanillate titers do not improve, motivating subsequent focus on conversion of protocatechuate to vanillate
Our findings indicate that limited availability of S-adenosylmethionine (SAM) represents a major hurdle to the improvement of vanillate titers from glucose as a sole carbon source in E. coli. To improve vanillate titers, we deregulate reactions in SAM biosynthesis and regeneration using two orthogonal strategies. Improvements in vanillate formation as a function of increased SAM availability have impact beyond the vanillin pathway given that SAM-dependent methyltransferases constitute a broad class of enzymes with potential for engineering numerous metabolic products [15, 16]. Our results increase the working knowledge of SAM biosynthesis engineering and provide a framework for improving titers of metabolic products dependent upon methylation reactions.
Results
Time-course experiments monitoring heterologous metabolites reveal limitation in conversion of protocatechuate to vanillate
As a starting point for understanding pathway limitations, we sought to increase carbon flux entering the heterologous portion of the vanillate pathway (Fig. 1a). We hypothesized that increasing flux would facilitate the identification of pathway bottlenecks based on changes in titer (units of mg/L) and specific yields [units of g/gDCW (grams of dry cell weight)] of intermediate metabolites. In our previous demonstration of de novo vanillin biosynthesis, titers of all measured heterologous metabolites (protocatechuate, vanillate, protocatechualdehyde, and vanillin) were low, each less than 60 mg/L [5]. Initially, we sought to achieve increased heterologous metabolite titers by targeting endogenous precursor biosynthesis genes for overexpression given that the first heterologous enzyme (AsbFBt) is reported to efficiently catalyze conversion of endogenous 3-dehydroshikimate into protocatechuate [17].
The heterologous vanillate pathway branches from endogenous aromatic amino acid biosynthesis, and efforts to improve aromatic amino acid biosynthesis in E. coli have been well-documented [18–20]. In our initial demonstration of vanillin synthesis, we harnessed one of these strategies by expressing a feedback-resistant variant of the enzyme that catalyzes the first committed step to aromatic amino acid biosynthesis (aroG*) [5]. Previous studies demonstrated two other strategies that were successfully used to increase titers of aromatic products, in these cases by increasing availability of two key aromatic precursor metabolites: phosphoenolpyruvate (PEP) from glycolysis and erythrose-4-phosphate (E4P) from the pentose phosphate pathway. PEP and E4P condense to form DAHP during the first committed step towards aromatic amino acid biosynthesis (Fig. 1a) [18–20]. One reported strategy to increase their availability is deletion of the phosphotransferase system (PTS), which is the primary means for glucose import and consumes 1 mole of PEP per mole of glucose. Growth of a PTS− strain on glucose as a sole carbon source can be made viable by upregulating the gene encoding galactose permease (galP), which allows glucose entry independent of PEP consumption (PTS− glu+). A second documented strategy to increase availability of PEP and E4P is to overexpress the genes encoding PEP synthase (ppsA) and transketolase (tktA) [18–20]. PEP synthase catalyzes the conversion of PEP into pyruvate, and transketolase catalyzes the reversible formation of E4P and xylulose 5-phosphate from fructose 6-phosphate and glyceraldehyde 3-phosphate.
To test these strategies, we engineered a PTS− glu+ variant of the RARE strain (PTS− glu+ RAREʹ), where the “RARE prime” designation indicates that this strain contains intact versions of two genes (dkgB and yeaE) that were targeted for deletion in the original RARE strain. Previous work found that these two genes did not substantially contribute to aldehyde reductase activity given their low native expression [5]. When the PTS− glu+ RAREʹ host overexpressed the aroG*, asbFBt, and OMTHs genes, 132 ± 14 mg/L of vanillate was produced 72 h after induction (Fig. 1b). When the same host was used to express the ppsA and tktA genes in addition to the previously mentioned pathway genes, vanillate titers slightly decreased but cumulative titers of heterologous metabolites increased. Specifically, protocatechuate titer increased from 200 to 300 mg/L in strains expressing the ppsA and tktA genes. In addition, the specific yield of protocatechuate noticeably increased around 24 h after pathway induction when ppsA and tktA were overexpressed (Additional file 1: Figure S1). In both experiments, protocatechuate was the dominant heterologous metabolite formed on a molar basis, and this suggested that the reaction catalyzed by the O-methyltransferase was limiting. It is possible that the decrease in vanillate titers resulted either from decreased OMTHs expression as two additional genes were being expressed, or from the decreased rate of biomass formation during expression of the two additional genes (Additional file 1: Figure S1), or from both contributions.
Although the gene encoding OMTHs was codon-optimized for expression in E. coli, we next wondered whether OMTHs may be expressing poorly or whether it may have low activity in E. coli. SDS-PAGE results suggested that OMTHs expresses well (Fig. 2a). In addition, cultures expressing the pathway were sampled for in vitro specific OMTHs activity. Activity measurements revealed that, while OMTHs activity decreases to 34 % of initially measured activity, the enzyme is still active 48 h after induction (Fig. 2b).
Fig. 2.
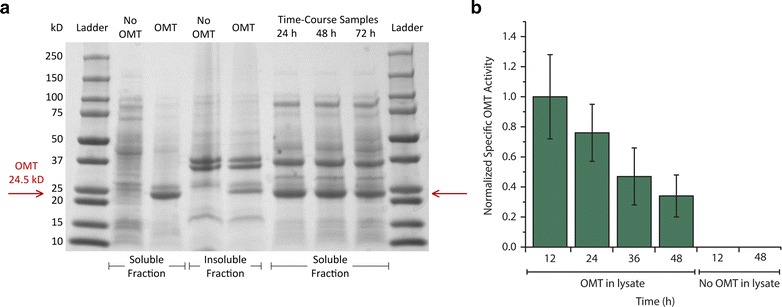
Expression and activity data suggests principal limitation is not heterologous O-methyltransferase. a SDS-PAGE gel suggesting robust OMTHs expression in soluble fractions of cells sampled from flask cultures. b In vitro specific activity measurements normalized to activity at 12 h time point. Activity data indicates that OMTHs activity declines from 12 to 48 h. However, notable activity even at 48 h strongly suggests that loss of OMTHs activity is not responsible for observed reduction in rate of vanillate formation
Supplementation of methionine and homocysteine improve vanillate titers, strongly suggesting that availability of the co-substrate S-adenosylmethionine (SAM) limits conversion of protocatechuate to vanillate
Given that OMTHs appeared to be expressed and active in cells at times when conversion of protocatechuate to vanillate was not occurring, we next considered that co-substrate availability may be limiting (Fig. 3a). The co-substrate SAM fulfills a variety of cellular roles, such as serving as the primary methyl donor for all cellular methylations, which compete directly with the engineered vanillin pathway. Methionine is endogenously converted to SAM by methionine adenosyltransferase (otherwise known as SAM synthetase) encoded by metK. Although exogenously supplied SAM does not enter E. coli, exogenously supplied methionine does enter cells and is known to indirectly perturb intracellular SAM availability in E. coli [6] and in yeast [21]. Importantly, in the sole previously reported effort to produce vanillate using E. coli, it was observed that methionine supplementation improved titers [6].
Fig. 3.
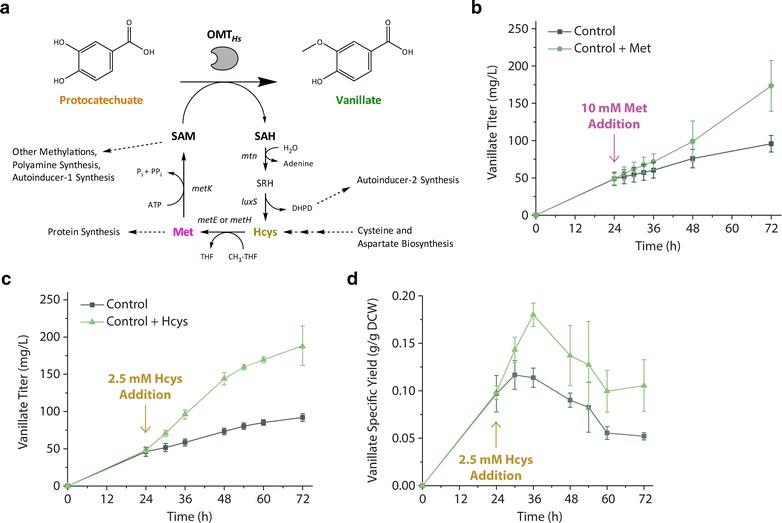
Supplementation experiments indicate limiting S-adenosylmethionine (SAM) availability and demonstrate SAM biosynthesis bottleneck upstream of homocysteine. a Metabolic pathway diagram illustrating co-substrate requirement for reaction catalyzed by O-methyltransferase. SAM is generated from methionine, which in turn is generated from homocysteine. b Vanillate titers resulting from the presence or absence of 10 mM methionine supplementation to cultures 24 h after induction. Cultures receiving methionine produced nearly twofold higher vanillate titers. c Vanillate titers resulting from the presence or absence of 2.5 mM homocysteine supplementation to cultures 24 h after induction. Lower concentrations of homocysteine were used relative to methionine given the potential for homocysteine toxicity. Once again, cultures receiving supplement produced nearly twofold higher vanillate titers. d Vanillate specific yields resulting from homocysteine supplementation experiment. Higher specific yield upon homocysteine supplementation demonstrates that increased vanillate production is due to greater output per cell and not because of additional biomass. These pathway experiments used the PTS− glu+ RAREʹ host and overexpression of aroG*,asbF Bt, OMT Hs , ppsA, and tktA
We monitored vanillate titers in cultures with and without supplementation of 10 mM l-methionine at peak vanillate productivity (24 h after induction) (Fig. 3b). Although methionine supplementation did not result in significant changes in vanillate titer immediately, final vanillate titer increased twofold 72 h after induction. In addition, vanillate synthesis occurred at a relatively constant rate until the final sampling time of 72 h upon methionine supplementation. This result suggests that potential depletion of SAM and methionine pools later in the culture may be responsible for limiting conversion of protocatechuate to vanillate.
