Abstract
Although microRNA-155 (miR-155) is known to play an important role in many cancers, its expression and function in oral squamous cell carcinoma (OSCC) was not fully understood. Thus, in the present study, we investigated the expression of miR-155 and also the role this miR plays in OSCC. We used the OSCC cell line (CAL27) and paired tumor and non-tumor tissue samples from patients with OSCC in order to detect the expression of miR-155. Cell proliferation, migration and invasion assays were then undertaken in order to determine the effect of miR-155 on the biological behavior of CAL27 cells following transient transfection with miR-155 mimic and antagomir. The regulatory effect of miR-155 on its target gene B-cell CLL/lymphoma 6 (BCL6) and downstream gene cyclin D2 (CCND2) was also analyzed. We found that miR-155 expression in OSCC cell and tumor tissues was significantly higher than that of the controls. We noted that the miR-155 mimic enhanced CAL27 cell proliferation, migration and invasion ability, downregulated BCL6 levels, and increased cyclin D2 expression. However, we noted that abrogating miR-155 with the miR-155 antagomir suppressed CAL27 cell proliferation, migration and invasion, upregulated BCL6 and reduced cyclin D2 expression. These results indicate that miR-155 plays a tumor-promoting role in OSCC by regulating the BCL6/cyclin D2 axis.
Keywords: miR-155, oral squamous cell carcinoma, proliferation, migration, invasion, B-cell CLL/lymphoma 6, cyclin D2
Introduction
Oral squamous cell carcinoma (OSCC) accounts for approximately 90% of all malignant oral tumors (1). OSCC often causes issues with chewing, swallowing and speaking and affects the physical appearance of patients. Although surgery, chemotherapy and radiotherapy cure the majority of cases of early stage OSCC, the outcome of most patients with advanced stage OSCC has not improved over the last three decades (2). The development of OSCC is a multi-step process involving the progressive acquisition and study of genetic and epigenetic alterations, such as deletions, point mutations and DNA methylation (3). Studying the key molecules that play important roles in the progression of OSCC will yield novel insights and assist in the search for new therapeutic targets.
MicroRNAs (miRNAs or miRs) are small non-coding RNAs that regulate mRNA transcription and translation in various cellular activities such as proliferation and differentiation (4,5). Previous research has demonstrated that aberrant miRNA expression is related to the development of several types of human cancer (6). MiR-155 maps within and is processed from an exon of a non-coding RNA transcribed from the B-cell Integration Cluster (BIC) located on chromosome 21 (7). Previous studies have reported that miR-155 plays an important role in many solid tumors, including laryngeal cell carcinoma, lung cancer, gliomas, colon cancer, hepatocellular carcinoma, pancreatic and breast cancer (8–15). In relation to laryngeal squamous cell carcinoma, it was noted that miR-155 increased cancer cell proliferation, migration and invasion through suppressor of cytokine signaling 1 (SOCS1) and signal transducer and activator of transcription 3 (STAT3) (9). In relation to colon cancer, Zhang et al (15) reported that the upregulation of miR-155 promotes tumor cell migration and invasion via claudin-1. Recently, a study revealed that miR-155 overexpression in OSCC promoted cell proliferation through cell division cycle 73 (CDC73) (16). However, it is not yet known whether miR-155 affects other aspects of the biological behavior of OSCC cells through other downstream target genes.
B-cell CLL/lymphoma 6 (BCL6) is a transcriptional repressor that has been proven to be a direct target gene of miR-155 in macrophages (17). BCL6 regulates a series of molecules that participate in regulating the cell cycle and inflammation, including cyclin D2 and various chemokines (18). Cyclin D2 plays an important role in the cell cycle, and the ectopic expression of cyclin D2 is closely related to invasion in OSCC cells (19). Moreover, BCL6 directly regulates cyclin D2 by binding with its promoter (20). Previous studies have revealed that the BCL6/cyclin D2 axis is involved in many disorders and that pro-heparin binding (HB)-epidermal growth factor (EGF) and forkhead box O3A (FOXO3A) regulate this axis (20,21). However, to the best of our knowledge, no previous study has discussed the pathological role of miR-155 in the BCL6/cyclin D2 axis in OSCC.
In the present study, we found that miR-155 was upregulated in the OSCC CAL27 cell line and in OSCC specimens. The overexpressed miR-155 enhanced the proliferative, migratory and invasive ability of CAL27 cells by decreasing BCL6 expression and increasing cyclin D2 levels. These results all suggest that miR-155 plays a tumor-promoting role during OSCC development, partly through the BCL6/cyclin D2 axis.
