Abstract
Microbial communities inhabit our entire planet and play a crucial role in biogeochemical processes, agriculture, biotechnology, and human health. Here, we argue that “in situ microbiome engineering” represents a new paradigm of community-scale genetic and microbial engineering. We discuss contemporary applications of this approach to directly add, remove, or modify specific sets of functions and alter community-level properties in terrestrial, aquatic, and host-associated microbial communities. Specifically, we highlight emerging in situ genome engineering approaches as tractable techniques to manipulate microbial communities with high specificity and efficacy. Finally, we describe opportunities for technological innovation and ways to bridge existing knowledge gaps to accelerate the development of in situ approaches for microbiome manipulations.
Keywords: microbiome engineering, genome engineering, microbial communities, prebiotics, probiotics, phage, xenobiotics, synthetic biology
Advances and roadblocks in microbiome research
Over the past decade, breakthroughs in high-throughput sequencing to read DNA from genomes have vastly outpaced our capabilities to edit genetic information. Developments in metagenomic and transcriptomic sequencing of mixed cell populations have enabled large-scale quantification of microbial community composition, function, and dynamics in a culture-independent manner [1]. However, we still lack a basic mechanistic understanding of the individual genetic factors that drive overall function and emergent ecological principles in these communities. To understand these complex communities and engineer them in useful ways, we must address critical technological and knowledge gaps in systems-level genetic manipulation [2].
Most microbial communities in nature exist in complex, dynamic consortia with highly interconnected networks of metabolic and ecological interactions that have yet to be unraveled. Consequently, the natural environments of most microbial communities are difficult, if not impossible, to recreate experimentally. In fact, a large majority of microbes have not been cultivated in the laboratory [3]. These microbes are not accessible for genetic studies, and their function and properties within their communities remain unknown. Even for culturable microbes, only a few genetic systems have been utilized in specific model strains [4]. Development of new genetic systems requires significant time and effort, and resulting tools are not necessarily transferrable between different microbes. Furthermore, studies of single model strains in laboratory conditions may not necessarily reflect behaviors relevant to natural environments. No general methods are available today to genetically engineer consortia of different organisms in situ. These challenges greatly limit the functional analysis and forward engineering of poly-microbial communities of any level of complexity.
Engineering microbial communities in situ
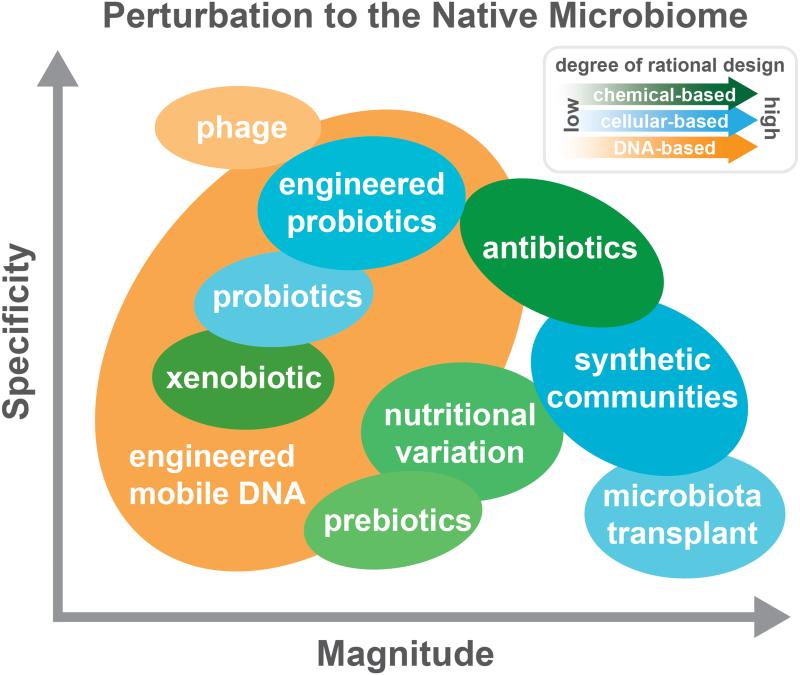
In situ microbiome engineering (see glossary terms) methods allow for manipulation and study of microbial communities in their native context without the need for individual laboratory domestication. These approaches can be classified by several characteristics: the magnitude of perturbation to community composition and function, specificity of perturbation to community members or processes, and degree of engineerability (Figure 1). While some approaches have low specificity and can lead to large-scale changes (e.g. microbiota transplants), others can be more easily designed to affect specific members while minimizing overall impact on the community (e.g. engineered probiotics). Here, we discuss contemporary in situ microbiome engineering approaches and offer in situ genome engineering as a new paradigm to directly manipulate communities with greater control of magnitude and specificity.
Figure 1. In situ microbiome perturbations vary in magnitude, specificity and degree of rational design required.
A variety of approaches, based on chemical (green), cellular (blue) and DNA (orange) methods can be applied to manipulate microbial communities in their native context. Each method can vary in its magnitude of perturbation to the native microbiome, shown increasing on the horizontal axis; its specificity of targeting to particular community members, shown increasing on the vertical axis; and its degree of required rational design, shown with increasing shading density. Particular combinations of magnitudes and specificities may be desirable for given target applications. Chemical-based approaches such as xenobiotics, prebiotics, and nutritional variation yield relatively broad spectrum changes, with varying magnitudes. Antibiotics, a class of xenobiotics, can yield larger magnitude changes with higher specificity. Cellular-based techniques such as probiotics and engineered probiotics can yield low magnitude, specific perturbations, while large-scale microbiota transplants or synthetic communities can yield to larger, but less specific changes. Finally, DNA-based methods such as phages can yield highly specific, albeit low magnitude perturbations, while engineered mobile DNA can yield perturbations over a large range of magnitudes and specificity. This flexible control of magnitude and specificity implies that engineered mobile DNA may be a desirable and tractable method for manipulating microbial communities in comparison to other methods.