To better understand the contributions of methionine biosynthesis to the vanillate pathway, a second supplementation experiment was performed, this time with the direct precursor to methionine, l-homocysteine (Fig. 3a). The methylation of homocysteine to form methionine is reported to be problematic under oxidative conditions due to the inactivation of catalytic residues in the cobalamin-independent methionine synthase (MetE) [22–25]. As before, cultures at peak vanillate productivity (24 h) were supplemented with and without l-homocysteine. Because l-homocysteine is reported to be toxic for E. coli [26, 27], we added 2.5 mM rather than 10 mM. Once again, supplemented cultures displayed an increase in vanillate titer (up to 200 mg/L) and an increase in duration of vanillate production consistent with what was observed for l-methionine addition (Fig. 3c). The average rate of vanillate synthesis doubled from 1.3 to 2.6 mg/L h. Importantly, vanillate specific yields showed that homocysteine supplementation led to higher specific production of vanillate rather than higher cell density (Fig. 3d). These results collectively suggested that reactions upstream of homocysteine synthesis in the methionine biosynthesis pathway need to be targeted to achieve increases in vanillate production from glucose as a sole carbon source.
Unlike aromatic amino acid biosynthesis, methionine biosynthesis in E. coli and other bacteria is not well understood and is regulated at multiple transcriptional as well as post-transcriptional levels (Fig. 4a). In fact, until recently, methionine was the only essential amino acid that was not commercially produced using fermentative processes [28, 29]. The protein encoded by metJ is the primary transcriptional regulator of several genes involved in methionine biosynthesis [30–32]. Methionine titers of 910 mg/L were previously achieved using an E. coli strain that was constructed by mutagenesis with nitrosoguanidine along with selection based on resistance to methionine analogs. This strain was observed to have a mutation in metJ that decreased repression of methionine biosynthesis [33]. However, genome sequencing was not performed and it is unlikely that metJ was the only gene containing a mutation. In addition to regulation by metJ, MetA (homoserine succinyltransferase) catalyzes the first committed step in methionine biosynthesis and is reported to be inhibited by both methionine and SAM [34]. l-cysteine is also a precursor to methionine. CysE (l-serine O-acetyltransferase) catalyzes the first step of cysteine biosynthesis and is reported to display significant inhibition by cysteine [35]. Fortunately, variants of MetA and CysE that have been engineered to display desensitization to feedback inhibition (MetA* and CysE*) have been reported [34, 35]. The combination of metA* expression, metJ deletion, and other modifications have previously led to a strain that accumulated 240 mg/L of methionine [34].
Fig. 4.
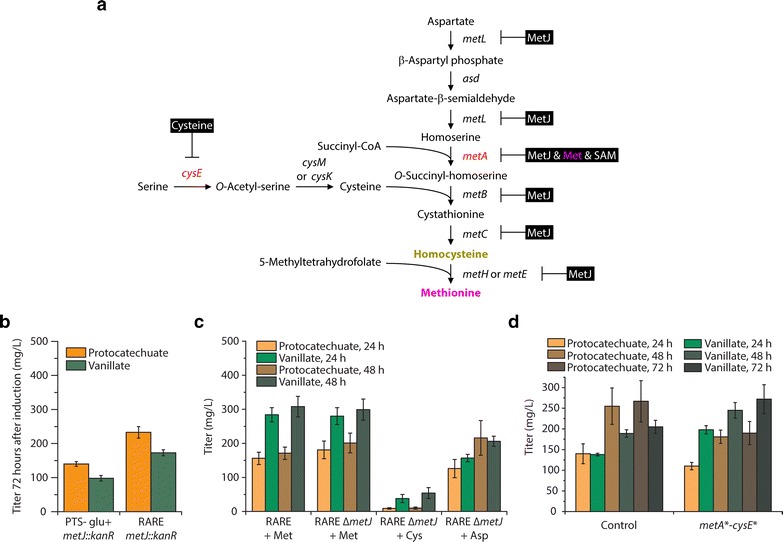
Effect of metJ deletion on protocatechuate and vanillate titers (a) in different host strains and (b) in the presence of amino acid supplementation. For these experiments, the following genes were overexpressed: aroG*,asbF Bt, OMT Hs , ppsA, and tktA. For the amino acid supplementation experiment (c), 10 mM of amino acid was added at induction. d Effect of overexpressing feedback-desensitized variants of metA and cysE along with aroG*,asbF Bt, OMT Hs , ppsA, and tktA in the RARE ∆metJ host. The control represents co-transformation with an empty pCOLADuet-1 plasmid
Deregulation of SAM biosynthesis through deletion of metJ and expression of metA* and cysE* improves vanillate titers
We hypothesized that vanillate titers might increase if methionine biosynthesis were modified by (i) deleting metJ; and (ii) expressing metA* and cysE* (Fig. 4a). Given our observation of slower growth in the PTS− glu+ RAREʹ strain relative to the RARE strain, we decided to delete metJ in both strains and compare performance. Upon overexpression of aroG*, asbF, OMT, ppsA, and tktA, the RARE ∆metJ host resulted in higher titers of protocatechuate and vanillate than the PTS− glu+ RAREʹ ∆metJ host (Fig. 4b). However, protocatechuate titers remained higher than vanillate titers in both cases. As previously noted, metJ is only one of many simultaneous modes of methionine biosynthesis regulation. Given feedback-resistance at the entrance to the pathway, one could envision the metJ deletion strain performing slightly worse because of increased expression of downstream genes with minimal flux entering the pathway. Given that the RARE ∆metJ host performed better than the PTS− glu+ RAREʹ ∆metJ host, we decided to continue with RARE ∆metJ for remaining experiments.
To further understand the limitations in methionine biosynthesis, we next supplemented the RARE ∆metJ host harboring the pathway with one of three amino acids: l-methionine, l-cysteine, and l-aspartate. Because cysteine and aspartate are precursors to methionine (Fig. 4a), our goal was to investigate whether reaction steps downstream of their biosynthesis were problematic. In this case, we added 10 mM of each amino acid at the time of induction (0 h) to see whether the time of supplementation would affect pathway kinetics. In this experiment, we also included the RARE host (with metJ intact) expressing the pathway in order to better understand the potential effect of metJ deletion. Performance of the RARE and RARE ∆metJ hosts supplemented with methionine was similar, with roughly 280 mg/L vanillate produced in just 24 h (Fig. 4c). Little additional vanillate formed after the first 24 h, suggesting that methionine supplementation at induction might be more effective than supplementation at 24 h. The decrease in the rate of vanillate formation suggested that all of the methionine added initially may have been depleted within 24 h. Addition of l-cysteine or l-aspartate did not improve vanillate titers, which supported the notion of next focusing on the reactions catalyzed by MetA and CysE. Interestingly, biomass formation occurred more rapidly than usual during l-aspartate supplementation, suggesting that uptake of this amino acid occurred readily but was diverted to other cellular purposes besides methionine biosynthesis (Additional file 1: Figure S2). On the other hand, addition of 10 mM l-cysteine significantly decreased titers and biomass formation. Cysteine has been shown to inhibit the growth of several E. coli strains by inhibiting threonine deaminase and therefore isoleucine synthesis [36].
We next explored whether expression of the metA* and cysE* genes would improve conversion of vanillate to protocatechuate without the addition of l-methionine to the culture medium (Fig. 4d). In the cases in which metA* and cysE* were overexpressed, average vanillate titer was observed to exceed average protocatechuate titer at all time points sampled. Expression of metA*−cysE* increased final vanillate titers by 33 % (from 205 ± 16 to 272 ± 35 mg/L). To gain insight into potential upper bounds of vanillate titer obtainable by increasing SAM availability, we supplemented cultures expressing metA*−cysE* with 10 mM methionine at both induction and at 24 h after induction (Additional file 1: Figure S3). In this case, vanillate titers continued to exceed protocatechuate titers at all time points sampled. Addition of methionine at induction led to 341 ± 25 mg/L of vanillate produced in the first 24 h. This suggested that further increased SAM availability within the first 24 h could still improve vanillate production. Although the addition of an extra 10 mM methionine (20 mM methionine total) at 24 h after induction resulted in a final vanillate titer of 419 ± 58 mg/L at 72 h after induction, protocatechuate titers remained above 100 mg/L. Thus, a strategy of additional exogenous methionine supplementation is insufficient to address the limitation of protocatechuate conversion, particularly once cultures have achieved late exponential or stationary phase. Given continued observations of a decreasing rate of vanillate formation after 24 h, we also investigated potential loss of the ampicillin-resistant plasmid harboring OMT (pET-AsbF-OMT). Based on the similar number of colonies appearing on plates taken at the same time points (Additional file 1: Figure S4), we could rule out plasmid loss as an explanation for the limited conversion observed.
