Abstract
ATAD3A is an integral mitochondrial membrane protein with unknown function, although we now show that high-level expression is associated with poor survival in breast cancer patients. Using a mass spectrometry approach we have demonstrated that ATAD3A interacts with the WASF3 metastasis promoting protein. Knockdown of ATAD3A leads to decreased WASF3 protein levels in breast and colon cancer cells. Silencing ATAD3A also results in loss of both cell anchorage-independent growth and invasion and suppression of tumor growth and metastasis in vivo using immuno-compromised mice. HSP70 is responsible for stabilizing WASF3 in the cytoplasm, but inactivation of HSP70 does not lead to loss of WASF3 stability at the mitochondrial membrane, where presumably it is protected through its interaction with ATAD3A. In response to endoplasmic reticulum stress, increases in the GRP78 protein level leads to increased WASF3 protein levels. We also show that ATAD3A was present in a WASF3-GRP78 complex and suppression of GRP78 led to destabilization of WASF3 at the mitochondrial membrane, which was ATAD3A dependent. Furthermore, ATAD3A-mediated suppression of CDH1/E-cadherin occurs through its regulation of GRP78-mediated WASF3 stability. Proteolysis experiments using isolated mitochondria demonstrates the presence of the N-terminal end of WASF3 within the mitochondria which is the interaction site with the N-terminal end of ATAD3A. It appears, therefore, that stabilization of WASF3 function occurs through its interaction with ATAD3A and GRP78, which may provide a bridge between the endoplasmic reticulum and mitochondria, allowing communication between the two organelles. These findings also suggest that pharmacologic inhibition of ATAD3A could be an effective therapeutic strategy to treat human cancer.
Keywords: ATAD3A, WASF3, GRP78, metastasis, stability, mitochondria
Introduction
WASF3 is a member of the Wiskott-Aldridge syndrome family of genes 1, which encode proteins with motifs that facilitate actin polymerization through recruitment of ARP2/3 complexes 2,3. WASF proteins are normally present in an inactive form, where the motifs that are responsible for actin polymerization are masked through conformational restraints imposed by associated protein complexes 4. Phosphorylation on a number of different tyrosine residues throughout the WASF3 protein, is required for its activation, which can be achieved through its interaction with kinases such as ABL, PI3K and JAK 5–7. Growth factor or cytokine stimulation leads to WASF3 phosphoactivation 5,7 and relocation to the inner cell membrane at the leading edges where actin reorganization is transferred into movement 5,7. In cancer cells, high level expression of WASF3 is correlated with more aggressive tumors 8 and knockdown of WASF3 in breast 9 and prostate 10 cancer cells leads to loss of both motility and invasion and metastasis, in part associated with a reduced ability to generate lamellipodia 5,10.
Gene expression analysis demonstrates that loss of WASF3 leads to dysregulation of a number of signaling pathways, notably by upregulating the KISS1 metastasis suppressor gene 11. KISS1 regulates NFκB activation by promoting its association with inhibitory IκBα, thereby sequestering NFκB in the cytoplasm. WASF3 overexpression induces down regulation of KISS1, promoting the movement of the p50/p65 components of NFκB into the nucleus, which activates a wide variety of genes including inflammatory cytokines such as IL6 and matrix metalloproteinases (MMP). Suppression of KISS1 also leads to activation of ZEB1, which leads to down regulation of the miRNA-200s that normally target WASF3 mRNA. This activation of ZEB1 leads to loss of CDH1/E-cadherin which promotes the epithelial-to-mesenchyme transition (EMT) 12. In response to IL6, WASF3 expression levels are up-regulated by activated STAT3 through a direct interaction with the WASF3 promoter 7. WASF3 is then phosphoactivated by JAK2 and the combined effects of the JAK2/STAT3 promotion of WASF3 function accounts for increased invasion 7. Inactivation of WASF3, regardless of the genetic background of the cancer cells leads to loss of invasion, suggesting it has a central role in mediating signaling to promote the metastatic phenotype.
In previous studies we have shown that proteins identified as part of the WASF3 immunocomplex have led to a better understanding of WASF3 function and its role in the promotion of metastasis 7,13. As part of this overall analysis we identified the ATPase family AAA domain containing 3A (ATAD3A) protein as another interacting partner. The function of ATAD3A is largely unknown but it is a nuclear encoded protein, which is located in mitochondria 14,15, and is a member of a family of proteins that includes ATAD3B which is a mitochondrial protein specific to embryonic stem cells that is re-expressed in cancer cells. ATAD3B has been shown to interact with ATAD3A in a dominant-negative manner suppressing its function 16. Knockdown of ATAD3A in normal cells leads to major changes in the mitochondrial profile, inhibits proliferation and modifies the functional interaction network between mitochondria and the endoplasmic reticulum (ER) 17,18. ATAD3A is amplified in a variety of cancer cell types and is associated with chemo- and radiation resistance 17,19. ATAD3A has been shown to be essential for the import of the apoptosis inducing factor (AIF) into the mitochondria from the endoplasmic reticulum (ER) via the mitochondrial membrane and import vesicles 20. Knockdown of ATAD3A using siRNA in uterine cervical cancer cells increases cell autophagy and apoptosis, and decreases resistance to anticancer drugs 21. In lung cancer, knockdown of ATAD3A increased mitochondrial fragmentation and cisplatin sensitivity 22. Although there has been some debate, it now appears that ATAD3A spans the mitochondrial membrane, since proteolysis studies demonstrate the N-terminal end is accessible, suggesting it is on the outside of the mitochondrial membrane but the C-terminal end is not accessible, suggesting it is in the mitochondria 15, 17.
In this report, we demonstrate that a novel WASF3-interacting protein, ATAD3A, acts as a crucial mediator to promote cell invasion in breast and colon cancer via regulating GPR78-mediated stabilization of WASF3.
Results
WASF3 interacts with ATAD3A
From an LTQ Orbitrap MS/MS analysis of tryptic peptides generated following immunoprecipitation (IP) from MDA-MB-231 breast cancer cells expressing an HA-tagged, exogenous, WASF3 construct (Figure S1A) 13, the ATAD3A protein was identified in the WASF3 immunocomplexes from each of two independent experiments (Figures S1A and S1B). ATAD3A was identified in immunoprecipitates using both HA (33% peptide coverage) and WASF3 antibodies (15% peptide coverage) (Figure S1C). IP analysis confirmed this interaction using a WASF3 antibody to immunoprecipitate the ATAD3A protein (Figure 1A). The interaction between ATAD3A and WASF3 was also demonstrated in SW620 colon cancer cells (Figure 1A), suggesting a wider association between these two proteins. Using the MitoTracker fluorescent dye, that stains the mitochondrial membrane, immunofluorescence (IF) analysis showed colocalization between the MitoTracker signal and that produced with an anti-ATAD3A antibody (Figure S2). When total cell lysates from MDA-MB-231 cells expressing an exogenous HA-tagged ATAD3A gene were compared with enriched mitochondria from these cells (Figure 1B), WASF3 was found in both fractions confirming its localization to the mitochondria. To determine whether WASF3 was binding to the N-terminal part of ATAD3A, we generated HA tagged constructs containing either the N-terminal 1–250 amino acids or the C-terminal 320–586 amino acids (Figure 1C). These constructs were introduced into parental MDA-MB-231 cells and IPs were generated using anti-HA antibodies. As shown in Figure 1C, WASF3 is present in the immunocomplex with the N-terminal protein and not the C-terminal protein.
