Abstract
Activation of the WASF3 protein by extracellular stimuli promotes actin cytoskeleton reorganization and facilitates cancer cell invasion, whereas WASF3 depletion suppresses invasion and metastasis. In quiescent cells, the interaction between WASF3 and a complex of proteins including CYFIP1 acts as a conformational restraint to prevent WASF3 activation. Therefore, we took advantage of this endogenous regulatory mechanism to investigate potential sites that disrupt WASF3 function. Here, we show that genetic knockdown of CYFIP1 in cancer cells led to the destabilization of the WASF3 complex, loss of WASF3 function, and suppressed invasion. Based on existing crystallographic data, we developed stapled peptides, referred to as WASF Helix Mimics (WAHM), that target an α-helical interface between WASF3 and CYFIP1. Treatment of highly invasive breast and prostate cancer cells with WAHM inhibitor peptides significantly reduced motility and invasion in vitro.
Mechanistic investigations revealed that these inhibitors suppressed the interaction between Rac and the WASF3 complex, which has been shown to promote cell migration. Furthermore, peptide-mediated inhibition of WASF3 also resulted in the dysregulation of known downstream targets such as MMP-9 and KISS1. Finally, we demonstrate that this invasive phenotype is specific to WASF3 as depletion of WASF1 and WASF2, which can also bind to CYFIP1, did not affect invasion. Collectively, our findings suggest that targeting WASF3 function with WAHM peptides could represent a promising therapeutic strategy for preventing tumor invasion and metastasis.
Keywords: CYFIP1, WASF3, stapled peptide, cancer invasion, metastasis, therapeutics
INTRODUCTION
WASF3 (1) is one of three genes in the Wiskott-Aldridge Syndrome family which have been implicated in the regulation of cell movement related to wound healing, neuronal migration, chemotaxis and immune cell activation, through control of membrane protrusions resulting from reorganization of the actin cytoskeleton (2–4). The protein C-termini carry motifs (VCA) that bind to monomeric actin and the ARP2/3 complex, to facilitate actin polymerization and cytoskeleton reorganization (1). In resting cells, WASF proteins are maintained in a conformation-restricted, inactive form as a result of binding the WASF regulatory complex (WRC) comprised of the CYFIP1/SRA1 (or the PIR121/CYFIP2 ortholog), NCKAP1/NAP1/HEM2 (or the HEM1 ortholog), ABI2 (or the ABI1 and ABI3 orthologs) and HSPC300/BRICK1 proteins (5–7). Activation of WASF proteins are facilitated by binding Rac proteins and phosphorylation of tyrosine residues, which leads to relaxation of the conformational constraints and, in the cases of WASF3, results in increased migration, invasion and metastasis (5).
WASF3 involvement in invasion/metastasis has largely been studied in model cell systems, but is supported by the observation that high-level WASF3 expression is associated with high-grade primary breast (5, 8) and prostate cancers (9). Knockdown of WASF3 in breast and prostate cancer cells leads to a reduction in cell invasion in vitro and metastasis in xenograft models in vivo (5, 9). Non-metastatic cells do not express WASF3 (10), but reexpression in these cells leads to acquisition of the invasion phenotype. Although primarily considered a protein that regulates actin cytoskeleton dynamics, WASF3 has also been shown to have a regulatory function that affects expression of genes involved in metastasis such as KISS1, ZEB1 and miRNA-200s (10–12) and its activity and expression is regulated by proteins such as JAK2, HSP70, ABL and HIF1 (13–16), which have also been implicated in metastasis. WASF3 also interacts with the ATAD3A mitochondrial protein, which regulates its stability at the mitochondrial membrane (17).
A relatively new class of inhibitors that provides the potential for much greater inhibition of protein function with high specificity has been developed, in which chemically stabilized peptides are used to target protein-protein interactions (PPIs). These “stapled peptides” (SP) are synthetically designed to stabilize and constrain an α-helical structure through macrocyclic ring formation using ring closing metathesis chemistry (18–21). Further, these locked peptides can exhibit drug-like properties including enhanced cell permeability and resistance to proteolytic degradation (22–24). SPs have only recently been considered as biological therapeutics and still face challenges of cost and delivery but several are currently being investigated in Phase I clinical trials (25,26).
The structure of the WASF proteins determines their function, which is regulated through interactions with two different subcomplexes (6,7) involving the CYFIP1-NCKAP1 dimer and the ABI2-HSPC300-WASF trimer. Regulation of the VCA domain, and hence actin polymerization, is facilitated by a complex structural interaction between CYFIP1/NCKAP1 and the WASF proteins that act allosterically to prevent actin polymerization. Analysis of the WASF1 crystal structure, and its association with the WRC proteins, demonstrates several critical interacting sites throughout the protein complex (6,7), identifying potential targeting sites to disrupt WASF3 function. In this report we describe the design of stapled peptides that target essential interactions between WASF3 and CYFIP1, and demonstrate that they can suppress WASF3 activation, thereby leading to loss of invasion potential in breast and prostate cancer cells without inhibiting cellular proliferation. As such, these inhibitor peptides offer an opportunity to investigate how suppression of WASF3 function can lead to suppression of invasion and metastasis.
MATERIALS AND METHODS
Stapled peptide synthesis
Peptides were prepared manually using standard Fmoc solid-phase peptide synthesis as described previously (27). The purified peptides were quantified using the Pierce HABA-Avidin microplate protocol by measuring absorbance at 500 nm using the Biotek Synergy 2 Microplate Reader. WAHM1 molecular weight = 2291.4 (expected = 2291.8), WAHM2 molecular weight= 2305.2 (expected = 2305.8), SCR1 molecular weight = 2291.4 (expected = 2291.8), SCR2 molecular weight 2305.8 (expected = 2305.8).
Molecular reagents and constructs
pLKO.1 lentiviral vectors harboring shRNAs targeting WASF1, WASF2, WASF3 or NCKAP1 were obtained from Open Biosystems and shCYFIP1 was from Sigma-Aldrich. WASF2 and WASF3 antibodies were purchased from Cell Signaling Technology. Antibodies against CYFIP1, NCKAP1, WASF1, Rac1 and Rac2 were from Abcam and KISS1 was from Santa Cruz Biotechnology. Antibodies against PY20 and β-Actin were from Sigma. HSP90 inhibitor 17-AAG was obtained from Selleckchem (Houston, TX).
