Abstract
Parturition in rodents is highly dependent on the engagement of the luteal prostaglandin F2 alpha receptor, which, through activation of the Gq/11 family of G proteins, increases the expression of Akr1c18, leading to an increase in progesterone catabolism. To further understand the involvement of Gq/11 on luteolysis and parturition, we used microarray analysis to compare the ovarian transcriptome of mice with a granulosa/luteal cell-specific deletion of Galphaq/11 with their control littermates on Day 18 of pregnancy, when mice from both genotypes are pregnant, and on Day 22, when mice with a granulosa/luteal cell-specific deletion of Galphaq/11 are still pregnant but their control littermates are 1–2 days postpartum. Ovarian genes up-regulated at the end of gestation in a Galphaq/11 -dependent fashion include genes involved in focal adhesion and extracellular matrix interactions. Genes down-regulated at the end of gestation in a Galphaq/11-dependent manner include Serpina6 (which encodes corticosteroid-binding globulin); Enpp2 (which encodes autotaxin, the enzyme responsible for the synthesis of lysophosphatidic acid); genes involved in protein processing and export; reproductive genes, such as Lhcgr; the three genes needed to convert progesterone to estradiol (Cyp17a1, Hsd17b7, and Cyp19a1); and Inha. Activation of ovarian Gq/11 by engagement of the prostaglandin F2 alpha receptor on Day 18 of pregnancy recapitulated the regulation of many but not all of these genes. Thus, although the ovarian transcriptome at the end of gestation is highly dependent on the activation of Gq/11, not all of these changes are dependent on the actions of prostaglandin F2 alpha.
Keywords: Gq/11, parturition, transcriptome
INTRODUCTION
It is now clear that at least two families of G proteins, Gs and Gq/11, are essential to maintain the functions of granulosa and luteal cells. Gs activation by the luteinizing hormone (LH) receptor in granulosa cells is required for initiating the events leading to the resumption of meiosis and the expansion of cumulus cells, but it is also required for the induction of the progesterone receptor (Pgr) and subsequent rupture of the ovarian follicles [1, 2]. The activation of Gq/11 by the LH receptor in granulosa cells is not required for the resumption of meiosis and the expansion of cumulus cells [3]. Together with the LH receptor-activated Gs, however, the LH receptor-activated Gq/11 is required for optimal induction of the granulosa cell Pgr, follicular rupture, and an optimal ovulatory response [1, 3]. Thus, genetic deletion of Gαq/11 from granulosa cells compromises the LH receptor-induced expression of the granulosa cell Pgr and ovulation [3].
Studies with mice harboring a granulosa/luteal cell-specific deletion of Gαq/11 have demonstrated that the differentiation of granulosa cells into luteal cells, as well as the establishment and maintenance of pregnancy, does not require Gq/11 [3, 4]. Parturition, on the other hand, is dependent on the prostaglandin F2α (PGF2α)-provoked activation of luteal Gq/11 [4]. A conditional deletion of Gαq/11 from granulosa/luteal cells disrupts the actions of PGF2α on luteal cells that normally occur toward the end of gestation [4]. The ovaries of pregnant mice with a conditional deletion of Gαq/11 from granulosa/luteal cells display a defect in the PGF2α-provoked induction of Ark1c18, the gene encoding for 20α-hydroxysteroid dehydrogenase, the enzyme that catabolizes progesterone to 20α-hydroxyprogesterone [4]. As such, the mice exhibit a delay or failure of parturition because they do not experience the reduction in progesterone levels that precipitate parturition [4].
The crucial role of luteal cell Gαq/11 in the expression of luteal Ark1c18 at the end of gestation prompted us to examine the hypothesis that luteal cell Gq/11 is also involved in the expression of other ovarian genes that may participate in luteolysis and parturition. This is an important question, because in addition to activating Gq/11, the PGF2α receptor also activates G12/13 [5], and because there are other hormones, such as oxytocin [6, 7] or lysophosphatidic acid [8, 9], that can activate luteal Gq/11. To test this hypothesis we compared the ovarian transcriptome of mice with a granulosa/luteal cell-specific deletion of Gαq/11 with the transcriptome of their control littermates on Days 18 and 22 of pregnancy. The results presented show that changes in the ovarian transcriptome that occur at the end of gestation (Day 22 of pregnancy, when mice lacking Gαq/11 still have not given birth but their control littermates have) are highly dependent on Gαq/11. Not all of the Gαq/11-dependent genes are dependent on the actions of PGF2α, however.
MATERIALS AND METHODS
Mice
A colony of Gαqf/f;Gα11−/−;Cre+ mice were generated by crossing Gαqf/f;Gα11−/− mice and Cyp19Cre transgenic mice as previously described [3]. The colony of Gαqf/f;Gα11−/−;Cre+ mice was maintained by crossing Gαqf/f;Gα11−/−;Cre+ males with Gαqf/f;Gα11−/−;Cre− females [4]. The resulting Gαqf/f;Gα11−/−;Cre+ and Gαqf/f;Gα11−/−;Cre− females were used as experimental and control animals, respectively. Genotyping was performed using tail genomic DNA and PCR amplification as described previously [10–14].
Adult females (ages 6–10 wk) were synchronized on estrus by injecting them with progesterone and cloprostenol [15], housed overnight with males of proven fertility, and checked the following morning for vaginal plugs. Day 1 of pregnancy (1 dpc) was counted as the morning when a vaginal plug was found, and the pregnant animals were killed in the morning on Days 18 or 22 of pregnancy as indicated. In some experiments pregnant mice (17 dpc) were injected subcutaneously with vehicle only or 1 μg of cloprostenol, a PGF2α agonist [16], and killed 24 h later for the preparation of ovarian RNA. All animal procedures were approved by the Institutional Animal Care and Use Committee for the University of Iowa.
cAMP Assays
Luteal cells were prepared from the ovaries of mice on 18 or 22 dpc exactly as described previously for pseudopregnant ovaries [4]. The freshly isolated luteal cells were immediately suspended in 500 μl of Dulbecco modified Eagle medium/F12 supplemented with 10 mM Hepes, bovine serum albumin (1 mg/ml), and 1 mM isobutyl methylxanthine and incubated without or with human chorionic gonadotropin (hCG; 100 ng/ml), for 30 min at 37°C. Total cAMP (cells + medium) was then extracted and quantitated as described earlier [17–19], except that we used a commercially available enzyme immunoassay kit from Cayman Chemical instead of a radioimmunoassay. All samples were assayed in a single assay and the intraassay coefficient of variation was 10%.
Microarray Analysis
Transcription profiling was performed in triplicate (n = 3) for each genotype on 18 dpc and 22 dpc, respectively. The profiling was done using ovarian RNA purified from the ovaries and the mouse WG-6 V2.0 Illumina platform. All steps of the transcription profiling (except for the isolation of the RNA) and analyses were performed by the Iowa Institute of Human Genetics. Gene enrichment analysis was carried out using publicly available software (http://bioinfo.vanderbilt.edu/webgestalt).
RNA Isolation and Real-Time PCR
Ovarian RNA was prepared as described earlier [4]. Target and control gene (Gapdh) expressions were measured by quantitative PCR (qPCR) using 5–50 ng of purified RNA, the iTaq Universal SYBR Green One Step Kit (Bio-Rad Laboratories), and a CFX90 real-time PCR detection system (Bio-Rad Laboratories). Each experimental group included a minimum of five replicate animals (see Figure legends), and each sample was run in duplicate on the qPCR reactions. Data were quantitated by the comparative Cq method as described previously [20], using Gapdh as control or normalizer gene, and are always expressed relative to a control group as indicated in each of the figures. There was no statistical difference among the average quantification cycles (Cq) for the control gene (Gapdh) for any group or condition.
The primers used were as follows: Lhcgr: forward, CCTTGTGGGTGTCAGCAGTTA; reverse, TTGTGACAGAGTGGA TTCCACAT; Cyp19a1: forward, GACGGGCCCTGGTCTTGT; reverse, CCGGTCCAAATGCTGCTT; Cyp11a1: forward, CCCGGAGCGGTTCCTT; reverse, CCAATGGGCCTCTGATAATACTG; Cyp17a1: forward, TGGAGGCCACTATCCGAGAA; reverse, CACATGTGTGTCCTTCGGGA; Inha: forward, TGTGGGTAAAGTGGGGGAGA; reverse, GTCTTTGGTGGTGTCTGCGA; Hsd17b7: forward, AGGCGTCGTGATGACCAATA; reverse, AAAAAGCGAAGGAGCCACATT; Enpp2: forward, GGGCTGCACCTGTGATGATA; reverse, GATGTCTCTCTTCTGTAGATCCTT; Serpina6: forward, TTCCAAGGCAGATGAACCTGTA; reverse, GTCTTTGGTGGTGTCTGCGA; Wfs1: forward, TCGTCAGCAGTGAATCCAAGAA; reverse, AATGATGCCCTTGGCGTACT; Jun: forward, CTTGTGCCCCAAGAACGTG; reverse, GTGACACTGGGAAGCGTGTT; Tnc: forward, CAATAACCACAGTCAGGGCGT; reverse, CCCTGGAATTTATGCCCGCT; Ssr4: forward, TGGCTCTTTATGCCGACGTT; reverse, AGGCTCCAGGACACCTGATA; and Gapdh: forward, GACGGCCGCATCTTCTTG; reverse, ACCGACCTTCACCATTTTGTCT.
Hormone Assays
Serum estradiol and Inhibin A concentrations were measured using ELISA by the Ligand Assay and Analysis Core of the University of Virginia. The measurable ranges for estradiol and inhibin A are 3–300 pg/ml and 10–950 pg/ml, respectively. The intraassay and interassay coefficients of variation for estradiol are 6.1% and 8.9%, respectively, and the corresponding values for Inhibin A are 1.5% and 4.1%.
