Abstract
The recent discovery of the potent regulatory nature of microRNAs (miRNAs), a relatively new class of approximately 22 nucleotide RNAs, has made them a primary focus in today’s biochemical and medical research. The relationship between miRNA expression patterns and the onset of cancer, as well as other diseases, has glimpsed the potential of miRNAs as disease biomarkers or drug targets, making them a primary research focus. Their promising future in medicine is hinged upon improving our scientific understanding of their intricate regulatory mechanisms. In the realm of analytical chemistry, the main challenge associated with miRNA is its detection. Their extremely small size and low cellular concentration poses many challenges for achieving reliable results. Current reviews in this area have focused on adaptations to microarray, PCR, and Northern blotting procedures to make them suitable for miRNA detection. While these are extremely powerful methods and accepted as the current standards, they are typically very laborious, semi-quantitative, and often require expensive imaging equipment and/or radioactive/toxic labels. This review aims to highlight emerging techniques in miRNA detection and quantification that exhibit superior flexibility and adaptability as well as matched or increased sensitivity in comparison to the current standards. Specifically, this review will cover colorimetric, fluorescence, bioluminescence, enzyme, and electrochemical based methods, which drastically reduce procedural complexity and overall expense of operation thereby increasing the accessibility of this field of research. The methods are presented and discussed as to their improvements over current standard methods as well as their potential complications preventing acceptance as standard procedures. These new methods have addressed the many of the problems associated with miRNA detection through the employment of enzyme-based signal amplification, enhanced hybridization conditions using PNA capture probes, highly sensitive and flexible forms of spectroscopy, and extremely responsive electrocatalytic nanosystems, among other approaches.
Introduction
The inadvertent discovery of the microRNA (miRNA) lin-4 in Caenorhabditis elegans (1993) by Lee and colleagues [1] was initially thought to be a sporadic anomaly of nature. However, the discovery of the miRNA let-7 and its ability to regulate lin-14 (a gene which controls the developmental stages of C. elegans) by Ruvkun and colleagues [2] just under a decade later (2000), set this new class of regulatory nucleic acids as the primary focus of a recent explosion of research. Since then, miRNAs have been studied at great length, and been found to control such functions as cell proliferation, apoptosis, and fat metabolism in humans; the distinct larval stages and neuronal patterning in developing nematodes; and leaf and flower growth in plants. [3] These tiny, approximately 22 nucleotide sequences achieve this incredible array of regulatory functions through the degradation and translational repression of messenger RNA (mRNA). [4]
The coding sequences for miRNAs are typically found in intergenic regions (spaces between genes in which there are few to no sequences coding for proteins) or within the introns (in eukaryotes, non-coding sequences which interrupt genes and are removed through mRNA processing before translation occurs) of protein-coding genes. Their initial template, known as primary miRNA (pri-miRNA) is transcribed by RNA polymerase II. These pri-miRNAs can be thousands of nucleotides long, and typically contain a variety of stem-loop moieties and a hairpin structure, making it pseudo-double-stranded. The miRNA sequence is further capped and polyadenylated in the nucleus before the RNase-III enzymes Drosha and Pasha excise a 60–80 nucleotide intermediate known as precursor miRNA (pre-miRNA). Following exportation to the cytoplasm by the Exportin-5 factor, the pre-miRNA is further cleaved into an 18–24 double-stranded oligonucleotide by the RNase-III enzyme Dicer. One of these strands is incorporated into a protein complex known as the RNA-induced silencing complex (RISC) and is now in the “mature” form capable of post-transcriptional regulation of mRNAs (Figure 1). [5; 6] This capacity for regulation allocates the mature form as the typical target for most miRNA assays.
Figure 1.
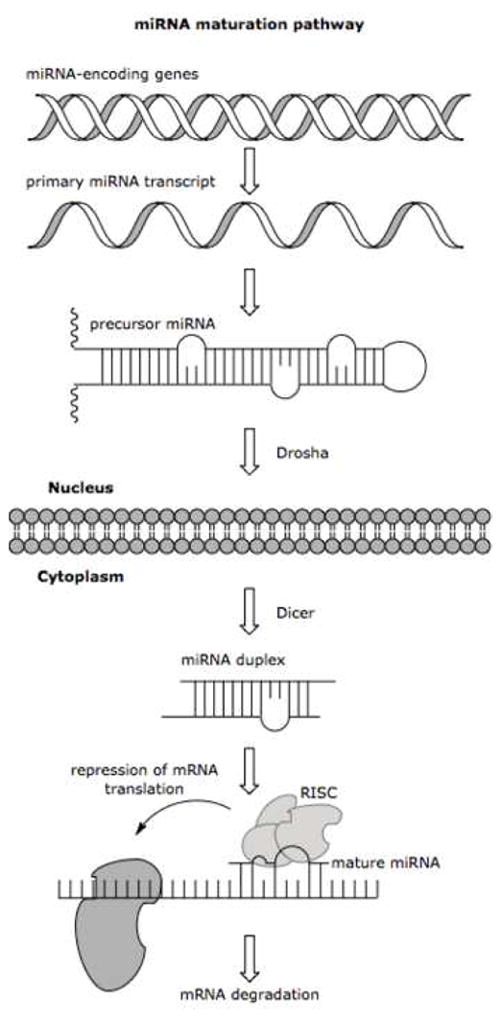
The genes coding for the primary miRNA transcript are typically found in intergenic regions or within the introns of eukaryotic protein-coding genes. The primary transcript is further processed and capped for nuclear export to the cytoplasm where the precursor form is cleaved into the 18–24 nucleotide mature sequence, which is incorporated into the RISC protein complex. Figure adapted from Macmillan Publishers Ltd: H. Grosshans, and W. Filipowicz, Molecular biology: the expanding world of small RNAs. Nature 451 (2008) 414–6.
According to miRBase (http://microrna.sanger.ac.uk/), a collective registry of currently known miRNA sequences and targets hosted by the Sanger Institute, there are currently 695 recognized miRNA sequences in the Homo sapiens genome, and the number is constantly growing as new sequences are discovered. [7] The rapid regulatory ability of these miRNAs has drastic implications on a cell’s health, especially when their expression pattern becomes erratic, and for this reason they are becoming an increasingly popular focus for research in the medical realm. Recent discoveries have shown that miRNA expression patterns are drastically altered in cancer cell lines, such that proliferation is up-regulated and apoptosis is considerably, if not entirely, repressed. [6; 8] Cellular miRNA expression levels can therefore be used as biomarkers for the onset of disease states. [9; 10] It has been shown that many miRNA sequences are oncogenic, and that their deregulation can force cells into tumorous states. [11; 12; 13] The causative nature of these types of disease factor miRNAs distinguishes them as possible drug targets. [14] Another major focus in medicinal research is centered on the possible usage of miRNAs in gene therapy for genetic disorders. If these miRNAs could be designed to target the mRNA of disease related genes, the phenotypic expression could be successfully silenced, preventing the onset of the diseased state. [8]
The characterization of such disease states requires accurate expression profiling of these miRNA sequences. The current standard methods, such as Northern blotting, microarrays, or RT-PCR, are laborious, semi-quantitative, and require expensive imaging equipment or radioactive/toxic labels. These requirements tend to make this area of research less accessible to academic or non-commercial environments. Current reviews in this area have introduced novel developments used to overcome the difficulties associated with miRNA detection using microarray platforms, and introduced some of the modifications made to PCR and Northern blotting procedures to permit their extension to miRNA detection. [15; 16] This review aims to explore emerging techniques in miRNA detection and quantification, which are not associated with the current standard PCR, Northern blotting, or microarray methods. These new assays demonstrate superior flexibility and adaptability as well as matched or increased sensitivity in comparison to the current standards. Additionally, their approaches drastically reduce the procedural complexity and overall expense of operation thereby increasing the accessibility of this field of research.
Colorimetric-Based miRNA Detection
Colorimetric-based detection methods are attractive because the setups are relatively simple. Most colorimetric experiments can be carried out with inexpensive imaging equipment such as a CCD camera and a personal computer for analysis. While these aspects are beneficial because they drastically reduce the cost and procedural complexity of the setup, colorimetric methods sometimes suffer from lower detection limits. This is not always the case, however, and there are many colorimetric assays which rival their fluorescent counterparts.
A simple solid phase assay developed by Yang and colleagues utilized adjacent complementary probes to capture a target sequence and register a corresponding fluorescent signal (Figure 2). [17] The capture probe is biotinylated, and the signal probe is directly conjugated to a gold nanoparticle via the chemical method developed by Mirkin and colleagues. [18] In this method, the oligonucleotide probes, modified with gold-reactive terminal thiols, are mixed with gold nanoparticles yielding the semi-spontaneous assembly of the probe. Each probe is complementary to half the target sequence. The two probes and miRNA target-containing mixture are combined under optimal conditions to form a hybridization complex. This complex may then be immobilized onto a streptavidin coated microtiter plate and silver enhanced to amplify the colorimetric signal from the absorbed gold nanoparticles. Silver enhancement is a reduction process which converts silver ions into their metallic form and ultimately coats the gold nanoparticles. The process is catalyzed by the gold nanoparticle, and is carried out by adding a solution of hydroquinone and silver nitrate in an acidic (pH 3.8) citrate buffer to the probe and incubating at room temperature. The miRNA is then quantified by measuring absorbance using a simple well plate reader.
Figure 2.