Cell lines and standard assays
MDA-MB-231 cells were obtained from ATCC (04/11) and have been verified using SNP-CGH (11) for characteristic cytogenetic changes. DU145 cells were obtained from ATCC on Feb 10, 2014 and passage <5 were used in this study. The ATCC Cell Authentication Testing service confirmed the identity of Hs578T and PC3 cells using STR DNA fingerprinting analysis (08/15/15). Lentiviral transduction, cell proliferation assays, wound healing assays, Transwell invasion assays, western blotting, flow cytometry and Real-time RT-PCR analysis were carried out as described previously (9–11,15–17).
Quantitation of filamentous actin
Cells were seeded at 2000 cells/well in a 96-well plate and treated with WAHM or scrambled peptides for 24 hours. Following incubation with Texas-red phalloidin (Life technologies) for 30 min and extensive washing, Phalloidin was solubilized with methanol and the supernatant transferred to a clear-bottom, black-walled microtiter plate, and the fluorescence was determined using the Infinite® M1000 PRO (Tecan). In parallel, cell number was determined using the Promega CellTiter-Glo® Luminescent Cell Viability kit.
Immunoprecipitation assays
Immunoprecipitation assays were carried out as described previously (15,16). To determine the physical interaction with individual peptides, MDA-MB-231 cells were pretreated with 10 µM biotin-labeled peptides for 24 hours and lysates were incubated with immobilized avidin resin overnight at 4 °C. The pull down samples were subjected to SDS-PAGE followed by Western blot analysis.
Detection of MMP-9 secretion in culture supernatant by ELISA assays
Cell supernatants were recovered and centrifuged at 1000 g at 4°C for 20 min and MMP9 levels were measured using the Human MMP9 Quantikine® ELISA kit (R&D Systems) and microplate reader at 450 nm.
Experimental metastasis assays
Mouse metastasis assays in SCID mice, approved by the GRU IACUC, were performed as described previously (5,9). Lungs were fixed in 10% neutral buffered formalin, embedded in paraffin blocks, sectioned at 5 µm, and subjected to hematoxylin and eosin staining.
RESULTS
Loss of CYFIP1 protein leads to WASF3 instability resulting in suppression of invasion
Analysis of the WASF complex suggested that the WRC proteins, and in particular the CYFIP1-NCKAP1 dimer, influence its functional activation (28,29). To evaluate the importance of this complex for the function of WASF3, we created shRNAs to knock down the CYFIP1 protein in MDA-MB-231 breast and PC3 prostate cancer cells. In both cases, two independent shRNAs were used and, as shown in Figure 1A, high-level suppression of protein expression was achieved in both cases. Coincident with the knockdown of CYFIP1, WASF3 protein levels were also significantly suppressed (Fig. 1A), whereas WASF3 transcript levels were not affected (Supplementary Figure S1), demonstrating that the stability of WASF3 protein is dependent on its interaction with CYFIP1. Invasion assays demonstrated that knockdown of CYFIP1 also led to a highly significant reduction in invasion potential (Fig. 1B), as seen for WASF3 knockdown in these cell lines (5,9,11). These data suggest that disrupting the protein-protein complex involving CYFIP1 could lead to suppression of invasion by affecting WASF3 function.
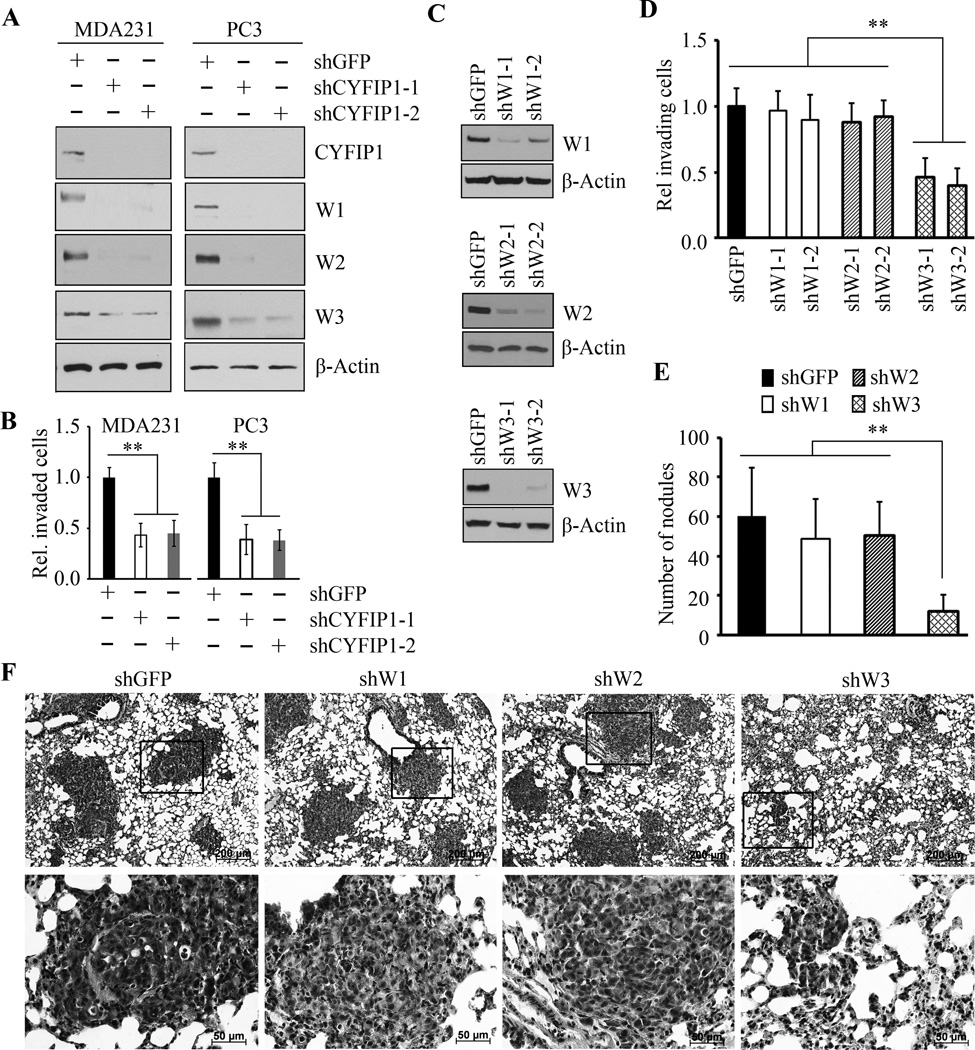
Figure 1. Knockdown of CYFIP1 suppresses cell invasion through destabilizing the WASF3 protein.