Materials and methods
Cell culture
The human OSCC cell line CAL27 was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA), and cultured in Dulbecco's modified Eagle's medium high-glucose medium (Life Technologies, Grand Island, NY, USA) supplemented with heat-inactivated 10% fetal bovine serum (HyClone Laboratories, Logan, UT, USA). The normal oral keratinocyte NOK-SI cell line was kindly provided by the National Institute of Health (Bethesda, MD, USA) and was grown in keratinocyte serum-free medium containing human recombinant epidermal growth factor and bovine pituitary extract (Life Technologies). Cells were cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Human tissue specimens
Primary OSCC tissues and matching adjacent non-tumor tissues were obtained from 12 patients who underwent OSCC resection without receiving preoperative chemotherapy or radiation at the Guanghua School of Stomatology, Hospital of Stomatology, Sun Yat-sen University (Guangzhou, China) between October 2011 and November 2012 (Table I). We obtained informed consent from all the patients. All tissue specimens were immediately frozen in liquid nitrogen and then stored at −80°C until the RNA was extracted. The samples were stained with hematoxylin and eosin and examined histopathologically (Table I). The Ethics Committee of Sun Yat-sen University approved the present study.
Table I.
Clinical data of patients with OSCC.
| Case no. | Gender | Age (years) | Tumor location | Pathological diagnosis |
|---|---|---|---|---|
| Patient 1 | Female | 44 | Tongue | Well differentiated |
| Patient 2 | Female | 68 | Gingiva | Well differentiated |
| Patient 3 | Female | 32 | Tongue | Well differentiated |
| Patient 4 | Female | 33 | Tongue | Well differentiated |
| Patient 5 | Male | 49 | Floor of the mouth | Poorly differentiated |
| Patient 6 | Male | 80 | Tongue | Poorly differentiated |
| Patient 7 | Female | 68 | Tongue | Well differentiated |
| Patient 8 | Male | 62 | Oropharynx | Moderately differentiated |
| Patient 9 | Male | 46 | Tongue | Moderately differentiated |
| Patient 10 | Male | 57 | Tongue | Poorly differentiated |
| Patient 11 | Female | 52 | Tongue | Moderately differentiated |
| Patient 12 | Male | 63 | Tongue | Poorly differentiated |
OSCC, oral squamous cell carcinoma.
Reverse transcription quantitative PCR (RT-qPCR)
In order to quantify miR-155, BCL6 and cyclin D2 (CCND2) mRNA expression using RT-qPCR, total RNA was extracted from cultured cells and clinical specimens using TRIzol (Life Technologies). miR-155 and its control U6 were stem-loop reverse transcribed using specific Bulge-Loop miRNA qRT-PCR Primer sets (Guangzhou RiboBio Co., Ltd., Guangzhou, China) and also a PrimeScript RT reagent kit (Takara, Shiga, Japan) in accordance with the manufacturers' instructions. BCL6, cyclin D2 and the control β-actin were reverse transcribed using a Transcriptor First Strand cDNA Synthesis kit (Roche, Mannheim, Germany). Subsequently, qPCR of miR-155 and U6 was performed using a LightCycler 480 Real-Time PCR system (Roche) and SYBR Premix Ex Taq II™ (Takara). The amplification profile involved initial denaturation at 95°C for 30 sec, followed by 40 cycles of denaturation at 95°C for 5 sec, annealing at 60°C for 20 sec and a final extension at 65°C for 15 sec. qPCR of BCL6, cyclin D2 and β-actin was performed using the LightCycler 480 Real-Time PCR system and LightCycler 480 SYBR-Green I Master Mix (Roche) with specific primers (Life Technologies) (Table II). The amplification profile involved initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 20 sec and a final extension at 72°C for 20 sec. miR-155, BCL6 and cyclin D2 expression relative to U6 or β-actin was determined as the respective comparative threshold cycle values (2−ΔΔCt).
Table II.
