Abstract
Repair of DNA double strand breaks (DSBs) by homologous recombination (HR) is critical for survival and genome stability of individual cells and organisms, but also contributes to the genetic diversity of species. A critical step in HR is MRN/CtIP-dependent end-resection that generates the 3′ single-stranded DNA overhangs required for the subsequent strand exchange reaction. Here, we identify EXD2 (EXDL2) as an exonuclease essential for DSB resection and efficient HR. EXD2 is recruited to chromatin in a damage-dependent manner and confers resistance to DSB-inducing agents. EXD2 functionally interacts with the MRN-complex to accelerate resection via its 3′-5′ exonuclease activity that efficiently processes dsDNA substrates containing nicks. Finally, we establish that EXD2 stimulates both short and long-range DSB resection, and thus together with MRE11 is required for efficient HR. This establishes a key role for EXD2 in controlling the initial steps of chromosomal break repair.
DNA double-strand breaks (DSBs) are extremely cytotoxic lesions that can arise during normal cellular processes or are induced by exogenous factors such as ionizing radiation as well as many commonly used anti-cancer drugs. The faithful repair of DSBs is essential for cell survival and organismal development, as defective repair can contribute to a plethora of inherited human syndromes with life-threatening symptoms including cancer, neurodegeneration or premature aging 1, 2. The two major pathways involved in the repair of DSBs in eukaryotic cells are non-homologous end-joining and homologous recombination (HR) 3-5. A key initial step in HR is resection of the DNA ends on either side of the break, which is carried out initially by the MRE11-RAD50-NBS1 complex (MRN) and CtIP to generate short stretches of ssDNA6-8. Subsequently, the EXO1 or DNA2 nucleases, in conjunction with the Bloom’s Syndrome helicase (BLM) extend these to generate longer 3′ ssDNA tails 9-15. These ssDNA strands are then bound by replication protein A (RPA) 10-12, 16-18 which is subsequently replaced by RAD51 in a BRCA2-dependent manner, leading to the formation of ssDNA-RAD51 nucleoprotein filaments essential for the strand exchange process 3, 19. In vitro, MRE11 displays a weak endo- and exonuclease activity, which may be due to the lack of accessory factors 16, 20. Accordingly, work from multiple laboratories has shown that CtIP, or its yeast homologue Sae2, can stimulate MRE11’s endonuclease activity 9, 16-18. Interestingly, MRE11 has been also shown to nick the DNA strand to be resected in multiple positions, as far as 300bp from the break itself, suggesting that resection could proceed from several entry points that are distal to the DSB 21, 22. However, it is unclear whether this would enhance MRE11-dependent nucleolytic processing of DNA ends, thus generating a better substrate for subsequent processing of the break by BLM-DNA2 and/or BLM-EXO1 complexes; or allow access for additional factors accelerating the initial strand processing. Indeed, the inhibition of MRE11’s endonuclease activity confers a stronger resection defect than inhibition of its exonuclease activity, suggesting perhaps that initial break processing might be also carried out by other exonucleases 23. Here we identify EXD2 as a cofactor of the MRN complex required for efficient DNA end-resection, recruitment of RPA, homologous recombination and suppression of genome instability.
EXD2 is required for repair of damage to DNA
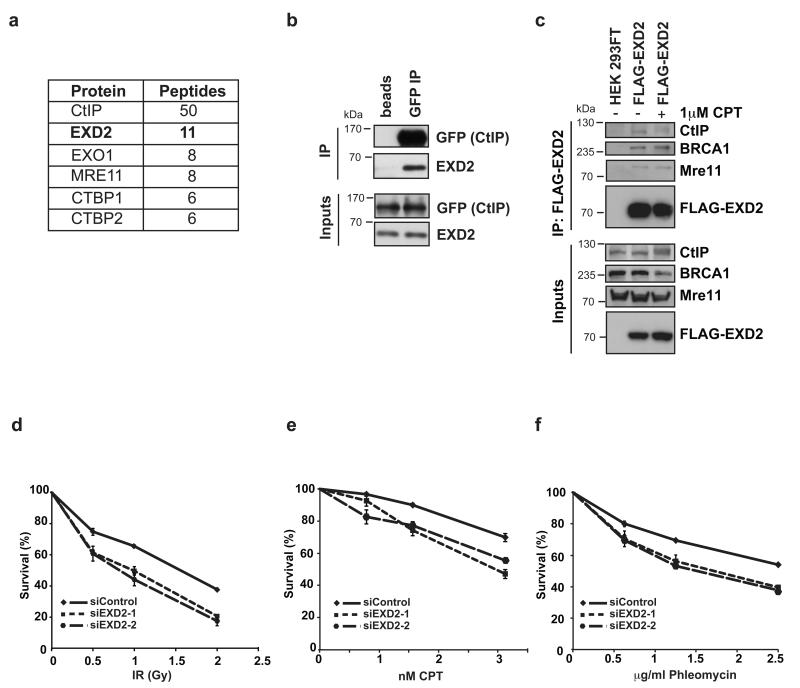
In an effort to identify factors required to promote HR, we carried out an unbiased proteomic approach to define the CtIP interactome. Here, we have identified EXD2, a largely uncharacterized protein with a putative exonuclease domain, as a candidate CtIP binding partner (Fig. 1a). We validated this interaction by co-immunoprecipitations from human cell extracts and found that we could readily detect endogenous EXD2 by western blotting of GFP-CtIP immunoprecipitates (Fig. 1b). Endogenous CtIP, as well as its known interactors MRE11 and BRCA1 were detected in a reciprocal FLAG-EXD2 immunoprecipitates (Fig. 1c; lysates were treated with benzonase to prevent DNA bridging). Therefore, we conclude that the two proteins likely exist in the same complex in cells.
Figure 1. EXD2 is a CtIP interactor and its depletion sensitizes cells to DNA damage.
a) Table showing proteins identified in IP/MS analysis of GFP-Trap purification from U2OS cells stably expressing GFP-CtIP.
b) Input (0.4% of total IP) and eluate fractions from immunoprecipitations of lysates prepared from U2OS cells stably expressing GFP-CtIP are shown with blocked agarose beads serving as a negative control (beads). Uncropped images of blots are shown in Supplementary Fig. 7. This experiment was carried out once as a validation of mass spectrometry data.
c) Input (0.4% of total IP) and eluate fractions of FLAG-HA-EXD2 pull downs from HEK293-FT cells transiently expressing this fusion protein treated with 1 μM CPT for 1h or left untreated as indicated. IP performed on mock-transfected HEK293FT cells serves as a negative control. This experiment was carried out two times independently. Uncropped images of blots are shown in Supplementary Fig. 7.
d, e and f) U2OS cells 72h post-transfection with control siRNA (siControl) or two independent siRNAs targeting EXD2 (siEXD2-1 or 2) were treated with the indicated doses of either ionizing radiation (IR), campthotecin (CPT) or phleomycin. Survival data represent mean +/− SEM, (n=3 independent experiments).
EXD2 is highly conserved across vertebrates (Supplementary Fig. 1) and was recently identified in the screen for suppression of sensitivity to mitomycin C 24. However, the biological and biochemical features of this protein are unknown. Since we identified EXD2 as an interactor of DBS-repair factors we tested its requirement in response to a range of DSBs-inducing agents namely, ionizing radiation (IR), campthotecin (CPT) and phleomycin. We found that depletion of EXD2 by two different siRNAs sensitized U2OS cells to these agents (Fig. 1d, e, f and Supplementary Fig. 2a and b). Taken together these results suggest a putative role for this protein in the repair of damaged DNA.