Western blot analysis following knockdown of CYFIP1 using two individual shRNAs (−1 and −2) in breast cancer MDA-MB-231 and prostate cancer PC3 cells shows that loss of the CYFIP1 protein leads to concomitant loss of all three WASF3 family members (A). As a result of CYFIP1 knockdown (B) both breast and prostate cancer cells show significant reduction in invasion potential. When two independent shRNAs were used to individually knockdown WASF1, WASF2 and WASF3 in MDA-MB-231 cells (C), Transwell invasion assays showed a marked suppression of invasion potential in WASF3 knockdown cells but not in WASF1 or WASF2 knockdown cells (D). Knockdown of WASF3, but not WASF1 or WASF2, significantly reduces the number of lung surface nodules in SCID mice (E). Histological analysis of lungs from mice with WASF3 knockdown MDA-MB-231 cells show fewer and smaller tumor foci in H & E-stained sections compared with knockdown of WASF1 and WASF2 (F). * p<0.05 and ** p<0.01.
Role of WASF family members in invasion and metastasis
As shown above, CYFIP1 interacts with WASF3 and loss of either protein leads to loss of invasion. CYFIP1, however, also interacts with WASF1 and WASF2, and so targeting the WASF3-CYFIP1 interaction may also affect the function of these proteins. As shown in Figure 1A, knockdown of CYFIP1 also leads to reduced protein levels of WASF1 and WASF2 without affecting their transcript levels (Supplementary Figure S1). Knock down of either WASF1 or WASF2 in MDA-MB-231 cells, however, using two independent shRNAs (Fig. 1C) shows no effect on proliferation (Supplementary Figure S2). Using Transwell invasion assays, there was also no significant suppression of invasion potential as a result of knockdown of WASF1 or WASF2 (Fig. 1D). In contrast, there was a significant reduction in invasion potential when WASF3 was knocked down (Fig. 1D). When MDA-MB-231 cells, in which the three WASF genes had been individually knocked down, were injected into the tail veins of SCID mice, the number of tumor nodules on the lung surface were reduced compared with parental cells. Analysis of lungs from mice injected with WASF3 knockdown MDA-MB-231 cells, showed a significant reduction in surface nodules compared with both parental cells and cells in which either WASF1 and WASF2 had been knocked down (Fig. 1E). In addition, histological analysis showed large tumor foci throughout the lungs in mice that had been injected with either WASF1 or WASF2 knockdown cells (Fig. 1F), compared with relatively few and significantly small tumor foci in cells in which WASF3 had been knocked down (Fig. 1F).
These observations suggest that, although the WASF proteins have been implicated in cell movement, only WASF3 is particularly and specifically associated with invasion and metastasis of cancer cells (Fig. 1). Therefore, it is unlikely that any specific invasion-suppressive effects produced by targeting the WASF3-CYFIP1 complex are a result of targeting functional protein-protein interactions involving the other WASF family members.
Targeting the CYFIP1-WASF interaction with stapled peptides
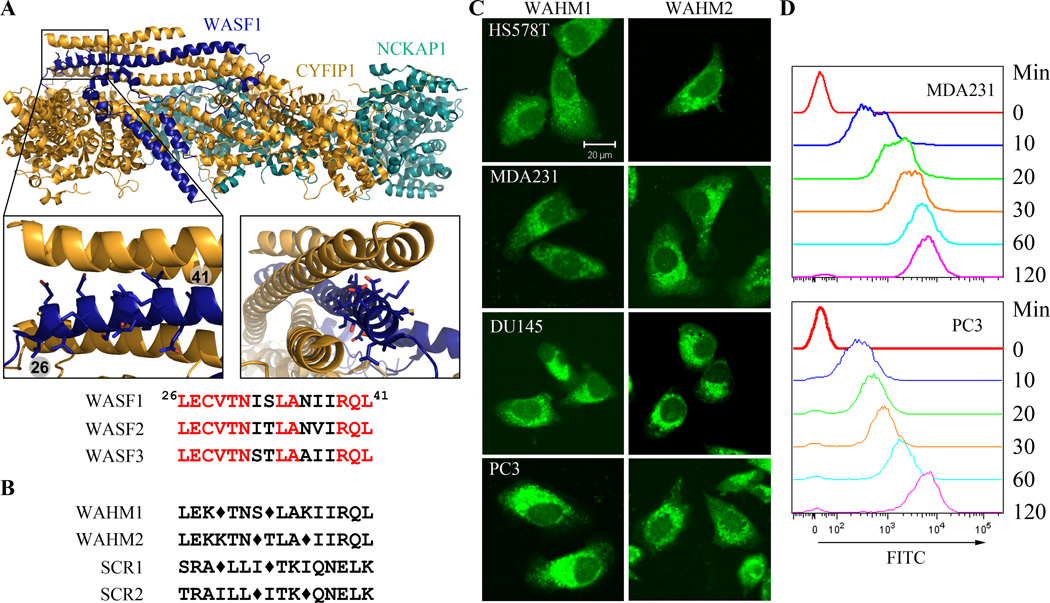
Defining α-helical interfaces between target proteins can only be derived from three-dimensional structural information. Although the crystal structure of WASF3 has not been established, related studies (6,7) identified a large α-helical interface between CYFIP1 and WASF1 at amino acid residues 26–41 (Fig. 2A), the amino acid sequence of which is virtually identical in WASF3 (Fig. 2A), suggesting that this region may perform the same function in both proteins. We therefore developed SP inhibitors derived from this sequence which are referred to as WASF Helix Mimics (WAHM) with sequences LEK*TNS*LAKIIRQL (WAHM1) and LEKKTN*TLA*IIRQL (WAHM2), where * indicates the position of the (S)-2-(4’-pentenyl)alanine that forms the staple (Fig. 2B). Two scrambled (SCR) peptides (SCR1: SRA*LLI*TKIQNELK for WAHM1, and SCR2: TRAILL*ITK*QNELK for WAHM2) were also designed as negative controls for the study. All peptides were modified to contain an N-terminal 5(6)-carboxyfluorescein label for studies involving cellular uptake and intracellular location.
