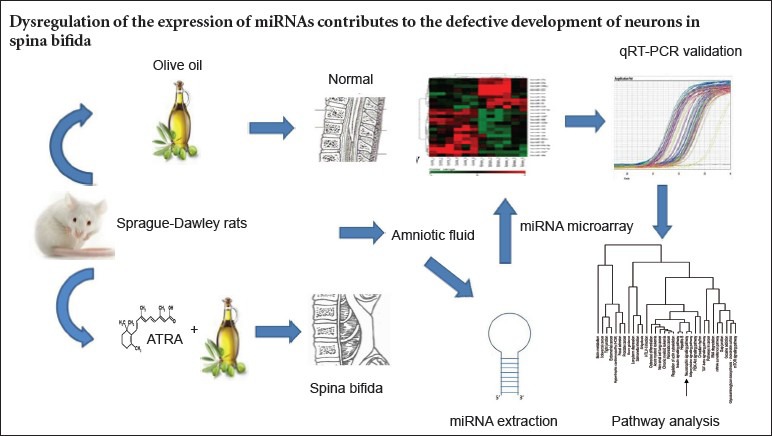
Keywords: nerve regeneration, spina bifida, amniotic fluid, all-trans retinoic acid, microarray, microRNA, reverse transcription-polymerase chain reaction, MAPK, neural regeneration
Abstract
MicroRNAs (miRNAs) are dynamically regulated during neurodevelopment, yet few reports have examined their role in spina bifida. In this study, we used an established fetal rat model of spina bifida induced by intragastrically administering olive oil-containing all-trans retinoic acid to dams on day 10 of pregnancy. Dams that received intragastric administration of all-trans retinoic acid-free olive oil served as controls. The miRNA expression profile in the amniotic fluid of rats at 20 days of pregnancy was analyzed using an miRNA microarray assay. Compared with that in control fetuses, the expression of miRNA-9, miRNA-124a, and miRNA-138 was significantly decreased (> 2-fold), whereas the expression of miRNA-134 was significantly increased (> 4-fold) in the amniotic fluid of rats with fetuses modeling spina bifida. These results were validated using real-time quantitative reverse-transcription polymerase chain reaction. Hierarchical clustering analysis of the microarray data showed that these differentially expressed miRNAs could distinguish fetuses modeling spina bifida from control fetuses. Our bioinformatics analysis suggested that these differentially expressed miRNAs were associated with many cytological pathways, including a nervous system development signaling pathway. These findings indicate that further studies are warranted examining the role of miRNAs through their regulation of a variety of cell functional pathways in the pathogenesis of spina bifida. Such studies may provide novel targets for the early diagnosis and treatment of spina bifida.
Introduction
Neural tube defects (NTDs) are a group of severe congenital malformations of the central nervous system resulting from failure of the neural tube to close during neurulation between 21 and 28 days post conception (Botto et al., 1999). NTDs are a significant cause of fetal mortality and morbidity globally, with an approximate incidence of 0.5–2 per 1,000 births (Greene et al., 2009). In the rural areas of the northern provinces of China, the prevalence of NTDs at birth is approximately 6 per 1,000 births (Moore et al., 1997). NTDs are multi-factorial in origin, arising from a combination of genetic and environmental factors (Frey and Hauser, 2003; Padmanabhan, 2006). One of the most common NTDs is spina bifida, a malformation of the spinal column characterized by herniation or exposure of the spinal cord through incompletely closed vertebrae. Even after fetal surgery to repair the defect, patients show neurological deficits of varying degrees, such as sensory and motor weakness in the legs as well as bowel and bladder incontinence, and they often require long-term care and assistance (Date et al., 1993). Despite years of epidemiologic and experimental studies, the mechanisms responsible for spina bifida remain largely unknown.
Endogenous, small non-coding RNAs approximately 22 nucleotides in length are called microRNAs (miRNAs). They constitute a class of negative regulators of gene expression, either inhibiting translation or inducing degradation of their target messenger RNA (mRNA), resulting in complete or incomplete binding to the 3′ untranslated region of their target mRNAs. This class of regulators reportedly plays an important role in a vast range of biological processes, such as proliferation, differentiation, and apoptosis (Esquela-Kerscher and Slack, 2006; Gu et al., 2009). A number of miRNA profiling studies have shown the expression of miRNAs during neural stem cell differentiation and morphological development of mammalian brain changes (Miska et al., 2004; Croce and Calin, 2005). However, few studies (Wei et al., 2013; Wu et al., 2013) have reported on the role of miRNA in spina bifida, and none have investigated a mechanism for the miRNA involvement. Additionally, these previous studies are limited to individual miRNA and, hence, lack a global level analysis.