Figure 1. ATAD3A is a novel WASF3-interacting protein.
(A) IP from MDA-MB-231 and SW620 cells using a WASF3 antibody identified ATAD3A in the immunocomplex (arrow). Smaller bands in the lower panel represent the IgG heavy chain. (B) Mitochondria from MDA-MB-231 cells expressing an HA-tagged ATAD3A gene were isolated. IP with HA shows that WASF3 was present in the immunocomplex of the whole cell lysate as expected as well as in isolated mitochondria. Purity of the mitochondrial fraction was noted by the COXIV mitochondrial specific protein and absence of the cytoplasmic GAPDH protein. (C) MDA-MB-231 cells were transfected with HA-tagged full length ATAD3A (HA-3A), N-terminal ATAD3A (amino acids 1–250, HA-3A-Nter) or C-terminal ATAD3A (amino acids 320–586, HA-3A-Cter). IP with HA shows that WASF3 is only present in the immunocomplexes where the N-terminal part of ATAD3A is present. (D) WASF3 is associated with isolated mitochondria where the purity of the fragment is demonstrated by the presence of the Cytochrome C mitochondrial-specific protein and absence of cytosolic GAPDH. Treatment of the isolated mitochondria with trypsin shows progressive reduction in overall WASF3 levels over 30 minutes but a resistant subpopulation of a smaller 40 kD WASF3 protein. (E) Following treatment with HSP90 inhibitors AUY922 or 17-AAG, IP of WASF3 from MDA-MB-231 cells shows decreased levels of phosphoactivated WASF3, which significantly reduces the specific binding to ATAD3A. In (A) and (B), preimmune IgG was used as a negative control.
While the IF and IP data demonstrates that WASF3 is associated with mitochondria, the topography of this interaction could not be determined from these studies. We suspected two potential alternatives where, either WASF3 resides on the outer mitochondrial surfaces and interacts with ATAD3A through its N-terminal end that protrudes out of the mitochondria, or that ATAD3A resides exclusively within the inter mitochondrial membrane space and WASF3 penetrates the mitochondrial membrane to facilitate the interaction. Using the same strategy that defined the location of ATAD3A at the mitochondrial membrane, we isolated mitochondria from MDA-MB-231 cells and treated them with trypsin for 30 minutes. Western blot analysis was then used to detect the presence of WASF3. As shown in Figure 1D, full length WASF3 is found within the purified mitochondrial fraction, which also showed the presence of the mitochondria-specific Cytochrome C protein. The absence of the common cytoplasmic GAPDH protein demonstrated the purity of the mitochondrial fraction. Following only 30 minutes of proteolysis, the presence of full-length WASF3 was substantially reduced in the mitochondrial fraction and a novel ~40kD truncated fragment was detected. More extensive digestion over 50 minutes led to complete loss of the full-length WASF3 protein but the 40 kD protein remained (Figure S3). Since the antibody used in this study identifies the N-terminal part of WASF3, as we have shown previously 6,13, this confirms that the interaction with ATAD3A is through the N-terminal region of WASF3. These observations suggest firstly that WASF3 can enter the mitochondrial membrane, since it is partially protected from proteolysis, and that it is the N-terminal end that is protected. From our results above, it is seen that the N-terminal end of ATAD3A associates with WASF3 and so it appears that these two proteins can interact within the interspace between the mitochondrial membranes (see discussion). The 40 kD fragment in the mitochondria accounts for the N-terminal ~350 amino acids of WASF3
Phosphoactivation of WASF3 is required for its regulation of cell invasion 5,6 which can be achieved by ABL kinase5, which is a client protein of HSP90. Inactivation of HSP90 in cancer cells suppresses WASF3 activation as a result of the destabilization of ABL13. To determine whether the association between ATAD3A and WASF3 is WASF3 phosphorylation-dependent, we treated MDA-MB-231 cells with the AUY922 and 17-AAG HSP90 inhibitors. IP analysis showed that inhibiting WASF3 phosphorylation in this way led to reduced binding to ATAD3A (Figure 1E), suggesting that ATAD3A preferentially binds to the activated form of WASF3.
ATAD3A knockdown significantly decreases cancer cell anchorage-independent growth and invasion in vitro
To determine the consequences of inactivating ATAD3A in cancer cells, we used shRNA strategies to knock it down in MDA-MB-231 and SW620 cells (Figure 2A) and assessed the effect on cell proliferation, anchorage-independent growth and invasion (Figure 2B-F). None of these cells lines showed a significant difference in either short-term (within 6 days) cell proliferation (Figure 2B) or cell viability (Figure S4), whether or not ATAD3A was expressed. When we analyzed anchorage-independent cell growth (over 3 weeks), shRNA-mediated depletion of ATAD3A led to both a reduction in the number and size of the colonies in both MDA-MB-231 breast cancer cells and SW620 colon cancer cells (Figure 2C and 2D). Furthermore, Matrigel invasion assays showed that knockdown of ATAD3A resulted in a significant reduction in cell invasion (Figure 2E and 2F).
Figure 2. ATAD3A knockdown inhibits cancer cell anchorage-independent growth and invasion in vitro.
(A) Western blot analysis shows that ATAD3A protein levels in MDA-MB-231 and SW620 cells are much lower in independently derived ATAD3A knockdown cells (sh3A-1, sh3A-2) compared with that in the knockdown control cells (shGFP). (B) Knockdown of ATAD3A does not affect cell proliferation over 6 days in both MDA-MB-231 and SW620 cells. Silencing ATAD3A inhibits the cell anchorage-independent growth (C) and invasion (E) in both MDA-MB-231 and SW620 cells. Quantification of the relative colony number and relatively invading cell number from the three independent experiments is shown in (D) and (F), respectively. * p<0.05, ** p<0.01.