Figure 2. Stapled peptide design and uptake in cancer cells.
The crystal structure of WASF1 in complex with CYFIP1-NCKAP1 shows the interaction surfaces derived from WASF3 (A) and defines an α-helical surface at amino acids 26–41 in CYFIP1 that provides contact points for the two proteins. Structures were rendered in PyMol using PDB 3P8C. The amino acid sequence between the three members of the WASF family of proteins is highly conserved (below). Using the WASF3 sequence, two stapled peptides were designed to target this interaction surface (B) where diamonds represent the position of the non-natural amino acids. Scrambled peptide controls were also generated for each WAHM peptide (B). MDA-MB-231 and HS578T breast cancer cells and DU145 and PC3 prostate cancer cells show cytoplasmic fluorescence labeling after 6 hours exposure to WAHM1/2 (C). A time course of peptide uptake using flow cytometry over the first 2 hours period of exposure (D) shows progressive fluorescein labeling in breast and prostate cancer cells.
Breast (HS578T and MDA-MB–231) and prostate (PC3 and DU145) cancer cells were exposed to WAHM1 and WAHM2 and their scrambled controls at a concentration of 10 µM, and cellular uptake was followed by assessing fluorescein levels in the cells using confocal microscopy and flow cytometry (Fig. 2C, D). After 6 hours, all four cell lines demonstrated high-level cytoplasmic fluorescence in > 80% of the cells (Fig. 2C). Flow cytometry showed a progressive increase in intracellular fluorescence over the first 2-hours (Fig. 2D). No obvious increase or decrease in fluorescence intensity was seen after 24 hours (data not shown). These data demonstrate a rapid and high-level uptake of these peptides by cancer cells, as we have seen for other SPs (27).
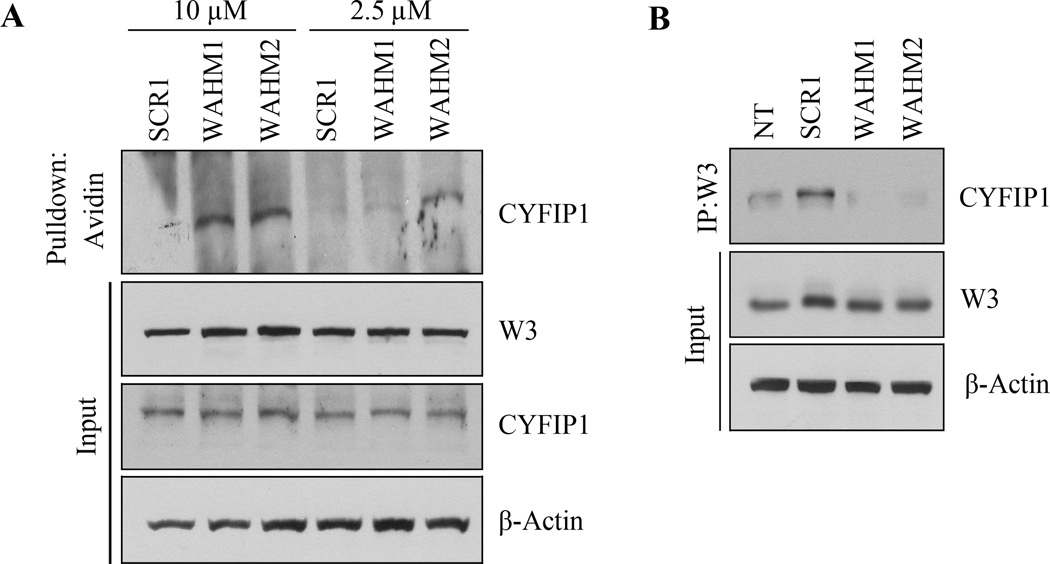
To determine whether the SPs formed a complex with WASF3 within the cells, MDA-MB-231 cells were exposed to biotinylated WAHM1 and WAHM2 for 6 hours, after which the cells were lysed and pull down assays were performed using avidin-coated beads. As shown in Figure 3a, WASF3 protein levels were not affected by the SPs, whether they were treated for short (2 hours) or longer (24 hours) periods (Supplementary Fig. S3). However, CYFIP1 was present in the pull down complex with both WAHM1 and WAHM2 but not the SCR peptides, in a concentration-dependent manner (2.5 µM versus 10 µM) (Fig. 3A). IP of WASF3 from these cells demonstrated that the CYFIP1 protein was largely absent in the immunocomplex from cells treated with the two SPs (Fig. 3B), further demonstrating that these peptides affect the assembly or stability of this protein complex.
Figure 3. The WAHM peptides lead to disruption of WASF3 complex.
In avidin-biotin pull down assays using biotinylated stapled peptides and a concentration of 10 µM, CYFIP1 was shown to interact with WAHM1/2 but not scrambled control peptide (SCR1) (A). At lower concentrations of peptides (2.5 µM), recovery of CYFIP1 was reduced. Treatment with WAHM1/2 for 24 hours did not affect intracellular levels of either WASF3 or CYFIP1. In an IP of WASF3, following treatment with WAHM1/2 (B), CYFIP1 was not present in the immunocomplex, demonstrating that WAHM1/2 leads to disruption of the complex without affecting the protein levels.
WAHM1/2 peptides suppress cancer cell motility and invasion
Since the stapled peptides are reasonably stable for at least 24 hours within the cell, we were able to investigate their effect on short-term cell motility, proliferation and invasion. Using MTS assays, there was no significant difference in cell proliferation in either breast or prostate cancer cells treated with either WAHM1 or WAHM2, or the scrambled peptides at a 10 µM concentration over 24 hours (Supplementary Fig. S4).