RESULTS
Microarray Analysis
Ovarian RNA was prepared from Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ females on 18 dpc (three mice of each genotype) and 22 dpc (three mice of each genotype). On 18 dpc, mice from both genotypes are pregnant. Because the Gαqf/f;Gα11−/−;Cre− mice give birth on 20–21 dpc, they are 1–2 days postpartum on 22 dpc. The Gαqf/f;Gα11−/−;Cre+ are still pregnant on 22 dpc because of the previously described delay or failure of parturition [4]. The 12 RNA samples were subjected to expression profiling using the mouse WG-6 V2.0 Illumina platform. Array data were imported using the Bead array software package (http://www.bioconductor.org/packages/release/bioc/html/beadarray.html) and were quantile normalized. Differential expression was determined using the Limma software package (http://bioconductor.org/packages/release/bioc/html/limma.html).
A multidimensional scale plot of the microarray data (Fig. 1) clearly shows that based on the similarity of gene expression, the 12 mice used for the microarray analysis can be segregated into 4 groups according to genotype and length of gestation. Ovarian gene expression was rather similar between the two genotypes on 18 dpc (when mice from both genotypes are pregnant) but was very different on 22 dpc, when the Gαqf/f;Gα11−/−;Cre− mice have given birth but the Gαqf/f;Gα11−/−;Cre+ mice have not. Also, the three mice of each length of gestation and genotype clustered very close to each other, but there was more variability in the group of Gαqf/f;Gα11−/−;Cre+ mice on 22 dpc than in any of the other three groups.
FIG. 1.
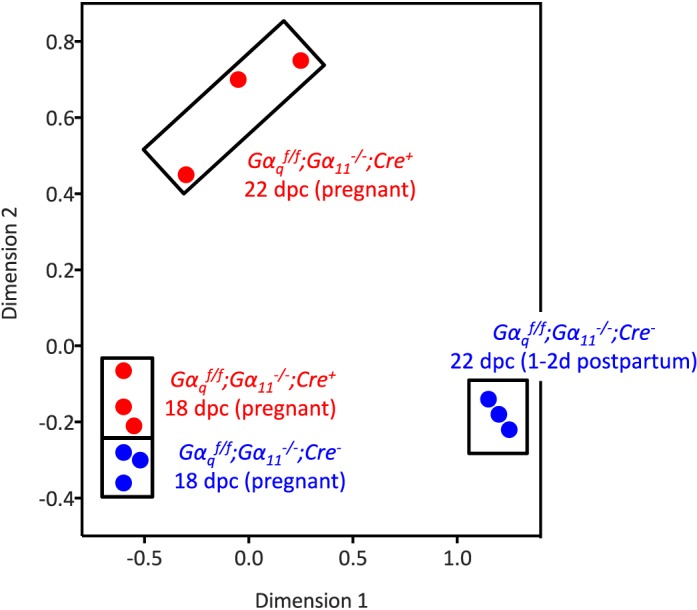
Multidimensional plot of ovarian gene expression in Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice on 18 and 22 dpc. The blue and red circles enclosed in the rectangles highlight the overall similarity of gene expression in the individual ovaries of the four groups of Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice analyzed on 18 and 22 dpc as indicated. On 18 dpc Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice are pregnant. On 22 dpc the Gαqf/f;Gα11−/−;Cre+ mice are still pregnant but the Gαqf/f;Gα11−/−;Cre− mice are 1–2 days postpartum.
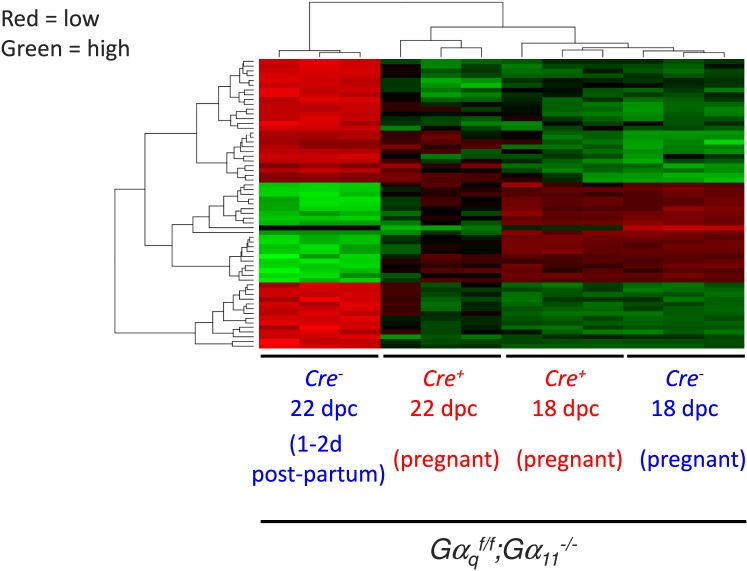
Because we used 12 mice and there are a large number of probes represented in the mouse WG-6 V2.0 Illumina chips (∼45 000), the results of the differential expression can be analyzed using rather stringent criteria for statistical significance (P < 0.001). A comparison of gene expression between the two genotypes on 18 dpc revealed no differentially expressed probes, either when using this criterion or when a more relaxed (P < 0.005) level of significance was used. A comparison of gene expression in Gαqf/f;Gα11−/−;Cre− mice on 22 and 18 dpc using P < 0.001 generated a list of 288 differentially expressed probes. This group of probes was then ranked to include only probes that were differentially expressed by at least 100-fold (2 log-fold), to generate the heat map presented in Figure 2. As expected from the above discussion, gene expression was generally similar in the ovaries of Gαqf/f;Gα11−/−;Cre+ and Gαqf/f;Gα11−/−;Cre− mice on 18 dpc. This map also clearly shows two clusters of probes (shown in red, about 40 probes) that were down-regulated in the ovaries of Gαqf/f;Gα11−/−;Cre− on 22 dpc, and one cluster of probes (shown in green, about 20 probes) that was up-regulated in the ovaries of Gαqf/f;Gα11−/−;Cre− mice on 22 dpc. The down- or up-regulation of these clusters of probes was not obvious in the ovaries of the Gαqf/f;Gα11−/−;Cre+ mice on 22 dpc, however.
FIG. 2.
Heat map of ovarian gene expression in Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice on 18 and 22 dpc. The probes included in this heat map were first selected by comparing gene expression in the Gαqf/f;Gα11−/−;Cre− mice on 22 and 18 dpc. On 18 dpc Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice are pregnant. On 22 dpc the Gαqf/f;Gα11−/−;Cre+ mice are still pregnant but the Gαqf/f;Gα11−/−;Cre− mice are 1–2 days postpartum. Only those probes that were differentially expressed with a P < 0.001 were chosen. This list was sorted again based on the extent of the differential expression, and only those probes that were differentially expressed by at least two orders of magnitude (100-fold) are shown. An increase in expression is shown by the different shades of green, and a decrease in expression is shown by the different shades of red. The clusters shown contain duplicate probes and probes for unknown genes. The identity of the up- and down-regulated genes (after removal of duplicate probes and probes for unknown genes) is shown in Tables 1 and 2.
Manually removing duplicate probes and probes for unknown genes further reduced the up-regulated cluster to the 11 genes shown in Table 1. This table shows that the magnitude of the increased expression of the 11 genes in the Gαqf/f;Gα11−/−;Cre− mice was much lower or absent compared with the Gαqf/f;Gα11−/−;Cre+ mice (compare the log of fold increase in the Cre−/Cre− and Cre+/Cre+ columns) and, with the exception of Akr1c18 and Chrna1, the up-regulation of the other genes in the Gαqf/f;Gα11−/−;Cre+ mice was no longer statistically significant (adjusted P value <0.001 in the Cre−/Cre− column and >0.001 in the Cre+/Cre+ column). The same type of analysis reduced the down-regulated clusters to the 28 genes shown in Table 2. Again, the decreased expression of these genes was much lower or absent (compare the log of fold decrease in the Cre−/Cre− and Cre+/Cre+ columns) and no longer statistically significant in the Gαqf/f;Gα11−/−;Cre+ mice (adjusted P value <0.001 in the Cre−/Cre− column and >0.001 in the Cre+/Cre+ column).
TABLE 1.
Ovarian genes up-regulated at the end of gestation in a Gαq/11-dependent fashion by at least two orders of magnitude and P < 0.001.

On 18 dpc Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice are pregnant. On 22 dpc the Gαqf/f;Gα11−/−;Cre+ mice are still pregnant but the Gαqf/f;Gα11−/−;Cre− mice are 1–2 days postpartum.
These genes were chosen for further analysis as discussed in the text.
TABLE 2.
Ovarian genes down-regulated at the end of gestation in a Gαq/11-dependent fashion by at least two orders of magnitude and P < 0.001.