Silencing ATAD3A expression suppresses tumor growth and metastasis in breast cancer cells in vivo
To relate the in vitro studies to clinical observations, we investigated whether ATAD3A expression was related to clinical parameters in human breast cancer patients. Survival analysis (Kaplan-Meier method, log-rank test) using the annotated GSE18229 data set from the GEO database, showed that high ATAD3A expression levels were associated with low overall survival of breast cancer patients (Figure 3A, p=0.002), supporting a role for ATAD3A in breast cancer progression. These observations were confirmed in an analysis of four additional data sets (Figure S5) correlating high ATAD3A with relapse free survival and disease free survival. We next used in vivo studies to investigate the importance of ATAD3A expression in breast cancer development and metastasis using human cell line xenografts. It has been shown that primary and metastatic tumors develop coincidently in immunocompromized NSG mice, precluding the need to remove the primary tumor during the experiment 23. We injected MDA-MB-231 cells into the mammary fat pad under the fourth (abdominal) nipple of NSG mice and monitored tumor development over 60 days, at which time the mice were sacrificed, the primary tumors removed, and the lungs examined for metastases. In the mice injected with the ATAD3A knockdown MDA-MB-231 cells, tumors were statistically significantly smaller as measured by tumor volume and weight, compared with mice that were injected with the knockdown control cells (Figure 3B). Since we did not observe an inhibitory effect on cell proliferation in vitro after knockdown of ATAD3A, we investigated tumor vasculature as a potential cause of reduced tumor growth using CD31 immunohistochemistry (IHC). In the tumors derived from ATADA3A knockdown cells, there was a significant reduction in the number of CD31-positive microvessels, compared with the tumors derived from the control cells (Figure 3C and 3D), indicating that ATAD3A affects neovascularization in vivo. In the analysis of metastasis, both lung weight and incidence of metastatic colonies observed on the lung surfaces were significantly reduced in the mice injected with the ATAD3A knockdown cells compared with the mice injected with the knockdown control cells (Figure 3E and 3F). This reduction in metastases between the two groups was confirmed by histological analysis of the lungs from these animals (Figure 3G). H&E-stained sections revealed that the mice injected with the knockdown control cells showed infiltrating tumors throughout the entire lung, whereas the mice injected with the cells in which ATAD3A had been knocked down showed only relatively few, small tumors in the lung (Figure 3G).
Figure 3. ATAD3A knockdown inhibits tumor growth and metastasis in vivo.
(A) Kaplan-Meier survival curves demonstrate that high ATAD3A expression is associated with low overall survival rate of breast cancer patients. (B) In NSG mice, knockdown of ATAD3A (sh3A) remarkably inhibits tumor growth, showing a smaller tumor size (upper panel) and reduced tumor weight (lower panel) compared with tumors derived from the knockdown control cells (shGFP). (C-D) In NSG mice, knockdown of ATAD3A inhibits tumor induced angiogenesis compared with tumors derived from the control cells. The representative images are shown in (C) and the CD31-positive microvessels are marked (arrows). Quantification of the CD31-positive microvessels is shown in (D). (E-F) Knockdown of ATAD3A significantly decreases lung weight (E) and the number of metastatic nodules on the lung surface (F). (G) Histological analysis shows that only a few, small-sized, metastatic tumors are observed in the lungs from the mice that were injected with ATAD3A-knockdown cells (arrow) compared with extensive infiltration seen in the knockdown control animals (arrow). Images on the right represent higher magnification of the boxed area in the left hand image. Error bars represent SD (n = 10), * p<0.05 and ** p<0.01.
ATAD3A is an important regulator of WASF3 protein stability at the mitochondrial membrane
In our previous studies, we demonstrated that several WASF3-interacting proteins affect WASF3 function through regulating either its expression level, protein stability or its protein phosphorylation status in cancer cells 7,13. We therefore determined how knockdown of ATAD3A affected these various aspects of WASF3 function. Suppressing ATAD3A effectively reduced WASF3 protein levels in MDA-MB-231 and SW620 cells (Figure 4A and 4B). Western blot analysis of tumors from the primary xenografts from ATAD3A knockdown MDA-MB-231 cells also demonstrated that WASF3 protein levels were significantly reduced (Figure 4C). Conversely, WASF3 protein levels were enhanced when full-length ATAD3A was overexpressed in MCF7 cells, which express low levels of WASF3, with a concomitant increase in cell invasion (Figure S6). Overexpressing ATAD3A in T47D cells which do not express WASF3, however, had no effect on invasion (Figure S6). The ability of ATAD3A to enhance invasion, therefore, appears to be dependent on the presence of the WASF3. Interestingly, overexpression of the N-terminal region of ATAD3A in MCF7 cells, also led to increased WASF3 protein levels and cell invasion (Figure S6). We did not see any effect when the C-terminal part of ATAD3A was overexpressed in either MCF7 or T47D cells (Figure S6). These observations suggested that the effect of ATAD3A on cancer progression is, at least in part, due to its control over the critical capacity of the WASF3 protein to influence invasion/metastasis.
Figure 4. ATAD3A regulates WASF3 protein stability associated with mitochondria.
(A) Western blot analysis shows that suppressing ATAD3A expression effectively reduces WASF3 and GRP78 protein levels in MDA-MB-231 and SW620 cells. No concomitant decrease in WASF3 mRNA levels, however, was detected in ATAD3A knockdown cells (below). Levels of other chaperone proteins are relatively unaffected. Quantification of the WASF3 protein levels from the three independent experiments is shown in (B), ** p<0.01. (C) Western blot analysis shows significantly reduced WASF3 protein levels in the tumors from the primary mouse xenografts from ATAD3A knockdown MDA-MB-231 cells. (D) Immunofluorescence assays show that the general distribution of the WASF3 protein in the cytoplasm is generally lost with the treatment of HSP70 inhibitor PES, except at a perinuclear location. (E) Western blot analysis shows a significant reduction of WASF3 in the cytoplasmic (Cyto) fraction but not in the isolated mitochondria (Mito) when treated with PES. ATAD3A protein levels, however, were unaffected in the mitochondrial fraction following PES treatment. GAPDH and COXIV serve as specific markers for the cytoplasm and mitochondria respectively, demonstrating the purity of the fractions. (F) In the presence of PES, Western blot analysis shows dramatic loss of WASF3 protein in ATAD3A knockdown cells compared with the knockdown control cells. (G) Immunofluorescence analysis shows a depletion of WASF3 in mitochondria in ATAD3A knockdown MDA-MB-231 cells. When the ATAD3A knockdown cells are treated with PES, WASF3 is undetectable in both mitochondria and cytoplasmic fraction. Mitochondria are labeled with MitoTracker dyes (red).
We did not, however, find a concomitant decrease in WASF3 mRNA levels in either MDA-MB-231 or SW620 cells in which ATAD3A had been knocked down (Figure 4A). The demonstration that reduced expression of ATAD3A leads to reduced WASF3 protein levels, suggested ATAD3A may be involved in stabilization of WASF3, as we described previously for HSP70, for example 13. To determine whether this stabilization is specifically related to ATAD3A, we analyzed HSP70 levels in the ATAD3A knockdown cells. In both MDA-MB-231 and SW620 cells, no significant changes in HSP70 levels were detected in the absence of the ATAD3A protein (Figure 4A). There was also no difference in HSP90 expression levels in the ATAD3A knockdown cells compared to the knockdown control cells (Figure 4A). The finding that ATAD3A binds to, and stabilizes WASF3 protein without affecting HSP70, suggests a novel potential mechanism for regulating WASF3 function. To explore this possibility further, we investigated the relationship between WASF3 protein stability and expression patterns of HSP70 and ATAD3A.