The congenital rat spina bifida model induced by administration of all-trans retinoic acid (ATRA), a metabolic product of vitamin A, is well established. In many respects, this model shows remarkable similarities to spina bifida generated by vitamin A deficiency in humans; thus, it is widely used in research of spina bifida. In the present study, we used miRNA microarray and real-time qRT-PCR analyses to characterize differentially expressed miRNAs in amniotic fluid samples obtained from rats with fetuses modeling spina bifida induced by ATRA and from control rats with normal fetuses. We aimed to clarify whether dysregulation of the expression of miRNAs contributes to the defective development of neurons in spina bifida.
Materials and Methods
Establishment of the spina bifida model
Adult Sprague-Dawley rats (10–12 weeks old; weighing 250–300 g) were purchased from Henan Provincial Laboratory Animal Center (license No. SYXK 2010-0001) in China. All animal experiments followed the policies and procedures recommended by the committee of animal research and ethics and approved by the ethics committee at Zhengzhou University, China. Female rats were mated with male rats overnight. The appearance of a vaginal plug the morning after mating was considered embryonic day 0 (E0). Spina bifida was induced with a single intragastric injection of ATRA (40 mg/mL in olive oil; 150 mg/kg body weight; Sigma, St. Louis, MO, USA) on E10 (n = 6 rats). Rats receiving the control treatment were injected with 4 mL/kg body weight of ATRA-free olive oil (Sigma) on the same day (n = 6 rats).
Sample collection
Pregnant rats were killed on E20 using an overdose of 10% chloral hydrate injected into the abdominal cavity, and the fetuses were harvested. After examining the fetuses for deformities under a microscope (Nikon SMZ1500 stereoscopic microscope, Nikon, Japan), amniotic fluid samples from both groups of pregnant rats were immediately centrifuged at 1,200 × g for 10 minutes, aliquoted in 1.5 mL Eppendorf tubes, and stored at −80°C for use in RNA extractions.
RNA isolation
Total amniotic fluid RNA was isolated using a mirVana PARIS miRNA isolation kit (Ambion, Foster City, CA, USA) according to the manufacturer's protocol. All RNAs were stored at −80°C. RNA purity was assessed using 260/280 nm and 260/230 nm absorbance ratios, with a 260/280 nm absorbance ratio > 1.8 and a 260/230 nm absorbance ratio > 1.5 considered high purity. RNA integrity was determined using the 28s:18s RNA ratio obtained from electrophoresis analysis with agarose gels. Total RNA from all samples demonstrated strong 28s and 18s bands (Figure 1), indicating sufficient quality of total RNA for subsequent miRNA microarray analysis.
Figure 1.
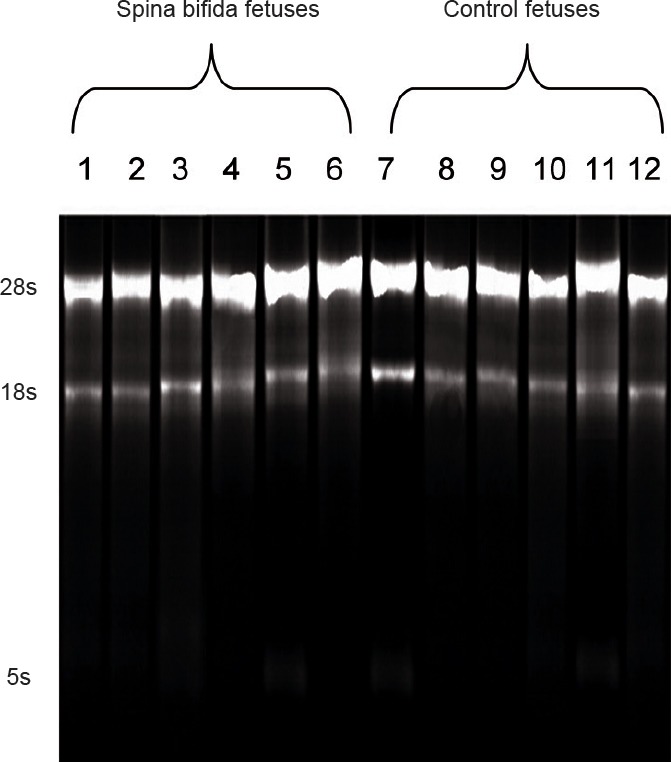
Total RNA electrophoresis.