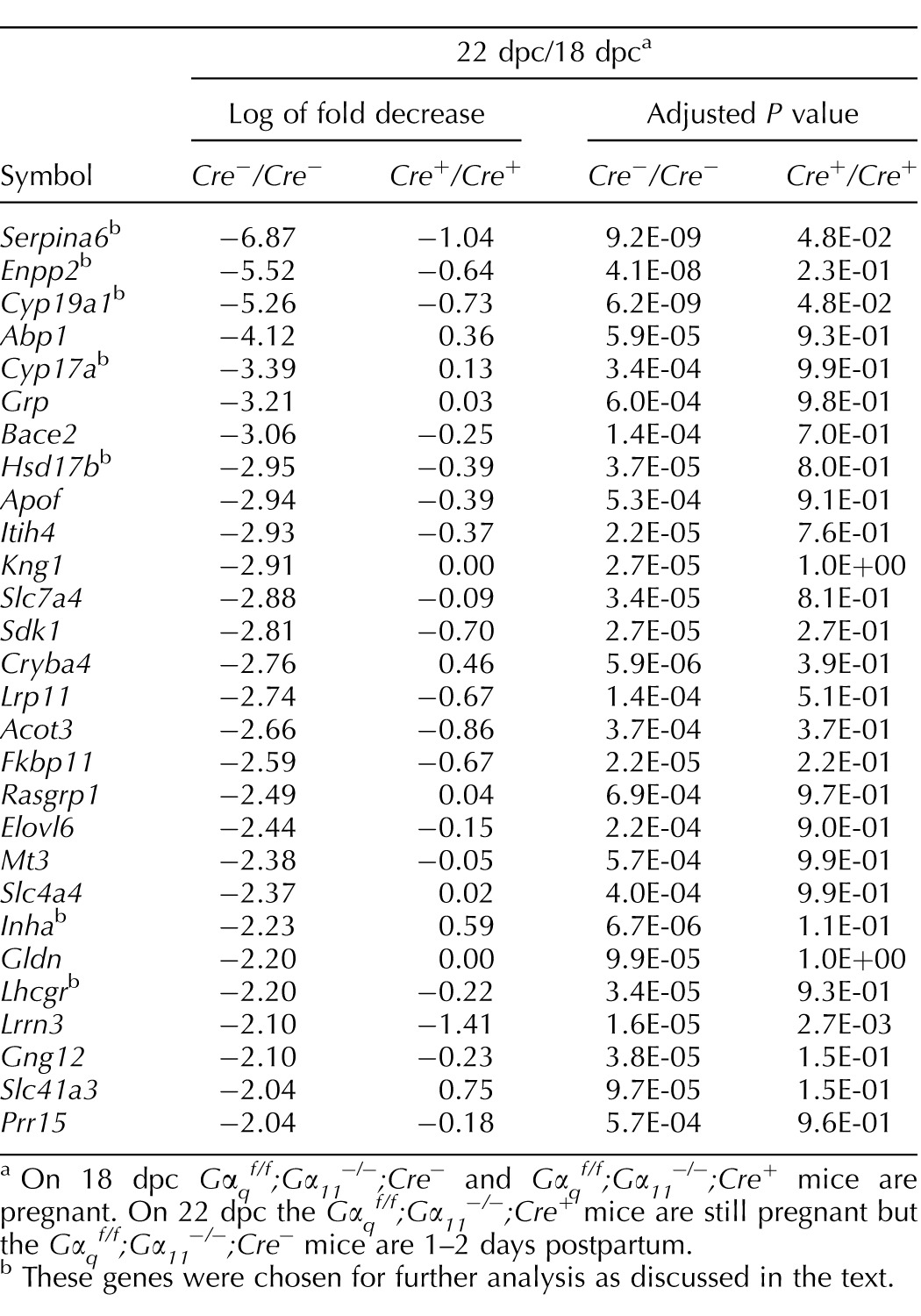
On 18 dpc Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice are pregnant. On 22 dpc the Gαqf/f;Gα11−/−;Cre+ mice are still pregnant but the Gαqf/f;Gα11−/−;Cre− mice are 1–2 days postpartum.
These genes were chosen for further analysis as discussed in the text.
We next used pathway enrichment software to categorize the 138 probes that were individually up-regulated as well as the 150 probes that were individually down-regulated (P < 0.001) into Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Using the default criteria for this analysis (at least two genes per group and P < 0.01 for the group) we identified 34 up-regulated genes that can be grouped into 8 KEGG pathways (Table 3) and 49 down-regulated genes that can be grouped in 7 KEGG pathways (Table 4).
TABLE 3.
Pathway enrichment analysis of ovarian genes that are up-regulated at the end of gestation in a Gαq/11-dependent fashion.
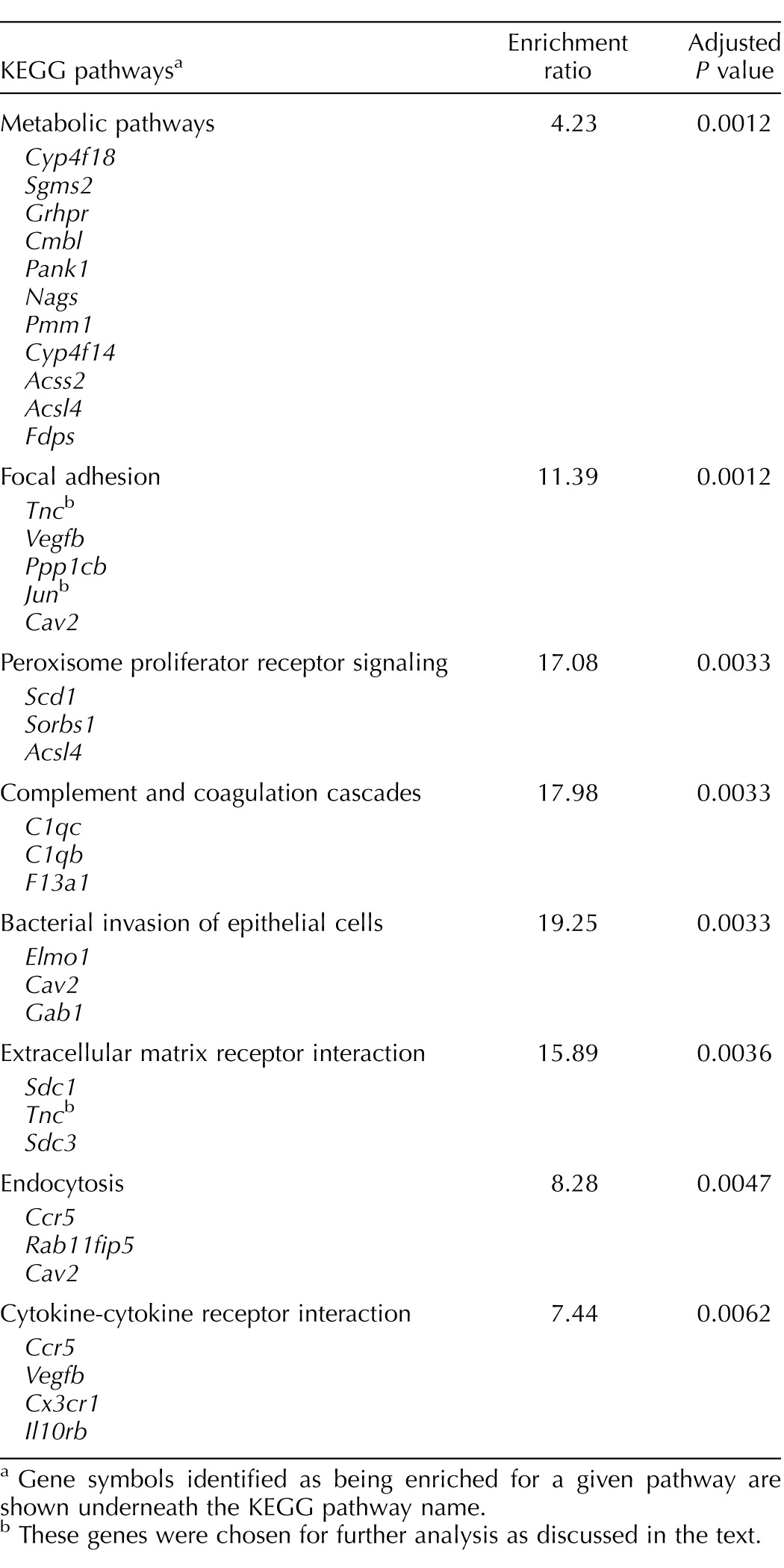
Gene symbols identified as being enriched for a given pathway are shown underneath the KEGG pathway name.
These genes were chosen for further analysis as discussed in the text.
TABLE 4.
Pathway enrichment analysis of ovarian genes that are down-regulated at the end of gestation in a Gαq/11-dependent fashion.

Gene symbols identified as being enriched for a given pathway are shown underneath the KEGG pathway name.
These genes were chosen for further analysis as discussed in the text.
The entire microarray data set is included herein (MS Excel file) as Supplemental Table S1 (available online at www.biolreprod.org).