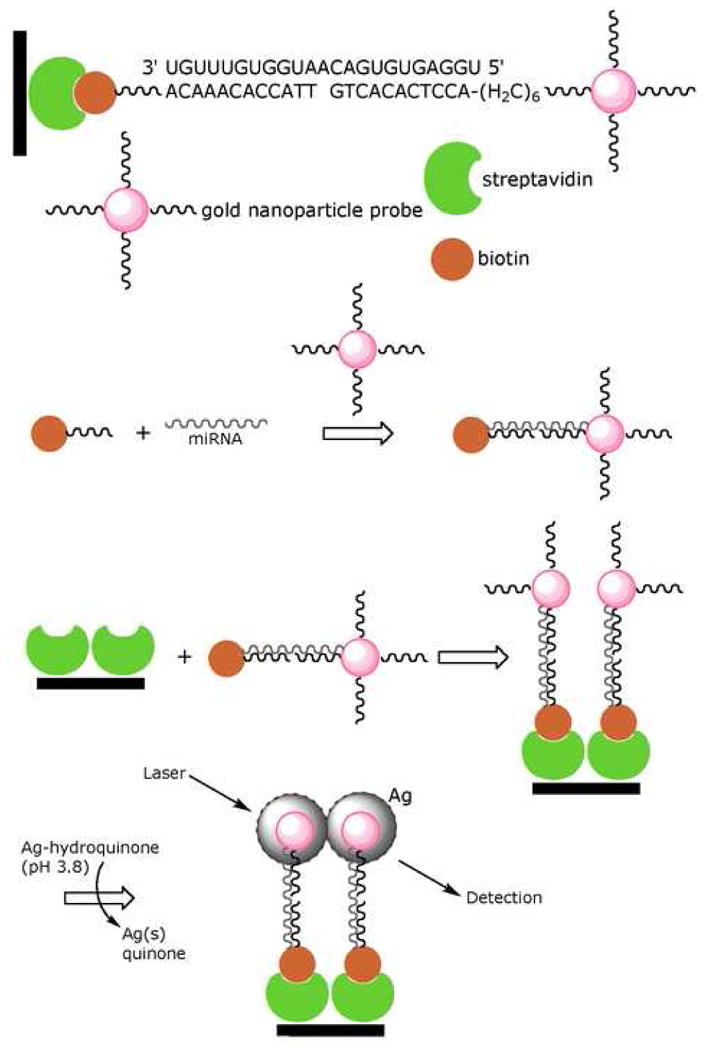
In the colorimetric assay designed by Yang and colleagues, the functionalized gold nanoparticle binds adjacent to the biotinylated probe on the miRNA target, thus allowing it to be immobilized on a streptavidin coated plate. The grayscale absorbance from the silver enhancement is used for colorimetric quantification of target. Figure adapted from Elsevier B.V.: W.J. Yang, X.B. Li, et al., Quantification of microRNA by gold nanoparticle probes. Analytical Biochemistry 376 (2008) 183–8.
A calibration study for the assay developed by Yang and colleagues was done by varying miR122a concentrations from 1 fM to 1 nM. The lower detection limit of this assay was found to be 10 fM for the miR-122a sequence extracted from mouse liver and brain tissues. The nanoparticle assay was also tested against a TaqMan miRNA RT-PCR assay. While the R2 value of this comparison was acceptable, 0.9249, the RT-PCR method has a slightly lower detection limit. Additionally, a specificity study was performed using probe sets containing single nucleotide polymorphisms. One set of miR-122a probes contained one point mutation each. The point mutation was detectable when it fell within the confines of the signal probe (gold-nanoparticle) sequence. In other words, mutations made in the capture probe sequence did not produce as noticeable a signal reduction as those made in the signal probe sequence. The assay was also performed using probes for the miR-128 sequence. The miR-128 target was only detected in mouse brain tissues, whereas the miR-122a target was only detected in liver tissues. The goal of these experimental variations was to show the relatively low susceptibility of the probe system to non-specific binding and/or cross-hybridization.
The gold-nanoparticle-based method discussed above is attractive in that it is relatively inexpensive and can be completed in a short time frame (approximately 1 h hybridization time). The high selectivity of this assay is hinged upon the optimization of the hybridization and rinse temperatures for each miRNA sequence targeted. When held in comparison to the amount of optimization required for a blotting assay, the adaptations required for this assay are quite manageable. There is currently no explanation as to why point mutations can only be detected in the signal probe domain. The authors suggest that this inconvenience may be avoided by using two sets of signal probes and swapping the molecules at the termini. This would require two assays to be performed per sample, but any discrepancy between the two assays would be indicative of a mutation and its general location on the target.
Fluorescence-Based miRNA Detection
Fluorescence is an extremely sensitive and adaptable form of spectroscopy, and for this reason it is frequently used for direct, in situ assays. There are many different types of fluorescent reporters available for these applications, from organic dyes to fluorescent proteins to inorganic nanostructures. Perhaps the most prominent fluorescent reporters in current research are inorganic nanostructures such as quantum dots or gold nanoparticles. These structures have outstanding fluorescent properties, such as broad excitation ranges complimented by extremely sharp emission bands, affording superior spectral resolution. Their high quantum yield makes them appealing for direct detection applications, removing the need for sample enrichment and amplification, which can be exceedingly inconvenient, and thereby reducing the procedural complexity, making the prescribed technique more versatile and accessible.
In a method developed by Neely and colleagues, a dual probe labeling system was used in conjunction with fluorescence correlation spectroscopy (FCS) to count and quantify single molecules of miRNA. [19] Two locked nucleic acid (LNA) DNA probes, each complementary to one half of the target sequence, were modified with fluorescent tags, one with Oyster 556 (green probe) and one with Oyster 656 (red probe). A Trilogy five laser (one DPSS 488 nm, one DPSS 532 nm, two HeNe 633 nm, and one 750 nm diode) confocal fluorescence detector capable of four color detection in a single microfluidic stream was used to acquire the data. The microfluidic setup consisted of a fused silica capillary sheathed by a needle for sample extraction from a sealed microtiter plate. An aperture oil-immersion objective was used to focus the beams to a 1 fl excitation volume within the capillary. Three lasers were used in this setup, the two HeNe 633nm (red 1 and 2) and the DPSS 532 nm (green). The red 2 laser was focused 12 μm downstream from the red 1 laser and was used to determine the flow velocity of the target molecules for cross-correlation (offset compensation) of the red 1 and green signals in the time domain. The green laser was focused 2 μm downstream from the red 1 laser, to avoid any spectral interference, and was used to detect coincident signals from targets bound to both fluorescent probes (excited by the red 1 and green lasers). To reduce background noise, free probes were arrested by hybridization with specially designed oligonucleotides containing fluorescent quenchers. (Figure 3)
Figure 3.
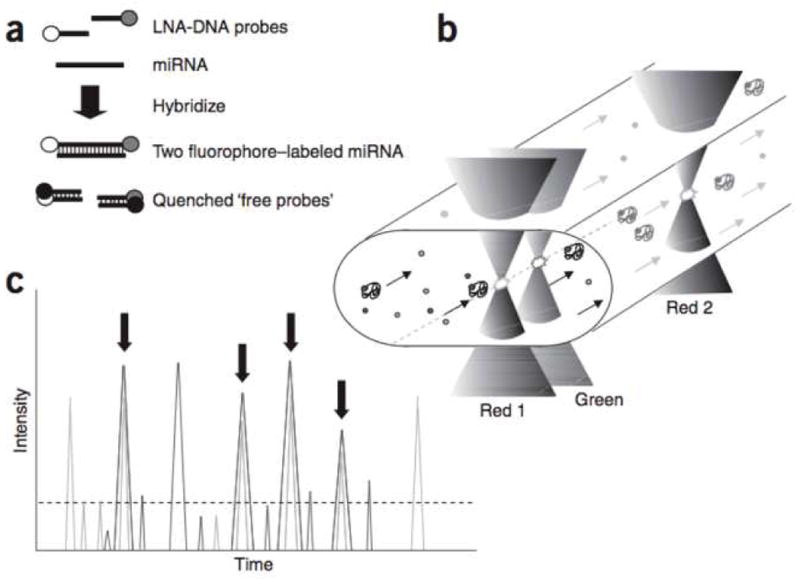
In the FCS based assay developed by Neely and colleagues, free probes were sequestered by specially designed quenching probes. The red 2 laser was used to correlate the red 1 and green signals. Reprinted by permission from Macmillan Publishers Ltd: L. Neely, S. Patel, et al., A single-molecule method for the quantitation of microRNA gene expression. Nat Meth 3 (2006) 41–6.
Photon bursts were recorded in 1 ms time intervals. The number of time intervals that exceeded a certain threshold were compared to three calibration curves prepared from a complex Escherichia coli RNA matrix spiked with 1, 2, 5, 10, 25, 50, 75, and 100 pM synthetic miRNA. All sequences tested were purchased pre-quantified by Northern blotting, and therefore acted as a control for comparison of this method to what is considered the current “gold-standard” for miRNA quantification. The results of the direct assay plotted against the Northern blotting results had an R2 value of 0.947, demonstrating that the assay is in general agreement with the current standard method. The assay had a lower detection limit of 15 fg per 1 μg of tissue RNA. The assay was used to profile the expression of 45 different human miRNAs in 16 different tissues, displaying superb selectivity and versatility.