EXD2 promotes DNA end resection and the generation of ssDNA
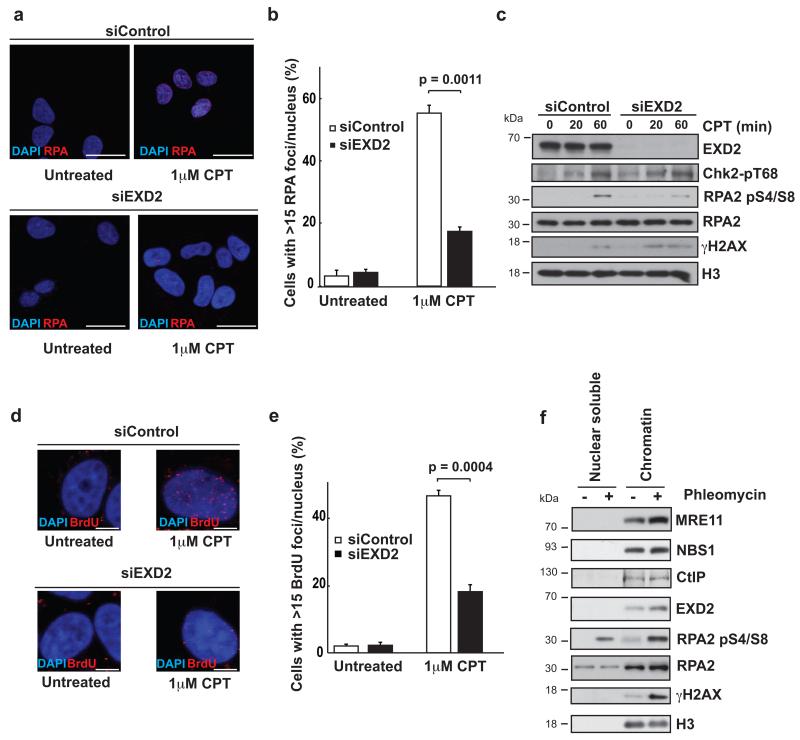
CtIP is essential for efficient DNA end processing during DSB repair, with cells depleted for this factor showing a defect in the generation of single stranded DNA (ssDNA) and the subsequent formation of RPA foci 16, 25, 26. Thus we hypothesized that EXD2 may promote DNA end resection. To test this, we analysed RPA focus formation in response to both CPT and IR in WT and EXD2-depleted cells. Strikingly, cells depleted for EXD2 showed severely impaired kinetics of RPA focus formation in response to both treatments (Fig. 2a and b and Supplementary Fig. 2c and d). RPA2 phosphorylation at S4 and S8 has been widely used as a marker for the generation of single-stranded DNA by DNA-end resection 27. Consistent with the data above, EXD2 depleted cells showed impaired RPA S4/S8 phosphorylation in response to DNA damage (Fig. 2c and Supplementary Fig. 2e). Treatment with both agents resulted in a robust phosphorylation of histone H2AX and CHK2 (Fig. 2c and Supplementary Fig. 2e), confirming induction of DNA damage in cells. Moreover, since these responses were intact in EXD2-depleted cells, EXD2 is most likely not required for initial sensing of the DNA damage. Failure to generate RPA foci could be associated with either a defect in exonucleolytic processing of the DSB into ssDNA, or with impaired recruitment of RPA itself to ssDNA. To distinguish between these two possibilities we tested the efficiency of ssDNA generation in EXD2-depleted cells exposed to DNA damage. To this end, we labelled cells with BrdU and then employed immunofluorescence microscopy using an anti-BrdU antibody under non-denaturing conditions to detect stretches of ssDNA. Depletion of EXD2 significantly reduced formation of ssDNA foci (Fig. 2d and e) suggesting impaired resection. This is reminiscent of the phenotype observed in cells depleted for essential components of DNA end resection machinery, such as MRE11 or CtIP 6, 16. Importantly, these phenotypes were not due to changes in the cell cycle, as EXD2-depletion had little effect on the overall cell-cycle distribution profile (Supplementary Fig. 2f). Despite multiple attempts, we were unable to visualise EXD2 recruitment to DNA damage foci. In this regard, we note that certain other proteins involved in DNA repair do not readily form cytologically discernible foci in mammalian cells i.e. Ku70, Ku80 or DNA2. However, EXD2 was recruited to chromatin in HeLa cells upon DNA damage as assayed by subcellular fractionation (Fig. 2f), supporting its putative role in the processing of DSBs. A similar recruitment was also observed for the key resection factor MRE11 (Fig. 2f). Taken together, these results indicate that EXD2 is a putative component of the resection machinery required for the efficient processing of DSBs.
Figure 2. EXD2 depletion impairs DNA end-resection following DSB induction.
a) Representative images of U2OS cells 72h post-transfection with control siRNA (siControl) or an siRNA oligo targeting EXD2 (siEXD2) treated with 1 μM CPT for 1h as indicated and stained for RPA and DAPI. Scale bar = 20μm.
b) Quantification of the percentage of U2OS cells treated as in (a). with greater than 15 RPA foci per nucleus. Bars represent +/− SEM. n=412 cells (siControl untreated), 358 cells (siEXD2 untreated), 396 cells (siControl 1μM CPT) and 403 cells (siEXD2 1μM CPT) respectively, grouped from three independent experiments. Statistical significance was determined by the Chi-square test.
c) Western blotting of various DDR proteins in U2OS cells 72h post-transfection with control siRNA (siControl) or an siRNA oligo targeting EXD2 (siEXD2) treated with 1 μM CPT for indicated amounts of time. Chk2-p T68 acts as a control for ATM activation, RPA2 pS4/S8 acts to indicate resection efficiency, γH2AX serves to indicate DSB induction with RPA and histone H3 acting as loading controls. This experiment was carried out two times independently. Uncropped images of blots are shown in Supplementary Fig. 7.
d) Representative images of U2OS cells 72h post-transfection with control siRNA (siControl) or an siRNA oligo targeting EXD2 (siEXD2) treated with 1 μM CPT for 1h as indicated and stained for BrdU and DAPI. BrdU staining was carried out under non-denaturing conditions, with foci indicating the presence of ssDNA. Scale bar = 5μm.
e) Quantification of the percentage of U2OS cells treated as in (d), exhibiting greater than 15 BrdU foci per nucleus. n=311 cells (siControl untreated), 300 cells (siEXD2 untreated), 300 cells (siControl 1μM CPT) and 300 cells (siEXD2 1μM CPT) respectively, grouped from three independent experiments. Error bars represent +/− SEM. The Chi-square test was used to determine statistical significance.
f) Chromatin fractionation of HeLa cells untreated or treated with 500 μM phleomycin for 1h as indicated. γH2AX and RPA2 pS4/S8 are used as markers of DNA damage and histone H3 (H3) acts as a loading control. This experiment was carried out two times independently. Uncropped images of blots are shown in Supplementary Fig. 7.
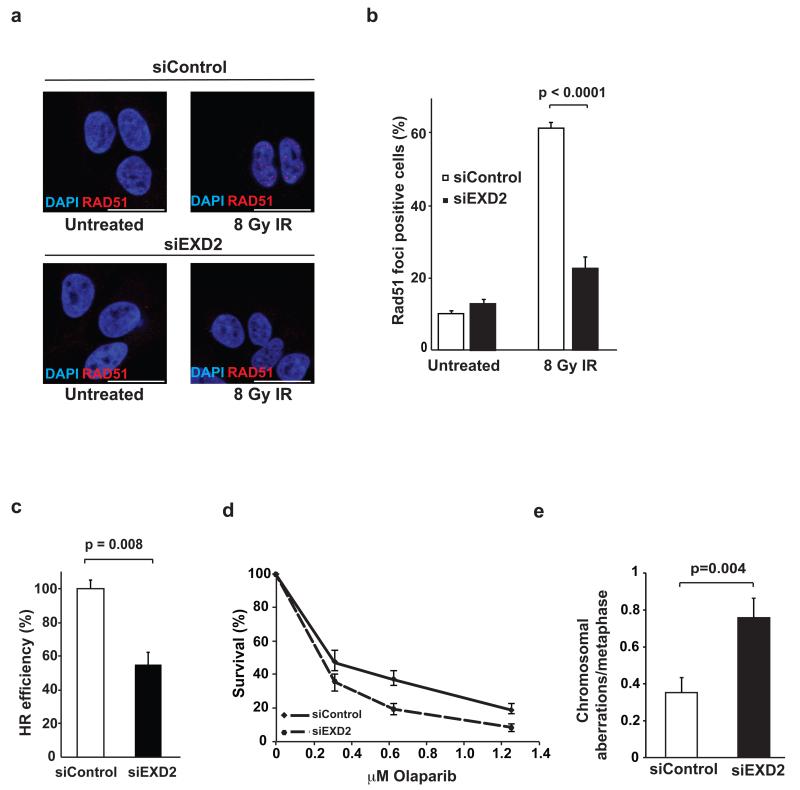
EXD2 promotes homologous recombination and suppresses genome instability
In vivo, RPA is required for RAD51 focus formation and in vitro it has been shown to promote RAD51-mediated strand exchange 28, 29. Consistent with this, treatment of U2OS cells with IR generated large numbers of RAD51 foci (Fig. 3a and b). In contrast, EXD2 depletion significantly impaired RAD51 focus formation (Fig. 3a and b). RAD51-ssDNA nucleoprotein filament formation is a crucial step in DSB repair by HR 30-32. To examine if EXD2 is also required for efficient HR we used a U2OS cell line carrying an integrated homologous recombination reporter transgene and an I-SceI recognition sequence 33. Transient expression of I-SceI endonuclease generates a DSB that, when repaired by HR, restores the expression of a functional GFP protein. We found that depletion of EXD2 significantly decreased the frequency of homologous recombination (Fig. 3c). Cells defective in HR are intrinsically sensitive to PARP inhibitors 34. Accordingly, EXD2-depletion sensitised cells to the PARP inhibitor olaparib (Fig. 3d). Failure to efficiently repair damaged DNA is associated with chromosomal instability and in line with this, we found a significantly greater number of chromosomal aberrations in EXD2-depleted U2OS cells as compared to the WT control (Fig. 3e). Taken together this data supports a role of EXD2 in promoting homologous recombination and genome stability.