Lanes 1–6 are results from rats with fetuses modeling spina bifida, and lanes 7–12 are from control rats with normal fetuses.
miRNA microarray
The expression profiles of miRNAs in amniotic fluid samples from six pregnant rats with fetuses modeling spina bifida were compared with those from six rats with control fetuses using TaqMan Low Density Array according to the manufacturer's protocols. Briefly, total RNA (350 ng) was reverse transcribed using a TaqMan MicroRNA RT kit and Multiplex RT rodent primer pool (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The cDNAs were added to Master Mix (Applied Biosystems) and applied to miRNA TaqMan low-density array rodent panels 2.0 (Applied Biosystems) according to the manufacturer's instructions for the simultaneous quantification of 375 miRNAs. The miRNA TaqMan assays were performed using a 7900 HT Fast Real-Time PCR system (Applied Biosystems). Each sample was screened with two arrays. Differentially expressed miRNA profiles were analyzed with DataAssist software v1.0 (Applied Biosystems). A value for P < 0.05 was considered statistically significant.
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
We used qRT-PCR to validate the miRNAs differentially expressed in the two groups. Briefly, 10 ng of total RNA was reverse transcribed to cDNA using microRNA specific primers and a TaqMan Reverse Transcription Kit (Applied Biosystems). Diluted cDNA was subjected to qRT-PCR using the TaqMan MicroRNA Assay and TaqMan Universal PCR Master Mix (ABI, Life Technologies, Foster City, CA, USA) with a 7500 Real-Time PCR system (Applied Biosystems). Relative quantification was performed using the ΔΔCt method (Yuan et al., 2006), and the data were normalized to U6 and RNU48 (Applied Biosystems), as endogenous controls. The PCR was performed in triplicate for each sample for both control and each miRNA simultaneously. Relative miRNA expression levels were quantified using the 2−ΔΔCt method.
Identification of miRNA related pathways
The DIANA miRPath web-based computational tool (http://diana.cslab.ece.ntua.gr/) was used to identify potential Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to the miRNAs differentially expressed in amniotic fluid samples from control rats and those with fetuses modeling spina bifida. A pathway assigned a value of P < 0.05 was considered significantly enriched.
Western blot analysis
The protein samples were extracted by using a ReadyPrep protein extraction kit (Bio-Rad, Hercules, CA, USA). The total protein sample was diluted using 5× sample buffer at a ratio of 4:1. The analytical sample was denatured at 100°C for 5 minutes and then underwent sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis. The proteins in the gel were subsequently transferred to polyvinylidene fluoride membranes, which were incubated in 5% fat-free milk at room temperature to block nonspecific proteins. The membranes were then incubated in mouse anti-rat MAPK monoclonal antibody (Cell Signaling Technology, Danvers, MA, USA) diluted to a ratio of 1:200 at 4°C overnight. After being washed with Tris-buffered saline containing Tween 20 (TBST: 50 mM Tris base, 0.9% NaCl, 0.05% Tween 20, pH 7.5), the membranes were incubated in fluorescein isothiocyanate-labeled goat anti-mouse IgG (for MAPK; ABI, Life Technologies) diluted to a ratio of 1:8,000 at 37°C for 1 hour. After being washed with TBST, the membranes were processed using enhanced chemiluminescence and exposed to X-ray film for 5 minutes. Beta-actin (mouse anti-rat monoclonal antibody, dilution 1:200, Cell Signaling Technology, Boston, MA, USA) was used as an internal reference. The expression levels of the proteins were compared with those of the β-actin control, based on the relative optical density of the bands. Band density was quantified using Quantity One v4.62 software (Bio-Rad, Hercules, CA, USA).
Statistical analysis
Data are expressed as the mean ± SD. Intergroup comparison was performed by a two-sided Student's t-test using SPSS v17.0 (SPSS Inc., Chicago, IL, USA), and a P value < 0.05 was considered statistically significant.
Results
miRNA expression altered in samples derived from rat fetuses modeling spina bifida
Hierarchical clustering analysis of the microarray data based on fold change and probability values revealed that there were 11 significantly altered miRNAs (Table 1). Only miRNAs showing expression level changes greater than 2-fold (log2 fold change) were further investigated. Among them, three significantly downregulated miRNAs (miRNA-9, miRNA-124a, and miRNA-138) that changed more than 2-fold (P < 0.05) and one significantly upregulated miRNA (miRNA-134) that changed more than 4-fold in expression (P < 0.05) were detected in the amniotic fluid of rats with fetuses modeling spina bifida as compared with control rats (Figure 2).