While the method developed by Neely and colleagues does require the usage of more sophisticated and expensive equipment, its ability to characterize and quantify miRNAs in conjunction with its versatility to automation makes it a competitive and highly useful assay, especially in profiling miRNA expression. Current miRNA expression profiles are carried out using microarrays, which are only moderately quantitative. The current standard for quantification of miRNAs is Northern blotting, but the huge time requirement and necessity for large RNA samples and potentially hazardous radioactive labels make it somewhat unsuitable for expression level studies. The method of Neely and colleagues is most attractive in its application to profiling studies. The simple procedural nature requires no sample enrichment or amplification, and the direct measurements eliminate the need for any subsequent clean-up steps. This is a very rapid, sensitive, and highly automatable method.
One of the most prominent biosensors in current research is the molecular beacon, which is essentially a highly unique oligonucleotide. Molecular beacons are a relatively new (their development was first reported in 1996) technology in bioanalytical fluorescence-based detection systems. [20] The beacons are designed as stem-loop DNA oligonucleotide probes containing the anti-target sequence flanked by two, usually five nucleotide, self-complementary sequences. The termini of the probe are conjugated to a fluorescent dye and a suitable quencher for that dye. In the absence of target, the flanking sequences anneal forming a stem-loop structure and bringing the fluorescent dye and quencher into close proximity. When target is present, it anneals to the probe, forcing it into a linear conformation, which separates the fluorescent dye and quencher. The benefit of this design is that the probe only elicits signal upon direct hybridization to the target sequence, thus eliminating the need to remove excess probe prior to taking a measurement. One unique approach for the detection and quantification of miRNA developed by Paiboonskuwong and colleagues utilized a molecular beacon probe and boasts the ability to solely detect the mature form of miRNA. [21] This is achieved by using a fluorescent dye, 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionic acid (BODIPY® FL), that is successfully quenched in the proximity of guanine. The probe is designed such that binding to the precursor form (not yet processed by Dicer) of the miRNA (pre-miRNA) target places the fluorescent dye in proximity with a guanine base, quenching its fluorescence. The mature miRNA is much shorter, however, and a binding event with this form of the target will no longer quench the fluorescent signal. (Figure 4)
Figure 4.
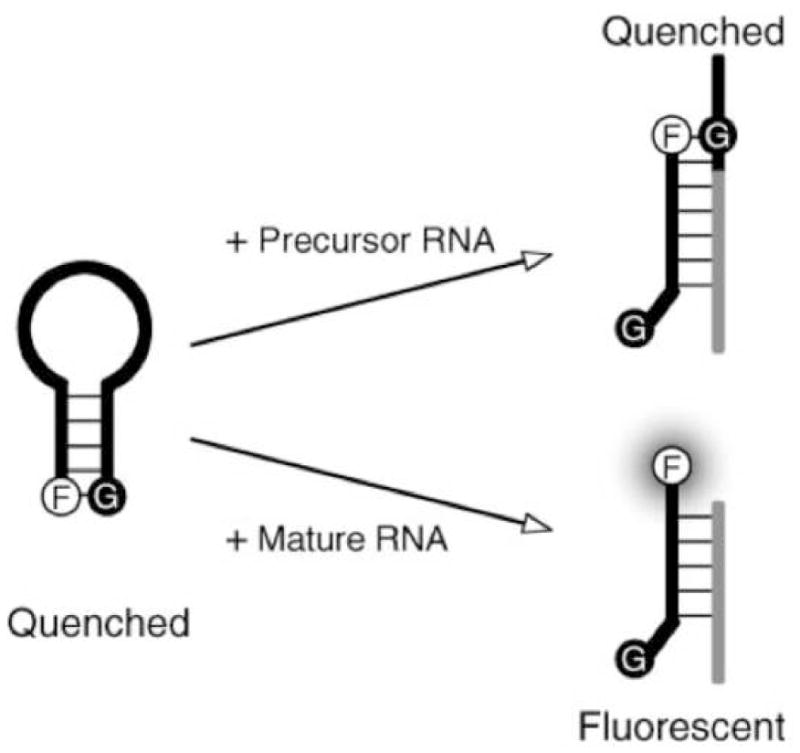
In the molecular beacon assay developed by Paiboonskuwong, adjacent guanine in the primary and precursor forms of the target successfully quenches the fluorophore and limits signal production to the mature form only. Reprinted by permission from Oxford University Press: K. Paiboonskuwong, and Y. Kato, Detection of the mature, but not precursor, RNA using a fluorescent DNA probe. Nucleic Acids Symposium Series (2006) 327–8.
This method could be highly useful in profiling cellular expression levels owing to the fact that both precursor and mature forms of the miRNA target are present in the cytosol. The chosen quenching method affords this unique selection capability, and also places stringent implications on the probe design. This molecular beacon is unique in that it does not require a conjugated quencher molecule. Instead, the addition of extra guanine beyond the flanking sequence will suffice to quench the signal. While this is easy to do, it must also be taken into consideration that the probe must be designed such that it hybridizes to the pre-miRNA in a location that places the fluorescent dye near a guanine. If this is not possible due to the pre-miRNA not containing guanine proximal to the targeted sequence, the molecular beacon will not be able to distinguish between precursor and mature forms of the target.
The ability to distinguish between the precursor and mature forms of an miRNA sequence is undoubtedly very useful. Another fluorescent method which is able to discriminate these differences was developed by Allawi and colleagues. [22] The developed assay is an application extension of the previously designed Invader mRNA assay. [23; 24] The mRNA Invader design utilized a 5′ nuclease (Cleavase) to release a generic 5′ flap from a linear probe annealed to the target scaffold but slightly displaced by a second invasive probe (Figure 5A). The released flap can then bind to a secondary scaffold and participate in another cleavage reaction to produce a fluorescent signal. [24] The small nature of miRNAs requires some modifications to the invader and probe oligonucleotides, which are each complementary to about half the miRNA target. In this setup, the invader and probe oligonucleotides are designed such that they contain self complementary sequences on their 5′ and 3′ ends, respectively (Figure 5C). These sequences form flanking hairpin structures on either side of the miRNA target specific region (TSR). These flanking structures help stabilize the miRNA target interaction with the probe system by promoting base stacking, and also increase the efficiency of the Cleavase (a structure-specific 5′ nuclease) reaction. Release of the probes from the miRNA target is regulated through temperature control. Higher temperatures make this exchange more rapid, thus improving enzymatic turnover and signal amplification. After cleavage, the assay proceeds just like the mRNA assay. The released 5′ flap anneals to a secondary reaction template (SRT) acting as an invasive oligonucleotide. The slight overlap of the FRET oligonucleotide causes Cleavase to act in the same manner and release the 5′ fluorescent dye (Figure 5B). Now out of close proximity of the quencher, the fluorescent signal may be read and quantified.
Figure 5.
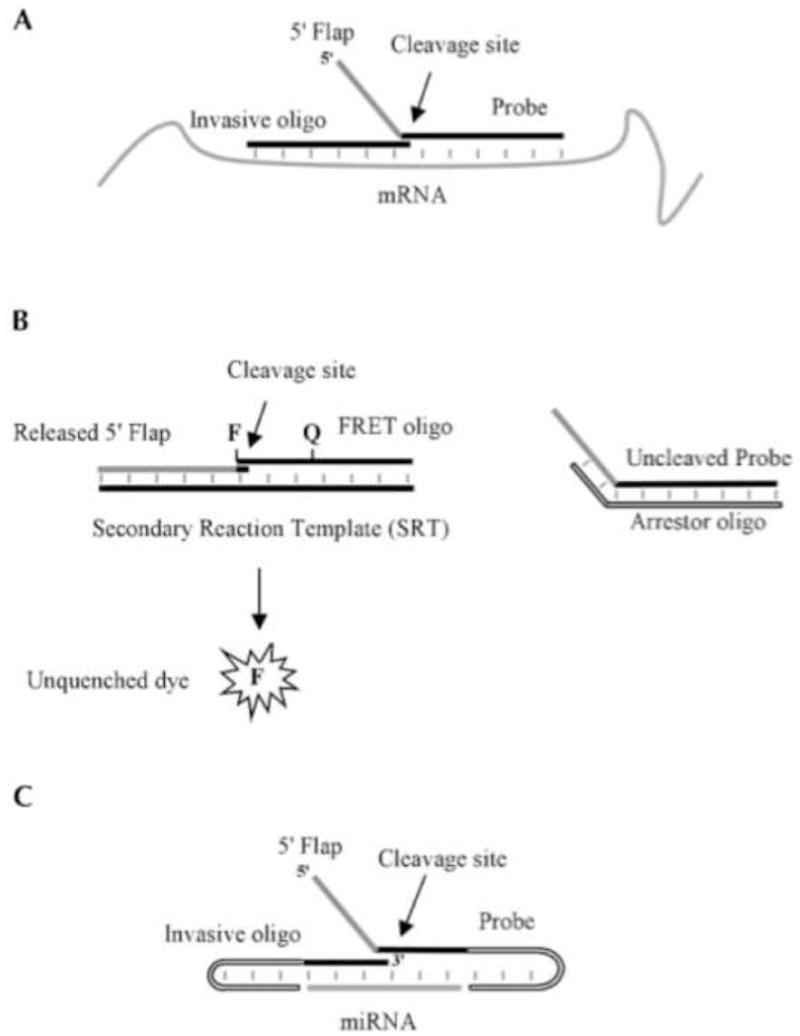
(A) The miRNA Invader assay developed by Allawi and colleagues is based on the same principles as the RNA Invader assay. (B) A general FRET oligo may be used for all sequences as it anneals to the SRT. (C) Self-complementary adaptations must be made to the invasive and probe oligos to compensate for the small size of the miRNA target. Reprinted by permission from the RNA Society: H.T. Allawi, J.E. Dahlberg, et al., Quantitation of microRNAs using a modified Invader assay. RNA 10 (2004) 1153–61.
