Abstract
Adult stem cells support tissue homeostasis and repair throughout the life of an individual. During ageing, numerous intrinsic and extrinsic changes occur that result in altered stem-cell behaviour and reduced tissue maintenance and regeneration. In the Drosophila testis, ageing results in a marked decrease in the self-renewal factor Unpaired (Upd), leading to a concomitant loss of germline stem cells. Here we demonstrate that IGF-II messenger RNA binding protein (Imp) counteracts endogenous small interfering RNAs to stabilize upd (also known as os) RNA. However, similar to upd, Imp expression decreases in the hub cells of older males, which is due to the targeting of Imp by the heterochronic microRNA let-7. In the absence of Imp, upd mRNA therefore becomes unprotected and susceptible to degradation. Understanding the mechanistic basis for ageing-related changes in stem-cell behaviour will lead to the development of strategies to treat age-onset diseases and facilitate stem-cell-based therapies in older individuals.
Many stem cells lose the capacity for self-renewal when removed from their local microenvironment (or niche), indicating that the niche has a major role in controlling stem-cell fate1. Changes to the local and systemic environments occur with age that result in altered stem-cell behaviour and reduced tissue maintenance and regeneration2,3. The stem-cell niche in the Drosophila testis is located at the apical tip, where both germline stem cells (GSCs) and somatic cyst stem cells are in direct contact with hub cells (Fig. 1a; reviewed in ref. 4). Hub cells express the self-renewal factor Upd, which activates the JAK-STAT signalling pathway to regulate the behaviour of adjacent stem cells5–7. Ageing results in a progressive and significant decrease in the levels of upd in hub cells (Fig. 1b). However, constitutive expression of upd in hub cells was sufficient to block the age-related loss of GSCs5, suggesting that mechanisms might be in place to regulate upd and maintain an active stem-cell niche.
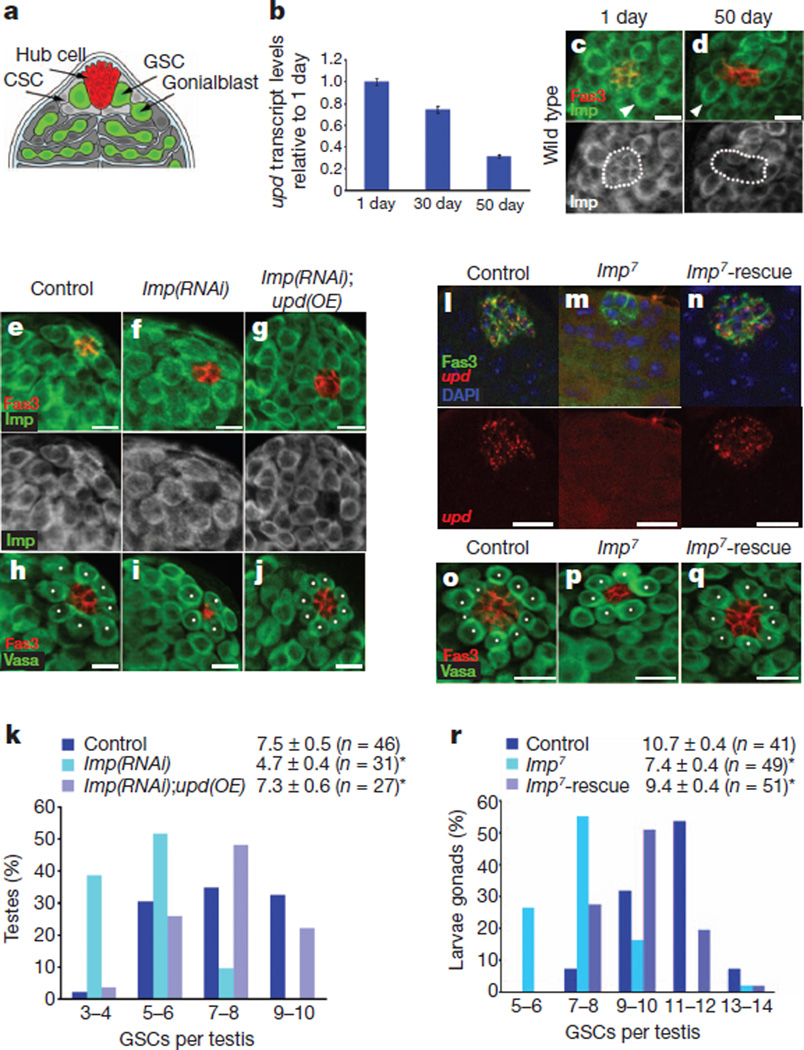
Figure 1. Imp regulates upd levels and GSC maintenance in the testis.
a, The apical tip of a Drosophila testis. CSC, cyst stem cell. b, upd mRNA decreases with age (qRT–PCR). One representative experiment is shown (n = 3); error bars denote s.d. of triplicate measurements. c, d, Testes from 1-day-old (c) or 50-day-old (d) flies immunostained for Imp (green) and Fas3 (red, hub). Note reduced Imp levels in hub cells of aged males (dotted lines, bottom panels) compared with a modest reduction in GSCs (arrowheads, top panels). e–j, Overexpression (OE) of upd suppresses the loss of GSCs owing to the loss of Imp. Staining for Imp (green) to confirm knockdown (e–g) or for the germline marker Vasa (green) (h–j). White dots in h–j denote GSCs. Genotypes: control, w−, upd-GAL4, UAS-gfp outcrossed to w1118; Imp(RNAi), w−, upd-GAL4, UAS-gfp; UAS-Imp(RNAi); Imp(RNAi);upd(OE), w−, upd-GAL4, UAS-gfp; UAS-Imp(RNAi); UAS-upd, TM2. k, GSC distribution as in h–j. l–q, L3 male gonads stained for Fas3 (green) and upd (red) (l–n) or Fas3 (red) and Vasa (green) (o–q). Nuclei in l–n were counterstained blue with 4′,6-diamidino-2-phenylindole (DAPI). White dots in o–q denote GSCs. Genotypes: control, EP(X)760; Imp7; Imp7-rescue, Imp7/Y; 8-156-GAL4; UAS-ImpT21. r, GSC distribution as in o–q. The mean ± 95% confidence interval are shown in k and r. *P < 0.01 compared with controls (Student’s t-test). Scale bars, 10 µm.
To identify potential regulators of upd, we screened a collection of transgenic flies carrying green fluorescent protein (GFP)-tagged proteins for expression in hub cells6. The Drosophila homologue of Imp protein is expressed throughout the testis tip in young flies (Fig. 1c and Supplementary Fig. 2a)7; however, antibody staining revealed a decrease (~50%) in Imp expression in the hub cells of aged males (Fig. 1d and Supplementary Fig. 2b). Imp is a member of a conserved family of RNA-binding proteins that regulate RNA stability, translation and localization8. Given the similarity in the ageing-related decline in Imp protein and upd mRNA in hub cells, we proposed that Imp could be a new regulator of upd.
Imp acts upstream to regulate upd mRNA
To address whether Imp acts in hub cells to regulate upd, we used the bipartite GAL4-UAS system9 in combination with RNA-mediated interference (RNAi) to reduce Imp expression exclusively in hub cells. Fluorescence in situ hybridization (FISH) to detect upd mRNA was used in combination with immunofluorescence microscopy to determine whether the loss of Imp expression affects upd levels. The loss of Imp specifically in hub cells resulted in reduced expression of upd (Fig. 1e, f (bottom), and Supplementary Fig. 2c, d), as well as a significant reduction in GSCs and hub cells (Fig. 1h, i, k and Supplementary Fig. 2e), when compared with controls. Consistent with a reduction in JAK-STAT signalling, decreased accumulation of STAT was observed when Imp levels were reduced by RNAi in hub cells (Supplementary Fig. 2f–h).