Contemporary methods for in situ microbiome engineering
Chemical, cellular and phage-based methods can be used to alter microbial communities in situ. Common examples of each approach are outlined in Table 1 and discussed in detail below.
Table 1.
Commonly utilized in situ microbiome engineering methods
| Method | Class (common formulations) |
Predominant Targets | Mechanism of Action |
|---|---|---|---|
| Prebiotics (chemical-based) |
Dietary fibers (Inulin), Polysaccharides (oligosaccharides) |
Firmicutes (Lactobacillus) Actinobacteria (Bifidobacteria) |
Promote bacterial growth; mechanism generally unknown |
| Antibiotics (chemical-based) |
β-lactams (Cephalosporins, Carbapenems) |
Clostridium, Staphylococcus, Streptococcus |
Block cell wall synthesis |
| Aminoglycosides (Kanamycin, Streptomycin) |
Klebsiella, Pseudomonas |
Block protein synthesis |
|
| Macrolides (Erythromycin, Azithromycin) |
Chlamydia, Legionella, Mycoplasma |
Block protein synthesis |
|
| Glycopeptides (Vancomycin) |
Enterococcus, Clostridium, Staphylococcus |
Block peptidoglycan synthesis |
|
| Quinolones (Ciprofloxacin, Levofloxacin) |
Neisseria, Pseudomonas, Streptococcus |
Block DNA replication | |
| Metronidazole | Bacteroides, Clostridium | Block DNA/RNA synthesis |
|
| Probiotics (cellular-based) |
Firmicutes (Lactobacillus), Actinobacteria (Bifidobacteria), Proteobacteria |
Variable, broadly targeting |
Compete for nutrients, produce antimicrobials, modulate environment |
| Microbiota transplants (cellular-based) |
Fecal microbiota transplants |
Variable, broadly targeting |
Replace native community; mechanism generally unknown |
| Bacteriophages (phage-based) |
Specific phage strains or cocktails of phages |
Variable, but strain- specific |
Cell lysis (lytic); genomic integration (lysogenic) |
Chemical modifiers of microbiomes
Biochemical availability can predictably affect microbiome composition and function [5]. Prebiotics are naturally occurring chemicals that selectively promote growth or activity in a community. Human-associated prebiotics are often non-digestible dietary polysaccharides that stimulate growth of commensal bacteria in the gut [6, 7]. Prebiotics can be used in other settings such as the food industry, where the polysaccharide β-glucan has been commercially sold to improve fish health and resistance to infection [8]. However, a major limitation of prebiotics is the inability to rationally predict or change the specificity of their manipulations. To address this shortcoming, approaches such as functional metagenomics, transcriptome and transposon sequencing have been used to identify gene-level fitness determinants of individual microbes in specific metabolic niches [9, 10]. These measurements have yielded more nuanced nutritional variation approaches that can be exploited to modulate the microbiome with higher precision [11]. For example, transposon sequencing of a model gut microbiome community revealed a specific dietary metabolite that could modulate the abundance of a single strain in the community [12]. Further, transcriptome sequencing of the gut bacterium E. lenta revealed specific genes that inactivate the cardiac drug digoxin and their transcriptional regulation, enabling design of a dietary modification to reduce inactivation in vivo [13].
Targeted biochemical modulation of microbiota can also utilize xenobiotics, compounds foreign to an environment designed to modulate microbial function or growth. For example, β-glucuronidase inhibitors have been used to reduce toxicity of a chemotherapeutic by inactivating bacterial enzymes that reactivate the drug [14]. Asparaginase can be delivered in vivo to protect against infection by degrading asparagine, a regulator of group A Streptococcus proliferation [15]. Finally, a small molecule structural analog of choline can inhibit TMA production by microbes, reducing levels of TMAO, a metabolite associated with cardiac disease [16].
Antibiotics are a widely used class of xenobiotics that modulate microbial growth by inhibiting essential cellular machinery. Most antibiotics target membrane integrity, protein synthesis or replication processes [17]. However, due to the widely conserved function of mechanisms targeted by antibiotics, they tend to affect a broad spectrum of bacteria, causing large and potentially undesirable changes in the community [18]. Antibiotic use can lead to persistent alterations in microbiota composition over time, and can further select for antibiotic resistance genes [19]. With the emergence of many antibiotic-resistant pathogens, there is a critical need for new classes of antibiotics that selectively target undesirable strains without promoting the spread of broad-spectrum resistance. Antimicrobial peptides and secondary metabolites such as bacteriocins are outstanding candidates for such novel antibiotics, as they display selective elimination of particular strains and a diverse set of compounds are present in natural communities [20, 21]. A better mechanistic understanding of specific biochemical processes and strain-level genetic factors will improve novel strategies to modulate microbiome growth and function with high specificity.