Orthogonal strategy of deregulating SAM regeneration through overexpression of mtn and luxS also achieves improvement in vanillate titers
To our knowledge, no previous study has sought to increase SAM availability by targeting genes downstream of SAM utilization, in what is known as the activated methyl cycle. S-adenosyl-l-homocysteine (SAH) is a co-product of the reaction along with vanillate, and SAH is a potent inhibitor of SAM-dependent methyltransferases [37, 38] (Fig. 5a). Homocysteine supplementation results presented earlier suggested that recycling of SAH back to homocysteine does not occur at a rate sufficient to maintain SAM availability. One of the two reactions required to recycle SAH is catalyzed by Mtn and converts SAH to S-ribosyl-l-homocysteine (SRH). The subsequent conversion of SRH to homocysteine is catalyzed by LuxS and coupled to the production of autoinducer AI-2, which is a quorum sensing molecule [39, 40]. In E. coli and other bacteria, AI-2 has been implicated as an inducer of biofilm formation [41, 42] and as an attractant for chemotaxis [43]. Because recycling of SAH is linked to the formation of AI-2, SAM regeneration is expected to be subject to immense regulation. A recent LC–MS study profiled the effect of deletions of mtn and luxS on intracellular concentrations of SAM, SAH, SRH, homocysteine, and methionine at different OD600 values for E. coli MG1655 [44]. However, overexpression of these genes and the possible effect on intracellular concentrations of the same metabolites was not tested. We hypothesized that overexpression of mtn and luxS may increase SAM availability and therefore improve vanillate titers.
Fig. 5.
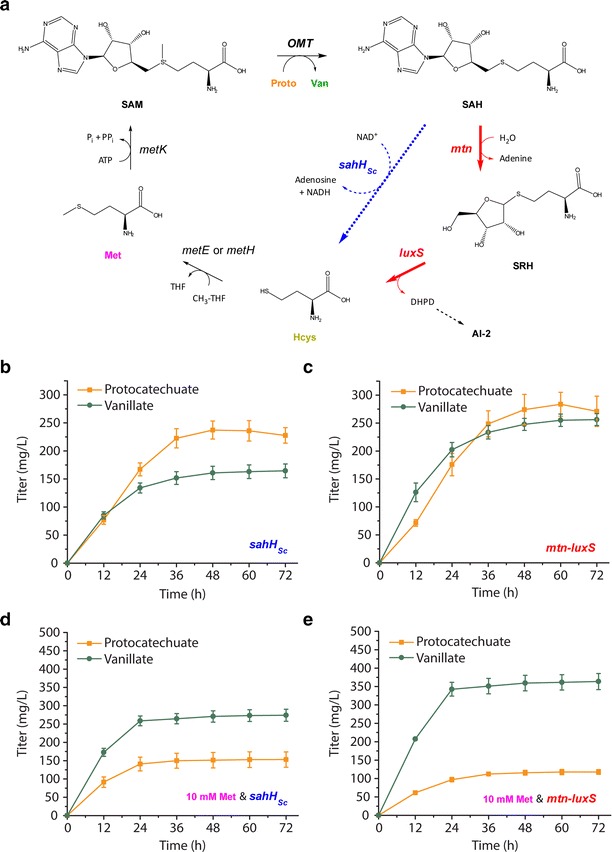
Orthogonal strategy of increasing S-adenosylhomocysteine (SAH) recycling through overexpression of mtn and luxS also improves vanillate titer. a The activated methyl cycle in E. coli (in black and red), along with an alternative SAH recycling route featuring a heterologous SAH hydrolase (sahH Sc, in blue). Native genes targeted for overexpression (mtn and luxS) are shown in red. b Effect of expressing sahH Sc with aroG*,asbF Bt, OMT Hs , ppsA, and tktA in the RARE ∆metJ host on protocatechuate and vanillate titers. c Effect of overexpressing mtn and luxS with aroG*,asbF Bt, OMT Hs , ppsA, and tktA in the RARE ∆metJ host on protocatechuate and vanillate titers. d Effect of 10 mM methionine supplementation at induction on protocatechuate and vanillate titers using strains expressing sahH Sc. e Effect of 10 mM methionine supplementation at induction on protocatechuate and vanillate titers using strains overexpressing mtn and luxS
In eukaryotes, archaea, and non-LuxS-containing bacteria, SAM regeneration occurs differently, with conversion of SAH directly to homocysteine and adenosine catalyzed by SAH hydrolase (SAHase) [40]. We also hypothesized that overexpression of a heterologous SAHase might provide an alternative way to improve protocatechuate conversion with minimal interference to native regulation (Fig. 5a). To explore this concept, we tested expression of a SAHase from S. cerevisiae (sahHSc). In all of the remaining experiments, neither metA* nor cysE* were expressed.
The two strategies intended to improve SAM regeneration had differing results. Final vanillate titers were 164 ± 12 mg/L when sahHSc was expressed (Fig. 5b), which is lower than the 205 ± 16 mg/L previously observed using a relevant control plasmid expressing no extra genes (Fig. 4d). Few conclusions should be drawn about the general concept of using a SAH hydrolase from the worsened pathway performance observed given that only one variant was tested and activity was not confirmed. Expression of sahHSc may have introduced a metabolic imbalance given its dependence on NAD+, or it may have simply added an expression burden if the gene product were inactive. However, when mtn and luxS were overexpressed, final vanillate titers were 256 ± 11 mg/L (Fig. 5c). The 25 % increase in average vanillate titers over the no expression control suggests that overexpression of the two native genes was effective at improving regeneration. A significant difference was also observed upon supplementing cultures containing each strain with 10 mM methionine at induction. Under these conditions, final vanillate titers were 274 ± 17 mg/L when sahHSc was expressed (Fig. 5d) and 364 ± 22 mg/L when mtn and luxS were overexpressed (Fig. 5e). For comparison, when 10 mM methionine was added to the control strain that did not overexpress these genes, final vanillate titers (at 48 h) were 299 ± 31 mg/L (Fig. 4c). Thus, overexpression of mtn and luxS improved vanillate titers in both the absence and presence of methionine supplementation. Table 1 contains a summary of protocatechuate and vanillate titers from key experiments, along with their cumulative titers.
Table 1.
Summary of titers from key experiments
| Number | Figure | Host | Genes overexpressed | Methionine added (mM) | Protocatechuate (P) Titer (mg/L) | Protocatechuate (P) Titer (mM) | Vanillate (V) Titer (mg/L) | Vanillate (V) Titer (mM) | Cumulative P+V Titer (mM) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1B | PTS-glu + RARE’ | aroG*, asbF, OMT | 0 | 216 | 1.4 | 135 | 0.8 | 2.2 |
| 2 | 1B | PTS-glu + RARE’ | aroG*, asbF, OMT, ppsA, tktA | 0 | 308 | 2.0 | 101 | 0.6 | 2.6 |
| 3 | 3B | PTS-glu + RARE’ | aroG*, asbF, OMT, ppsA, tktA | 0 | 323 | 2.1 | 101 | 0.6 | 2.7 |
| 4 | 4D | RARE ∆metJ | aroG*, asbF, OMT, ppsA, tktA | 0 | 262 | 1.7 | 202 | 1.2 | 2.9 |
| 5 | 4D | RARE ∆ metJ | aroG*, asbF, OMT, ppsA, tktA, metA, cysE | 0 | 185 | 1.2 | 269 | 1.6 | 2.8 |
| 6 | 5B | RARE ∆metJ | aroG*, asbF, OMT, ppsA, tktA, sahH | 0 | 231 | 1.5 | 168 | 1.0 | 2.5 |
| 7 | 5C | RARE ∆ metJ | aroG*, asbF, OMT, ppsA, tktA, mtn, luxS | 0 | 277 | 1.8 | 252 | 1.5 | 3.3 |
| 8 | 3B | PTS-glu + RARE’ | aroG*, asbF, OMT, ppsA, tktA | 10 | 246 | 1.6 | 168 | 1.0 | 2.6 |
| 9* | 4C | RARE ∆metJ | aroG*, asbF, OMT, ppsA, tktA | 10 | 200 | 1.3 | 303 | 1.8 | 3.1 |
| 10 | 5D | RARE ∆metJ | aroG*, asbF, OMT, ppsA, tktA, sahH | 10 | 154 | 1.0 | 269 | 1.6 | 2.6 |
| 11 | 5E | RARE ∆ metJ | aroG*, asbF, OMT, ppsA, tktA, mtn, luxS | 10 | 123 | 0.8 | 370 | 2.2 | 3.0 |
| 12 | S3 | RARE ∆ metJ | aroG*, asbF, OMT, ppsA, tktA, metA, cysE | 20 | 139 | 0.9 | 420 | 2.5 | 3.4 |
Rows in bold formatting are highlighted in “Discussion” section
* Titers shown for experiment 9 correspond to samples obtained 48 h after induction, which was the final sampling time. All other titers correspond to samples obtained 72 h after induction
Discussion
The results of this study present a detailed perspective on de novo vanillate biosynthesis in engineered E. coli. First, genetic perturbations were made upstream of the heterologous pathway in order to improve titers and better determine which reaction steps may be limiting. Upon identification of a limitation in the reaction catalyzed by OMTHs, experiments were performed that provided several lines of evidence that SAM availability was the dominant cause of the limitation. Given that conversion of exogenously supplied homocysteine improved vanillate titers, attention was next focused on deregulating methionine biosynthesis by targeting the global regulator MetJ and the feedback-sensitive enzymes MetA and CysE. When deletion of metJ was coupled with expression of feedback-desensitized variants of metA* and cysE*, average de novo vanillate titers improved by 33 %. The orthogonal strategy of deregulating SAM regeneration by overexpressing mtn and luxS improved average de novo vanillate titers by 25 %. Overall, de novo vanillate titers improved roughly twofold over the course of experiments described in this manuscript.
