Abstract
Introduction
Aspirin, clopidogrel, prasugrel and ticagrelor are antiplatelet agents for the prevention of ischemic events in patients with acute coronary syndromes (ACS), percutaneous coronary intervention (PCI), and other indications. Variability in response is observed to different degrees with these agents, which can translate to increased risks for adverse cardiovascular events. As such, potential pharmacogenetic determinants of antiplatelet pharmacokinetics, pharmacodynamics and clinical outcomes have been actively studied.
Areas covered
This article provides an overview of the available antiplatelet pharmacogenetics literature. Evidence supporting the significance of candidate genes and their potential influence on antiplatelet response and clinical outcomes are summarized and evaluated. Additional focus is directed at CYP2C19 and clopidogrel response, including the availability of clinical testing and genotype-directed antiplatelet therapy.
Expert opinion
The reported aspirin response candidate genes have not been adequately replicated and few candidate genes have thus far been implicated in prasugrel or ticagrelor response. However, abundant data supports the clinical validity of CYP2C19 and clopidogrel response variability among ACS/PCI patients. Although limited prospective trial data are available to support the utility of routine CYP2C19 testing, the increased risks for reduced clopidogrel efficacy among ACS/PCI patients that carry CYP2C19 loss-of-function alleles should be considered when genotype results are available.
Keywords: Antiplatelet agents, aspirin, clopidogrel, prasugrel, ticagrelor, candidate genes, CYP2C19, pharmacogenetics, pharmacogenomics
1. INTRODUCTION
Platelets are activated in response to vascular injury and/or atherosclerotic plaque rupture through a complex network of intra- and intercellular pathways [1]. Activated platelets facilitate cell adhesion, initiate the arachidonic acid (AA) pathway to produce thromboxane A2 (TXA2), and excrete adenosine diphosphate (ADP), serotonin and other proteins from their granules, ultimately to form a platelet clot and eventually a thrombus. Given this fundamental role that platelets have in blood loss prevention and vasculature integrity, they are inherently implicated in cardiovascular diseases such as atherosclerosis, coronary artery disease (CAD) and myocardial infarction (MI), as well as cerebrovascular disease and stroke.
Since platelet activation is influenced, in part, by TXA2, ADP, serotonin, thrombin, epinephrine and collagen [2], these biological pathways have been leveraged as potential targets for antiplatelet therapies. The currently approved oral antiplatelet agents include aspirin, clopidogrel, prasugrel and ticagrelor, which are prescribed for prevention of ischemic events among patients with ischemic stroke and symptomatic peripheral artery disease (PAD), and as dual antiplatelet therapy (DAPT) for patients with acute coronary syndromes (ACS). However, variability in patient response to these agents are observed, which can translate to increased risks for adverse cardiovascular events [3, 4]. Potential pharmacogenetic determinants of response variability have been actively studied for all the antiplatelet agents, but none more so than clopidogrel.
The identification of a biologically relevant candidate gene for clopidogrel responsiveness (i.e., cytochrome P450-2C19) and the availability of alternative antiplatelet therapies have provided the opportunity for genotype-directed antiplatelet therapy in selected patient populations. However, despite the enthusiasm for personalized antiplatelet therapy from advocates of this paradigm, the uncertain clinical utility and cost-effectiveness of this approach has made the field of antiplatelet pharmacogenetics highly studied and frequently debated. The current status of antiplatelet pharmacogenetics, with an emphasis on antiplatelet candidate genes and clinical pharmacogenetic testing implementation, is the focus of this review and is detailed below.
2. ASPIRIN PHARMACOGENETICS
2.1. ASPIRIN RESPONSE
Given its benefit in reducing arterial thrombosis and recurrent cardiovascular events, aspirin (or acetylsalicylic acid) has remained a mainstay in antiplatelet therapy with indications including coronary, cerebral, and peripheral vascular disease. However, despite its general effectiveness in most patients, a considerable number of individuals experience sub-optimal aspirin response, assessed by either laboratory platelet reactivity testing or through its ability to prevent cardiovascular events. This interindividual variability in platelet response to aspirin has been well documented, and patients with high on-treatment platelet reactivity (HTPR) to AA have increased risk of ischemic events [5–7]. Although estimates of aspirin non-responsiveness are controversial given the lack of standardized definitions, the use of multiple platelet function tests and evaluation of different surrogate endpoints suggest that 5–60% of patients do not adequately respond to aspirin [8].
Platelet response to aspirin is influenced by several clinical variables (e.g., age, gender, smoking, and non-adherence) and coexisting comorbidities including obesity, diabetes, and hyperlipidemia [9]; however, these factors only explain ~15% of the variability in on-treatment ex vivo platelet aggregation [10]. Heritability estimates suggest that 14–39% of the variability in platelet responsiveness to aspirin can be attributed to genetic factors, and potentially through variants that influence both cyclooxygenase-1 (COX1)-dependent and COX1-independent platelet activation pathways [10].
Aspirin inhibits platelet aggregation primarily by the irreversible acetylation of COX1, which prevents the conversion of AA to TXA2, a potent platelet agonist. As such, most traditional tests of aspirin response have focused on the COX1 pathway through measurement of AA-stimulated platelet aggregation or circulating thromboxane B2 levels, the stable inactive metabolite of TXA2. Using such assays, aspirin leads to near complete inhibition of COX1 in approximately 95% of individuals [11, 12] suggesting that a substantial proportion of the variability in response is mediated by factors outside of the COX1 pathway. While COX1 inhibition is nearly complete, the effect of aspirin on other platelet activation pathways (e.g., collagen, epinephrine, and ADP) is more heterogeneous and may explain, in part, the observed variability in response. Recent studies using collagen-stimulated platelet aggregation have identified novel circulating biomarkers and genetic risk loci associated with response variability [13–15]. Consequently, while COX1 dependent platelet function assays are the most specific test of aspirin’s canonical mechanism of action, recent studies have increasingly used non-COX1-dependent assays to more comprehensively define aspirin response and to identify novel genetic determinants of on-treatment platelet aggregation and cardiovascular outcomes.
2.2. ASPIRIN CANDIDATE GENES
Most of the initial pharmacogenetic studies of aspirin response variability consisted of relatively underpowered candidate gene studies with different designs, participant selection (i.e., healthy vs. CAD/ACS patients), and primary outcome (i.e., platelet aggregation vs. cardiovascular events). Furthermore, these studies used different aspirin response phenotypes and platelet function tests [e.g., light transmission aggregometry, platelet function analyzer-100 (PFA-100), and VerifyNow® Aspirin], which subsequently have been shown to poorly correlate given the lack of standard definitions of aspirin responsiveness and the fact that these assays measure different platelet activation pathways (e.g., AA, epinephrine, and collagen) [16, 17]. Although variability in platelet function testing has been previously reviewed [9, 18], it is important to consider these limitations when assessing the potential roles of the following candidate genes in aspirin response variability.
2.2.1. Cyclooxygenase-1 (COX1)
Given that COX1 is the molecular target of aspirin, multiple studies have evaluated the effect of genetic variants in the COX1 gene [also known as prostaglandin synthase 1 (PTGS-1)] on aspirin response, most commonly involving the linked c.-842A>G (rs10306114) and c.50C>T (rs3842787) variants [19]. Although it was initially reported that healthy individuals with the minor haplotype (c.[-842G;50T]) had better on-treatment inhibition of prostaglandin H2 and AA-induced platelet aggregation compared to those with the common haplotype (c.[-842A;50C]) [19], subsequent studies on stable CAD patients identified the c.-842G allele to actually be associated with aspirin resistance and non-responsiveness based on AA-induced platelet aggregation and serum TXB2 levels [20]. More recent studies on the COX1 c.-842A>G and c.50C>T variants using several different aspirin response phenotypes and platelet function tests observed no significant association between these variants and TBX2 levels, platelet aggregation, or cardiovascular outcomes [21–28], including a recent systematic review [29]. Consequently, the available evidence does not support a clinically relevant role for COX1 variants in aspirin response.
2.2.2. Glycoprotein IIIa (GPIIIa)
The glycoprotein IIb/IIIa complex (GPIIb/IIIa) is a critical regulator of thrombosis formation through its ability to bind fibrinogen resulting in platelet-platelet crosslinks. The PIA1/A2 (c.176T>C, p.L59P, rs5918) variant in the ITGB3 gene that encodes the GPIIIa subunit has been extensively studied as a risk factor for cardiovascular disease and drug response to both aspirin and the GPIIb/IIIa inhibitor abciximab. A thorough review of ITGB3 PIA1/A2, including its potential effect on aspirin response, has been previously reported [18]. Although there is evidence suggesting that the PIA2 allele contributes to MI, stent thrombosis, unstable angina and sudden cardiac death, studies measuring the effect of this variant on aspirin response have been less conclusive. Collectively, using different platelet function tests and aspirin response definitions, these studies have reported that the PIA2 allele results in increased, decreased, or no change in on-treatment platelet reactivity [9]. A recent systematic review has highlighted the inconsistency in PIA1/A2 study results, most likely due to differing platelet function tests and/or study cohorts [29]. As a result, while ITGB3 PIA1/A2 likely influences coronary thrombosis and the occurrence of stent thrombosis under DAPT [30], its role in aspirin response variability remains undetermined.
2.2.3. Glycoproteins VI (GPVI), GPIa/IIa, and GPIbα
Given that collagen stimulates platelet aggregation by binding to glycoprotein VI (GPVI) and the glycoprotein Ia/IIa (GPIa/IIa) receptor complex on the platelet surface, these genes have been considered as candidates for aspirin response variability. Pharmacogenetic studies of GPVI and aspirin response have led to mixed results. Specifically, the common GPVI c.655C>T variant (p.P219S, rs1613662) has been associated with on-treatment platelet function variability in CAD patients [20]; however, subsequent studies have not replicated this finding [24, 26]. The commonly studied c.759C>T variant of the GPIa gene (ITGA2; rs1126643) has been associated with increased risk of stroke, MI, and cardiovascular death [31–33]; however, studies on platelet aggregation variability after DAPT with aspirin and clopidogrel have also been conflicting and largely not supportive of a clinically meaningful effect on drug response [34–37]. Given that most of these studies were small and generally underpowered, larger scale replication efforts will be needed to determine the precise roles of these variants on aspirin response.
The GPIbα c.-5T>C variant (rs2243093) of the Von Willebrand receptor has also been studied as a candidate for aspirin response variability. Initial studies suggested that this variant altered GPIbα mRNA translation [38] and was associated with increased platelet reactivity (as measured by reduced PFA-100 closure time) and increased risk of MI among aspirin-treated CAD patients [39, 40]. In contrast, subsequent studies reported no evidence of association between c.-5T>C and cardiovascular outcomes or aspirin response (including TXB2 levels, collagen-stimulated platelet aggregation and PFA-100 closure time) [26, 35, 41, 42]. Taken together, the available evidence does not support a role for the GPIbα c.-5T>C variant in aspirin efficacy.
2.2.4. Platelet Endothelial Aggregation Receptor 1 (PEAR1)
The platelet endothelial aggregation receptor 1 (PEAR1) is a type 1 transmembrane receptor that is involved in platelet aggregation through GPIIb/IIIa [43] as well as altered megakaryopoiesis and thrombopoiesis via the PI3K/PTEN pathways [44]. Early genetic studies identified several PEAR1 variants significantly associated with platelet aggregation in response to multiple agonists before [45, 46] and after [47–50] aspirin exposure. The most notable PEAR1 association has been between the intronic rs12041331 variant and ex vivo platelet aggregation in response to several platelet agonists (ADP, collagen, epinephrine) as well as pre- and post-antiplatelet therapy treatment (i.e., aspirin and prasugrel) [46, 48–51]. Furthermore, PEAR1 rs12041331 significantly reduced 1-year survival in aspirin-treated patients undergoing percutaneous coronary intervention (PCI) and increased rates of MI in an independent cohort of aspirin-treated patients with stable CAD [49]. Paradoxically, the allele that was associated with improved aspirin response, as defined by ex vivo platelet aggregometry, was the same allele that resulted in an increased risk of experiencing a thrombotic event. In addition, a recent study did not detect any association between PEAR1 rs12041331 and clinical outcomes in CAD patients [26], indicating that additional clinical studies on PEAR1 and aspirin response are still warranted.
2.3. Other Aspirin Candidate Genes
Other commonly investigated aspirin response candidate genes include the TXA2 receptor (TBXA2R), ADP receptors (P2RY1 and P2RY12), coagulation factor XIII (F13A1), and UDP-glucuronosyltransferase 1A6 (UGT1A6) [18, 52]; however, their inconsistent results make it difficult to form any firm conclusions.
3. CLOPIDOGREL PHARMACOGENETICS
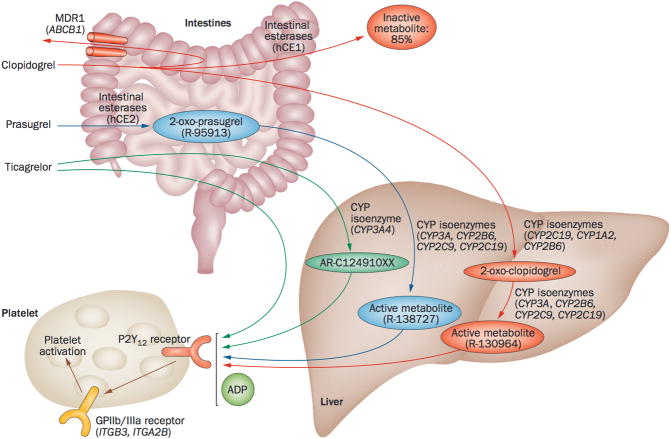
Clopidogrel is a second generation thienopyridine that undergoes hepatic biotransformation to an active metabolite, which binds irreversibly to the P2Y12 receptor and inhibits ADP-mediated platelet activation and aggregation (Figure 1). The majority of clopidogrel (~85%) is hydrolyzed to inactive metabolites by esterases, including carboxylesterase 1 (CES1), leaving only ~15% available for transformation to the active metabolite [53]. Two sequential oxidative reactions by the cytochrome P450 (CYP450) system form the active metabolite: the first involving CYP1A2, CYP2B6 and CYP2C19, and the second involving CYP2B6, CYP2C9, CYP2C19, CYP3A4 and CYP3A5 [53, 54]. Clopidogrel and aspirin administered as DAPT reduces cardiovascular death and ischemic events in ACS patients and those undergoing PCI [55–57]. However, wide interindividual variability in ex vivo platelet aggregation is common among DAPT-treated patients, and some still experience thrombotic events [58–60]. Importantly, patients with persistent HTPR to ADP are at increased risk for adverse cardiovascular events [3]. Other clinical factors implicated in clopidogrel response variability include age, co-medications, diabetes, disease activity, renal failure, and cardiac failure.