Figure 3. EXD2 promotes homologous recombination and suppresses genome instability.
a) RAD51 foci in U2OS cells 72h following transfection with control siRNA (siControl) or an siRNA oligo targeting EXD2 (siEXD2). Cells were either untreated or were exposed to 8 Gy IR and left to recover for 6h prior to fixation and staining for RAD51 and DAPI as indicated. Scale bar = 20μm.
b) Quantification of the percentage of cells treated as in (a), exhibiting RAD51 foci; n=334 cells (siControl untreated), 368 cells (siEXD2 untreated), 378 cells (siControl 8Gy IR) and 376 cells (siEXD2 8gy IR) respectively, grouped from three independent experiments. Error bars represent +/− SEM. The Chi-square test was used to determine statistical significance.
c) Quantification of the relative efficiency of HR as measured by the DR-GFP assay (see methods for details) in cells treated with control siRNA (siControl) or an siRNA oligo targeting EXD2 (siEXD2). Efficiency of HR following transient expression of I-SceI enzyme was analysed by FACS and siEXD2 samples compared to the siControl (normalized to 100%). Data represent the mean +/− SEM (n=3 independent experiments). Statistical significance was determined using the student’s t-test.
d) siRNA-treatment was carried out using either control siRNA (siControl) or siRNA targeting EXD2 (siEXD2) in cells treated with the indicated doses of Olaparib. Survival data represent mean +/− SEM, (n=3 independent experiments).
e) Quantification of the frequency of chromosomal aberrations from mitotic spreads prepared from U2OS cells treated with control siRNA (siControl) or an siRNA smartpool targeting EXD2 (siEXD2). Error bars represent +/− SEM, n= 75metaphase spreads pooled from 3 independent experiments. Statistical significance was determined using the student’s t-test.
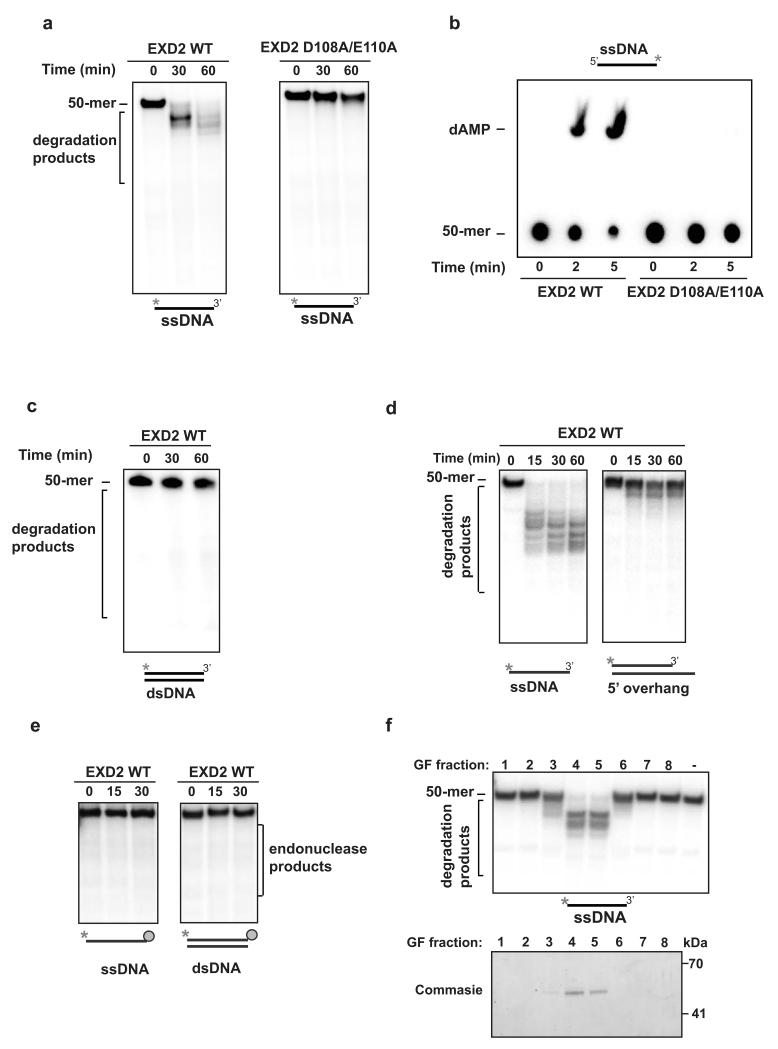
EXD2’s exonuclease activity facilitates the generation of ssDNA
The MRN complex processes DSBs to generate ssDNA, which requires MRE11’s 3′-5′ exonuclease activity 20, 35. Interestingly, EXD2 has a predicted exonuclease fold, which has sequence homology to the 3′-5′ exonuclease domain of the Werner syndrome protein (WRN). Analysis of the alignment between EXD2 and WRN identified two key amino acids (D108 and E110) within the putative exonuclease domain of EXD2, which are also highly conserved in other DnaQ type exonucleases, including WRN, that coordinate the binding of metal ions within the active site 36 (Supplementary Fig. 3a). Mutation of the equivalent residues in WRN (D82 and E84) renders the protein devoid of nuclease activity 37. Therefore, we hypothesized that the equivalent residues in EXD2 may be also required for its putative nuclease activity. To test this, we expressed the full length GST-tagged EXD2 and the D108A and E110A mutant protein in bacteria, and purified them to apparent homogeneity (Supplementary Fig. 3b). Next, we tested the activity of these purified proteins on single stranded DNA radiolabeled on the 3′ or 5′ end (Fig. 4a and b). We found that purified EXD2, but not the D108A and E110A mutant, exhibited a robust nuclease activity on short 5′ labeled ssDNA (Fig. 4a). Furthermore, a time course of the 3′ labelled substrate digestion indicates that EXD2 degrades the labelled DNA strand from the 3′ end, as evidenced by the release of the single labelled nucleotide (Fig. 4b). This data shows that EXD2 displays a 3′-5′ exonuclease activity in vitro. Moreover, under these conditions the WT protein exhibited only weak activity towards blunt end double stranded DNA (Fig. 4c). To verify this data, we also identified a highly-soluble truncated form of EXD2 (spanning residues lysine 76 through to valine 564, containing the predicted exonuclease domain) that can be produced at very high yields and purity in a three-step procedure (Supplementary Fig. 3c-e). This version of EXD2, and its D108A E110A variant, behaved indistinguishably from full-length EXD2 (Supplementary Fig. 3f). In addition, the protein showed only a weak activity towards dsDNA with resected 3’end (Fig. 4d), and did not display any endonuclease activity on ssDNA or dsDNA with biotin/streptavidin blocked 3′ end (Fig. 4e). Importantly, purified EXD2 displayed a robust exonuclease activity, which co-elutes with the protein (Fig. 4f). Thus our data identify EXD2 as a bone fide exonuclease with a 3′-5′ polarity.
Figure 4. EXD2 displays 3′ – 5′ exonuclease activity in vitro.
a) 5′ radiolabeled ssDNA 50-mer substrate (10 nM molecules) was incubated for the indicated amounts of time with EXD2 WT or EXD2 D108A E110A mutant protein (70 nM). Samples were resolved on a 20% TBE-Urea polyacrylamide gel and visualised by phosphorimaging. This experiment was carried out two times independently.
b) 3′ radiolabeled ssDNA 50-mer substrate (0.25 μM molecules) was incubated for the indicated amounts of time with EXD2 WT or EXD2 D108A E110A (70 nM) mutant protein. Samples were resolved by TLC in 1M sodium formate pH 3.4 and visualised by phosphorimaging. This experiment was carried out two times independently.
c) 5′ dsDNA 50-mer substrates (10 nM molecules) were incubated for the indicated amounts of time with EXD2 WT protein (70 nM). Samples were resolved on a 20% TBE-Urea polyacrylamide gel and visualised by phosphorimaging. This experiment was carried out two times independently.
d) 5′ radiolabeled ssDNA or dsDNA with 5’overhang substrate (3 nM molecules) was incubated for indicated amounts of time with EXD2 WT (K76 - V564) protein (25 nM). Samples were resolved on a 20% TBE-Urea polyacrylamide gel and visualised by phosphorimaging. This experiment was carried out two times independently.
e) 5′ radiolabeled ssDNA or dsDNA (3 nM molecules) with 3′ end blocked by biotin – streptavidin was incubated for indicated time with EXD2 WT (K76 - V564) protein (25 nM) in buffer supplemented with 1 mM ATP. Samples were resolved on a 20% TBE-Urea polyacrylamide gel and visualised by phosphorimaging. This experiment was carried out two times independently.
f) (upper panel) EXD2 WT (K76 - V564) gel-filtration fractions were tested for nuclease activity against 5′ radiolabeled ssDNA (10 nM molecules). Reactions were incubated for 30 min and resolved on a 15% TBE-Urea polyacrylamide gel and visualised by phosphorimaging; (lower panel) Coomassie blue–stained gel depicting the EXD2 protein in gel-filtration fractions analysed in the upper panel. This experiment was carried out two times independently.
