Successful nerve regeneration requires not only that neurons reconstruct new axons distal to the site of injury, but also those growing axons must navigate through the neuropil to make appropriate synaptic connections with target cells. While this is an imposing task for the thousands of axons that may occupy a regenerating nerve in the peripheral nervous system or a tract in the central nervous system, the billions of neurons in the developing brain must accomplish similar tasks making connections that number in the trillions. How do neurons do this?
One of the ways researchers have studied these questions is to introduce radiolabeled amino acids into cell bodies of neurons in anaesthetized, experimental animals during embryonic development or nerve regeneration. This in vivo approach labels newly synthesized proteins in the neuron, some of which are transported into the elongating axon. Researchers can then ask what is unique or changed in the biochemistry of neurons with growing axons compared with those in which axons are intact. Using these techniques, the growth associated protein, neuromodulin (GAP43), was shown to be up-regulated and transported axonally in greatly increased abundance during regeneration and development, when compared with mature neurons (Kalil and Skene, 1986).
A similar approach has been used to investigate the possibility that RNA is transported axonally, substituting radiolabeled RNA precursors for amino acids. The original goal of these experiments was to explore the possibility that protein synthesis could occur locally in axons, the logic being that if proteins were being synthesized in axons, then the major stable RNAs (ribosomal and transfer RNAs) should be produced in the neuronal cell body and transported into the axon. These studies resulted in the surprising finding that in vertebrate neurons only a single species of RNA, 4S RNA (that co-migrated on SDS PAGE with 76nt, tRNA, markers), could be demonstrated to be transported axonally, and only during axon growth (in regenerating optic axons of goldfish and sciatic nerves of rats), and during elongation of optic axons in developing rat and chick brains (reviewed in Ingoglia, 1982). Thus, the axonal transport of 76nt RNAs, parallels that of GAP-43.
Perhaps the most remarkable finding in the experiments examining RNA in growing axons was the electron microscopic autoradiography data that showed high levels of 76nt RNAs in distal axons and axonal growth cones of goldfish optic nerves as they regenerated towards their targets in the optic tectum (Gambetti et al., 1978). These tantalizing results raised the question of the function(s) 76nt RNAs played in axonal growth, guidance and reconnection.
At the time these findings were reported, the only roles known for a 76nt RNA was as tRNA in classical protein synthesis and as tRNAarg that functioned as an arginine donor in the posttranslational N-terminal arginylation of proteins. In squid axoplasm, some of the axonal 4S RNA had the properties of tRNA, but the disproportionate levels of 76nt RNA in squid axoplasm and in regenerating axons, suggested an additional role for 4S RNA in axons (reviewed in Ingoglia, 1982). Thus, we began a series of experiments to test the hypothesis that the 76nt RNA in growing axons functioned as an Arg donor in the posttranslational modification of axonal proteins.
Following protocols in which sciatic nerves of anaesthetized rats were crushed and allowed to regenerate in vivo and N-terminal arginylation was assayed in extracts of nerve segments in vitro, we found that protein arginylation increased dramatically following injury to rat sciatic nerves. The greatest increases occurred within hours of a crush injury in proximal nerve segments that included the site of injury, and days later, in the distal portion of nerve that included regenerating axons (reviewed in Ingoglia, 2015). In subsequent experiments, we tested the hypothesis that the function of posttranslational arginylation was to modify damaged proteins marking them for ubiquitin mediated degradation. The results of those experiments were inconclusive. We also had limited success in identifying endogenous protein targets for arginylation in the sciatic nerve, regenerating goldfish optic nerves, or in squid axoplasm.
Thus, by the mid 1980s we knew that crushing the optic nerves of goldfish induced the synthesis of 76nt RNAs in retinal ganglion cells and the axonal transport of those RNAs in large amounts along regenerating axons, into axonal growth cones as they approached their targets in the goldfish brain. But, we had no understanding of the function these RNAs played in axon regeneration.