In principle, the strategies of targeting biosynthesis or regeneration for deregulation provide different tradeoffs. Increasing SAM regeneration by increasing flux through the activated methyl cycle should be more efficient on a carbon basis than increasing flux through SAM biosynthesis because it depletes SAH, which is a toxic molecule and is known to inhibit O-methyltransferases, rather than potentially unbalancing central metabolic precursors. However, a potential disadvantage of increasing SAM regeneration is that the native E. coli pathway is coupled to biosynthesis of quorum sensing molecules that have widespread regulatory effects. The use of a yeast SAH hydrolase was motivated by the potential ability to decouple SAM regeneration and quorum sensing. The effectiveness of our implementation of these strategies can be determined from the titers shown in Table 1. Although overexpression of metA* and cysE* (Table 1, row #5) led to slightly higher vanillate titers than overexpression of mtn and luxS (Table 1, row #7), the latter strategy resulted in greater cumulative titers of protocatechuate and vanillate. In all cases, methionine supplementation led to a greater molar ratio of vanillate to protocatechuate (of note are rows #11–12). These results suggest that further efforts towards increasing SAM availability would be worthwhile for improving de novo production.
When compared with results from previously reported vanillin pathway optimization studies performed in yeast, our observations highlight the context dependence of engineered metabolic pathways. Two reports published after the initial demonstration of the engineered yeast vanillin pathway describe improvements obtained using flux balance analysis (FBA) model-guided optimization [8, 9]. In the first report, OptGene was used to identify target reactions that, if deleted, would increase vanillin production. Different deletion targets were suggested depending on the reference flux distribution in the minimization of metabolic adjustment (MOMA) biological objective function. In general, the predicted benefit from these modifications was increased availability of the co-factors ATP and NADPH required by CarNi. Genes related to pyruvate metabolism, ammonium metabolism, the pentose phosphate pathway, and central carbon metabolism were identified. The only three modifications tested experimentally were deletion of one of the pyruvate decarboxylases (PDC1), deletion of the most active glutamate dehydrogenase (GDH1), and overexpression of GDH2 to ensure sufficient nitrogen uptake in the absence of GDH1. These modifications led to an overproducer strain that, when cultivated in a low dilution rate continuous fermentation, resulted in vanillin-β-d-glucoside titers of 500 mg/L [8]. As of that study, identification and overexpression of a potential rate-limiting enzyme in the pathway had not yet been described. In a second report, OMTHs and CarNi were overexpressed in the highest-producing strain obtained from the previous study [9] based on the observed accumulation of two heterologous intermediates (protocatechuate and protocatechualdehyde). CarNi overexpression did not lead to an increase in production, whereas OMTHs overexpression led to a 30 % increase in titer (now 380 mg/L vanillin-β-d-glucoside compared to a different baseline than referenced above). However, OMTHs overexpression in the parental strain from the first study did not alter titer. The authors concluded that this was likely because CarNi was limiting due to low availability of ATP and NADPH without the model-guided modifications made in the first study.
Given the many potential applications of methyltransferases in metabolic engineering [15, 16], the small number of reported examples in E. coli may reflect the limited host methylation capacity that we have encountered. In one report, a novel bacterial fatty acid methyltransferase was used to catalyze the formation of fatty acid methyl esters using free fatty acids and SAM [45]. The authors of that study note that SAM availability strongly regulated methyl ester production. By deleting the metJ gene mentioned earlier, and by overexpressing a gene encoding methionine adenosyltransferase from rat, the authors achieved an improvement in methyl ester production. However, the normalized titers of methyl esters in the supernatant achieved in their study increased from below 1 µM/OD to roughly 2.5 µM/OD. The corresponding amount of SAM required to achieve such conversion is orders of magnitude below what drives production of hundreds of milligrams per liter of vanillate.
Other reported examples of SAM-dependent methyltransferase reactions engineered in E. coli include several biotransformations to produce valuable methylated flavonoids, such as conversion of quercetin to rhamnetin [46], naringenin to 3-O-methylkaempferol [47], coumaric acid to 7-methoxyapigenin [48, 49], and coumaric acid to 7-O-methylaromadendrin [50]. Several of these methylated flavonoids are reported to have pharmacological properties. The largest titer reported in any of these examples was 111 mg/L of rhamnetin upon overexpression of E. coli SAM synthetase (metK) [46]. Results obtained in this study, such as those from methionine and homocysteine supplementation experiments, strongly suggest that conversion of methionine to SAM is not a primary limitation under native conditions of SAM biosynthesis. Rather, steps in upstream methionine biosynthesis and in downstream SAM regeneration appear to be limiting SAM utilization by heterologous methyltransferases, and these steps ought to receive greater initial focus for targeted deregulation. Strategies described in this work would likely further improve production of these valuable methylated flavonoids and other desired methylated compounds. In addition, based on the ease of detecting protocatechuate and vanillate, we propose that the vanillate pathway could serve as a convenient system in future studies that aim to understand and improve SAM biosynthesis and regeneration in E. coli.
Conclusion
In this report, we have demonstrated that the de novo vanillin biosynthesis pathway engineered in E. coli suffers from a limitation in the conversion of protocatechuate to vanillate. Using several experimental approaches, we established that limited availability of the co-substrate SAM is the primary cause of the limitation. We then implemented two orthogonal strategies intended to increase SAM availability. One approach focused on deregulating SAM biosynthesis through metJ deletion and overexpression of feedback desensitized metA* and cysE*. The other approach aimed at deregulating SAM regeneration through overexpression of mtn and luxS. Both approaches led to increases in vanillate titer, and further increases were obtained through methionine supplementation. These results provide useful insights for improving production from pathways that involve methylation in E. coli, such as the vanillin pathway.
Methods
Strains and plasmids
Escherichia coli strains and plasmids used in this study are listed in Table 2. Molecular biology techniques were performed according to standard practices [51] unless otherwise stated. Molecular cloning and vector propagation were performed in DH5α. All host strains used for production experiments were derived from E. coli K-12 MG1655(DE3). In order to construct new host strains, two methods were used. The first was P1 transduction [52] using donor strains from the Keio collection [53] and P1 bacteriophage from ATCC (25404-B1). P1 transduction was used for all deletions of single genes. The second method was recombineering using the λ Red system [54]. Recombineering was used to delete the yqhC-dkgA operon and to upregulate galP expression by promoter substitution, as previously described [5]. Oligonucleotides were purchased from Sigma. Q5 High Fidelity DNA Polymerase (New England Biolabs, MA) was used for DNA amplification. In all cases of host strain modifications, pCP20 was used to cure the kanamycin resistance cassette [54].
Table 2.