Table 1.
miRNA differentially expressed in the amniotic fluid of rats with fetuses modeling spina bifida and control rats with normal fetuses
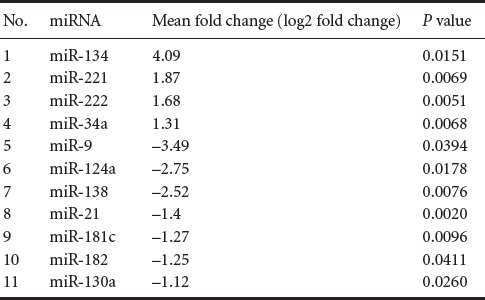
Figure 2.
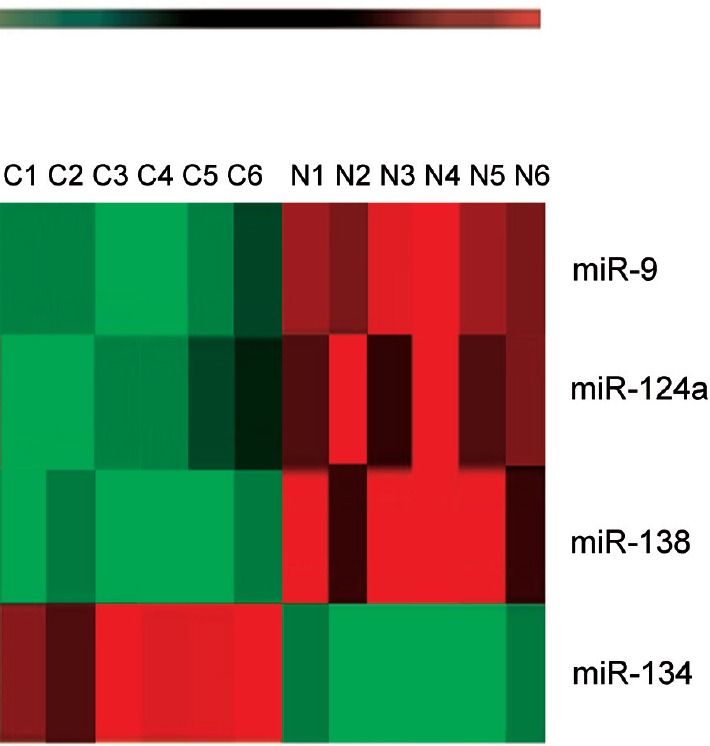
Hierarchical clustering of miRNAs differentially expressed in the amniotic fluid of rats with fetuses modeling spina bifida and control rats with normal fetuses based on results of a microarray analysis.
Each row represents an miRNA, and each column represents a sample. The miRNA clustering tree is shown on the left, and the sample clustering tree appears across the top. The color scale shown at the top illustrates the relative expression level of an miRNA, in which red represents a high expression level and green represents a low expression level. C1–C6 identify rats with fetuses modeling spina bifida and N1–N6 identify control rats with normal fetuses.
Real-time qRT-PCR validation
Retinoic acid induces spina bifida manifesta 20 days post conception. To validate the results from the microarray platform, we assessed the expression of four differentially expressed miRNAs using qRT-PCR. As shown in Figure 3, there were significant differences in the expression of these miRNAs. In particular, the levels of expression for miRNA-9, miRNA-124a, and miRNA-138 were significantly downregulated, whereas expression of miRNA-134 was significantly upregulated in the amniotic fluid of rats with fetuses modeling spina bifida compared with those in rats with normal fetuses. These data were consistent with the microarray findings.
Figure 3.
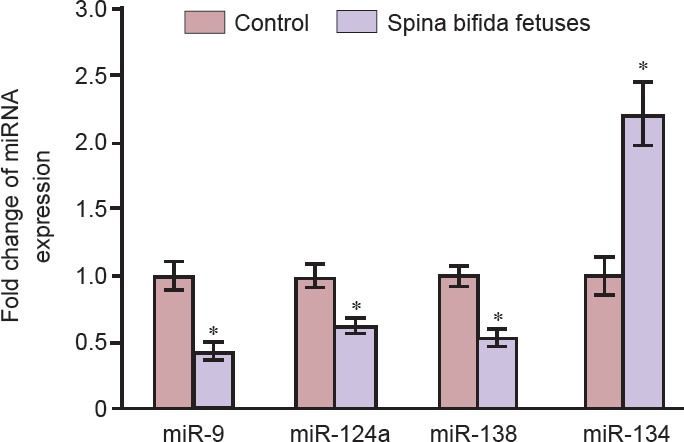
Expression levels of miRNA-9, miRNA-124a, miRNA-138, and miRNA-134 in the amniotic fluid of rats with fetuses modeling spina bifida and control rats with normal fetuses as determined using real-time quantitative reverse transcription-polymerase chain reaction.