In the mid 1990s a new role was described for a class of small (~21nt) RNAs. Several labs using a variety of biological systems showed that these small RNAs can function as post-transcriptional regulatory mechanisms to degrade mRNAs (siRNA) or temporally suppress translation (miRNA) by binding to specific mRNA sequences. This new role for small RNAs, termed RNA interference (RNAi), has been shown to be a universal mechanism for regulating translation in a wide range of cells, including neurons. Subsequent experiments showed that these small RNAs are processed from higher molecular weight precursors. The immediate precursor of miRNAs is a double stranded, 76nt, stem-loop RNA, termed pre-miRNA (reviewed in Bartel, 2009). Pre-miRNA co-migrates with tRNA markers on SDS PAGE, raising a third possibility for the function of 76nt RNAs in growing axons, i.e., it might contain pre-miRNAs which can be cleaved into miRNAs that then locally regulate mRNA translation in the growing axon and growth cone.
Research over the past decade supports each of the possibilities raised above. First, it is now well accepted that a select group of mRNAs are present and translated in growing axons and axonal growth cones (reviewed in Holt and Schuman, 2013). Thus, some of the 76nt RNA transported in growing axons is likely to be tRNA used in axonal protein synthesis. Regarding the origin of the other major stable RNAs, recent experiments demonstrate that ribosomes can be imported into axons from adjacent Schwann cells (Court et al., 2008), accounting for the paucity of evidence for axonally transported ribosomal RNAs in vertebrate axons. Another traditional argument against protein synthesis in axons, stems from the failure to observe ribosomal structures in axonal domains. This appears to be due to the localization of axonal ribosomes to periaxolemmal regions where they might be overlooked using standard microscopic techniques (reviewed in Holt and Shuman, 2013). Thus, it is now generally accepted that during axonal growth, a select group of mRNAs are present and can be translated into proteins in the axonal compartment of a neuron. It is likely that the 76nt RNAs transported during axonal growth contain, in part, tRNAs serving as amino acid donors in protein synthesis.
The regulation of protein synthesis in axons is not well understood. But a large body of recent evidence indicates that the RNAi pathway is operative in growing axons and is a likely mechanism for regulating translation. Thus, Dicer, the ribonuclease that cleaves 21nt RNAs from premiRNA, (but not Drosher, the enzyme responsible for the production of pre-miRNA), is present in developing axons and growth cones (Hengst et al., 2006; Kim et al., 2015). Other studies have shown that a variety of micro RNAs are also present in distal regions of elongating axons and axonal growth cones (reviewed by Iyer et al., 2014). These findings strongly support the concept that the 76nt RNA, that was shown to be transported axonally in growing axons more than 30 years ago, is made up, at least in part, of pre-miRNAs.
Finally, experiments reported in the past decade raise the possibility that tRNAarg may play a role as the Arg donor in the posttranslational modification of actin in growing axons. Actin, an abundant cytoskeletal protein, exists in multiple isoforms. In fibroblasts, beta (but not gamma) actin is a target for posttranslational arginylation. Other experiments have shown that arginylation of beta actin is required for lamellae formation and process extension. The arginylation of beta-actin appears to keep the actin monomers in a depolymerized state, allowing elongation to occur. Based on these and other observations it has been proposed that arginylation is a general mechanism for regulating actin isoform interactions (Kashina, 2006). Since beta-actin mRNA is present in growth cones and is essential for axon growth and turning behavior (reviewed in Holt and Schuman, 2013), we speculate that some of the axonally transported growth cone 76nt RNA is tRNAarg that arginylates beta actin, allowing it to maintain a depolymerized, and therefore flexible, configuration as the axon extends and the growth cone searches for its target (Figure 1).
Figure 1.
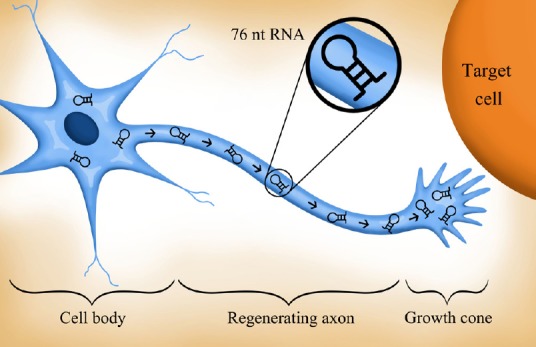
Neurons with axons regenerating after injury (or elongating during development), synthesize and transport axonally large amounts of ~76nt RNAs.