RNA-binding proteins characteristically target several RNAs; therefore, we wanted to determine whether upd is a physiologically relevant target of Imp. Expression of upd together with an Imp RNAi construct was sufficient to completely rescue the defects caused by reduced Imp expression in hub cells (Fig. 1e–k and Supplementary Fig. 2e), suggesting that Upd acts downstream of Imp to maintain GSCs and niche integrity. Importantly, the constitutive expression of upd alone in hub cells did not lead to an increase in GSCs in testes from 1-day-old males (w, upd-GAL4; 8.3 ± 0.7 (mean ± 95% confidence interval), n = 21, and upd-GAL4; UAS-upd, TM2; 8.3 ± 0.8, n = 32). These data suggest that Imp acts in hub cells to promote niche integrity and GSC maintenance, at least in part, by positively regulating upd.
If Imp acts in hub cells in adult testes to regulate upd mRNA, we speculated that the loss of Imp function during development might lead to a decrease in upd and a subsequent reduction in GSCs. Null mutations in Imp result in lethality at the pharate adult stage; therefore, we examined testes from third instar larvae (L3) carrying Imp null alleles, Imp7 and Imp8 (ref. 10). Deletion of the Imp locus was verified by PCR of genomic DNA (Supplementary Fig. 3a). Combined immunofluorescence and FISH showed that although Fas3+ hub cells were easily detected, the expression of upd was significantly reduced: 24% of Imp7 mutants (n = 67) and 15% of Imp8 mutants (n = 41) had no detectable upd at this stage (Fig. 1l, m). In addition, the average number of GSCs and hub cells in testes from Imp mutants was significantly reduced when compared with control L3 testes (Fig. 1o–p, r and Supplementary Fig. 3c–e). Notably, the re-expression of Imp in somatic niche cells was sufficient to rescue upd expression in Imp mutants to comparable levels to controls (Fig. 1l–n), and the reduction in the average number of GSCs and hub cells in Imp mutants was also reversed (Fig. 1o–q, r and Supplementary Fig. 3c–e).
Imp binds to upd in vivo and in vitro
Imp family members contain conserved KH domains that mediate direct binding to RNA targets. To determine whether Imp could associate directly with upd mRNA in vivo, testes were dissected from young flies expressing GFP-tagged Imp (Supplementary Fig. 2a)6. Immunoprecipitation of Imp with anti-GFP antibodies, followed by quantitative reverse transcriptase PCR (qRT–PCR) analysis, showed a significant enrichment (~208-fold) of associated upd mRNA relative to control antibodies (Fig. 2a). Minimal enrichment for the ubiquitously expressed RNAs rp49 (also known as RpL32; ~4-fold) and GapDH (also known as Gapdh1; ~8 fold) or for the negative control med23 (~4-fold; see Methods), was observed after Imp immunoprecipitation, indicating that the interaction between Imp and upd mRNA in hub cells is specific (Fig. 2a). Consistent with these observations, Imp protein and upd RNA co-localized in hub cells within perinuclear foci, probably ribonucleoprotein particles (Supplementary Fig. 4a).
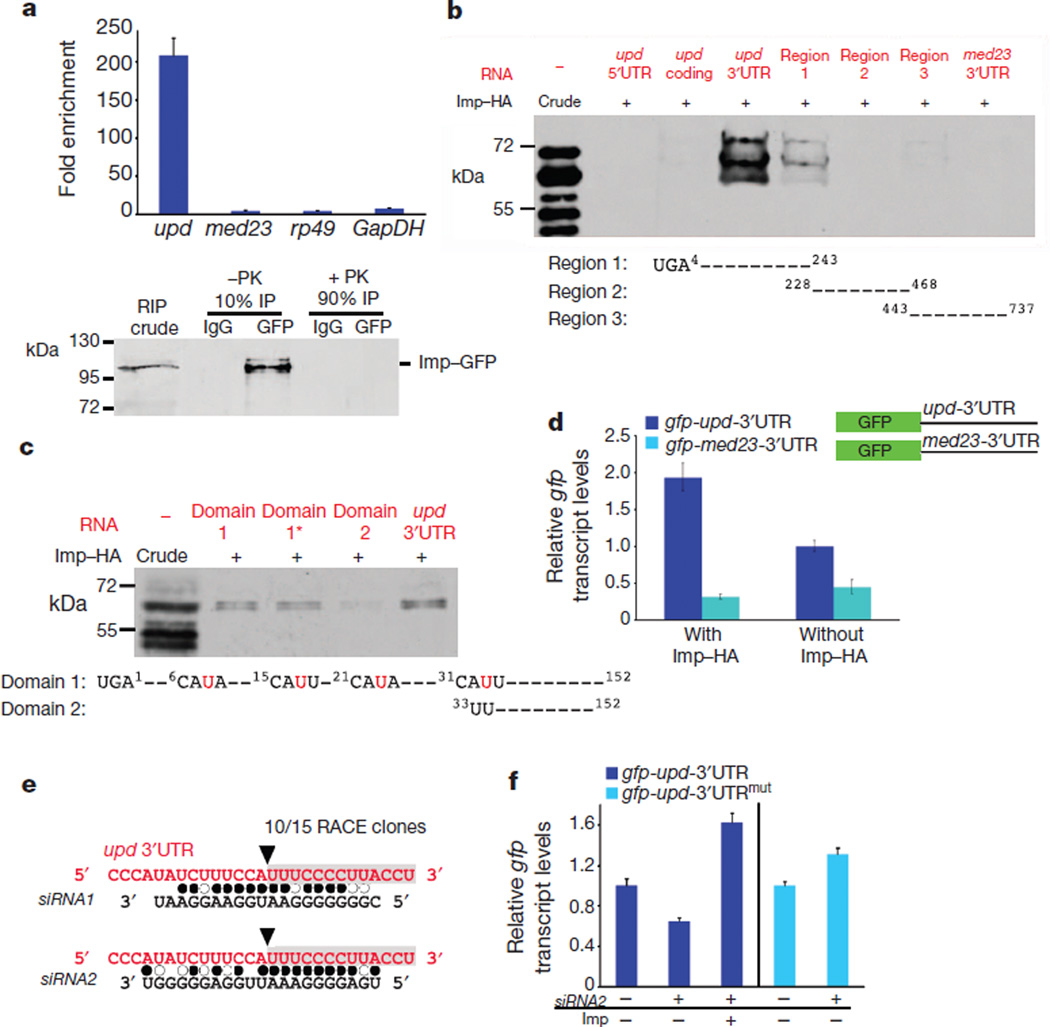
Figure 2. Imp binds to upd mRNA and counteracts siRNA-mediated degradation.
a, Top, fold-enrichment of the indicated mRNAs after RNA immunoprecipitation (RIP) with anti-GFP antibodies relative to control IgG from the testes of young Imp–GFP flies. Bottom, immunoblot for Imp–GFP. Ten per cent of the immunoprecipitate was used to confirm immunoprecipitation efficiency; the remaining 90% was treated with proteinase K (PK) and used for qRT–PCR. b, c, In vitro protein–RNA binding assay. Top panels, immunoblots for Imp–HA binding to designated RNA fragments. Imp–HA binds to region 1 of the upd 3′UTR (b), and to domain 1 and 1* within region 1 of the upd 3′UTR, but not to domain 2 (c). Mutations in domain 1* of the upd 3′UTR are indicated in red (c). d, qRT–PCR showing relative gfp mRNA levels in cells co-transfected with actin-GAL4 and the indicated gfp reporter constructs. Expression of gfp is presented relative to cells without Imp–HA and is normalized toGAL4. Note that gfp-upd-3′UTR mRNA is sixfold higher than gfp-med23-3′UTR in cells expressing Imp–HA. e, Sequence of cleavage product from aged testes identified a fragment starting at nucleotide 33 within the upd 3′UTR (black arrowhead, grey box). Alignment of the upd 3′UTR with endo-siRNAs found in aged testis library. Filled circles, canonical base pairing; open circles, GU pairs. f, qRT–PCR showing gfp fromS2 cells transfected with gfp-upd-3′UTR or with gfp-upd-3′UTRmut (32AUU = CGG), with or without Imp and siRNA2. Levels normalized to rp49 and relative to control gfp levels without siRNA2. Error bars in a, d and f denote s.d. of triplicate measurements.