Relative miRNA expression is shown as the mean ± SD (n = 6 rats per group). *P < 0.05, vs. control fetuses (two-sided Student's t-test).
Pathway enrichment analysis
Single miRNA can bind to many target genes and function as multiple pathway regulators. Using the web-based pathway analysis system DIANA miRPath, we interpreted the biological and signaling pathways of all altered miRNA. The KEGG results showed these miRNA were involved in multiple significant pathways (Figure 4). The neurotrophin signaling pathway was of special interest for spina bifida.
Figure 4.
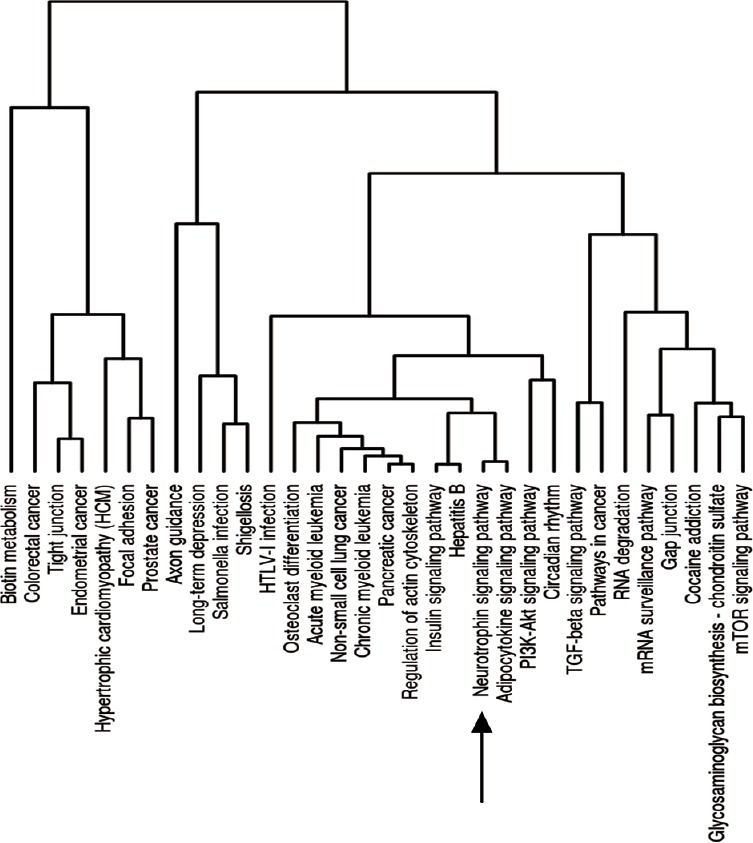
Significantly enriched signaling pathways associated with differentially expressed miRNAs as determined using KEGG pathway cluster analysis.
The neurotrophin signaling pathway is indicated with an arrow.
Association of miRNA-124a and MAPK expression and protein levels
Based on the DIANA miRPath analysis results, miRNA-124a had a relationship with the neurotrophin signaling pathway, which involves the MAPK gene (Longo and Massa, 2013). We thus explored the role of miRNA-124a in the regulation of MAPK gene expression. The downregulation of miRNA-124a paralleled the reduction in MAPK expression in the amniotic fluid of rats with fetuses modeling spina bifida compared with controls (Figure 5).
Figure 5.
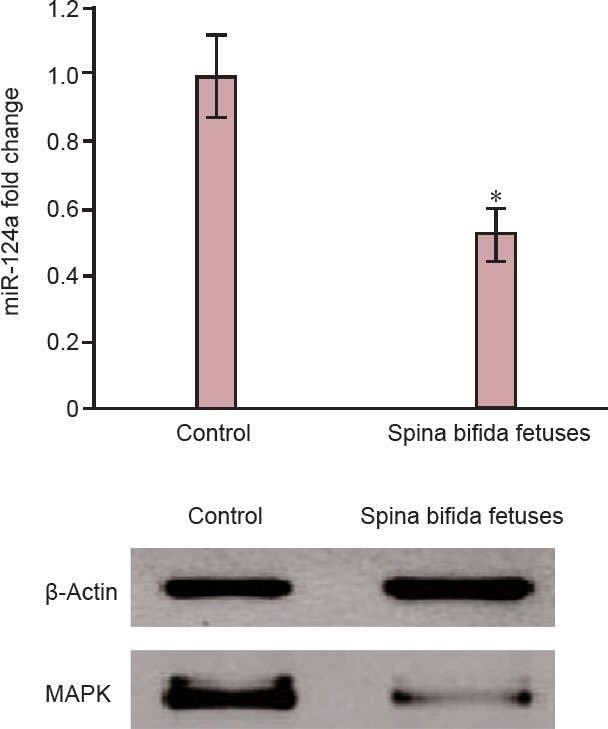
Parallel reductions in miRNA-124a and MAPK expression in the amniotic fluid of rats with fetuses modeling spina bifida fetuses as determined using western blot analysis.