2-phenylethynesulfonamide (PES) disrupts the interaction between HSP70 and its client proteins 13,24, and has been shown to lead to reduced cellular WASF3 protein levels, as well as disruption of its association with WASF3 13. To determine whether the intracellular localization of ATAD3A affects the ability of HSP70 to stabilize WASF3, we used confocal microscopy to analyze the cells treated with PES. In MDA-MB-231 cells, the WASF3 protein is distributed generally throughout the cytoplasm (Figure 4D). Following PES treatment, however, cytoplasmic WASF3 is largely lost, except at a perinuclear location (Figure 4D), suggesting a colocalization with mitochondria. We then isolated cytoplasmic and mitochondrial fractions and determined that PES treatment caused a significant reduction in the cytoplasmic WASF3 fraction but not in the isolated mitochondria (Figure 4E). PES treatment did not affect the levels of ATAD3A in the mitochondria (Figure 4E). These data demonstrate an association between WASF3 and mitochondria, possibly through an association with the ATAD3A protein. When the same analysis was performed in cells in which ATAD3A had been knocked down, there was no WASF3 protein in mitochondria and complete loss of WASF3 protein following PES treatment compared with the knockdown control cells (Figure 4F). Western blot analysis showed a more dramatic reduction in WASF3 protein levels in ATAD3A knockdown MDA-MB-231 cells following PES treatment compared with the knockdown control cells. Immunofluorescence analysis using MitoTracker shows no WASF3 signal associated with mitochondria when ATAD3A was knocked down (Figure 4G). Moreover, in the presence of PES, WASF3 was barely detectable in ATAD3A knockdown cells, but was clearly seen associated with mitochondria in the knockdown control cells (Figure 4G), which is consistent with the Western blot results (Figure 4F). It appears, therefore, that WASF3 is stabilized in association with mitochondria through its interaction with ATAD3A.
GRP78 is required for ATAD3A-mediated WASF3 stability
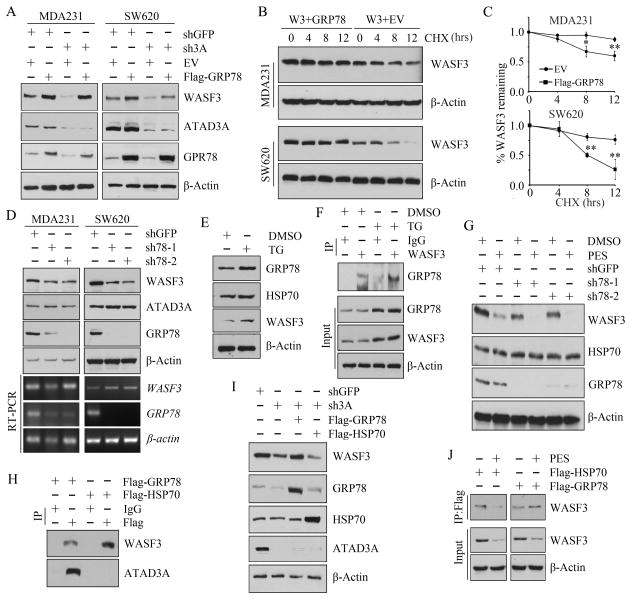
Since HSP70 and HSP90 protein levels did not change in ATAD3A knockdown cells, we extended our analysis to investigate other chaperone proteins: GRP78/BiP, GRP94 and HSP60. Only GRP78 protein levels were decreased in both ATAD3A knockdown MDA-MB-231 and SW620 cells, in which reduced WASF3 protein levels were seen compared with the knockdown control cells (Figure 4A). GRP78 is an ER resident chaperone involved in protein degradation 25,26. Under certain circumstances, it can be retargeted to, and may be involved in, coordinating the unfolded protein response (UPR) signaling between mitochondria and the ER 27,28. These data suggest that GRP78 may be involved in ATAD3A-mediated WASF3 stability. To examine this possibility, we overexpressed Flag-tagged GRP78 in the ATAD3A-deficient MDA-MB-231 and SW620 cells. In this analysis, we found that knockdown of ATAD3A in both cell lines led to reduced WASF3 protein levels but this suppression was ameliorated when GRP78 was overexpressed in these cells (Figure 5A). These data indicate that GRP78 is required for ATAD3A-mediated WASF3 stability. We then overexpressed Flag-tagged GRP78 in cells overexpressing WASF3 and treated them with cycloheximide (CHX) to prevent de novo protein synthesis. In both cell lines, analysis over 12 hours showed that the half-life of the WASF3 protein was significantly extended in the presence of excess GRP78 (Figure 5B and 5C). When GRP78 was knocked down in MDA-MB-231 and SW620 cells, a reduction in WASF3, but not ADAT3A, protein levels was observed (Figure 5D). This effect was shown to act at the protein stability level, since gene expression levels of WASF3 were unaffected as a result of GRP78 knockdown (Figure 5D).
Figure 5. GRP78 is required for ATAD3A-mediated WASF3 protein stability.
(A) Western blot analysis shows that overexpression of GRP78 rescues WASF3 protein levels in ATAD3A knockdown MDA-MB-231 and SW620 cells. (B) Inhibition of protein synthesis by CHX followed by Western blot at the indicated times shows that the half-life of WASF3 is increased in the MDA-MB-231 cells when co-transfected with pCDH-HA-WASF3 (W3) and pCMV-Flag-GRP78 (GRP78) vectors, compared with that in the cells which co-transfected with pCDH-HA-WASF3 (W3) and pCMV-Flag (EV) vectors. The same results were observed from SW620 cells. (C) WASF3 protein levels are normalized to β-Actin and the WASF3 half-life from three independent experiments was quantified (lower panel). * p<0.05 and ** p<0.01. (D) Western blot shows that knockdown of GRP78 (sh78-1, sh78-2) in MDA-MB-231 or SW620 cells leads to a decrease in WASF3 protein levels compared with the knockdown control cells (shGFP). RT-PCR shows that mRNA expression levels of WASF3 were not affected as a result of GRP78 knockdown. (E) Western blot analysis shows increased protein levels of both GRP78 and WASF3, but not HSP70, when MDA-MB-231 cells were treated with the TG ER stress inducer. (F) IP analysis shows more GRP78 protein in the WASF3 immunocomplex following the treatment with TG. (G) Western blot analysis shows a more dramatic reduction in WASF3 protein levels in GRP78 knockdown MDA-MB-231 cells when compared with the knockdown control cells. (H) IP analysis shows that ATAD3A binds to GRP78, but not to HSP70 in MDA-MB-231 cells. (I) Western blot shows that the loss of WASF3 in ATAD3A knockdown cells cannot be rescued by overexpression of HSP70. (J) IP analysis shows that binding of WASF3 to HSP70 is impaired, but does not affect WASF3 binding to GRP78 following PES treatment.