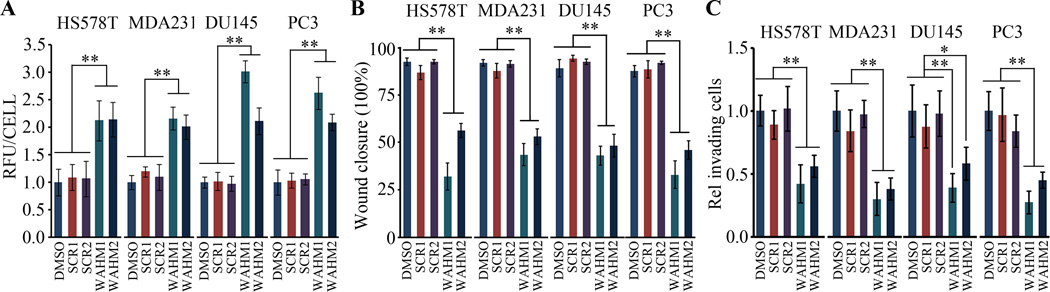
Decreased cell motility has been associated with increased stress fiber formation (9,13). To investigate whether the SPs affected organization of the microfilament network in cells treated with WHAM peptides, we used Phalloidin-binding assays to quantify polymerized F-actin in four different cell lines treated with either WAHM1 or WAHM2. In all cases the actin cytoskeleton became more organized, with increased numbers and thickness of the actin stress fibers (not shown) which led to increased fluorescence intensity (Fig. 4A), compared with cells treated with DMSO or scrambled SPs. Thus, disrupting the CYFIP1-WASF3 interaction increases intracellular actin bundles, which is also a feature of WASF3 knockdown cells (30) and is associated with reduced cell migration and invasion. Using the scratch wound assay, treatment of breast and prostate cell lines with SPs led to reduced migration potential (Fig. 4B and Supplementary Fig. S5A), which is consistent with previous observations following shRNA knockdown of WASF3 (5,9,11). Transwell invasion analysis over 24 hours following treatment with 10 µM WAHM1 or WAHM2 resulted in reduced (~50–75%) invasion potential, compared with cells treated with either the SCR peptides or DMSO (Fig. 4C and Supplementary Fig. S5B). The equivalency of this effect between DMSO and SCR treatment demonstrates that neither SCR control peptide had any significant effect on the invasion phenotype. High dose treatment (10 µM) suppressed cell invasion more efficiently than low dose treatment (2.5 µM), indicating a dose-dependent effect of WAHM1 and WAHM2 on cell invasion (Supplementary Fig. S6). Thus, targeting the CYFIP1-WASF3 interaction with SPs affects cancer cell cytoskeleton organization, motility and invasion, consistent with knockdown of WASF3 function achieved by other means.
Figure 4. WAHM peptides suppress cancer cell invasion.
Fluorescence intensity of phalloidin stained cells (A) demonstrates that the cells treated with WAHM1/2 show increased intensity indicative of increased levels of stress fibers, in contrast to cells treated with either the DMSO vehicle or scrambled peptides. When prostate and breast cancer cells were treated with WAHM1/2 there was a significant reduction in cell motility compared with DMSO and scrambled peptide treatment (B). Similarly, treatment with WAHM1/2 significantly suppresses invasion in breast and prostate cells (C). * p<0.05 and ** p<0.01.
WAHM treatment leads to loss of WASF3 activation
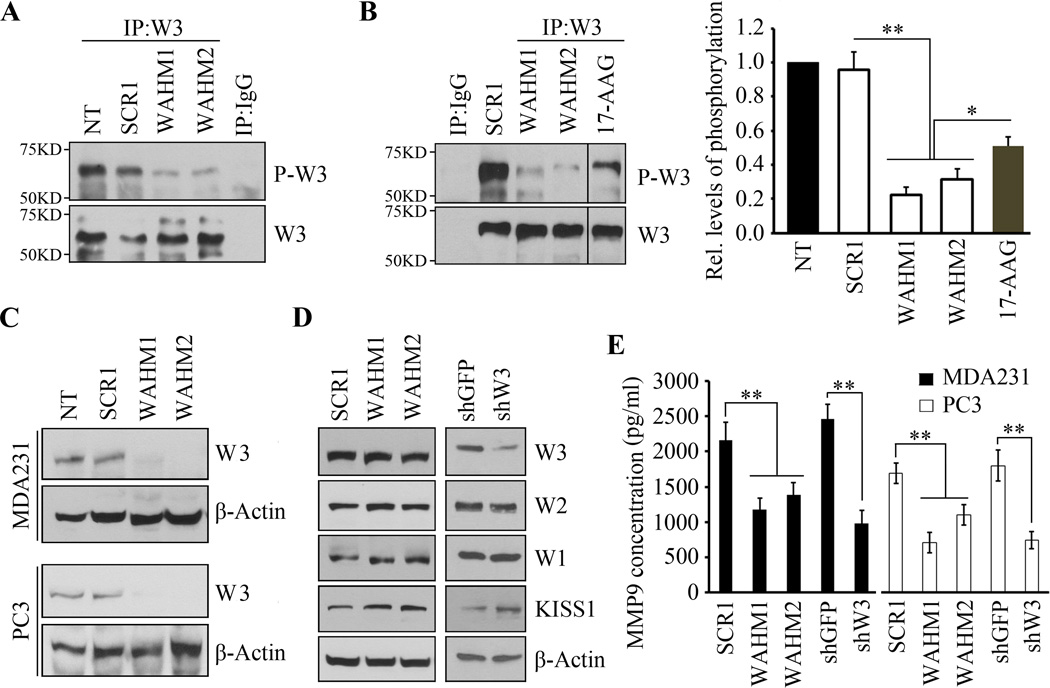
ShRNA knockdown of CYFIP1 leads to WASF3 protein destabilization (Fig. 1). In the SP treated cells, however, there was no change in WASF3 protein levels compared with control treated cells (Fig. 3 and Supplementary Fig. S3). Thus, targeting the WASF3 complex with WAHM1/2 does not lead to its degradation as seen following genetic knockdown of any of the three WASF proteins, even though invasion and migration are dramatically affected. Similarly, SP treatment did not affect WASF1 or WASF2 protein levels (Fig. 5D). Since WASF3 function depends on its phosphoactivation (13,15,16), we investigated the phosphorylation status of WASF3 following treatment with WAHM1 or WAHM2. Treatment of MDA-MB-231 cells with either WAHM1 or WAHM2 led to a dramatic decrease in WASF3 phosphorylation levels (Fig. 5A, B), without a reduction in WASF3 protein levels. The same effect was observed in PC3 prostate cancer cells (data not shown). Thus, loss of WASF3 activation resulting from SP treatment accounts for the suppression of invasion. We previously demonstrated that inactivation of HSP90 with 17-AAG also led to reduced WASF3 phosphoactivation due to suppression of ABL kinase function (15). Treatment with the SPs, however, showed that suppression of WASF3 activation was far more significant (Fig. 5B) than that seen following 17-AAG treatment.