(A) miRNA-124a expression in rats with fetuses modeling spina bifida and control rats with normal fetuses. Data are expressed as the mean ± SD (n = 6 rats per group). *P < 0.05, vs. control fetuses (two-sided Student's t-test). (B) MAPK protein and loading control (β-actin) expression levels in amniotic fluid obtained from rats with fetuses modeling spina bifida and control rats with normal fetuses.
Discussion
Despite the clinical application of fetal surgical treatments available for spina bifida, current therapeutics for spina bifida are still unsatisfactorily compounded by lasting neurological complications. Thus, understanding the mechanisms of neurological dysfunction is essential for developing effective treatments for spina bifida.
A metabolic product of vitamin A, ATRA, is required for normal neuronal differentiation, with the spinal cord being the most active site of ATRA signaling during development (Maden, 2000, 2006). Excessive ATRA is known to cause NTDs in pregnant mammals, spina bifida in particular, and can induce lesions similar to those found in human vitamin A deficiency (Zhao et al., 2008). Maternal administration of ATRA has long been used to induce an experimental model of spina bifida in the fetal rat (Diez-Pardo et al., 1995). No previous studies have provided direct evidence to show which doses of ATRA significantly modify miRNA expression levels. However, the results of some studies showed significantly changed gene expression in vivo after treatment with 150 mg/kg ATRA and increased apoptosis percentage in mouse embryo cells induced by 100 and 150 mg/kg ATRT treatment (Stopera and Bird, 1993; Shum et al., 1999). Hence, we used the higher dose, 150 mg/kg, in the present study to induce miRNA expression alteration.
A number of studies have established that miRNAs are critical regulators of cell and tissue differentiation during embryogenesis (Mineno et al., 2006). In mammals, certain miRNAs have been implicated in the maintenance of the pluripotent cell state during early embryogenesis (Houbaviy et al., 2003), and other miRNAs mediate tissue-specific and organ-specific development (Lagos-Quintana et al., 2002). Maintenance and regulation of endogenous miRNA levels are necessary for proper mammalian neurogenesis and neurodevelopment. A previous study of miRNA expression during brain development and function of miRNA biogenesis in neuronal tissues clearly demonstrated their importance in healthy neuronal development and in the pathogenesis of CNS diseases (Vreugdenhil and Berezikov, 2010). Wu et al. (2013) reported the alteration of miRNA in spina bifida and concluded that overexpression of miRNA-451 and miRNA-375, and the consequent upregulation of p53, may further promote apoptosis in spina bifida. With key roles in the regulation of both neural stem cell differentiation and brain development, miRNAs are candidate contributors to the etiopathology of spina bifida. The analysis of miRNA expression based on genome-wide microarrays has been widely used to rapidly and systematically identify new miRNA markers in human cancers (Lu et al., 2005). Using miRNA microarray analysis in the present study, we demonstrated that the expression levels of miRNA-9, miRNA-124a, and miRNA-138 were significantly downregulated, whereas miRNA-134 expression was significantly upregulated in the amniotic fluid of rats with fetuses modeling spina bifida compared with rats having healthy fetuses. The microarray analysis-identified differentially expressed miRNAs were validated using qRT-PCR. Our results are the first to reveal a unique miRNA expression profile in the amniotic fluid of a rat model of spina bifida and may help identify potential therapeutic targets for spina bifida.
The highly conserved, brain-enriched miRNA-9 is strongly expressed at the fifth stage of embryogenesis, which is a critical period in neural differentiation. Repressed expression of miRNA-9 in neural progenitors could reduce the number of neurons by 30%. Another brain-specific miRNA, miRNA-124a, is enriched in neural cells, including embryonic stem cell-derived neurospheres. It has been reported that expression levels of miRNA-9 and miRNA-124a are reduced in human anencephaly (Zhang et al., 2010, 2014) and in E19 rat embryos modeling spina bifida (Wei et al., 2013). Significantly reduced expression of miRNA-124a in the spinal cord of a rat model of spina bifida occurred at an earlier developmental stage (from E12 to E18) compared with the expression in control rats, and downregulation of miRNA-9 was observed on E12, suggesting that the change in miRNA expression occurred during an early stage of embryonic development in a rat model of spina bifida (Wei et al., 2013). Both miRNA-9 and miRNA-124a have a binding site for repressor element silencing transcription factor (REST), which suppresses their expression in neuronal cells. As the effector of REST, miRNA-124a has been reported to suppress the repression of BAF53a and therefore participate in neuronal differentiation in the developing neural tube (Conaco et al., 2006). It is thus reasonable to postulate that the reduced expression of these two microRNAs observed in the amniotic fluid of rats with fetuses modeling spina bifida in our study is due to an increase in REST expression. Indeed, it has been shown that in the defective spinal cords of a rat model of spina bifida with downregulated miRNAs, the expression of REST was upregulated at all stages (Wei et al., 2013).