Validation of the Microarray Analysis by qPCR
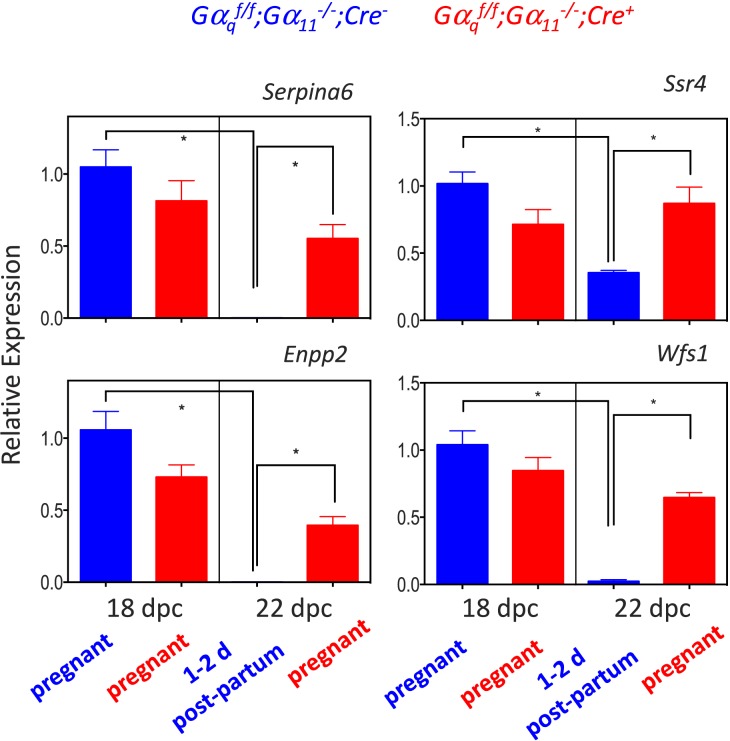
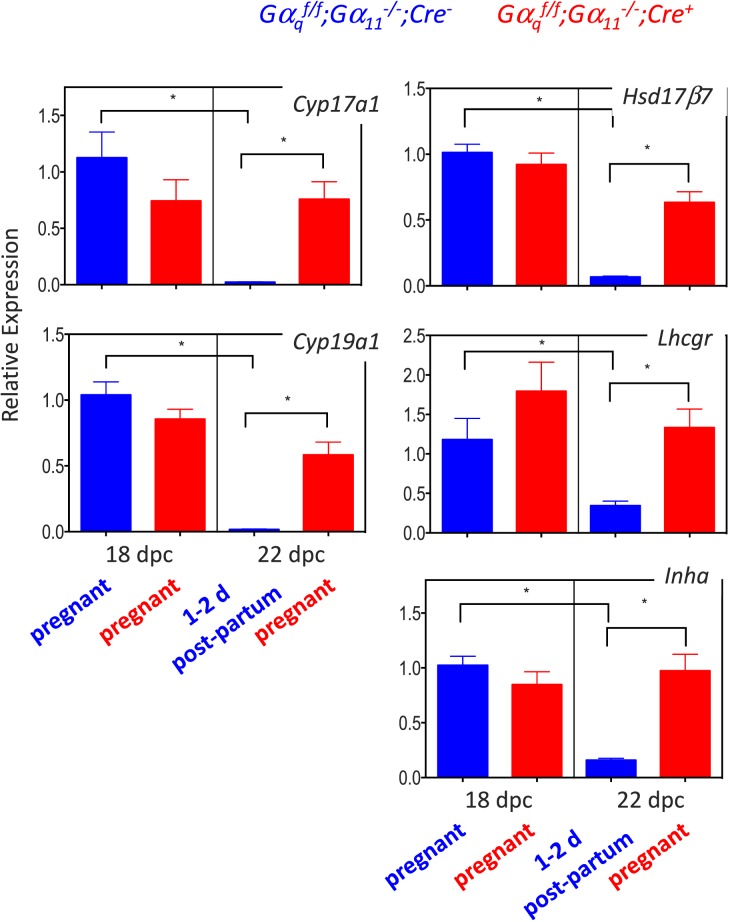
Akr1c18 was the gene with the highest magnitude of Gαq/11-dependent up-regulation (Table 1), and the validation of this result by qPCR has already been accomplished elsewhere [4]. We chose to further analyze two up-regulated genes, Tnc and Jun, based on a combination of the magnitude of up-regulation (Table 1) and their grouping into the “focal adhesion” KEGG pathway (Table 3). We also chose to further analyze nine down-regulated genes based on the magnitude of down-regulation (Serpina6 and Enpp2; Table 2), their known involvement in reproductive functions (Cyp19a1, Cyp17a1, Hsd17b7, Lhcgr, and Inha; Table 2), their grouping into a KEGG pathway (Wfs1 and Ssr4 in the “protein processing in endoplasmic reticulum” KEGG pathway in Table 4), or a combination of all three criteria—magnitude of down-regulation, their known involvement in reproductive functions, and their grouping into the “steroid hormone biosynthesis” KEGG pathway (Cyp19a1, Cyp17a1, and Hsd17b7; Tables 2 and 4).
Quantitative PCR validation of the microarray data for the up-regulated (Fig. 3) and down-regulated (Figs. 4 and 5) genes on 18 and 22 dpc clearly recapitulated the microarray data. Their expression increased (Fig. 3) or decreased (Figs. 4 and 5) at the end of gestation in the Gαqf/f;Gα11−/−;Cre−, mice but the magnitude of this change was blunted or absent in the Gαqf/f;Gα11−/−;Cre+ mice.
FIG. 3.
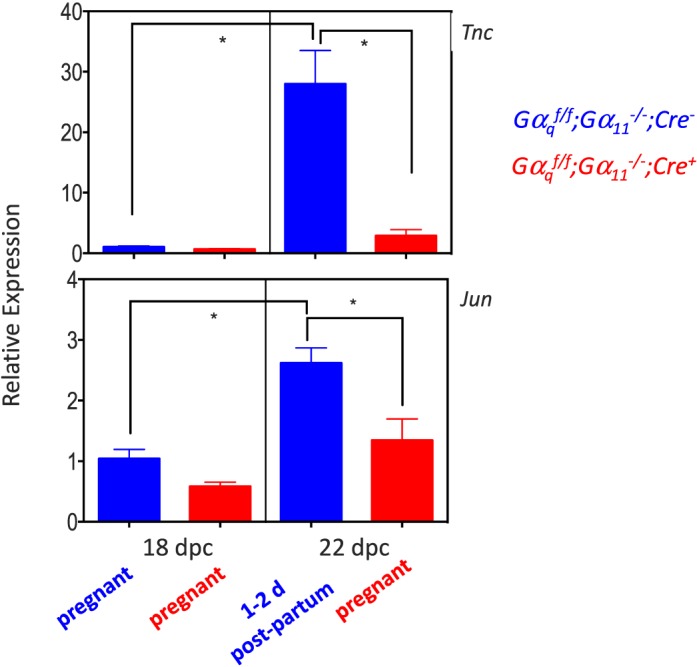
Quantitative PCR validation of the changes in expression of ovarian Tnc and Jun at the end of gestation in Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice. For each gene, data are expressed relative to the gene's expression in the ovaries of Gαqf/f;Gα11−/−;Cre− mice on 18 dpc (blue bar on the left panel of each graph). Each bar is the mean ± SEM of five different animals analyzed on the indicated days. Statistically significant differences (P < 0.05) were calculated using ANOVA and are shown by the brackets and asterisks. On 18 dpc Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice are pregnant. On 22 dpc the Gαqf/f;Gα11−/−;Cre+ mice are still pregnant but the Gαqf/f;Gα11−/−;Cre− mice are 1–2 days postpartum.
FIG. 4.
Quantitative PCR validation of the changes in expression of ovarian Serpina6, Enpp2, Wfs1, and Ssr4 at the end of gestation in Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice. For each gene, data are expressed relative to the gene's expression in the ovaries of Gαqf/f;Gα11−/−;Cre− mice on 18 dpc (blue bar on the left panel of each graph). Each bar is the mean ± SEM of 5–10 different animals analyzed on the indicated days. Statistically significant differences (P < 0.05) were calculated using ANOVA and are shown by the brackets and asterisks. Some bars are not visible because of the scale used. On 18 dpc Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice are pregnant. On 22 dpc the Gαqf/f;Gα11−/−;Cre+ mice are still pregnant but the Gαqf/f;Gα11−/−;Cre− mice are 1–2 days postpartum.
FIG. 5.
Quantitative PCR validation of the changes in expression of the genes known to be involved in reproductive functions at the end of gestation in Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice. For each gene, data are expressed relative to the gene's expression in the ovaries of Gαqf/f;Gα11−/−;Cre− mice on 18 dpc (blue bar on the left panel of each graph). Each bar is the mean ± SEM of 8–10 different animals analyzed on the indicated days. Statistically significant differences (P < 0.05) were calculated using ANOVA and are shown by the brackets and asterisks. On 18 dpc Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice are pregnant. On 22 dpc the Gαqf/f;Gα11−/−;Cre+ mice are still pregnant but the Gαqf/f;Gα11−/−;Cre− mice are 1–2 days postpartum.
Effect of a PGF2α Receptor Agonist on the Expression of Luteal Gαq/11-Dependent Genes
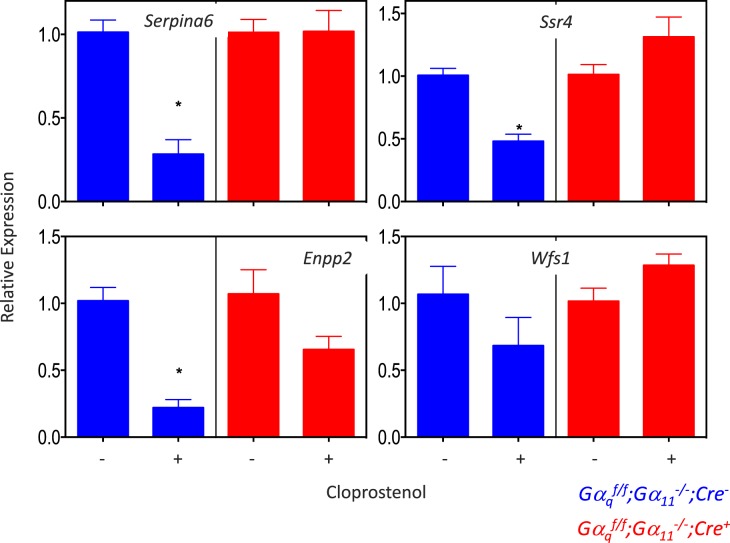
The engagement of the PGF2α receptor by uterine-derived PGF2α is recognized as the main determinant of luteolysis and parturition in rodents [4, 21–29], and the increased expression of one of the genes found in the microarray (Akr1c18; Table 1) has already been shown to be dependent on the activation of Gq/11 by the PGF2α receptor [4, 26, 27]. To determine whether PGF2α also influenced the expression of the 11 Gαq/11-dependent genes shown in Figures 3–5, we injected mice of both genotypes with cloprostenol (a PGF2α agonist) on 17 dpc and analyzed ovarian gene expression 24 h later. Figure 6 shows that the expression of only one (Jun) of the two chosen up-regulated genes was increased when pregnant Gαqf/f;Gα11−/−;Cre− mice were injected with cloprostenol, and that this response was absent in the Gαqf/f;Gα11−/−;Cre+ mice. The expression of ovarian Tnc was not affected by cloprostenol in either genotype.
FIG. 6.