Primers used for RT-qPCR.
| Gene | Primer | 5′→3′ |
|---|---|---|
| BCL6 | Forward | ACACATCTCGGCTCAATTTGC |
| Reverse | AGTGTCCACAACATGCTCCAT | |
| Cyclin D2 | Forward | TTTGCCATGTACCCACCGTC |
| Reverse | AGGGCATCACAAGTGAGCG | |
| β-actin | Forward | CTCTGGCCGTACCACTGGC |
| Reverse | GTGAAGCTGTAGCCGCGC |
BCL6, B-cell CLL/lymphoma 6.
miRNA transfection
Homo sapiens (hsa)-miR-155-5p mimic (miR-155 mimic), miRNA mimic negative control (mimic-NC) (both from Life Technologies), hsa-miR-155-5p antagomir (miR-155 antagomir), and miRNA antagomir negative control (antagomir-NC) (both from Guangzhou RiboBio) were used for transfection. Cells were incubated in plates 24 h before transfection. When cells reached 30–40% confluence, 5 nM miR-155 mimic, 5 nM mimic-NC, 100 nM miR-155 antagomir, or 100 nM antagomir-NC mixed with Lipofectamine RNAiMAX transfection reagent according to the manufacturer's instructions (Life Technologies) was added. Six hours after transfection, the culture medium was replaced with fresh medium. We also constructed a mock group, in which we added Lipofectamine RNAiMAX and culture medium to cells but no synthetic miRNA sequence.
Cell proliferation assay
CAL27 cells were seeded in 96-well plates at 3,500 cells/well and cultured at 37°C. Twenty-four hours after seeding, the cells were transfected with the synthetic miRNA sequences. Every 24 h after transfection, 10 μl Cell Counting Kit-8 solution (Dojindo Laboratories, Kumamoto, Japan) was added to each well, which contained 100 μl culture medium, and the plates were further incubated for 2 h at 37°C. Absorbance was measured using an automated microplate reader (Thermo Fisher Scientific, Waltham, MA, USA) at 450 nm.
Migration and invasion assay
The cell migration and invasion assays were performed in cell culture inserts packaged in companion plates (Becton-Dickinson, Franklin Lakes, NJ, USA) with 8-μm polycarbonate nuclepore filters. Transfected CAL27 cells were plated at 7×104 cells/insert suspended in 100 μl serum-free culture medium. For the invasion assay, each insert was coated with Matrigel matrix (BD) before the cells were seeded. After 24 h incubation, cells on the apical side of the inserts were removed with cotton swabs, and the inserts were then fixed with 4% paraformaldehyde for 30 min and stained with 0.4% crystal violet for 15 min. The number of migrated cells in 10 random visual fields was directly counted under a light microscope (Zeiss, Jena, Germany). The cells were also counted indirectly by eluting the membrane-binding crystal violet with 500 μl 10% glacial acetic acid, and then measuring absorbance at 590 nm using an automated microplate reader (Thermo Fisher Scientific).
Western blot analysis
To isolate the proteins, cells were washed twice with cold phosphate-buffered saline and then lysed with RIPA and PSFM (Beyotime Institute of Biotechnology, Nanjing, China). Protein concentrations were determined using a BCA protein assay kit (Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China). The proteins were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and were transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). After blocking in 5% milk, the membranes were incubated with rabbit monoclonal anti-BCL6 antibody (D65C10, diluted 1:1,000), rabbit monoclonal anti-cyclin D2 antibody (D52F9, diluted 1:1,000) (both from Cell Signaling Technology, Boston, MA, USA), mouse monoclonal anti-GAPDH antibody (ab125247, diluted 1:2,000), and rabbit polyclonal anti-H2A.X antibody (ab10475, diluted 1:500) (both from Abcam, Cambridge, MA, USA) at 4°C overnight. The membranes were then incubated with the secondary antibodies goat polyclonal anti-rabbit IgG (ab6721, diluted 1:5,000) and goat polyclonal anti-mouse IgG (ab85760, diluted 1:2,000) (both from Abcam) for 2 h. The bands were then visualized using an enhanced chemiluminescence detection system (Alpha Innotech Cοrp., San Leandro, CA, USA) and quantified using ImageJ software.
Statistical analysis
All statistical analyses were carried out using SPSS version 16.0 (SPSS, Inc., Chicago, IL, USA). Data are expressed as the means ± standard deviation. Statistical analysis was performed using Student's t-test, or one-way ANOVA or the Kruskal-Wallis test. A P-value <0.05 was considered to indicate a statistically significant difference.