The transcriptional activation of GRP78 responds to stress conditions that target the ER 28–30. To mimic increased levels of GRP78 in response to ER stress, we treated MDA-MB-231 cells with thapsigargin (TG), a tumor promoter and ER Ca2+-ATPase inhibitor, which resulted in increased protein levels for both GRP78 and WASF3, but not HSP70 (Figure 5E). The same phenotype was observed when cells were treated with other ER stress inducers, Brefeldin A (BFA) and tunicamycin (TA) (data not shown). IP analysis showed that GRP78 binds to WASF3, and increased GRP78 protein levels were found in the WASF3 immunocomplex when treated with TG (Figure 5F), demonstrating that GRP78 promotes WASF3 protein stability through enhancing the interaction with WASF3 under ER stress. To further investigate the regulation of GRP78 on WASF3 stability, we treated the GPR78 knockdown and knockdown control MDA-MB-231 cells with PES. Following PES treatment, there was a remarkable reduction in WASF3 protein levels, which was even more dramatic in GRP78 knockdown cells compared with the knockdown control cells (Figure 5G).
Since WASF3 binds to both ATAD3A and GRP78, we investigated whether these three protein were in the same complex. To investigate this, we overexpressed either Flag-tagged GRP78 or HSP70 in MDA-MB-231 cells and immunoprecipitated these proteins using a Flag antibody. IP analysis shows that ATAD3A also interacts with GRP78, but it does not bind to HSP70, even though WASF3 binds to both GRP78 and HSP70 (Figure 5H), suggesting that ATAD3A only exists in the WASF3-GRP78 complex. Western blot analysis further indicates that overexpression of HSP70 cannot rescue WASF3 protein levels in ATAD3A knockdown cells (Figure 5I). Moreover, PES severely disrupted binding between WASF3 and HSP70, but did not impair the integrity of the WASF3-GRP78 complex (Figure 5J). Taken together, these observations suggest that GRP78 and HSP70 function cooperatively to mediate WASF3 protein stability in different cell compartments, and support the idea that GRP78 is required for ATAD3A-mediated stabilization of WASF3 at the mitochondrial membrane.
ATAD3A downregulates CDH1 through WASF3 in cancer cells
Knockdown of ATAD3A in MDA-MB-231 cells leads to a profound change in cell invasion and metastasis. To determine how this affects gene expression in these cells, we performed mRNA expression studies using Affymetrix (U133A Plus 2.0) microarrays. A significant change in the gene expression profile was detected in the ATAD3A knockdown cells with 308 genes showing increased expression and 217 genes showing down regulation (fold change>2 and p<0.05, Figure 6A). Interestingly, from the four different probes of VEGF on the array, the fold reduction in the ATAD3A knockdown cells was -7, -4.4, -3.75 and -3.4 respectively, which could account for the reduced vascularization in the tumors (Figure 3C). Gene ontology (GO) analysis further shows that most of the dysregulated genes in the ATAD3A knockdown cells are involved in the cancer cell motility and metastasis phenotypes (Figure 6B), such as CDH, where the expression levels were upregulated 34.9 fold compared with that in the knockdown control cells (Figure 6C). Microarray expression analysis also showed that WASF3 levels were not altered as a result of ATAD3A knockdown (Figure 6C), which is consistent with our results above. Quantitative reverse transcription-PCR (QRT-PCR) analysis of 6 genes (CDH1, CHRDL1, GRP110, HMGN5, PARM1 and TLR4) associated with cell movement showed the clearly altered gene expression patterns when ATAD3A was knocked down, which confirmed the observations from the microarray assays (Figure 6C).
Figure 6. WASF3 is involved in ATAD3A-mediated alteration of CDH1 expression.
(A) Heatmap analysis of microarray data shows differential gene expression in MDA-MB-231 as a result of ATAD3A knockdown. Red square: up-regulated genes (fold change more 2.0); blue square: down-regulated genes (fold change less than 0.5). (B) GO analysis for the biological functions of differentially expressed genes shows most of the dysregulated genes in the ATAD3A knockdown cells are involved in the cancer cell motility and metastasis phenotypes. (C) QRT-PCR analysis of selected gene expression shows their altered expression levels in ATAD3A knockdown cells, which is consistent with those seen in the microarray experiments. Relative expression levels are presented as a percentage of control expression levels. Error bars represent SD (n = 3). (D) Western blot shows increased CDH1 protein levels in ATAD3A knockdown MDA-MB-231 cells. 1, 2 and 3 indicate three independent repeats. (E) Western blot analysis shows that overexpressing GRP78 in the ATAD3A knockdown MDA-MD-231 cells leads to a suppression of CDH1 upregulation mediated by loss of ATAD3A. However, CDH1 levels did not change in WASF3 knockdown cells whether or not GRP78 was overexpressed. (F) Matrigel invasion assays show a significant increase in invasion potential when GRP78 was overexpressed in ATAD3A knockdown cells, but there was no change in invasion potential when GRP78 was overexpressed in WASF3 knockdown cells. Error bars represent SD (n = 3), * p<0.05.
CDH1 is critical in maintaining cell-cell adhesion and loss of its expression is associated with transition to a mesenchymal phenotype associated with metastasis 31,32. Western blot analysis confirmed the significant elevation of CDH1 in the ATAD3A knockdown cells (Figure 6D). As we reported previously, WASF3 downregulates CDH1 in breast cancer cells while promoting cancer invasion and metastasis 12. To investigate whether WASF3 is required for the ATAD3A-mediated alteration in CDH1 expression, we overexpressed GRP78 in either in ATAD3A or WASF3 deficient MDA-MD-231 cells. In the WASF3 knockdown cells, overexpressing GRP78 did not change CDH1 levels (Figure 6E). In ATAD3A knockdown cells overexpressing GRP78, however, CDH1 levels were below the level of detection. These observations indicate that ATAD3A exerts its effect on CDH1 through a WASF3 dependent mechanism. Matrigel invasion assays demonstrated a large increase in invasion potential when GRP78 was overexpressed in ATAD3A knockdown cells, but there was no change in invasion potential when GRP78 was overexpressed in WASF3-silenced cells (Figure 6F). These results demonstrate that the loss of invasion seen in ATAD3A knockdown cells can be rescued by restoring WASF3 protein function. Together, our data suggest that ATAD3A has a WASF3-dependent effect on gene expression involved in cell invasion and metastasis.