Figure 5. WAHM peptides lead to loss of WASF3 phosphoactivation and suppression of downstream signaling.
MDA-MB-231 cells, treated with WAHM1/2, show no reduction in WASF3 protein levels but, unlike cells treated with the scrambled control (A), WASF3 phosphorylation is reduced. IgG IP was used as a negative control. WAHM1/2 is more efficient than the 17-AAG HSP90 inhibitor in suppressing WASF3 phosphorylation (B). When MDA-MB-231 and PC3 cells were starved overnight and then treated with WAHM1/2, WASF3 levels were reduced below detectable levels (C) in contrast to untreated cells (NT) and cells treated with scrambled control peptides (SCR1). Stapled peptides do not affect the protein levels of any of the WASF family members (D). KISS1 levels increase when cells are treated with WAHM1/2 (D, left), compared with scrambled peptides. Knockdown of WASF3 (shW3) leads to increased KISS1 protein levels (D, right) compared with control shRNA treatment (shGFP). Following treatment of MDA-MB-231 and PC3 cells with WAHM1/2 (E) downregulation of MMP9 levels are comparable to those seen in WASF3 knockdown cells (shW3). In contrast, cells treated with the scrambled peptide or control shRNA (shGFP) show no affect on MMP-9 activity.
Growth factors and cytokines induce phosphoactivation of WASF3 (16,30). Thus, in cells cultured in serum, there is a consistent subpopulation of the WASF3 protein that is activated as demonstrated by its relocation to the leading edge of invading cells. In starved cells, however, WASF3 activation is virtually undetectable (Supplementary Fig. S7). To determine whether treatment with SPs specifically affects activated WASF3, we starved MDA-MB-231 and PC3 cells overnight and then treated them for 4 hours with WHAM1 and WHAM2. As shown in Figure 5C, in the absence of serum, WASF3 levels are reduced below detectable levels following SP treatment compared with non-treatment or scrambled peptide treatment. These data suggest that the inactive form of WASF3 is more susceptible to degradation following disruption of the CYFIP1-WASF3 complex.
WAHM1/2 suppresses MMP9 secretion through inactivation of WASF3
WASF3-mediated invasion is associated with downregulation of KISS1 (10,11) but this effect was not seen in WASF1- or WASF2-depleted cells (10). Knockdown of WASF3 also leads to downregulation of MMP9 expression and secretion (10). Western blot analysis of MDA-MB-231 cells treated with WAHM1 or WAHM2 showed increased KISS1 levels (Fig. 5D) and in MDA-MB-231 and PC3 cells, there was a remarkable reduction in secreted MMP9 levels compared with treatment with scrambled controls (Fig. 5E). Collectively, these results indicate that disruption of the CYFIP1-WASF complex leads to suppression of MMP-9 secretion through inhibition of the WASF3-KISS1 signaling cascade.
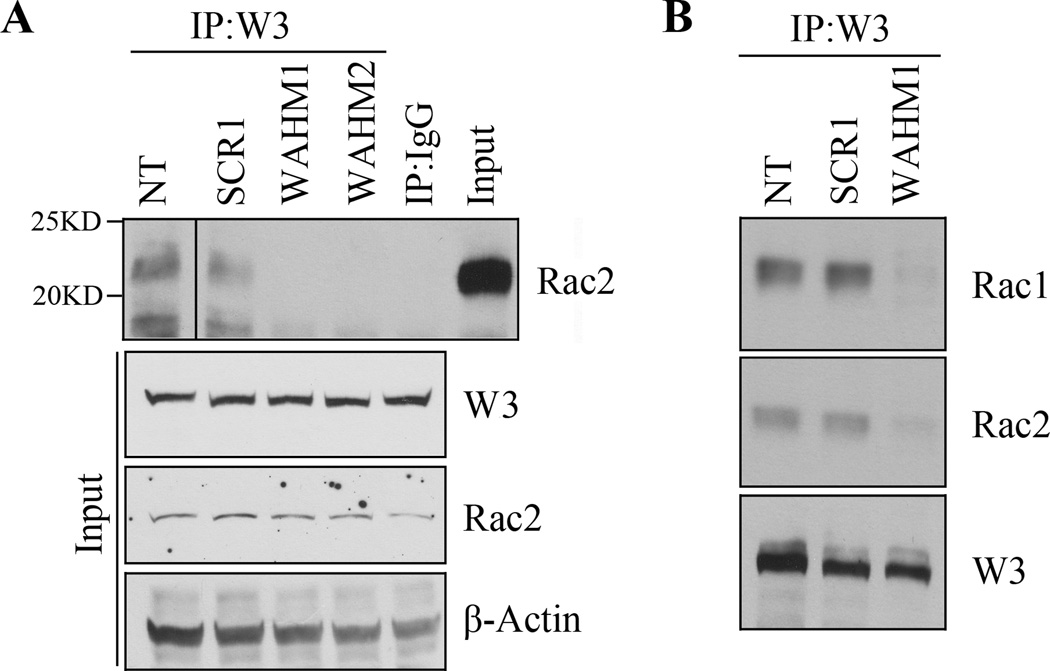
WAHM peptides disrupt the binding of Rac1/2 to the WASF3 complex
Activated Rac1 induces lamellipodia formation and is suggested to bind the WASF regulatory complex including CYFIP1/NCKAP. Although Rac1 has been implicated in this function, our MS analysis of PC3 cells for example (15) identified Rac2 in the immunocomplex as well (Supplementary Fig. S8), which was confirmed by IP and western blotting (Fig. 6A). To determine whether the WHAM1/2 effect on the CYFIP-WASF3 complex also involved Rac binding, we generated IP using WASF3 antibodies and then western blot analysis of MDA-MB-231 cells following treatment with either WAHM1/2 or SCR1 control peptides. As seen in Figure 6A, Rac2, while present in the untreated cells and scramble control treated cells, could not be detected in the WASF3 IP from WAHM treated cells. IP analysis further demonstrated that both Rac1 and Rac2 are present in the WASF3 immunocomplex (Fig. 6B).
Figure 6. WAHM peptides prevent Rac1/2 recruitment to the WASF3 complex.
Treatment of MDA-MB-231 cells with WAHM1/2 leads to loss of Rac2 in the immunocomplex (A) compared with cells either untreated (NT) or treated with the scrambled control peptide (SCR1). Both Rac1 and Rac2 were absent from the WASF3 immunocomplex in the presence of WAHM peptide (B).