Results
miR-155 is highly expressed in the OSCC cell line and OSCC tissue samples
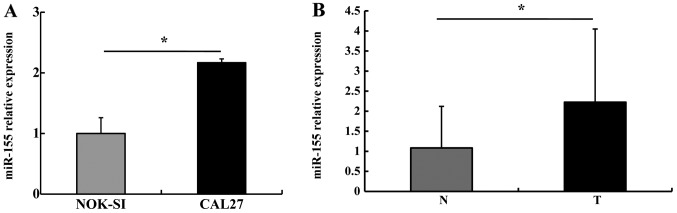
We first detected miR-155 expression in NOK-SI and CAL27 cells using stem-loop RT-qPCR. miR-155 expression in the CAL27 cells was 2.17±0.26 times higher than that in the NOK-SI cells (P<0.05) (Fig. 1A). We then detected miR-155 levels in the OSCC specimens: miR-155 expression in the OSCC tumor tissues was higher than that in the paired adjacent non-tumor tissues (P<0.05) (Fig. 1B). These results suggest that miR-155 is typically upregulated in OSCC.
Figure 1.
miR-155 expression in oral squamous cell carcinoma (OSCC) cells and tissues. (A) Stem-loop RT-qPCR was used to detect miR-155 expression, and U6 was used to normalize data. miR-155 expression in CAL27 cells was higher than in NOK-SI cells. (B) miR-155 levels were higher in tumor tissues (T) than in non-tumor tissues (N). *P<0.05.
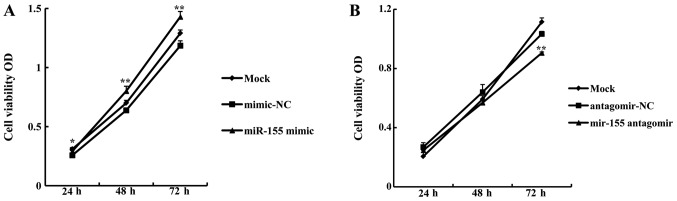
miR-155 promotes CAL27 cell proliferation
In order to examine whether miR-155 affected OSCC cell growth, a proliferation assay was carried out using CAL27 cells following transfection with miR-155 mimic or antagomir. The growth rate of CAL27 cells transfected with 5 nM miR-155 mimic was significantly higher than that of cells transfected with mimic-NC (miR-155 mimic OD/mimic-NC OD) at 24 h (113.14±2.75%, P<0.05), 48 h (125.39±6.49%, P<0.01), and 72 h (120.6±0.41%, P<0.01) (Fig. 2A). By contrast, the growth rate of cells transfected with 100 nM miR-155 antagomir was significantly lower compared with antagomir-NC at 72 h (87.61±1.67%, P<0.01) (Fig. 2B). These data indicate that miR-155 promotes OSCC cell proliferation.
Figure 2.
miR-155 induces increased proliferation of oral squamous cell carcinoma (OSCC) cells. (A) The cell growth rate was calculated as optical density (OD) values, measured using a CCK-8 assay. miR-155 mimic promoted cell proliferation compared with mimic-NC at 24, 48 and 72 h. (B) miR-155 antagomir inhibited CAL27 cell proliferation compared with antagomir-NC at 72 h. *P<0.05, **P<0.01 compared with mimic-NC or antagomir-NC.
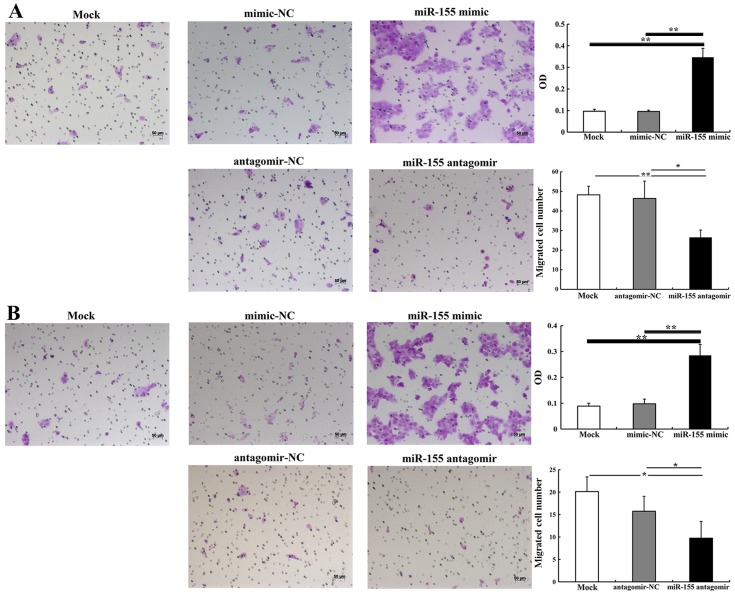
miR-155 increases CAL27 cell migration and invasion
The effect of miR-155 on CAL27 cell migration and invasion was detected using cell migration and invasion assays. At 24 h after transfection, the number of migrated cells in the group transfected with 5 nM miR-155 mimic was significantly higher than in the group treated with mimic-NC (P<0.01) (Fig. 3A). However, there were significantly fewer migrated cells in the group transfected with 100 nM miR-155 antagomir compared with the group treated with antagomir-NC (P<0.05) (Fig. 3B). The invasion assay demonstrated that 5 nM miR-155 mimic significantly increased the number of invasive cells when compared with mimic-NC (P<0.01). There were significantly fewer invasive cells in the group treated with 100 nM miR-155 antagomir than in the group treated with antagomir-NC (P<0.05) (Fig. 3B). Thus, we suggest that miR-155 effectively enhances the migratory and invasive abilities of OSCC cells.
