Keywords: nerve regeneration, GIT1, hippocampal neurons, neurite outgrowth, tubulin, microtubule-associated proteins, neural regeneration
Abstract
GIT1, a G-protein-coupled receptor kinase interacting protein, has been reported to be involved in neurite outgrowth. However, the neurobiological functions of the protein remain unclear. In this study, we found that GIT1 was highly expressed in the nervous system, and its expression was maintained throughout all stages of neuritogenesis in the brain. In primary cultured mouse hippocampal neurons from GIT1 knockout mice, there was a significant reduction in total neurite length per neuron, as well as in the average length of axon-like structures, which could not be prevented by nerve growth factor treatment. Overexpression of GIT1 significantly promoted axon growth and fully rescued the axon outgrowth defect in the primary hippocampal neuron cultures from GIT1 knockout mice. The GIT1 N terminal region, including the ADP ribosylation factor-GTPase activating protein domain, the ankyrin domains and the Spa2 homology domain, were sufficient to enhance axonal extension. Importantly, GIT1 bound to many tubulin proteins and microtubule-associated proteins, and it accelerated microtubule assembly in vitro. Collectively, our findings suggest that GIT1 promotes neurite outgrowth, at least partially by stimulating microtubule assembly. This study provides new insight into the cellular and molecular pathogenesis of GIT1-associated neurological diseases.
Introduction
GIT1 is a member of the GIT subfamily of ADP ribosylation factor (ARF)-GTPase activating protein (GAP) family of proteins (Turner et al., 2001; Kahn et al., 2008; Myers and Casanova, 2008), which are characterized by an ARFGAP domain that stimulates the GTPase activity of proteins. GIT1 also contains additional domains, including three ankyrin domains, a Spa2 homology domain (SHD), a synaptic localization domain (SLD), and a paxillin binding domain (PBD) (Zhang et al., 2003). These diverse domains enable GIT1 to interact with various proteins, and as a result, GIT1 has the capacity to play a role in multiple biological processes (Hoefen and Berk, 2006). For example, GIT1 has been reported to play a role in endocytosis (Bhanot et al., 2010; Nakaya et al., 2013; Podufall et al., 2014), adhesion (Wilson et al., 2014; Hammer et al., 2015), cell migration (Hsu et al., 2014; Penela et al., 2014; Podufall et al., 2014), proliferation (Wu et al., 2014; Xiao et al., 2014) and apoptosis (Zhang et al., 2009, 2015).
Histological studies have demonstrated that GIT1 is mainly localized to brain tissues (Schmalzigaug et al., 2007), and accordingly, the biological function of GIT1 in the nervous system has been extensively studied. Several GIT1 knockout mouse models have been developed to investigate the association between GIT1 and brain development (Schmalzigaug et al., 2009; Menon et al., 2010; Won et al., 2011). Although brain developmental defects have been observed in these mouse models, the phenotypes are not consistent. A recent report found that GIT1 deletion leads to a microcephaly-like small brain phenotype (Hong and Mah, 2015). Therefore, the functions of GIT1 in brain development are unclear and require further study.
The function of GIT1 in the development of spines and synapses has been relatively well studied, and interactions between GIT1 and specific proteins, particularly cytoskeleton-related proteins, are the foundation of neural plasticity (Zhang et al., 2005, 2013; Segura et al., 2007; Richier et al., 2010; Koles et al., 2012; Rocca et al., 2013; Lim et al., 2014; Smith et al., 2014). Neurite outgrowth plays a pivotal role in neuronal development and regeneration (Beller and Snow, 2014; Stiles et al., 2014; Villarroel-Campos et al., 2014; Baldwin et al., 2015; Dayer et al., 2015; Madl and Heilshorn, 2015). Studies on the association between GIT1 and neurite outgrowth are emerging, but inconclusive. For example, GIT1 knockout leads to reduced neurite length in mice (Menon et al., 2010), while overexpression of full-length GIT1 in vitro does not affect neuritic morphology (Albertinazzi et al., 2003; Za et al., 2006; Totaro et al., 2012).
To better understand the role of GIT1 in neurite outgrowth, we investigated GIT1 expression at different developmental stages in the mouse brain. We examined the relation between GIT1 expression levels and neurite length in primary cultures of mouse hippocampal neurons. In addition, we assessed whether nerve growth factor (NGF), a well-known neurotrophin that promotes neurite outgrowth (O’Keeffe et al., 2008; Yammine et al., 2014; Sarma et al., 2015), could upregulate GIT1 expression and rescue the axonal growth defect caused by GIT1 knockout. Furthermore, using immunoprecipitation (IP) coupled with mass spectrometry, we identified a number of novel binding partners for GIT1. Potential interactions were subsequently confirmed by co-IP and colocalization experiments. Subsequently, in vitro polymerization assay was performed to examine whether GIT1 directly promotes microtubule assembly.
Materials and Methods
Experimental animals
Breeding pairs of GIT1-lacZ+/- mice, C.129S4(B6)-Git1Gt(FHCRC-GT-S10-12C1)Sor/WeisJ were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Neither GIT1 heterozygote (HET) nor GIT1 knockout (KO) animals statistically differed in body weight or organ size from wild type (WT) mice. All experimental procedures were performed in accordance with the Guideline for the Care and Use of Laboratory Animals of the Animal Research Ethics Committee of Renji Hospital, Shanghai Jiao Tong University School of Medicine, China.