Discussion
ATAD3A is a mitochondrial protein that has been associated with a number of mitochondrial functions including fragmentation and fission, protein transport and mitochondrial physiology 14–18. In cancer cells, ATAD3A has mostly been associated with overexpression in gliomas, lung adenocarcinomas, uterine cervical cancer and head and neck tumors, and in some cases this is related to early recurrence and poor overall survival 18–22. Despite these phenotypic associations, the molecular function of ATAD3A is still largely unknown. In this report we demonstrate that ATAD3A is associated with the WASF3 metastasis promoting protein and, while not significantly affecting cancer cell proliferation, is required to maintain the invasion and metastasis phenotypes in breast and colon cancer cell lines, through a WASF3-dependent mechanism. Knockdown of ATAD3A also leads to suppression of tumor development by breast cancer cells at the primary xenograft site as demonstrated previously for knockdown of WASF3 in breast cancer cells 9, although this is not due to reduced proliferation potential but rather to an effect on the development of vascularization of the tumors. Tumor cells often overexpress VEGF, which provides autocrine stimulation of neovascularization in the tumors that develop. The expression levels of VEGF were suppressed in the ATAD3A knockdown cells, suggesting one possible mechanism for the reduced vascularization and smaller tumor size. It appears, therefore, that the WASF3-ATAD3A axis may not only be a target for suppressing invasion but also for suppressing tumor growth.
In our studies, the major effect of the ATAD3A interaction with WASF3 is on cell movement and invasion, since cell proliferation over 6–7 days appears to be unaffected. Knockout of ATAD3 in C. elegans, Drosophila melanogaster and mouse development 33–35, however, leads to an embryonic lethality which is suggested to result from compromised mitochondrial function at critical stages of development. It should be noted that knockdown of WASF proteins also leads to developmental lethality due to inability to transition through specific developmental stages which possibly rely on early development and implantation processes 36,37. Indeed, in the C. elegans studies, although development was retarded, the overall life span of the mutant and wild type animals was the same, suggesting cell viability was not the critical defect. In our studies over 6–7 days there were no cell viability issues either, which may reflect that we were using cancer cells where, unlike normal development, the dependence on energy derived from mitochondrial function has switched to glycolysis making a dependence on mitochondria less critical, at least in short term assays.
Although there has been some controversy about the exact location of ATAD3A, it now seems clear that it spans the inner and outer mitochondrial membranes, with the C-terminal end located in the mitochondrial matrix, possibly associating with the nucleoid complexes 15. The N-terminal end is largely located between the inner and outer mitochondrial membranes with one interpretation favoring an exclusively intra-mitochondrial location while another 15,17 suggests the potential of the N–terminal end protruding through the membrane and into the cytoplasm of the cell. Using truncated forms of ATAD3A, we showed that WASF3 interacts exclusively with the N-terminal end, and that this interaction is essential for the stability of the WASF3 protein, since loss of ATAD3A leads to reduction in WASF3 protein levels but not mRNA levels. The demonstration that the N-terminal end of WASF3 penetrated the outer mitochondrial membrane, where it is protected from proteolysis, provides an explanation for the interaction with ATAD3A, regardless of where it is positioned in the mitochondrial. The 40 kD protected fragment includes the ~350 N-terminal amino acids, which includes a coiled coil region within the WASF3 protein. The N-terminal end of ATAD3A also includes a coiled coil domain, which is potentially the site of the interaction between the two proteins. In this model, the C-terminal end of WASF3 remains in the cytoplasm. This model does not discriminate whether there are two specific forms of WASF3, one exclusively on the outside of the mitochondria and the other within the mitochondria. These subtleties, however, do not impact on the importance of the interaction between these two proteins in affecting the invasion phenotype, which is the main focus of this study. The association of WASF3 with mitochondria is also consistent with observations that other family members show a similar association in a different context. , for example, has been shown to bind to PKA and a number of other kinases, suggesting it functions as an A kinase associated protein (AKAP) delivering functional protein complexes to specific locations within the cell, including the mitochondria 38,39. This protein complex is associated with the BAD-mediated mechanism of apoptosis although the mechanism is not known 40,41. A study of interacting kinases using MS demonstrated the presence of PKA and PKC in the WASF3 complex 13. Nonetheless, the suggestion that WASF proteins can deliver kinase cargoes to mitochondria, provides additional circumstantial evidence for the functional interaction between this protein family and the mitochondria, which merits further investigation.
Since we have repeatedly shown that loss of the WASF3 protein leads to loss of invasion and metastasis, it is likely that the effect of ATAD3A on metastasis is mediated through its control of WASF3 stability. To determine whether the effects of ATAD3A knockdown on gene expression profiles were due to loss of ATAD3A per se, or whether this reflected the consequence of reduced WASF3 levels, we compared the gene expression profiles described here with those derived from WASF3 knockdown cells 11. Although there was variation between the genes identified in the two experiments, there was a significant concordance between the two datasets with 12/20 of the highest upregulated genes present in both data sets (data not shown). CDH1, for example, was upregulated in both datasets which may suggest that the effects on cell movement in the ATAD3A knockdown cells are, in part, due to reduced levels of WASF3. It should be noted that knockdown of ATAD3A only results in a 50% knockdown of WASF3, and so it would be expected that the profiles were different since the WASF3 knockdown cells showed a much higher level of inactivation.
We have shown previously that WASF3 is stabilized in the cytoplasm by HSP70 13, and that inactivation of HSP70 leads to significant loss of the WASF3 protein. We have now extended these studies and demonstrated that even though inactivation of HSP70 leads to a significant reduction in cytoplasmic WASF3, mitochondrial associated WASF3 is protected through its interaction with ATAD3A. The GRP78 ER stress chaperone promotes tumor proliferation, survival, metastasis, and resistance to a wide variety of therapies 42,43. The stabilization of WASF3 appears to be due to an interaction with GRP78, which is required for the ATAD3A stabilization of WASF3. GRP78 is a well-recognized resident in the ER, but is also found on the mitochondrial-associated membrane of the ER as well as inside mitochondria, although the transport mechanism is not known 27–29. It has been shown previously that knockdown of ATAD3A leads to mitochondrial fragmentation and decreased co-localization with the ER 14–17, suggesting a possible role in communicating with the ER. Many other proteins are involved in the transport of cargo into the mitochondria and it is possible that WASF3 may be involved in this transport mechanism through creating a bridge between the ER and the mitochondria through its interaction with the GRP78 and ATAD3A proteins. Alternatively, the interaction with GRP78 occurs exclusively within the mitochondria which contributes to the stability of this complex.
In summary, our studies have shown that loss of WASF3 function invariably leads to suppression of invasion and motility in a wide variety of cancer cell types, and inactivation of ATAD3A provides another mechanism of inactivating WASF3 in breast and colon cancer cells. ATAD3A, therefore, may represent a novel therapeutic target for human cancers.
Materials and methods
Cell culture and standard assays
MDA-MB-231 and SW620 cells were purchased from the American Type Culture Collection (ATCC; Rockville, MD, USA) and maintained according to the supplier’s instructions. RT-PCR, QRT-PCR, lentiviral transduction, soft agar colony formation assays, Matrigel invasion assays, Western blot, IP, IF, IHC and CHX chase assay were carried out as described previously 10–13.