Strains and plasmids used in this study
| Name | Relevant genotype | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1 | Invitrogen |
| DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139Δ(ara, leu)7697 galU galK λ− rpsL nupG | Invitrogen |
| MG1655 | F− λ− ilvG- rfb-50 rph-1 | ATCC 700926 |
| MG1655(DE3) | F− λ− ilvG- rfb-50 rph-1 (DE3) | Ref. [55] |
| RARE | MG1655(DE3) ∆dkgB ∆yeaE ∆(yqhC-dkgA) ∆yahK ∆yjgB | Ref. [5] |
| RARE ∆metJ | MG1655(DE3) ∆dkgB ∆yeaE ∆(yqhC-dkgA) ∆yahK ∆yjgB ∆metJ | This study |
| PTS− glu+ | MG1655(DE3) ∆ptsHIcrr Pglk::Pcon* galPq | This study, but based on Ref. [56] |
| PTS− glu+ RARE’ | MG1655(DE3) ∆ptsHIcrr Pglk::Pcon* galPq ∆(yqhC-dkgA) ∆yahK ∆yjgB | This study |
| PTS− glu+ RARE’ ∆metJ | MG1655(DE3) ∆ptsHIcrr Pglk::Pcon* galPq ∆(yqhC-dkgA) ∆yahK ∆yjgB ∆metJ | This study |
| E. coli plasmids | ||
| pCP20 | λ cI857 (ts), λ pr Repts, AmpR, CmR, λ pr FLP | CGSC 7629 |
| pKD13 | oriRγ, AmpR, kan | CGSC 7633 |
| pKD46 | oriR101, repA101 ts, AmpR, araC, araBp-λ γ-λ β-λexo | CGSC 7739 |
| pETDuet-1 | AmpR, lacI, T7lac | Novagen |
| pACYCDuet-1 | CmR, lacI, T7lac | Novagen |
| pCOLADuet-1 | KanR, lacI, T7lac | Novagen |
| pCDFDuet-1 | StrR, lacI, T7lac | Novagen |
| pACYC-car-sfp | pACYCDuet-1 harboring car opt (carboxylic acid reductase from Nocardia iowensis, codon-optimized for expression in E. coli) and sfp opt (phosphopantetheinyl transferase from Bacillus subtilis, codon optimized for expression in E. coli) | Ref. [5] |
| pET-OMT-asbF | pETDuet-1 harboring Hs-S-COMT opt (catechol O-methyltransferase from Homo sapiens, codon-optimized for expression in E. coli) and asbF opt (dehydroshikimate dehydratase from Bacillus thuringiensis, codon optimized for expression in E. coli) | Ref. [5] |
| pS4 | Plasmid containing the shikimate module, version 4, kindly provided by the Keasling Lab at UC Berkeley. (Source of aroG*-ppsA-tktA artificial operon) | Ref. [57] |
| pACYC-aroG* | pACYCDuet-1 harboring the feedback-resistant aroG* from E. coli | This study |
| pACYC-aroG*-ppsA-tktA | pACYCDuet-1 harboring three E. coli genes in an artificial operon: aroG*, ppsA, and tktA | This study |
| pCOLA-metK | pCOLADuet-1 harboring the E. coli metK gene | This study |
| pET-OMT | pETDuet-1 harboring Hs-S-COMT opt | This study |
| pCOLA-metA* | pCOLADuet-1 harboring a feedback-desensitized version of E. coli metA (27_Arg- > Cys, 296_Ile- > Ser, and 298_Pro- > Leu, “metA*”) | This study |
| pCOLA-cysE* | pCOLADuet-1 harboring a feedback-desensitized version of E. coli cysE (95_Val- > Arg, and 96_Asp- > Pro, “cysE*”) | This study |
| pCOLA-metA*-cysE* | pCOLADuet-1 harboring an artificial operon consisting of the metA* and cysE* genes | This study |
| pCDF-car-sfp | pCDFDuet-1 harboring an artificial operon containing car opt and sfp opt | This study |
| pCOLA-mtn | pCOLADuet-1 harboring the E. coli mtn gene | This study |
| pCOLA-mtn-luxS | pCOLADuet-1 harboring an artificial operon consisting of the E. coli mtn and luxS genes | This study |
| pCOLA-sahH | pCOLADuet-1 harboring sahH Sc (S-adenosylhomocysteine hydrolase from S. cerevisiae, codon-optimized for expression in E. coli) | This study |
The aroG*, ppsA, and tktA genes were kindly provided by Professor Jay D. Keasling at the University of California, Berkeley (USA). The genes encoding metA* and cysE* were synthesized as gBlocks (IDT, CA) and their sequences are included in Table 3. The gene encoding sahH from S. cerevisiae (UniProt #P39954) was codon-optimized for expression in E. coli and also synthesized as a gBlock (Table 3). The E. coli metK, mtn, and luxS genes were amplified from MG1655(DE3) genomic DNA using PCR amplication and the oligonucleotides shown in Table 4. All genes of interest were cloned into the Duet vector system (Novagen, WI) using restriction digest-based cloning. Restriction enzymes used to clone each gene are either shown in Table 3 (if the gene was synthesized) or in Table 4 (if the gene was amplified). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. Propagated constructs were purified using a QIAprep Miniprep Kit (Qiagen, CA) and agarose gel fragments were purified using a Zymoclean Gel DNA Recovery Kit (Zymo Research, CA). All constructs were confirmed to be correct by nucleotide sequencing (Genewiz, NJ).
Table 3.
Synthesized gene sequences used in this study
| Gene (and restriction enzymes) | DNA sequence. Restriction enzyme sites are underlined and start/stop codons are in bold |
|---|---|
| metA* (NdeI/AatII) | AAAAAACAT ATGCCGATTCGTGTGCCGGACGAGCTACCCGCCGTCAATTTCTTGCGTGAAGAAAACGTCTTTGTGATGACAACTTCTTGTGCGTCTGGTCAGGAAATTCGTCCACTTAAGGTTCTGATCCTTAACCTGATGCCGAAGAAGATTGAAACTGAAAATCAGTTTCTGCGCCTGCTTTCAAACTCACCTTTGCAGGTCGATATTCAGCTGTTGCGCATCGATTCCCGTGAATCGCGCAACACGCCCGCAGAGCATCTGAACAACTTCTACTGTAACTTTGAAGATATTCAGGATCAGAACTTTGACGGTTTGATTGTAACTGGTGCGCCGCTGGGCCTGGTGGAGTTTAATGATGTCGCTTACTGGCCGCAGATCAAACAGGTGCTGGAGTGGTCGAAAGATCACGTCACCTCGACGCTGTTTGTCTGCTGGGCGGTACAGGCCGCGCTCAATATCCTCTACGGCATTCCTAAGCAAACTCGCACCGAAAAACTCTCTGGCGTTTACGAGCATCATATTCTCCATCCTCATGCGCTTCTGACGCGTGGCTTTGATGATTCATTCCTGGCACCGCATTCGCGCTATGCTGACTTTCCGGCAGCGTTGATTCGTGATTACACCGATCTGGAAATTCTGGCAGAGACGGAAGAAGGGGATGCATATCTGTTTGCCAGTAAAGATAAGCGCATTGCCTTTGTGACGGGCCATCCCGAATATGATGCGCAAACGCTGGCGCAGGAATTTTTCCGCGATGTGGAAGCCGGACTAGACCCGGATGTACCGTATAACTATTTCCCGCACAATGATCCGCAAAATACACCGCGAGCGAGCTGGCGTAGTCACGGTAATTTACTGTTTACCAACTGGCTCAACTATTACGTCTACCAGAGCACGCTATACGATCTACGGCACATGAATCCAACGCTGGATTAA GACGTCAAAAAA |
| cysE* (AatII/XhoI) | AAAAAAGACGTCTAATAAAAGGAGATATACCATGTCGTGTGAAGAACTGGAAATTGTCTGGAACAATATTAAAGCCGAAGCCAGAACGCTGGCGGACTGTGAGCCAATGCTGGCCAGTTTTTACCACGCGACGCTACTCAAGCACGAAAACCTTGGCAGTGCACTGAGCTACATGCTGGCGAACAAGCTGTCATCGCCAATTATGCCTGCTATTGCTATCCGTGAAGTGGTGGAAGAAGCCTACGCCGCTGACCCGGAAATGATCGCCTCTGCGGCCTGTGATATTCAGGCGGTGCGTACCCGCGACCCGGCAAGACCCAAATACTCAACCCCGTTGTTATACCTGAAGGGTTTTCATGCCTTGCAGGCCTATCGCATCGGTCACTGGTTGTGGAATCAGGGGCGTCGCGCACTGGCAATCTTTCTGCAAAACCAGGTTTCTGTGACGTTCCAGGTCGATATTCACCCGGCAGCAAAAATTGGTCGCGGTATCATGCTTGACCACGCGACAGGCATCGTCGTTGGTGAAACGGCGGTGATTGAAAACGACGTATCGATTCTGCAATCTGTGACGCTTGGCGGTACGGGTAAATCTGGTGGTGACCGTCACCCGAAAATTCGTGAAGGTGTGATGATTGGCGCGGGCGCGAAAATCCTCGGCAATATTGAAGTTGGGCGCGGCGCGAAGATTGGCGCAGGTTCCGTGGTGCTGCAACCGGTGCCGCCGCATACCACCGCCGCTGGCGTTCCGGCTCGTATTGTCGGTAAACCAGACAGCGATAAGCCATCAATGGATATGGACCAGCATTTCAACGGTATTAACCATACATTTGAGTATGGGGATGGGATCTAA CTCGAGAAAAAA |
| sahH Sc (NcoI/NotI) | ATATATCC ATGGGCGCCCCTGCTCAGAATTATAAAATTGCAGATATCTCACTGGCAGCGTTCGGTCGTAAGGAAATTGAATTGGCAGAGCACGAAATGCCAGGCCTGATGGCAATTCGTAAAGCCTATGGCGACGTTCAACCGCTGAAGGGCGCACGTATCGCGGGTTGCTTACACATGACGATCCAAACCGCCGTGCTGATTGAGACATTAGTCGCTTTGGGAGCCGAGGTTACGTGGTCGTCATGCAATATCTACTCGACGCAAGATCATGCGGCAGCGGCAATCGCTGCAAGCGGCGTGCCTGTGTTTGCTTGGAAAGGTGAAACCGAAGAGGAATACCTGTGGTGCATTGAACAGCAGCTGTTTGCATTCAAGGATAACAAAAAATTGAACCTGATTTTGGACGACGGCGGGGATTTAACAACCTTAGTTCACGAGAAACATCCTGAAATGCTGGAGGATTGTTTTGGCCTTTCCGAAGAAACCACCACCGGTGTCCACCACCTCTACCGCATGGTTAAAGAGGGTAAACTTAAAGTCCCGGCTATCAATGTGAACGACTCAGTAACCAAATCCAAATTTGATAACTTATATGGTTGCCGTGAAAGCCTGGTTGACGGGATTAAGCGCGCCACGGATGTTATGCTTGCCGGAAAGGTGGCTGTTGTGGCCGGTTACGGTGACGTGGGTAAAGGATGCGCAGCCGCGTTACGTGGCATGGGTGCACGCGTGTTAGTTACGGAAATTGATCCTATCAATGCCCTGCAGGCCGCGATGGAAGGCTATCAAGTGGTAACAATGGAAGATGCCTCACATATCGGTCAGGTGTTCGTTACGACCACGGGTTGTCGTGATATCATCAACGGGGAGCATTTCATTAACATGCCGGAAGATGCAATTGTTTGTAACATCGGGCATTTCGATATTGAGATCGACGTGGCGTGGCTCAAAGCCAATGCCAAAGAGTGTATTAACATTAAACCGCAGGTAGATCGTTATTTGCTGAGCTCTGGCCGTCATGTCATCCTCCTGGCAAATGGACGCCTTGTGAATCTGGGTTGTGCGACGGGTCACTCGTCCTTCGTCATGAGCTGCTCTTTTTCCAACCAGGTTCTGGCACAAATCGCCCTGTTTAAATCTAATGACAAATCTTTTCGCGAAAAACACATCGAGTTCCAGAAGACCGGTCCCTTCGAAGTCGGGGTTCATGTGTTGCCTAAAATCTTAGATGAGGCAGTCGCTAAATTTCATCTTGGCAACCTGGGTGTCCGTCTGACCAAACTGAGTAAAGTCCAGAGCGAGTACCTGGGTATTCCGGAGGAAGGACCTTTCAAAGCGGATCATTATCGCTATTAA GCGGCCGCAAATTT |
Table 4.