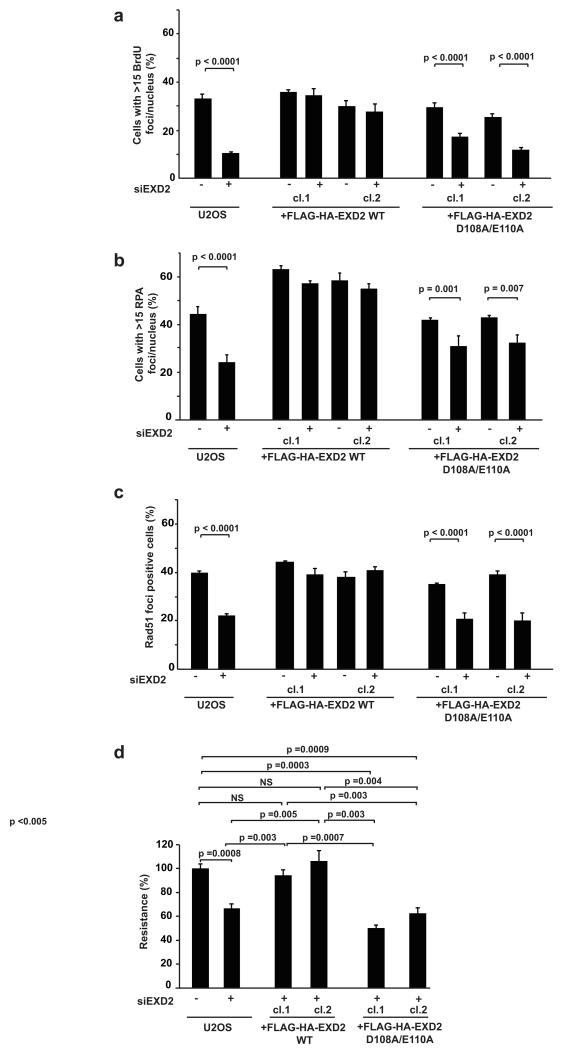
To address the potential biological significance of EXD2’s exonuclease activity, we tested whether this activity was required to promote DNA-end resection in vivo. To this end, we examined the phenotypes of two independently derived U2OS clones stably expressing wild-type or the nuclease-dead (D108A and E110A) EXD2 mutant. The endogenous protein was depleted with siRNA targeting the 3′ un-translated region (UTR) of EXD2 (Supplementary Fig. 4a). Notably, cells expressing the nuclease-dead protein did not correct phenotypes associated with EXD2 deficiency as compared to cells expressing wild-type EXD2 (Fig. 5a-d). This result is consistent with our data showing a requirement for EXD2 in the processing of DSBs into ssDNA. Cells overexpressing WT EXD2 displayed elevated resection. In this regard we note that overexpression of WT MRE11 also increases resection efficiency resulting in elevated levels of RPA foci formation and RPA phosphorylation 38.
Figure 5. EXD2’s nuclease activity is required for DSB repair in vivo.
a) U2OS control cells or cells stably expressing Flag–HA–EXD2 WT or D108A–E110A mutant fusion proteins 72 h post transfection with an siRNA targeting EXD2 3′ UTR or without siRNA were treated with 1 μM CPT for 1 h. Quantification of the percentage of cells with more than 15 BrdU foci per nucleus is represented. n = 223 cells (U2OS – EXD2 siRNA), 235 cells (U2OS + EXD2 siRNA), 209 cells (WT clone 1 – EXD2 siRNA), 170 cells (WT clone 1 + EXD2 siRNA), 183 cells (WT clone 2 – EXD2 siRNA), 203 cells (WT clone 2 + EXD2 siRNA), 200 cells (D108A–E110A clone 1 – EXD2 siRNA), 190 cells (D108A–E110A clone 1 + EXD2 siRNA), 282 cells (D108A–E110A clone 2 – EXD2 siRNA) and 223 cells (D108A–E110A clone 2 + EXD2 siRNA), pooled from three independent experiments.
(b) U2OS control cells or cells stably expressing Flag–HA–EXD2 WT or D108A–E110A mutant fusion proteins 72 h post transfection with an siRNA targeting EXD2’s 3′ UTR or without siRNA were treated with 1 μM CPT for 1 has indicated. Cells were fixed and stained for RPA by immunofluorescence with quantification of the percentage of cells with more than 15 RPA foci per nucleus represented. n = 308 cells (U2OS – EXD2 siRNA), 308 cells (U2OS + EXD2 siRNA), 327 cells (WT clone 1 – EXD2 siRNA), 320 cells (WT clone 1 + EXD2 siRNA), 308 cells (WT clone 2 – EXD2 siRNA), 353 cells (WT clone 2 + EXD2 siRNA), 321 cells (D108A–E110A clone 1 – EXD2 siRNA), 368 cells (D108A–E110A clone 1 + EXD2 siRNA), 370 cells (D108A–E110A clone 2 – EXD2 siRNA) and 337 cells (D108A–E110A clone 2 + EXD2 siRNA), pooled from three independent experiments. At least 100 cells were scored for each experiment.
(c) U2OS control cells or cells stably expressing Flag–HA–EXD2 WT or D108A–E110A mutant fusion proteins 72 h post transfection with an siRNA targeting EXD2 3′ UTR or without siRNA were irradiated with 8 Gy. Cells were fixed and stained for RAD51 6 h post treatment and the percentage of RAD51-positive cells quantified. n = 213 cells (U2OS – EXD2 siRNA), 254 cells (U2OS + EXD2 siRNA), 169 cells (WT clone 1 – EXD2 siRNA), 176 cells (WT clone 1 + EXD2 siRNA), 191 cells (WT clone 2 – EXD2 siRNA), 184 cells (WT clone 2 + EXD2 siRNA), 195 cells (D108A–E110A clone 1 – EXD2 siRNA), 224 cells (D108A–E110A clone 1), 191 cells (D108A–E110A clone 2 – EXD2 siRNA) and 198 cells (D108A–E110A clone 2 + EXD2 siRNA), pooled from three independent experiments.
(d) Survival assay in U2OS cells complemented with WT or D108A–E110A mutant EXD2 protein transfected with control siRNA (sample 1) or siRNA targeting 3′ UTR of EXD2 (samples 2–6). Cells were treated with 5 μg ml–1 phleomycin, and the resistance of cells transfected with control siRNA was set at 100% (n = 4 independent experiments). For all panels, bars represent mean ± s.e.m. Statistical significance was determined using the Chi-square test (a–c) or Student’s t-test (d).
EXD2 cooperates with MRE11 in the repair of DSBs
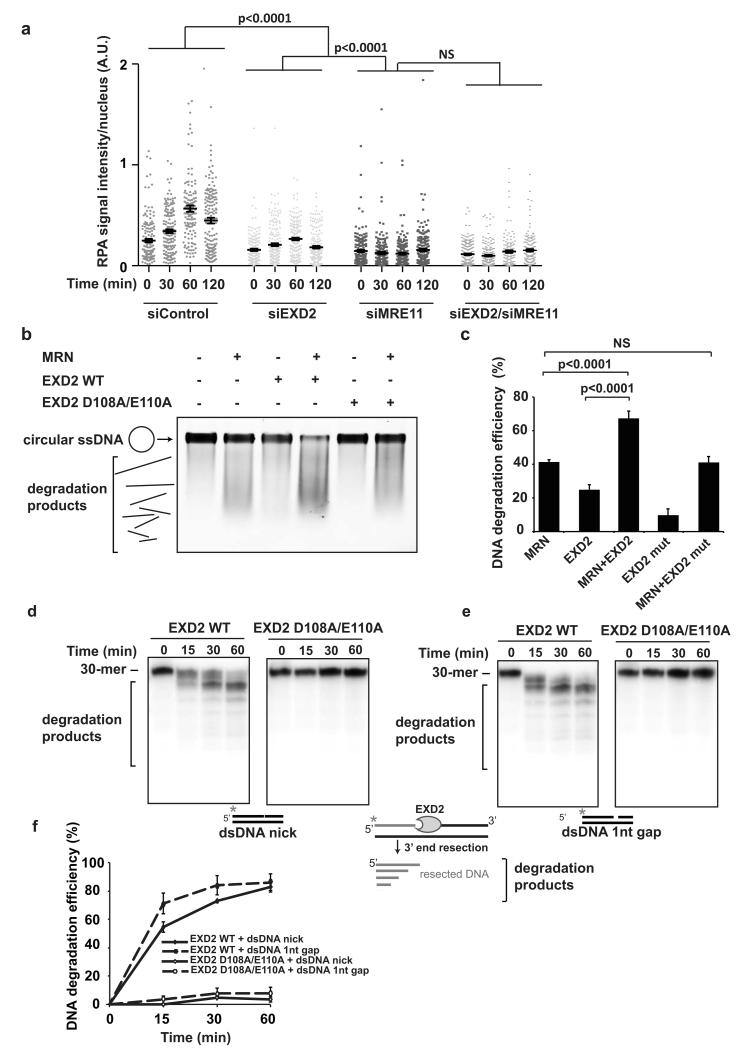
The in vivo resection that initiates DSB repair is catalysed by the MRN complex 6, 7, 14, 39. To test whether or not EXD2 collaborates in this process with MRE11 we analysed the kinetics of RPA foci formation (a marker of resection) in cells depleted for either of these proteins or concomitantly depleted for both EXD2 and MRE11. We found that combined depletion resulted in a comparable inhibition of resection as observed for depletion of MRE11 alone (Fig. 6a). A similar relationship was also observed for RAD51 foci (Supplementary Fig. 4b and c). Interestingly, the defect observed in EXD2-depleted cells was slightly weaker than that observed in MRE11 alone, suggesting that MRE11 functions upstream of EXD2 in DNA resection, perhaps initiating resection through its endonuclease activity. Indeed, it has been suggested recently that MRE11 may create multiple nicks on the strand being resected that could serve as additional exonuclease entry sites to further enhance nucleolytic processing 21, 22. We tested this notion in several ways. First, we analysed if MRN-dependent DNA resection is accelerated in the presence of EXD2. Purified EXD2 and the MRN complex (MRE11, RAD50, NBS1) were incubated together with circular single-stranded PhiX174 DNA. This substrate requires initial endonuclease-dependent nicking by the MRN complex in order to undergo resection. As previously reported, the MRN complex exhibited nuclease activity under these conditions 16 (Fig. 6b). Importantly, combining EXD2 with the MRN complex resulted in increased ssDNA degradation in vitro (Fig. 6b and c). As expected, addition of exonuclease-dead EXD2 protein to the MRN complex resulted in DNA degradation similar to what we observed for MRN alone (Fig. 6b and c). Secondly, we predicted that EXD2 should be able to initiate resection from a nicked and a gapped duplex substrate designed to mimic the substrates generated by MRE11 endonuclease activity during the initial stage of DNA-end resection. Strikingly, EXD2 exhibited robust exonuclease activity on both the nicked and gapped substrates (Fig. 6d, e and f and Supplementary Fig. 4d and e). Taken together, these data show that EXD2 functionally collaborate with the MRN complex in promoting DNA degradation.