DNA constructs, antibodies and other reagents
pLKO.1 lentiviral vectors harboring hRNAs targeting ATAD3A, WASF3 or GRP78 were obtained from Open Biosystems (Huntsville, AL, USA). Lentiviral pCDH-CMV-MCS-EF1-PURO-HA-WASF3 (pCDH-HA-WASF3) and pcDNA-Flag-HSP70 constructs were generated as described previously 13. To construct the GPR78 expression vector, human GRP78 was PCR amplified from full-length cDNA (Open Biosystems) and subcloned into the pCMV-Flag vector (XhoI and NotI). The full-length (HA-3A) and truncated human ATAD3A (HA-3A-Nter, HA-3A-Cter) with HA tags were subcloned into pCDH-CMV-MCS-EF1-PURO (System Biosciences, Mountain View, CA) using XbaI and NotI sites. MitoTracker® Red was purchased from Invitrogen (Carlsbad, CA, USA), PES was obtained from Calbiochem (San Diego, CA, USA), and CHX, TG, BFA and TA were purchased from Sigma (St Louis, MO, USA). 17-AAG and AUY922 were obtained from Selleckchem (Houston, TX, USA). The primers used for RT-PCR or QRT-PCR studies are listed in Table S1, and the primary antibodies used for Western blot or IP assays are listed in Table S2.
Cell proliferation and viability
Cell proliferation and viability at the indicated incubation time was determined by CellTiter 96® AQueous One Solution Cell Proliferation kit and CellTiter-Glo® Luminescent Cell Viability kit from Promega (Madison, WI, USA), respectively.
MS and protein identification
MDA-MB-231 cells expressing either the pCDH-HA-WASF3 or the pCDH empty vector were harvested and the cleared lysates were incubated with either anti-HA, anti-WASF3 antibodies or control rabbit IgG. Detailed procedures for protein preparation for MS analysis were provided previously 7,13. All MS/MS data were analyzed using Sequest (Thermo Fisher Scientific, San Jose, CA, USA; version 1.2.0.208) and X!Tandem (GPM, version 2007.01.01.1). Scaffold (version Scaffold_3.1.4.1, Proteome Software, Inc., Portland, OR, USA) was used to validate MS/MS-based peptide and protein identifications as described previously 7,13.
Mitochondria isolation and trypsin proteolysis
Mitochondrial fractionation was performed using the ProteoExtract® Mitochondria/Cytosol Fractionation Kit (Millpore, Billerica, MA, USA) according to the manufacturer’s instructions. The highly enriched mitochondrial and cytoplasmic fractions were confirmed by Western blot analysis using the COXIV mitochondrial-specific marker. Trypsin treatment of mitochondria was performed as described previously 44. Briefly, crude preparations of mitochondria were exposed to 50 μg/ml trypsin for 0–50 minutes and were then pelleted, lysed, and separated by SDS-PAGE. The protein binding to the organelle was analyzed using the antibodies against WASF3 and a mitochondrial inner membrane protein Cytochrome C.
In vivo tumor growth and metastasis analysis
All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Georgia Regents University. Six-week-old female NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and maintained in accordance with IACUC guidelines. The animal experiments were performed using the NSG mouse model as described previously 22. Briefly, 2×106 cells, in 100 μl of Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), were injected subcutaneously into the fourth abdominal fat pad at the base of the nipple. Tumor growth was measured externally every 3 to 5 days using vernier calipers and the tumor volume was calculated as length×width2×0.52. The mice were sacrificed on day 60, and the lungs were removed and processed for histological analyses. H&E staining was used to examine tumor metastasis in the lung.
Gene expression microarray
Total RNA was extracted from stable ATAD3A knockdown and the knockdown control MDA-MB-231 cells using Trizol reagent (Invitrogen). For gene expression profiling, labeled RNA was hybridized to a GeneChip® Human Genome U133A Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA) probing 18,400 transcripts and variants. The microarray data were analyzed as described previously 13 and deposited in the Gene Expression Omnibus with accession number GSE56084. GO analysis was performed with DAVID 45 for all differentially expressed genes (>2-fold, p < 0.05).
Statistical analysis
To assess the role of ATAD3A for the survival of breast cancer patients, we investigated the gene expression in microarray data of breast cancer patients obtained from Gene Expression Omnibus (GEO) database (GSE18229, GSE42568, GES7390, GSE4922 and GSE21653). All percentiles between the lower and upper quartiles of gene expression were computed for the Kaplan-Meier method and the best performing threshold was used as a cut-off point for high and low groups of ATAD3A gene expression. Associations between gene expression and clinical features were investigated using Pearson’s chi-square tests. In univariate survival analyses, the Kaplan-Meier method and the log-rank test were used to compare overall survival curves between high and low gene expression groups.
Supplementary Material
Acknowledgments
This work was supported in part by grant CA120510 from the National Institutes of Health. We would like to thank Yun Mei, James Ross and the staff of Cancer Center Shared Resources at GRU for the technical assistance.
Footnotes
Conflict of interest: The authors declare no conflicts of interest related to this work.
References
- 1.Sossey-Alaoui K, Su G, Malaj E, Roe B, Cowell JK. WAVE3, an actin-polymerization gene, is truncated and inactivated as a result of a constitutional t(1;13)(q21;q12) chromosome translocation in a patient with ganglioneuroblastoma. Oncogene. 2002;21:5967–5974. doi: 10.1038/sj.onc.1205734. [DOI] [PubMed] [Google Scholar]
- 2.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 3.Sossey-Alaoui K, Head K, Nowak N, Cowell JK. Genomic organization and expression profile of the human and mouse WAVE gene family. Mamm Genome. 2003;14:314–322. doi: 10.1007/s00335-002-2247-7. [DOI] [PubMed] [Google Scholar]
- 4.Bisi S, Disanza A, Malinverno C, Frittoli E, Palamidessi A, Scita G. Membrane and actin dynamics interplay at lamellipodia leading edge. Curr Opin Cell Biol. 2013;25:565–573. doi: 10.1016/j.ceb.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Sossey-Alaoui K, Li X, Ranalli TA, Cowell JK. WAVE3-mediated cell migration and lamellipodia formation are regulated downstream of phosphatidylinositol 3-kinase. J Biol Chem. 2005;280:21748–21755. doi: 10.1074/jbc.M500503200. [DOI] [PubMed] [Google Scholar]
- 6.Sossey-Alaoui K, Li X, Cowell JK. c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J Biol Chem. 2007;282:26257–26265. doi: 10.1074/jbc.M701484200. [DOI] [PubMed] [Google Scholar]