Oligonucleotides used in this study
| Name | Sequence (5′– >3′) |
|---|---|
| MetJ-verify-f | TCTTTAGCAATCACCACG |
| MetJ-verify-r | GGAATATTCTTGCCGTAAC |
| PtsHICrr-verify-f | GAAAGGCGCAATCCAA |
| PtsHICrr-verify-r | CGATTTGACTGCCAGAAT |
| AroG*-f (BglII) | AAAAAAAGATCTGATGAATTATCAGAACGACGATTTAC |
| AroG*-r (AvrII) | AAAAAACCTAGGCCTCCTTTAGATCCTTACCC |
| AroG*-PpsA-TktA-f (BglII) | AAAAAAAGATCTGATGAATTATCAGAACGACGATTTAC |
| AroG*-PpsA-TktA-r (AvrII) | AAAAAACCTAGGTTACAGCAGTTCTTTTGCTTTC |
| MetK-f (NdeI) | AAAAAACATATGGCAAAACACCTTTTTAC |
| MetK-r (AvrII) | AAAAAACCTAGGTTACTTCAGACCGGCAG |
| Mtn-f (NcoI) | AAAAAACCATGGGCAAAATCGGCATCATTGG |
| Mtn-r (NotI) | AAAAAAGCGGCCGCTTAGCCATGTGCAAGTTTCT |
| LuxS-f (NotI) | AAAAAAGCGGCCGCTAATAAAGGAGATATACCATGCCGTTGTTAGATAGCTT |
| LuxS-r (AflII) | AAAAAACTTAAGCTAGATGTGCAGTTCCTGC |
Chemicals
The following compounds were purchased from Sigma: vanillic acid, protocatechuic acid (otherwise known as 3,4-dihydroxybenzoic acid), l-methionine, l-homocysteine, l-cysteine, and l-aspartate. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was purchased from Denville Scientific. Ampicillin sodium salt, chloramphenicol, streptomycin sulfate, and kanamycin sulfate were purchased from Affymetrix.
Culture conditions
A 1X M9 salt medium (Sigma) containing 6.78 g/L Na2HPO4∙7H2O, 3 g/L KH2PO4, 1 g/L NH4CI, and 0.5 g/L NaCl, supplemented with 2 mM MgSO4, 0.1 mM CaCl2, glucose, trace elements, and antibiotics was used as the culture medium for experiments in this study. The trace element solution (100×) used contained 5 g/L EDTA, 0.83 g/L FeCl3∙6H2O, 84 mg/L ZnCl2, 10 mg/L CoCl2∙6H2O, 13 mg/L CuCI2∙2H2O, 1.6 mg/L MnCl2∙2H2O and 10 mg/L H3BO3 dissolved in water. This was added to a concentration of 1× to supplement the M9-glucose medium. This medium will be henceforth referred to as “M9-glu-trace.” For all experiments except the experiment corresponding to Fig. 1b, the initial glucose concentration was 1.8 %. The initial glucose concentration was 1.2 % for the experiment corresponding to Fig. 1b.
All experiments were performed in 250 ml baffled PYREX shake flasks that contained 50 ml culture volumes. Overnight cultures were grown in 3 ml in 14 ml round-bottom tubes (Corning). Experimental cultures were initiated as follows: 1 % (v/v) inoculum volumes of overnight culture in LB medium were first transferred into overnight culture in M9-glu-trace medium, and then 1 % (v/v) inoculum volumes of overnight culture in M9-glu-trace were transferred into 50 mL M9-glu-trace medium, incubated at 30 °C, and agitated at 250 rpm. The OD600 was measured regularly during exponential growth using a DU800 UV/Vis spectrophotometer (Beckman Coulter). Cultures were induced with 0.5 mM IPTG at OD600 values between 0.8 and 1.0. Depending on the experiment, culture medium was supplemented with either 50 mg/L ampicillin, 17 mg/L chloramphenicol, 25 mg/L streptomycin, 25 mg/L kanamycin, or combinations of the previous antibiotics to provide selective pressure for plasmid maintenance. In some experiments (Figs. 4, 5; Additional file 1: Figures S2–S4), 50 mg/L carbenicillin was used in place of ampicillin in liquid cultures to provide more stringent pressure for plasmid maintenance. All experiments were performed in biological triplicate, and results are presented as averages with error bars representing one standard deviation.
For experiments in which S-adenosyl-l-methionine precursors were supplemented, flask cultures were set up as mentioned before but with culture volumes adjusted to achieve final concentrations as follows: 10 mM l-methionine, 2.5 mM l-homocysteine, 10 mM l-cysteine, or 10 mM l-aspartate. Stocks of supplemented metabolites were pH-neutralized and sterile filtered. In these experiments, control cultures received an equal volume of sterile deionized water instead of the metabolic precursors at the time of supplementation. Supplementation times varied from occurring at induction (0 h), occurring 24 h after induction, and occurring at both induction and 24 h after induction.
Metabolite analysis
Culture samples were pelleted by centrifugation and aqueous supernatant was collected for HPLC analysis using an Agilent 1100 series instrument equipped with a diode array detector. Heterologous compounds produced in vanillin experiments were separated using a Zorbax Eclipse XDB-C18 column (Agilent) and detected using a wavelength of 280 nm. A gradient method used the following solvents: (A) 50 % acetonitrile+ 0.1 % trifluoroacetic acid (TFA); (B) water+ 0.1 % TFA. The gradient began with 5 % Solvent A and 95 % Solvent B. The setting at 20 min was 60 % Solvent A and 40 % Solvent B. The program restored the original ratio at 22 min and ended at 25 min. The flow rate was 1.0 ml/min and all vanillin pathway compounds of interest eluted within 15 min. Column temperature was maintained at 30 °C.
SDS–PAGE analysis
To determine qualitative protein expression level of OMT in the absence of other pathway gene overexpression, E. coli MG1655(DE3) was transformed with empty pETDuet-1 or pET-OMT. Single colonies from plates of each transformation were grown overnight in 3 ml of LB with 50 mg/L ampicillin. Cells were passaged into second overnight cultures by inoculating 3 ml of M9+ 1.8 % glucose with 100 μL of the overnight LB cultures. Shake flask cultures containing 50 ml M9+ 1.8 % glucose were inoculated at 1 % inoculum from overnight M9 cultures and incubated with agitation at 30 °C and 250 rpm. Cultures were induced with 0.5 mM IPTG at OD600 values between 0.8 and 1.0. Twenty-four hours after induction, 5 ml of each culture were sampled and pelleted by centrifugation. Cell pellets were resuspended in 1 ml of 10 mM Tris–HCl at pH 8.0 and lysed using sonication. After lysis, samples were pelleted by centrifugation (16,000g, 4 °C, 20 min) and supernatant was collected as soluble lysate. The remaining pellet was resuspended in 10 mM Tris–HCl and deemed the insoluble fraction.