RNA extraction and semi-quantitative RT-PCR
Total RNA was extracted from various tissues, including heart, brain, spine, lung, muscle, stomach, intestine, liver, spleen, thymus, kidney, prostate, and testis of adult mice using TRI Reagent (Sigma-Aldrich, St. Louis, MO, USA) and reverse transcribed using the First Strand cDNA Synthesis Kit (Takara, Dalian, China). The Git1 cDNA was amplified with 5′-TGTGGACGAAAGCCTGATC-3′ and 5′-ATCCACCTCATCATACACATCC-3′ primers, Git2 with 5′-CAACAATGGTGCTAACTC-3′ and 5′-CGAACGCTAACATCTGATAC-3′ primers, and Gapdh with 5′-ATTCAACGGCACAGTCAA-3′ and 5′-CTTCTGGGTGGCAGTGAT-3′ primers. PCR was performed using rTaq (Takara), with 25 cycles of amplification with 30 seconds denaturation at 95°C, 30 seconds annealing at 58°C and 60 seconds extension at 72°C.
Cell culture and transfection
Primary hippocampal neuron cultures were prepared as described previously (Beaudoin et al., 2012). Briefly, the hippocampus was dissected out from postnatal day 0 (P0) mouse brain and cut into small pieces, followed by 20 minutes digestion in 0.25% trypsin (Worthington, Lakewood, NJ, USA) at 37°C. After terminating the reaction with 0.1% trypsin inhibitor (Sigma-Aldrich), the digested tissue was triturated and allowed to sit undisturbed for 5 minutes, which allowed non-dissociated tissue to settle at the bottom. The upper fraction containing dissociated cells was removed to another tube, and the non-dissociated tissue was incubated with 0.1% DNase (BD Bioscience, Shanghai, China) for 5 minutes at room temperature and triturated again. The upper fraction was removed again, and this procedure was repeated three times. All the upper fractions containing dissociated cells were combined and centrifuged at 500 × g for 5 minutes at room temperature. The collected cells were resuspended in Neurobasal medium containing B27 supplement (GIBCO, New York, NY, USA), 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were then plated and cultured in a 37°C/5% CO2 incubator. After attaching, cells were transfected using Lipofectamine® 3000 (Life Technologies, Carlsbad, CA, USA) and cultured for an additional 5 days. NGF-β (50 ng/mL; Peprotech, Rocky Hill, NJ, USA) was used as an inducer of neurite growth. To investigate the effect of NGF-β on GIT1 expression, three different concentrations of the neurotrophin were used, including 0, 25 and 50 ng/mL.
Plasmid construction
Full-length human GIT1 cDNA was obtained from the pEGFP-GIT1 plasmid (Addgene, Cambridge, MA, USA). GIT1, cGIT1, CDΔAS/SLD, nGIT1 and GIT1ΔSLD were amplified as described previously (Zhang et al., 2003), and were inserted into modified pIRESneo vectors (Takara-Clontech, Mountain View, CA, USA) with a Flag/HA tag added at the N terminal using Infusion-HD (Takara).
Immunocytochemistry
Neurons were fixed in 4% paraformaldehyde in PBS and permeabilized with 0.1% Triton X-100. The fixed cells were blocked with 10% donkey serum, and then incubated in primary antibody against GIT1 (goat, 1:50; Santa Cruz Biotechnology, Santa Cruz, CA, USA), Clasp2 (rabbit, 1:25; Biorbyt, San Francisco, CA, USA), microtubule-associated protein (MAP) 2 (rabbit, 1:300; Cell Signaling Technology, Danvers, MA, USA), Tuj1 (rabbit, 1:300; Sigma-Aldrich) or Flag (mouse, 1:500; Sigma-Aldrich) at 4°C overnight. After three washes in PBS, cells were incubated with the corresponding anti-mouse, anti-rabbit or anti-goat secondary antibody (Alexa Fluor 488, 594, 640; 1:600; Life Technologies) and DAPI (1:2,000; Sigma-Aldrich) away from light at room temperature for 1 hour. Images were acquired on a LSM-710 Zeiss confocal microscope (Carl Zeiss, Oberkochen, Germany). Neurite length was analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA) as described by Schmidt et al. (1997). Briefly, the straight-line distance from the tip of the neurite to the junction between the cell body and neurite base or to the neurite branch point was measured as an individual neurite length. The sum of all individual neurite lengths of a neuron was taken as the total neurite length per neuron, and the process with the longest individual neurite length in each neuron was considered an axon-like process.
Western blot analysis
Tissues or cells were lysed in RIPA Lysis and Extraction Buffer (Thermo, Waltham, MA, USA). The proteins were then separated on a polyacrylamide gel (Bio-Rad, Shanghai, China) and transferred onto nitrocellulose membranes (Bio-Rad). After blocking in milk, the blots were incubated in primary antibody against GIT1 (goat, 1:800; Santa Cruz Biotechnology), Clasp2 (rabbit, 1:600; Biorbyt), CRMP2 (rabbit, 1:1,000; Proteintech, Chicago IL, USA), MAP2 (rabbit, 1:1,000; Cell Signaling Technology), Tuj1 (rabbit, 1:1,000; Sigma-Aldrich), TUBGCP3 (rabbit, 1:1,000; Proteintech), MAP1B (rabbit, 1:1,000; Proteintech), Tau (rabbit, 1:1,000; Sigma-Aldrich) or beta-actin (mouse, 1:2,000; Millipore, Darmstadt, Germany) at 4°C overnight. After washing, the blots were incubated with IRDye-conjugated secondary antibody against mouse, rabbit or goat (1:10,000; Licor, Lincoln, NE, USA) away from light at room temperature for 1 hour, and visualized with an Odyssey imaging system (Licor).