Figure 3.
Transwell migration and invasion assays. (A) miR-155 mimic enhanced CAL27 cell migration and miR-155 antagomir-inhibited migration. (B) Invasion of CAL27 was enhanced by miR-155 mimic and inhibited by miR-155 antagomir. *P<0.05, **P<0.01. OD, optical density.
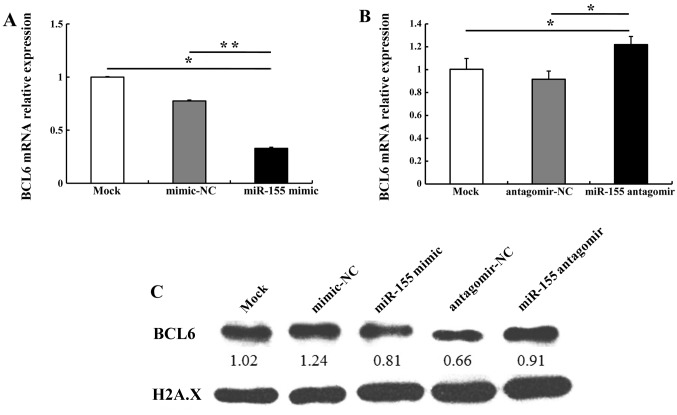
miR-155 regulates BCL6 and cyclin D2 expression in OSCC cells
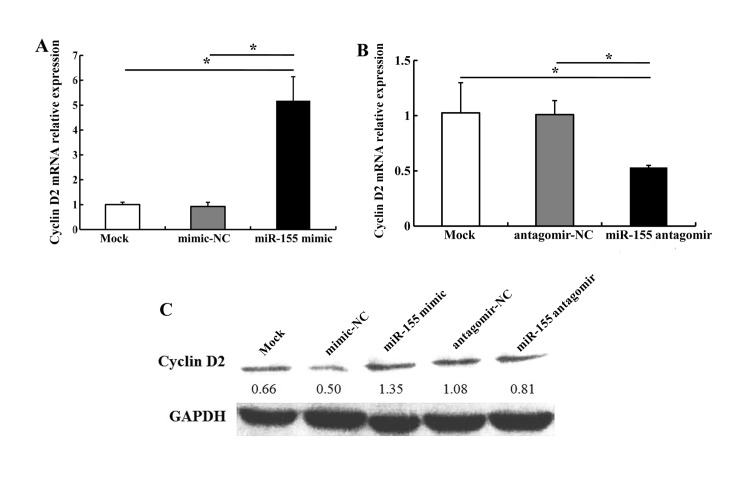
It has previously been reported that miR-155 directly regulates BCL6 expression (17). In the present study, bioinformatics analyses using miRanda (http://www.microrna.org/microrna/home.do) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) revealed that the 3′ untranslated region (3′UTR) of BCL6 mRNA contained a highly conserved miR-155 binding site (data not shown). Therefore, in order to validate whether miR-155 regulates BCL6 and its downstream gene cyclin D2 in OSCC cells, we compared the alteration in BCL6 and cyclin D2 expression in CAL27 cells transfected with miR-155 mimic, mimic-NC, miR-155 antagomir or antagomir-NC. miR-155 mimic (5 nM) downregulated BCL6 mRNA and protein levels; 100 nM miR-155 antagomir upregulated BCL6 mRNA and protein levels as compared with antagomir-NC (Fig. 4). We noted that transfection with 5 nM miR-155 mimic increased cyclin D2 mRNA and protein levels; 100 nM miR-155 antagomir markedly decreased cyclin D2 expression as compared with antagomir-NC (Fig. 5). These results indicate that miR-155 regulates the expression of BCL6 and cyclin D2.