Cellular modifiers of microbiomes
Beyond biochemical approaches, live bacterial strains or communities can be used to manipulate microbial ecosystems. In contrast to molecular modulators, these cellular approaches can yield more nuanced interaction and function over space and time. Probiotics are bacteria which can confer a benefit to a particular host environment [22]. For example, probiotic Lactobacilli have been used in livestock to decrease incidence of pathogenic infections [23]. In humans, probiotic bacteria alter the gut microbiota by competing for nutrients, producing antimicrobial compounds, or modulating host immunity [24, 25]. However, our fledgling understanding of probiotic mechanistic function has limited their value as a tool for predictive microbiome manipulation.
Genetically engineered probiotics have potential for more targeted community manipulations. For example, in the poultry industry, genetically modified Salmonella lacking virulence factors have been used to vaccinate chickens against infection [26]. In humans, synthetic biology tools can be applied to engineer probiotics with precise and novel functions. For example, bacteria have been modified to modulate microbiota or host physiology by secreting chemicals or proteins, including human interleukin-10 to reduce inflammation [27], NAPEs to reduce food intake and obesity [28], and bacterial quorum signals to modulate microbiota composition [29]. Wholly new functions can also be engineered; Danino et al. engineered an E. coli probiotic as an orally administered diagnostic of liver metastasis in mice through production of a detectable signal in urine [30]. More complex synthetic biology circuits such as combinatorial logic [31] or memory [32] circuits can be layered upon these simple designs to improve the precision, specificity and controllability of desired perturbations.
Mixtures of bacteria can also be utilized to manipulate microbiomes. Microbiota transplantation is the beneficial transfer of live bacteria from one environment into another. This approach has been recently popularized through successful clinical trials of fecal microbiota transplants to treat recurrent C. difficile infections [33]. It is hypothesized that transplantation may replace the existing microbial community with a more infection-resistant community from a healthy donor, but the mechanisms of this process remain largely unclear [34]. A more refined approach to transplantation is the transfer of synthetic communities, which could replicate the functions of complex consortia, but contain defined members that are amenable to detailed genetic and biochemical studies [35-37]. We envision that novel experimental tools to delineate interspecies and host-microbe interactions and improved metabolic and ecological modeling [38], combined with functional studies in gnotobiotic animals [39], will enable better design of microbial consortia to precisely manipulate microbial communities.
Phage-based modifiers of microbiomes
Bacteriophages (phages) are the most abundant, diverse, and rapidly replicating life forms on earth [40]. Phages infect a host microbe, hijack its replication machinery to reproduce, and then replicate via stable genomic integration (lysogenic cycle) or lysis of the host and dissemination (lytic cycle). This life cycle makes them ideal genetic engineering candidates to selectively eliminate strains or transfer specific genes in microbial populations. Indeed, natural phages have been used to limit growth of undesirable or pathogenic bacteria in humans [41], agriculture [42], food processing [43], and aquaculture [44]. Phages have been further genetically engineered to deliver specific DNA payloads or to alter host specificity. For example, phages have been designed to deliver biofilm dispersal enzymes [45] or genes that increase antibiotic sensitivity [46]. By delivering the CRISPR-Cas RNA-guided nuclease system with phages, designated strains can be selectively eliminated based on their genetic content [47]. Furthermore, phage host ranges can be modularly engineered by swapping phage tail components [48]. Altering phage populations represents another avenue for microbiome modulation; exposing the gut microbiome to antibiotics alters its associated virome and ecological networks [49]. Quantitative characterization of phage ecology, combined with advances in forward genetic engineering of phage function, will allow for more complex manipulations of microbial communities in situ.
In situ genome engineering: an emerging frontier in microbiome modulation
Despite much progress, contemporary methods for modifying microbiomes have not seen widespread success in achieving desired manipulations. We attribute these shortcomings to two major knowledge and technical barriers. First, we lack a fine-scale understanding of how individual microbial species function in the context of their natural environments, and a subsequent large-scale understanding of emergent ecosystem function. Second, we lack techniques to efficiently manipulate microbial communities over a large range of magnitudes and specificities. These barriers have limited the effective design and physical implementation of manipulations.
Current methods face a variety of outstanding engineering challenges. For example, chemical manipulations, such as transformation, are specific to particular microbial strains and biochemical processes and cannot be broadly applied. Cellular approaches require colonization of a foreign strain into an ecologically competitive environment, which may be difficult to engineer or lead to unwanted consequences.
Rather than targeting specific strains or functions, one could instead directly modify the metagenomic content of a community to achieve a desired manipulation. While genomes vary greatly between microbes, the metagenome of a community is more constant [50], and governs its biochemical and cellular function. Compared to currently available techniques, direct genomic manipulation could enable perturbations with magnitude and specificity tunable over a much greater range. For example, a metabolic pathway could be added directly to the genome of a native microbe, rather than introducing a foreign strain containing the pathway, thus reducing off-target effects and achieving a high-specificity manipulation. Alternatively, the same pathway could be targeted to a broad range of native organisms to achieve a large-magnitude manipulation. Advances in materials science to manipulate and polymerize chemical building blocks enabled the proliferative use of plastics in the 20th century; we analogously envision that direct, tunable manipulations of the genetic building blocks of microbial communities will enable novel bioengineering applications. We advocate for the development of in situ genome engineering approaches, or techniques to directly manipulate genetic information and engineer new functions in complex microbial communities (Figure 2).