Figure 1.
Metabolic pathway of P2Y12-receptor inhibitors. CYP: cytochrome P450; GP: glycoprotein; hCE: human carboxylesterase; MDR1: multidrug resistance protein 1. Reprinted by permission from Macmillan Publishers Ltd: [199] © 2014.
In order to identify variants that influence clopidogrel response variability, a number of candidate genes in the clopidogrel pharmacokinetic and pharmacodynamic pathways have been studied. Among them, the most robust association has been with the common CYP2C19*2 loss-of-function allele (c.681G>A; rs4244285), which was initially reported in 2006 to be significantly associated with HTPR in healthy subjects [61]. As detailed below, since this initial observation, numerous studies have confirmed CYP2C19 as the major genetic determinant of clopidogrel metabolite levels, on-treatment platelet reactivity, and adverse cardiovascular event risks.
A single genome-wide association study (GWAS) on clopidogrel response has been reported, which identified variants in the CYP2C18-CYP2C19-CYP2C9-CYP2C8 gene cluster on chromosome 10q24 to be significantly associated with ADP-induced platelet aggregation in a healthy Amish cohort [62]. The most significant variant (rs12777823) was in strong linkage disequilibrium with CYP2C19*2, accounting for ~12% of the variation in ADP-induced platelet aggregation [62]. Although no other variants reached genome-wide significance, this agnostic GWAS validated previous CYP2C19 candidate gene studies and confirmed its role as the major genetic determinant of interindividual clopidogrel response variability. This important study also determined that on-treatment platelet response is a highly heritable trait (h2=0.73) [62].
3.1. CYTOCHROME P450-2C19 (CYP2C19)
The CYP2C19 enzyme metabolizes a large number of clinically relevant drugs (e.g., antidepressants, proton pump inhibitors) and has over 30 reported variant star (*) alleles, many of which encode reduced or complete loss-of-function enzyme variants [63–65]. Like many other CYP450 genes, CYP2C19*1 is considered the wild-type allele encoding normal enzyme activity. The CYP2C19*2 variant allele frequencies are ~15% in Caucasians and Africans, and 29–35% in Asians while the *3 loss-of-function allele (c.636G>A; rs4986893) is typically only found in Asians (2–9%) [66, 67]. The *4 – *8 alleles are rare in the general population (<1%) but have extensive in vitro evidence for loss-of-function [63]. In contrast, the CYP2C19*17 allele (c.-806C>T; rs12248560) results in enhanced transcription and increased activity, and has multi-ethnic allele frequencies ranging from ~3–21% [66]. Based on CYP2C19 genotype, individuals can be categorized as ultrarapid (*1/*17, *17/*17), extensive (*1/*1), intermediate (e.g., *1/*2, *1/*3, *2/*17), or poor (e.g., *2/*2, *2/*3) metabolizers [63].
3.2. CYP2C19 CLINICAL VALIDITY
3.2.1. Clopidogrel Pharmacokinetics and Pharmacodynamics
Given that CYP2C19 is directly involved in both steps of clopidogrel bioactivation, impaired CYP2C19 activity due to germline loss-of-function alleles results in reduced formation of the active metabolite in healthy subjects [68–74] and cardiac patients [75, 76]. Consistent with the association between CYP2C19 and clopidogrel pharmacokinetics, numerous studies have confirmed the important role of CYP2C19 loss-of-function alleles in HTPR, typically measured by ex vivo ADP-induced platelet aggregometry, in both healthy subjects [61, 62, 68, 71, 72, 77] and CAD patients [62, 71, 76, 78–84]. Although the *2 – *8 alleles are all loss-of-function variants, the *3 allele has been reported to have a greater effect on on-treatment platelet reactivity among East Asian PCI patients [85]; however, other studies have not detected any difference in effect between the CYP2C19 loss-of-function alleles on clopidogrel response [86–88]. In addition, some studies have also found that the CYP2C19*17 increased activity allele results in enhanced platelet inhibition and possibly an increased bleeding risk [71, 89–91]; however, this potential effect has not been replicated in all studies [62, 92–95].
3.2.2. Clinical Outcomes and Indication
In addition to the association between CYP2C19 loss-of-function alleles and reduced active clopidogrel metabolites and HTPR, evidence exists linking CYP2C19 genotype with clinical outcomes among clopidogrel-treated ACS patients, particularly those undergoing PCI. Since the initial 2008 report suggesting a relationship between CYP2C19*2, HTPR, and 1-year incidence of death and MI among clopidogrel-treated ACS/PCI patients [96], many clinical studies have detected a significant association between CYP2C19 loss-of-function alleles and adverse cardiovascular events (e.g., cardiovascular death, MI, stroke) including an increased risk for stent thrombosis [30, 62, 71, 78, 93, 97–99]. However, a significant effect of CYP2C19 on clinical outcomes has not been detected among lower risk CAD patient cohorts [e.g., those with low frequencies (<20%) of PCI] or patients with other indications (e.g., atrial fibrillation) [100–102], underscoring the importance of indication when considering CYP2C19 genetic testing for antiplatelet management [103]. This is supported by data from several studies that found the effect of CYP2C19 on clinical outcomes among patients treated with clopidogrel to be dependent on the indication for antiplatelet therapy [30, 62, 71, 78, 93, 97, 99, 100, 104, 105]. Lower risk indications, such as medical management of ACS and PAD, derive a lesser overall benefit from clopidogrel therapy compared to higher risk indications such as PCI [100, 102, 104]. Consequently, the influence of CYP2C19 on clinical outcomes has been most evident among PCI patient cohorts.
Reported meta-analyses suggest that clopidogrel-treated ACS/PCI patients who are CYP2C19*2 carriers have an increased risk compared to wild-type patients for both major adverse cardiovascular events (MACE) [hazard ratio (HR) 1.55, 95% confidence interval (CI) 1.11–2.17 for heterozygotes; HR 1.76, 95% CI 1.24–2.50 for homozygotes] and stent thrombosis (HR 2.67, 95% CI 1.69–4.22 for heterozygotes; HR 3.97, 95% CI 1.75–9.02 for homozygotes) [105]. The significance of CYP2C19*2 in stent thrombosis risk among ACS/PCI patients has been the most notable, with reported odds ratios for CYP2C19*2 carriers ranging from 1.75 to 4.68 [106–113]. Given the relationship between CYP2C19 and clopidogrel indication detailed above, meta-analyses that include studies with low frequencies of PCI or patients without coronary disease have not supported a major role for CYP2C19 in clopidogrel response variability in these specific patient populations [104].
3.3. CYP2C19 CLINICAL UTILITY AND PRACTICE GUIDELINES
3.3.1. Randomized Controlled Trials
Clinical utility is commonly established by prospective randomized controlled trials (RCTs); however, they can be challenging to implement for pharmacogenetic interventions for a number of reasons (e.g., ethnical considerations, rapid turnaround time genotyping, adequate power). Despite the difficulties with pharmacogenetic RCTs, CYP2C19 genotype-directed antiplatelet therapy trials have been reported (Table 1). For example, RAPID GENE concluded that point-of-care genetic testing after PCI can be done effectively at the bedside and that treatment of CYP2C19*2 carriers with prasugrel can reduce HTPR [114], which subsequently was expanded (with similar conclusions) to an ST-elevation MI patient cohort in the RAPID STEMI trial [115]. Notably, RAPID STEMI also showed no differences in risk for HTPR between prasugrel-treated CYP2C19*2 carriers and clopidogrel-treated non-carriers, suggesting that this strategy may have clinical utility. These pharmacodynamic end-point trials are further supported by a Korean RCT, which also concluded that genotype-directed antiplatelet therapy can reduce HTPR among ACS patients [116].
TABLE 1.
Reported randomized controlled trials testing CYP2C19 genotype-directed antiplatelet therapy
| Trial | Primary Endpoint | Sample Size | Clinicaltrials.gov Identifier | Major Finding | Reference |
|---|---|---|---|---|---|
| RAPID GENE | The proportion of CYP2C19*2 carriers with HTPR after 1 week of treatment | 187 | NCT01184300 | No CYP2C19*2 carriers treated with 10 mg prasugrel daily in the rapid genotyping group had HTPR at day 7, compared to 30% given standard treatment (75 mg clopidogrel daily) (p=0.0092). | [114] |
| RAPID STEMI | The proportion of CYP2C19*2 or ABCB1 TT carriers with HTPR after 1 month of treatment | 102 | NCT01452139 | Among carriers of at-risk genotypes, treatment with prasugrel was superior to an augmented dosing strategy of clopidogrel in reducing HTPR. | [115] |
| Ahn SG, et al. | The proportion of patients with HTPR after 30 days of treatment | 65 | NA | Tailored antiplatelet therapy according to point-of-care genetic and phenotypic testing reduced HTPR after 30 days. | [116] |
| Xie X, et al. | Composite of major adverse cardiac or cerebrovascular events, including death from any cause, MI, stroke and ischemia-driven target-vessel revascularization, for the 180-day period after randomization | 600 | ChiCTR-TRC-11001807 a | Personalized antiplatelet therapy according to CYP2C19 genotype after PCI can significantly decrease the incidence of MACE and the risk of 180-day ST in Chinese population. | [117] |
| GIANT | Death, MI and ST between genetically resistant patients (*2 genotype) with adapted treatment versus non-resistant patients (*1 genotype) | 1445 | NCT01134380 | CYP2C19 genotype-directed antiplatelet therapy post-PCI may reduce ischemic events at one year. | [118] |
| TAILOR-PCI | Occurrence of MACE one year after PCI | 5945 | NCT01742117 | Estimated primary completion: June 2016 | NA |
HTPR: high on-treatment platelet reactivity; MACE: major adverse cardiovascular events; MI: myocardial infarction; NA: not available; PCI: percutaneous coronary intervention; ST: stent thrombosis.
Chinese Clinical Trial Registry.
Although the reported pharmacodynamic RCTs support CYP2C19 genotype-directed antiplatelet therapy, RCTs powered for actual clinical outcomes are ultimately more likely to definitively establish or refute the clinical utility of CYP2C19 genotyping. Notably, a prospective RCT with a Chinese PCI patient cohort recently reported that CYP2C19 genotype-directed antiplatelet therapy significantly decreased the incidence of MACE and the risk of 180-day stent thrombosis [117]. In addition to this encouraging RCT, preliminary results from the Genotyping Infarct Patients to Adjust and Normalize Thienopyridine Treatment (GIANT) trial, presented at the 2013 Transcatheter Cardiovascular Therapeutics (TCT) annual meeting, suggest that CYP2C19 genotype-directed antiplatelet therapy post-PCI may reduce ischemic events at one year [118]. The influence of CYP2C19 genotype-directed antiplatelet therapy on clinical outcomes will also be evaluated by the much larger and ongoing Tailored Antiplatelet Therapy Following PCI (TAILOR-PCI) trial.
3.3.2. Professional Practice Guidelines
The U.S. Food and Drug Administration (FDA) added a boxed warning to the clopidogrel label in 2010 highlighting the role of CYP2C19 in clopidogrel response. The American College of Cardiology Foundation (ACCF) and the American Heart Association (AHA) with endorsement from the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons, subsequently issued a ‘clinical alert’ in response to the boxed warning [119]. This expert committee urged clinicians to be aware of CYP2C19, impaired clopidogrel metabolism and reduced platelet inhibition, but did not recommend routine genetic testing prior to clopidogrel initiation, citing a lack of sufficient evidence. However, they did suggest consideration of genetic testing for patients at moderate to high risk for poor outcomes (e.g., those undergoing elective high-risk PCI procedures), and that CYP2C19 poor metabolizers should be prescribed an alternative antiplatelet regimen [119]. Similarly, the 2012 ACCF/AHA ‘Focused Update’ guideline for management of unstable angina/non-ST-elevation MI did not recommend routine CYP2C19 genetic testing, but stated that it ‘might be considered if results of testing may alter management’ [120].
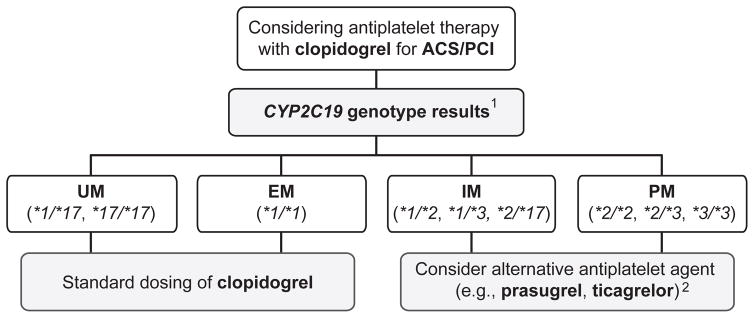
Additional practice guidelines are available for clinicians when the patient genotype is already known from previous and/or unrelated testing. These include the Royal Dutch Association for the Advancement of Pharmacy Pharmacogenetics Working Group (KNMP-PWG) [121], and the Clinical Pharmacogenetics Implementation Consortium (CPIC) [66], which have recommended consideration of an alternative antiplatelet agent (i.e., prasugrel or ticagrelor) for CAD patients who are either CYP2C19 intermediate (e.g., *1/*2) or poor metabolizers (e.g., *2/*2) (Figure 2). The CPIC recommendation is restricted to PCI patients as current evidence does not support CYP2C19 genotype-directed prescribing for patients with lower risk for adverse cardiovascular events (e.g., stroke, PAD; Figure 2) [103]. However, CYP2C19 genotype has recently been shown to influence clopidogrel efficacy and prognosis in ischemic stroke [122, 123] and subcortical stroke [124] patients, suggesting that CYP2C19 genotype-directed therapy for stroke patients might potentially be considered in the future.