These RNAs may function: 1) as tRNAs in classical protein synthesis, 2) as pre-miRNAs, precursors of 22nt miRNAs, that regulate axonal translation, 3) as tRNAarg in posttranslational modification, perhaps of beta-actin, and/or 4) in a role yet to be described. The figure was drawn by Stephanie Karena Jones.
Many questions remain regarding the function of 76nt RNAs in growing axons. Among these are: which pre-miRNAs are found in elongating axons? What roles do they play in axon elongation, axon guidance in response to external cues, and in synaptogenesis? How does the neuron ‘decide’ which pre-miRNAs are exported into the axon? What environmental cues lead to the activation of the axonal RNAi pathway? Is the mRNA for arginyl transferase (ATE1), the enzyme responsible for arginylation of beta actin, present in axons and does the RNAi pathway regulate its translation? Do miRNAs regulate the translation of beta-actin mRNA? What other axonal mRNAs does the RNAi pathway regulate? Finally, mutations in many genes encoding proteins that interact with and regulate RNAs, including the ZC3H14 polyadenosine RNA binding protein, have been shown to be associated with heritable forms of human intellectual disability. In the case of ZC3H14, its Drosophila ortholog, dNab2 is required for axon projection patterns in brain neurons (Kelly et al., 2015) and its overexpression results in axon projection deficits in the optic lobes (Jalloh et al., unpublished). It will be interesting to see if any of these RNA binding proteins, or ZC3H14/dNab2 in particular, play a role in targeting mRNAs or components of the RNAi complex to growing axons and growth cones.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court FA, Midha R, Cisterna BA, Grochmal J, Shakhbazau A, Hendriks WT, Van Minnen J. Morphological evidence for a transport of ribosomes from Schwann cells to regenerating axons. Glia. 2011;59:1529–1539. doi: 10.1002/glia.21196. [DOI] [PubMed] [Google Scholar]
- Gambetti P, Ingoglia NA, Autilio-Gambetti L, Weis P. Distribution of [3H] RNA in goldfish optic tectum following intraocular or intracranial injection of [3 H] uridine. Evidence of axonal migration of RNA in regenerating optic fibers. Brain Res. 1978;154:285–300. doi: 10.1016/0006-8993(78)90701-1. [DOI] [PubMed] [Google Scholar]
- Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingoglia NA. 4S RNA in regenerating optic axons of goldfish. J Neurosci. 1982;3:331–338. doi: 10.1523/JNEUROSCI.02-03-00331.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingoglia NA. Arginylation in a partially purified fraction of 150k × g supernatants of axoplasm and injured vertebrate nerves. Methods Mol Biol. 2015;1337:25–32. doi: 10.1007/978-1-4939-2935-1_4. [DOI] [PubMed] [Google Scholar]
- Iyer AN, Bellon A, Baudet ML. Micro RNAs in axon guidance. Front Cell Neurosci. 2014;8:78. doi: 10.3389/fncel.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil K, Skene JH. Elevated synthesis of an axonally transported protein correlates with axon outgrowth in normal and injured pyramidal tracts. J Neurosci. 1986;9:2563–2570. doi: 10.1523/JNEUROSCI.06-09-02563.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS. Differential arginylation of actin isoforms: the mystery of the actin N-terminus. Trends Cell Biol. 2006;16:610–615. doi: 10.1016/j.tcb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Kelly SM, Bienkowski R, Banerjee A, Melicharek DJ, Brewer ZA, Marenda DR, Corbett AH, Moberg KH. The Drosophila ortholog of the Zc3h14 RNA binding protein acts within neurons to pattern axon projection in the developing brain. Dev Neurobiol. 2015 doi: 10.1002/dneu.22301. doi: 10.1002/dneu.22301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kim P, Phay M, Soonmooo Y. Identification of precursor microRNAs within distal axons of sensory neuron. J Neurochem. 2015;134:193–199. doi: 10.1111/jnc.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]