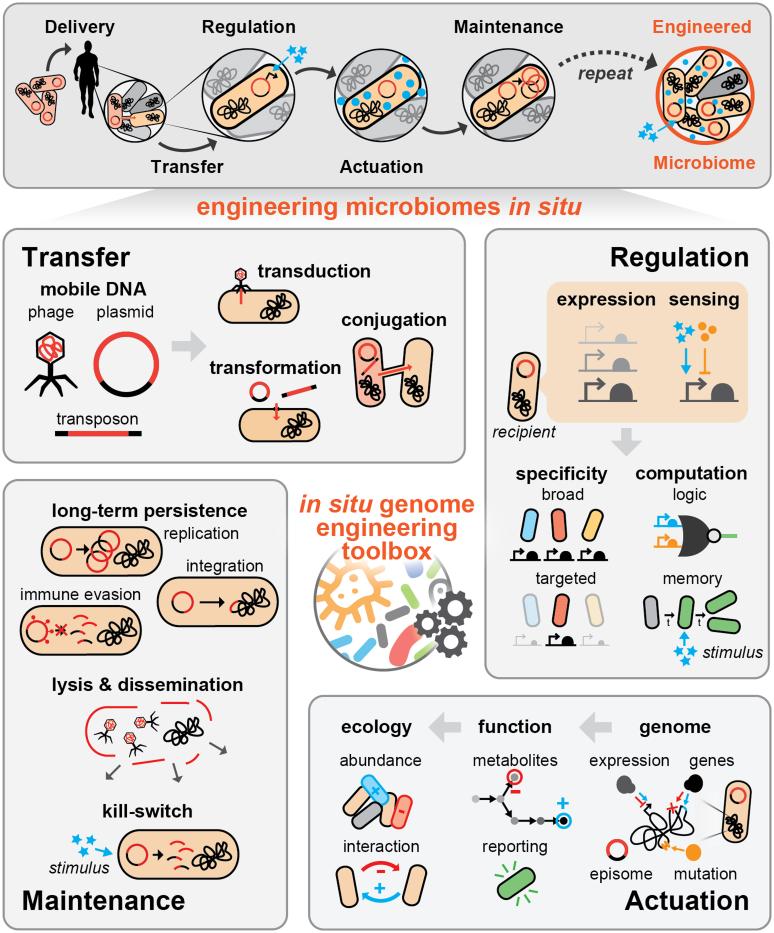
Figure 2. The in situ genome engineering toolbox.
The genomic content of native microbial communities can be directly engineered via in situ genome engineering. As we depict in the top panel, mobile genetic elements can be delivered and transferred to an endogenous microbiome, where they elicit desired function via a combination of regulation and actuation strategies; these elements can then maintain themselves over time to achieve long term desired function. A toolbox of existing and novel genetic tools will enable engineering of mobile genetic elements for in situ genome engineering methods. Transfer methods such as phages, plasmids and transposons can be used to deliver and circulate engineered DNA sequences to microbial communities, via processes such as transduction, transformation and conjugation. Regulatory parts, including transcription and translation parts and sensors of endogenous and exogenous chemical ligands will enable the construction of more complex genetic devices to tune host-range and endow higher order functions such as logic and memory. Actuation of genomes, including addition and removal of genes, modulation of expression, targeted mutations, and episomal modifications will allow for changes to underlying community metabolic function, or introduction of wholly new functions such as reporting on the state of an environment. These functional manipulations can further alter communities at the ecological level by altering the abundances of specific strains or introducing competitive or cooperative interactions. Maintenance of these mobile elements allows for dynamic and long-term control of engineered genetic content; with replication, integration and optimized immune evasion, vectors can be stably propagated over time. Alternatively, lysis can quickly disseminate phages across a community, or engineered circuits such as kill-switches could be used to eliminate circuits as a safe-guard.
Because complex communities are difficult to recapitulate in the laboratory, new approaches are needed for genome engineering in situ. In nature, microbial genomes are in constant flux as a result of abundant horizontal gene transfer events mediated by mobile genetic elements [51]. These horizontal gene transfer events have been increasingly recognized for the important roles they play in the evolution of individual genomes [52] and of entire microbial communities [49]. Furthermore, these events occur rapidly, on timescales less than a week between bacterial species in the gut microbiome [53]. Thus, natural horizontal gene transfer processes, including conjugation, natural transformation, and phage delivery are viable entry points for in situ genome engineering.
Conjugation via plasmids or transposable elements enables transfer of genetic material through direct cell-to-cell contact. An in situ conjugative system could utilize a transiently introduced donor cell carrying an engineered mobile plasmid or transposable element that would be transferred to native microbiota. Secondary conjugations could be engineered to occur following the first transfer to promote further propagation. Bacteria can also uptake extracellular DNA through natural transformation machinery [54] or chemical and physical transformation processes, such as abrasion with mineral particles [55], which creates small membrane pores that allow DNA into the cytoplasm [56]. Phages can also be utilized to transfer and insert genes into bacterial genomes.