DISCUSSION
One of the challenges in exploring strategies to increase survival in cancer patients is to devise a means of suppressing the most lethal aspect of the disease that results from metastatic progression. Overexpression of WASF3 is now clearly correlated with high-grade breast and prostate cancer (8,9,31), and experimental evidence to date suggests that its inactivation leads to suppression of invasion and metastasis in several model cell systems (5,9–11), raising the possibility that targeting its function may provide a means of suppressing metastasis. The present study supports this idea, where specific targeting of WASF3 function, using stapled peptides, affecting the WASF3-CYFIP1 protein-protein interaction disrupts WASF3 activity and suppresses the invasion phenotype.
Genetic knockdown of CYFIP1 leads to an almost complete absence of the protein, with a concomitant reduction in WASF3 protein levels. As expected, this leads to suppression of invasion, demonstrating a functional consequence of disrupting the WRC complex. In the absence of CYFIP1, WASF3 protein levels are almost completely lost, demonstrating the importance of these interactions in maintaining the stability of members of the complex and underscoring the importance of the CYFIP1-NCKAP1 dimer in maintaining the function of the WASF3 protein. In contrast, however, targeting the interface between CYFIP1 and WASF3 (amino acids 21–46) with the stapled peptides, does not affect WASF3 protein levels, but rather prevents phosphoactivation, which we have shown is absolutely required for invasion (13,15,16). Activation of the WASF3 proteins can be achieved by binding to GTP-loaded Rac1 (32) and, as we show here, Rac2 is also identified in the immunocomplex with WASF3 suggesting a possible role for this family member in WASF3 activation as well. It has been shown that Rac binding, in contrast to the shRNA knockdown, does not disrupt the WRC but triggers conformational changes that allow inhibition of the VCA domain to be removed, allowing actin reorganization at the leading edges of cells (33,34). When CYFIP1 is absent from the complex, WASF proteins cannot respond to Rac1 activation and subsequent cytoskeletal remodeling, supporting our observation that knockdown of CYFIP1 also leads to loss of invasion. These observations suggest that the WAHM SPs interfere with the WRC conformation sufficiently to prevent Rac binding, but not enough to disrupt stability of WASF3. As a consequence, however, WASF3 phosphoactivation is prevented, which accounts for the loss of invasion. These observations are consistent with previous reports that preventing phosphoactivation of WASF3, either by targeting critical activating kinases, or destabilizing them, leads to the same loss of the invasion phenotype (13,15,16). This phosphoactivation of WASF3 appears to have a role in maintaining WASF3 integrity since, in the absence of serum, where WASF3 exists in an unphosphorylated, inactive form, WAHM1/2 treatment leads to loss of WASF3 protein, whereas in the presence of serum where WASF3 is activated, the WASF3 protein is protected from degradation, even in the absence of bound CYFIP1. These data further support the idea that WASF3 function is highly dependent on the overall structural conformation imposed on the WRC by the constituent proteins and that WAHM peptides are sufficient to disrupt the complex to prevent Rac binding and WASF3 activation.
WASF1 and WASF2 have also been implicated in cell movement through regulation of other membrane structures such as membrane ruffles (35). While it might be expected that targeting the CYFIP1-WASF3 interface may also affect the function of the other two WASF family members, we now show, in a side-by-side comparison of the same cell types that respond to WASF3 loss, neither of these family members are required for cancer cell invasion. The SPs therefore, specifically target this phenotype through regulation of WASF3 function. These observations further support a specific role for the WASF3 family member in the regulation of invasion and metastasis.
Our studies indicate that knockdown of CYFIP1 in highly invasive cancer cells leads to suppression of invasion due to its regulatory role over WASF3. It was suggested in a previous study (36), however, that CYFIP1 may have a tumor suppressor function, although these studies were largely performed in normal MCF10A breast cells, which may provide a different context for the function of CYFIP1 and in its interactions with other members of the WASF family, in particular WASF2, which we have shown does not influence invasion in highly metastatic cells. It is possible, therefore, that CYFIP1 may have different effects in normal cells that result from a disruption of the functional interaction with WASF1 and WASF2, but our studies clearly show that CYFIP1 is required to engage in the WASF3 complex to promote cancer cell invasion in different cell types.
Here we show that targeting the CYFIP1-WASF3 interaction leads to suppression of invasion as well as downregulation of invasion-related signaling cascades that were previously reported to be WASF3-dependent, including regulation of KISS1 and MMP-9 levels as a result of WASF3 phosphoactivation. Thus, from a molecular and functional standpoint, the SPs can effectively suppress invasion by suppressing WASF3 signaling, although the levels of suppression of invasion were only ~50–75%, compared with an 80–90% knockdown achieved with the best performing shRNAs. We feel, however, that, as with different shRNAs that have variable efficiencies, achieving effective suppression of function using SPs depends on optimizing the target sequence and peptide design, since the position of the staple and the sequence being targeted can have significant effects on efficacy for optimal suppression of function (37). In the current proof-of-principle series of experiments, however, we demonstrated that the WAHM inhibitors can suppress WASF3 function by targeting a particular PPI. Using this as a starting point, more comprehensive SP libraries can be designed for optimization of suppression of WASF3 function and support targeting CYFIP1-WASF3 in the future to suppress invasion in vivo once robust delivery systems can be developed.
The selected target for the WASF3 SPs was based on the trimeric crystallographic structure of CYFIP1-NCKAP1-WASF1. This interaction is highly conserved between the WASF1 and WASF2 proteins, which might suggest that these SPs could also affect the function of the other two family members. While there may be unknown effects as a result of affecting WASF1/2 function, these do not manifest into changes in cell proliferation or invasion, since knockdown of either WASF1 or WASF2 individually did not lead to a change in invasion in the cancer cell types studied. It appears, therefore, that WASF3 function is specifically related to invasion and that this relies on an interaction with the CYFIP1-NCKAP1 complex. WASF1 and WASF2 have also been implicated in regulating cell movement through influencing other membrane structures such as ruffles and filopodia (38,39) but it is clear from the knockdown studies that these membrane structures do not have a significant effect on invasion or metastasis. As a result, targeting the CYFIP1-NCKAP1-WASF3 complex may provide some specificity for approaches designed to inactivate WASF3 in order to achieve suppression of invasion and metastasis.