Figure 2.
Clinical Pharmacogenetics Implementation Consortium (CPIC) algorithm for suggested clinical actions based on CYP2C19 genotype when considering clopidogrel treatment for ACS/PCI patients. ACS: acute coronary syndromes; PCI: percutaneous coronary intervention; UM: ultrarapid metabolizer; EM: extensive metabolizer; IM: intermediate metabolizer; PM: poor metabolizer. 1 Other possible CYP2C19 genotypes with rare loss-of-function alleles exist beyond those illustrated (see [66]). 2 Note that prasugrel and ticagrelor are only recommended when not contraindicated clinically. Reprinted from [66] by permission of John Wiley and Sons © 2013 American Society for Clinical Pharmacology and Therapeutics.
3.4. OTHER CLOPIDOGREL CANDIDATE GENES
3.4.1. ATP-Binding Cassette, Sub-family B (MDR/TAP), Member 1 (ABCB1)
Multidrug resistance protein 1 (MDR1), also known as P-glycoprotein 1 (P-gp), is an ATP-binding cassette (ABC) efflux transporter encoded by ABCB1 that is involved in the intestinal absorption of clopidogrel. The most commonly studied ABCB1 variant is the synonymous c.3435C>T (p.Ile1145=, rs1045642), which has been associated with HTPR [125] and an increased risk of cardiovascular events [30, 93, 126–128]; however, other studies have not detected any effect [72, 82, 129] or found an opposite effect [101], and meta-analyses of ABCB1 c.3435C>T among CAD patients concluded that there was no significant association between c.3435C>T and HTPR or adverse cardiovascular events [130]. Given the conflicting results surrounding ABCB1 c.3435C>T, further studies are warranted to better understand the relationship between ABCB1 and clinical outcomes during clopidogrel treatment.
3.4.2. Carboxylesterase 1 (CES1)
As noted above, 85% of clopidogrel is metabolized into inactive carboxylic acid derivatives by hepatic carboxylesterases, primarily CES1. As such, genetic variability in CES1 has considerable potential to significantly influence active metabolite formation, on-treatment platelet reactivity, and possibly cardiovascular event risk. A recent healthy Amish study showed that carriers of the CES1 p.G143E loss-of-function variant (c.428G>A, rs71647871) had ~1.6-fold higher circulating active metabolite levels compared to c.428G/G homozygotes (30.3 vs. 19.0 ng/ml, p=0.001) [131]. This translated to variant allele carriers having greater inhibition of on-treatment ex vivo ADP-stimulated platelet aggregation in both healthy subjects (43 vs. 29% of baseline, p=0.003) and CAD patients undergoing PCI (45 and 25%, p=0.03) compared to wild-type homozygotes. Although the CAD patient cohort was underpowered to test for cardiovascular events, consistent with the pharmacokinetic and pharmacodynamic data, no patients who were homozygous for the variant allele experienced a cardiovascular event after one year compared to 13.7% of wild-type homozygotes (p=0.44) [131]. Consistent with these data, a recent study in human liver s9 fractions revealed that the CES1 p.G143E variant completely inhibited the hydrolysis of both clopidogrel and 2-oxo-clopidogrel [132]. Despite the clinical validity of CES1 p.G143E for clopidogrel pharmacokinetics and response variability, additional studies that comprehensively interrogate the CES1 gene and that are powered for clinical outcomes are still warranted.
3.4.3. Other CYP450 Genes
Other CYP450 enzymes are directly involved in the hepatic bioactivation of clopidogrel [53], which prompted their interrogation as candidate genes for clopidogrel response. In addition to CYP2C19, several other CYP450 enzymes (i.e. CYP1A2, CYP2B6, CYP2C9, CYP3A4 and CYP3A5) are also necessary for active clopidogrel metabolite formation, and all of these genes have known variant alleles. Some candidate gene studies identified significant effects on clopidogrel response (pharmacodynamic or clinical outcomes) with germline variants in CYP1A2 (with possible smoking interaction) [133, 134], CYP2C9 [68, 98, 135–137], CYP3A4 [138, 139], and CYP3A5 [140, 141]; however, the majority of clinical clopidogrel pharmacogenetic studies have not confirmed a significant independent role for these other CYP450 genes [61, 71, 72, 77, 80, 93, 142–145].
3.4.4. Purinergic Receptor P2Y, G-protein Coupled, 12 (P2RY12)
The platelet P2Y12 purinergic receptor is encoded by P2RY12 and two functional haplotypes (H1 and H2) have been identified [146]. The minor H2 haplotype has been associated with increased ADP-induced platelet aggregation in healthy subjects [146]; however, recent studies of these alleles have concluded that the H2 haplotype has no influence on platelet function among clopidogrel-treated patients undergoing PCI [147–149] [150]. Based on these and other conflicting reports [72, 80, 93, 151], a clinically relevant effect of common P2RY12 variants on clopidogrel efficacy is unlikely.
3.4.5. Paraoxonase 1 (PON1)
Paraoxonase 1 (PON1) is an aromatic esterase responsible for hydrolyzing endogenous and xenobiotic compounds in the liver, and in 2011, the PON1 p.Q192R variant (c.575A>G, rs662) was reported to significantly impact clopidogrel pharmacokinetics, pharmacodynamics, and the occurrence of both MACE (OR=3.9, 95% CI 2.1–7.2, p<0.001) and stent thrombosis (HR=10.2, 95% CI 4.3–71.4, p<0.001) [152]. These dramatic data suggested that PON1 p.Q192R had a greater effect on clopidogrel efficacy than the well-described CYP2C19*2 allele. The PON1 association was identified using in vitro metabolomic profiling of HEK293 cell microsomal preparations, which suggested that PON1 hydrolyzed the γ-thiobutyrolactone ring of 2-oxo-clopidogrel into the active H4 thiol metabolite. However, subsequent clopidogrel kinetic studies concluded that PON1 cannot generate H4 in cell-based systems, but mediates the formation of another thiol metabolite (Endo), which does not correlate with antiplatelet response [73]. In addition, the reported association between PON1 p.Q192R and clopidogrel pharmacokinetics, pharmacodynamics and clinical outcomes has been refuted by multiple pharmacogenetic studies [127, 153–158], including a systematic meta-analysis [159]. As such, the current body of evidence does not support a role for PON1 in clopidogrel pharmacogenetics.
4. PRASUGREL PHARMACOGENETICS
Prasugrel is a third generation thienopyridine administered with aspirin as DAPT for the management of ACS patients undergoing PCI. Prasugrel is hydrolyzed by carboxylesterases to yield thiolactone (R-95913), which undergoes hepatic bioactivation by CYP3A4, CYP2B6, CYP2C9, CYP2C19 and CYP2D6, to generate its active metabolite (R-138727) (Figure 1). Like clopidogrel, the prasugrel active metabolite antagonizes the P2Y12 receptor and impairs ADP-mediated activation [160]. Prasugrel is rapid-acting and generates a higher level of active metabolite compared to clopidogrel, resulting in a more potent and effective platelet inhibition [161–164]; however, the increased efficacy is counterbalanced by an increased risk for major bleeding [165]. Despite the advantages of prasugrel over clopidogrel, HTPR has also been reported among prasugrel-treated PCI patients, which was associated with higher rates of thrombotic events [166].
Given that prasugrel undergoes CYP450-mediated hepatic bioactivation, initial pharmacogenetic studies on prasugrel response focused on CYP450 variant alleles [68, 74, 75, 164]. Prasugrel pharmacokinetics and pharmacodynamics were initially tested for association with CYP450 variants among healthy subjects; however, no significant relationship was detected for either active metabolite exposure or pharmacodynamic response [68]. A small study of CAD patients also failed to detect a significant difference in prasugrel active metabolite exposure or pharmacodynamic responses based on CYP2C19 genotype status [75]. Notably, the large TRITON-TIMI 38 trial included a pharmacogenetic substudy of prasugrel-treated ACS patients with planned PCI and genotyped 54 alleles in six CYP450 genes. No significant effects on prasugrel pharmacokinetics or pharmacodynamics were identified, nor were any CYP450 variants associated with clinical outcomes [164]. Considering most studies use ADP-induced platelet aggregation as a measure of platelet function, a subsequent clinical study was performed to assess the influence of CYP2C19 alleles on prasugrel response as determined by platelet reactivity index (PRI) from vasodilator-stimulated phosphoprotein (VASP) analysis. Interestingly, similar to clopidogrel, CYP2C19*2 carriers had a significantly higher PRI and risk of HTPR than noncarriers [167], which remained consistent with a subsequent study on prasugrel maintenance therapy response by PRI VASP [168]. Although not conclusive as these results have yet to be replicated by an independent research group, these data do underscore the lack of concordance between different platelet function tests and the challenges with interpreting data across studies that measure antiplatelet therapy response by light transmission aggregometry, VASP and/or the VerifyNow® platelet function assay.
In addition to the CYP450 genes, a few other candidate genes have been interrogated for association with prasugrel response. For example, the ABCB1 gene was also genotyped in the TRITON-TIMI 38 pharmacogenetic substudy; however, it was not significantly associated with any outcomes in prasugrel-treated ACS/PCI patients [126]. Interestingly, the PEAR1 gene was genotyped in a small study of healthy Han Chinese subjects, which reported a significant association between selected PEAR1 variants and ADP-induced platelet aggregation; however, the extremely small sample size of the study (n=36) indicates that these findings are preliminary.
5. TICAGRELOR PHARMACOGENETICS
Ticagrelor is a cyclopentyl-triazolo-pyrimidine agent that is an allosteric ADP antagonist that does not require hepatic bioactivation to generate an active metabolite; however, after oral administration and absorption, it is degraded to its primary active (ARC124910XX) and inactive (AR-C133913XX) metabolites through CYP3A4/5-mediated metabolism (Figure 1) [169, 170]. Consequently, the ticagrelor label recommends avoiding coadministration with strong CYP3A inhibitors and inducers among patients with ACS. Although this also suggests that CYP3A4 and/or CYP3A5 variant alleles may potentially influence ticagrelor efficacy, pharmacogenetic studies have yet to prove this hypothesis. Additionally, the role of CYP3A4 in generating the active ticagrelor metabolite may also be responsible for the reported drug interaction between ticagrelor and statins. Since ticagrelor is both a CYP3A4 substrate and inhibitor, its use results in higher serum concentrations of simvastatin and lovastatin when coadministered, as these drugs are also metabolized by CYP3A4. This interaction may be responsible, in part, for the mortality benefit observed with ticagrelor compared to clopidogrel in the PLATO trial, as ticagrelor significantly increases the potency of CYP3A4-metabolized statins, which in turn may increase the vascular benefit derived from the statin [171].
Ticagrelor has a faster onset and offset of action and achieves a more pronounced and consistent antiplatelet response than clopidogrel [172], which has translated to superior efficacy among ACS patients, including reductions in stent thrombosis and all-cause mortality [173]. A subset of patients in the PLATO trial were genotyped for CYP2C19 loss-of-function and increased-function alleles and the common ABCB1 c.3435C>T variant; however, unlike clopidogrel, no significant association was observed between either gene and the primary composite outcome of cardiovascular death, MI, or stroke at 12 months [101]. In addition, a GWAS was also performed with this cohort in an effort to identify variants associated with ticagrelor plasma and major metabolite (AR-C124910XX) levels [174]. Although only reported to date in abstract form, one variant in SLCO1B1 (rs113681054) and two independent variants (rs62471956, and rs56324128) were significantly associated with ticagrelor plasma levels. The SLCO1B1 rs113681054 variant and an additional variant in UGT2B7 (rs61361928) were also significantly associated with metabolite levels; however, both of these pharmacogenetic effects were limited to ticagrelor pharmacokinetics, as the variants did not associate with efficacy or safety of ticagrelor treatment [174].
6. ANTIPLATELET PHARMACOGENETIC TESTING AND CLINICAL IMPLEMENTATION
6.1. CYP2C19 GENETIC TESTING
Although the landscape of U.S. FDA regulatory oversight over clinical genetic testing is potentially about to undergo a major restructuring [175, 176], clinical laboratories currently can offer genetic tests that are either approved for in vitro diagnostic testing by the U.S. FDA or as in-house validated laboratory-developed tests. Most U.S. FDA-approved CYP2C19 tests interrogate *2, *3 and *17, but do not include the low frequency *4 – *8 alleles despite their established loss-of-function (Table 2). In contrast, these alleles are frequently included in most CYP2C19 laboratory-developed tests. Clinical laboratories that interrogate CYP2C19 by Sanger or next-generation sequencing would identify these rare alleles as well as other novel coding variants of uncertain clinical significance. In addition to surveying the testing menus of local CLIA-certified laboratories for CYP2C19 genetic testing availability, the National Institutes of Health (NIH) Genetic Testing Registry (GTR) is a central location for voluntary submission of genetic test information by laboratory providers [177].
TABLE 2.