Total protein was quantified by the Bradford assay method [58] using Bio-Rad Protein Assay Dye Reagent (Cat #500-0006) and a bovine serum album (BSA) standard. A Bio-Rad 10 % Mini-PROTEAN TGX gel (Cat #456-1034) was run using the Mini-PROTEAN Tetra Cell electrophoresis apparatus. Bio-Rad Precision Plus Protein All Blue Standard (Cat #161-0373) and 10 μg of total protein for each sample was loaded on the gel. After running at 200 volts for 33 min, the gel was washed with deionized water before staining with Bio-Rad Bio-Safe Coomassie Stain (Cat #161-0786).
O-methyltransferase in vitro specific activity
To determine the specific activity of the O-methyltransferase as a function of time in the context of the pathway, E. coli MG1655(DE3) was transformed with pET-OMT-AsbF and pACYC-AroG*-PpsA-TktA plasmids. Single colonies from plates of each transformation were grown overnight in 3 ml of LB with 50 mg/L ampicillin and 17 mg/L chloramphenicol. Cells were passaged into second overnight cultures by inoculating 3 ml of M9+ 1.8 % glucose+ antibiotics with 100 μL of the overnight LB cultures. Shake flask cultures containing 50 ml M9+ 1.8 % glucose+ antibiotics were inoculated at 1 % inoculum from overnight M9 cultures and incubated with agitation at 30 °C and 250 rpm. Cultures were induced with 0.5 mM IPTG at OD600 values between 0.8 and 1.0. At 12 h time point increments after induction, 5 ml of each culture were sampled and pelleted by centrifugation. Cell pellets were resuspended in 1 ml of 100 mM sodium phosphate buffer at pH 7.5 and lysed using sonication. After lysis, samples were pelleted by centrifugation (16,000g, 4 °C, 20 min) and supernatant containing OMT was collected.
The OMT activity assay contained the following final concentrations to a final volume of 600 µL per sample: 100 mM sodium phosphate buffer (pH 7.5), 2 mM protocatechuate, 2 mM S-adenosylmethionine, 5 mM MgCl2, and roughly 0.1 mg/mL BSA protein equivalent of lysate (60 µL). Lysate was always added last, and immediately upon addition, the assay solution was split into two 300 µL volumes (ti and tf). The ti volume was quenched with 1 % TFA, and the tf was quenched in the same manner after 1 h. Both volumes for each sample were maintained at 30 °C. Potential debris from quenched volumes were pelleted by centrifugation (16,000g, 4 °C, 3 min) and supernatant was analyzed by HPLC. Concentrations of protocatechuate and vanillate were measured as described before. Specific OMT activity was calculated by dividing differences in vanillate concentration by total protein concentration in corresponding lysate. Specific activities were subsequently normalized to the activity at the first time point sampled (12 h). Lysate from a strain transformed with empty pET was used as a control in otherwise identical assays to ensure there was no background conversion of protocatechuate.
Calculation of maximum theoretical pathway yield
Maximum theoretical yields presented for vanillin and vanillate were calculated using an in silico genome-scale metabolic model of E. coli (iJO1366) [59] and the constraint-based reconstruction and analysis (COBRA) toolbox version 2.0 [60, 61] within the framework of MATLAB 2013a. Heterologous vanillin pathway reactions were added to the model, and uptake rates for glucose and oxygen were set to −10 mmol/gDW-h and −1000 mmol/gDW-h, respectively [62].
Additional file
10.1186/s12934-016-0459-x Supplementary material
Authors’ contributions
AMK and KLJP conceived of the study. AMK performed experiments and data analysis. JCH assisted with sampling of time-course experiments and with OMT activity assays. AMK and KLJP drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by the National Science Foundation through the Synthetic Biology Engineering Research Center (SynBERC, Grant No. EEC-0540879) and through a Graduate Research Fellowship to A.M.K.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Aditya M. Kunjapur, Email: aditya_kunjapur@hms.harvard.edu
Jason C. Hyun, Email: jhyun95@mit.edu
Kristala L. J. Prather, Phone: 617.253.1950, Email: kljp@mit.edu
References
- 1.Krings U, Berger RG. Biotechnological production of flavours and fragrances. Appl Microbiol Biotechnol. 1998;49(1):1–8. doi: 10.1007/s002530051129. [DOI] [PubMed] [Google Scholar]
- 2.Feron G, Wache Y. 1.16 microbial biotechnology of food flavor production. In: Pometto A, Shetty K, Paliyath G, Levin R, editors. Food Biotechnology, 2nd ed. Boca Raton: CRC Press; 2006. p. 407–442.
- 3.Walton NJ, Mayer MJ, Narbad A. Vanillin. Phytochemistry. 2003;63:505–515. doi: 10.1016/S0031-9422(03)00149-3. [DOI] [PubMed] [Google Scholar]
- 4.Hansen EH, Møller BL, Kock GR, Bünner CM, Kristensen C, Jensen OR, Okkels FT, Olsen CE, Motawia MS, Hansen J. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae) Appl Environ Microbiol. 2009;75:2765–2774. doi: 10.1128/AEM.02681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunjapur AM, Tarasova Y, Prather KLJ. Synthesis and accumulation of aromatic aldehydes in an engineered strain of Escherichia coli. J Am Chem Soc. 2014;136:11644–11654. doi: 10.1021/ja506664a. [DOI] [PubMed] [Google Scholar]
- 6.Li K, Frost JW. Synthesis of vanillin from glucose. J Am Chem Soc. 1998;120:10545–10546. doi: 10.1021/ja9817747. [DOI] [Google Scholar]
- 7.Kunjapur AM, Prather KLJ. Microbial engineering for aldehyde synthesis. Appl Environ Microbiol. 2015;81:1892–1901. doi: 10.1128/AEM.03319-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brochado A, Matos C, Moller B, Hansen J, Mortensen U, Patil K. Improved vanillin production in baker’s yeast through in silico design. Microb Cell Fact. 2010;9:84. doi: 10.1186/1475-2859-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brochado AR, Patil KR. Overexpression of O-methyltransferase leads to improved vanillin production in baker’s yeast only when complemented with model-guided network engineering. Biotechnol Bioeng. 2013;110:656–659. doi: 10.1002/bit.24731. [DOI] [PubMed] [Google Scholar]
- 10.Hayden EC. Synthetic-biology firms shift focus. Nature. 2014;505:598. doi: 10.1038/505598a. [DOI] [PubMed] [Google Scholar]
- 11.Fischer C, Klein-Marcuschamer D, Stephanopoulos G. Selection and optimization of microbial hosts for biofuels production. Metab Eng. 2008;10:295–304. doi: 10.1016/j.ymben.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Muntendam R, Melillo E, Ryden A, Kayser O. Perspectives and limits of engineering the isoprenoid metabolism in heterologous hosts. Appl Microbiol Biotechnol. 2009;84:1003–1019. doi: 10.1007/s00253-009-2150-1. [DOI] [PubMed] [Google Scholar]
- 13.Jarboe LR, Zhang X, Wang X, Moore JC, Shanmugam KT, Ingram LO. Metabolic engineering for production of biorenewable fuels and chemicals: contributions of synthetic biology. BioMed Res Int. 2010;2010:761042. doi: 10.1155/2010/761042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaldivar J, Martinez A, Ingram LO. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng. 1999;65:24–33. doi: 10.1002/(SICI)1097-0290(19991005)65:1<24::AID-BIT4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Struck A-W, Thompson ML, Wong LS, Micklefield J. S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis biosynthesis and other biotechnological applications. ChemBioChem. 2012;13:2642–2655. doi: 10.1002/cbic.201200556. [DOI] [PubMed] [Google Scholar]
- 16.Wessjohann L, Dippe M, Tengg M, Gruber-Khadjawi M. Methyltransferases in biocatalysis, in cascade biocatalysis: integrating stereoselective and environmentally friendly reactions. In: Riva S, W-D Fessner, editors, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim; 2014.