Immunoprecipitation assays and mass spectrometry
Whole brains were excised from 8-week-old mice, homogenized in IP buffer (250 mM hydroxyethyl piperazine ethanesulfonic acid (pH 7.4), 100 mM NaCl, 10 mM KCl, 10 mM NaF, 1 mM Na3VO4, 10 mM β-glycerophosphate, 10% glycerine, 0.5% CHAPS and 0.5% Triton X-100) supplemented with protease inhibitor cocktail tablets (Roche, Shanghai, China), and centrifuged for 20 minutes at 12,000 × g at 4°C The supernatant, containing 1 mg total protein, was incubated with 10 μg anti-GIT antibody (Santa Cruz Biotechnology). The target antibody complex was immunoprecipitated with Dynabeads® Protein G (Life Technologies) and eluted with elution buffer in the immunoprecipitation kit (Life Technologies). The eluted supernatant was subjected to polyacrylamide gel electrophoresis (PAGE) for mass spectrometric detection or protein immunoblotting. For mass spectrometry, the proteins were excised from the stacking gel and digested with trypsin as previously described (Koles et al., 2012). The peptides were separated using a high performance liquid chromatography system (Agilent, Palo Alto, CA, USA) and detected with a ThermoElectron Finnigan LCQ DECA ion trap mass spectrometer (Thermo).
Tubulin polymerization assay
Purified tubulins (Cytoskeleton, Denver, CO, USA) were reconstituted in tubulin buffer (80 mM piperazine-1,4-bisethanesulfonic acid (pH 6.9), 2 mM MgCl2, 0.5 mM ethylene glycol tetraacetic acid, 1 mM guanosine triphosphate and 5% glycerol), placed in a 24-well plate and pre-warmed to 37°C. Immediately, 1 or 5 μM GIT1 protein (Proteintech) diluted in tubulin buffer was added to the wells. As a positive control, 10 μM paclitaxel (Cytoskeleton), a well-known microtubule stabilizer (Yu et al., 2015) was added instead of GIT1. The blank control well contained only tubulin buffer. The tubulin polymerization curve was recorded every minute, with absorbance readings at 340 nm and 37°C. The optical density is proportional to the quantity of microtubules formed. All assays were done in triplicate.
Statistical analysis
Data are expressed as the mean ± SEM. Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA). Inter-group comparisons were analyzed by one-way analysis of variance followed by Tukey's test. P < 0.05 was considered statistically significant.
Results
GIT1 was more abundantly expressed in the developing brain than other tissues
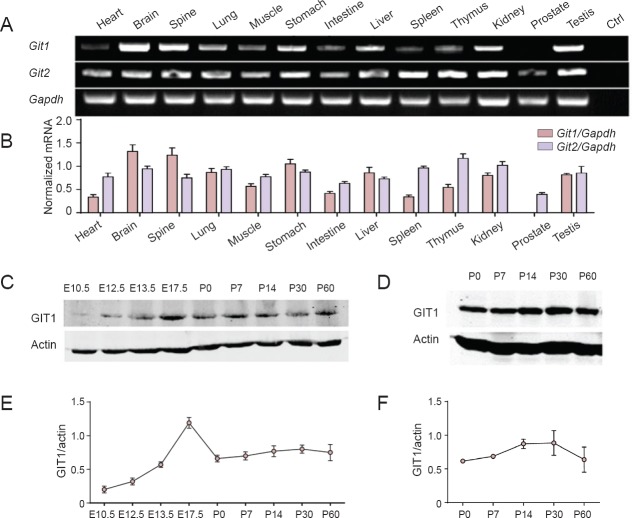
To investigate the relative GIT1 abundance in various tissues, we first performed semi-quantitative RT-PCR to obtain Git1 and Git2 mRNA expression profiles. We found that Git1 was expressed most abundantly in the brain and spine, weakly in the heart and spleen, and was almost undetectable in the prostate; however, Git2 was expressed ubiquitously in all 13 examined tissues (Figure 1A, B). This suggests that GIT1 likely has very important functions in the nervous system. Therefore, we subsequently examined GIT1 protein expression levels at different stages of brain development by western blot assay (Figure 1C, E). GIT1 could be detected at embryonic day 10.5 (E10.5), and thereafter, expression levels rapidly increased, reaching a maximum at E17.5. At P0, the expression level decreased to roughly half of the peak amount, and was maintained until P60 with little fluctuation. In the hippocampus, GIT1 expression levels increased starting at P7, peaking at P30, and then tended to decrease by P60, consistent with the time window of hippocampal development (Gu et al., 2012) (Figure 1D, F).
Figure 1.
GIT1 was highly expressed in the nervous system.
(A) Expression profiles of Git1 and Git2 in 2-month-old mice, as assessed by RT-PCR. Ctrl: Negative control. (B) The relative mRNA expression levels of Git1 and Git2 in 2-month-old mice, normalized to Gapdh. The data are expressed as optical density ratio of Git/Gapdh (n = 3; data are given as mean ± SEM). (C) GIT1 protein expression in the whole brain from embryonic day (E) 10.5 to postnatal day (P) 60. (D) GIT1 expression in the mouse hippocampus at P0, 7, 14, 30 and 60. Quantification of GIT1 expression in the whole brain (E) and hippocampus (F) were normalized to β-actin. Data represent the mean ± SEM of three independent observations.