An in vitro protein–RNA binding assay11 showed that Imp associates with the upd 3′ untranslated region (UTR), specifically the first 250 base pairs (region 1), as no substantial binding to other portions of the upd 3′UTR was detected (Fig. 2b). Moreover, Imp did not bind the 5′ untranslated or coding regions of upd or to the med23 3′UTR (Fig. 2b). Notably, a putative consensus binding sequence CAUH (in which Hdenotes A, U or C) for the mammalian IMP homologues (IGF2BP1–3)12 occurs 22 times within the upd 3′UTR, including a cluster of four tandem repeats within the first 35 nucleotides of region 1 (Fig. 2c). To test whether this motif mediates binding between Imp and upd, we removed the first 33 nucleotides to generate a sequence excluding the CAUH repeats, which resulted in a reduction in binding (Fig. 2c, compare domain 1 with domain 2). Point mutations in the third nucleotide of each motif (U = G) did not abolish the binding (Fig. 2c, domain 1*); however, point mutations in the consensus motif of MRPL9 RNA, a target of mammalian IGF2BPs, also did not abolish binding12, suggesting that secondary structures probably mediate the association between IGFBP family members and their target RNAs. Altogether, our data identify the first 33 base pairs of the upd 3′UTR as a putative target sequence for Imp, and support our observations that Imp associates specifically with upd in vivo (Fig. 2a, c).
To gain further insight into the mechanism by which Imp regulates upd, a GFP reporter was constructed that contained the 3′UTR from either upd or med23 (Fig. 2d). Transcript levels for gfp were fivefold higher in Drosophila Schneider (S2) cells that co-expressed Imp with the gfp-upd-3′UTR reporter than in cells that co-expressed Imp with the gfp-med23-3′UTR reporter (Fig. 2d). The significant increase in reporter mRNA levels indicates that it is likely that Imp regulates upd mRNA stability.
Imp counters targeting of upd by endo-siRNAs
RNA-binding proteins, including mammalian IGF2BP1 (refs 13, 14), have been shown to counter microRNA (miRNA)-mediated targeting of mRNAs. However, no consensus miRNA seeds were located within the first 34 base pairs of domain 1 of the upd 3′UTR (Fig. 2c). We speculated that if Imp binding blocks small RNA-mediated degradation of upd, polyadenylated, cleaved upd degradation intermediates would be detected in the testes of older males, when Imp expression in hub cells is reduced (Fig. 1d). Using a modified rapid amplification of complementary DNA ends (RACE) technique we identified a specific cleavage product starting at nucleotide 33 of the upd 3′UTR in the testes of 30-day-old flies (Fig. 2e), but not in RNA extracts from the testes of 1-day-old males. Importantly, the same degradation product of upd was also detected in the testes of young flies when Imp was specifically depleted from hub cells using RNAi-mediated knockdown. As a positive control, we detected the esi-2-mediated cleavage product of mus308 (ref. 15) in testes from both 1- and 30-day-old flies.
To test whether small RNAs might mediate upd cleavage, we cloned and deep-sequenced small RNA libraries generated from the testes of 1- and 30-day-old flies. Although no small RNAs with exact pairing to the upd degradation product were identified, two short interfering RNAs (siRNAs; termed siRNA1 and siRNA2) with high sequence complementarity to the predicted target site in the upd 3′UTR were present in the testis library generated from 30-day-old males (Fig. 2e and Supplementary Table 1). Using qRT–PCR for mature small RNAs, we found that the siRNA2 levels in the testes, relative to the levels of the control small RNAs bantam and mir-184, were similar in young and old males (deep sequencing analysis demonstrated that expression of these two control miRNAs did not change with age). The source of siRNA2 is the gypsy5 transposon, which is inserted at several loci throughout the fly genome and is conserved in numerous Drosophila species.
To gain further insight into the mechanism by which Imp and siRNA2 regulate upd, we investigated the levels of the upd GFP reporter (gfp-upd-3′UTR) in the presence or absence of Imp and siRNA2 in S2 cells. To generate a reporter that should not be susceptible to siRNA-mediated degradation, we mutated the cleavage site in the upd 3′UTR that was identified by RACE (32AUU = CGG; gfp-upd-3′UTRmut). Cells were transfected with either of the GFP reporter constructs, with or without haemagglutinin-tagged Imp (Imp–HA), and subsequently transfected with siRNA2; gfp expression was quantified by qRT–PCR.
The co-expression of siRNA2 and the gfp-upd-3′UTR reporter resulted in a significant decrease in gfp transcript levels (Fig. 2f). Conversely, the co-expression of Imp blocked siRNA2-mediated reduction of gfp mRNA such that gfp levels were higher than in control cells (Fig. 2f). Furthermore, mutation of the putative cleavage site rendered the upd 3′UTR resistant to siRNA2-mediated degradation (Fig. 2f). These data, in combination with the in vitro binding data, suggest that Imp binds to and protects the upd 3′UTR from endogenous and exogenous siRNA2 in S2 cells. Thus, endo-siRNA2 is a bona fide candidate that could direct upd degradation when Imp is absent or its levels are reduced, although targeting by other small RNAs cannot be excluded.
In Drosophila, Argonaute-1 (AGO1) is the principle acceptor of miRNAs and primarily regulates targets in a cleavage-independent mode, whereas AGO2 is preferentially loaded with siRNAs and typically regulates targets by mRNA cleavage16 (reviewed in refs 17, 18). AGO2 expression was detected throughout the tip of the testis, as verified by immunostaining of testes from transgenic flies expressing 3×Flag-HA-tagged AGO2 (ref. 15; Fig. 3a, b). To test whether AGO2 binds to upd mRNA in vivo, thereby potentially regulating upd levels directly, testes were dissected from aged (30-day-old) 3×Flag–HA–AGO2 males. Immunoprecipitation of AGO2, followed by qRT–PCR, showed significant enrichment (~102-fold) of upd mRNA bound to AGO2 (Fig. 3c). Negligible binding of a negative control, rp49, to AGO2 was detected, suggesting specific association of AGO2 with upd mRNA in vivo and supporting our previous findings that upd is probably targeted by the siRNA pathway (Fig. 3c).