A calibration curve was prepared from 5 fM to 5 pM samples of synthetic miRNA, and assay results were quantified by comparison to this standard curve. The authors report a lower detection limit of 20,000 molecules of a single miRNA in a tissue sample size of 50–100 ng. The let-7a miRNA sequence assay was compared against three other miRNA sequences (let-7c, let-7e, and let-7f), each containing a single nucleotide polymorphism. A 400 fold (or greater) difference in fluorescence signal was observed between the let-7a and its polymorphic variants, thereby indicating that this assay can be applied in the detection of SNPs. Additionally, the hairpin structures of the invader and probe oligonucleotides make this system sensitive to the distinctive forms of an miRNA sequence. In other words, it is able to discriminate between the mature and precursor forms of the target.
The redundant bind and release nature of this assay provides superb signal amplification, which affords the benefit of requiring only a small sample size, 50–100 ng, of cellular RNA. The more common Northern blotting techniques require a 2–10 μg sample. [25] While the optimal temperature for the primary reaction must be determined for each miRNA sequence assayed, the procedure is drastically simplified by the fact that secondary reaction conditions and probes remain the same in all cases. In other words, the secondary reaction design is independent from the primary reaction and therefore remains constant no matter what target sequence is selected. Perhaps the only concern with this assay is the variation in results when compared to Northern blotting and previous literature reported standards. However, as the authors mention, these results are made upon the assumption of total RNA mass per cell. If their assumption varied from those made for the standard techniques (i.e. Northern blotting or RT-PCR amplification) it would explain the observed variance. In addition to being sensitive, and specific, this assay also avoids the use of radioactive labels and can be completed in 2–3 h. It is also directly compatible with cellular detergent lysates, making sample enrichment and/or amplification unnecessary.
Bioluminescence-Based miRNA Detection
Bioluminescent proteins emit light via a chemical reaction therefore these proteins offer the advantage of minimizing background signal by eliminating the use of an external excitation light source. The methods described in this section for miRNA detection utilize the bioluminescent protein, Renilla luciferase, either alone or as a bioluminescent resonance energy transfer (BRET) partner with fluorescent nanoparticles to create competitive assays. Renilla luciferase is a 38 kDa protein which, in the presence of the coelenterazine substrate, emits bioluminescence at 485 nm. The assays described below have, continued to expand the realm of nucleic acid detection/quantification with reported detection levels in the fM range. Additionally these assays offer simple one step nucleic acid quantification both in buffer and cellular extract, removing many of the additional preparation and isolation steps previously required for miRNA detection. As seen for the electrochemical assays described in the previous section these also require minimal amounts of total RNA for detection, in the ng or μg range.
The work reported by Cissell et al. on nucleic acid detection and quantification utilized a BRET based competitive assay. [26] This assay relied upon the signal generated by an energy transfer between Renilla luciferase (Rluc) and an acceptor quantum dot (QD705) upon nucleic acid hybridization. Rluc was selected as the BRET partner for the QD705 due to Rluc’s emission at 485 nm, a good compliment for the 300–500 nm range of the QD excitation. The QDs themselves offered advantages to this assay, including high quantum yield, large molar extinction coefficient, and photo stability.
For this assay an amine-modified CT1 oligonucleotide probe (5′ TCAACATCAGTCTGATAAGCTA-(CH2)6-NH2-3′) corresponding to miR21, was labeled with a carboxy-activated QD705 using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) chemistry. The competitive, thiol-modified T1 probe (5′-SH-(CH2)6-TAGCTTATCAGACTGATGTTGA-3′) was labeled with Rluc via sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Sulfo-SMCC). The two probes when mixed together can hybridize to each other resulting in a measurable signal at 710 nm, in the presence of coelenterazine, caused by the energy transfer from the Rluc to the QD705 (Figure 6). This same system, in the presence of the target nucleic acid, shows a marked decrease in emission signal at 710 nm with an increase in the concentration of the target. This decrease was measured as a BRET ratio, of the signal at 485 nm to that at 710 nm. To create a calibration curve, the concentration of the target nucleic acid was plotted against this ratio of the two emission peaks (I485/I710). Luminescent studies on both labeled probes showed that the conjugation had no effect on luminescent profiles of the protein or QD. The energy transfer from Rluc to QD710 upon adding labeled probes in a cell extract demonstrated that the cellular matrix had no effect on the assay, suggesting its application in sensing samples without purification or isolation steps.
Figure 6.
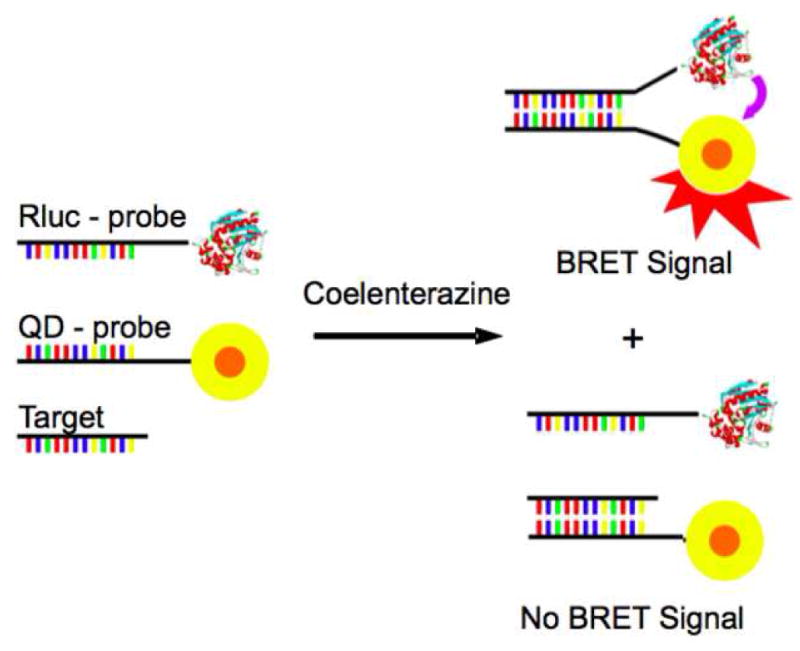
In the absence of the target, utilized in the BRET-based assay developed by Cissell et al., the Rluc-probe and the QD-probe hybridize creating a BRET signal. In the presence of the target, there is a competition between the target and the Rluc-probe, decreasing the BRET signal. With kind permission from Springer Science+Business Media: Bioanal Chem, Rapid single-step nucleic acid detection, 319, 2008, 2577, K.A. Cissell, Y. Rahimi, S. Shrestha, E.A. Hunt, and S.K. Deo, fig. 1.
While this BRET study was designed to detect and quantify nucleic acids, it could easily be adapted to specific miRNA studies. The assay as presented here showed detection limits of 4 pmol (20 nM), within 30 min hybridization. Since this assay is designed as mix and measure it demonstrates a simplification over solid phase assays by removing wash steps. Additionally the assay is useful for nucleic acid detection in cell extract and does not suffer from background interference due to non-specific hybridization.
Further work in the field of miRNA detection and quantification has utilized the bioluminescence of proteins, alone, as tags for miRNA detection. One such assay, reported by Cissell and colleagues, focused on the use of Rluc as a label in a solid phase competitive hybridization assay. [27] Rluc was selected as the tag for this work since it does not require an external light source to catalyze the bioluminescence emission, instead relying on the chemical reaction with coelenterazine. This increases the signal to noise ratio, allowing for highly sensitive miRNA detection. Luciferase tagged miR21 was used in competition with untagged “free” miR21 from a total RNA sample. The two miR21 samples, Rluc labeled and free, were hybridized in solution to a biotinylated miR21-complementary oligonucleotide probe. These hybridized miRNAs were then immobilized onto a neutravidin-coated microtiter plate via the biotinylated oligonucleotide probe (Figure 7). The bioluminescence of the wells was measured, and the decrease in bioluminescence correlated to the concentration of the target, “free”, miRNA via a dose-response curve. This assay was used to compare miR21 expression levels from MCF-7 cancer cell lines, which is tumorgenic and MCF-10A non-tumorgenic cells.
Figure 7.
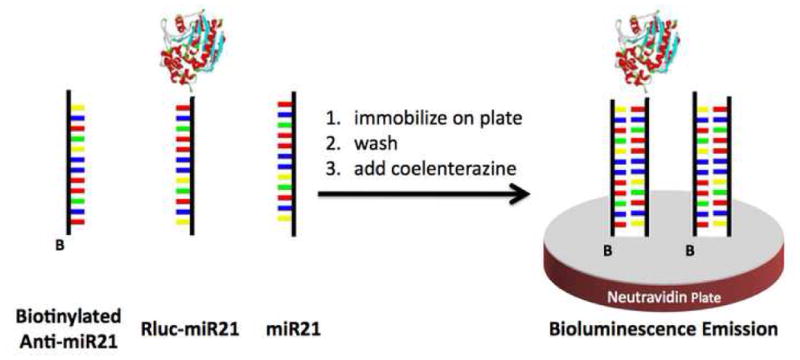
The miRNA (both labeled and unlabeled) hybridize to the biotinylated probe, in the solid phase assay described by Cissell et al. The hybridized probe is immobilized onto a neutravidin plate, washed to remove unbound miRNA, and the emission signal recorded. Figure adapted from the American Chemical Society: K.A. Cissell, Y. Rahimi, et al., Bioluminescence-based detection of microRNA mir21 in breast cancer cells. Anal Chem 80 (2008) 2319–25.