GIT1 KO inhibited neurited extension
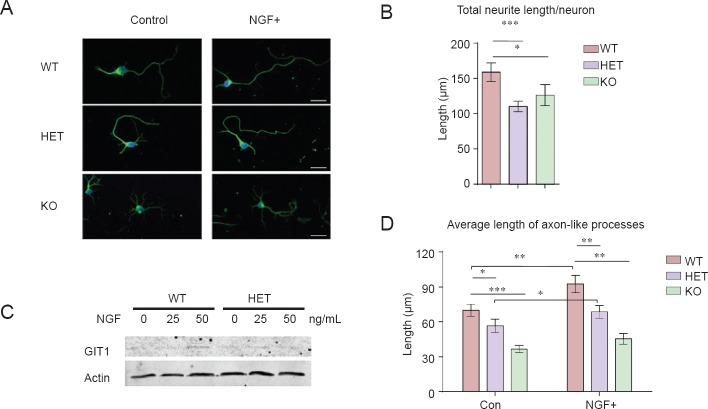
To evaluate GIT1's role in neurite outgrowth, primary hippocampal neurons were isolated and cultured in vitro for 5 days. Neurons were immunolabeled for Tuj1, a neuron-specific marker (Figure 2A). Compared with WT neurons, both GIT1 KO and GIT1 HET neurons showed a significant reduction in total neurite length per neuron (Figure 2B). The neurite outgrowth defect was more obvious when another important morphological characteristic was measured, axonal length (Figure 2D). Because NGF enhances the expression of GIT1 (Figure 2C), we examined whether NGF could rescue the defects in axonal growth in the GIT1 mutant. As shown in Figure 2D, NGF treatment caused a significant increase in the average length of axon-like processes in WT and HET, but not GIT1 KO, neurons. Interestingly, there was a greater increase in WT neurons than in HET neurons, indicating that the increase in the length of axon-like processes induced by NGF treatment is dependent on Git1 gene dosage.
Figure 2.
Neurite growth was impaired in GIT1 knockout neurons.
(A) Hippocampal neurons isolated from wild type (WT), GIT1 heterozygote (HET) and GIT1 knockout (KO) mice at postnatal day (P) 0 were cultured for 5 days in control medium (left) or medium containing 50 ng/mL nerve growth factor (NGF) (right). Neurons were immunostained with anti-Tuj1 (green) antibody and DAPI (blue). Scale bar: 20 μm. Quantification of total neurite length per neuron (B) and average length of axon-like processes (D) of WT, HET and KO hippocampal neurons in vitro. The effects of NGF treatment are shown in Figure D. n = 30–50 cells; data are expressed as the mean ± SEM. The experiments were repeated three times. (C) Western blot of lysates from P0 WT and HET hippocampal neurons cultured for 5 days in the presence of 0, 25 or 50 ng/mL NGF. Anti-GIT1 and anti-actin antibodies were used to detect target protein expression. *P < 0.05, **P < 0.01, ***P < 0.001. One-way analysis of variance followed by Tukey's test.
GIT1 overexpression rescued the axonal growth defects in GIT1 KO neurons, and the N terminal was sufficient for this effect
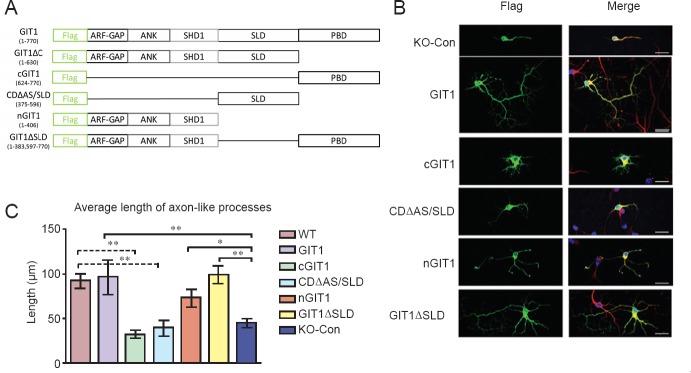
To further examine the neurite outgrowth-promoting effect of GIT1, we performed GIT1 overexpression experiments in the hippocampal neuronal cultures. As shown in Figure 3B and C, overexpression of GIT1 in primary hippocampal neurons isolated from GIT1 KO mice resulted in a complete rescue of the axon growth defect. The length of axon-like processes in these cells was comparable to that in WT neurons after 5 days in culture. To determine which domains of GIT1 mediate this effect, we made various constructs of the different functional domains. These GIT1 constructs were designated cGIT1, CDΔAS/SLD, nGIT1 and GIT1ΔSLD, each with a Flag tag at the N terminal (Figure 3A). Each construct was transfected into GIT1 KO neurons, followed by immunostaining for Tuj1 (red) and Flag (green) (Figure 3B). Similar to GIT1 overexpression, transfection of nGIT1 or GIT1Δ SLD (GIT1 with the SLD domain deleted) effectively rescued the axon outgrowth defect in GIT1 KO neurons. In sharp contrast, all other GIT1 fragments failed to do so (Figure 3C). These results demonstrate that nGIT1 is sufficient for the axon growth promoting effect of GIT1.
Figure 3.
Axonal growth in GIT1 knockout (KO) hippocampal neurons and the effects of overexpression of GIT1 or GIT1 deletion constructs.
(A) Schematic diagram of the full-length GIT1 and different GIT1 deletion constructs: cGIT1, CDΔAS/SLD, nGIT1 and GIT1ΔSLD. The Flag-tag was attached to the N terminal of the constructs. (B) KO hippocampal neurons were cultured for 5 days after transfection with the constructs, and then stained with anti-Flag (green) and anti-Tuj1 (red) antibodies and DAPI (blue). Scale bars: 20 μm. (C) Average length of axon-like processes in KO hippocampal neurons cultured for 5 days after transfection with the GIT1 deletion constructs. Cells were analyzed using ImageJ software. n = 3 cells; data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. One-way analysis of variance followed by Tukey's test. The experiments were repeated three times. Con: Control.