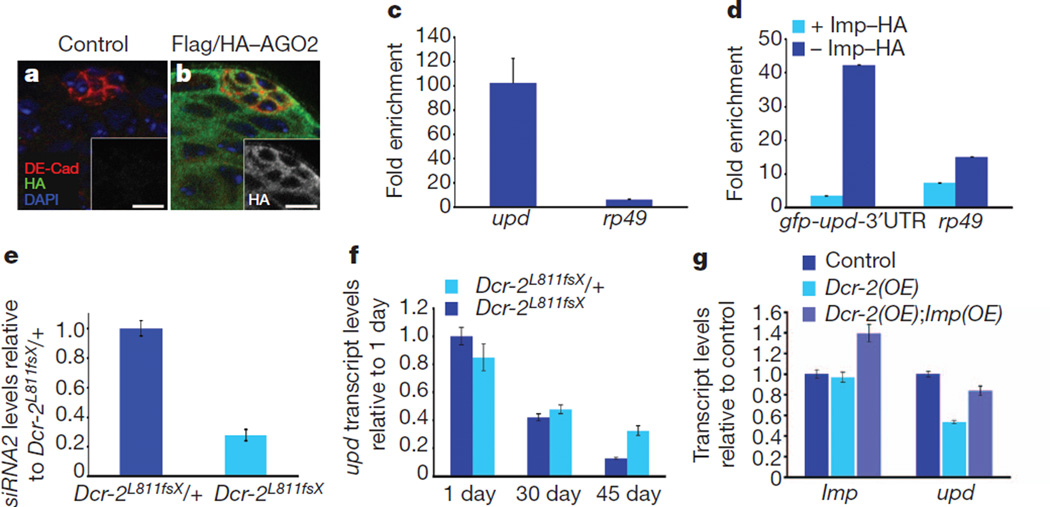
Figure 3. Imp counteracts AGO2 and Dicer-2 to regulate upd levels and stem-cell maintenance.
a, b, Testes from control (a) or Flag/HA–AGO2 (b) males showing AGO2 expression throughout the testis tip. DE-Cad, Drosophila E-cadherin. Scale bars, 10 µm. c, Flag/HA–AGO2 RNA immunoprecipitation from the testes of 30-day-old males. The fold enrichment of upd or rp49 RNA bound to mouse anti-Flag relative to control IgG is shown. d, qRT–PCR after Flag–AGO2 RNA immunoprecipitation from S2 cells, showing fold enrichment of gfp-upd-3′UTR or rp49 RNA bound to anti-Flag antibodies relative to control IgG, in the presence (light blue) or absence (dark blue) of Imp–HA. e, qRT–PCR showing mature siRNA2 levels relative to bantam in testes from Dcr-2L811fsX/+ heterozygous and Dcr-2L811fsX homozygous flies. Expression normalized to heterozygotes. Note the 72% reduction in siRNA2 levels in Dcr-2L811fsX homozygous flies. f, qRT–PCR of upd levels in testes from Dcr-2L811fsX/+ and Dcr-2L811fsX flies measured in testes from 1-, 30- and 45-day-old males; shown are upd levels relative to levels in 1-day-old Dcr-2 heterozygotes. g, qRT–PCR showing mRNA abundance for Imp and upd relative to controls. Note reduction of upd transcript levels in flies expressing ectopic Dcr-2, which is suppressed by co-expression of Imp. mRNA levels in f and g were normalized to GapDH. Error bars in c–g denote s.d. of triplicate measurements.
To test whether Imp can impede the binding of AGO2 to the upd 3′UTR, S2 cells stably expressing Flag-tagged AGO2 (ref. 19) were transfected with the gfp-upd-3′UTR reporter. Consistent with our previous observations, transcript levels of gfp-upd-3′UTR increased ~18-fold when Imp was co-expressed (Fig. 2d and Supplementary Fig. 5b). Despite increases in the overall levels of gfp mRNA, the presence of Imp markedly reduced the association of AGO2 with the upd 3′UTR (Fig. 3d), indicating that Imp antagonizes the ability of AGO2 to bind the upd 3′UTR.
Similar to the AGO family, Drosophila encodes two Dicer proteins that seem to have distinct roles in small RNA biogenesis. Dicer-1 (Dcr-1) is essential for the generation of miRNAs, and Dcr-2 is required for siRNA production from exogenous and endogenous sources16 (reviewed in refs 17, 18). If siRNAs were involved in upd degradation in older males, we predicted that the loss of Dcr-2 would suppress the ageing-related decline in upd and GSCs. Consistent with a role for Dcr-2 in the generation of siRNAs, siRNA2 levels were significantly reduced in Dcr-2 homozygous mutants relative to heterozygous controls (Fig. 3e). Testes from 30- and 45-day-old Dcr-2 mutant flies showed increased levels of upd by qRT–PCR when compared with controls. Whereas a ~90% reduction of upd is observed in the testes from aged Dcr-2 heterozygous controls, we observed only a ~45% reduction in upd in testes from age-matched, Dcr-2 homozygous mutants (Fig. 3f), indicating that upd levels are higher when Dcr-2 function is compromised. Furthermore, the testes from aged Dcr-2 mutants contained more GSCs, on average, when compared with controls (Supplementary Fig. 5c–h). Conversely, the forced expression of Dcr-2 in hub cells resulted in a reduction in the average number of GSCs (Supplementary Fig. 5i–j, l) and led to a significant reduction in upd levels, as detected using qRT–PCR and combined immunofluorescence and FISH, which seemed to be specific, as no significant change in Imp transcript levels was observed (Fig. 3g and Supplementary Fig. 5m, n). Expression of Imp in combination with Dcr-2 resulted in a significant increase in upd levels (Fig. 3g), with a concomitant increase in the average number of GSCs (Supplementary Fig. 5i–l). These observations indicate that Imp can counter the decrease in upd levels resulting from forced Dcr-2 expression, providing further evidence that Imp protects upd from targeted degradation by the siRNA pathway.
let-7 targets Imp in the Drosophila testis
Our data suggest that Imp has a role in stabilizing upd in hub cells; therefore, the ageing-related decline in Imp would be a major contributing factor to the decrease in upd mRNA in the hub cells of aged males. To investigate the mechanism that leads to the decline in Imp expression with age, we examined the Imp 3′UTR for potential instability elements. Within the first 160 base pairs there is a canonical seed sequence for the heterochronic miRNA let-7 (Fig. 4a). Expression of a reporter gene under the control of the let-7 promoter showed that let-7 expression increases in hub cells of ageing males (Fig. 4b, c), which was confirmed by let-7 FISH of testes from aged males (Supplementary Fig. 6a–d). In addition, mature let-7 miRNA was enriched twofold in the testes from 30-day-old flies, relative to 1-day-old males (Fig. 4d). Therefore, an age-related increase in let-7 is one mechanism by which Imp expression could be regulated in an ageing-dependent manner in testes from older males.