CYP2C19 genotyping tests approved by the U.S. FDA for in vitro diagnostic (IVD) usea
| Assay | Alleles Interrogated | Company |
|---|---|---|
| INFINITI® CYP2C19 Assay | *2, *3, *17 | Autogenomics, Inc. |
| Verigene® CYP2C19 Test | *2, *3, *17 | Nanosphere, Inc. |
| Spartan RX CYP2C19 Assay | *2, *3, *17 | Spartan Bioscience, Inc. |
| xTAG® CYP2C19 Kit v3 | *2, *3, *17 | Luminex Molecular Diagnostics, Inc. |
As listed on the U.S. Food and Drug Administration in vitro Diagnostic Product Database [198].
6.2. BARRIERS TO CYP2C19 IMPLEMENTATION
The ongoing publication of genome-directed practice guidelines and the availability of high-throughput multiplexed genotyping and next-generation sequencing technologies are increasing the accessibility of clinical pharmacogenetic testing, both theoretically and practically. However, physician adoption of clinical CYP2C19 testing has not been widespread, which is likely due to a number of barriers [178, 179], including testing logistics, clinician education and acceptance, pharmacogenetic testing reimbursement, and uncertain cost-effectiveness.
6.2.1. CYP2C19 Testing Logistics
One of the frequently cited barriers to implementing CYP2C19 genetic testing for antiplatelet therapy is the need for a rapid turnaround time of results to the patient’s medical record to enable drug selection by clinicians prior to patient discharge. Although genotyping platforms have been developed that can be completed within a few hours from receipt of a specimen, clinical genetic testing laboratories also need to have dedicated sample accessioning, technologist and director effort, and electronic report returning capabilities to efficiently execute same-day testing. Additionally, cardiac catheterization laboratories also need dedicated effort to consent their patients for genetic testing, which would likely translate to unpredictable daily specimen volumes being sent to the genetic testing laboratory. The need for rapid results combined with this irregular receipt of specimens together contribute to significant challenges when implementing real-world CYP2C19 genotype-directed antiplatelet therapy. Despite these difficult testing logistics, prospective clinical CYP2C19 genetic testing has been successfully accomplished at selected medical centers [180, 181].
Another testing strategy that can circumvent the issue of rapid turnaround time genotyping is pre-emptive pharmacogenetic testing [182]. This approach deposits CYP2C19 genotype data into patient electronic medical records through prospective or biobank patient recruitment and CLIA-certified genetic testing, and alerts prescribers through clinical decision support at the point-of-care if and when clopidogrel is ordered and the patient carries an at-risk CYP2C19 genotype. Although this model has inherent challenges and significant costs for effective clinical implementation, pre-emptive CYP2C19 genetic testing has recently been deployed at several academic medical centers [183–185].
6.2.2. CYP2C19 and HTPR Association
An important barrier to clinical implementation of CYP2C19 genotype-directed antiplatelet therapy is the association between CYP2C19 and HTPR. CYP2C19 genetic testing has a relatively low estimated positive predictive value for HTPR (~20%) [186], which has driven the ongoing search for additional germline variants implicated in clopidogrel response variability. Similarly, the summarized sensitivity and specificity of CYP2C19*2 for predicting HTPR has been reported to be 38% and 80%, respectively [187], indicating that a significant number of patients who are not *2 carriers will still have HTPR and potentially be overlooked following a negative genotype result. As such, these data suggest that in addition to CYP2C19 genotype, the available phenotype and clinical data should also be incorporated to guide antiplatelet therapy.
6.2.3. Clinician Awareness, Education and Acceptance
An ongoing effort towards the application of clinical pharmacogenetics is increasing clinician education and acceptance. Continuing education efforts in genomics for practicing physicians are becoming more available given that most survey studies have consistently concluded that the physician workforce is unprepared for any large-scale application of genomic medicine [188, 189]. Education in pharmacogenetics is inherently a part of those efforts, and enhancing the professional curricula for all relevant healthcare professionals (e.g., physicians, physician assistants, pharmacists, nurses and genetic counselors) is going to be necessary for proper implementation of genotype-guided pharmacotherapy [190]. Clinician acceptance of clinical pharmacogenetics currently varies widely, and is undoubtedly tied to their general understanding and perception of the field. Ongoing professional education in pharmacogenetics will hopefully facilitate not only a greater acceptance and understanding of the field, but a more informed and rational implementation of clinical pharmacogenetic testing, including genotype-guided antiplatelet therapy.
6.2.4. CYP2C19 Testing Reimbursement and Cost-effectiveness
Insurance coverage for CYP2C19 genetic testing is available from selected providers; however, this issue is continually evolving. A recent proposed draft for local coverage determination by the Centers for Medicare and Medicaid Services (CMS; Palmetto GBA, Virginia) determined that CYP2C19 genetic testing (CPT 81225) is medically necessary for patients with ACS undergoing PCI initiating or reinitiating clopidogrel therapy, but not for medical management of ACS without PCI, stroke, or PAD. How, or if, CMS will be implementing this draft more broadly is not yet determined. Notably, the University of Florida Health Personalized Medicine Program recently reported that ~85% of third-party payers (including Medicare) reimbursed CYP2C19 genotyping for PCI patients [191].
Cost-effectiveness studies on CYP2C19-guided antiplatelet therapy have been inconclusive, but have suggested that this strategy may be a more cost-effective approach [192–194]. However, some studies also support widespread use of ticagrelor regardless of CYP2C19 status [192, 195]. Real-world application of these approaches will undoubtedly be influenced by factors that are difficult to model, including clinician preference, treatment indication and contraindications, and personal insurance coverage and policy.
7. EXPERT OPINION
Interpatient variability in pharmacokinetics, pharmacodynamics and/or clinical outcomes when treated with antiplatelet agents has prompted extensive studies on potential pharmacogenetic determinants of antiplatelet response. This has been further driven, in part, by the fact that CAD patients with HTPR have increased risks for ischemic events. However, discordant results across aspirin pharmacogenetic studies have hampered the ability to identify true aspirin response genes and variants, likely due to differences in study designs, response definitions, and assays used to measure platelet function. As such, the available data do not support any implementation of clinical genetic testing for aspirin response at this time. Similarly, although there is limited data available, no candidate genes have been reported for prasugrel and ticagrelor that have been adequately replicated with pharmacokinetic or pharmacodynamic response measurements, nor have any genes been convincingly associated with any clinical outcomes using these potent antiplatelet agents.
The major genetic determinant of clopidogrel metabolite levels, on-treatment platelet reactivity, and adverse cardiovascular event risks among ACS/PCI patients are CYP2C19 loss-of-function alleles. In contrast, the clinical validity of other candidate clopidogrel response genes (ABCB1, CES1, other CYP450 genes, and P2RY12) is uncertain due to the absence of adequate replication at this time. The effect of reduced CYP2C19 activity on clopidogrel response has prompted the availability of clinical CYP2C19 genotyping and the implementation of genotype-directed antiplatelet therapy at some institutions. However, given that the only reported prospective trials testing CYP2C19 genotype-guided antiplatelet therapy had pharmacodynamic primary endpoints (i.e., platelet reactivity) and not clinical outcomes, the utility of this approach is frequently debated. Cost-effectiveness studies have also been inconclusive with respect to pharmacogenetic guided antiplatelet therapy and cardiology society guidelines do not currently recommended routine CYP2C19 genotyping, together ultimately leaving the decision to test ACS/PCI patients up to the individual clinician when clopidogrel is being considered.
Consistent with the ACCF/AHA guideline statements, CYP2C19 genotyping should be considered when treating patients at moderate to high risk for poor outcomes (including those undergoing PCI) with clopidogrel. In addition, CYP2C19 poor metabolizers should be prescribed an alternative antiplatelet regimen [119] following physician consideration of all available clinical information. The debate regarding whether or when to perform CYP2C19 genetic testing is complicated and ongoing, and will hopefully be better informed by the ongoing prospective trials evaluating CYP2C19-directed antiplatelet therapy, clarity regarding third-party payer policies, and more convincing cost-effectiveness data. However, the increasing availability of direct-to-consumer genetic testing, other sequencing programs and general public awareness/interest in genomics is resulting in patients already having personal genetic data available, which will likely only increase in the near and ongoing future. In this context, CYP2C19 genotype data, and potentially other future candidate gene variants, can be used to inform antiplatelet therapy, and recommendations on how to incorporate these pharmacogenetic variables can be found by CPIC [66] and other professional guidelines. A personalized strategy for ACS/PCI patients has the potential to target the more potent antiplatelet agents to at-risk patients (i.e., CYP2C19 loss-of-function allele carriers), while sparing the remaining patients from the increased expense of non-generic medication and associated increased risks for bleeding.
With respect to the ongoing discovery efforts in antiplatelet pharmacogenetics, it is becoming increasingly appreciated that research studies need to utilize more multidisciplinary and integrative systems biology designs to more comprehensively evaluate the effect of antiplatelet agents on platelet reactivity and cardiovascular events. For example, recent studies have successfully used a composite aspirin platelet function score that included both non-COX1-dependent platelet reactivity in response to multiple agonists (ADP, collagen, and epinephrine) and canonical measures of aspirin response (AA-stimulated platelet aggregation) to identify novel determinants of aspirin responsiveness [196, 197]. Moreover, the Pharmacogenomics Research Network (PGRN) and Pharmacometabolomics Research Network (PMRN) have shown that integration of pharmacogenomics data with high-resolution metabolomic profiling can increase our understanding of the effects of aspirin on metabolism, identify novel metabolites, signaling pathways, and gene variants that influence response to therapy [13–15]. In addition to highlighting their utility for the antiplatelet pharmacogenomics field, the success of these study design approaches also suggest that they should be considered for other drug response discovery research.
ARTICLE HIGHLIGHTS BOX.
Oral antiplatelet agents are indicated for prevention of ischemic events resulting from acute coronary syndromes, ischemic stroke, and symptomatic peripheral artery disease; however, variability in patient response has been observed, which can translate to increased risks for adverse cardiovascular events.
Aspirin pharmacogenetic studies have identified a number of candidate aspirin response genes and variants; however, heterogeneity in study design, aspirin response definition, and platelet function assays have contributed to limited replication between studies.
The major genetic determinant of clopidogrel metabolite levels, on-treatment platelet reactivity, and adverse cardiovascular event risks are CYP2C19 loss-of-function alleles, which has prompted the recent implementation of clinical CYP2C19 genotype-directed antiplatelet therapy at selected institutions.
Prasugrel and ticagrelor result in a more potent and effective platelet inhibition than clopidogrel and the limited pharmacogenetic data available have not identified any genetic variants with strong effects implicated in response variability for these agents.
Pharmacogenomics discovery research needs to embrace multidisciplinary and integrative systems biology study designs to more comprehensively evaluate interindividual variability in drug response and to facilitate the identification of more robust candidate genes and variants.
Footnotes
Financial and competing interests disclosure
JP Lewis receives support from NIH for antiplatelet pharmacogenomics research. J-S Hulot has received research grant support from Biotronik, and consulting fees from Daiichi Sankyo. SA Scott receives support from NIH for antiplatelet pharmacogenomics research and is an Associate Director of a clinical laboratory that performs CYP2C19 testing. This research was supported in part by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH), through grants K23 GM102678 (J.P.L.) and K23 GM104401 (S.A.S.). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed
References
Papers of special note have been highlighted as either of interest (*) or of considerable interest (**) to readers.