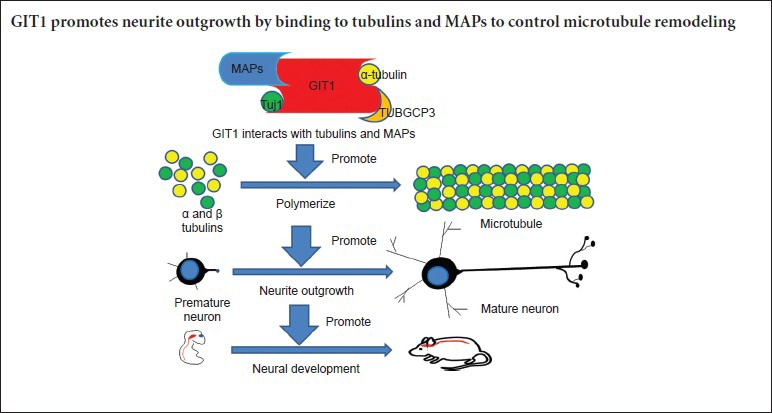
GIT1 binded to tubulins and MAPs and promoted microtubule assembly
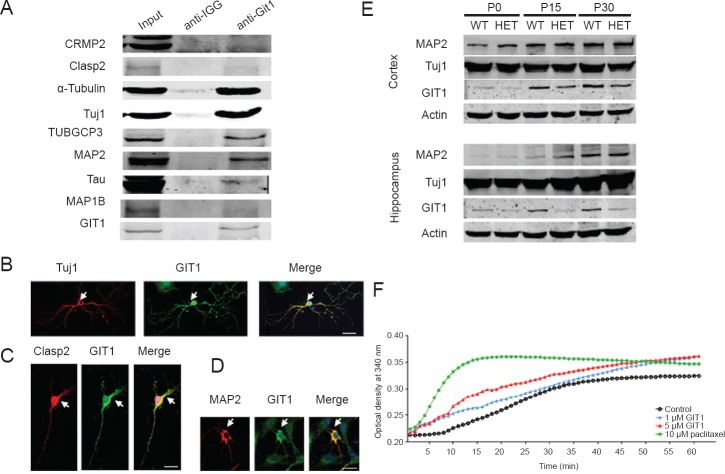
To screen for proteins that interact with GIT1, GIT1 was immunoprecipitated from brain extracts and mass spectrometry was performed. Many tubulins and MAPs were identified (Supplementary Table 1 (24.3KB, pdf) online). We verified these results by co-IP experiments. As shown in Figure 4A, tubulins, including α-tubulin, Tuj1 and TUBGCP3 (a member of the γ-tubulin complex), and MAPs (Clasp2, Tau, MAP1B and MAP2) were co-precipitated by the GIT1 antibody. Some of the interactions were further confirmed by immunofluorescence colocalization experiments. In hippocampal neurons, GIT1 was clearly colocalized with Tuj1 (Figure 4B), Clasp2 (Figure 4C) and MAP2 (Figure 4D). Furthermore, western blot assay was performed to examine the expression of Tuj1 and MAP2 in the cortex and hippocampus in both HET and WT mice at P0, P15 and P30 (Figure 4E). Notably, lower GIT1 protein levels were detected in the cortex and hippocampus of HET mice compared with WT animals at the different developmental time points. In contrast, no substantial difference was seen in expression levels of Tuj1 or MAP2 between WT and HET mice, indicating that GIT1 does not affect levels of these proteins. These results suggest that GIT1 may affect neurite outgrowth by impacting tubulin polymerization. We, therefore, evaluated the capacity of purified tubulin to form microtubules in the presence of GIT1 in vitro (Figure 4F). When incubated with 1 μM GIT1, tubulin started to polymerize by as early as 2 minutes and displayed rapid polymerization during the first 15 minutes. In comparison, in the control, the start of tubulin polymerization was delayed to ~6 minutes. In the presence of 5 μM GIT1, tubulin polymerization was increased significantly. As expected, the positive control paclitaxel dramatically accelerated tubulin polymerization. These data indicate that GIT1 promotes microtubule assembly in vitro in a concentration-dependent manner.
Figure 4.
GIT1 interacted with tubulins and microtubule-associated proteins, and stimulated microtubule assembly.
(A) Brain extract from 8-week-old mice was immunoprecipitated with the GIT1 antibody, and western blots were probed by antibodies against CRMP2, Clasp2, α-tubulin, Tuj1, TUBGCP3, MAP2, Tau, MAP1B and GIT1. The images show the colocalization of Tuj1 (red) and GIT1 (green) (B), Clasp2 (red) and GIT1 (green) (C), and MAP2 (red) and GIT1 (green) (D) in primary cultured hippocampal neurons on postnatal day (P) 0. Scale bars: 20 μm. (E) Expression levels of Tuj1, MAP2 and GIT1 in the cortex and hippocampus of wild-type (WT) and GIT1 heterozygote (HET) mice on postnatal day (P) 0, P15 and P30. (F) GIT1 activates tubulin polymerization in vitro. Result represents means of three independent observations. min: Minutes.
List of tubulins and microtubule associated protein MAPs co-immunoprecipitated with GIT1, as detected by mass spectrometry
Discussion
Although GIT1 has been reported to stimulate neurite outgrowth, the observations have been inconsistent (Za et al., 2006; Totaro et al., 2012). Our present findings strongly suggest that GIT1 promotes neurite outgrowth, especially of axon-like processes.
We found relatively high expression levels of GIT1 in the nervous system compared with other tissues, which correlates well with neuritogenesis in the brain. Although GIT1 is detected in most tissues at various levels, its expression is predominantly in the brain, indicating that GIT1 has important functions in this organ. This hypothesis is further supported by the fact that GIT1 expression dramatically increases from E12.5 and remains at a high level throughout embryogenesis, during which neuronal migration and axon and dendrite growth and sprouting occur. Hence, GIT1 is strongly correlated with neuritogenesis. Furthermore, GIT1 is highly expressed in the hippocampus, which peaks at around P30, consistent with a critical role in synaptogenesis. These findings provide additional support for an important role of GIT1 in spine and synapse development. Compared with GIT1, GIT2, a homolog, has a quite different expression pattern. GIT2 is ubiquitously expressed in all tissues, without any specificity. GIT2 expression in the hippocampus is consistently low and is not correlated with synapse formation (data not shown). Therefore, it is unlikely that GIT2 expression and synaptic plasticity are closely related.