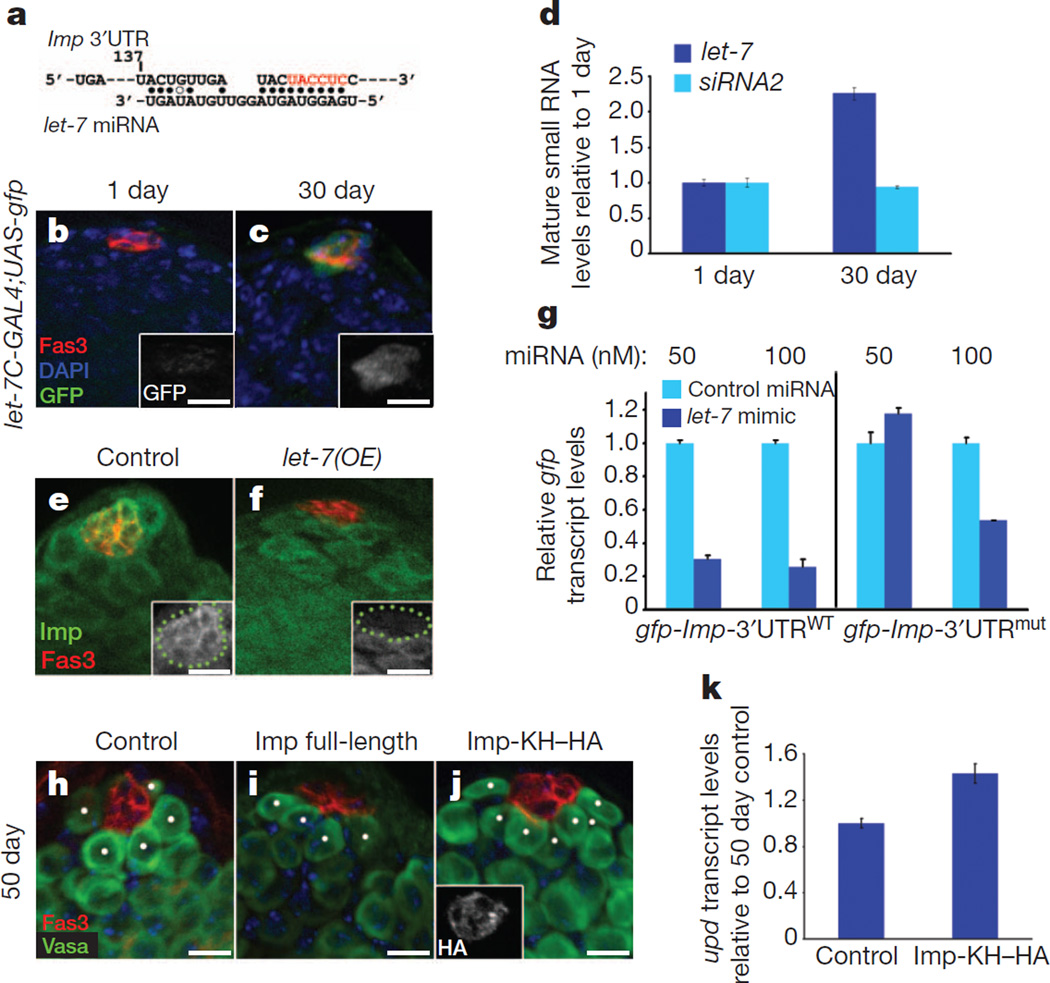
Figure 4. Imp is targeted by let-7 miRNA in the testis.
a, Canonical let-7 seed sequence within the Imp 3′UTR. Start site at position 137. Filled circles, canonical base pairs; open circles, GU base pairs. Note perfect seed pairing of nucleotides 2–7 with the 5′end of let-7 (red). b, c, Testes from1-day-old (b) and 30-day old (c) males immunostained for GFP (green, insets) to follow let-7C expression. d, qRT–PCR for mature let-7 and siRNA2 relative to bantam in the testes of 1- and 30-day-old males. Levels normalized to 1-day-old adults. e, f, Overexpression of let-7 in hub cells reduces Imp expression. Larval testes immunostained as indicated. Green dots in insets outline hub cells. Genotypes: control, upd-GAL4−;TM3, kr-GAL4, UAS-gfp (e); let-7(OE), upd-GAL4; UASlet-7701.12.9 (f). g, let-7 targets a gfp-Imp-3′UTR reporter in S2 cells. qRT–PCR for gfp transcript levels relative to control miRNA at the same concentration. Transfection of 50 nM let-7 mimic results in a 70% reduction in the gfp transcript of gfp-Imp-3′UTRWT but not of gfp-Imp-3′UTRmut (mutated bases to disrupt let-7 binding are coloured red in a). Higher concentrations of let-7 target the Imp 3′UTR through additional sites. h–j, Immunofluorescence of testes from 50-day-old control (h), Imp full-length (i) or Imp with a truncated 3′UTR (Imp-KH–HA; j). Genotypes: control, w, upd-GAL4, UAS-gfp outcrossed to w1118 (h); Imp full-length, w–, upd-GAL4, UAS-gfp; UASImpT21 (i); Imp-KH–HA, w, upd-GAL4; UAS-ImpHA-KH1-3-3 (j). Scale bars, 10 µm. k, qRT–PCR showing relative upd levels in testes from 50-day-old males. Control and Imp-KH–HA genotypes are as in h and j, respectively. Note the 1.5-fold increase in upd levels in aged flies overexpressing the Imp-KH–HA transgene. gfp and upd mRNA expression in g and k are shown relative to controls and normalized to GapDH.
Consistent with these observations, the forced expression of let-7 specifically in hub cells led to a decrease in Imp (Fig. 4e, f, insets). In addition, let-7 expression in S2 cells reduced the levels of a heterologous gfp-Imp-3′UTRWT reporter. S2 cells were transfected with a let-7 mimic or with negative control miRNA, and gfp expression was quantified by qRT–PCR. There was a 70% reduction in gfp-Imp-3′UTRWT expression in the presence of let-7, relative to control miRNA (Fig. 4g). A gfp-Imp-3′UTRmut reporter with mutations in the canonical seed for let-7 (at nucleotide 137) was unaffected by let-7 expression, indicating that mutation of the let-7 seed rendered the RNA resistant to degradation. These data confirm that let-7 can destabilize Imp through sequences in the 3′UTR (Fig. 4g). However, further increasing the levels of let-7 resulted in a decrease in gfp expression from the mutated 3′UTR, indicating that other, putative let-7 seeds in the Imp 3′UTR can be targeted by let-7 (Fig. 4g and Supplementary Fig. 6e).
If the age-related decrease in Imp contributes to a decline in upd and subsequent loss of GSCs, we proposed that re-expression of Imp in hub cells would rescue the ageing-related decrease in upd. Therefore, flies in which Imp was constitutively expressed in hub cells were aged, and upd levels were quantified by qRT–PCR. The expression of an Imp construct containing a truncated 3′UTR (Imp-KH–HA) lacking let-7 target sequences specifically in hub cells was sufficient to suppress the ageing-related decline in upd, with concomitant maintenance of GSCs (Fig. 4h, j, k and Supplementary Fig. 6f), similar to what was observed by re-expressing upd in the hub cells of aged males5,33. Maintenance of Imp-KH–HA expression in aged males was verified by staining with an anti-HA antibody (Fig. 4j, inset). Conversely, the expression of an Imp construct that is susceptible to degradation by let-7 (ImpT21) did not lead to an accumulation of Imp in the testes of 30- and 50-day-old flies, as levels were similar to the levels of endogenous Imp at later time points (Supplementary Fig. 6g)20. Consequently, the expression of this construct was not sufficient to block the ageing-related decline in GSCs (Fig. 4h, i and Supplementary Fig. 6f). These data indicate that let-7-mediated regulation of Imp contributes to the decline in Imp protein in older flies, and supports a model in which an ageing-related decline in Imp, mediated by let-7, exposes upd to degradation by siRNAs (Supplementary Fig. 1). Thus, both the miRNA and siRNA pathways act upstream to regulate the ageing of the testis stem-cell niche by generating let-7 and siRNA2, which target Imp and upd, respectively.