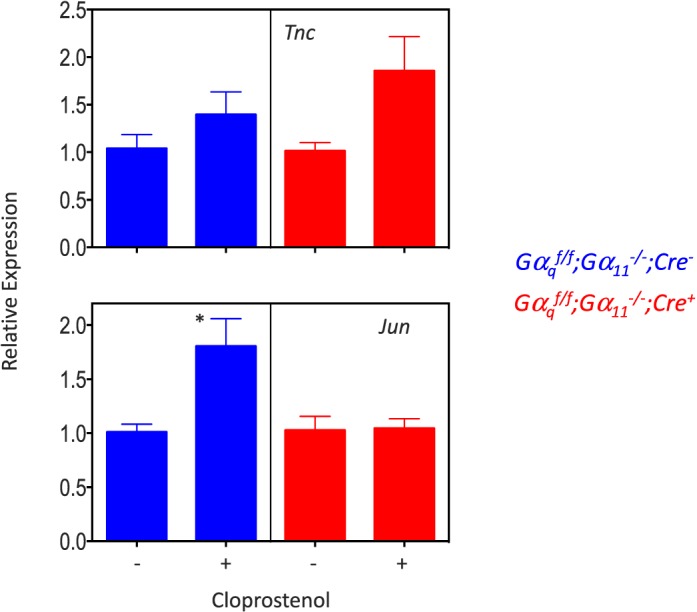
Effect of cloprostenol on the expression of Tnc and Jun in the ovaries of Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice on 17 dpc. Mice were injected with vehicle or 1 μg of cloprostenol on 17 dpc and killed 24 h later. For each gene, data are expressed relative to the gene's expression in the ovaries of mice of the same genotype injected with vehicle only. Each bar is the mean ± SEM of five different animals. Statistically significant differences (P < 0.05) were calculated only by comparing expression in each genotype (t-test) and are shown by asterisks.
The effect of cloprostenol on the down-regulated genes was also mixed (Figs. 7 and 8). This PGF2α agonist decreased the expression of ovarian Serpina6, Ssr4, Enpp2, Lhcgr, Hsd17b7, and Cyp19a1 in Gαqf/f;Gα11−/−;Cre− mice but not in Gαqf/f;Gα11−/−;Cre+ mice. Cloprostenol had no effect on the expression of ovarian Wfts1, Cyp17a1, or Inha in either mouse genotype.
FIG. 7.
Effect of cloprostenol on the expression of Serpina6, Enpp2, Wfs1, and Ssr4 in the ovaries of Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice on 17 dpc. Mice were injected with vehicle or 1 μg of cloprostenol on 17 dpc and killed 24 h later. For each gene, data are expressed relative to the gene's expression in the ovaries of mice of the same genotype injected with vehicle only. Each bar is the mean ± SEM of 5–10 different animals. Statistically significant differences (P < 0.05) were calculated only by comparing expression in each genotype (t-test) and are shown by asterisks.
FIG. 8.
Effect of cloprostenol on the expression of the genes known to be involved in reproductive functions in the ovaries of Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice on 17 dpc. Mice were injected with vehicle or 1 μg of cloprostenol on 17 dpc and killed 24 h later. For each gene, data are expressed relative to the gene's expression in the ovaries of mice of the same genotype injected with vehicle only. Each bar is the mean ± SEM of 5–10 different animals. Statistically significant differences (P < 0.05) were calculated only by comparing expression in each genotype (t-test) and are shown by asterisks.
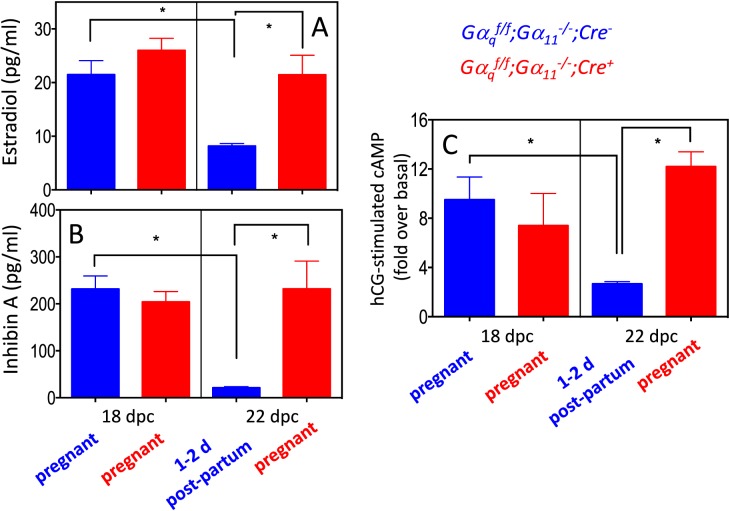
Physiological Correlates of the Gαq/11-Dependent Changes in Reproductive Gene Expression That Occur at the End of Gestation
The decrease in expression of steroidogenic enzyme genes and the Lhcgr that occurred at the end of gestation (Table 2 and Fig. 5) implies that the decline in progesterone and estradiol levels that occur during this time period [27, 30] is coordinately regulated in a Gαq/11-dependent fashion. The dependency of the decline in progesterone on Gαq/11 has already been documented [4]. Figure 9A shows that estradiol levels in Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice on 18 dpc were similar and that they declined on 22 dpc in Gαqf/f;Gα11−/−;Cre− mice but remained elevated in Gαqf/f;Gα11−/−;Cre+ mice. Figure 9B similarly shows that the responsiveness of luteal cells to LH/CG was similar in Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice on 18 dpc, and that it declined on 22 dpc in Gαqf/f;Gα11−/−;Cre− mice but remained elevated in Gαqf/f;Gα11−/−;Cre+ mice. Lastly, the data in Figure 9C show that circulating levels of Inhibin A, another ovarian product, follow the same pattern predicted by the changes in the ovarian expression of Inha documented in Table 2 and Figure 5.
FIG. 9.
Physiological correlates of the changes in ovarian reproductive gene expression at the end of gestation in Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice. For A and B, each bar shows the mean ± SEM of five to eight different animals analyzed on the indicated days. C) The effects of hCG on cAMP accumulation in freshly isolated luteal cells (see Materials and Methods for details). These data are presented as fold over basal level for ease of comparison. The basal levels of cAMP among the four groups shown varied between 30 and 50 pmol/mg protein. Each bar shows the mean ± SEM of three to four different animals analyzed on the indicated days. Statistically significant differences (P < 0.05) were calculated using ANOVA and are shown by the brackets and asterisks. On 18 dpc Gαqf/f;Gα11−/−;Cre− and Gαqf/f;Gα11−/−;Cre+ mice are pregnant. On 22 dpc the Gαqf/f;Gα11−/−;Cre+ mice are still pregnant but the Gαqf/f;Gα11−/−;Cre− mice are 1–2 days postpartum.
DISCUSSION
The murine parturition pathway is initiated by the increased expression of the Ark1c18 gene in luteal cells. This gene encodes for 20α-hydroxysteroid dehydrogenase an enzyme that inactivates progesterone by converting it to 20α-hydroxyprogesterone. In pregnant rodents the expression of luteal Akr1c18 is initially silenced by the actions of prolactin and placental lactogens [29, 31–38]. As pregnancy progresses, expression of the luteal PGF2α receptor increases [21–24], and it is engaged by uterine-derived PGF2α [25], which, through the activation of Gq/11 [4], increases the expression of the Ark1c18 gene and the conversion of progesterone to 20α-hydroxyprogesterone [26–29]. The resulting drop in progesterone ultimately allows uterine contractions and parturition to begin (reviewed in Davis and Rueda [25], Ratajczak and Muglia [39], Stocco et al. [40], and Henkes et al. [41]). We cannot assume, however, that all actions of PGF2α are mediated by Gq/11, because the PGF2α receptor also activates G12/13 [5]. In addition, luteal Gq/11 can also be activated by other hormones, such as oxytocin [6, 7] or lysophosphatidic acid [8, 9], and these could be involved in luteolysis and parturition as well.
The results of the microarray analysis reported here show that Ark1c18 is the ovarian gene that is most robustly up-regulated at the end of gestation in a Gαq/11-dependent fashion. Because we recently showed that the increased expression of ovarian Akr1c18 provoked by activation of the PGF2α receptor or observed at the end of gestation is dependent on Gαq/11 [4], the finding of Akr1c18 as the ovarian gene that is most robustly up-regulated in our microarray is a reassuring finding that serves as a positive control. The data presented here and in a previous publication [4] also show that the up-regulation of this gene is greatly reduced in magnitude but not abolished in the Gαqf/f;Gα11−/−;Cre+ mice. Nevertheless, this residual up-regulation detected in Gαqf/f;Gα11−/−;Cre+ mice does not appear to be physiologically significant because these mice experience a delay or failure of parturition [4]. When grouped by the magnitude of the difference in expression, the other up-regulated genes code for proteins involved in a variety of cellular functions, including components of the extracellular matrix (Tnc), control of the cell cycle (Cdkn2b), solute transport (Slc46a3), and nicotinic receptor signaling (Chrna1). When grouped by their presumed involvement in cellular functions, they segregate into some categories, such as focal adhesion, extracellular matrix, endocytosis, and cytokine signaling, that may be involved in the regression of the corpus luteum and other ovarian remodeling that occurs at the end of gestation and the reentry into the estrous cycle. Although the “metabolic pathway” KEGG grouping has the largest number of genes and the highest level of statistical significance we chose not to further consider genes belonging to this KEGG grouping because of its broadness. We instead validated the microarray results of the up-regulated genes with Tnc and Jun, two up-regulated genes placed in the focal adhesion KEGG pathway. Tnc codes for tenascin C (Tn-C), a nonstructural extracellular matrix protein that is highly up-regulated during active tissue remodeling [42]. Tn-C is expressed in several ovarian compartments, including degenerating luteal cells [43–45]. Jun was also placed in this KEGG pathway, but because it is a component of the AP-1 transcription factor [46], it can modulate the expression of many genes and participate in many pathways. Jun is present in the porcine corpus luteum, and under some conditions it can be induced by PGF2α [47].