Another major finding of the present study is that both GIT1 KO and GIT1 overexpression significantly affect neurite length. Our results show that GIT1 is required for axonal extension and that NGF enhances axonal growth in neurons from WT and HET, but not KO, mice. Furthermore, we found that the expression of GIT1 can be stimulated by NGF, a well-known neurite outgrowth promoter. This suggests that the neurite outgrowth stimulating effect of NGF may be mediated by GIT1, although the underlying mechanisms are unknown.
Albertinazzi et al. (2003) found that transfection of full-length p95-APP1 (GIT1) did not have an obvious effect on neurite outgrowth, although some deletion constructs did. p95-APP1 with an SHD domain deletion (p95-ΔSHD) and p95-APP1 with a C-terminal truncation after the SHD domain (P95-C) tended to stimulate neurite extension. However, an N-terminal construct truncated after the SHD domain (p95-N) and a C-terminal construct truncated before the SHD domain (p95-C2) completely blocked neurite outgrowth. In the current study, we investigated the function of various GIT1 deletion constructs on neurite outgrowth. We found that the SLD domain (CDΔAS/SLD) and the PBD domain (cGIT1) alone had no effect on neurite extension. In contrast, nGIT1 (corresponding to p95-N) and GIT1-ΔSLD (GIT1 with an SLD domain deletion) significantly stimulated axon-like neurite extension. Therefore, we conclude that the GIT1 N-terminal is sufficient to stimulate axonal extension, and that the effect can be enhanced by the PBD domain and weakened by the SLD domain.
The molecular mechanisms by which GIT1 stimulates neurite outgrowth remain unclear. Albertinazzi et al. (2003) proposed that activation of the GIT1-Pix-Rac1B-PAK signaling pathway triggers growth cone actin remodeling to drive neurite extension. Our current results suggest the presence of a novel mechanism, in which GIT1 promotes neurite extension by inducing microtubule remodeling. By mass spectrometry, we found that many tubulins and MAPs were co-immunoprecipitated by the GIT1 antibody, and these interactions were confirmed by co-IP and colocalization experiments. The tubulin polymerization assay revealed that GIT1 facilitates tubulin polymerization in a concentration-dependent manner, without directly affecting expression levels of Tuj1 or MAP2. Taken together, our findings support the concept that GIT1 affects neurite growth by modulating both actin and microtubule dynamics. Our working model is that GIT1 binds to tubulins and MAPs to control microtubule remodeling, thereby promoting neurite outgrowth.
Acknowledgments
The authors thank Dr. Guang-lei Zhuang from Renji-Med X Stem Cell Research Center, Ren Ji Hospital, China for proofreading.
Footnotes
Funding: This study was supported by the grants to HLS from the National Natural Science Foundation of China (81371507), Medicine and Engineering Cross-talking Funds of Shanghai Jiao Tong University (YG2013MS40), Science and Technology Projects of Shanghai Jiao Tong University Medical School (13XJ10016) and the National Basic Research Program of China (973 Program; 2013CB945600); and by the grants to WQG from the Chinese Ministry of Science and Technology (2012CB966800 and 2013CB945600), the National Natural Science Foundation of China (81130038 and 81372189), the Science and Technology Commission of Shanghai Municipality (Pujiang Program), the Shanghai Health Bureau Key Disciplines and Specialties Foundation, the Shanghai Education Committee Key Discipline and Specialties Foundation (J50208) and KC Wong Foundation.
Conflicts of interest: None declared.
Supplementary data: Supplementary information is available at http://www.nrronline.org.
Plagiarism check: This paper was screened twice using Cross-Check to verify originality before publication.
Peer review: This paper was double-blinded and stringently reviewed by international expert reviewers.
Copyedited by Yokoyama S, Zimmer G, Li CH, Wang L, Song LP, Zhao M
References
- Albertinazzi C, Za L, Paris S, de Curtis I. ADP-ribosylation factor 6 and a functional PIX/p95-APP1 complex are required for Rac1B-mediated neurite outgrowth. Mol Biol Cell. 2003;14:1295–1307. doi: 10.1091/mbc.E02-07-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin KT, Carbajal KS, Segal BM, Giger RJ. Neuroinflammation triggered by beta-glucan/dectin-1 signaling enables CNS axon regeneration. Proc Natl Acad Sci U S A. 2015;112:2581–2586. doi: 10.1073/pnas.1423221112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin GM, 3rd, Lee SH, Singh D, Yuan Y, Ng YG, Reichardt LF, Arikkath J. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat Protoc. 2012;7:1741–1754. doi: 10.1038/nprot.2012.099. [DOI] [PubMed] [Google Scholar]
- Beller JA, Snow DM. Proteoglycans: road signs for neurite outgrowth. Neural Regen Res. 2014;9:343–355. doi: 10.4103/1673-5374.128235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanot H, Young AM, Overmeyer JH, Maltese WA. Induction of nonapoptotic cell death by activated Ras requires inverse regulation of Rac1 and Arf6. Mol Cancer Res. 2010;8:1358–1374. doi: 10.1158/1541-7786.MCR-10-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayer AG, Jacobshagen M, Chaumont-Dubel S, Marin P. 5-HT6 receptor: A new player controlling the development of neural circuits. ACS Chem Neurosci. 2015;6:951–960. doi: 10.1021/cn500326z. [DOI] [PubMed] [Google Scholar]