Figure 4.
miR-155 regulation of BCL6 expression in CAL27 cells. (A) miR-155 mimic decreased BCL6 mRNA levels. (B) miR-155 antagomir upregulated BCL6 mRNA levels. (C) BCL6 protein levels were downregulated by miR-155 mimic and upregulated by miR-155 antagomir. *P<0.05, **P<0.01.
Figure 5.
miR-155 regulation of cyclin D2 expression in CAL27 cells. (A) miR-155 mimic upregulated cyclin D2 mRNA levels. (B) miR-155 antagomir downregulated cyclin D2 mRNA levels. (C) miR-155 mimic increased cyclin D2 protein levels in CAL27 cells; miR-155 antagomir decreased it. *P<0.05.
Discussion
It has been shown in previous studies that miR-155 is an important regulatory gene which is involved in the carcinogenesis of various epithelial tumors, and is also involved in tumor cell proliferation, invasion and other biological behaviors. miR-155 is always upregulated in laryngeal squamous cell carcinoma, cervical and colon cancer (9,10,23). To the best of our knowledge, there are but few studies on the role of miR-155 in OSCC (16,24,25). In the present study, we detected miR-155 expression in the OSCC cell line CAL27 and in NOK-SI, the control cell line. We found that miR-155 expression was higher in CAL27 cells than in NOK-SI cells. We also compared miR-155 expression in paired tumor and non-tumor tissues from patients with OSCC, and found that miR-155 was upregulated in the tumor tissues. These results are consistent with the findings of previous studies (16,24,25). Our results and those of others indicate that miR-155 participates in the development of OSCC, and thus our understanding of the pathogenesis of OSCC has been improved.
Cell proliferation is required for tumor growth and acts as the foundation for cell invasion and migration. miR-155 upregulates cell proliferation in several types of cancer, including laryngeal squamous cell carcinoma, breast and pancreatic cancer (9,12,13). To date, relatively few studies on the effect of miR-155 on OSCC cell proliferation have been undertaken. Recently, Rather et al (16) reported that antagomir-155 decreased the proliferation of the OSCC cell line KB through CDC73. In the present study, to explore whether miR-155 promotes proliferation in OSCC, we transfected CAL27 cells with miR-155 mimic and antagomir. miR-155 mimic was used to imitate miR-155 function, and miR-155 antagomir was used to abolish the effect of miR-155. We demonstrated that miR-155 mimic increased CAL27 cell proliferation, whereas miR-155 antagomir reduced it, indicating that miR-155 plays a proliferation-promoting role in OSCC.
Cell motility is critical for the spread of tumor cells to the lymph nodes and blood vessels. miR-155 functions as a migration- and invasion-promoting gene in many cancers: e.g., it promotes migration and invasion in laryngeal squamous cell carcinoma by targeting the SOCS1-STAT3 pathway (9). In breast cancer, miR-155 mediates the loss of CCAAT/enhancer binding protein β (C/EBPβ) and shifts the response to transforming growth factor-β (TGF-β) from growth inhibition to invasion and metastasis (26). However, to the best of our knowledge, no study on the effect of miR-155 on OSCC cell migratory and invasive ability had previously been undertaken. In the present study, by examining the gain or loss of function of miR-155, we discovered that miR-155 enhances OSCC cell migration and invasion. These findings demonstrate that miR-155 plays an important role in the progression of OSCC by enhancing tumor cell migratory and invasive abilities.
Typically, miR-155 exerts its regulatory functions by binding to the 3′UTR of the target gene. miR-155 targets FOXO3, SOCS1, claudin-1 and other genes in tumors (9,14,15). Moreover, it has been reported that the transcriptional repressor BCL6, which inhibits cell growth and induces apoptosis, is a target gene of miR-155, and it has been confirmed using a luciferase assay that miR-155 binds to the conserved binding site in the 3′UTR of BLC6 mRNA and regulates the expression of BCL6 (17,27). In the present study, we found that miR-155 mimic significantly decreased BCL6, while miR-155 antagomir significantly increased it. Thus, we deduced that miR-155 regulated BCL6. Considering that the aforementioned previous studies have verified that miR-155 binds to the conserved binding site in the 3′UTR of BCL6 through luciferase assays, we did not undertake the same assay in our study.