Serpina6 and Enpp2 are the genes that exhibit the largest Gαq/11-dependent decrease in expression at the end of gestation. The finding that these two genes are expressed in the ovary and down-regulated at the end of gestation is novel and could have potential implications in reproductive functions. Serpina6 codes for corticosteroid-binding globulin (CBG), a circulating protein that binds cortisol and progesterone [48]. In the rat, the liver is the main source of circulating CBG, and its expression can be stimulated with estrogens [49, 50]. Circulating CBG levels increase during the second and third trimesters in pregnant women and influence progesterone levels in the blood and at the maternal-fetal interface [51, 52]. Our data show that Serpina6 is expressed in the pregnant mouse ovary and that it declines at the end of gestation in a Gαq/11-dependent fashion. The functions of ovarian Serpina6 and its possible contribution to circulating CBG remain to be determined, however. Enpp2 codes for autotaxin [53–55], an extracellular enzyme that synthesizes lysophosphatidic acid (LPA). Lysophosphatidic acid is an extracellular phospholipid that activates many cellular functions by engaging one or more of the six known LPA receptors [8, 9], but its role in reproduction in general and ovarian physiology in particular are poorly understood [9]. The human ovary was initially reported as a site of high Enpp2 expression [56], but this was not confirmed by the expression data available on public databases [57]. Autotaxin expression was shown in the rat corpus luteum but not in developing or mature follicles [58]. In addition, LPA is present in very high concentrations in human follicular fluid isolated from women undergoing in vitro fertilization, and circulating LPA as well as circulating autotaxin increase during human pregnancy [59, 60]. Lastly, several LPA receptor subtypes are expressed in the ovary [9, 58], and addition of LPA to luteal cells impacts steroid and interleukin biosynthesis [61, 62]. When considered together, these data suggest that ovarian-derived LPA is an autocrine/paracrine regulator of ovarian functions. This latter possibility is currently being investigated.
When grouped by their presumed involvement in cellular functions (Table 4), the genes that are down-regulated at the end of gestation in a Gαq/11-dependent fashion segregate into several KEGG pathways, including three—“protein processing in endoplasmic reticulum,” “N-glycan biosynthesis,” and “protein export”—that are involved in protein synthesis, processing, and glycosylation. We validated the microarray results of the down-regulated genes with Wfs1 and Ssr4, two down-regulated genes placed in the “protein processing in endoplasmic reticulum” KEGG pathway, the down-regulated pathway with the highest level of statistical significance. Wfs1 codes for an endoplasmic reticulum protein called WFS1 that is involved in calcium homeostasis and the unfolded protein response. In humans, mutations of the WFS1 gene result in a disorder (called Wolfram syndrome) characterized by neurodegeneration and diabetes mellitus [63]. Ssr4 codes for SSR4, one of the four signal sequence receptor proteins that are part of a complex of membrane proteins needed for N-glycosylation [64] and to translocate secreted proteins across the endoplasmic reticulum membrane [65]. There is no available information about the expression or possible functions of these two genes in the ovary, but because of the secretory nature of luteal cells [66], the down-regulation of these ovarian genes could also be involved in the transition that occurs as the mice resume the estrous cycle after delivery.
The group of down-regulated genes in the “steroid hormone biosynthesis” KEGG pathway is also of interest because it contains the three enzymes required to convert progesterone to estradiol (Cyp17a1, Hsd17b7, and Cyp19a1). These three genes, as well as two other ovarian genes known to be involved in reproductive functions (Lhcgr and Inha), are also among the genes with the largest Gαq/11-dependent decrease in expression at the end of gestation. Thus, we also validated the microarray results with these five additional genes. Two of these (Cyp19a1 and Hsd17b7) were previously shown to decrease at the end of gestation [30, 67], but our finding that they are dependent on Gαq/11 is new. The changes in expression of these five reproductive genes have clear physiological correlates. The decrease in Lhcgr reduces the responsiveness of luteal cells to hCG, the decrease in Inha results in a reduction in the circulating levels of Inhibin A, and the decrease in the three steroidogenic enzymes is associated with a decrease in circulating estradiol levels. We hypothesize that the Gαq/11-dependent decrease in expression of Lhcgr, Cyp17a1, Cyp19a1, and Hsd17b7, and the increase in expression of Akr1c18 coordinate the drop in circulating progesterone and estradiol that occurs at the end of gestation [4, 25–29, 39–41]. The decrease in Lhcgr reduces the stimulus for progesterone synthesis, and the increase in Akr1c18 enhances progesterone metabolism. Because the magnitude of progesterone production is several orders of magnitude higher than that of estradiol (ng/ml levels of circulating progesterone and pg/ml levels for circulating estradiol), a decline in progesterone production alone (such as that induced by the changes in Lhcgr and Akr1c18) would have to be very large to translate into a significant drop in estradiol production. Thus, the additional decrease in expression of Cyp17a1, Cyp19a1, and Hsd17b7 is likely to contribute to a reduction in estradiol synthesis even if the substrate (progesterone) was still relatively high.
The decreased expression of Inha and circulating Inhibin A predicts that follicle-stimulating hormone (FSH) levels will rebound at the end of gestation in the Gαqf/f;Gα11−/−;Cre− mice but remain low in the Gαqf/f;Gα11−/−;Cre+ mice. We attempted to test this hypothesis by measuring FSH in both groups of mice on 22 dpc, but most of the samples collected had undetectable levels of FSH.
The failure of parturition and lack of progesterone withdrawal observed in the Ptgfr null mice shows that activation of the luteal PGF2α receptor is the trigger for parturition in mice (reviewed in Davis and Rueda [25], Ratajczak and Muglia [39], Stocco et al. [40], and Henkes et al. [41]). Because we have recently shown that the phenotype of Gαqf/f;Gα11−/−;Cre+ mice is very similar to that of the Ptgfr null mice, and because luteal cells derived from Gαqf/f;Gα11−/−;Cre+ mice fail to respond to PGF2α with an increase in Akr1c18 expression, we concluded that the effects of the luteal PGF2α receptor on the induction of Akr1c18 are mediated by the activation of Gq/11 [4]. These results, together with the Gαq/11 dependency of the changes in gene expression that occur at the end of gestation, suggest that they are induced by PGF2α. Indeed, (in addition to Akr1c18) some of the genes identified here have already been shown to be targets of PGF2α. The decreased expression of ovarian Hsd17b7 and Cyp19a1 that occurs at the end of gestation was shown to be blunted in Ptgfr null mice [67], but the data reported here show for the first time that this effect is dependent on the granulosa/luteal Gαq/11. Also, a previous study has shown that administration of PGF2α to pseudopregnant rats decreases the binding of 125I-hCG to the corpus luteum and the ability of hCG to stimulate progesterone synthesis [68]. Our findings that Lhcgr declines at the end of gestation in a Gαq/11-dependent fashion are also new. When we tested the effects of a PGF2α agonist on the expression of 11 of the Gαq/11-dependent genes characterized here, we found that only 7 of them (Jun, Serpina6, Ssr4, Enpp2, Lhcgr, Hsd17b7, and Cyp19a1) are regulated by PGF2α, whereas the other 4 (Tnc, Wfs1, Cyp17a1, and Inha) are not. It is possible, however, that some of the PGF2-insensitive mRNAs have a relatively long half-life, and therefore changes in expression may not be detectable within 24 h. In contrast, the microarray and the qPCR experiments done at the end of gestation were done by comparing animals on 18 and 22 dpc (a 96-h difference). Another possibility is that the expression of these genes is dependent on the actions of endocrine or paracrine/autocrine hormones that also activate luteal Gq/11 (such as oxytocin, see Blanks and Thornton [6] and Mitchell et al. [7]) or LPA [8, 9]. When engaged by oxytocin, the oxytocin receptor is known to activate Gq/11 [6, 7], and oxytocin has been shown to induce progesterone withdrawal and parturition in Ptgr null mice [69]. The actions of LPA in the corpus luteum need to be characterized, but some of the LPA receptors are known to activate Gq/11 [8, 9]. The changes in the expression of Gαq/11-dependent but PGF2α-independent genes may also not be a direct effect of the activation of Gq/11. It may be secondary to the decrease in estradiol levels that occurs as a consequence of Gq/11 activation. Lastly, the dependency of luteal Cyp17a1 expression on Gαq/11 is also curious because Cyp17a1 is expressed in theca cells, not granulosa cells [70], and the deletion of Gαq/11 took place initially in granulosa cells [3, 4]. We therefore hypothesize that the regulation of luteal Cyp17a1 expression is due to Gαq/11-dependent changes in a granulosa/luteal cell-specific product that acts on theca/luteal cells to inhibit Cyp17a1 expression.
In summary, the results presented here have identified 138 and 150 ovarian genes that are up-regulated or down-regulated, respectively, at the end of gestation in a Gαq/11-dependent fashion. Genes that participate in the structure/function of the extracellular matrix are among those that are up-regulated, whereas some of the down-regulated genes participate in the processing and transport of proteins in the endoplasmic reticulum. Several ovarian genes known to be involved in reproductive functions and two novel ovarian genes that may participate in reproductive functions were also down-regulated in a Gαq/11-dependent fashion. These changes in ovarian gene expression are likely to be involved in the transition that occurs at the end of gestation and in the reentry into the estrous cycle. PGF2α, acting thought the luteal prostaglandin F2α receptor, a Gq/11-coupled receptor, is a prominent regulator of luteolysis, but not all of the genes that are regulated at the end of gestation in a Gαq/11-dependent fashion are targets of PGF2α
Supplementary Material
ACKNOWLEDGMENT
The estradiol and Inhibin A assays were performed by the Ligand Assay and Analysis Core of the University of Virginia.