- Gu Y, Arruda-Carvalho M, Wang J, Janoschka SR, Josselyn SA, Frankland PW, Ge S. Optical controlling reveals time-dependent roles for adult-born dentate granule cells. Nat Neurosci. 2012;15:1700–1706. doi: 10.1038/nn.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer A, Oladimeji P, De Las Casas LE, Diakonova M. Phosphorylation of tyrosine 285 of PAK1 facilitates betaPIX/GIT1 binding and adhesion turnover. FASEB J. 2015;29:943–959. doi: 10.1096/fj.14-259366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefen RJ, Berk BC. The multifunctional GIT family of proteins. J Cell Sci. 2006;119:1469–1475. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- Hong ST, Mah W. A critical role of GIT1 in vertebrate and invertebrate brain development. Exp Neurobiol. 2015;24:8–16. doi: 10.5607/en.2015.24.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu RM, Hsieh YJ, Yang TH, Chiang YC, Kan CY, Lin YT, Chen JT, Yu JS. Binding of the extreme carboxyl-terminus of PAK-interacting exchange factor beta (betaPIX) to myosin 18A (MYO18A) is required for epithelial cell migration. Biochim Biophys Acta. 2014;1843:2513–2527. doi: 10.1016/j.bbamcr.2014.06.023. [DOI] [PubMed] [Google Scholar]
- Kahn RA, Bruford E, Inoue H, Logsdon JM, Jr, Nie Z, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, Cassel D. Consensus nomenclature for the human ArfGAP domain-containing proteins. J Cell Biol. 2008;182:1039–1044. doi: 10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koles K, Nunnari J, Korkut C, Barria R, Brewer C, Li Y, Leszyk J, Zhang B, Budnik V. Mechanism of evenness interrupted (Evi)-exosome release at synaptic boutons. J Biol Chem. 2012;287:16820–16834. doi: 10.1074/jbc.M112.342667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Ritt DA, Zhou M, Morrison DK. The CNK2 scaffold interacts with vilse and modulates Rac cycling during spine morphogenesis in hippocampal neurons. Curr Biol. 2014;24:786–792. doi: 10.1016/j.cub.2014.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl CM, Heilshorn SC. Matrix interactions modulate neurotrophin-mediated neurite outgrowth and pathfinding. Neural Regen Res. 2015;10:514–517. doi: 10.4103/1673-5374.155426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon P, Deane R, Sagare A, Lane SM, Zarcone TJ, O’Dell MR, Yan C, Zlokovic BV, Berk BC. Impaired spine formation and learning in GPCR kinase 2 interacting protein-1 (GIT1) knockout mice. Brain Res. 2010;1317:218–226. doi: 10.1016/j.brainres.2009.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008;18:184–192. doi: 10.1016/j.tcb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M, Tajima M, Kosako H, Nakaya T, Hashimoto A, Watari K, Nishihara H, Ohba M, Komiya S, Tani N, Nishida M, Taniguchi H, Sato Y, Matsumoto M, Tsuda M, Kuroda M, Inoue K, Kurose H. GRK6 deficiency in mice causes autoimmune disease due to impaired apoptotic cell clearance. Nat Commun. 2013;4:1532. doi: 10.1038/ncomms2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keeffe GW, Gutierrez H, Pandolfi PP, Riccardi C, Davies AM. NGF-promoted axon growth and target innervation requires GITRL-GITR signaling. Nat Neurosci. 2008;11:135–142. doi: 10.1038/nn2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penela P, Nogués L, Mayor F., Jr Role of G protein-coupled receptor kinases in cell migration. Curr Opin Cell Biol. 2014;27:10–17. doi: 10.1016/j.ceb.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Podufall J, Tian R, Knoche E, Puchkov D, Walter AM, Rosa S, Quentin C, Vukoja A, Jung N, Lampe A, Wichmann C, Bohme M, Depner H, Zhang YQ, Schmoranzer J, Sigrist SJ, Haucke V. A presynaptic role for the cytomatrix protein GIT in synaptic vesicle recycling. Cell Rep. 2014;7:1417–1425. doi: 10.1016/j.celrep.2014.04.051. [DOI] [PubMed] [Google Scholar]
- Richier L, Williton K, Clattenburg L, Colwill K, O’Brien M, Tsang C, Kolar A, Zinck N, Metalnikov P, Trimble WS, Krueger SR, Pawson T, Fawcett JP. NOS1AP associates with Scribble and regulates dendritic spine development. J Neurosci. 2010;30:4796–4805. doi: 10.1523/JNEUROSCI.3726-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca DL, Amici M, Antoniou A, Blanco Suarez E, Halemani N, Murk K, McGarvey J, Jaafari N, Mellor JR, Collingridge GL, Hanley JG. The small GTPase Arf1 modulates Arp2/3-mediated actin polymerization via PICK1 to regulate synaptic plasticity. Neuron. 2013;79:293–307. doi: 10.1016/j.neuron.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma T, Koutsouris A, Yu JZ, Krbanjevic A, Hope TJ, Rasenick MM. Activation of microtubule dynamics increases neuronal growth via the nerve growth factor (NGF) - and Galphas-mediated signaling pathways. J Biol Chem. 2015;290:10045–10056. doi: 10.1074/jbc.M114.630632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalzigaug R, Phee H, Davidson CE, Weiss A, Premont RT. Differential expression of the ARF GAP genes GIT1 and GIT2 in mouse tissues. J Histochem Cytochem. 2007;55:1039–1048. doi: 10.1369/jhc.7A7207.2007. [DOI] [PubMed] [Google Scholar]