Figure 6. EXD2 and MRE11 promote resection through a common mechanism with MRE11.
a) Quantification of the signal intensity of RPA foci in U2OS cells depleted for EXD2, MRE11 or both by siRNA (as indicated) at various time-points following treatment with 8Gy IR. ImageJ was used to quantify signal intensity per nucleus (using RPA as a marker of resection, DAPI staining marks the nucleus). n= 154, 155, 150, and 161 cells for siControl 0, 30, 60 and 120 min post-treatment, respectively. n= 177, 187, 189, and 173 cells for siEXD2 0, 30, 60 and 120 min post-treatment, respectively. n= 182, 167, 184, and 187 cells for siMRE11 0, 30, 60 and 120 min post-treatment, respectively. n= 214, 182, 150 and 163 cells for siEXD2/siMRE11 0, 30, 60 and 120 min post-treatment, respectively. In all cases cells were pooled from three independent experiments. Error bars represent +/− SEM. Statistical significance was determined using the Mann-Whitney test.
b) EXD2 stimulates MRN complex–dependent nuclease activity. MRN complex (MRE11-RAD50-NBS1) (50 nM) was incubated with PhiX174 substrate DNA in the presence or absence of EXD2 WT (K76 - V564) or corresponding mutant protein (350 nM). Reaction products were resolved on agarose gel, stained with SYBR Gold and visualized.
c) Quantification of the percentage of intact DNA from (b) representing DNA degradation efficiency. The percentage of intact DNA was obtained relative to the no-protein control sample. Error bars represent +/− SEM, n= 4 independent experiments, student’s t-test was used to determine statistical significance.
d) and e) 5′ radiolabeled dsDNA substrate (10 nM molecules) containing a nick (d) or a 1 nucleotide gap (e) was incubated for the indicated amounts of time with EXD2 WT (K76 - V564) or EXD2 (K76 - V564) D108A E110A protein (100 nM). Samples were resolved on a 20% TBE-Urea polyacrylamide gel and visualised by phosphorimaging. These experiments were carried out three times independently.
f) Quantification of the efficiency of DNA degradation from (d) and (e). Error bars represent +/− SEM, n= 3 independent experiments.
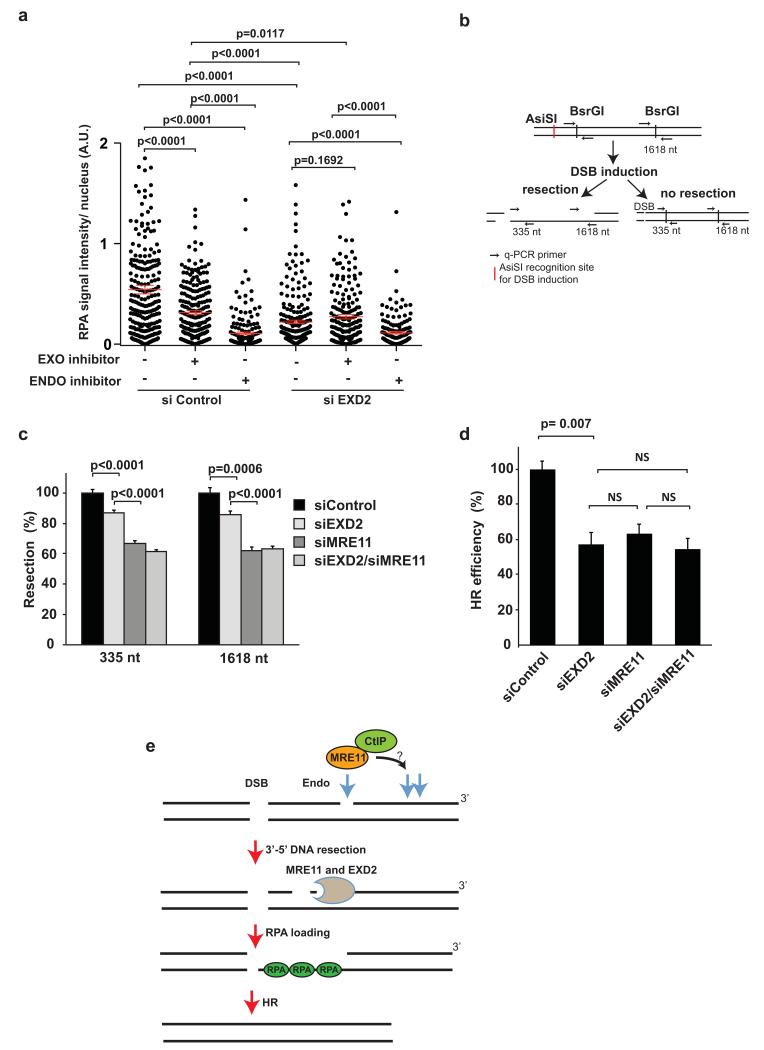
To gain more functional insight into the role of EXD2’s exonuclease activity in DNA-end resection in vivo, we took advantage of the recently developed small molecule inhibitors targeting the exo- or endonuclease activity of MRE11 23. Inhibition of MRE11 endonuclease activity resulted in almost total inhibition of resection, whereas cells treated with the MRE11 exonuclease inhibitor showed milder resection defect (Fig. 7a) as reported previously 23. Knockdown of EXD2 alone resulted in a resection defect significantly stronger (p<0.0001) than the one observed in cells treated with the MRE11 exonuclease inhibitor alone. Depletion of EXD2 in the presence of the MRE11 exonuclease inhibitor did not decrease efficiency of resection further than what was achieved with EXD2 depletion alone (Fig. 7a). Interestingly, some residual resection was still observed in cells concomitantly depleted for EXD2 and incubated with the MRE11 exonuclease inhibitor, indicating either an involvement of another exonuclease in this step of DNA-end processing 23 or that EXD2 knockdown or MRE11 exonuclease inhibition was not complete. Nevertheless, this data suggests that both the exonuclease activity of EXD2 and that of MRE11 function within the same pathway and are required for efficient DNA end-resection. Knockdown of EXD2 in cells treated with an inhibitor targeting MRE11’s endonuclease activity showed a similar resection defect when compared to the inhibitor alone (Fig. 7a), supporting the notion that EXD2 functions downstream of MRE11’s endonuclease activity.
Figure 7. EXD2 is required for efficient homologous recombination.
a) Quantification of the signal intensity of RPA foci in U2OS cells treated with DMSO or small molecule inhibitors specifically targeting MRE11’s exo- or endonuclease activity (EXO or ENDO inhibitors, as indicated) 72h post-transfection with siRNA targeting EXD2 (siEXD2) or control siRNA (siControl). Cells were pre-treated for 30 min with 100 μM PFM39 (EXO inhibitor) or 100 μM PFM01 (ENDO inhibitor) before irradiation with 3Gy ionizing radiation. Cells were harvested 2h post-irradiation and stained for RPA foci. The intensity of RPA signal per cell nucleus was analysed using ImageJ n = 231 cells (siControl EXO/ENDO −), 226 cells (siControl, EXO +), 177 cells (siControl, ENDO +), 233 cells (siEXD2 EXO/ENDO −), 201 cells (siEXD2 EXO +), 158 cells (siEXD2, ENDO +), respectively, pooled from three independent experiments. Error bars represent +/− SEM. The Mann-Whitney test was used to determine statistical significance.
b) Schematic diagram of the resection assay in human cells using the ER-AsiSI system. Arrows indicate q-PCR primers for measurement of resection efficiency following induction of the DSB.
c) ER-AsiSI U2OS cells were treated with 300 nM 4-OHT for 1 h, genomic DNA was extracted and digested or mock digested with BsrGI overnight. DNA-end resection adjacent to the DSB was measured by qPCR. The percentage of ssDNA was calculated and related to the siControl treated sample, which was set as 100%. Bars represent mean values +/− SEM (n= 5 independent experiments). Student’s t-test was used to determine statistical significance.
d) Quantification of the relative efficiency of HR in U2OS cells carrying the DR-GFP reporter construct with concomitant depletion of EXD2 and MRE11 by siRNA following transient expression of the I-SceI enzyme (see Methods and text for details). Efficiency of HR was measured by FACS and all knockdowns compared to siControl (normalized to 100%). Data represents the mean +/− SEM (n= 3 independent experiments). Student’s t-test was employed to determine statistical significance.
e) Schematic model of EXD2’s putative role in DNA-end resection. DSB induction leads to recruitment of MRE11, whose endonuclease activity results in nicking of the DNA strand proximal to the DSB. EXD2 can then process this substrate, either alone or in conjunction with the MRN complex, thereby promoting DNA-end resection.