Footnotes
R.M. was partially supported by a K12 grant from the National Institutes of Health (NIH; HD063117). This core is supported by NIH grant P50-HD28934.
These authors contributed equally to this work.
REFERENCES
- Richards JS, Liu Z, Shimada M. Plant TM, Zeleznik AJ, editors. Knobil and Neill's Physiology of Reproduction, vol. 1, 4th ed. 2015:997–1021. Ovulation In: (eds.) San Diego, CA: Academic Press; [Google Scholar]
- Conti M, Hsieh M. Musa Zamah A, Oh JS. Novel signaling mechanisms in the ovary during oocyte maturation and ovulation. Mol Cell Endocrinol. 2012;356:65–73. doi: 10.1016/j.mce.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen SM, Andric N, Ping T, Xie F, Offermans S, Gossen JA, Ascoli M. Ovulation involves the luteinizing hormone-dependent activation of Gq/11 in granulosa cells. Mol Endocrinol. 2013;27:1483–1491. doi: 10.1210/me.2013-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia R, Waite C, Ascoli M. Activation of Gq/11 in the mouse corpus luteum is required for parturition. Mol Endocrinol. 2015;29:238–246. doi: 10.1210/me.2014-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goupil E, Tassy D, Bourguet C, Quiniou C, Wisehart V, Petrin D, Le Gouill C, Devost D, Zingg HH, Bouvier M, Saragovi HU, Chemtob S, et al. A novel biased allosteric compound inhibitor of parturition selectively impedes the prostaglandin F2alpha-mediated Rho/ROCK signaling pathway. J Biol Chem. 2010;285:25624–25636. doi: 10.1074/jbc.M110.115196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanks AM, Thornton S. The role of oxytocin in parturition. BJOG. 2003;110((suppl 20)):46–51. doi: 10.1016/s1470-0328(03)00024-7. [DOI] [PubMed] [Google Scholar]
- Mitchell BF, Fang X, Wong S. Oxytocin: a paracrine hormone in the regulation of parturition? Rev Reprod. 1998;3:113–122. doi: 10.1530/ror.0.0030113. [DOI] [PubMed] [Google Scholar]
- Yung YC, Stoddard NC, Chun J. LPA receptor signaling: pharmacology, physiology, and pathophysiology. J Lipid Res. 2014;55:1192–1214. doi: 10.1194/jlr.R046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Chun J. Lysophosphatidic acid (LPA) signaling in vertebrate reproduction. Trends Endocrinol Metab. 2010;21:17–24. doi: 10.1016/j.tem.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermanns S, Zhao LP, Gohla A, Sarosi I, Simon MI, Wilkie TM. Embryonic cardiomyocyte hypoplasia and craniofacial defects in Galpha q/Galpha 11-mutant mice. EMBO J. 1998;17:4304–4312. doi: 10.1093/emboj/17.15.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettschureck N, Lee E, Libutti SK, Offermanns S, Robey PG, Spiegel AM. Parathyroid-specific double knockout of Gq and G11 alpha-subunits leads to a phenotype resembling germline knockout of the extracellular Ca2+-sensing receptor. Mol Endocrinol. 2007;21:274–280. doi: 10.1210/me.2006-0110. [DOI] [PubMed] [Google Scholar]
- Kero J, Ahmed K, Wettschureck N, Tunaru S, Wintermantel T, Greiner E, Schutz G, Offermanns S. Thyrocyte-specific Gq/G11 deficiency impairs thyroid function and prevents goiter development. J Clin Invest. 2007;117:2399–2407. doi: 10.1172/JCI30380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Liu Z, Shimada M, Sterneck E, Johnson PF, Hedrick SM, Richards JS. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science. 2009;324:938–941. doi: 10.1126/science.1171396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Shimada M, Liu Z, Cahill N, Noma N, Wu Y, Gossen J, Richards J. Selective expression of KrasG12D in granulosa cells of the mouse ovary causes defects in follicle development and ovulation. Development. 2008;135:2127–2137. doi: 10.1242/dev.020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallares P, Gonzalez-Bulnes A. A new method for induction and synchronization of oestrus and fertile ovulations in mice by using exogenous hormones. Lab Anim. 2009;43:295–299. doi: 10.1258/la.2008.008056. [DOI] [PubMed] [Google Scholar]
- Dukes M, Russell W, Walpole AL. Potent luteolytic agents related to prostaglandin F2alpha. Nature. 1974;250:330–331. doi: 10.1038/250330a0. [DOI] [PubMed] [Google Scholar]
- Donadeu FX, Ascoli M. The differential effects of the gonadotropin receptors on aromatase expression in primary cultures of immature rat granulosa cells are highly dependent on the density of receptors expressed and the activation of the inositol phosphate cascade. Endocrinology. 2005;146:3907–3916. doi: 10.1210/en.2005-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipkin RW, Liu X, Ascoli M. Truncation of the C-terminal tail of the follitropin (FSH) receptor does not impair the agonist- or phorbol ester-induced receptor phosphorylation and uncoupling. J Biol Chem. 1995;270:26683–26689. doi: 10.1074/jbc.270.44.26683. [DOI] [PubMed] [Google Scholar]
- Hipkin RW, Wang Z, Ascoli M. Human chorionic gonadotropin- and phorbol ester stimulated phosphorylation of the LH/CG receptor maps to serines 635, 639, 645 and 652 in the C-terminal cytoplasmic tail. Mol Endocrinol. 1995;9:151–158. doi: 10.1210/mend.9.2.7776965. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Anderson LE, Wu YL, Tsai SJ, Wiltbank MC. Prostaglandin F(2alpha) receptor in the corpus luteum: recent information on the gene, messenger ribonucleic acid, and protein. Biol Reprod. 2001;64:1041–1047. doi: 10.1095/biolreprod64.4.1041. [DOI] [PubMed] [Google Scholar]
- Sugatani J, Masu Y, Nishizawa M, Sakamoto K, Houtani T, Sugimoto T, Ito S. Hormonal regulation of prostaglandin F2 alpha receptor gene expression in mouse ovary. Am J Physiol. 1996;271:E686–E693. doi: 10.1152/ajpendo.1996.271.4.E686. [DOI] [PubMed] [Google Scholar]
- Hasumoto K, Sugimoto Y, Yamasaki A, Morimoto K, Kakizuka A, Negishi M, Ichikawa A. Association of expression of mRNA encoding the PGF2 alpha receptor with luteal cell apoptosis in ovaries of pseudopregnant mice. J Reprod Fertil. 1997;109:45–51. doi: 10.1530/jrf.0.1090045. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Hasumoto K, Namba T, Irie A, Katsuyama M, Negishi M, Kakizuka A, Narumiya S, Ichikawa A. Cloning and expression of a cDNA for mouse prostaglandin F receptor. J Biol Chem. 1994;269:1356–1360. [PubMed] [Google Scholar]
- Davis JS, Rueda BR. The corpus luteum: an ovarian structure with maternal instincts and suicidal tendencies. Front Biosci. 2002;7:d1949–1978. doi: 10.2741/davis1. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Yamasaki A, Segi E, Tsuboi K, Aze Y, Nishimura T, Oida H, Yoshida N, Tanaka T, Katsuyama M, Hasumoto K, Murata T, et al. Failure of parturition in mice lacking the prostaglandin F receptor. Science. 1997;277:681–683. doi: 10.1126/science.277.5326.681. [DOI] [PubMed] [Google Scholar]
- Stocco CO, Zhong L, Sugimoto Y, Ichikawa A, Lau LF, Gibori G. Prostaglandin F2alpha-induced expression of 20alpha-hydroxysteroid dehydrogenase involves the transcription factor NUR77. J Biol Chem. 2000;275:37202–37211. doi: 10.1074/jbc.M006016200. [DOI] [PubMed] [Google Scholar]
- Stocco CO, Lau LF, Gibori G. A calcium/calmodulin-dependent activation of ERK1/2 mediates JunD phosphorylation and induction of nur77 and 20alpha-hsd genes by prostaglandin F2alpha in ovarian cells. J Biol Chem. 2002;277:3293–3302. doi: 10.1074/jbc.M110936200. [DOI] [PubMed] [Google Scholar]
- Piekorz RP, Gingras S, Hoffmeyer A, Ihle JN, Weinstein Y. Regulation of progesterone levels during pregnancy and parturition by signal transducer and activator of transcription 5 and 20alpha-hydroxysteroid dehydrogenase. Mol Endocrinol. 2005;19:431–440. doi: 10.1210/me.2004-0302. [DOI] [PubMed] [Google Scholar]
- Stocco C. In vivo and in vitro inhibition of cyp19 gene expression by prostaglandin F2alpha in murine luteal cells: implication of GATA-4. Endocrinology. 2004;145:4957–4966. doi: 10.1210/en.2004-0625. [DOI] [PubMed] [Google Scholar]
- Bast JD, Melampy RM. Luteinizing hormone, prolactin and ovarian 20 -hydroxysteroid dehydrogenase levels during pregnancy and pseudopregnancy in the rat. Endocrinology. 1972;91:1499–1505. doi: 10.1210/endo-91-6-1499. [DOI] [PubMed] [Google Scholar]
- Akinola LA, Poutanen M, Vihko R, Vihko P. Expression of 17beta-hydroxysteroid dehydrogenase type 1 and type 2, P450 aromatase, and 20alpha-hydroxysteroid dehydrogenase enzymes in immature, mature, and pregnant rats. Endocrinology. 1997;138:2886–2892. doi: 10.1210/endo.138.7.5258. [DOI] [PubMed] [Google Scholar]
- Albarracin CT, Parmer TG, Duan WR, Nelson SE, Gibori G. Identification of a major prolactin-regulated protein as 20 alpha-hydroxysteroid dehydrogenase: coordinate regulation of its activity, protein content, and messenger ribonucleic acid expression. Endocrinology. 1994;134:2453–2460. doi: 10.1210/endo.134.6.8194472. [DOI] [PubMed] [Google Scholar]