Drosophila has proven to be a valuable model system for investigating ageing-related changes in stem-cell behaviour21. Cell autonomous and extrinsic changes contribute to altered stem-cell activity; thus, determining the mechanisms underlying the ageing-related decline of self-renewal factors, such as the cytokine-like factor Upd, may provide insight into strategies to maintain optimal niche function.
Our data indicate that Imp can regulate gene expression by promoting the stability of selected RNA targets by countering inhibitory small RNAs. Therefore, rescue of the aged niche by Imp expression may be a consequence of effects on Imp targets, in addition to upd, in somatic niche cells. Furthermore, as Imp is expressed in germ cells, it could also act in an autonomous manner to regulate the maintenance of GSCs. The canonical let-7 seed in the Imp 3′UTR is conserved in closely related species, and reports have predicted that the let-7 family of miRNAs target mammalian Imp homologues (IGF2BP1–3)22. Given the broad role of the let-7 family in ageing23, stem cells24–28, cancer29,30 and metabolism31, the regulation of Imp by let-7 may be an important, conserved mechanism in numerous physiological processes.
Non-coding RNAs can ensure biological robustness and provide a buffer against relatively small fluctuations in a system32. However, after a considerable change, a molecular switch is flipped, which allows a biological event to proceed unimpeded. In our model, Imp preserves niche function in young flies until a time at which miRNAs and siRNAs act together to trigger an ‘ageing’ switch that leads to a definitive decline in upd and, ultimately, in stem-cell maintenance (Supplementary Fig. 1). Therefore, targeting signalling pathways at several levels using RNA-based mechanisms will probably prove to be a prevalent theme to ensure robustness in complex biological systems.
METHODS
Drosophila stocks and genotypes
Wild-type flies were Oregon R. Additional strains used were: upd-GAL4, UAS-gfp (E. Bach); upd-GAL4 (T. Xie); w−;UAS-upd, TM2 (D. Harrison); w; let-7CGK1/CyO and UAS-let7701.12.9 (N. Sokol35); 3×Flag/HA–AGO2 (G. Hannon15); ImpCB04573 (A. Spradling6); 8-156-GAL4 (U. Heberlein36); UAS-ImpT21 (P Macdonald20); UAS-Imp HA-KH1 3-3 (from T. Hays33; UAS-Imp(RNAi) (Vienna Drosophila RNAi Center (VDRC) stock number 20322); and UAS-gfpnls (Bloomington Drosophila Stock Center (BDSC) number 4775).
Imp7 and Imp8 were gifts from D. St Johnston10. EP(X)760 (w1118, P{w+mc = EP}ImpEP760; BDSC number 17294) was used as a control for Imp7 and Imp8 alleles. Imp7 and Imp8 hemizygous larvae (L3) were obtained by crossing Imp7/FM0 or Imp8/FM0 females with FM7,KrGAL4, UAS-gfp males (BDSC). L3 mutants were selected against GFP. Specifically, Imp7/FM7, KrGAL4, UAS-gfp; 8-156-GAL4 or Imp8/FM7, KrGAL4, UAS-gfp; 8-156-GAL4 females were crossed to w−/Y;+; UAS-ImpT21 males, and non-GFP L3 male progeny were selected.
UAS-Dcr-2 (VDRC number 60008) and yw, eyFLP; FRT42D, Dcr-2L811fsX were gifts from R.W. Carthew16. Heterozygous flies were obtained by out-crossing to w; FRT42D (BDSC number 1802). An independent Dcr-2 mutant background was obtained by crossing Df(2R)BSC45w+mC/SM6a (Dcr-2 deficiency chromosome; BDSC number 7441) with y, w, eyFLP; FRT42D, Dcr-2L811fsX. Heterozygous flies were obtained by out-crossing Df(2R)BSC45, w+ mC/SM6a to w; FRT42D.
Antibodies
Primary antibodies for immunofluorescence were as follows: rabbit anti-Imp (1:600, gift from P. M. Macdonald20), mouse anti-HA (1:800, Covance), rabbit anti β-galactosidase (1:2,000, Cappel), rabbit anti-GFP (1:5,000, Invitrogen), rabbit anti-Vasa (1:3,000, gift from P. Lasko), rabbit anti-STAT (1:800, gift from D. Montell); mouse anti-Fas3 (1:50), rat anti-DEcad (1:20) and rat anti-DNcad (1:20) were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences. Secondary antibodies were obtained from Invitrogen.
Combined FISH and immunofluorescence
For let-7 miRNA detection, testes were dissected, squashed onto slides37, and incubated with primary antibodies using PBT (diethylpyrocarbonate PBS, 0.1% Tween 20) and PBTHR solutions34. After washes and secondary antibodies, slides were treated with proteinase K for 3 min, rinsed and post-fixed in 10% formaldehyde. Slides were then treated according to the manufacturer’s recommendations for miRNA in situ (Exiqon). Slides were hybridized with 40 nM let-7 LNA-digoxigenin (DIG) probe for 1 h at 50 °C with Exiqon hybridization buffer in a humid chamber. let-7 FISH detection signal and mounting was performed similar to upd.
Generation of DNA constructs
The nucleotide numbers of the following sequences refer to their numbers in the 3′UTRs/5′UTRs or open reading frames (ORF), as shown on FlyBase. All point mutations described were generated by site-directed mutagenesis (Stratagene). pBS-upd was a gift from D.A. Harrison38. pAc5.1-Imp-HA and pBS-Imp-HA were produced by PCR of Imp-RB ORF using Imp cDNA (expressed sequence tag (EST) clone SD07045) as a template. pBS-upd-3′UTR or pBS-med23-3′UTR was produced by PCR of the upd 3′UTR (nucleotides 73–707) or med23 3′UTR (nucleotides 791–1391) and ligated into pBS. To generate pAc5.1-gfp-upd-3′UTR and pAc5.1-gfp-upd-3′UTRmut (32ATT = CGG), nucleotides 1–737 of the upd 3′UTR were ligated into pAc5.1 following the gfp ORF. pBS-upd-5′UTR and pBS-upd-coding were produced by PCR of the upd 5′UTR (nucleotides 1–224 of os-RA) or upd coding region (nucleotides 29–1191 of os ORF) and ligated into pBS. Region 1 (nucleotides 4–243), region 2 (nucleotides 228–468), region 3 (nucleotides 443–737), domain 1 (nucleotides 1–152) and domain 2 (nucleotides 33–152) of the upd 3′UTR were amplified by PCR and ligated into pBS. Domain 1* of the upd 3′UTR was generated by four point mutations (U8G, U17G, U23G and U33G). To generate pAc5.1-gfp-Imp-3′UTRWT, Imp-RB 3′UTR (nucleotides 45–781) was amplified by PCR and then ligated into pGEM T-Easy (Promega). In pAc5.1-gfp-Imp-3′UTRMUT, the let-7 seed was disrupted by five tandem point mutations (149TACCT = CGTTC). pGEM T-Easy-Imp 3′UTR wild-type and mutant were then digested with NotI and ligated downstream of the gfp coding region in pAc5.1-gfp. The sequence of all DNA constructs described above was verified by DNA sequencing.
RNA–protein binding assay and western blotting
Biotin-labelled RNA was transcribed in vitro from cDNAs cloned into pBS using T7 RNA polymerase. The RNA concentration was measured, and an equal number of RNA molecules (according to size) was assayed for protein binding. Imp–HA was translated in vitro using the TNT Coupled Reticulocyte Lysate System (Promega), and 5 µl of the translation mixture was mixed with biotin-labelled RNA. Binding assays were performed as described12. Associated protein complexes were separated on 10% SDS–PAGE gels, followed by western blotting according to standard procedures. Antibodies for western analysis (1:1,000) were: mouse anti-Flag and mouse anti-α-tubulin (Sigma), mouse anti-HA (Covance) and mouse anti-GFP (Clontech).