- 1.Sangkuhl K, Shuldiner AR, Klein TE, et al. Platelet aggregation pathway. Pharmacogenetics and genomics. 2011 Aug;21(8):516–21. doi: 10.1097/FPC.0b013e3283406323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jennings LK. Mechanisms of platelet activation: need for new strategies to protect against platelet-mediated atherothrombosis. Thrombosis and haemostasis. 2009 Aug;102(2):248–57. doi: 10.1160/TH09-03-0192. [DOI] [PubMed] [Google Scholar]
- 3.Buonamici P, Marcucci R, Migliorini A, et al. Impact of platelet reactivity after clopidogrel administration on drug-eluting stent thrombosis. J Am Coll Cardiol. 2007 Jun 19;49(24):2312–7. doi: 10.1016/j.jacc.2007.01.094. [DOI] [PubMed] [Google Scholar]
- 4.Tantry US, Bonello L, Aradi D, et al. Consensus and update on the definition of on-treatment platelet reactivity to adenosine diphosphate associated with ischemia and bleeding. J Am Coll Cardiol. 2013 Dec 17;62(24):2261–73. doi: 10.1016/j.jacc.2013.07.101. [DOI] [PubMed] [Google Scholar]
- 5.Tantry US, Mahla E, Gurbel PA. Aspirin resistance. Prog Cardiovasc Dis. 2009 Sep-Oct;52(2):141–52. doi: 10.1016/j.pcad.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Michelson AD, Cattaneo M, Eikelboom JW, et al. Aspirin resistance: position paper of the Working Group on Aspirin Resistance. Journal of thrombosis and haemostasis : JTH. 2005 Jun;3(6):1309–11. doi: 10.1111/j.1538-7836.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 7.Cattaneo M. Aspirin and clopidogrel: efficacy, safety, and the issue of drug resistance. Arteriosclerosis, thrombosis, and vascular biology. 2004 Nov;24(11):1980–7. doi: 10.1161/01.ATV.0000145980.39477.a9. [DOI] [PubMed] [Google Scholar]
- 8.Mansour K, Taher AT, Musallam KM, et al. Aspirin resistance. Adv Hematol. 2009;2009:937352. doi: 10.1155/2009/937352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faraday N, Becker DM, Becker LC. Pharmacogenomics of platelet responsiveness to aspirin. Pharmacogenomics. 2007 Oct;8(10):1413–25. doi: 10.2217/14622416.8.10.1413. [DOI] [PubMed] [Google Scholar]
- 10.Faraday N, Yanek LR, Mathias R, et al. Heritability of platelet responsiveness to aspirin in activation pathways directly and indirectly related to cyclooxygenase-1. Circulation. 2007 May 15;115(19):2490–6. doi: 10.1161/CIRCULATIONAHA.106.667584. [DOI] [PubMed] [Google Scholar]
- 11.Becker DM, Segal J, Vaidya D, et al. Sex differences in platelet reactivity and response to low-dose aspirin therapy. JAMA : the journal of the American Medical Association. 2006 Mar 22;295(12):1420–7. doi: 10.1001/jama.295.12.1420. [DOI] [PubMed] [Google Scholar]
- 12.Grosser T, Fries S, Lawson JA, et al. Drug resistance and pseudoresistance: an unintended consequence of enteric coating aspirin. Circulation. 2013 Jan 22;127(3):377–85. doi: 10.1161/CIRCULATIONAHA.112.117283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yerges-Armstrong LM, Ellero-Simatos S, Georgiades A, et al. Purine pathway implicated in mechanism of resistance to aspirin therapy: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2013 Oct;94(4):525–32. doi: 10.1038/clpt.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis JP, Yerges-Armstrong LM, Ellero-Simatos S, et al. Integration of pharmacometabolomic and pharmacogenomic approaches reveals novel insights into antiplatelet therapy. Clin Pharmacol Ther. 2013 Nov;94(5):570–3. doi: 10.1038/clpt.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellero-Simatos S, Lewis JP, Georgiades A, et al. Pharmacometabolomics reveals that serotonin is implicated in aspirin response variability. CPT Pharmacometrics Syst Pharmacol. 2014;3:e125. doi: 10.1038/psp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lordkipanidze M, Pharand C, Schampaert E, et al. A comparison of six major platelet function tests to determine the prevalence of aspirin resistance in patients with stable coronary artery disease. Eur Heart J. 2007 Jul;28(14):1702–8. doi: 10.1093/eurheartj/ehm226. [DOI] [PubMed] [Google Scholar]
- 17.Fontana P, Nolli S, Reber G, et al. Biological effects of aspirin and clopidogrel in a randomized cross-over study in 96 healthy volunteers. Journal of thrombosis and haemostasis : JTH. 2006 Apr;4(4):813–9. doi: 10.1111/j.1538-7836.2006.01867.x. [DOI] [PubMed] [Google Scholar]
- 18.Wurtz M, Kristensen SD, Hvas AM, et al. Pharmacogenetics of the antiplatelet effect of aspirin. Curr Pharm Des. 2012;18(33):5294–308. doi: 10.2174/138161212803251907. [DOI] [PubMed] [Google Scholar]
- 19.Halushka MK, Walker LP, Halushka PV. Genetic variation in cyclooxygenase 1: effects on response to aspirin. Clin Pharmacol Ther. 2003 Jan;73(1):122–30. doi: 10.1067/mcp.2003.1. [DOI] [PubMed] [Google Scholar]
- 20.Lepantalo A, Mikkelsson J, Resendiz JC, et al. Polymorphisms of COX-1 and GPVI associate with the antiplatelet effect of aspirin in coronary artery disease patients. Thrombosis and haemostasis. 2006 Feb;95(2):253–9. doi: 10.1160/TH05-07-0516. [DOI] [PubMed] [Google Scholar]
- 21.Li Q, Chen BL, Ozdemir V, et al. Frequency of genetic polymorphisms of COX1, GPIIIa and P2Y1 in a Chinese population and association with attenuated response to aspirin. Pharmacogenomics. 2007 Jun;8(6):577–86. doi: 10.2217/14622416.8.6.577. [DOI] [PubMed] [Google Scholar]
- 22.Clappers N, van Oijen MG, Sundaresan S, et al. The C50T polymorphism of the cyclooxygenase-1 gene and the risk of thrombotic events during low-dose therapy with acetyl salicylic acid. Thrombosis and haemostasis. 2008 Jul;100(1):70–5. doi: 10.1160/TH08-03-0172. [DOI] [PubMed] [Google Scholar]
- 23.Pettinella C, Romano M, Stuppia L, et al. Cyclooxygenase-1 haplotype C50T/A-842G does not affect platelet response to aspirin. Thrombosis and haemostasis. 2009 Apr;101(4):687–90. [PubMed] [Google Scholar]
- 24.Kunicki TJ, Williams SA, Nugent DJ, et al. Lack of association between aspirin responsiveness and seven candidate gene haplotypes in patients with symptomatic vascular disease. Thrombosis and haemostasis. 2009 Jan;101(1):123–33. [PubMed] [Google Scholar]
- 25.Chakroun T, Addad F, Yacoub S, et al. The cyclooxygenase-1 C50T polymorphism is not associated with aspirin responsiveness status in stable coronary artery disease in Tunisian patients. Genet Test Mol Biomarkers. 2011 Jul-Aug;15(7–8):513–6. doi: 10.1089/gtmb.2010.0225. [DOI] [PubMed] [Google Scholar]
- 26.Voora D, Horton J, Shah SH, et al. Polymorphisms associated with in vitro aspirin resistance are not associated with clinical outcomes in patients with coronary artery disease who report regular aspirin use. Am Heart J. 2011 Jul;162(1):166–72. e1. doi: 10.1016/j.ahj.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 27.Yi X, Zhou Q, Lin J, et al. Platelet response to aspirin in Chinese stroke patients is independent of genetic polymorphisms of COX-1 C50T and COX-2 G765C. J Atheroscler Thromb. 2013;20(1):65–72. doi: 10.5551/jat.14092. [DOI] [PubMed] [Google Scholar]
- 28.Postula M, Janicki PK, Rosiak M, et al. Effect of common single nucleotide polymorphisms in COX-1 gene on related metabolic activity in diabetic patients treated with acetylsalicylic acid. Arch Med Sci. 2014 Dec 22;10(6):1198–205. doi: 10.5114/aoms.2013.35442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodman T, Ferro A, Sharma P. Pharmacogenetics of aspirin resistance: a comprehensive systematic review. Br J Clin Pharmacol. 2008 Aug;66(2):222–32. doi: 10.1111/j.1365-2125.2008.03183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Cayla G, Hulot JS, O’Connor SA, et al. Clinical, angiographic, and genetic factors associated with early coronary stent thrombosis. JAMA : the journal of the American Medical Association. 2011 Oct 26;306(16):1765–74. doi: 10.1001/jama.2011.1529. Case/control study on early stent thrombosis. [DOI] [PubMed] [Google Scholar]
- 31.Carlsson LE, Santoso S, Spitzer C, et al. The alpha2 gene coding sequence T807/A873 of the platelet collagen receptor integrin alpha2beta1 might be a genetic risk factor for the development of stroke in younger patients. Blood. 1999 Jun 1;93(11):3583–6. [PubMed] [Google Scholar]
- 32.Santoso S, Kunicki TJ, Kroll H, et al. Association of the platelet glycoprotein Ia C807T gene polymorphism with nonfatal myocardial infarction in younger patients. Blood. 1999 Apr 15;93(8):2449–53. [PubMed] [Google Scholar]
- 33.Roest M, Banga JD, Grobbee DE, et al. Homozygosity for 807 T polymorphism in alpha(2) subunit of platelet alpha(2)beta(1) is associated with increased risk of cardiovascular mortality in high-risk women. Circulation. 2000 Oct 3;102(14):1645–50. doi: 10.1161/01.cir.102.14.1645. [DOI] [PubMed] [Google Scholar]
- 34.Lim E, Carballo S, Cornelissen J, et al. Dose-related efficacy of aspirin after coronary surgery in patients With Pl(A2) polymorphism ( NCT00262275) Ann Thorac Surg. 2007 Jan;83(1):134–8. doi: 10.1016/j.athoracsur.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Macchi L, Christiaens L, Brabant S, et al. Resistance in vitro to low-dose aspirin is associated with platelet PlA1 (GP IIIa) polymorphism but not with C807T(GP Ia/IIa) and C-5T Kozak (GP Ibalpha) polymorphisms. J Am Coll Cardiol. 2003 Sep 17;42(6):1115–9. doi: 10.1016/s0735-1097(03)00921-5. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Conejero R, Rivera J, Corral J, et al. Biological assessment of aspirin efficacy on healthy individuals: heterogeneous response or aspirin failure? Stroke. 2005 Feb;36(2):276–80. doi: 10.1161/01.STR.0000151362.65339.f9. [DOI] [PubMed] [Google Scholar]
- 37.Bernardo E, Angiolillo DJ, Ramirez C, et al. Lack of association between gene sequence variations of platelet membrane receptors and aspirin responsiveness detected by the PFA-100 system in patients with coronary artery disease. Platelets. 2006 Dec;17(8):586–90. doi: 10.1080/09537100600881412. [DOI] [PubMed] [Google Scholar]
- 38.Afshar-Kharghan V, Li CQ, Khoshnevis-Asl M, et al. Kozak sequence polymorphism of the glycoprotein (GP) Ibalpha gene is a major determinant of the plasma membrane levels of the platelet GP Ib-IX-V complex. Blood. 1999 Jul 1;94(1):186–91. [PubMed] [Google Scholar]
- 39.Douglas H, Michaelides K, Gorog DA, et al. Platelet membrane glycoprotein Ibalpha gene -5T/C Kozak sequence polymorphism as an independent risk factor for the occurrence of coronary thrombosis. Heart. 2002 Jan;87(1):70–4. doi: 10.1136/heart.87.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Douglas H, Davies GJ, Michaelides K, et al. Detection of functional differences between different platelet membrane glycoprotein Ibalpha variable number tandem repeat and Kozak genotypes as shown by the PFA-100 system. Heart. 2006 May;92(5):676–8. doi: 10.1136/hrt.2004.058545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiwara T, Ikeda M, Esumi K, et al. Exploratory aspirin resistance trial in healthy Japanese volunteers (J-ART) using platelet aggregation as a measure of thrombogenicity. Pharmacogenomics J. 2007 Dec;7(6):395–403. doi: 10.1038/sj.tpj.6500435. [DOI] [PubMed] [Google Scholar]
- 42.Croft SA, Hampton KK, Daly ME, et al. Kozak sequence polymorphism in the platelet GPIbalpha gene is not associated with risk of myocardial infarction. Blood. 2000 Mar 15;95(6):2183–4. [PubMed] [Google Scholar]
- 43.Kauskot A, Di Michele M, Loyen S, et al. A novel mechanism of sustained platelet alphaIIbbeta3 activation via PEAR1. Blood. 2012 Apr 26;119(17):4056–65. doi: 10.1182/blood-2011-11-392787. [DOI] [PubMed] [Google Scholar]
- 44.Kauskot A, Vandenbriele C, Louwette S, et al. PEAR1 attenuates megakaryopoiesis via control of the PI3K/PTEN pathway. Blood. 2013 Jun 27;121(26):5208–17. doi: 10.1182/blood-2012-10-462887. [DOI] [PubMed] [Google Scholar]
- 45.Jones CI, Bray S, Garner SF, et al. A functional genomics approach reveals novel quantitative trait loci associated with platelet signaling pathways. Blood. 2009 Aug 13;114(7):1405–16. doi: 10.1182/blood-2009-02-202614. [DOI] [PubMed] [Google Scholar]
- 46.Johnson AD, Yanek LR, Chen MH, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010 Jul;42(7):608–13. doi: 10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrera-Galeano JE, Becker DM, Wilson AF, et al. A novel variant in the platelet endothelial aggregation receptor-1 gene is associated with increased platelet aggregability. Arteriosclerosis, thrombosis, and vascular biology. 2008 Aug;28(8):1484–90. doi: 10.1161/ATVBAHA.108.168971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faraday N, Yanek LR, Yang XP, et al. Identification of a specific intronic PEAR1 gene variant associated with greater platelet aggregability and protein expression. Blood. 2011 Sep 22;118(12):3367–75. doi: 10.1182/blood-2010-11-320788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Lewis JP, Ryan K, O’Connell JR, et al. Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circulation Cardiovascular genetics. 2013 Apr;6(2):184–92. doi: 10.1161/CIRCGENETICS.111.964627. Identification of PEAR1 association with platelet aggregation and clinical outcomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y, Suktitipat B, Yanek LR, et al. Targeted deep resequencing identifies coding variants in the PEAR1 gene that play a role in platelet aggregation. PLoS One. 2013;8(5):e64179. doi: 10.1371/journal.pone.0064179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang Q, Cui Y, Zhao X, et al. Identification of PEAR1 SNPs and their influences on the variation in prasugrel pharmacodynamics. Pharmacogenomics. 2013 Jul;14(10):1179–89. doi: 10.2217/pgs.13.108. [DOI] [PubMed] [Google Scholar]