- Richards JS, Williams JJ. Luteal cell receptor content for prolactin (PRL) and luteinizing hormone (LH): regulation by LH and Endocrinology PRL. 1976. 99 1571 1581 [DOI] [PubMed] [Google Scholar]
- Bachelot A, Beaufaron J, Servel N, Kedzia C, Monget P, Kelly PA, Gibori G, Binart N. Prolactin independent rescue of mouse corpus luteum life span: identification of prolactin and luteinizing hormone target genes. Am J Physiol Endocrinol Metab. 2009;297:E676–E684. doi: 10.1152/ajpendo.91020.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdemouge I, Bachelot A, Lucas A, Baran N, Kelly PA, Binart N. Effects of deletion of the prolactin receptor on ovarian gene expression. Reprod Biol Endocrinol. 2003;1:12. doi: 10.1186/1477-7827-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- Ratajczak CK, Muglia LJ. Insights into parturition biology from genetically altered mice. Pediatr Res. 2008;64:581–589. doi: 10.1203/PDR.0b013e31818718d2. [DOI] [PubMed] [Google Scholar]
- Stocco C, Telleria C, Gibori G. The molecular control of corpus luteum formation, function, and regression. Endocr Rev. 2007;28:117–149. doi: 10.1210/er.2006-0022. [DOI] [PubMed] [Google Scholar]
- Henkes LE, Davis JS, Rueda BR. Mutant mouse models and their contribution to our knowledge of corpus luteum development, function and regression. Reprod Biol Endocrinol. 2003;1:87. doi: 10.1186/1477-7827-1-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka-Yoshida K, Aoki H. Tenascin-C and mechanotransduction in the development and diseases of cardiovascular system. Front Physiol. 2014;5:283. doi: 10.3389/fphys.2014.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagavandoss P. Temporal expression of tenascin-C and type I collagen in response to gonadotropins in the immature rat ovary. Acta Histochem. 2014;116:1125–1133. doi: 10.1016/j.acthis.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Hagiwara E, Takeuchi A, Mukai C, Matsui C, Sakai A, Tamotsu S. Changes in the distribution of tenascin and fibronectin in the mouse ovary during folliculogenesis, atresia, corpus luteum formation and luteolysis. Zoolog Sci. 2005;22:237–245. doi: 10.2108/zsj.22.237. [DOI] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol Endocrinol. 2006;20:1300–1321. doi: 10.1210/me.2005-0420. [DOI] [PubMed] [Google Scholar]
- Shaulian E. AP-1–the Jun proteins: oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22:894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Diaz FJ, Luo W, Wiltbank MC. Prostaglandin F2alpha regulation of mRNA for activating protein 1 transcriptional factors in porcine corpora lutea (CL): lack of induction of JUN and JUND in CL without luteolytic capacity. Domest Anim Endocrinol. 2013;44:98–108. doi: 10.1016/j.domaniend.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HY, Muller YA, Hammond GL. Molecular and structural basis of steroid hormone binding and release from corticosteroid-binding globulin. Mol Cell Endocrinol. 2010;316:3–12. doi: 10.1016/j.mce.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Feldman D, Mondon CE, Horner JA, Weiser JN. Glucocorticoid and estrogen regulation of corticosteroid-binding globulin production by rat liver. Am J Physiol. 1979;237:E493–E499. doi: 10.1152/ajpendo.1979.237.6.E493. [DOI] [PubMed] [Google Scholar]
- Smith CL, Hammond GL. Hormonal regulation of corticosteroid-binding globulin biosynthesis in the male rat. Endocrinology. 1992;130:2245–2251. doi: 10.1210/endo.130.4.1547738. [DOI] [PubMed] [Google Scholar]
- O'Leary P, Boyne P, Flett P, Beilby J, James I. Longitudinal assessment of changes in reproductive hormones during normal pregnancy. Clin Chem. 1991;37:667–672. [PubMed] [Google Scholar]
- Lei JH, Yang X, Peng S, Li Y, Underhill C, Zhu C, Lin HY, Wang H, Hammond GL. Impact of corticosteroid-binding globulin deficiency on pregnancy and neonatal sex. J Clin Endocrinol Metab. 2015;100:1819–1827. doi: 10.1210/jc.2014-4254. [DOI] [PubMed] [Google Scholar]
- Barbayianni E, Kaffe E, Aidinis V, Kokotos G. Autotaxin, a secreted lysophospholipase D, as a promising therapeutic target in chronic inflammation and cancer. Prog Lipid Res. 2015;58:76–96. doi: 10.1016/j.plipres.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Moolenaar WH. Autotaxin: structure-function and signaling. J Lipid Res. 2014;55:1010–1018. doi: 10.1194/jlr.R046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico L, Jeong KJ, Vellano CP, Mills GB. Autotaxin, a lysophospholipase D with pleomorphic effects in oncogenesis and cancer progression. J Lipid Res. 2016;57:25–35. doi: 10.1194/jlr.R060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Murata J, Clair T, Polymeropoulos MH, Torres R, Manrow RE, Liotta LA, Stracke ML. Cloning, chromosomal localization, and tissue expression of autotaxin from human teratocarcinoma cells. Biochem Biophys Res Commun. 1996;218:714–719. doi: 10.1006/bbrc.1996.0127. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge C, Haase J, Janes J, Huss J, Su A. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Haruta S, Orino K, Kawaminami M, Kurusu S. Autotaxin as a novel, tissue-remodeling-related factor in regressing corpora lutea of cycling rats. FEBS J. 2013;280:6600–6612. doi: 10.1111/febs.12565. [DOI] [PubMed] [Google Scholar]
- Tokumura A, Miyake M, Nishioka Y, Yamano S, Aono T, Fukuzawa K. Production of lysophosphatidic acids by lysophospholipase D in human follicular fluids of In vitro fertilization patients. Biol Reprod. 1999;61:195–199. doi: 10.1095/biolreprod61.1.195. [DOI] [PubMed] [Google Scholar]
- Tokumura A, Kanaya Y, Miyake M, Yamano S, Irahara M, Fukuzawa K. Increased production of bioactive lysophosphatidic acid by serum lysophospholipase D in human pregnancy. Biol Reprod. 2002;67:1386–1392. doi: 10.1095/biolreprod.102.004051. [DOI] [PubMed] [Google Scholar]
- Chen SU, Chou CH, Lee H, Ho CH, Lin CW, Yang YS. Lysophosphatidic acid up-regulates expression of interleukin-8 and −6 in granulosa-lutein cells through its receptors and nuclear factor-kappaB dependent pathways: implications for angiogenesis of corpus luteum and ovarian hyperstimulation syndrome. J Clin Endocrinol Metab. 2008;93:935–943. doi: 10.1210/jc.2007-1512. [DOI] [PubMed] [Google Scholar]
- Budnik LT, Brunswig-Spickenheier B. Differential effects of lysolipids on steroid synthesis in cells expressing endogenous LPA2 receptor. J Lipid Res. 2005;46:930–941. doi: 10.1194/jlr.M400423-JLR200. [DOI] [PubMed] [Google Scholar]
- Inoue H, Tanizawa Y, Wasson J, Behn P, Kalidas K, Bernal-Mizrachi E, Mueckler M, Marshall H, Donis-Keller H, Crock P, Rogers D, Mikuni M, et al. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome) Nat Genet. 1998;20:143–148. doi: 10.1038/2441. [DOI] [PubMed] [Google Scholar]
- Losfeld ME, Ng BG, Kircher M, Buckingham KJ, Turner EH, Eroshkin A, Smith JD, Shendure J, Nickerson DA, Bamshad MJ; University of Washington Center for Mendelian Genomics, Freeze HH. A new congenital disorder of glycosylation caused by a mutation in SSR4, the signal sequence receptor 4 protein of the TRAP complex. Hum Mol Genet. 2014;23:1602–1605. doi: 10.1093/hmg/ddt550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E, Gorlich D, Kostka S, Otto A, Kraft R, Knespel S, Burger E, Rapoport TA, Prehn S. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur J Biochem. 1993;214:375–381. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- Stouffer RL, Hennebold JD. Structure, function, and regulation of the corpus luteum. Plant TM, Zeleznik AJ, editors. Knobil and Neill's Physiology of Reproduction, vol. 1, 4th ed. 2015:1023–1076. In: (eds.) San Diego, CA: Academic Press; [Google Scholar]
- Foyouzi N, Cai Z, Sugimoto Y, Stocco C. Changes in the expression of steroidogenic and antioxidant genes in the mouse corpus luteum during luteolysis. Biol Reprod. 2005;72:1134–1141. doi: 10.1095/biolreprod.104.037598. [DOI] [PubMed] [Google Scholar]
- Grinwich DL, Hichens M, Behrman HR. Control of the LH receptor by prolactin and prostaglandin F2alpha in rat corpora lutea. Biol Reprod. 1976;14:212–218. doi: 10.1095/biolreprod14.2.212. [DOI] [PubMed] [Google Scholar]
- Kawamata M, Yoshida M, Sugimoto Y, Kimura T, Tonomura Y, Takayanagi Y, Yanagisawa T, Nishimori K. Infusion of oxytocin induces successful delivery in prostanoid FP-receptor-deficient mice. Mol Cell Endocrinol. 2008;283:32–37. doi: 10.1016/j.mce.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Auchus RJ. Human steroid biosynthesis. Plant TM, Zeleznik AJ, editors. Knobil and Neill's Physiology of Reproduction, 4th ed. 2015:295–312. In: (eds.) San Diego, CA: Academic Press; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.