Building an expansive in situ genome engineering toolbox
We envision a suite of genetic tools that will significantly expand our ability to activate stable and controllable synthetic gene circuits in complex natural microbiomes. First, natural horizontal gene transfer vectors could be engineered with tunable host ranges and dynamics. These vectors could then be augmented with existing and emerging synthetic biology tools, such as transcriptional and translational regulatory parts, logic gates, and genome editing tools to add, remove, or modify particular functions and ecologies. Finally, maintenance of these engineered vectors could be precisely controlled, resulting in propagation across specific members of a community or destruction via controlled kill switches. Recent studies suggest that such mobile genetic element-mediated transfer may be a tractable approach to manipulate diverse communities. Plasmids can be broadly mobilized into naturally occurring soil bacteria, transferring to bacteria from 11 different phyla [57] with efficiencies of up to 1 in 10,000 cells within a few days [58, 59]. Furthermore, viral tagging experiments have revealed that in nature, a single bacterial host can harbor dozens of different phage populations, suggesting that viruses can be isolated and engineered for a broad range of hosts [60]. Here, we highlight key components that need to be developed to form the foundation for this new in situ genome engineering toolbox.
New replicative or integrative plasmids are needed for stable propagation of exogenous DNA in the microbiome. Recent sequencing efforts have demonstrated that plasmids are prevalent in microbial communities [61]. However, the host ranges of most plasmids remain unknown, and their associated proteins and modes of regulation are poorly characterized [62]. Few, if any, new plasmids are being developed, as almost all current vectors are based on plasmids isolated during the pre-genomic era. Characterizing more natural plasmids with various host ranges will elucidate the principles underlying their host specificity, transfer dynamics, and stability, and will aid in the construction of new synthetic vectors [63].
Tunable gene regulation systems with varying strengths and host-specificities are needed to better control the activity of synthetic circuits across microbial communities. A repository of characterized regulatory parts such as promoters and ribosome binding sites for diverse microbes does not yet exist and needs to be developed. Furthermore, basic measurement techniques to assess genetic circuit function across many species in parallel have not been developed. New strategies will be needed to engineer genetic circuits with broad and defined host ranges [64]. Existing and novel genetic parts, combined with community-level measurement strategies, will yield design principles for building genetic circuits with predictable performance in different hosts.
Recent advances in genome editing have enabled programmable modification of microbial genomes. RNA-guided CRISPR-Cas9 systems have been used to site-specifically edit bacterial genomes [65], while Ll.LtrB group II introns, retrotransposons that undergo RNA intermediary steps, have been successfully repurposed for targeted gene editing of multiple bacterial species [66]. While these techniques enable powerful interrogation of the genome, new tools to enable strain-and site-specific genomic integration of large synthetic constructs are still required, as current approaches are often inefficient or difficult to target to desired genomic sites. Additionally, increased knowledge and functional annotation of microbial genomes, coupled with advances in modeling techniques to predict the effects of particular genomic modifications, will be necessary to maximize the utility of existing genome editing tools.
More sophisticated genetic devices are also needed to perform higher function processes. Programmable transcriptional and post-transcriptional regulators [67, 68], combined with new chemical sensing pathways, will enable sophisticated genetic circuitry such as memory devices and kill switches to control engineered functions in complex communities [69]. Other strategies, such as a gene drive to propagate engineered function in higher order organisms [70], conceptually similar to bacterial in situ genome engineering, may be further developed for microbial populations.
A better understanding of the function and dynamics of natural mobile genetic elements will enable strategies for long-term persistence of mobile genetic elements. A major challenge in utilizing these natural elements is ensuring efficient delivery, transfer, and stability of the system over time. Delivery of mobile genetic elements and subsequent transfer between endogenous microbiota in situ could minimize perturbations to the overall structure of a given community. Mobile elements found in the wild employ elegant strategies to ensure their long-term presence; for example, an E. faecalis conjugative plasmid expressing a bacteriocin enhances niche colonization of its host, facilitating transfer to other E. faecalis strains in the mammalian gut [53]. Similar strategies which couple a niche or metabolic advantage to a mobile element could be employed to engineer selection and stability over time.
Finally, many bacterial immune systems such as CRISPRs and restriction endonucleases prevent foreign DNA from infiltrating the cell. Active immune evasion is required to enable efficient transfer and propagation of engineered DNA, which may require sequence recoding to avoid restriction enzyme digestion or modification of DNA methylation patterns to match that of the recipient cells. In fact, methylation-matching has been shown to increase gene transfer rates by several orders of magnitude [71]. Experimental and design tools for bacterial immune evasion are needed to predictably manipulate gene transfer efficiencies in situ.
Design principles, knowledge gaps and applications of in situ microbiome engineering
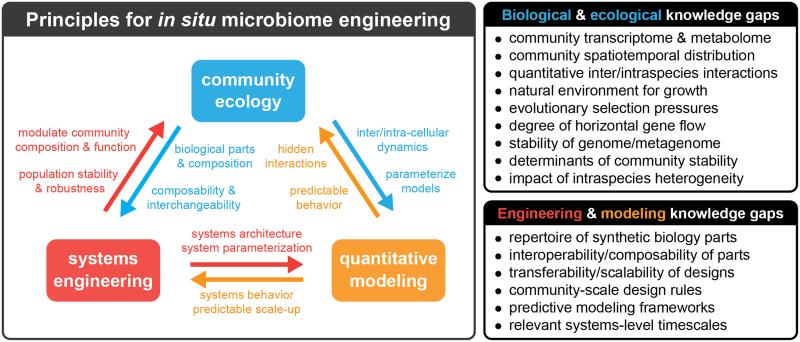
Successful in situ microbiome engineering will require an expanded understanding of basic ecological principles, new systems measurement methods and genetic parts, and the application of existing and new quantitative modeling frameworks (Figure 3, Key Figure). These three major knowledge areas directly inform and influence each other; for example, underlying ecology provides a starting point for determining new genetic parts for systems engineering. The performance of these parts can then be used to parameterize quantitative models of complex microbial systems, which could ultimately reveal hidden or underlying ecological interactions. However, as outlined in Figure 3, key knowledge gaps still remain to be addressed.