Supplementary Material
Acknowledgements
We are grateful to Irina Georgieva for technical assistance.
Financial support: This work was supported in part by grants from the National Institutes of Health, CA120510 (JKC) and 1K22CA154600 (EJK).
Footnotes
Disclosure of Potential Conflicts of Interest The authors declare no competing financial interests.
REFERENCES
- 1.Sossey-Alaoui K, Su G, Malaj E, Roe B, Cowell JK. WAVE3, an actin-polymerization gene, is truncated and inactivated as a result of a constitutional t(1;13)(q21;q12) chromosome translocation in a patient with ganglioneuroblastoma. Oncogene. 2002;38:5967–5974. doi: 10.1038/sj.onc.1205734. [DOI] [PubMed] [Google Scholar]
- 2.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 3.Kurisu S, Takenawa T. WASP and WAVE family proteins: friends or foes in cancer invasion? Cancer Sci. 2010;101:2093–2104. doi: 10.1111/j.1349-7006.2010.01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendoza MC. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin Cell Dev Biol. 2013;24:272–279. doi: 10.1016/j.semcdb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sossey-Alaoui K, Safina A, Li X, Vaughan MM, Hicks DG, Bakin AV, et al. Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am J Pathol. 2007;170:211–221. doi: 10.2353/ajpath.2007.060975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, et al. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulkarni S, Augoff K, Rivera L, McCue B, Khoury T, Groman A, et al. Increased expression levels of WAVE3 are associated with the progression and metastasis of triple negative breast cancer. PLoS One. 2012;7:e42895. doi: 10.1371/journal.pone.0042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teng Y, Ren M, Cheney R, Sharma S, Cowell JK. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br J Cancer. 2010;103:1066–1075. doi: 10.1038/sj.bjc.6605850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teng Y, Mei Y, Hawthorn LA, Cowell JK. WASF3 regulates miR-200 inactivation by ZEB1 through suppression of KISS1 leading to increased invasiveness in breast cancer cells. Oncogene. 2014;33:203–211. doi: 10.1038/onc.2012.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng Y, Liu M, Cowell JK. Functional interrelationship between the WASF3 and KISS1 metastasis-associated genes in breast cancer cells. Int J Cancer. 2011;129:2825–2835. doi: 10.1002/ijc.25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teng Y, Ross JL, Cowell JK. The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT. 2014;3:e28086. doi: 10.4161/jkst.28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sossey-Alaoui K, Li X, Cowell JK. c-ABL-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J Biol Chem. 2007;82:26257–26265. doi: 10.1074/jbc.M701484200. [DOI] [PubMed] [Google Scholar]
- 14.Ghoshal P, Teng Y, Lesoon L, Cowell JK. HIF1A induces expression of the WASF3 metastasis associated gene under hypoxic conditions. Int J Cancer. 2012;131:E905–E915. doi: 10.1002/ijc.27631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teng Y, Ngoka L, Mei Y, Lesoon L, Cowell JK. HSP90 and HSP70 are essential for stabilization and activation of the WASF3 metastasis promoting protein. J Biol Chem. 2012;287:10051–10059. doi: 10.1074/jbc.M111.335000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teng Y, Ghoshal P, Ngoka L, Mei Y, Cowell JK. Critical role of the WASF3 gene in JAK2/STAT3 regulation of cancer cell invasion. Carcinogenesis. 2013;4:1994–1999. doi: 10.1093/carcin/bgt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng Y, Ren X, Li H, Shull A, Kim J, Cowell JK. Mitochondrial ATAD3A combines with GRP78 to regulate the WASF3 metastasis-promoting protein. Oncogene. 2015 doi: 10.1038/onc.2015.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schafmeister CE, Po J, Verdine GL. An all-hydrocarbon crosslinking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- 19.Blackwell HE, Sadowsky JD, Howard RJ, Sampson JN, Chao JA, Steinmetz WE, et al. Ring-closing metathesis of olefinic peptides: design, synthesis, and structural characterization of macrocyclic helical peptides. J Org Chem. 2001;66:5291–5302. doi: 10.1021/jo015533k. [DOI] [PubMed] [Google Scholar]
- 20.Walensky LD. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higueruelo AP, Jubb H, Blundell TL. Protein-protein interactions as druggable targets: recent technological advances. Curr Opin Pharmacol. 2013;13:791–796. doi: 10.1016/j.coph.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 23.Wittrup KD, Verdine GL. Protein engineering for therapeutics, part B. Preface. Methods Enzymol. 2012;503:xiii–xiv. doi: 10.1016/B978-0-12-396962-0.00013-6. [DOI] [PubMed] [Google Scholar]
- 24.Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To KH, et al. Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci U S A. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grigoryev Y. Stapled peptide to enter human testing, but affinity questions remain. Nat Med. 2013;19:120. doi: 10.1038/nm0213-120a. [DOI] [PubMed] [Google Scholar]
- 26.Cromm PM, Spiegel J, Grossmann TN. Hydrocarbon stapled peptides as modulators of biological function. ACS Chem Biol. 2015;10:1362–1375. doi: 10.1021/cb501020r. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Ho TG, Bertinetti D, Neddermann M, Franz E, Mo GC, et al. Isoform-Selective Disruption of AKAP-Localized PKA Using Hydrocarbon Stapled Peptides. ACS Chem Biol. 2014;9:635–642. doi: 10.1021/cb400900r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, et al. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 30.Sossey-Alaoui K, Ranalli TA, Li X, Cowell JK. WAVE3 promotes cell motility and invasion through the regulation of MMP-1, MMP-3 and MMP-9- expression. Exp Cell Res. 2005;308:135–145. doi: 10.1016/j.yexcr.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, et al. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009;16:561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lebensohn AM1, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 36.Silva JM, Ezhkova E, Silva J, Heart S, Castillo M, Campos Y, et al. Cyfip1 is a putative invasion suppressor in epithelial cancers. Cell. 2009;137:1047–1061. doi: 10.1016/j.cell.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu Q, Moellering R, Hilinski G, Kim Y, Grossmann T, Johannes T, et al. Towards understanding cell penetration by stapled peptides. Med Chem Commun. 2015;6:111–119. [Google Scholar]
- 38.Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5:2595–2609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- 39.Beli P, Mascheroni D, Xu D, Innocenti M. WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat Cell Biol. 2008;10:849–857. doi: 10.1038/ncb1745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.