BCL6 plays a role in cell growth and other biological behaviors by regulating downstream genes such as CCND2, and various chemokines (18). Previous research has reported that cyclin D2 plays an important role in G1-to-S phase shifting in the cell cycle (28). Moreover, cyclin D2 overexpression results in excessive proliferation in many cell types and enhances OSCC cell invasive ability in vitro and in vivo (19). It has also been reported that BCL6 directly regulates the expression of cyclin D2 by binding with the conserved site in its promoter, as evidenced by luciferase reporter assay, and BCL6 protein bound specifically to cyclin D2, as determined using electrophoretic mobility shift DNA binding assay (20). Several studies have demonstrated that BCL6/cyclin D2 are involved in many disorders. The BCL6/cyclin D2 axis affects the cell cycle and proliferation by regulating pro-HB-EGF and FOXO (21,22,29). In the present study, cyclin D2 levels were increased, and BCL6 was repressed, by miR-155 mimic, and these levels were decreased when miR-155 antagomir decreased BCL6 repression. These data indicate that ectopic expression of miR-155 downregulates BCL6 expression and increases cyclin D2 expression, and subsequently facilitates cell proliferation, migration and invasion. Moreover, as miR-155 binds to the 3′UTR of BCL6 mRNA to regulate BCL6 expression, and as CCND2 is a target gene of BCL6, we infer that miR-155 promotes OSCC carcinogenesis and development partly through the BCL6/cyclin D2 axis. In future research, we will explore the critical roles of BCL6/cyclin D2 in miR-155-induced proliferation, migration and invasion in OSCC cells.
In conclusion, miR-155 overexpression plays a promoting role in the proliferative, migratory and invasive behavior of OSCC cells. Its effects on OSCC are possibly associated with its regulation of the BCL6/cyclin D2 axis. Collectively, our results are of great significance for understanding OSCC development. However, further research is required to fully elucidate the regulatory mechanisms of BCL6/cyclin D2 mediating the miR-155-induced promoting effects in OSCC.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (nos. 81272948 and 81000446), the Specialized Research Fund for the Doctoral Program of Higher Education of China (201001711201050), the Provincial Natural Scientific Foundation of Guangdong (10451008901005050), the Guangdong Department of Science and Technology Translational Medicine Center Grant (2011A080300002) and the Priming Scientific Research Foundation for Junior Teachers of Medicine in Sun Yat-sen University (10ykpy17).
References
- 1.Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008;87:14–32. doi: 10.1177/154405910808700104. [DOI] [PubMed] [Google Scholar]
- 2.Gibson MK, Forastiere AA. Multidisciplinary approaches in the management of advanced head and neck tumors: state of the art. Curr Opin Oncol. 2004;16:220–224. doi: 10.1097/00001622-200405000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Sasahira T, Kirita T, Kuniyasu H. Update of molecular pathobiology in oral cancer: a review. Int J Clin Oncol. 2014;19:431–436. doi: 10.1007/s10147-014-0684-4. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Kluiver J, van den Berg A, de Jong D, Blokzijl T, Harms G, Bouwman E, Jacobs S, Poppema S, Kroesen BJ. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–3776. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- 8.Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, Harris CC. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer. 2012;130:1378–1386. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao XD, Zhang W, Liang HJ, Ji WY. Overexpression of miR-155 promotes proliferation and invasion of human laryngeal squamous cell carcinoma via targeting SOCS1 and STAT3. PLoS One. 2013;8:e56395. doi: 10.1371/journal.pone.0056395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakirtzi K, Hatziapostolou M, Karagiannides I, Polytarchou C, Jaeger S, Iliopoulos D, Pothoulakis C. Neurotensin signaling activates microRNAs-21 and -155 and Akt, promotes tumor growth in mice, and is increased in human colon tumors. Gastroenterology. 2011;141:1749–61.e1. doi: 10.1053/j.gastro.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan XL, Jia YL, Chen L, Zeng Q, Zhou JN, Fu CJ, Chen HX, Yuan HF, Li ZW, Shi L, et al. Hepatocellular carcinoma-associated mesenchymal stem cells promote hepatocarcinoma progression: role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology. 2013;57:2274–2286. doi: 10.1002/hep.26257. [DOI] [PubMed] [Google Scholar]