- 52.Wurtz M, Lordkipanidze M, Grove EL. Pharmacogenomics in cardiovascular disease: focus on aspirin and ADP receptor antagonists. Journal of thrombosis and haemostasis : JTH. 2013 Sep;11(9):1627–39. doi: 10.1111/jth.12318. [DOI] [PubMed] [Google Scholar]
- 53.Sangkuhl K, Klein TE, Altman RB. Clopidogrel pathway. Pharmacogenetics and genomics. 2010 Jul;20(7):463–5. doi: 10.1097/FPC.0b013e3283385420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kazui M, Nishiya Y, Ishizuka T, et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos. 2010 Jan;38(1):92–9. doi: 10.1124/dmd.109.029132. [DOI] [PubMed] [Google Scholar]
- 55.Mehta SR, Yusuf S, Peters RJ, et al. Effects of pretreatment with clopidogrel and aspirin followed by long-term therapy in patients undergoing percutaneous coronary intervention: the PCI-CURE study. Lancet. 2001 Aug 18;358(9281):527–33. doi: 10.1016/s0140-6736(01)05701-4. [DOI] [PubMed] [Google Scholar]
- 56.Sabatine MS, Cannon CP, Gibson CM, et al. Effect of clopidogrel pretreatment before percutaneous coronary intervention in patients with ST-elevation myocardial infarction treated with fibrinolytics: the PCI-CLARITY study. JAMA : the journal of the American Medical Association. 2005 Sep 14;294(10):1224–32. doi: 10.1001/jama.294.10.1224. [DOI] [PubMed] [Google Scholar]
- 57.Steinhubl SR, Berger PB, Mann JT, 3rd, et al. Early and sustained dual oral antiplatelet therapy following percutaneous coronary intervention: a randomized controlled trial. JAMA : the journal of the American Medical Association. 2002 Nov 20;288(19):2411–20. doi: 10.1001/jama.288.19.2411. [DOI] [PubMed] [Google Scholar]
- 58.Gurbel PA, Bliden KP, Hiatt BL, et al. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation. 2003 Jun 17;107(23):2908–13. doi: 10.1161/01.CIR.0000072771.11429.83. [DOI] [PubMed] [Google Scholar]
- 59.Jaremo P, Lindahl TL, Fransson SG, et al. Individual variations of platelet inhibition after loading doses of clopidogrel. Journal of internal medicine. 2002 Sep;252(3):233–8. doi: 10.1046/j.1365-2796.2002.01027.x. [DOI] [PubMed] [Google Scholar]
- 60.Muller I, Besta F, Schulz C, et al. Prevalence of clopidogrel non-responders among patients with stable angina pectoris scheduled for elective coronary stent placement. Thrombosis and haemostasis. 2003 May;89(5):783–7. [PubMed] [Google Scholar]
- 61**.Hulot JS, Bura A, Villard E, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006 Oct 1;108(7):2244–7. doi: 10.1182/blood-2006-04-013052. Identification of CYP2C19 association with platelet aggregation by a candidate gene study. [DOI] [PubMed] [Google Scholar]
- 62**.Shuldiner AR, O’Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA : the journal of the American Medical Association. 2009 Aug 26;302(8):849–57. doi: 10.1001/jama.2009.1232. Identification of CYP2C19 association with platelet aggregation by a GWAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scott SA, Sangkuhl K, Shuldiner AR, et al. PharmGKB summary: very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 19. Pharmacogenetics and genomics. 2012 Feb;22(2):159–65. doi: 10.1097/FPC.0b013e32834d4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sim SC, Ingelman-Sundberg M. The Human Cytochrome P450 (CYP) Allele Nomenclature website: a peer-reviewed database of CYP variants and their associated effects. Hum Genomics. 2010 Apr;4(4):278–81. doi: 10.1186/1479-7364-4-4-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martis S, Peter I, Hulot JS, et al. Multi-ethnic distribution of clinically relevant CYP2C genotypes and haplotypes. Pharmacogenomics J. 2012 Apr 10; doi: 10.1038/tpj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013 Sep;94(3):317–23. doi: 10.1038/clpt.2013.105. CPIC guidelines on CYP2C19-directed antiplatelet therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto K, Hokimoto S, Chitose T, et al. Impact of CYP2C19 polymorphism on residual platelet reactivity in patients with coronary heart disease during antiplatelet therapy. J Cardiol. 2011 Mar;57(2):194–201. doi: 10.1016/j.jjcc.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Brandt JT, Close SL, Iturria SJ, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. Journal of thrombosis and haemostasis : JTH. 2007 Dec;5(12):2429–36. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 69.Kim KA, Park PW, Hong SJ, et al. The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: a possible mechanism for clopidogrel resistance. Clin Pharmacol Ther. 2008 Aug;84(2):236–42. doi: 10.1038/clpt.2008.20. [DOI] [PubMed] [Google Scholar]
- 70.Umemura K, Furuta T, Kondo K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. Journal of thrombosis and haemostasis : JTH. 2008 Aug;6(8):1439–41. doi: 10.1111/j.1538-7836.2008.03050.x. [DOI] [PubMed] [Google Scholar]
- 71.Mega JL, Close SL, Wiviott SD, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009 Jan 22;360(4):354–62. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 72.Simon T, Bhatt DL, Bergougnan L, et al. Genetic polymorphisms and the impact of a higher clopidogrel dose regimen on active metabolite exposure and antiplatelet response in healthy subjects. Clin Pharmacol Ther. 2011 Aug;90(2):287–95. doi: 10.1038/clpt.2011.127. [DOI] [PubMed] [Google Scholar]
- 73.Gong IY, Crown N, Suen CM, et al. Clarifying the importance of CYP2C19 and PON1 in the mechanism of clopidogrel bioactivation and in vivo antiplatelet response. Eur Heart J. 2012 Nov;33(22):2856–64. doi: 10.1093/eurheartj/ehs042. [DOI] [PubMed] [Google Scholar]
- 74.Kelly RP, Close SL, Farid NA, et al. Pharmacokinetics and pharmacodynamics following maintenance doses of prasugrel and clopidogrel in Chinese carriers of CYP2C19 variants. Br J Clin Pharmacol. 2012 Jan;73(1):93–105. doi: 10.1111/j.1365-2125.2011.04049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Varenhorst C, James S, Erlinge D, et al. Genetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery disease. Eur Heart J. 2009 Jul;30(14):1744–52. doi: 10.1093/eurheartj/ehp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Collet JP, Hulot JS, Anzaha G, et al. High doses of clopidogrel to overcome genetic resistance: the randomized crossover CLOVIS-2 (Clopidogrel and Response Variability Investigation Study 2) JACC Cardiovasc Interv. 2011 Apr;4(4):392–402. doi: 10.1016/j.jcin.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 77.Fontana P, Hulot JS, De Moerloose P, et al. Influence of CYP2C19 and CYP3A4 gene polymorphisms on clopidogrel responsiveness in healthy subjects. Journal of thrombosis and haemostasis : JTH. 2007 Oct;5(10):2153–5. doi: 10.1111/j.1538-7836.2007.02722.x. [DOI] [PubMed] [Google Scholar]
- 78.Collet JP, Hulot JS, Pena A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009 Jan 24;373(9660):309–17. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 79.Frere C, Cuisset T, Morange PE, et al. Effect of cytochrome p450 polymorphisms on platelet reactivity after treatment with clopidogrel in acute coronary syndrome. Am J Cardiol. 2008 Apr 15;101(8):1088–93. doi: 10.1016/j.amjcard.2007.11.065. [DOI] [PubMed] [Google Scholar]
- 80.Giusti B, Gori AM, Marcucci R, et al. Cytochrome P450 2C19 loss-of-function polymorphism, but not CYP3A4 IVS10 + 12G/A and P2Y12 T744C polymorphisms, is associated with response variability to dual antiplatelet treatment in high-risk vascular patients. Pharmacogenetics and genomics. 2007 Dec;17(12):1057–64. doi: 10.1097/FPC.0b013e3282f1b2be. [DOI] [PubMed] [Google Scholar]
- 81.Gurbel PA, Shuldiner AR, Bliden KP, et al. The relation between CYP2C19 genotype and phenotype in stented patients on maintenance dual antiplatelet therapy. Am Heart J. 2011 Mar;161(3):598–604. doi: 10.1016/j.ahj.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Price MJ, Murray SS, Angiolillo DJ, et al. Influence of genetic polymorphisms on the effect of high- and standard-dose clopidogrel after percutaneous coronary intervention: the GIFT (Genotype Information and Functional Testing) study. J Am Coll Cardiol. 2012 May 29;59(22):1928–37. doi: 10.1016/j.jacc.2011.11.068. [DOI] [PubMed] [Google Scholar]
- 83.Hokimoto S, Mizobe M, Akasaka T, et al. Impact of CYP2C19 polymorphism and proton pump inhibitors on platelet reactivity to clopidogrel and clinical outcomes following stent implantation. Thromb Res. 2014 Apr;133(4):599–605. doi: 10.1016/j.thromres.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 84.Tabata N, Hokimoto S, Akasaka T, et al. Chronic kidney disease status modifies the association of CYP2C19 polymorphism in predicting clinical outcomes following coronary stent implantation. Thromb Res. 2014 Nov;134(5):939–44. doi: 10.1016/j.thromres.2014.07.039. [DOI] [PubMed] [Google Scholar]
- 85.Jeong YH, Abadilla KA, Tantry US, et al. Influence of CYP2C19*2 and *3 loss-of-function alleles on the pharmacodynamic effects of standard- and high-dose clopidogrel in East Asians undergoing percutaneous coronary intervention: the results of the ACCEL-DOUBLE-2N3 study. Journal of thrombosis and haemostasis : JTH. 2013 Jun;11(6):1194–7. doi: 10.1111/jth.12200. [DOI] [PubMed] [Google Scholar]
- 86.Jeong YH, Tantry US, Kim IS, et al. Effect of CYP2C19*2 and *3 loss-of-function alleles on platelet reactivity and adverse clinical events in East Asian acute myocardial infarction survivors treated with clopidogrel and aspirin. Circ Cardiovasc Interv. 2011 Dec 1;4(6):585–94. doi: 10.1161/CIRCINTERVENTIONS.111.962555. [DOI] [PubMed] [Google Scholar]
- 87.Kim HS, Chang K, Koh YS, et al. CYP2C19 poor metabolizer is associated with clinical outcome of clopidogrel therapy in acute myocardial infarction but not stable angina. Circulation Cardiovascular genetics. 2013 Oct;6(5):514–21. doi: 10.1161/CIRCGENETICS.113.000109. [DOI] [PubMed] [Google Scholar]
- 88.Sorich MJ, Rowland A, McKinnon RA, et al. CYP2C19 genotype has a greater effect on adverse cardiovascular outcomes following percutaneous coronary intervention and in Asian populations treated with clopidogrel: a meta-analysis. Circulation Cardiovascular genetics. 2014 Dec;7(6):895–902. doi: 10.1161/CIRCGENETICS.114.000669. [DOI] [PubMed] [Google Scholar]
- 89.Frere C, Cuisset T, Gaborit B, et al. The CYP2C19*17 allele is associated with better platelet response to clopidogrel in patients admitted for non-ST acute coronary syndrome. Journal of thrombosis and haemostasis : JTH. 2009 Aug;7(8):1409–11. doi: 10.1111/j.1538-7836.2009.03500.x. [DOI] [PubMed] [Google Scholar]
- 90.Sibbing D, Gebhard D, Koch W, et al. Isolated and interactive impact of common CYP2C19 genetic variants on the antiplatelet effect of chronic clopidogrel therapy. Journal of thrombosis and haemostasis : JTH. 2010 May 21; doi: 10.1111/j.1538-7836.2010.03921.x. [DOI] [PubMed] [Google Scholar]
- 91.Tiroch KA, Sibbing D, Koch W, et al. Protective effect of the CYP2C19*17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J. 2010 Sep;160(3):506–12. doi: 10.1016/j.ahj.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 92.Geisler T, Grass D, Bigalke B, et al. The Residual Platelet Aggregation after Deployment of Intracoronary Stent (PREDICT) score. Journal of thrombosis and haemostasis : JTH. 2008 Jan;6(1):54–61. doi: 10.1111/j.1538-7836.2007.02812.x. [DOI] [PubMed] [Google Scholar]
- 93.Simon T, Verstuyft C, Mary-Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009 Jan 22;360(4):363–75. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 94.Sorich MJ, Polasek TM, Wiese MD. Systematic review and meta-analysis of the association between cytochrome P450 2C19 genotype and bleeding. Thrombosis and haemostasis. 2012 Jul;108(1):199–200. doi: 10.1160/TH12-02-0095. [DOI] [PubMed] [Google Scholar]
- 95.Lewis JP, Stephens SH, Horenstein RB, et al. The CYP2C19*17 variant is not independently associated with clopidogrel response. Journal of thrombosis and haemostasis : JTH. 2013 Sep;11(9):1640–6. doi: 10.1111/jth.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trenk D, Hochholzer W, Fromm MF, et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol. 2008 May 20;51(20):1925–34. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 97**.Giusti B, Gori AM, Marcucci R, et al. Relation of cytochrome P450 2C19 loss-of-function polymorphism to occurrence of drug-eluting coronary stent thrombosis. Am J Cardiol. 2009 Mar 15;103(6):806–11. doi: 10.1016/j.amjcard.2008.11.048. Meta-analysis of CYP2C19 association with adverse clinical outcomes among predominantly PCI patients. [DOI] [PubMed] [Google Scholar]
- 98.Harmsze A, van Werkum JW, Bouman HJ, et al. Besides CYP2C19*2, the variant allele CYP2C9*3 is associated with higher on-clopidogrel platelet reactivity in patients on dual antiplatelet therapy undergoing elective coronary stent implantation. Pharmacogenetics and genomics. 2010 Jan;20(1):18–25. doi: 10.1097/FPC.0b013e328333dafe. [DOI] [PubMed] [Google Scholar]
- 99.Sibbing D, Stegherr J, Latz W, et al. Cytochrome P450 2C19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. Eur Heart J. 2009 Apr;30(8):916–22. doi: 10.1093/eurheartj/ehp041. [DOI] [PubMed] [Google Scholar]