Next, we asked which part of the resection process i.e. short-range vs. long-range resection is affected in cells depleted for EXD2. To test this, we depleted EXD2 in a cell line allowing for the induction of a DSB in a specific genomic locus in vivo 40. Then we analysed the efficiency of DNA resection at this DSB by q-PCR at two positions: one located close to the break (335bp downstream of the break - short range resection) and the other located at 1618bp from the break (long range resection) 40. Interestingly, we found that EXD2-depletion affected both short range as well as long-range resection (Fig. 7b and c and Supplementary Fig. 4f). Knockdown of MRE11 resulted in a similar albeit stronger resection phenotype thus providing further evidence to support its upstream function in this process. Importantly, concomitant depletion of both proteins did not further potentiate the resection defect.
Given that both EXD2 and MRE11 regulate DSB resection, we tested the effect of their combined depletion on homologous recombination using the DR-GFP assay. In support of their role in this process, we observed that combined depletion of EXD2 and MRE11 did not decrease homologous recombination efficiency further than what we observed in the single knockdowns (Fig. 6d and Supplementary Fig. 4g).
These findings therefore show that EXD2 promotes DNA end resection and homologous recombination by enhancing the generation of ssDNA through a common mechanism with the MRN complex. Furthermore, it seems likely that MRE11 functions upstream of EXD2 in this process, likely initiating resection through its endonuclease activity.
To verify and extend the above conclusions, we used CRISPR-Cas9 nickase based gene editing 41 in HeLa cells to generate EXD2−/− clones (Supplementary Fig. 5a). The use of Cas9 nickase has been recently shown to minimize any off-target effects 42. Comparable to siRNA treated U2OS cells, EXD2−/− HeLa cells showed dramatically decreased RPA focus formation in response to CPT (Supplementary Fig. 5b and c), diminished RPA2 phosphorylation on S4/S8 (Supplementary Fig. 5d) and decreased survival in response to CPT (Supplementary Fig. 5e). We also tested if EXD2 depletion affects the MRE11 or CtIP protein stability and/or their recruitment to DSBs. We found this not to be the case, as cells lacking EXD2 had similar level of endogenous MRE11 or CtIP as the WT control (Supplementary Fig. 5f). Likewise, MRE11 or GFP-CtIP localization to DSBs induced by microirradiation 43 was not affected (Supplementary Fig. 6a, b and c). These finding therefore establish EXD2 as an important regulator of DSBs resection.
Discussion
We have shown that EXD2 facilitates DSB resection thus promoting recruitment of RPA and homologous recombination. Accordingly, cells depleted for EXD2 show spontaneous chromosomal instability and are sensitive to DNA damage induced by agents that generate DSBs. Furthermore, we establish that EXD2 functionally interacts with the MRN complex utilizing its 3′-5′ exonuclease activity to accelerate DSB resection and promote efficient HR. In line with this, complementation experiments showed that exonuclease-dead mutant protein failed to complement these phenotypes. Interestingly, EXD2 seems to be dispensable for the initial sensing of the break as evidenced by efficient γH2AX and CHK2 phosphorylation, and most likely acts downstream of MRE11. Finally we reveal that both, EXD2 and MRE11 function in the same pathway for DSB resection and HR. It is unclear at present why cells would need two exonucleases with the same polarity. However, a paradigm for such a requirement is evident from the fact that cells have two alternate machineries, consisting of BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN that carry out long-range resection9, 10, 13, 44. Thus, by analogy EXD2 may function together with MRE11 to accelerate resection in the 3′-5′ direction in order to efficiently produce short 3′ ssDNA overhangs. This could promote faster generation of longer stretches of ssDNA, which in turn may serve as a better substrate for BLM–DNA2 or BLM-EXO1 to initiate long-range resection. Accordingly, depletion of EXD2 adversely impacts on this process. Ultimately, efficient generation of ssDNA with minimal homology length required for productive HR would supress unscheduled deleterious recombination events. This may be particularly important in vertebrates, as they require significantly longer stretches of ssDNA (200-500 bp) to initiate productive HR 45, which is in contrast to yeast, where as little as 60 bp of 3′ ssDNA is sufficient to support HR 46. Not mutually exclusive is the possibility that EXD2 could also augment resection efficiency under specific circumstances, for instance in the presence of modifications to the damaged DNA and/or polypeptides bound at the 5′ ends. In line with this, we show that in vivo EXD2 depletion impairs short-range resection. Recently, it has been proposed that MRE11 may create multiple incisions on the DNA strand undergoing resection up to 300 bp distal to the break, which could allow for more efficient resection 21, 22. Indeed, inhibition of MRE11’s endonuclease activity seems to be dominant in promoting the generation of ssDNA over its exonuclease activity 23. Thus, we postulate a model whereby EXD2 functionally collaborates with the resection machinery, most likely utilizing DNA nicks generated by MRE11’s endonuclease activity 3′ of the DSB. This would enhance the generation of ssDNA tails required for efficient homologous recombination (model Fig. 6d).
In summary, our work identifies EXD2 as a critical factor in the maintenance of genome stability through homologous recombination dependent repair of DSBs, including those induced by commonly used anti-cancer agents, such as IR or CPT. This highlights EXD2 itself and/or its enzymatic activity as a potential candidate for development of anti-cancer drugs.
METHODS
Cell lines
HEK 293FT cells were a kind gift from Dr G. Stewart while HeLa and U2OS cells were a generous gift from Dr F. Esashi. These cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and standard antibiotics. U2OS cells stably expressing GFP-CtIP and U2OS cells harbouring the HR reporter DR-GFP were a generous gift from Prof. S.P. Jackson and were maintained in media supplemented with 500 μg ml−1 G-418. The ER-AsiSI U2OS cell line 47, a kind gift from Prof. G. Legube, was maintained in DMEM media without phenol red supplemented with 10% dialysed FBS (Life Technologies) and 1 μg ml−1 Puromycin. Cell lines stably expressing FLAG-HA EXD2 WT or D108A/E110A fusion proteins were generated by transfection of U2OS cells with these plasmid constructs followed by clonal selection of cells grown in media containing 500 μg ml−1 G418 (Life Technologies). All cell lines have been verified mycoplasma free by PCR based test (Takara).
Plasmids
The open reading frame (ORF) of human EXD2 was purchased as a gateway entry clone in the pDONR221 plasmid backbone from DNASU Plasmid Repository (HsCD00295838). Discrepancies in the amino-acid sequence in comparison to the reference sequence for human EXD2 (NM_001193360.1) were corrected by site-directed mutagenesis. Site-directed mutagenesis was then employed to generate EXD2 D108A–E110A in pDONR221.
Flag–HA–EXD2 WT and D108A–E110A as well as GST–EXD2 WT and D108A–E110A plasmid constructs were generated by recombination of the WT or D108A–E110A EXD2 ORF in pDONR221 by LR Clonase reaction into either the pHAGE-N-Flag–HA destination vector (a gift from R. Chapman, The Wellcome Trust Centre for Human Genetics, University of Oxford, UK), or the pDEST-pGEX6P-1 destination vector (a gift from C. Green, The Wellcome Trust Centre for Human Genetics, University of Oxford, UK), respectively. LR Clonase reactions were carried out using the Gateway LR Clonase II enzyme mix according to the instructions of the manufacturer (Life Technologies). The pCMV-I-Sce1 plasmid was a gift from V. Macaulay (Department of Oncology, University of Oxford, UK). pmCherry-C1 was obtained from Clontech. pX335-GFP plasmid (pX335 vector48 containing PGK-EGFP-P2A-Neo-pA) was a gift from J. Riepsaame and M. de Bruijn (MRC Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, University of Oxford, UK).
His–EXD2 (Lys76–Ser589) construct was generated by cloning of truncated human EXD2 (Lys76–Ser589) in the expression vector pNIC28-Bsa4 (ref. 49), containing an amino-terminal His tag followed by a tobacco etch virus protease cleavage site. The construct was subsequently subjected to site-directed mutagenesis to introduce the D108A–E110A mutations. Plasmids were transfected into human cells using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions.
Immunoblotting
Cell extracts were prepared by lysing cells in urea buffer (9 M urea, 50 mM Tris HCL, pH 7.3, 150 mM β-mercaptoethanol) followed by sonication using a soniprep 150 (MSE) probe sonicator. Proteins were resolved by SDS-PAGE and transferred to PVDF. Immunoblots were carried out using the indicated antibodies (See Supplementary Table 1)
Cell Survival Assay
Alamar Blue survival assays were performed in accordance with the manufacturer’s recommendations (Life Technologies). Briefly, 300 cells per well in 96-well plates were untreated or treated with indicated doses of camptothecin, ionising radiation, phleomycin or olaparib and incubated for 7 days. Alamar blue reagent (Life Technologies) was added to each well and fluorometric measurements taken after 2h incubation at 37°C.
RNAi treatment
siRNAs used in this study are listed in Supplementary Table 2. ON-TARGETplus Non-targeting Pool or siRNA targeting luciferase 50 were used as control siRNAs where appropriate.