The work described above, demonstrates a rapid, highly sensitive competitive miRNA hybridization assay. Detection limits of 1 fmol were reported for this work, making it comparable to the electrochemical methods described in the previous section. The MCF-10A cells showed 50 fmol level of miR21, which is comparable to previously reported in vitro assays, demonstrating that the cellular extract has no effect on the overall assay. miR21 spiked samples of MCF-7 were used to determine the accuracy of the above method, with 97–103% recovery reported. Both of these suggest that there is no non-specific binding from other miRNA within the sample. Overall this work offers a simple, expedient assay that requires no sample enrichment, which can be used to determine miRNA concentrations in crude samples, including saliva and blood. Additionally since this work is done in a micro titer plate, it can easily be expanded to high through put work.
Both of these studies offer some advancements over the more often favored optical, Northern blotting, or PCR-based quantification methods. They use simple solution and solid phase assays that can be used to evaluate crude samples and require only small quantities of total RNA for detection of a target. They offer detection limits in the pmol - fmol range, which are sensitive enough for expression level studies in total cellular extract. In addition both demonstrate selectivity for the target small molecule in a total RNA sample, which simplifies the assays by removing isolation and amplification steps, previously required, and demonstrates a lack of non-specific binding.
Enzyme-Based miRNA Detection
As demonstrated with the Invader assay approach (which combined fluorescence-based and enzyme-based techniques), the utilization of enzymes to generate a reporter signal can provide immense signal amplification if substrate turnover is substantial. For the direct or in situ detection of miRNAs, enzyme based approaches can be a simple solution to the necessity of sample enrichment or amplification procedures. This means higher assay sensitivity may be obtained without drastically heightening the procedural complexity.
The enzyme based, colorimetric assay developed by Su and colleagues utilized immobilized peptide nucleic acid (PNA) probes to capture target DNA or RNA molecules. [25] While their work in RNA detection specifically focused on miRNAs, the assay can theoretically be applied to any form of RNA. PNA resembles DNA in structure, but its entire sugar-phosphate backbone is replaced by a neutral peptide sequence. This neutral charge reduces electrostatic repulsions between the capture probe and the target, thus drastically improving hybridization. The PNA probe is complementary to the target sequence, terminally conjugated to biotin, and immobilized on a streptavidin coated microtiter plate (Figure 8). It is a somewhat recent discovery that horseradish peroxidase (HRP) can be absorbed onto DNA (or RNA) through electrostatic interaction with the negatively charged sugar-phosphate backbone. [28] Hybridization of the PNA capture probe to the miRNA target spatially localizes the negative charge for HRP adsorption. This unique technique removes the need for a target labeling step and thereby increases assay sensitivity. Treatment with 3,3′,5,5′-tetramethylbenzidine (TMB) and hydrogen peroxide, which are substrates for HRP, yields a blue oxidation product that can be further converted to a yellow diamine by treatment with strong acid. The absorbance was then read at 450 nm.
Figure 8.
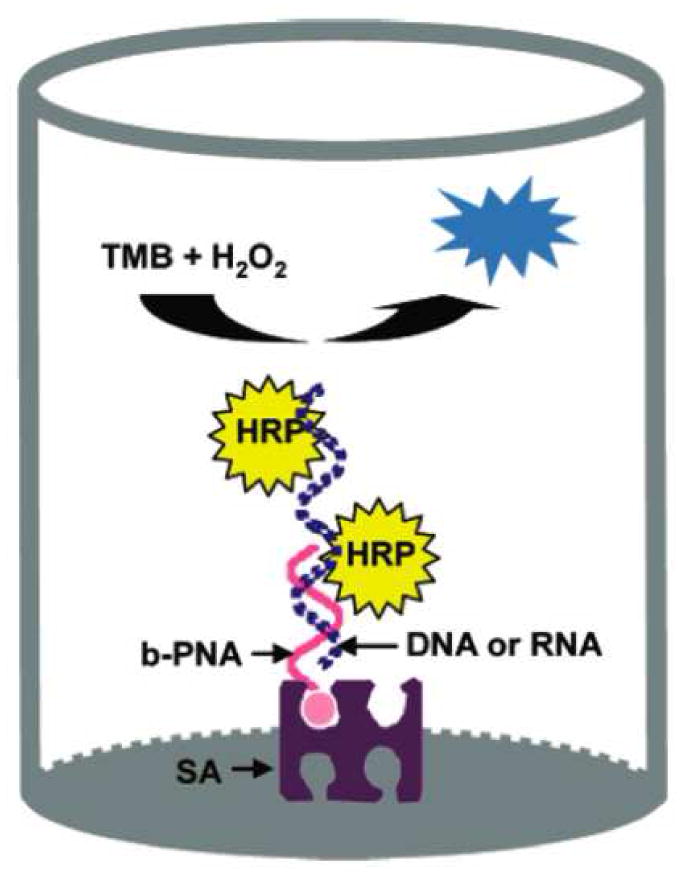
In the enzymatic/colorimetric assay developed by Su and colleagues, the PNA capture probe improves hybridization conditions and localizes negative charge for HRP adsorption. Reprinted by permission from the American Chemical Society: X. Su, H.F. Teh, et al., Enzyme-based colorimetric detection of nucleic acids using peptide nucleic acid-immobilized microwell plates. Anal Chem 79 (2007) 7192–7.
A 22 bp PNA probe was used to detect let-7b miRNA in HeLa and A549 lung cancer cells. All assays were done on total RNA extractions varying from 1–58 μg. Samples were quantified against calibration curves created using varying concentrations (0.1–100 nM) of a synthetic 22 bp DNA oligonucleotide. It was determined that the lowest amount of total RNA required for this assay was around 2 μg. While the Invader miRNA assay is much more sensitive, requiring only 50 ng, the colorimetric assay sample size is still comparable to Northern blotting techniques. In many ways, the lower sensitivity of this colorimetric method is outweighed by its versatility. Not only is the assay rapid, requiring only 1 h of incubation for hybridization, and fairly inexpensive, but it can also be adapted to other solid support formats such as membranes or glass for microarray or gene profiling applications. There are some minor concerns of minimal HRP adsorption to unhybridized PNA capture probe, and a variable dependence of HRP adsorption on target length. The authors propose valid explanations for these issues, such as increased negative charge due to more phosphate groups present on longer targets. However, it appears that the problems have little to no effect on the reliability or accuracy of this assay.
The relatively low sensitivity of direct detection methods is commonly overcome through the employment of some kind of signal amplification. The novel method developed by Hartig and colleagues utilized an intermolecular ribozyme to detect miRNA sequences with superior signal amplification. [29] Ribozymes are RNA molecules with special physical features that give them catalytic properties. The hairpin ribozyme used in this assay contains two helical domains (A and B). The catalytic nature of this type of ribozyme is directly related to the conformational flexibility of the overall structure, and cleavage of the external substrate nucleotide can only occur upon the docking of the A and B domains. [30] (Figure 9) The ribozyme setup used by Hartig and colleagues is somewhat special in that a third, stem-loop domain (C) has been engineered into the ribozyme such that its conformational flexibility can be controlled in the presence or absence of a target sequence (Figure 10). The C domain is complementary to the target miRNA sequence and to the substrate binding regions of the A domain (termed H1 and H2 in Figure 7), thus restricting conformational flexibility by being in a native loop structure in the absence of target and hence preventing substrate nucleotide from binding to the A domain. In this case, the substrate is a specially designed FRET oligonucleotide. In the presence of target sequence, the C domain becomes helical, the substrate binding regions of the A domain become available for substrate hybridization, and the docking of domains A and B becomes conformationally permissible, which enables the catalytic activity of the ribozyme. The cleavage of the FRET oligonucleotide separates the fluorescent dye from the quencher and gives rise to a fluorescent signal.
Figure 9.
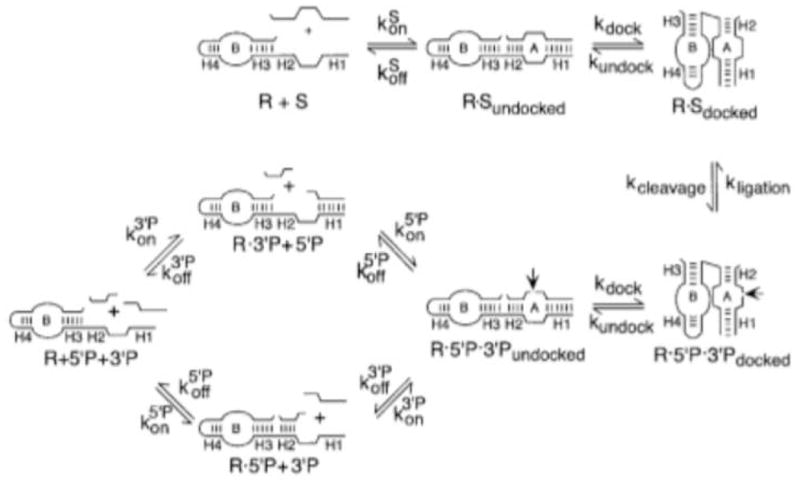
The catalytic activity of a ribozyme is directly related to its structural conformation. Reprinted by permission from Elsevier B.V.: M.J. Fedor, Structure and function of the hairpin ribozyme. J Mol Biol 297 (2000) 269–91.