In silico examination of the upd 3′UTR, 5′UTR and coding regions showed that the upd 3′UTR contains an abundance of AU-rich sequences. Annotated Drosophila 3′UTRs were surveyed for the AU/CG ratio, and identified the med23 3′UTR as one that should not be recognized by Imp, owing to equal AU/CG ratios. Statistical analysis of possible Imp-binding sequences was done in MATLAB (Mathwork). All sequences that bound Imp in vitro were analysed by Meme39 to search for sequence motifs. Alignment based on locally stable RNA structure was performed by LocARNA (the Vienna package: http://rna.tbi.univie.ac.at/).
S2 cell culture and transfection
S2 adherent cells were cultured in Schneider’s Drosophila medium and supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen). Each experiment was done at least three times and each transfection was performed in triplicate. Plasmids were transfected according to the manufacturer’s recommendations (Fugene HD, Roche). For GFP-3′UTR dependent assays, cells were transfected with pBS-actin-GAL4 (obtained from T. Volk) and with either UAS-gfp-upd-3′UTR or UAS-gfp-med23-3′UTR and with 1 µg pAC 5.1 empty vector or pAC 5.1-ImpHA. Cells were collected 20–26 h after transfection and RNA was extracted with the RNeasy kit (Qiagen) and used for qRT–PCR. gfp levels were quantified relative to GAL4 to account for transfection efficiency.
To test the activity of siRNA2 functionally, S2 cells were transfected with pAc5.1-gfp-upd-3′UTR, with either pAC 5.1 empty vector or pAc5.1-Imp-HA, or with pAc5.1-gfp-upd-3′UTRmut (32ATT = CGG). Thirty-six hours after transfection, each plate was split into two. One plate was transfected with 100 nM annealed oligonucleotides for siRNA2 (guide sequence: 5′-UGAGGGGAAAU UGGAGGGGGU-3′, passenger sequence: 5′-CCCCUCCAAUUUCCCUUUU UU-3′) using HighPerfect (Qiagen). Cells were collected 20–26 h after second transfection and used for qRT–PCR. gfp levels were quantified relative to rp49 RNA levels. Each transfection was performed in triplicate.
For the gfp or let-7 reporter assay, cells were transfected with reporter construct of gfp-Imp-3′UTRWT or gfp-Imp-3′UTRmut. Twenty-four hours after transfection, cells from each transfection were reseeded in 6-cm plates and transfected with 50 or 100 nM of synthetic pre-miRNAs to gain expression of dme-miR-let-7 or negative control miRNA (Applied Biosystems) using HiPerfect (Qiagen). Cells were collected 48 h after miRNA transfection and total RNA was extracted with TRIzol (Invitrogen). For each sample we used qRT–PCR to measure let-7 and bantam miRNA expression with Taqman probes for mature miRNA and gfp reporter levels using primers for gfp and GapDH.
RNA–protein co-immunoprecipitation
Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) was used to precipitate GFP-tagged Imp (Imp–GFP) or 3×Flag/HA-tagged AGO2 and associated RNA from testes. Testes from ~1,500 males were dissected on dry ice, and homogenized in RNA immunoprecipitation lysis buffer. Each lysate was divided into two samples; one was used for immunoprecipitation with either rabbit anti-GFP (Clontech) or mouse anti-Flag (Sigma) antibodies, and the second was used with control rabbit or mouse IgG according to the manufacturer’s instructions. RNA was phenol–chloroform precipitated from samples and from input (10% of control immunoprecipitate input before washes). Input RNA was used to establish a standard curve to calculate the percentage of precipitated RNA. Complementary DNA was prepared with random hexamer primers, followed by qRT–PCR.
For AGO2 RNA immunoprecipitation, S2 cells stably expressing Flag–AGO2 (gift from N. Perrimon19) were transfected with pAc5.1-gfp-upd-3′UTR with and without pAc5.1-Imp. Thirty-six hours after transfection, protein extracts from each plate were split into two samples; mouse anti-Flag antibodies (Sigma) or anti-mouse IgG were used for immunoprecipitation. To quantify bound RNAs, qRT–PCR was performed with primers for gfp, rp49 and Imp (primers sequence below). Each transfection was performed in triplicate.
qRT–PCR
S2 cells were washed with PBS and testes from ~125 males were dissected in PBS and immediately frozen on dry ice. Cells or testes were homogenized in RLT buffer from the RNeasy kit (Qiagen), and RNA was extracted according to kit instructions. RNA was quantified and 1 µg was treated with DNaseI (Promega) and reverse-transcribed with random hexamer mixture and SuperScript III (Invitrogen), according to instructions. Quantitative PCR was carried out in 7900HT Fast Real-Time PCR System using SYBR Green PCR Master mix (Applied Biosystems). We used the ΔΔCT method40, and the efficiency of the target and the reference amplification was approximately equal. Specific primers for qRT–PCR of testes were: upd forward, 5′-CCTCCACACGCACAACTACAAG-3′, reverse, 5′-AGCTGGCCACGTAAGTTTGC-3′; Imp forward, 5′-GGTGGGCCGTATCATTGG-3′, reverse, 5′-TCACGCGCTGCAATTCC-3′; med23 forward, 5′-TCAGCGTGGTGACCGAGTAC-3′, reverse, 5′-CCGATCAGGTGCTGGTTGT-3′; rp49 forward, 5′-ATCGATATGCTAAGCTGTCGC-3′, reverse, 5′-TGTCGATACCCTTGGGCTTG-3′. See ref. 41 for GapDH primer. For S2 cells, specific primers were: GAL4 forward, 5′-GCAACGGTCCGAACCTCA-3′, reverse, 5′-GAGGCAATTGGTTGTGAAAGC-3′; gfp forward, 5′-TCCGCCCTGAGCAAAGAC-3′, reverse, 5′-GAACTCCAGCAGGACCATGTG-3′. Reactions were performed in triplicate and averaged, and the transcript expression level was quantified using the relative CT method40 and normalized to the level of GapDH transcript in testes and to the level of GAL4 in S2 cells (to correct for transfection efficiency). To quantify small RNAs, total RNA from S2 cells and from testes was extracted using TRIzol (Invitrogen), and cDNA was prepared with small RNA-specific reverse-transcription primers for let-7, siRNA2 or bantam (Applied Biosystems). Small RNA levels were measured using a Taqman-specific microRNA probe. Both let-7 and siRNA2 levels were normalized relative to bantam, which did not change with age according to the deep sequencing data.
Statistics
For quantification of cell number (GSC or hub cells) and for densitometric analysis of pixel intensity, the mean ± 95% confidence interval and the number (n) of testes examined are shown. P values were generated after a two-tailed Student’s t-test.
For qRT–PCR, one of at least three representative experiments is shown (mean ± s.d. from triplicate measurements). P values were generated after a two-tailed Student’s t-test was used to compare ΔCT between time points or genotypes across three independent biological replicates42.