Immunofluorescence microscopy
Antibodies employed for immunofluorescence are listed in Supplementary Table 1. For visualisation of RPA foci, cells were pre-extracted on ice for 2 min (10 mM PIPES, pH 6.8, 300 mM Sucrose, 100 mM NaCl, 1.5 mM MgCl2 and 0.5% TritonX-100) and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Coverslips were washed 3 × in PBS and blocked in 10% FBS in PBS for 30 min before incubation with primary antibody in 0.1% FBS in PBS for 1h at room temperature. Unbound primary antibody was removed by washing 4 × 5 min in PBS at room temperature followed by incubation with the indicated secondary antibody for 45 min at room temperature. Slides were then washed 4 × 5 min in PBS before mounting with Vectashield mounting medium (Vector Laboratories) with DAPI.
In order to visualise ssDNA foci, the same protocol for RPA focus staining was used, preceded by treatment of cells growing on coverslips with 10 μM BrdU for 24 hours before fixation.
In order to visualise RAD51 foci, cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature followed by permeabilisation with 0.5% TritonX-100 in PBS for 10 min at room temperature. Cells were then blocked, incubated with primary and secondary antibodies and mounted for analysis as described above. Confocal microscopy was carried out using a Zeiss LSM 510 laser scanning confocal microscope with Zen 2009 software using a 63x objective. Image analysis was carried out with FIJI (ImageJ) software.
Microirradiation experiments
Induction of localised DSBs in human cells was carried out as described previously 43. Briefly, cells were grown on coverslips and pre-treated for 24h with 10 μM BrdU before microirradiation. To induce localised DSBs, the media was removed and cells were washed once in PBS, with subsequent removal of excess PBS. Isopore membrane filters (Millipore TMTP02500, 0.5 μM pore size) were placed on top of the coverslips and cells were exposed to 30 J m−2 UVC using a Stratagene UV Stratalinker 2400. Membrane filters were removed and media placed back on the cells which were allowed to recover for the indicated times before fixation. Cells were fixed, permeabilised and blocked as per RAD51 focus staining (described above). Cells expressing GFP-CtIP were stained for GFP (using the GFP-Booster reagent, Chromotek, 1:200,) and γH2AX using the indicated secondary antibody. Images of microirradiated cells were acquired using a DeltaVision DV Elite microscope using either a 40x objective. Image analysis was carried out with FIJI (ImageJ) and Huygens Professional (Scientific Volume Imaging) software. U2OS cells were stained for MRE11 and γH2AX and visualised on a Zeiss LSM 510 confocal microscope using a 40x objective. Image analysis was carried out using FIJI (ImageJ).
Immunoprecipitation experiments
Lysates for co-immunoprecipitation experiments were prepared as follows; cells were washed twice in PBS and then lysed in IP buffer (100 mM NaCl, 0.2% Igepal CA-630, 1 mM MgCl2, 10% glycerol, 5 mM NaF, 50 mM Tris-HCl, pH 7.5), supplemented with complete EDTA-free protease inhibitor cocktail (Roche) and 25 U ml−1 Benzonase (Novagen). After Benzonase digestion, the NaCl and EDTA concentrations were adjusted to 200 mM and 2 mM, respectively, and lysates cleared by centrifugation (16,000 × g for 25 min). Lysates were then incubated with 20 μl of GFP-Trap agarose beads (ChromoTek) blocked with 5% BSA in IP lysis buffer for 1h at 4 °C in the case of GFP trap IPs or with 20 μl of anti-FLAG M2 affinity gel in the case of FLAG IPs for 2 hours with end-to-end mixing at 4 °C. Complexes were washed extensively in IP buffer (including 200mM NaCl and 2 mM EDTA) before elution. In the case of GFP trap IPs, beads were resuspended in 2X SDS sample buffer and boiled for 3 min before centrifugation at 5000 × g for 5 min. The resultant supernatant fraction was retained as the eluate. In the case of FLAG IPs, beads were incubated for 30 min with gentle agitation at 4 °C in IP buffer supplemented with 400 μM 3x FLAG peptide (Sigma) followed by centrifugation at 5000 × g for 5 min. The resultant supernatant fraction was collected as eluate.
For mass spectrometry analyses eluates from immunoprecipitation experiments were analysed by the Mass Spectrometry Laboratory (IBB PAS, Warsaw, Poland) using the Thermo Orbitrap Velos system and protein hits were identified by MASCOT.
Chromosomal aberrations
Cells were prepared for analyses of chromosomal aberrations as described previously 51. Biriefly, Colcemid (0.1 μg/ml) was added 4 hours prior to cell harvesting. Cells were trypsinized and incubated in 0.075 M KCl for 20 min. After fixing in methanol:acetic acid (3:1) for 30 min, cells were dropped onto slides and stained with Leishman’s solution for 2 min. Slides were then coded and scored blind to the observer.
Homologous recombination DR-GFP assay
48 hours after siRNA transfection, U2OS DR-GFP cells 16 were co-transfected using Amaxa nucleofection with an I-SceI expression vector (pCMV-I-SceI) and a vector expressing mCherry fluorescent protein (pmCherry-C1). 24 hours after I-SceI transfection cells were harvested and analysed by flow cytometry (CyAn ADP Analyzer, Beckman Coulter). The percentage of GFP-positive cells among transfected cells (mCherry-positive cells) was determined using Summit 4.3 software. siControl treated sample was set as 100%. Statistical significance was determined with the Student’s t-test.
Recombinant protein purification
Glutathione S-transferase (GST) tagged proteins were purified as described 51 with some modifications. Briefly, GST protein expression was induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (Sigma-Aldrich) at 16°C for 18 hours. Bacteria were harvested by centrifugation and resuspended in lysis buffer containing 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 1 mM DTT, 1% Triton X-100, and protease inhibitors. Lysates were sonicated and cleared by centrifugation. Supernatants were incubated with Glutathione HiCap Matrix (Qiagen) for 2 h with rotation at 4°C. Beads were washed with lysis buffer containing increasing NaCl concentration, elution buffer (50 mM Tris-HCl pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.2% Triton X-100) and resuspended in elution buffer supplemented with PreScission Protease (50 units/ml) (GE Healthcare) and incubated for 18 h with rotation at 4°C. Eluates were dialysed to buffer containing 20 mM Hepes-KOH pH7.2, 100 mM NaCl, 1 mM DTT, 10% glycerol, aliquoted and stored at −80°C. His-EXD2 (K76-S589) protein and corresponding D108A/E110A mutant protein were expressed in E. coli BL21(DE3)-R3-pRARE2cells 49 grown in TB medium and induced with 0.5 mM IPTG at 18°C overnight. Cells were harvested by centrifugation and resuspended in a lysis buffer containing 50mM HEPES, pH 7.5, 500mM NaCl, 10mM imidazole, 5% glycerol, 1 mM TCEP, 0.5% Triton, supplemented with a protease inhibitor mixture (Roche Applied Science). The cells were sonicated, polyethyleneimine was added to 0.15% (w v−1) from a 5% pH7.5 stock solution, and lysates cleared by centrifugation. The supernatant was applied to a Ni-sepharose resin, washed with 50 mM HEPES, pH 7.5, 300 mM NaCl, 45 mM imidazole, 5% glycerol, 1 mM TCEP, 0.5% Triton, 1 mM PMSF and 2 mM benzamidine, and eluted in 50 mM HEPES, pH 7.5, 300 mM NaCl, 300 mM imidazole, 5% glycerol, 1 mM TCEP, 1 mM PMSF and 2 mM benzamidine. The eluate was further purified on two sequential Superdex S200 gel filtration columns in GF buffer (50 mM HEPES, pH 7.5, 500 mM NaCl, 5% glycerol, 1 mM TCEP, 0.1% Triton, 1 mM PMSF and 2 mM benzamidine). At each stage the presence of protein was confirmed on an InstantBlue-stained SDS-PAGE gel, and the identity of the final preparation was confirmed using electrospray ionisation-TOF mass spectrometry. Mass spectrometry indicated that both the WT and D108A, E110A mutant proteins had a shorter mass than predicted. Analysis using PAWS software (Genomic Solutions) suggests the proteins were lacking amino acids 565-589 at the C-terminus, probably due to proteolysis resulting in an EXD2 protein (WT or mutant) that consisted of amino acids K76-V564.
In vitro nuclease assay
Sequences of DNA oligos used are listed in Supplementary Table 3. In order to generate 3′ end labeled substrates, the indicated ssDNA oligo was labeled using [α-32P] dATP and TdT enzyme (New England Biolabs). To generate 5′ end labeled substrates, the indicated ssDNA oligo was labeled using [γ-32P] dATP and PNK enzyme (New England Biolabs). To obtain dsDNA substrates, complementary ssDNA oligos (as indicated in Supplementary Table 3) were mixed in an equimolar ratio and annealed by heating at 100 °C for 5 min followed by gradual cooling to room temperature. Where indicated DNA substrates with biotin label at 3′ end were used and pre-incubated 5 min at room temperature with 10-fold molar excess of streptavidin (Sigma).