- 100.Pare G, Mehta SR, Yusuf S, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010 Oct 28;363(18):1704–14. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 101.Wallentin L, James S, Storey RF, et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010 Oct 16;376(9749):1320–8. doi: 10.1016/S0140-6736(10)61274-3. [DOI] [PubMed] [Google Scholar]
- 102.Hulot JS, Collet JP, Montalescot G. Genetic substudy of the PLATO trial. Lancet. 2011 Feb 19;377(9766):637. doi: 10.1016/S0140-6736(11)60227-4. author reply 37–8. [DOI] [PubMed] [Google Scholar]
- 103.Johnson JA, Roden DM, Lesko LJ, et al. Clopidogrel: a case for indication-specific pharmacogenetics. Clin Pharmacol Ther. 2012 May;91(5):774–6. doi: 10.1038/clpt.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Holmes MV, Perel P, Shah T, et al. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA : the journal of the American Medical Association. 2011 Dec 28;306(24):2704–14. doi: 10.1001/jama.2011.1880. [DOI] [PubMed] [Google Scholar]
- 105.Mega JL, Simon T, Collet JP, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA : the journal of the American Medical Association. 2010 Oct 27;304(16):1821–30. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bauer T, Bouman HJ, van Werkum JW, et al. Impact of CYP2C19 variant genotypes on clinical efficacy of antiplatelet treatment with clopidogrel: systematic review and meta-analysis. BMJ. 2011;343:d4588. doi: 10.1136/bmj.d4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hulot JS, Collet JP, Silvain J, et al. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration: a systematic meta-analysis. J Am Coll Cardiol. 2010 Jul 6;56(2):134–43. doi: 10.1016/j.jacc.2009.12.071. [DOI] [PubMed] [Google Scholar]
- 108.Jang JS, Cho KI, Jin HY, et al. Meta-analysis of cytochrome P450 2C19 polymorphism and risk of adverse clinical outcomes among coronary artery disease patients of different ethnic groups treated with clopidogrel. Am J Cardiol. 2012 Aug 15;110(4):502–8. doi: 10.1016/j.amjcard.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 109.Jin B, Ni HC, Shen W, et al. Cytochrome P450 2C19 polymorphism is associated with poor clinical outcomes in coronary artery disease patients treated with clopidogrel. Mol Biol Rep. 2011 Mar;38(3):1697–702. doi: 10.1007/s11033-010-0282-0. [DOI] [PubMed] [Google Scholar]
- 110.Singh M, Shah T, Adigopula S, et al. CYP2C19*2/ABCB1-C3435T polymorphism and risk of cardiovascular events in coronary artery disease patients on clopidogrel: Is clinical testing helpful? Indian Heart J. 2012 Jul-Aug;64(4):341–52. doi: 10.1016/j.ihj.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sofi F, Giusti B, Marcucci R, et al. Cytochrome P450 2C19*2 polymorphism and cardiovascular recurrences in patients taking clopidogrel: a meta-analysis. Pharmacogenomics J. 2011 Jun;11(3):199–206. doi: 10.1038/tpj.2010.21. [DOI] [PubMed] [Google Scholar]
- 112.Yamaguchi Y, Abe T, Sato Y, et al. Effects of VerifyNow P2Y12 test and CYP2C19*2 testing on clinical outcomes of patients with cardiovascular disease: A systematic review and meta-analysis. Platelets. 2012 Jul 3; doi: 10.3109/09537104.2012.700969. [DOI] [PubMed] [Google Scholar]
- 113.Zabalza M, Subirana I, Sala J, et al. Meta-analyses of the association between cytochrome CYP2C19 loss- and gain-of-function polymorphisms and cardiovascular outcomes in patients with coronary artery disease treated with clopidogrel. Heart. 2012 Jan;98(2):100–8. doi: 10.1136/hrt.2011.227652. [DOI] [PubMed] [Google Scholar]
- 114.Roberts JD, Wells GA, Le May MR, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet. 2012 May 5;379(9827):1705–11. doi: 10.1016/S0140-6736(12)60161-5. [DOI] [PubMed] [Google Scholar]
- 115.So DY, Wells GA, McPherson R, et al. A prospective randomized evaluation of a pharmacogenomic approach to antiplatelet therapy among patients with ST-elevation myocardial infarction: the RAPID STEMI study. Pharmacogenomics J. 2015 Apr 7; doi: 10.1038/tpj.2015.17. [DOI] [PubMed] [Google Scholar]
- 116.Ahn SG, Yoon J, Kim J, et al. Genotype- and Phenotype-Directed Personalization of Antiplatelet Treatment in Patients with Non-ST Elevation Acute Coronary Syndromes Undergoing Coronary Stenting. Korean circulation journal. 2013 Aug;43(8):541–9. doi: 10.4070/kcj.2013.43.8.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xie X, Ma YT, Yang YN, et al. Personalized antiplatelet therapy according to CYP2C19 genotype after percutaneous coronary intervention: a randomized control trial. International journal of cardiology. 2013 Oct 9;168(4):3736–40. doi: 10.1016/j.ijcard.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 118.Chevalier B. Foundation CR, editor. GIANT: Evaluation of the genetic profile of CYP2C19 in patients undergoing primary angioplasty. J Am Coll Cardiol; Twenty-Fifth Annual Transcatheter Cardiovascular Therapeutics Symposium; 2013; San Francisco, California: Moscone Center; 2013. [Google Scholar]
- 119.Holmes DR, Jr, Dehmer GJ, Kaul S, et al. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J Am Coll Cardiol. 2010 Jul 20;56(4):321–41. doi: 10.1016/j.jacc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 120.Anderson JL, Adams CD, Antman EM, et al. 2012 ACCF/AHA focused update incorporated into the ACCF/AHA 2007 guidelines for the management of patients with unstable angina/non-ST-elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013 Jun 11;61(23):e179–347. doi: 10.1016/j.jacc.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 121.Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011 May;89(5):662–73. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 122.Jia DM, Chen ZB, Zhang MJ, et al. CYP2C19 polymorphisms and antiplatelet effects of clopidogrel in acute ischemic stroke in China. Stroke. 2013 Jun;44(6):1717–9. doi: 10.1161/STROKEAHA.113.000823. [DOI] [PubMed] [Google Scholar]
- 123.Sun W, Li Y, Li J, et al. Variant recurrent risk among stroke patients with different CYP2C19 phenotypes and treated with clopidogrel. Platelets. 2014 Sep;10:1–5. doi: 10.3109/09537104.2014.953044. [DOI] [PubMed] [Google Scholar]
- 124.McDonough CW, McClure LA, Mitchell BD, et al. CYP2C19 Metabolizer Status and Clopidogrel Efficacy in the Secondary Prevention of Small Subcortical Strokes (SPS3) Study. Journal of the American Heart Association. 2015;4(6) doi: 10.1161/JAHA.114.001652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Campo G, Parrinello G, Ferraresi P, et al. Prospective evaluation of on-clopidogrel platelet reactivity over time in patients treated with percutaneous coronary intervention relationship with gene polymorphisms and clinical outcome. J Am Coll Cardiol. 2011 Jun 21;57(25):2474–83. doi: 10.1016/j.jacc.2010.12.047. [DOI] [PubMed] [Google Scholar]
- 126.Mega JL, Close SL, Wiviott SD, et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010 Oct 16;376(9749):1312–9. doi: 10.1016/S0140-6736(10)61273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Simon T, Steg PG, Becquemont L, et al. Effect of paraoxonase-1 polymorphism on clinical outcomes in patients treated with clopidogrel after an acute myocardial infarction. Clin Pharmacol Ther. 2011 Oct;90(4):561–7. doi: 10.1038/clpt.2011.193. [DOI] [PubMed] [Google Scholar]
- 128.Carlquist JF, Knight S, Horne BD, et al. Cardiovascular risk among patients on clopidogrel anti-platelet therapy after placement of drug-eluting stents is modified by genetic variants in both the CYP2C19 and ABCB1 genes. Thrombosis and haemostasis. 2013 Jan 31;109(4) doi: 10.1160/TH12-05-0336. [DOI] [PubMed] [Google Scholar]
- 129.Verschuren JJ, Boden H, Wessels JA, et al. Value of platelet pharmacogenetics in common clinical practice of patients with ST-segment elevation myocardial infarction. International journal of cardiology. 2012 Aug 29; doi: 10.1016/j.ijcard.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 130.Su J, Xu J, Li X, et al. ABCB1 C3435T polymorphism and response to clopidogrel treatment in coronary artery disease (CAD) patients: a meta-analysis. PLoS One. 2012;7(10):e46366. doi: 10.1371/journal.pone.0046366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lewis JP, Horenstein RB, Ryan K, et al. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenetics and genomics. 2013 Jan;23(1):1–8. doi: 10.1097/FPC.0b013e32835aa8a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhu HJ, Wang X, Gawronski BE, et al. Carboxylesterase 1 as a determinant of clopidogrel metabolism and activation. J Pharmacol Exp Ther. 2013 Mar;344(3):665–72. doi: 10.1124/jpet.112.201640. [DOI] [PubMed] [Google Scholar]
- 133.Park KW, Park JJ, Jeon KH, et al. Enhanced clopidogrel responsiveness in smokers: smokers’ paradox is dependent on cytochrome P450 CYP1A2 status. Arteriosclerosis, thrombosis, and vascular biology. 2011 Mar;31(3):665–71. doi: 10.1161/ATVBAHA.110.217182. [DOI] [PubMed] [Google Scholar]
- 134.Cresci S, Depta JP, Lenzini PA, et al. Cytochrome p450 gene variants, race, and mortality among clopidogrel-treated patients after acute myocardial infarction. Circulation Cardiovascular genetics. 2014 Jun;7(3):277–86. doi: 10.1161/CIRCGENETICS.113.000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Harmsze AM, van Werkum JW, Ten Berg JM, et al. CYP2C19*2 and CYP2C9*3 alleles are associated with stent thrombosis: a case-control study. Eur Heart J. 2010 Dec;31(24):3046–53. doi: 10.1093/eurheartj/ehq321. [DOI] [PubMed] [Google Scholar]
- 136.Gremmel T, Kopp CW, Seidinger D, et al. Differential impact of cytochrome 2C9 allelic variants on clopidogrel-mediated platelet inhibition determined by five different platelet function tests. International journal of cardiology. 2013 Jun 5;166(1):126–31. doi: 10.1016/j.ijcard.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 137.Gladding P, White H, Voss J, et al. Pharmacogenetic testing for clopidogrel using the rapid INFINITI analyzer: a dose-escalation study. JACC Cardiovasc Interv. 2009 Nov;2(11):1095–101. doi: 10.1016/j.jcin.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 138.Lau WC, Gurbel PA, Watkins PB, et al. Contribution of hepatic cytochrome P450 3A4 metabolic activity to the phenomenon of clopidogrel resistance. Circulation. 2004 Jan 20;109(2):166–71. doi: 10.1161/01.CIR.0000112378.09325.F9. [DOI] [PubMed] [Google Scholar]
- 139.Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, et al. Contribution of gene sequence variations of the hepatic cytochrome P450 3A4 enzyme to variability in individual responsiveness to clopidogrel. Arteriosclerosis, thrombosis, and vascular biology. 2006 Aug;26(8):1895–900. doi: 10.1161/01.ATV.0000223867.25324.1a. [DOI] [PubMed] [Google Scholar]
- 140.Suh JW, Koo BK, Zhang SY, et al. Increased risk of atherothrombotic events associated with cytochrome P450 3A5 polymorphism in patients taking clopidogrel. CMAJ : Canadian Medical Association journal = journal de l’Association medicale canadienne. 2006 Jun 6;174(12):1715–22. doi: 10.1503/cmaj.060664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim KA, Park PW, Park JY. Effect of CYP3A5*3 genotype on the pharmacokinetics and antiplatelet effect of clopidogrel in healthy subjects. Eur J Clin Pharmacol. 2008 Jun;64(6):589–97. doi: 10.1007/s00228-008-0471-0. [DOI] [PubMed] [Google Scholar]
- 142.Wu H, Qian J, Xu J, et al. Effects of CYP2C19 variant alleles on postclopidogrel platelet reactivity and clinical outcomes in an actual clinical setting in China. Pharmacogenetics and genomics. 2012 Dec;22(12):887–90. doi: 10.1097/FPC.0b013e328359253a. [DOI] [PubMed] [Google Scholar]
- 143.Kreutz RP, Owens J, Jin Y, et al. Cytochrome P450 3A4*22, PPAR-alpha, and ARNT polymorphisms and clopidogrel response. Clinical pharmacology : advances and applications. 2013;5:185–92. doi: 10.2147/CPAA.S53151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Park JJ, Park KW, Kang J, et al. Genetic determinants of clopidogrel responsiveness in Koreans treated with drug-eluting stents. International journal of cardiology. 2013 Feb 10;163(1):79–86. doi: 10.1016/j.ijcard.2012.09.075. [DOI] [PubMed] [Google Scholar]
- 145.Viviani Anselmi C, Briguori C, Roncarati R, et al. Routine assessment of on-clopidogrel platelet reactivity and gene polymorphisms in predicting clinical outcome following drug-eluting stent implantation in patients with stable coronary artery disease. JACC Cardiovasc Interv. 2013 Nov;6(11):1166–75. doi: 10.1016/j.jcin.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 146.Fontana P, Dupont A, Gandrille S, et al. Adenosine diphosphate-induced platelet aggregation is associated with P2Y12 gene sequence variations in healthy subjects. Circulation. 2003 Aug 26;108(8):989–95. doi: 10.1161/01.CIR.0000085073.69189.88. [DOI] [PubMed] [Google Scholar]
- 147.von Beckerath N, von Beckerath O, Koch W, et al. P2Y12 gene H2 haplotype is not associated with increased adenosine diphosphate-induced platelet aggregation after initiation of clopidogrel therapy with a high loading dose. Blood coagulation & fibrinolysis : an international journal in haemostasis and thrombosis. 2005 Apr;16(3):199–204. doi: 10.1097/01.mbc.0000164429.21040.0a. [DOI] [PubMed] [Google Scholar]