- Schmalzigaug R, Rodriguiz RM, Bonner PE, Davidson CE, Wetsel WC, Premont RT. Impaired fear response in mice lacking GIT1. Neurosci Lett. 2009;458:79–83. doi: 10.1016/j.neulet.2009.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CE, Shastri VR, Vacanti JP, Langer R. Stimulation of neurite outgrowth using an electrically conducting polymer. Proc Natl Acad Sci U S A. 1997;94:6. doi: 10.1073/pnas.94.17.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura I, Essmann CL, Weinges S, Acker-Palmer A. Grb4 and GIT1 transduce ephrinB reverse signals modulating spine morphogenesis and synapse formation. Nat Neurosci. 2007;10:301–310. doi: 10.1038/nn1858. [DOI] [PubMed] [Google Scholar]
- Smith KR, Davenport EC, Wei J, Li X, Pathania M, Vaccaro V, Yan Z, Kittler JT. GIT1 and betaPIX are essential for GABA(A receptor synaptic stability and inhibitory neurotransmission. Cell Rep. 2014;9:298–310. doi: 10.1016/j.celrep.2014.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles TL, Kapiloff MS, Goldberg JL. The role of soluble adenylyl cyclase in neurite outgrowth. Biochim Biophys Acta. 2014;1842:2561–2568. doi: 10.1016/j.bbadis.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A, Tavano S, Filosa G, Gartner A, Pennucci R, Santambrogio P, Bachi A, Dotti CG, de Curtis I. Biochemical and functional characterisation of alphaPIX, a specific regulator of axonal and dendritic branching in hippocampal neurons. Biol Cell. 2012;104:533–552. doi: 10.1111/boc.201100060. [DOI] [PubMed] [Google Scholar]
- Turner CE, West KA, Brown MC. Paxillin-ARF GAP signaling and the cytoskeleton. Curr Opin Cell Biol. 2001;13:593–599. doi: 10.1016/s0955-0674(00)00256-8. [DOI] [PubMed] [Google Scholar]
- Villarroel-Campos D, Gastaldi L, Conde C, Caceres A, Gonzalez-Billault C. Rab-mediated trafficking role in neurite formation. J Neurochem. 2014;129:240–248. doi: 10.1111/jnc.12676. [DOI] [PubMed] [Google Scholar]
- Wilson E, Leszczynska K, Poulter NS, Edelmann F, Salisbury VA, Noy PJ, Bacon A, Rappoport JZ, Heath JK, Bicknell R, Heath VL. RhoJ interacts with the GIT-PIX complex and regulates focal adhesion disassembly. J Cell Sci. 2014;127:3039–3051. doi: 10.1242/jcs.140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Mah W, Kim E, Kim JW, Hahm EK, Kim MH, Cho S, Kim J, Jang H, Cho SC, Kim BN, Shin MS, Seo J, Jeong J, Choi SY, Kim D, Kang C. GIT1 is associated with ADHD in humans and ADHD-like behaviors in mice. Nat Med. 2011;17:566–572. doi: 10.1038/nm.2330. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang Y, Yin Q, Xia H, Wang J. Platelet-derived growth factor promotes osteoblast proliferation by activating G-protein-coupled receptor kinase interactor-1. Mol Med Rep. 2014;10:1349–1354. doi: 10.3892/mmr.2014.2374. [DOI] [PubMed] [Google Scholar]
- Xiao J, Chen X, Xu L, Zhang Y, Yin Q, Wang F. PDGF regulates chondrocyte proliferation through activation of the GIT1- and PLCgamma1-mediated ERK1/2 signaling pathway. Mol Med Rep. 2014;10:2409–2414. doi: 10.3892/mmr.2014.2506. [DOI] [PubMed] [Google Scholar]
- Yammine M, Saade M, Chauvet S, Nguyen C. Spatial gene's (Tbata) implication in neurite outgrowth and dendrite patterning in hippocampal neurons. Mol Cell Neurosci. 2014;59:1–9. doi: 10.1016/j.mcn.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Yu Y, Gaillard S, Phillip JM, Huang TC, Pinto SM, Tessarollo NG, Zhang Z, Pandey A, Wirtz D, Ayhan A, Davidson B, Wang TL, Shih IeM. Inhibition of Spleen Tyrosine Kinase Potentiates Paclitaxel-Induced Cytotoxicity in Ovarian Cancer Cells by Stabilizing Microtubules. Cancer Cell. 2015;28:82–96. doi: 10.1016/j.ccell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Za L, Albertinazzi C, Paris S, Gagliani M, Tacchetti C, de Curtis I. betaPIX controls cell motility and neurite extension by regulating the distribution of GIT1. J Cell Sci. 2006;119:2654–2666. doi: 10.1242/jcs.02996. [DOI] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Horwitz AF. Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol. 2003;161:131–142. doi: 10.1083/jcb.200211002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LQ, Zhao GZ, Xu XY, Fang J, Chen JM, Li JW, Gao XJ, Hao LJ, Chen YZ. Integrin-beta1 regulates chondrocyte proliferation and apoptosis through the upregulation of GIT1 expression. Int J Mol Med. 2015;35:1074–1080. doi: 10.3892/ijmm.2015.2114. [DOI] [PubMed] [Google Scholar]
- Zhang S, Hisatsune C, Matsu-Ura T, Mikoshiba K. G-protein-coupled receptor kinase-interacting proteins inhibit apoptosis by inositol 1,4,5-triphosphate receptor-mediated Ca2+ signal regulation. J Biol Chem. 2009;284:29158–29169. doi: 10.1074/jbc.M109.041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhou W, Fan J, Ren Y, Yin G. G-protein-coupled receptor kinase interactor-1 serine 419 accelerates premature synapse formation in cortical neurons by interacting with Ca(2+)/calmodulin-dependent protein kinase IIbeta. Brain Res Bull. 2013;95:70–77. doi: 10.1016/j.brainresbull.2013.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of tubulins and microtubule associated protein MAPs co-immunoprecipitated with GIT1, as detected by mass spectrometry