Exonuclease assays were performed as described 52 with some modifications. Briefly, reactions were carried out in a buffer containing 20 mM HEPES-KOH, pH 7.5, 50 mM KCl, 0.5 mM DTT, 10 mM MnCl2, 0.05% Triton-X, 0.1 mg ml−1 BSA, 5% glycerol, and 50 ng of EXD2 protein and initiated by adding the indicated amount of substrate and incubated at 37 °C for the indicated amounts of time. Reactions were stopped by addition of EDTA to a final concentration of 20 mM and 1/5 volume of formamide. The samples were resolved on denaturing 20% polyacrylamide TBE-Urea gels. Gels were fixed, dried and visualised using a Typhoon FLA 9500 instrument (GE Healthcare).
Thin layer chromatography (TLC) was performed as described 53. Briefly, exonuclease reactions were terminated by addition of stop solution (2% SDS, 120 mM EDTA), 1 μl of reaction mixtures was spotted on PEI cellulose thin layer plates (Merck). Plates were developed in 1.0 M Sodium formate pH 3.4 and subsequently visualised using Typhoon FLA 9500 instrument (GE Healthcare).
The PhiX174 circular single-stranded substrate (30 μM nucleotides) from New England Biolabs was incubated with MRN complex (50 nM) in the presence or absence of His-EXD2 (K76-S589) (350 nM) or corresponding mutant protein in buffer containing (20 mM HEPES-KOH, pH 7.5, 50 mM KCl, 0.5 mM DTT, 5 mM MnCl2, 0.05% Triton-X, 0.1 mg ml−1 BSA, 5% glycerol, 1 mM ATP). After 2 hours the reaction was stopped by adding 1/5 volume of Stop solution (2% SDS, 50 mM EDTA). Reactions were resolved on agarose gels, stained with Sybr Gold and visualised using a Typhoon FLA 9500 instrument (GE Healthcare).
ER-AsiSI resection assay
The level of resection adjacent to specific DSB (Chr 1: 89231183) was measured as described 40 with some modifications. Briefly ER-AsiSI U2OS cells were treated with 300 nM of 4-hydroxytamoxifen (4-OHT, Sigma) for 1 hour to allow the AsiSI enzyme to enter the nucleus and induce DSBs. Cells were then harvested and genomic DNA was extracted as previously described 40. Genomic DNA was then digested with the BsrGI enzyme or mock digested and was used as a template for qPCR performed using iTaq Universal Sybr Green Mix (Bio-Rad) and Rotor-Gene (Corbett Research) qPCR system. Primers used are listed in Supplementary Table 4 40. The percentage of ssDNA was calculated as previously described 40. All data were then related to the siControl treated sample, which was set as 100%. Statistical significance was determined with the Student’s t-test.
Generation of EXD2−/− cells by CRISP/Cas9
The following guide RNA (gRNA) sequences targeting first exon of EXD2 were selected using Optimized CRISPR Design tool (http://crispr.mit.edu; 54: gRNA1 AAGGCATCCAGCGCCGCCGA, gRNA2 CCCTACAGCCACACCCAGAA.
DNA oligonucleotides were purchased from IDT and cloned into pX335-GFP vector 48 to generate targeting constructs that were subsequently co-transfected in an equimolar ratio into HeLa cells using Lipofectamine. 24 hours after transfection, cells were sorted using a MoFlo cell sorter (Beckman Coulter) for cells expressing Cas9 nickase (GFP-positive cells) and left to recover for 6 days before sorting for single cells and allowing colonies to form. EXD2 expression was analysed by western blotting. Two clones showing loss of all detectable EXD2 were selected for subsequent analysis.
Chromatin fractionation
HeLa cells were treated with 500 μM phleomycin for 1 hour, washed with ice cold PBS, scraped into PBS and the chromatin fractionation was performed as described 55, 56. Briefly, cells were resuspended in buffer A (10 mM Hepes-KOH pH7.9, 10 mM KCl, 1.5 MgCl2, 340 mM Sucrose, 10% glycerol, 1 mM DTT, protease inhibitors) and Triton X-100 was added to final concentration 0.1 %. After 5 min incubation on ice, nuclei were spun down at 1300 × g for 4 min. Pelleted nuclei were washed with buffer A, resuspended in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, protease inhibitors) and lysed for 20 min on ice before centrifugation at 1700 × g for 5 min. Supernatant (nuclear soluble fraction) was saved, and pellet (chromatin fraction) was washed with buffer B, resuspended in urea buffer (9 M urea, 50 mM Tris-HCl, pH 7.3) and sonicated.
Statistics and reproducibility
Microsoft Excel or Prism 6 software were used to perform statistical analyses. Detailed information (statistical tests used, number of independent experiments, P values) are listed in individual figure legends. All experiments were repeated at least twice unless stated otherwise.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Jackson, M. de Bruijn, J. Riepsaame, V. Macaulay, G. Legube, G. Stewart, F. Esashi, C. Green and R. Chapman for cell lines, plasmids and antibodies; J. A. Tainer for the provision of MRE11 exo- and endonuclease inhibitors; the Mass Spectrometry Laboratory (IBB PAS, Warsaw, Poland) for their work on analyses of GFP–CtIP IP experiments; J. A. Newman for advice on protein purification and D. Waithe for help with image analysis. We also thank G. Ira for critical reading of the manuscript. R.B., J.N. and W.N. are funded by Worldwide Cancer Research and MRC Senior Non-Clinical Fellowships awarded to W.N., P.J.M. and O.G. are funded by an MRC Grant, H.T.B. is funded by a Cancer Research UK Studentship and R.A.D. and T.T.P. are funded by the Cancer Research and Prevention Institute of Texas (CPRIT) grant RP110465-P4.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annual review of genetics. 2012;46:455–473. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 3.Krogh BO, Symington LS. Recombination proteins in yeast. Annual review of genetics. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 4.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annual review of genetics. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 5.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mimitou EP, Symington LS. DNA end resection--unraveling the tail. DNA repair. 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nature reviews. Molecular cell biology. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes & development. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paull TT. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA repair. 2010;9:1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackwood JK, et al. End-resection at DNA double-strand breaks in the three domains of life. Biochemical Society transactions. 2013;41:314–320. doi: 10.1042/BST20120307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicolette ML, et al. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nature structural & molecular biology. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- 19.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nature structural & molecular biology. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 20.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Molecular cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 21.Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. The EMBO journal. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata A, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Molecular cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Molecular cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Molecular cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kousholt AN, et al. CtIP-dependent DNA resection is required for DNA damage checkpoint maintenance but not initiation. The Journal of cell biology. 2012;197:869–876. doi: 10.1083/jcb.201111065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eggler AL, Inman RB, Cox MM. The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. The Journal of biological chemistry. 2002;277:39280–39288. doi: 10.1074/jbc.M204328200. [DOI] [PubMed] [Google Scholar]
- 29.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. The Journal of biological chemistry. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 30.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 31.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 32.Filippone S, et al. On the mechanism of the thermal retrocycloaddition of pyrrolidinofullerenes (retro-Prato reaction) Chemistry. 2008;14:5198–5206. doi: 10.1002/chem.200800096. [DOI] [PubMed] [Google Scholar]
- 33.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes & development. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCabe N, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer research. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 35.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi JM, et al. Probing the roles of active site residues in the 3′-5′ exonuclease of the Werner syndrome protein. The Journal of biological chemistry. 2007;282:9941–9951. doi: 10.1074/jbc.M609657200. [DOI] [PubMed] [Google Scholar]
- 37.Li B, Reddy S, Comai L. Sequence-specific processing of telomeric 3′ overhangs by the Werner syndrome protein exonuclease activity. Aging. 2009;1:289–302. doi: 10.18632/aging.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rass E, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nature structural & molecular biology. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 39.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nature structural & molecular biology. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, Caron P, Legube G, Paull TT. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic acids research. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen B, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nature methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki K, Yamauchi M, Oka Y, Suzuki M, Yamashita S. Creating localized DNA double-strand breaks with microirradiation. Nature protocols. 2011;6:134–139. doi: 10.1038/nprot.2010.183. [DOI] [PubMed] [Google Scholar]
- 44.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes & development. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubnitz J, Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Molecular and cellular biology. 1984;4:2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Molecular and cellular biology. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iacovoni JS, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. The EMBO journal. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savitsky P, et al. High-throughput production of human proteins for crystallization: The SGC experience. Journal of Structural Biology. 2010;172:3–13. doi: 10.1016/j.jsb.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuschl T. Cotransfection of Luciferase Reporter Plasmids with siRNA Duplexes. CSH protocols. 2006;2006 doi: 10.1101/pdb.prot4342. [DOI] [PubMed] [Google Scholar]
- 51.Blackford AN, et al. TopBP1 Interacts with BLM to Maintain Genome Stability but Is Dispensable for Preventing BLM Degradation. Molecular cell. 2015;57:1133–1141. doi: 10.1016/j.molcel.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sengerova B, et al. Characterization of the human SNM1A and SNM1B/Apollo DNA repair exonucleases. The Journal of biological chemistry. 2012;287:26254–26267. doi: 10.1074/jbc.M112.367243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu L, Halligan BD. V(D)J recombinational signal sequence DNA binding activities expressed by fetal bovine thymus. Veterinary immunology and immunopathology. 1999;71:277–289. doi: 10.1016/s0165-2427(99)00099-9. [DOI] [PubMed] [Google Scholar]
- 54.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Molecular and cellular biology. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, et al. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. The EMBO journal. 2014;33:862–877. doi: 10.1002/embj.201386064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.