- 148.Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, et al. Lack of association between the P2Y12 receptor gene polymorphism and platelet response to clopidogrel in patients with coronary artery disease. Thromb Res. 2005;116(6):491–7. doi: 10.1016/j.thromres.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 149.Lev EI, Patel RT, Guthikonda S, et al. Genetic polymorphisms of the platelet receptors P2Y(12), P2Y(1) and GP IIIa and response to aspirin and clopidogrel. Thromb Res. 2007;119(3):355–60. doi: 10.1016/j.thromres.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 150.Smith SM, Judge HM, Peters G, et al. Common sequence variations in the P2Y12 and CYP3A5 genes do not explain the variability in the inhibitory effects of clopidogrel therapy. Platelets. 2006 Jun;17(4):250–8. doi: 10.1080/09537100500475844. [DOI] [PubMed] [Google Scholar]
- 151.Namazi S, Kojuri J, Khalili A, et al. The impact of genetic polymorphisms of P2Y12, CYP3A5 and CYP2C19 on clopidogrel response variability in Iranian patients. Biochemical pharmacology. 2012 Apr 1;83(7):903–8. doi: 10.1016/j.bcp.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 152.Bouman HJ, Schomig E, van Werkum JW, et al. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat Med. 2011 Jan;17(1):110–6. doi: 10.1038/nm.2281. [DOI] [PubMed] [Google Scholar]
- 153.Hulot JS, Collet JP, Cayla G, et al. CYP2C19 but not PON1 genetic variants influence clopidogrel pharmacokinetics, pharmacodynamics, and clinical efficacy in post-myocardial infarction patients. Circ Cardiovasc Interv. 2011 Oct 1;4(5):422–8. doi: 10.1161/CIRCINTERVENTIONS.111.963025. [DOI] [PubMed] [Google Scholar]
- 154.Lewis JP, Fisch AS, Ryan K, et al. Paraoxonase 1 (PON1) gene variants are not associated with clopidogrel response. Clin Pharmacol Ther. 2011 Oct;90(4):568–74. doi: 10.1038/clpt.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sibbing D, Koch W, Massberg S, et al. No association of paraoxonase-1 Q192R genotypes with platelet response to clopidogrel and risk of stent thrombosis after coronary stenting. Eur Heart J. 2011 Jul;32(13):1605–13. doi: 10.1093/eurheartj/ehr155. [DOI] [PubMed] [Google Scholar]
- 156.Trenk D, Hochholzer W, Fromm MF, et al. Paraoxonase-1 Q192R polymorphism and antiplatelet effects of clopidogrel in patients undergoing elective coronary stent placement. Circulation Cardiovascular genetics. 2011 Aug 1;4(4):429–36. doi: 10.1161/CIRCGENETICS.111.960112. [DOI] [PubMed] [Google Scholar]
- 157.Ancrenaz V, Desmeules J, James R, et al. The paraoxonase-1 pathway is not a major bioactivation pathway of clopidogrel in vitro. Br J Pharmacol. 2012 Aug;166(8):2362–70. doi: 10.1111/j.1476-5381.2012.01946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Reny JL, Combescure C, Daali Y, et al. Influence of the paraoxonase-1 Q192R genetic variant on clopidogrel responsiveness and recurrent cardiovascular events: a systematic review and meta-analysis. Journal of thrombosis and haemostasis : JTH. 2012 Jul;10(7):1242–51. doi: 10.1111/j.1538-7836.2012.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reny JL, Combescure C, Daali Y, et al. Influence of the paraoxonase-1 Q192R genetic variant on clopidogrel responsiveness and recurrent cardiovascular events: a systematic review and meta-analysis. Journal of thrombosis and haemostasis : JTH. 2012 Jul;10(7):1242–51. doi: 10.1111/j.1538-7836.2012.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rehmel JL, Eckstein JA, Farid NA, et al. Interactions of two major metabolites of prasugrel, a thienopyridine antiplatelet agent, with the cytochromes P450. Drug Metab Dispos. 2006 Apr;34(4):600–7. doi: 10.1124/dmd.105.007989. [DOI] [PubMed] [Google Scholar]
- 161.Brandt JT, Payne CD, Wiviott SD, et al. A comparison of prasugrel and clopidogrel loading doses on platelet function: magnitude of platelet inhibition is related to active metabolite formation. Am Heart J. 2007 Jan;153(1):66e9–16. doi: 10.1016/j.ahj.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 162.Payne CD, Li YG, Small DS, et al. Increased active metabolite formation explains the greater platelet inhibition with prasugrel compared to high-dose clopidogrel. Journal of cardiovascular pharmacology. 2007 Nov;50(5):555–62. doi: 10.1097/FJC.0b013e3181492209. [DOI] [PubMed] [Google Scholar]
- 163.Wallentin L, Varenhorst C, James S, et al. Prasugrel achieves greater and faster P2Y12receptor-mediated platelet inhibition than clopidogrel due to more efficient generation of its active metabolite in aspirin-treated patients with coronary artery disease. Eur Heart J. 2008 Jan;29(1):21–30. doi: 10.1093/eurheartj/ehm545. [DOI] [PubMed] [Google Scholar]
- 164.Mega JL, Close SL, Wiviott SD, et al. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation. 2009 May 19;119(19):2553–60. doi: 10.1161/CIRCULATIONAHA.109.851949. [DOI] [PubMed] [Google Scholar]
- 165.Wiviott SD, Trenk D, Frelinger AL, et al. Prasugrel compared with high loading- and maintenance-dose clopidogrel in patients with planned percutaneous coronary intervention: the Prasugrel in Comparison to Clopidogrel for Inhibition of Platelet Activation and Aggregation-Thrombolysis in Myocardial Infarction 44 trial. Circulation. 2007 Dec 18;116(25):2923–32. doi: 10.1161/CIRCULATIONAHA.107.740324. [DOI] [PubMed] [Google Scholar]
- 166.Bonello L, Pansieri M, Mancini J, et al. High on-treatment platelet reactivity after prasugrel loading dose and cardiovascular events after percutaneous coronary intervention in acute coronary syndromes. J Am Coll Cardiol. 2011 Jul 26;58(5):467–73. doi: 10.1016/j.jacc.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 167.Cuisset T, Loosveld M, Morange PE, et al. CYP2C19*2 and *17 alleles have a significant impact on platelet response and bleeding risk in patients treated with prasugrel after acute coronary syndrome. JACC Cardiovasc Interv. 2012 Dec;5(12):1280–7. doi: 10.1016/j.jcin.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 168.Grosdidier C, Quilici J, Loosveld M, et al. Effect of CYP2C19*2 and *17 genetic variants on platelet response to clopidogrel and prasugrel maintenance dose and relation to bleeding complications. Am J Cardiol. 2013 Apr 1;111(7):985–90. doi: 10.1016/j.amjcard.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 169.Htun WW, Steinhubl SR. Ticagrelor: the first novel reversible P2Y(12) inhibitor. Expert opinion on pharmacotherapy. 2013 Feb;14(2):237–45. doi: 10.1517/14656566.2013.757303. [DOI] [PubMed] [Google Scholar]
- 170.Teng R. Pharmacokinetic, pharmacodynamic and pharmacogenetic profile of the oral antiplatelet agent ticagrelor. Clin Pharmacokinet. 2012 May 1;51(5):305–18. doi: 10.2165/11630960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 171.Dinicolantonio JJ, Serebruany VL. Exploring the ticagrelor-statin interplay in the PLATO trial. Cardiology. 2013;124(2):105–7. doi: 10.1159/000346151. [DOI] [PubMed] [Google Scholar]
- 172.Gurbel PA, Bliden KP, Butler K, et al. Randomized double-blind assessment of the ONSET and OFFSET of the antiplatelet effects of ticagrelor versus clopidogrel in patients with stable coronary artery disease: the ONSET/OFFSET study. Circulation. 2009 Dec 22;120(25):2577–85. doi: 10.1161/CIRCULATIONAHA.109.912550. [DOI] [PubMed] [Google Scholar]
- 173.Wallentin L, Becker RC, Budaj A, et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2009 Sep 10;361(11):1045–57. doi: 10.1056/NEJMoa0904327. [DOI] [PubMed] [Google Scholar]
- 174.Varenhorst CEN, Johansson A, Barratt B, Akerblom EH, Syvanen A, et al. Ticagrelor plasma levels but not clinical outcomes are associated with transporter and metabolism enzyme genetic polymorphisms. J Am Coll Cardiol. 2014 Apr;63(12_S) [Google Scholar]
- 175.Sharfstein J. FDA Regulation of Laboratory-Developed Diagnostic Tests: Protect the Public, Advance the Science. JAMA : the journal of the American Medical Association. 2015 Jan 5; doi: 10.1001/jama.2014.18135. [DOI] [PubMed] [Google Scholar]
- 176.Evans JP, Watson MS. Genetic Testing and FDA Regulation: Overregulation Threatens the Emergence of Genomic Medicine. JAMA : the journal of the American Medical Association. 2015 Jan 5; doi: 10.1001/jama.2014.18145. [DOI] [PubMed] [Google Scholar]
- 177.Rubinstein WS, Maglott DR, Lee JM, et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic acids research. 2013 Jan;41(Database issue):D925–35. doi: 10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Scott SA. Personalizing medicine with clinical pharmacogenetics. Genetics in medicine : official journal of the American College of Medical Genetics. 2011 Dec;13(12):987–95. doi: 10.1097/GIM.0b013e318238b38c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Abul-Husn NS, Owusu Obeng A, Sanderson SC, et al. Implementation and utilization of genetic testing in personalized medicine. Pharmacogenomics and personalized medicine. 2014;7:227–40. doi: 10.2147/PGPM.S48887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Johnson JA, Elsey AR, Clare-Salzler MJ, et al. Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics. 2013 May;14(7):723–6. doi: 10.2217/pgs.13.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shuldiner AR, Palmer K, Pakyz RE, et al. Implementation of pharmacogenetics: the University of Maryland Personalized Anti-platelet Pharmacogenetics Program. American journal of medical genetics Part C, Seminars in medical genetics. 2014 Mar;166C(1):76–84. doi: 10.1002/ajmg.c.31396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Haga SB, Moaddeb J. Comparison of delivery strategies for pharmacogenetic testing services. Pharmacogenetics and genomics. 2014 Mar;24(3):139–45. doi: 10.1097/FPC.0000000000000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gottesman O, Scott SA, Ellis SB, et al. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther. 2013 Aug;94(2):214–7. doi: 10.1038/clpt.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pulley JM, Denny JC, Peterson JF, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012 Jul;92(1):87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther. 2014 Oct;96(4):482–9. doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hulot JS, Fuster V. Antiplatelet therapy: Personalized medicine for clopidogrel resistance? Nature reviews Cardiology. 2009 May;6(5):334–6. doi: 10.1038/nrcardio.2009.28. [DOI] [PubMed] [Google Scholar]
- 187.Fontana P, Cattaneo M, Combescure C, et al. Tailored Thienopyridine therapy: no urgency for CYP2C19 genotyping. Journal of the American Heart Association. 2013 Apr;2(2):e000131. doi: 10.1161/JAHA.112.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Burke W, Emery J. Genetics education for primary-care providers. Nature reviews Genetics. 2002 Jul;3(7):561–6. doi: 10.1038/nrg845. [DOI] [PubMed] [Google Scholar]
- 189.Murray MF. Educating physicians in the era of genomic medicine. Genome medicine. 2014;6(6):45. doi: 10.1186/gm564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nickola TJ, Green JS, Harralson AF, et al. The current and future state of pharmacogenomics medical education in the USA. Pharmacogenomics. 2012 Sep;13(12):1419–25. doi: 10.2217/pgs.12.113. [DOI] [PubMed] [Google Scholar]
- 191.Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. American journal of medical genetics Part C, Seminars in medical genetics. 2014 Mar;166C(1):56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kazi DS, Garber AM, Shah RU, et al. Cost-effectiveness of genotype-guided and dual antiplatelet therapies in acute coronary syndrome. Annals of internal medicine. 2014 Feb 18;160(4):221–32. doi: 10.7326/M13-1999. [DOI] [PubMed] [Google Scholar]
- 193.Reese ES, Daniel Mullins C, Beitelshees AL, et al. Cost-effectiveness of cytochrome P450 2C19 genotype screening for selection of antiplatelet therapy with clopidogrel or prasugrel. Pharmacotherapy. 2012 Apr;32(4):323–32. doi: 10.1002/PHAR.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lala A, Berger JS, Sharma G, et al. Genetic testing in patients with acute coronary syndrome undergoing percutaneous coronary intervention: a cost-effectiveness analysis. Journal of thrombosis and haemostasis : JTH. 2013 Jan;11(1):81–91. doi: 10.1111/jth.12059. [DOI] [PubMed] [Google Scholar]
- 195.Sorich MJ, Horowitz JD, Sorich W, et al. Cost-effectiveness of using CYP2C19 genotype to guide selection of clopidogrel or ticagrelor in Australia. Pharmacogenomics. 2013 Dec;14(16):2013–21. doi: 10.2217/pgs.13.164. [DOI] [PubMed] [Google Scholar]
- 196.Voora D, Ortel TL, Lucas JE, et al. Time-dependent changes in non-COX-1-dependent platelet function with daily aspirin therapy. J Thromb Thrombolysis. 2012 Apr;33(3):246–57. doi: 10.1007/s11239-012-0683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Voora D, Cyr D, Lucas J, et al. Aspirin exposure reveals novel genes associated with platelet function and cardiovascular events. J Am Coll Cardiol. 2013 Oct 1;62(14):1267–76. doi: 10.1016/j.jacc.2013.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.U.S. Food and Drug Administration. [last accessed June 30 2015];In Vitro Diagnostics. Available from: http://www.accessdata.fda.gov/scripts/cdrh/devicesatfda/index.cfm?
- 199.Levine GN, Jeong YH, Goto S, et al. Expert consensus document: World Heart Federation expert consensus statement on antiplatelet therapy in East Asian patients with ACS or undergoing PCI. Nature reviews Cardiology. 2014 Oct;11(10):597–606. doi: 10.1038/nrcardio.2014.104. [DOI] [PubMed] [Google Scholar]