Figure 10.
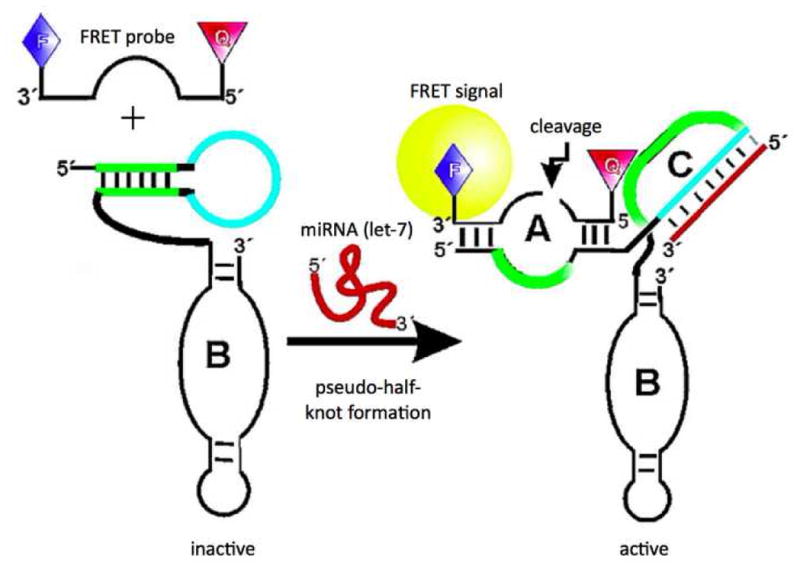
In the ribozyme based assay developed by Hartig and colleagues, the stem-loop domain (C) prevents cleavage of the FRET oligo when not bound to target by restricting the ribozyme to an inactive conformation and blocking the substrate binding domain (A). Figure adapted from the American Chemical Society: J.S. Hartig, I. Grüne, et al., Sequence-specific detection of microRNAs by signal-amplifying ribozymes. J Am Chem Soc 126 (2004) 722–3.
There are several benefits to this ribozyme design strategy. For instance, the intermolecular ribozyme design is capable of rapid substrate turnover, which provides an amplification of the signal and thereby improves the assay sensitivity. Additionally, the C domain provides excellent target specificity. There were a total of nine miRNA sequences from Drosophila detected using this assay. There was no noticeable difference in signal output between reactions containing only the target sequence and reactions containing the target plus a matrix of the other eight miRNA sequences. This stringent discrimination was further verified when signal outputs for samples containing either no sequences or the eight non-target sequences were virtually the same. In order to verify that this assay exhibited higher sensitivity than other direct, in situ detection methods, it was compared side by side to a molecular beacon assay. In all cases, the ribozyme assay yielded a 20-fold higher signal and a much smaller standard deviation than the molecular beacon assay. This is because the molecular beacon signal is directly dependent upon the target concentration and lacks any signal amplification step. While the signal amplification afforded by the ribozyme approach does give a much better signal to noise ratio, it tends to complicate the use of this assay for quantitative measurements. The reaction time and assay conditions must be carefully regulated to ensure the substrate turnover rate remains relatively the same for all trials. While it is true that the entire ribozyme structure must be synthesized specifically for each target, the same FRET oligonucleotide can be used in all cases. One concern with this assay is that the ribozyme will always exhibit some small amount of catalytic activity, even in the absence of target. However, upon the addition of target the ribozyme reaction rate increases 10-fold. Therefore, with prompt application of the assay this anomaly will cause negligible background noise. Though no lower detection limit was reported for this specific assay, it appears by statistical analysis of the plots provided in the supporting information that it is sequence dependent and varies from about 3 to 5 nM.
Electrochemical-Based miRNA Detection
Another detection principle employed in quantitative detection of miRNA is based on electrochemical measurements. Previous direct electrochemical detection methods, of small molecules, have been limited by the low readout signals produced by these systems and the poor binding of small oligonucleotide probes to the miRNA. Gao and colleagues, in a series of articles, have addressed these issues; these articles employ electrocatalytic moieties to enhance the readout from redox reactions upon binding miRNA. Gao’s work has focused on designing simple assays, which require very little sample preparation, small amounts of total RNA sample, and inexpensive equipment. These assays use electrocatalytic moieties, which, under mild conditions, are chemically ligated directly to the miRNA. The attached electrocatalytic moieties then assist an oxidation reaction, which is recorded amperometrically. Detection limits for these assays have been reported in the femtomolar scale, which is comparable to the more common Northern blotting and PCR-based assays.
Two such articles by Gao and colleagues utilized direct chemical ligation to tag miRNAs, in a total RNA, with an electrocatalytic moiety. [31; 32] The first of these studies used the electrocatalytic properties of an isoniazid-substituted osmium complex, (Os(dmpy)2(IN)Cl+) on the oxidation of ascorbic acid (AA). [32] Total RNA samples were extracted from HeLa cells. The miRNA within this sample was ligated, via a covalent bond, to (Os(dmpy)2(IN)Cl+) through a simple condensation reaction (Figure 11). The ligated and tagged miRNA, from the total RNA sample, was extracted via ethanol precipitation. The tagged miRNAs were hybridized to complementary oligonucleotide primers immobilized onto a gold electrode, and washed. The electro-oxidation current, of the AA, was measured in a PBS buffer containing 10 mM AA at 0.15 V. A further series of experiments by Gao and colleagues utilizing the same principal focused on the electrocatalytic moiety, Ru(PD)2Cl2 [PD=1,10-phenanthroline-5,6-dione], to monitor the oxidation of hydrazine. [31] The electrocatalytic moiety was attached to the miRNA, again, in a total RNA sample through coordinative bonds with the miRNA purine bases. The tagged miRNA was hybridized to immobilized complementary oligonucleotides, washed, and the oxidation signal was recorded after the addition of hydrazine (Figure 12). Additionally, this work used an indium tin oxide (ITO) electrode rather than the gold used for the previously described work.
Figure 11.
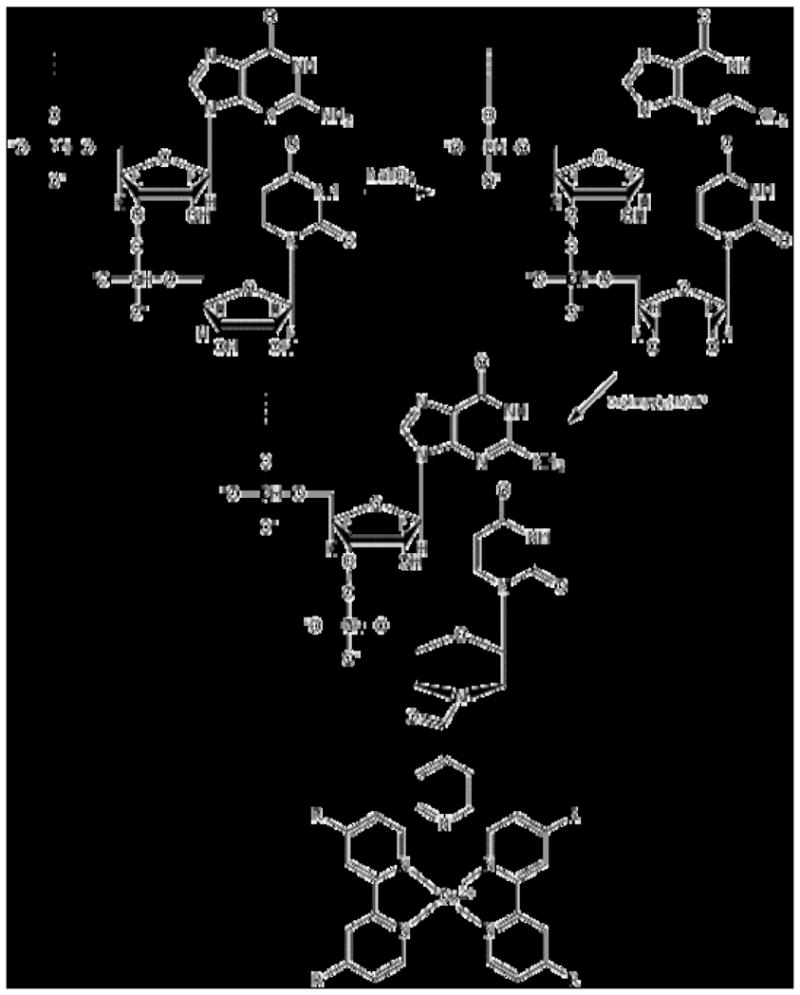
In the electrochemical method developed by Gao et al, the miRNA was ligated, via a covalent bond, to (Os(dmpy)2(IN)Cl+) through a simple condensation reaction. Figure adapted from Elsevier B.V.: Z. Gao, and Y.H. Yu, A microRNA biosensor based on direct chemical ligation and electrochemically amplified detection. Sensors and Actuators B: Chemical 121 (2007) 552–9.
Figure 12.
For the electrochemical work by Gao et al., the labeled miRNA was hybridized to the immobilized probes; the oxidation signal was recorded upon the addition of hydrazine. Figure adapted from Elsevier B.V.: Z. Gao, and Y.H. Yu, Direct labeling microRNA with an electrocatalytic moiety and its application in ultrasensitive microRNA assays. Biosensors and Bioelectronics 22 (2007) 933–40.