- 12.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang CM, Zhao J, Deng HY. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J Biomed Sci. 2013;20:79–88. doi: 10.1186/1423-0127-20-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling N, Gu J, Lei Z, Li M, Zhao J, Zhang HT, Li X. microRNA-155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncol Rep. 2013;30:2111–2118. doi: 10.3892/or.2013.2685. [DOI] [PubMed] [Google Scholar]
- 15.Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med. 2013;31:1375–1380. doi: 10.3892/ijmm.2013.1348. [DOI] [PubMed] [Google Scholar]
- 16.Rather MI, Nagashri MN, Swamy SS, Gopinath KS, Kumar A. Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: implications for cancer therapeutics. J Biol Chem. 2013;288:608–618. doi: 10.1074/jbc.M112.425736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nazari-Jahantigh M, Wei Y, Noels H, Akhtar S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest. 2012;122:4190–4202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dent AL, Vasanwala FH, Toney LM. Regulation of gene expression by the proto-oncogene BCL-6. Crit Rev Oncol Hematol. 2002;41:1–9. doi: 10.1016/S1040-8428(01)00164-0. [DOI] [PubMed] [Google Scholar]
- 19.Liu SC, Bassi DE, Zhang SY, Holoran D, Conti CJ, Klein-Szanto AJ. Overexpression of cyclin D2 is associated with increased in vivo invasiveness of human squamous carcinoma cells. Mol Carcinog. 2002;34:131–139. doi: 10.1002/mc.10057. [DOI] [PubMed] [Google Scholar]
- 20.Fernández de Mattos S, Essafi A, Soeiro I, Pietersen AM, Birkenkamp KU, Edwards CS, Martino A, Nelson BH, Francis JM, Jones MC, et al. FoxO3a and BCR-ABL regulate cyclin D2 transcription through a STAT5/BCL6-dependent mechanism. Mol Cell Biol. 2004;24:10058–10071. doi: 10.1128/MCB.24.22.10058-10071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirata Y, Ogasawara N, Sasaki M, Mizushima T, Shimura T, Mizoshita T, Mori Y, Kubota E, Wada T, Tanida S, et al. BCL6 degradation caused by the interaction with the C-terminus of pro-HB-EGF induces cyclin D2 expression in gastric cancers. Br J Cancer. 2009;100:1320–1329. doi: 10.1038/sj.bjc.6605010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glauser DA, Schlegel W. The FoxO/Bcl-6/cyclin D2 pathway mediates metabolic and growth factor stimulation of proliferation in Min6 pancreatic beta-cells. J Recept Signal Transduct Res. 2009;29:293–298. doi: 10.3109/10799890903241824. [DOI] [PubMed] [Google Scholar]
- 23.Lao G, Liu P, Wu Q, Zhang W, Liu Y, Yang L, Ma C. Mir-155 promotes cervical cancer cell proliferation through suppression of its target gene LKB1. Tumour Biol. 2014;35:11933–11938. doi: 10.1007/s13277-014-2479-7. [DOI] [PubMed] [Google Scholar]
- 24.Shi LJ, Zhang CY, Zhou ZT, Ma JY, Liu Y, Bao ZX, Jiang WW. MicroRNA-155 in oral squamous cell carcinoma: Overexpression, localization, and prognostic potential. Head Neck. 2014;37:970–976. doi: 10.1002/hed.23700. [DOI] [PubMed] [Google Scholar]
- 25.Gombos K, Horváth R, Szele E, Juhász K, Gocze K, Somlai K, Pajkos G, Ember I, Olasz L. miRNA expression profiles of oral squamous cell carcinomas. Anticancer Res. 2013;33:1511–1517. [PubMed] [Google Scholar]
- 26.Johansson J, Berg T, Kurzejamska E, Pang MF, Tabor V, Jansson M, Roswall P, Pietras K, Sund M, Religa P, Fuxe J. MiR-155-mediated loss of C/EBPβ shifts the TGF-β response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene. 2013;32:5614–5624. doi: 10.1038/onc.2013.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandhu SK, Volinia S, Costinean S, Galasso M, Neinast R, Santhanam R, Parthun MR, Perrotti D, Marcucci G, Garzon R, Croce CM. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eμ-miR-155 transgenic mouse model. Proc Natl Acad Sci USA. 2012;109:20047–20052. doi: 10.1073/pnas.1213764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponzio G, Loubat A, Rochet N, Turchi L, Rezzonico R, Farahi Far D, Dulic V, Rossi B. Early G1 growth arrest of hybridoma B cells by DMSO involves cyclin D2 inhibition and p21(CIP1) induction. Oncogene. 1998;17:1159–1166. doi: 10.1038/sj.onc.1202040. [DOI] [PubMed] [Google Scholar]
- 29.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/S1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]