Figure 3. Key Figure. Principles and knowledge gaps in in situ genome engineering.
Key principles for in situ microbiome engineering include community ecology (blue), systems engineering (red), and quantitative modeling (orange). On the left, we illustrate the direct interplay between each of the three principles; these directional interactions and knowledge that inform one-another are denoted via color coded arrows and text. On the right, we detail specific biological and ecological and engineering and modeling knowledge gaps.
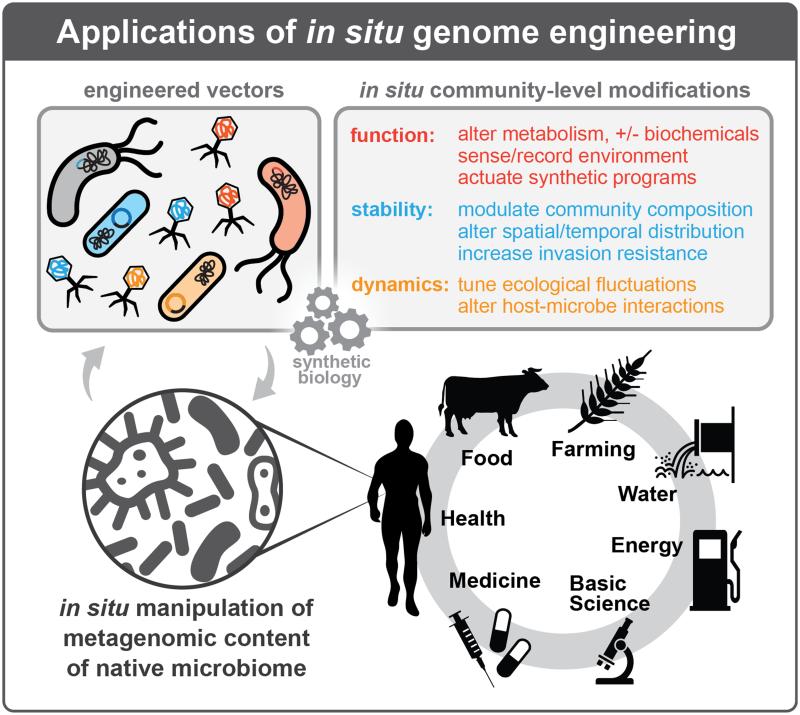
The application of in situ microbiome engineering to basic science questions will enable a new class of experiments to elucidate the determinants of individual and community function. In individual strains, these new tools will allow for genetic studies of unculturable microbes and the mechanistic basis of microbial fitness and function. In communities, these tools will enable large-scale perturbations of community composition and interactions at unprecedented resolution and scale. Further, in situ microbiome engineering has numerous applications to the addition, deletion or modification of community-level functions and properties (Figure 4). In human-associated communities, microbiome manipulations could improve health and nutrition via removal of detrimental host interactions (e.g., chronic inflammation) and addition of beneficial processes (e.g., producing essential nutrients). Altering community-level properties such as resilience to infection could yield novel ecology-based treatments for infectious diseases. In an agricultural setting, targeted manipulations could increase crop yield by accelerating nutritional absorption or enhance bioremediation by removing toxins. In human-made environments such as buildings, pipelines, and ship hulls, engineered communities could enable exclusion of strains with undesirable properties (e.g. pathogenic or biofouling) and augmentation of materials with desirable “smart” properties such as self-healing, chemical production, and recording exposure to biochemical compounds. Finally, synthetic communities with defined functions could replace natural communities in certain settings to enable predictability and control over particular biochemical processes. Such communities could potentially be used to colonize environments lacking endogenous microbiota, such as other planets, to improve habitability for humans.
Figure 4. Applications of in situ genome engineering.
In situ genome engineering approaches could be utilized to address fundamental basic science questions and key applications in medicine, health, food, farming, water and energy. Engineered vectors introduced into native environments could allow for manipulation of metagenomic content, and would propagate within the community over time. These engineered vectors could then endow these communities with desirable alterations to community-level function, stability, and dynamics, detailed in the upper right panel.
Safety and regulation
The manipulation and engineering of microbial ecosystems in natural environments will require significant advances in our ability to reliably predict engineering outcomes and safeguard against undesirable events. Current manipulations of microbial ecosystems, intended or not, are widespread and subject to a complex litany of regulatory policies with varying stringency. New policies and regulatory frameworks will be required to appropriately evaluate the safety and implementation of emerging approaches such as genetically engineered probiotics and mobile vectors. Analogous to conversations around the use of gene drives to cause forced inheritance of engineered traits in a population [72], or CRISPR-Cas9 for gene editing purposes [73], the societal implications of microbiome engineering technologies and their regulation must be carefully considered, evaluated, and communicated to the public in order for these open-environment engineering approaches to be rationally evaluated and safely adopted.
Concluding remarks
In situ microbiome and genome engineering offer exciting opportunities at the frontier of population and ecological engineering with applications in basic science, human health, agriculture, and beyond. As we move from understanding and engineering individual organisms to entire ecosystems, we envision that these emerging techniques will reveal a vast diversity and elegance underlying natural microbial ecosystems, and will correspondingly suggest wholly new strategies to manipulate microbial communities (See Outstanding Questions box).