- 7.Teng Y, Ghoshal P, Ngoka L, Mei Y, Cowell JK. Critical role of the WASF3 gene in JAK2/STAT3 regulation of cancer cell motility. Carcinogenesis. 2013;34:1994–1999. doi: 10.1093/carcin/bgt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sossey-Alaoui K, Safina A, Li X, Vaughan MM, Hicks DG, Bakin AV, et al. Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am J Pathol. 2007;170:2112–2121. doi: 10.2353/ajpath.2007.060975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teng Y, Ren MQ, Cheney R, Sharma S, Cowell JK. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br J Cancer. 2010;103:1066–1075. doi: 10.1038/sj.bjc.6605850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng Y, Liu M, Cowell JK. Functional interrelationship between the WASF3 and KISS1 metastasis-associated genes in breast cancer cells. Int J Cancer. 2011;129:2825–2835. doi: 10.1002/ijc.25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teng Y, Mei Y, Hawthorn L, Cowell JK. WASF3 regulates miR-200 inactivation by ZEB1 through suppression of KISS1 leading to increased invasiveness in breast cancer cells. Oncogene. 2014;33:203–211. doi: 10.1038/onc.2012.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teng Y, Ngoka L, Mei Y, Lesoon L, Cowell JK. HSP90 and HSP70 proteins are essential for stabilization and activation of WASF3 metastasis-promoting protein. J Biol Chem. 2012;287:10051–10059. doi: 10.1074/jbc.M111.335000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 16.Merle N, Féraud O, Gilquin B, Hubstenberger A, Kieffer-Jacquinot S, Assard N, et al. ATAD3B is a human embryonic stem cell specific mitochondrial protein, re-expressed in cancer cells, that functions as dominant negative for the ubiquitous ATAD3A. Mitochondrion. 2012;12:441–448. doi: 10.1016/j.mito.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Gilquin B, Taillebourg E, Cherradi N, Hubstenberger A, Gay O, Merle N, et al. The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol Cell Biol. 2010;30:1984–1996. doi: 10.1128/MCB.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubstenberger A, Labourdette G, Baudier J, Rousseau D. ATAD 3A and ATAD 3B are distal 1p-located genes differentially expressed in human glioma cell lines and present in vitro anti-oncogenic and chemoresistant properties. Exp Cell Res. 2008;314:2870–2883. doi: 10.1016/j.yexcr.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 19.You WC, Chiou SH, Huang CY, Chiang SF, Yang CL, Sudhakar JN, et al. Mitochondrial protein ATPase family, AAA domain containing 3A correlates with radioresistance in glioblastoma. Neuro Oncol. 2013;15:1342–1352. doi: 10.1093/neuonc/not077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang SF, Huang CY, Lin TY, Chiou SH, Chow KC. An alternative import pathway of AIF to the mitochondria. Int J Mol Med. 2012;29:365–372. doi: 10.3892/ijmm.2011.849. [DOI] [PubMed] [Google Scholar]
- 21.Chen TC, Hung YC, Lin TY, Chang HW, Chiang IP, Chen YY, et al. Human papillomavirus infection and expression of ATPase family AAA domain containing 3A, a novel anti-autophagy factor, in uterine cervical cancer. Int J Mol Med. 2011;28:689–696. doi: 10.3892/ijmm.2011.743. [DOI] [PubMed] [Google Scholar]
- 22.Fang HY, Chang CL, Hsu SH, Huang CY, Chiang SF, Chiou SH, et al. ATPase family AAA domain-containing 3A is a novel anti-apoptotic factor in lung adenocarcinoma cells. J Cell Sci. 2010;123:1171–1180. doi: 10.1242/jcs.062034. [DOI] [PubMed] [Google Scholar]
- 23.Iorns E, Drews-Elger K, Ward TM, Dean S, Clarke J, Berry D, et al. A new mouse model for the study of human breast cancer metastasis. PLoS One. 2012;7:e47995. doi: 10.1371/journal.pone.0047995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 26.Grkovic S, O’Reilly VC, Han S, Hong M, Baxter RC, Firth SM. IGFBP-3 binds GRP78, stimulates autophagy and promotes the survival of breast cancer cells exposed to adverse microenvironments. Oncogene. 2013;32:2412–2420. doi: 10.1038/onc.2012.264. [DOI] [PubMed] [Google Scholar]
- 27.Li N, Zoubeidi A, Beraldi E, Gleave ME. GRP78 regulates clusterin stability, retrotranslocation and mitochondrial localization under ER stress in prostate cancer. Oncogene. 2013;32:1933–1942. doi: 10.1038/onc.2012.212. [DOI] [PubMed] [Google Scholar]
- 28.Sun FC, Wei S, Li CW, Chang YS, Chao CC, Lai YK. Localization of GRP78 to mitochondria under the unfolded protein response. Biochem J. 2006;396:31–39. doi: 10.1042/BJ20051916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 30.Dong D, Stapleton C, Luo B, Xiong S, Ye W, Zhang Y, et al. A critical role for GRP78/BiP in the tumor microenvironment for neovascularization during tumor growth and metastasis. Cancer Res. 2011;71:2848–2857. doi: 10.1158/0008-5472.CAN-10-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maître JL, Heisenberg CP. Three functions of cadherins in cell adhesion. Curr Biol. 2013;23:R626–R633. doi: 10.1016/j.cub.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sotomayor M, Gaudet R, Corey DP. Sorting out a promiscuous superfamily: towards cadherin connectomics. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.03.007. pii:S0962–8924(14)00060–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann M, Bellance N, Rossignol R, Koopman WJ, Willems PH, Mayatepek E, et al. C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS One. 2009;4:e7644. doi: 10.1371/journal.pone.0007644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goller T, Seibold UK, Kremmer E, Voos W, Kolanus W. Atad3 function is essential for early post-implantation development in the mouse. PLoS One. 2013;8:e54799. doi: 10.1371/journal.pone.0054799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogaeri T, Eto K, Otsu M, Ema H, Nakauchi H. The actin polymerization regulator WAVE2 is required for early bone marrow repopulation by hematopoietic stem cells. Stem Cells. 2009;27:1120–1129. doi: 10.1002/stem.42. [DOI] [PubMed] [Google Scholar]
- 37.Dahl JP, Wang-Dunlop J, Gonzales C, Goad ME, Mark RJ, Kwak SP. Characterization of the WAVE1 knock-out mouse: implications for CNS development. J Neurosci. 2003;23:3343–3352. doi: 10.1523/JNEUROSCI.23-08-03343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawe VY, Ramalho-Santos J, Payne C, Chemes HE, Schatten G. WAVE1, an A-kinase anchoring protein, during mammalian spermatogenesis. Hum Reprod. 2004;19:2594–2604. doi: 10.1093/humrep/deh513. [DOI] [PubMed] [Google Scholar]
- 39.Beene DL, Scott JD. A-kinase anchoring proteins take shape. Curr Opin Cell Biol. 2007;19:192–198. doi: 10.1016/j.ceb.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Li JH, Wang J, Zhang CY. Molecular modeling of BAD complex resided in a mitochondrion integrating glycolysis and apoptosis. J Theor Biol. 2010;266:231–241. doi: 10.1016/j.jtbi.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 43.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–818. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellot G, Cartron PF, Er E, Oliver L, Juin P, Armstrong LC, et al. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 2007;14:785–794. doi: 10.1038/sj.cdd.4402055. [DOI] [PubMed] [Google Scholar]
- 45.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.