The quantitation of the hybridized miRNA in both of these studies was determined by the creation of a calibration curve correlating recorded current (μA) to miRNA concentration. In order for a linear relationship to be recorded the rate-determining step must be the catalytic process. This was achieved by increasing the substrate concentration, which increases the mass-transport rates. Additionally, higher electron-transfer rates were achieved by working with a more positive applied potential. For these studies, an ideal set of parameters was deemed to be 10 mM AA at 0.15 V and 5 mM hydrazine at 0.10 V. The selectivity of the assays was also evaluated, each showing clear discrimination between single base-mismatched miRNA and probe. The recorded current of the AA and hydrazine oxidation from tagged let-7b, let-7c, and mir-106 (mir-92 for hydrazine) hybridized to let-7b complementary oligonucleotide probes were compared. These showed clear differentiation between the complementary and non-complementary miRNAs, including a greater than 75% decrease in signal for the let-7c, which has only a single base mismatch with let-7b.
These assays demonstrated a simple, efficient, and low-cost way to quantitatively determine the presence of miRNA. Both proved to be ultrasensitive and selective with total RNA sample sizes as small as 10 ng, showing an improvement over previous methods which have been limited by the need for extensive sample preparation and/or miRNA extension (or amplification). Detection limits of 500 femtomolar (using AA) and 0.20 picomolar (using hydrazine) have been reported for these assays, offering an improvement over optical methods, with much less time. The assays reported here can yield results within hours, post extraction the miRNA is reacted with the tag for 3 h and the hybridization to complementary probes needs an additional hour. These methods could also easily be adapted to low-density electrochemical arrays for miRNA expression profiling. While the authors do state that the total miRNA recovered from the cell extract was quantified for both assays by UV-Vis neither article gives any indication of the efficiency of their extraction protocols. Additionally the authors are less than clear about the efficiency of their tagging method, stating that gel electrophoresis indicated that the miRNA was tagged but not how complete this tagging was.
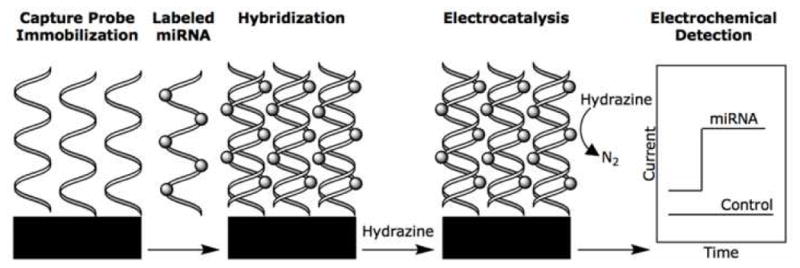
Further work by Gao and colleagues continued to focus on electrochemical detection of miRNA, however this work utilized electrocatalytic nanoparticle tags rather than chemical moieties used for the previous studies. [33] For this work OsO2 nanoparticles, which catalyze the oxidation of hydrazine, were attached to the miRNAs following their hybridization to immobilized capture probes, on an ITO electrode. Immobilized oligonucleotide probes were incubated with the total RNA from extracted HeLa cells to allow the miRNA to hybridize to the probes. The hybridized miRNA was reacted with OsO2 nanoparticles, in a hydrazine buffer, and washed with buffer to remove any unbound OsO2, and the oxidation potential of the hydrazine recorded. The nanoparticles were chemically ligated to the miRNA through a condensation reaction between isoniazid molecules, grafted onto the nanoparticles, and the 3′-end dialdhydes of the miRNA (Figure 13). As described for the previous studies, mass-transport and electron transfer parameters were evaluated and an ideal set of conditions determined, −0.10 V and 30 mM hydrazine in PBS buffer. Once these parameters had been stabilized, the read out potential was directly correlated to the concentration of the hybridized miRNA.
Figure 13.
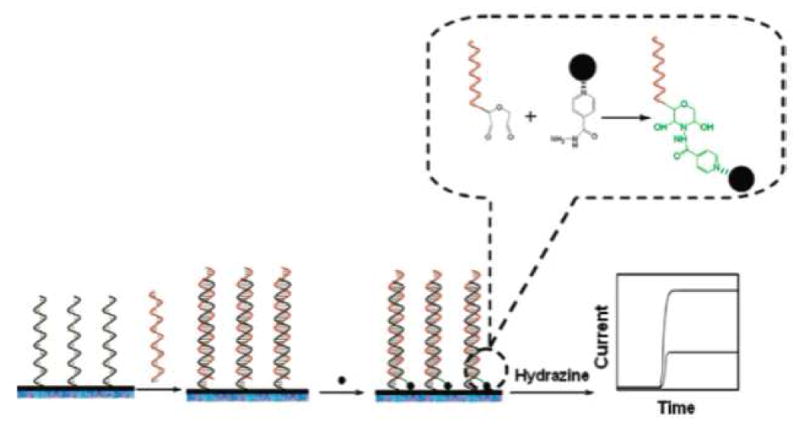
Nanoparticles were chemically ligated to the hybridized miRNAs, in the electrocatalytic assays developed by Gao et al., through a condensation reaction. Reprinted by permission from the American Chemical Society: Z. Gao, and Z. Yang, Detection of microRNAs using electrocatalytic nanoparticle tags. Anal Chem 78 (2006) 1470–7.
The assay reported a linear relationship between current and concentration from 0.3 – 200 pM miRNA with a measureable signal reported for as low as 80 fM miRNA. The specificity of the assay was evaluated by comparison of the amperometric readings of let-7b, mir-106, and mir-139 hybridized to let-7b complementary probes. Additionally, comparison between let-7b and let-7c, showed that the assay can easily distinguish between a single base mismatch, with a signal detected for fully matched miRNA and less than 25% signal reported for mismatched miRNA. These results are comparable to the previous electrochemical work by this group, and offer many of the same advantages over more conventional methods, such as PCR-based and Northern blot techniques. The use of these nanoparticles, over the electrocatalytic moieties presented previously in this section offer additional advantages for the electrocatalytic quantification of miRNA. These advantages include control over the choice of the capping groups on the nanoparticle, which simplifies their ligation to the miRNA and the improved catalytic effect on the oxidation of the hydrazine that results in improved signal. However the authors do not address the efficiency and reliability of the conjugation of the nanoparticle tag to the miRNA.
An electrochemical assay was developed by Fan and colleagues, which utilized conducting polymer nanowires for miRNA quantification. [34] For this work PNA capture probes were immobilized onto a nano-gapped biosensor array made up of 100 pairs of interlocking comb-like microelectrodes. These electrodes have 150–200 fingers, each 700 nm wide and 200 μm long, and separated by a 300 nm gap. These fingers allow for simple multiplexing, if different probes were immobilized on each. A total RNA sample from Hela and lung cancer cells was hybridized to these capture probes, creating a net negative charge. The hybridized miRNA were then incubated with a mixture of aniline/HRP/H2O2 at pH 4.0, the protonated aniline molecules align around the negatively charged, hybridized miRNA, forming polyaniline polymers. These conducting polymer polyaniline (PAn) nanowires were then expanded, by doping with HCl vapors, creating an electrically conducting network bridging the gaps of the biosensor array (Figure 14). Since the total deposition of these nanowires, and their electrically conducting network, is dependent upon the amount of hybridized miRNA, a calibration curve was created correlating the recorded conductance to the concentration of miRNA present.
Figure 14.
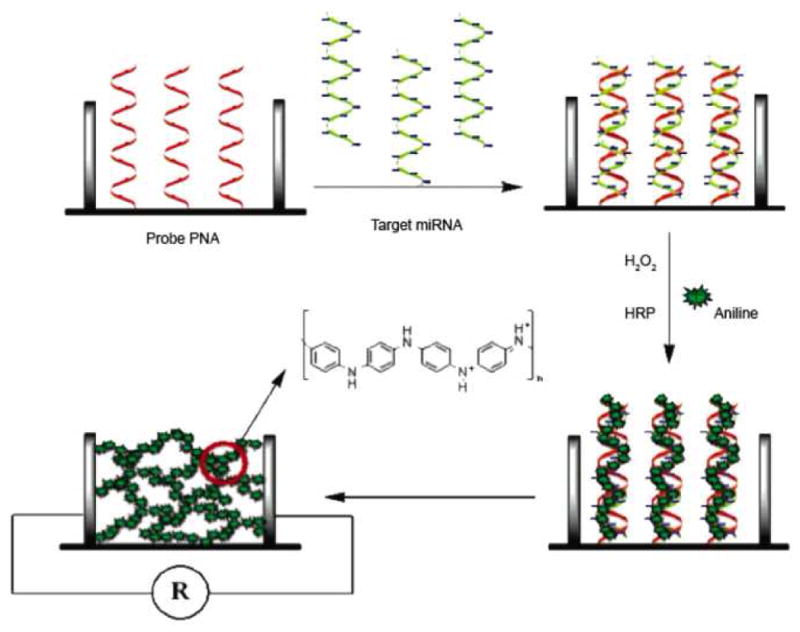
For the electrochemical assay developed by Fan and colleagues, the total RNA sample was hybridized to the capture probes, creating a net negative charge. The hybridized miRNA was then incubated with an aniline mixture. The protonated aniline molecules align around the hybridized miRNA forming polyaniline polymers. The nanowires were then expanded by doping with HCl vapors, creating an electrically conducting network bridging the gaps of the biosensor array. Reprinted by permission from the American Chemical Society: Y. Fan, X. Chen, et al., Detection of microRNAs using target-guided formation of conducting polymer nanowires in nanogaps. J Am Chem Soc 129 (2007) 5437–43.