OUTSTANDING QUESTIONS.
How do specific biological factors such as genetic content or biochemical interactions lead to emergent ecological properties, including spatial variation, dynamics and stability?
What is the extent of horizontal gene transfer in microbiomes, and how does this impact intraspecies heterogeneity, genomic stability, evolutionary selection and community function?
How can standardized synthetic biology genetic parts for genetic transfer, regulation, and genome editing be characterized and designed across many different bacteria?
What systems engineering principles can be leveraged to increase transferability and composability of these parts into more complex genetic devices across many different hosts?
What are appropriate quantitative modeling frameworks to predict specific microbiome manipulations?
TRENDS BOX.
High-throughput sequencing advances have provided a detailed survey of microbial community composition and prevalence, but a functional and mechanistic understanding of microbial ecology is lacking.
Manipulating microbiome composition and function is of great interest for basic science and engineering applications; contemporary methods for manipulating microbial communities in situ yield perturbations limited to particular specificities and magnitudes.
Emerging in situ genome engineering tools can precisely alter the metagenomic content of microbial communities over a large range of specificities and magnitudes.
Acknowledgements
We thank members of the Wang lab for helpful discussions and feedback that help shape the content of this work. H.H.W. acknowledges funding support from the NIH (1DP5OD009172-02), NSF (MCB-1453219), Sloan Foundation (FR-2015-65795), DARPA (W911NF-15-2-0065), and ONR (N00014-15-1-2704). R.U.S. is supported by a Fannie and John Hertz Foundation Graduate Fellowship and an NSF Graduate Research Fellowship (DGE-11-44155). S.P.C. is supported by a NIH MSTP training grant (NIH T32GM007367).
GLOSSARY
- Bacteriophage/phage
a virus that infects and hijacks a bacteria’s machinery to reproduce; can integrate stably into the bacterial genome.
- Conjugation
a mechanism of genetic material transfer via direct cell-cell contact.
- Genome engineering
technologies or approaches to alter genetically inheritable information in a targeted or specific manner.
- In situ microbiome engineering
manipulation of microbial communities in their native environment.
- Metagenome
the collection of genes, genomes and inheritable information present in a given environment.
- Microbiome
encompassing term referring to the microbiota, metagenome, and surrounding environment of a microbial community.
- Microbiota
the set of microorganisms present in a given environment.
- Mobile genetic elements
genetic information that can be transferred between cells; includes conjugative plasmids, transposons, bacteriophages.
- Prebiotics
naturally-occurring chemicals that can promote growth or activity in a community in a selective manner.
- Probiotics
bacteria that can confer a benefit to a particular host environment.
- Xenobiotics
biochemical compounds unnatural to an environment that can promote or limit the growth or function of specific microbial community members.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449(7164):804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esvelt KM, Wang HH. Genome-scale engineering for systems and synthetic biology. Mol Syst Biol. 2013;9:641. doi: 10.1038/msb.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart EJ. Growing unculturable bacteria. J Bacteriol. 2012;194(16):4151–60. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yaung SJ, Church GM, Wang HH. Recent progress in engineering human-associated microbiomes. Methods Mol Biol. 2014;1151:3–25. doi: 10.1007/978-1-4939-0554-6_1. [DOI] [PubMed] [Google Scholar]
- 5.Faith JJ, et al. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333(6038):101–4. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouhnik Y, et al. The capacity of nondigestible carbohydrates to stimulate fecal bifidobacteria in healthy humans: a double-blind, randomized, placebo-controlled, parallel-group, dose-response relation study. Am J Clin Nutr. 2004;80(6):1658–64. doi: 10.1093/ajcn/80.6.1658. [DOI] [PubMed] [Google Scholar]
- 7.Roberfroid M, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr. 2010;104 Suppl2:S1–63. doi: 10.1017/S0007114510003363. [DOI] [PubMed] [Google Scholar]
- 8.Song SK, et al. Prebiotics as immunostimulants in aquaculture: a review. Fish Shellfish Immunol. 2014;40(1):40–8. doi: 10.1016/j.fsi.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Yaung SJ, et al. Improving microbial fitness in the mammalian gut by in vivo temporal functional metagenomics. Mol Syst Biol. 2015;11(1):788. doi: 10.15252/msb.20145866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol. 2013;11(7):435–42. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M, et al. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science. 2015;350(6256):aac5992. doi: 10.1126/science.aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haiser HJ, et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341(6143):295–8. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace BD, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330(6005):831–5. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baruch M, et al. An extracellular bacterial pathogen modulates host metabolism to regulate its own sensing and proliferation. Cell. 2014;156(1-2):97–108. doi: 10.1016/j.cell.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, et al. Non-lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell. 2015;163(7):1585–95. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8(6):423–35. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson CJ, Young VB. Antibiotic administration alters the community structure of the gastrointestinal micobiota. Gut Microbes. 2010;1(4):279–284. doi: 10.4161/gmic.1.4.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson MK, Crofts TS, Dantas G. Antibiotics and the developing infant gut microbiota and resistome. Curr Opin Microbiol. 2015;27:51–6. doi: 10.1016/j.mib.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–50. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 21.Cullen TW, et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347(6218):170–5. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Toole PW, Cooney JC. Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdiscip Perspect Infect Dis. 2008;2008:175285. doi: 10.1155/2008/175285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mappley LJ, et al. Oral treatment of chickens with Lactobacillus reuteri LM1 reduces Brachyspira pilosicoli-induced pathology. J Med Microbiol. 2013;62(Pt 2):287–96. doi: 10.1099/jmm.0.051862-0. [DOI] [PubMed] [Google Scholar]