- 17.Fox DT, Hotta K, Kim C-Y, Koppisch AT. The missing link in petrobactin biosynthesis: asbF encodes a (-)-3-dehydroshikimate dehydratase. Biochemistry. 2008;47:12251–12253. doi: 10.1021/bi801876q. [DOI] [PubMed] [Google Scholar]
- 18.Berry A. Improving production of aromatic compounds in Escherichia coli by metabolic engineering. Trends Biotechnol. 1996;14:250–256. doi: 10.1016/0167-7799(96)10033-0. [DOI] [PubMed] [Google Scholar]
- 19.Draths KM, Pompliano DL, Conley DL, Frost JW, Berry A, Disbrow GL, Staversky RJ, Lievense JC. Biocatalytic synthesis of aromatics from d-glucose: the role of transketolase. J Am Chem Soc. 1992;114:3956–3962. doi: 10.1021/ja00036a050. [DOI] [Google Scholar]
- 20.Snell KD, Draths KM, Frost JW. Synthetic modification of the Escherichia coli chromosome: enhancing the biocatalytic conversion of glucose into aromatic chemicals. J Am Chem Soc. 1996;118:5605–5614. doi: 10.1021/ja9538041. [DOI] [Google Scholar]
- 21.Holcomb ER, Shapiro SK. Assay and regulation of S-adenosylmethionine synthetase in Saccharomyces cerevisiae and Candida utilis. J Bacteriol. 1975;121:267–271. doi: 10.1128/jb.121.1.267-271.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hondorp ER, Matthews RG. Oxidative Stress Inactivates Cobalamin-Independent Methionine Synthase (MetE) in Escherichia coli. PLoS Biol. 2004;2:e336. doi: 10.1371/journal.pbio.0020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004;2:e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hondorp ER, Matthews RG. Oxidation of cysteine 645 of cobalamin-independent methionine synthase causes a methionine limitation in Escherichia coli. J Bacteriol. 2009;191:3407–3410. doi: 10.1128/JB.01722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mordukhova EA, Pan J-G. Evolved cobalamin-independent methionine synthase (MetE) improves the acetate and thermal tolerance of Escherichia coli. Appl Environ Microbiol. 2013;79:7905–7915. doi: 10.1128/AEM.01952-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roe AJ, O’Byrne C, McLaggan D, Booth IR. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology. 2002;148:2215–2222. doi: 10.1099/00221287-148-7-2215. [DOI] [PubMed] [Google Scholar]
- 27.Tuite NL, Fraser KR, O’Byrne CP. Homocysteine toxicity in Escherichia coli is caused by a perturbation of branched-chain amino acid biosynthesis. J Bacteriol. 2005;187:4362–4371. doi: 10.1128/JB.187.13.4362-4371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar D, Gomes J. Methionine production by fermentation. Biotechnol Adv. 2005;23:41–61. doi: 10.1016/j.biotechadv.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Willke T. Methionine production—a critical review. Appl Microbiol Biotechnol. 2014;98:9893–9914. doi: 10.1007/s00253-014-6156-y. [DOI] [PubMed] [Google Scholar]
- 30.Su C-H, Greene RC. Regulation of methionine biosynthesis in Escherichia coli: mapping of the metJ locus and properties of a metJ +/metJ- diploid. Proc Natl Acad Sci. 1971;68:367–371. doi: 10.1073/pnas.68.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saint-Girons I, Duchange N, Cohen GN, Zakin MM. Structure and autoregulation of the metJ regulatory gene in Escherichia coli. J Biol Chem. 1984;259:14282–14285. [PubMed] [Google Scholar]
- 32.Shoeman R, Redfield B, Coleman T, Greene RC, Smith AA, Brot N, Weissbach H. Regulation of methionine synthesis in Escherichia coli: effect of metJ gene product and S-adenosylmethionine on the expression of the metF gene. Proc Natl Acad Sci. 1985;82:3601–3605. doi: 10.1073/pnas.82.11.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamori S, Kobayashi S, Nishimura T, Takagi H. Mechanism of l-methionine overproduction by Escherichia coli: the replacement of Ser-54 by Asn in the MetJ protein causes the derepression of l-methionine biosynthetic enzymes. Appl Microbiol Biotechnol. 1999;52:179–185. doi: 10.1007/s002530051506. [DOI] [PubMed] [Google Scholar]
- 34.Usuda Y, Kurahashi O. Effects of deregulation of methionine biosynthesis on methionine excretion in Escherichia coli. Appl Environ Microbiol. 2005;71:3228–3234. doi: 10.1128/AEM.71.6.3228-3234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris C. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J Bacteriol. 1981;145:1031–1035. doi: 10.1128/jb.145.2.1031-1035.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kai Y, Kashiwagi T, Ishikawa K, Ziyatdinov MK, Redkina EI, Kiriukhin MY, Gusyatiner MM, Kobayashi S, Takagi H, Suzuki E. Engineering of Escherichia colil-serine O-acetyltransferase on the basis of crystal structure: desensitization to feedback inhibition by l-cysteine. Protein Eng Des Sel. 2006;19:163–167. doi: 10.1093/protein/gzj015. [DOI] [PubMed] [Google Scholar]
- 37.Coward JK, D’Urso-Scott M, Sweet WD. Inhibition of catechol-O-methyltransferase by S-adenosylhomocysteine and S-adenosylhomocysteine sulfoxide, a potential transition-state analog. Biochem Pharmacol. 1972;21:1200–1203. doi: 10.1016/0006-2952(72)90114-1. [DOI] [PubMed] [Google Scholar]
- 38.Coward JK, Slisz EP, Wu FYH. Kinetic studies on catechol O-methyltransferase, product inhibition and the nature of the catechol binding site. Biochemistry. 1973;12:2291–2297. doi: 10.1021/bi00736a017. [DOI] [PubMed] [Google Scholar]
- 39.Parveen N, Cornell KA. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol. 2011;79:7–20. doi: 10.1111/j.1365-2958.2010.07455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6:191–197. doi: 10.1016/S1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 41.Barrios AFG, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Attila C, Wang L, Wood TK, Valdes JJ, Bentley WE. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J Bacteriol. 2007;189:6011–6020. doi: 10.1128/JB.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegde M, Englert DL, Schrock S, Cohn WB, Vogt C, Wood TK, Manson MD, Jayaraman A. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J Bacteriol. 2011;193:768–773. doi: 10.1128/JB.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halliday NM, Hardie KR, Williams P, Winzer K, Barrett DA. Quantitative liquid chromatography–tandem mass spectrometry profiling of activated methyl cycle metabolites involved in LuxS-dependent quorum sensing in Escherichia coli. Anal Biochem. 2010;403:20–29. doi: 10.1016/j.ab.2010.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Nawabi P, Bauer S, Kyrpides N, Lykidis A. Engineering E. coli for biodiesel production utilizing a bacterial fatty acid methyltransferase. Appl Environ Microbiol. 2011;77:8052–8061. doi: 10.1128/AEM.05046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sung SH, Ahn JH. Optimization of rhamnetin production in Escherichia coli. J Microbiol Biotechnol. 2011;21:854–857. doi: 10.4014/jmb.1104.04048. [DOI] [PubMed] [Google Scholar]
- 47.Kim BG, Joe EJ, Ahn JH. Molecular characterization of flavonol synthase from poplar and its application to the synthesis of 3-O-methylkaempferol. Biotechnol Lett. 2010;32:579–584. doi: 10.1007/s10529-009-0188-x. [DOI] [PubMed] [Google Scholar]
- 48.Leonard E, Chemler J, Lim KH, Koffas MA. Expression of a soluble flavone synthase allows the biosynthesis of phytoestrogen derivatives in Escherichia coli. Appl Microbiol Biotechnol. 2006;70:85–91. doi: 10.1007/s00253-005-0059-x. [DOI] [PubMed] [Google Scholar]
- 49.Jeon YM, Kim BG, Ahn JH. Biological synthesis of 7-O-methyl apigenin from naringenin using Escherichia coli expressing two genes. J Microbiol Biotechnol. 2009;19:491–494. doi: 10.4014/jmb.0807.402. [DOI] [PubMed] [Google Scholar]
- 50.Malla S, Koffas MA, Kazlauskas RJ, Kim BG. Production of 7-O-methyl aromadendrin, a medicinally valuable flavonoid, in Escherichia coli. Appl Environ Microbiol. 2012;78:684–694. doi: 10.1128/AEM.06274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 52.Thomason LC, Costantino N, Court DL. E. coli genome manipulation by P1 transduction. In: Current protocols in molecular biology. Wiley; 2001. [DOI] [PubMed]
- 53.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2006(2):0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tseng H-C, Martin CH, Nielsen DR, Prather KLJ. Metabolic engineering of Escherichia coli for enhanced production of (R)- and (S)-3-hydroxybutyrate. Appl Environ Microbiol. 2009;75:3137–3145. doi: 10.1128/AEM.02667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solomon KV, Moon TS, Ma B, Sanders TM, Prather KLJ. Tuning primary metabolism for heterologous pathway productivity. ACS Synth Biol. 2013;2:126–135. doi: 10.1021/sb300055e. [DOI] [PubMed] [Google Scholar]
- 57.Juminaga D, Baidoo EEK, Redding-Johanson AM, Batth TS, Burd H, Mukhopadhyay A, Petzold CJ, Keasling JD. Modular engineering of l-tyrosine production in Escherichia coli. Appl Environ Microbiol. 2012;78:89–98. doi: 10.1128/AEM.06017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 59.Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, Palsson BØ. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol Syst Biol. 2011;7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA toolbox. Nat Protoc. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 61.Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinksi DC, Bordbar A, Lewis NE, Rahmanian S, Kang J. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
10.1186/s12934-016-0459-x Supplementary material