Similar to miRNA-124a, miRNA-134 is confined to brain cells, where it positively regulates the development of dendrites and spines as well as synaptic integration (Christensen et al., 2010). The upregulation of miRNA-134 has been shown to reduce cell migration in neural progenitors (Gaughwin et al., 2011), a process that is crucial for neural tube closure. Due to the observed upregulation of miRNA-134 in amniotic fluid of rats with fetuses modeling spina bifida in our study, miRNA-134 overexpression may be considered a high risk factor for NTDs. To date, no functional studies have reported the role of miRNA-138 in brain development and in NTDs. Thus, the biological effects of miRNA-138 should be further explored.
The neurotrophin pathway has been identified as a target of miRNA-124a. Neurotrophins are a family of closely related proteins that were identified initially as survival factors for sensory and sympathetic neurons (Reichardt, 2006; Wang et al., 2012). Previous studies (Bernd, 2008) reported the effects of neurotrophins during the early developmental stage. One of the component genes in this pathway is MAPK, a downstream signal in neural tube development (Chi et al., 2005; Krupp et al., 2012). Our results revealed a parallel decrease in miRNA-124a and MAPK protein expression, with miRNA-124a previously shown to have a regulatory function in spina bifida pathology.
In summary, we provided an overview of differentially expressed miRNAs in the amniotic fluid of rats with fetuses modeling spina bifida compared with control rats. However, the result of this investigation into the mechanisms associated with this defect are preliminary, and future studies with larger sample sizes are needed to validate the screened differentially expressed miRNAs and to investigate their potential target genes, which may contribute to therapeutic strategies with wide clinical implications for the treatment or prevention of spina bifida.
Footnotes
Conflicts of interest: None declared.
Plagiarism check: This paper was screened twice using Cross-Check to verify originality before publication.
Peer review: This paper was double-blinded and stringently reviewed by international expert reviewers.
Copyedited by Smith T, Robens J, Yu J, Li CH, Song LP, Zhao M
References
- Bernd P. The role of neurotrophins during early development. Gene Expr. 2008;14:241–250. doi: 10.3727/105221608786883799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, Moore CA, Khoury MJ, Erickson JD. Neural-tube defects. New Engl J Med. 1999;341:1509–1519. doi: 10.1056/NEJM199911113412006. [DOI] [PubMed] [Google Scholar]
- Chi H, Sarkisian MR, Rakic P, Flavell RA. Loss of mitogen-activated protein kinase kinase kinase 4 (MEKK4) results in enhanced apoptosis and defective neural tube development. Proc Natl Acad Sci U S A. 2005;102:3846–3851. doi: 10.1073/pnas.0500026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M, Larsen LA, Kauppinen S, Schratt G. Recombinant adeno-associated virus-mediated microRNA delivery into the postnatal mouse brain reveals a role for miR-134 in dendritogenesis in vivo. Front Neural Circuits. 2010;3:16. doi: 10.3389/neuro.04.016.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han J-J, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Date I, Yagyu Y, Asari S, Ohmoto T. Long-term outcome in surgically treated spina bifida cystica. Surg Neurol. 1993;40:471–475. doi: 10.1016/0090-3019(93)90049-7. [DOI] [PubMed] [Google Scholar]
- Diez-Pardo JA, Mariño JM, Baoquan Q, Delgado-Baeza E, Fernáneez A, Morales MC, Tovar JA. Neural tube defects: an experimental model in the foetal rat. Eur J Pediatr Surg. 1995;5:198–202. doi: 10.1055/s-2008-1066204. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Frey L, Hauser WA. Epidemiology of neural tube defects. Epilepsia 44 Suppl. 2003;3:4–13. doi: 10.1046/j.1528-1157.44.s3.2.x. [DOI] [PubMed] [Google Scholar]
- Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21:1857–1869. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- Greene ND, Stanier P, Copp AJ. Genetics of human neural tube defects. Human molecular genetics. 2009;18:R113–129. doi: 10.1093/hmg/ddp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3[prime] untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Krupp DR, Xu PT, Thomas S, Dellinger A, Etchevers HC, Vekemans M, Gilbert JR, Speer MC, Ashley-Koch AE, Gregory SG. National Birth Defects Prevention Study (2012) Transcriptome profiling of genes involved in neural tube closure during human embryonic development using long serial analysis of gene expression (long-SAGE) Birth Defects Res A Clin Mol Teratol. 94:683–692. doi: 10.1002/bdra.23040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Longo FM, Massa SM. Small-molecule modulation of neurotrophin receptors: a strategy for the treatment of neurological disease. Nat Rev Drug Discov. 2013;12:507–525. doi: 10.1038/nrd4024. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet- Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Maden M. The role of retinoic acid in embryonic and post-embryonic development. Proc Nutr Soc. 2000;59:65–73. doi: 10.1017/s0029665100000082. [DOI] [PubMed] [Google Scholar]
- Maden M. Retinoids and spinal cord development. J Neurobiol. 2006;66:726–738. doi: 10.1002/neu.20248. [DOI] [PubMed] [Google Scholar]
- Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, Kato I. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CA, Li S, Li Z, Hong SX, Gu HQ, Berry RJ, Mulinare J, Erickson JD. Elevated rates of severe neural tube defects in a high-prevalence area in northern China. Am J Med Genet. 1997;73:113–118. [PubMed] [Google Scholar]
- Padmanabhan R. Etiology, pathogenesis and prevention of neural tube defects. Congenit Anom (Kyoto) 2006;46:55–67. doi: 10.1111/j.1741-4520.2006.00104.x. [DOI] [PubMed] [Google Scholar]
- Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum AS, Poon LL, Tang WW, Koide T, Chan BW, Leung YC, Shiroishi T, Copp AJ. Retinoic acid induces down-regulation of Wnt-3a, apoptosis and diversion of tail bud cells to a neural fate in the mouse embryo. Mech Develop. 1999;84:17–30. doi: 10.1016/s0925-4773(99)00059-3. [DOI] [PubMed] [Google Scholar]
- Stopera SA, Bird RP. Effects of all-trans retinoic acid as a potential chemopreventive agent on the formation of azoxymethane-induced aberrant crypt foci: differential expression of c-myc and c-fos MRNA and protein. Int J Cancer. 1993;53:798–803. doi: 10.1002/ijc.2910530516. [DOI] [PubMed] [Google Scholar]
- Vreugdenhil E, Berezikov E. Fine-tuning the brain: MicroRNAs. Front Neuroendocrinol. 2010;31:128–133. doi: 10.1016/j.yfrne.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Wang W, Kwon EJ, Tsai L-H. MicroRNAs in learning, memory, and neurological diseases. Learn Memory. 2012;19:359–368. doi: 10.1101/lm.026492.112. [DOI] [PubMed] [Google Scholar]
- Wei X, Li H, Miao J, Liu B, Zhan Y, Wu D, Zhang Y, Wang L, Fan Y, Gu H, Wang W, Yuan Z. miR-9*- and miR-124a-Mediated switching of chromatin remodelling complexes is altered in rat spina bifida aperta. Neurochem Res. 2013;38:1605–1615. doi: 10.1007/s11064-013-1062-8. [DOI] [PubMed] [Google Scholar]
- Wu LN, Wei XW, Fan Y, Miao JN, Wang LL, Zhang Y, Wu D, Yuan ZW. Altered expression of 14-3-3æ protein in spinal cords of rat fetuses with spina bifida aperta. PLoS One. 2013;8:e70457. doi: 10.1371/journal.pone.0070457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WD, Yu X, Fu X, Huang S, Jin SJ, Ning Q, Luo XP. MicroRNAs function primarily in the pathogenesis of human anencephaly via the mitogen-activated protein kinase signaling pathway. Genet Mol Res. 2014;13:1015–1029. doi: 10.4238/2014.February.20.3. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chang H, Li Y, Zhang T, Zou J, Zheng X, Wu J. MicroRNAs: Potential regulators involved in human anencephaly. Int J Biochem Cell Biol. 2010;42:367–374. doi: 10.1016/j.biocel.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Sun DG, Wang J, Liu SR, Zhang CY, Zhu MX, Ma X. Retinoic acid downregulates microRNAs to induce abnormal development of spinal cord in spina bifida rat model. Childs Nerv Syst. 2008;24:485–492. doi: 10.1007/s00381-007-0520-5. [DOI] [PubMed] [Google Scholar]