- 24.Spinler JK, et al. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe. 2008;14(3):166–71. doi: 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–8. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassan JO, Curtiss R., 3rd Development and evaluation of an experimental vaccination program using a live avirulent Salmonella typhimurium strain to protect immunized chickens against challenge with homologous and heterologous Salmonella serotypes. Infect Immun. 1994;62(12):5519–27. doi: 10.1128/iai.62.12.5519-5527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steidler L, et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289(5483):1352–5. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J Clin Invest. 2014;124(8):3391–406. doi: 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson JA, et al. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10(11):1861–71. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 30.Danino T, et al. Programmable probiotics for detection of cancer in urine. Sci Transl Med. 2015;7(289):289ra84. doi: 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mimee M, et al. Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Systems. 2015;1(1):62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotula JW, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc Natl Acad Sci U S A. 2014;111(13):4838–43. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–15. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 34.Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9(2):88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- 35.Faith JJ, et al. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010;4(9):1094–8. doi: 10.1038/ismej.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tvede M, Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet. 1989;1(8648):1156–60. doi: 10.1016/s0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- 37.Petrof EO, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: 'RePOOPulating' the gut. Microbiome. 2013;1(1):3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoaie S, et al. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab. 2015;22(2):320–31. doi: 10.1016/j.cmet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Seedorf H, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159(2):253–66. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28(2):127–81. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Summers WC. Bacteriophage therapy. Annu Rev Microbiol. 2001;55:437–51. doi: 10.1146/annurev.micro.55.1.437. [DOI] [PubMed] [Google Scholar]
- 42.Jones JB, et al. Considerations for using bacteriophages for plant disease control. Bacteriophage. 2012;2(4):208–214. doi: 10.4161/bact.23857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perera MN, et al. Bacteriophage cocktail significantly reduces or eliminates Listeria monocytogenes contamination on lettuce, apples, cheese, smoked salmon and frozen foods. Food Microbiol. 2015;52:42–8. doi: 10.1016/j.fm.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Silva YJ, et al. Influence of environmental variables in the efficiency of phage therapy in aquaculture. Microb Biotechnol. 2014;7(5):401–13. doi: 10.1111/1751-7915.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007;104(27):11197–202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009;106(12):4629–34. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bikard D, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32(11):1146–50. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ando H, et al. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Systems. 2015;1(3):187–196. doi: 10.1016/j.cels.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Modi SR, et al. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499(7457):219–22. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smillie CS, et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480(7376):241–4. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 52.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 53.Kommineni S, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526(7575):719–22. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2(3):241–9. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez-Beltran J, et al. The animal food supplement sepiolite promotes a direct horizontal transfer of antibiotic resistance plasmids between bacterial species. Antimicrob Agents Chemother. 2013;57(6):2651–3. doi: 10.1128/AAC.02363-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilharm G, et al. A simple and rapid method of bacterial transformation. J Microbiol Methods. 2010;80(2):215–6. doi: 10.1016/j.mimet.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 57.Klumper U, et al. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2015;9(4):934–45. doi: 10.1038/ismej.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Musovic S, et al. Long-term manure exposure increases soil bacterial community potential for. plasmid uptake. Environ Microbiol Rep. 2014;6(2):125–30. doi: 10.1111/1758-2229.12138. [DOI] [PubMed] [Google Scholar]
- 59.Klumper U, et al. Novel assay to measure the plasmid mobilizing potential of mixed microbial communities. Front Microbiol. 2014;5:730. doi: 10.3389/fmicb.2014.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng L, et al. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature. 2014;513(7517):242–5. doi: 10.1038/nature13459. [DOI] [PubMed] [Google Scholar]
- 61.Shintani M, Sanchez ZK, Kimbara K. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol. 2015;6:242. doi: 10.3389/fmicb.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frost LS, et al. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3(9):722–32. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 63.Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat Methods. 2007;4(1):55–61. doi: 10.1038/nmeth964. [DOI] [PubMed] [Google Scholar]
- 64.Kushwaha M, Salis HM. A portable expression resource for engineering cross-species genetic circuits and pathways. Nat Commun. 2015;6:7832. doi: 10.1038/ncomms8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang W, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–9. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karberg M, et al. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol. 2001;19(12):1162–7. doi: 10.1038/nbt1201-1162. [DOI] [PubMed] [Google Scholar]
- 67.Na D, et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol. 2013;31(2):170–4. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 68.Nielsen AA, Voigt CA. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol Syst Biol. 2014;10:763. doi: 10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farzadfard F, Lu TK. Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science. 2014;346(6211):1256272. doi: 10.1126/science.1256272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Esvelt KM, et al. Concerning RNA-guided gene drives for the alteration of wild populations. Elife. 2014:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Butler CA, Gotschlich EC. High-frequency mobilization of broad-host-range plasmids into Neisseria gonorrhoeae requires methylation in the donor. J Bacteriol. 1991;173(18):5793–9. doi: 10.1128/jb.173.18.5793-5799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oye KA, et al. Biotechnology. Regulating gene drives. Science. 2014;345(6197):626–8. doi: 10.1126/science.1254287. [DOI] [PubMed] [Google Scholar]
- 73.Baltimore D, et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348(6230):36–8. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]