Cleavage site mapping for endo-siRNA targets
Testes from 1- or 30-day-old OreR or 10-day-old Imp(RNAi) (w−, upd-GAL4, UAS-gfp; UAS-Imp(RNAi)) or control (w−, upd-GAL4, UAS-gfp outcrossed to w1118) males (~600 testes for each time point) were collected on dry ice. Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s protocol. Fifteen micrograms of total RNA were used as starting material. PolyA RNA was isolated with the Dynabeads mRNA purification kit (Invitrogen). Ligation of an RNA adaptor, reverse transcription using the GeneRacer oligo (dT) primer and 5′ RACE-PCR were performed according to the manufacturer’s instructions (GeneRacer kit, Invitrogen). upd 5′ RACE-PCR was carried out using the GeneRacer 5′ primer (5′-CGACTGGAGCACGAGGACACTGA-3′) and an upd gene-specific reverse primer (5′-TAGTACTCGATGCGGGTGCGGAATG-3′), followed by one round of nested PCR using the GeneRacer 5′ nested primer (5′-GGACACTGACATGGACTGAAGGAGTA-3′) and a nested primer specific to upd (5′-CGGCAACTGCAGATTGTGGTTTCGT-3′). PCR products were gel purified and cloned into pCR4Blunt-TOPO (Invitrogen). Fifteen clones were sequenced with T7 primer and subjected to further analysis. The esi-2-mediated cleavage product of mus308 was detected as described15.
Small RNA libraries
Total RNA from the testes of 1,005 (1-day-old) and 1,018 (30-day-old) wild-type (OreR) males was isolated using TRIzol (Invitrogen). Small RNAs (19–28 nucleotides) were cloned as described43. Libraries were sequenced in-house using the Illumina GAII sequencing platform. Obtained sequences were deposited in the Gene Expression Omnibus database under accession GSE3704.
The analysis of small RNA libraries was performed as described20. Illumina reads were stripped of the 3′ linker, collapsed, and the resulting small RNA sequences were matched without mismatches to the Drosophila release 5 genome. Only reads that met these conditions were subjected to further analyses. All reads between 20 and 22 nucleotides in size were extracted bioinformatically regardless of their annotations. Using a custom Perl script, these sequences were mapped against the predicted cleavage site at nucleotide 33 within the 3′UTR of upd, allowing up to six mismatches between the target site and nucleotides 2 to 18 of the small RNA (all mapping results are shown in Supplementary Table 1). Our search was focused on nucleotides 2–18 because previous studies suggested that pairing of the 5′ terminal nucleotide of the small RNA with its target is not required for efficient silencing. Furthermore, several mismatches between the 3′ end of the small RNA and the target site can be tolerated for target regulation44,45.
Supplementary Material
Acknowledgments
We are grateful to D. St Johnston, W. Chia, P. Macdonald, R. Carthew, E. Bach, D. Harrison, T. Volk, U. Heberlein, A. Spradling, J. Kadonaga, G. Hannon, T. Hays, N. Sokol, H. Siomi and P. Lasko for reagents and fly stocks, to C. Doe, R. Hans, G. Volohonsky, T. Juven-Gershon, A. Pasquinelli, R. Zhou and S. Aigner for guidance on methods used in this manuscript, to O. Tam for computational support, and to C. Koehler for technical assistance. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation, the Ellison Medical Foundation, the Emerald Foundation, the American Federation for Aging Research, and the National Institutes of Health (to D.L.J.). B.C. is supported by a PhD fellowship from the Boehringer Ingelheim Fonds. E.L. is supported by the National Science Foundation.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions H.T., C.D. and D.L.J. designed experiments. H.T. and C.D. performed experiments. B.C. generated and analysed small RNA libraries. E.L. performed bioinformatic analysis to identify Imp-binding sequences. H.T., C.D., B.C. and D.L.J. evaluated the data and wrote the manuscript.
The small RNA libraries from the testes of 1-day-old and 30-day-old flies have been deposited in the Gene Expression Omnibus database under accession GSE37041.
The authors declare no competing financial interests.
References
- 1.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- 2.Jones DL, Rando TA. Emerging models and paradigms for stem cell ageing. Nature Cell Biol. 2011;13:506–512. doi: 10.1038/ncb0511-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voog J, Jones DL. Stem cells and the Niche: a dynamic duo. Cell Stem Cell. 2010;6:103–115. doi: 10.1016/j.stem.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuller MT. In: The Development of Drosophila Melanogaster. Bate M, Martinez-Arias A, editors. Cold Spring Harbor Laboratory Press; 1993. pp. 71–147. [Google Scholar]
- 5.Boyle M, Wong C, Rocha M, Jones DL. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1:470–478. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Buszczak M, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabrizio JJ, et al. Imp (IGF-II mRNA-binding protein) is expressed during spermatogenesis in Drosophila melanogaster. Fly. 2008;2:47–48. doi: 10.4161/fly.5659. [DOI] [PubMed] [Google Scholar]
- 8.Yisraeli JK. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol. Cell. 2005;97:87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]
- 9.Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- 10.Munro TP, Kwon S, Schnapp BJ, St Johnston D. A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J. Cell Biol. 2006;172:577–588. doi: 10.1083/jcb.200510044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabel-Rosen H, Dorevitch N, Reuveny A, Volk T. The balance between two isoforms of the Drosophila RNA-binding protein how controls tendon cell differentiation. Mol. Cell. 1999;4:573–584. doi: 10.1016/s1097-2765(00)80208-7. [DOI] [PubMed] [Google Scholar]
- 12.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 14.Elcheva I, Goswami S, Noubissi FK, Spiegelman V. SCRD-BP protects the coding region of βTrCP1 mRNA from miR-183-mediated degradation. Mol. Cell. 2009;35:240–246. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 17.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golden DE, Gerbasi VR, Sontheimer EJ. An inside job for siRNAs. Mol. Cell. 2008;31:309–312. doi: 10.1016/j.molcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czech B, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol. Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng C, Macdonald P. MImp associates with squid and Hrp48 and contributes to localized expression of gurken in the oocyte. Mol. Cell. Biol. 2006;26:9508–9516. doi: 10.1128/MCB.01136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Jones DL. The effects of aging on stem cell behavior in Drosophila. Exp. Gerontol. 2010;46:340–344. doi: 10.1016/j.exger.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyerinas B, et al. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 23.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducingp16Ink4a andp19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao C, et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl Acad. Sci. USA. 2010;107:1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, et al. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm. Genome. 2007;18:316–327. doi: 10.1007/s00335-007-9032-6. [DOI] [PubMed] [Google Scholar]
- 27.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 28.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-κB, Lin28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 31.Zhu H, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boylan KL, et al. Motility screen identifies Drosophila IGF-II mRNA-binding protein–zipcode-binding protein acting in oogenesis and synaptogenesis. PLoS Genet. 2008;4:e36. doi: 10.1371/journal.pgen.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitadate Y, et al. Boss/Sev signaling from germline to soma restricts germline-stem-cell-niche formation in the anterior region of Drosophila male gonads. Dev. Cell. 2007;13:151–159. doi: 10.1016/j.devcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
References
- 35.Sokol NS, et al. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldwell JC, Fineberg SK, Eberl DF. reduced ocelli encodes the leucine rich repeat protein Pray For Elves in Drosophila melanogaster. Fly. 2007;1:146–152. doi: 10.4161/fly.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 1996;109:2779–2788. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- 38.Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Min KJ, Yamamoto R, Buch S, Pankratz M, Tatar M. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell. 2008;7:199–206. doi: 10.1111/j.1474-9726.2008.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 44.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nature Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 45.Haley B, Foys B, Levine M. Vectors and parameters that enhance the efficacy of RNAi-mediated gene disruption in transgenic Drosophila. Proc. Natl Acad. Sci. USA. 2010;107:11435–11440. doi: 10.1073/pnas.1006689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.