The assay showed a dynamic range from 20 pM to 10 fM and a detection limit of 5.0 fM, which is more sensitive than the previously reported electrocatalytic methods of Gao and colleagues. The selectivity of this assay was examined, by comparing the output of perfectly matched and single base mismatched miRNAs, let-7b and let-7c respectively; with let-7b capture probes. As seen for the previous assays reported by Gao et al., the single polymorphism of let-7c can easily be discriminated, however this assay showed an even greater signal increase of 20:1 for let-7b over let-7c. Finally the blank sample, without target miRNA, demonstrated that the non-hybridization related signal is extremely low. While this assay offers many advantages over the previously discussed electrochemical methods, the major improvement is due to the PNA capture probes. These probes have a much higher affinity for miRNA, due to the absence of repulsion, normally caused by electrostatic charges along the backbones of the probe and the miRNA. Additionally, the neutral backbone of these probes prevents adsorption of the PAn, decreasing the background noise.
The work described above by Gao and Fan offers many advantages over optical detection methods. These methods yield sensitive and selective results on a scale not previously seen for electrochemical detection. Gao and Fan have demonstrated detection limits in the fM range, which can be obtained with as little as 10 ng of total RNA. These detection limits showed a three orders of magnitude sensitivity improvement over direct voltammetry. Expression level studies indicated good agreement with the more common Northern blotting, while being more efficient and more sensitive. These also offer the advantage of requiring mostly inexpensive simple electronics, little to no sample preparation, and smaller samples. Since most of these methods label the miRNA itself rather than the oligonucleotide probe, and these attachments are made under mild conditions, the issues seen previously with labeling short sequences is reduced. A further improvement is the labeling of the target post hybridization, to prevent the label/tag from interfering with the hybridization.
Summary
In this review we have covered several unique miRNA detection and quantification techniques that may be applied to direct or in situ screening assays. While these novel designs hold immense promise for the future of miRNA detection, there are still several issues which must be addressed before they are accepted among the current standard PCR or Northern blotting methods. Generally, the methods must be more adaptable. For instance, the molecular beacon design by Paiboonskuwong is currently limited to miRNA targets that are flanked by guanines in their primary transcripts. Without the guanine in appropriate proximity to the quencher, detection is not limited to the mature form of the target. In the methods developed by Gao and Fan, the efficiency of tagging of miRNAs with electrocatalytic moieties and nanoparticles must be further evaluated before their methods can be considered mainstream. Of equal concern is that many of the methods described in this review are not adaptable to multiplex miRNA detection platforms, which is essential for miRNA expression profiling. While many of the discussed assays demonstrated exceptional adaptability, the assay developed by Neely for instance (which tested 45 human miRNAs in 16 different tissues), the robustness of these methods must be further explored before they can be used as standard procedures. This is especially true of the solution phase assays, which may be applied directly to a variety of complex matrices and hopefully display acceptable background tolerance. These are the kinds of challenges the discussed assays will face as they are further developed.
As we continue to discover the vast impact that miRNAs have on the course of diseases, such as cancer, and the immense promise they hold in the area of gene therapy, rapid and sensitive methods for their direct detection and quantification are of utmost importance. A simple approach to direct detection is to monitor some type of hybridization event. However, the small size and low in situ concentration of miRNA molecules typically foist signal or target amplification as a prerequisite and make the direct detection approach less viable through procedural complication. To address these problems, novel techniques have been developed which employ one or more of the following: signal amplification through enzyme substrate turnover, highly favorable hybridization conditions using LNA or PNA capture probes, highly sensitive forms of spectroscopy such as FCS or bioluminescence, optimized reporter molecules such as gold nanoparticles, or highly attuned and responsive electrocatalytic/amperometric monitoring.
These discussed methods stand out above the rest because they have achieved comparable or improved sensitivity and specificity with respect to the current “gold standards” without the necessity for potentially hazardous materials, such as radioactive labels or toxic dyes, and highly expensive imaging equipment. These factors are becoming increasingly important in today’s environmentally conscious society, and by drastically reducing the procedural complexity and overall expense of such assays, it opens the doors to academic environments making research in the field of miRNAs much more accessible.
Acknowledgments
We appreciate funding from the National Institutes of Health (EB008231-01) and the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006 doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–12. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 6.Stahlhut Espinosa CE, Slack FJ. The role of microRNAs in cancer. Yale J Biol Med. 2006;79:131–40. [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grosshans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451:414–6. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 10.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 11.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 12.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 13.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;442:82–5. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- 15.Cissell KA, Shrestha S, Deo SKD. MicroRNA Detection: Challenges for the Analytical Chemist. Anal Chem. 2007;79:4754–4761. [Google Scholar]
- 16.Wark AW, Lee HJ, Corn RM. Multiplexed detection methods for profiling microRNA expression in biological samples. Angew Chem Int Ed Engl. 2008;47:644–52. doi: 10.1002/anie.200702450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang WJ, Li XB, Li YY, Zhao LF, He WL, Gao YQ, Wan YJ, Xia W, Chen T, Zheng H, Li M, Xu SQ. Quantification of microRNA by gold nanoparticle probes. Anal Biochem. 2008;376:183–8. doi: 10.1016/j.ab.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–9. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 19.Neely L, Patel S, Garver J, Gallo M, Hackett M, Mclaughlin S, Nadel M, Harris J, Gullans S, Rooke J. A single-molecule method for the quantitation of microRNA gene expression. Nat Methods. 2006;3:41–6. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- 20.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–8. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 21.Paiboonskuwong K, Kato Y. Detection of the mature, but not precursor, RNA using a fluorescent DNA probe. Nucleic Acids Symp Ser. 2006:327–8. doi: 10.1093/nass/nrl163. [DOI] [PubMed] [Google Scholar]
- 22.Allawi HT, Dahlberg JE, Olson S, Lund E, Olson M, Ma WP, Takova T, Neri BP, Lyamichev VI. Quantitation of microRNAs using a modified Invader assay. RNA. 2004;10:1153–61. doi: 10.1261/rna.5250604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Arruda M, Lyamichev VI, Eis PS, Iszczyszyn W, Kwiatkowski RW, Law SM, Olson MC, Rasmussen EB. Invader technology for DNA and RNA analysis: principles and applications. Expert Rev Mol Diagn. 2002;2:487–96. doi: 10.1586/14737159.2.5.487. [DOI] [PubMed] [Google Scholar]
- 24.Eis PS, Olson MC, Takova T, Curtis ML, Olson SM, Vener TI, Ip HS, Vedvik KL, Bartholomay CT, Allawi HT, Ma WP, Hall JG, Morin MD, Rushmore TH, Lyamichev VI, Kwiatkowski RW. An invasive cleavage assay for direct quantitation of specific RNAs. Nat Biotechnol. 2001;19:673–6. doi: 10.1038/90290. [DOI] [PubMed] [Google Scholar]
- 25.Su X, Teh HF, Lieu X, Gao Z. Enzyme-based colorimetric detection of nucleic acids using peptide nucleic acid-immobilized microwell plates. Anal Chem. 2007;79:7192–7. doi: 10.1021/ac0709403. [DOI] [PubMed] [Google Scholar]
- 26.Cissell KA, Campbell S, Deo SK. Rapid, single-step nucleic acid detection. Anal Bioanal Chem. 2008;391:2577–81. doi: 10.1007/s00216-008-2215-5. [DOI] [PubMed] [Google Scholar]
- 27.Cissell KA, Rahimi Y, Shrestha S, Hunt EA, Deo SKD. Bioluminescence-based detection of microRNA, miR21 in breast cancer cells. Anal Chem. 2008;80:2319–25. doi: 10.1021/ac702577a. [DOI] [PubMed] [Google Scholar]
- 28.Tong Z, Yuan R, Chai Y, Chen S, Xie Y. Direct electrochemistry of horseradish peroxidase immobilized on DNA/electrodeposited zirconium dioxide modified, gold disk electrode. Biotechnol Lett. 2007;29:791–5. doi: 10.1007/s10529-007-9319-4. [DOI] [PubMed] [Google Scholar]
- 29.Hartig JS, Grüne I, Najafi-Shoushtari SH, Famulok M. Sequence-specific detection of MicroRNAs by signal-amplifying ribozymes. J Am Chem Soc. 2004;126:722–3. doi: 10.1021/ja038822u. [DOI] [PubMed] [Google Scholar]
- 30.Fedor MJ. Structure and function of the hairpin ribozyme. J Mol Biol. 2000;297:269–91. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- 31.Gao Z, Yu YH. Direct labeling microRNA with an electrocatalytic moiety and its application in ultrasensitive microRNA assays. Biosens Bioelectron. 2007;22:933–40. doi: 10.1016/j.bios.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Gao Z, Yu YH. A microRNA biosensor based on direct chemical ligation and electrochemically amplified detection. Sens Actuators, B. 2007;121:552–9. [Google Scholar]
- 33.Gao Z, Yang Z. Detection of microRNAs using electrocatalytic nanoparticle tags. Anal Chem. 2006;78:1470–7. doi: 10.1021/ac051726m. [DOI] [PubMed] [Google Scholar]
- 34.Fan Y, Chen X, Trigg AD, Tung CH, Kong J, Gao Z. Detection of MicroRNAs using target-guided formation of conducting polymer nanowires in nanogaps. J Am Chem Soc. 2007;129:5437–43. doi: 10.1021/ja067477g. [DOI] [PubMed] [Google Scholar]


