Abstract
Microtubule-based kinesin motors have many cellular functions, including the transport of a variety of cargos. However, unconventional roles have recently emerged, and kinesins have also been reported to act as scaffolding proteins and signaling molecules. In this work, we further extend the notion of unconventional functions for kinesin motor proteins, and we propose that Kif13b kinesin acts as a signaling molecule regulating peripheral nervous system (PNS) and central nervous system (CNS) myelination. In this process, positive and negative signals must be tightly coordinated in time and space to orchestrate myelin biogenesis. Here, we report that in Schwann cells Kif13b positively regulates myelination by promoting p38γ mitogen-activated protein kinase (MAPK)-mediated phosphorylation and ubiquitination of Discs large 1 (Dlg1), a known brake on myelination, which downregulates the phosphatidylinositol 3-kinase (PI3K)/v-AKT murine thymoma viral oncogene homolog (AKT) pathway. Interestingly, Kif13b also negatively regulates Dlg1 stability in oligodendrocytes, in which Dlg1, in contrast to Schwann cells, enhances AKT activation and promotes myelination. Thus, our data indicate that Kif13b is a negative regulator of CNS myelination. In summary, we propose a novel function for the Kif13b kinesin in glial cells as a key component of the PI3K/AKT signaling pathway, which controls myelination in both PNS and CNS.
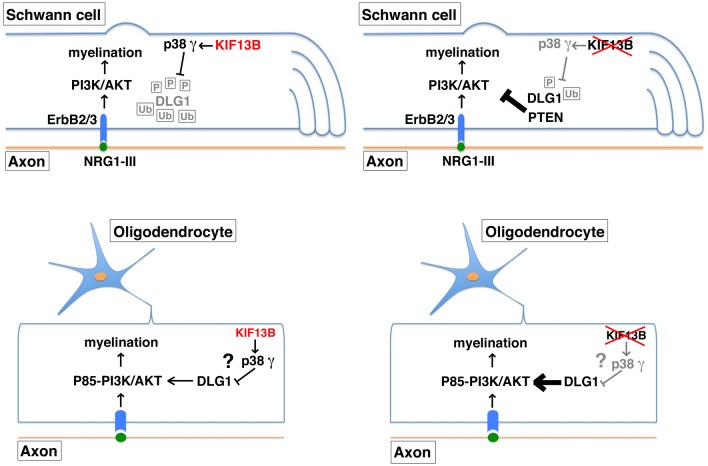
Kif13b is an unconventional kinesin that acts as a signaling molecule, regulating myelination via the Dlg1 scaffold in both Schwann cells (in the peripheral nervous system) and oligodendrocytes (in the central nervous system).
Author Summary
Myelin is a multilayered extension of the Schwann and oligodendrocyte cell membranes, which wraps around neuronal axons to facilitate propagation of electric signals and to support axonal metabolism. However, the signals regulating myelin formation and how they are integrated and controlled to achieve homeostasis are still poorly understood.
In Schwann cells, the Discs large 1 (Dlg1) protein is a known brake of myelination, which negatively regulates the amount of myelin produced so that myelin thickness is proportional to axonal diameter. In this paper, we report that in Schwann cells Dlg1 itself is tightly regulated to ensure proper myelination. We propose that Dlg1 function is further controlled by the Kif13b kinesin motor protein, which acts as a "brake of the brake" by downregulating Dlg1 activity. Surprisingly, we found that in oligodendrocytes Dlg1 is a positive and not a negative regulator of myelination. Thus, Kif13b-mediated negative regulation of Dlg1 ensures appropriate myelin production and thickness in the central nervous system. Our data further extend recently emerged unconventional roles for kinesins, which are usually implicated in cargo transport rather than in the modulation of signaling pathways. The elucidation of mechanisms regulating myelination may help to design specific approaches to favor re-myelination in demyelinating disorders in which this process is severely impaired.
Introduction
Myelination is a multistep process that includes axon recognition and contact, ensheathment, and myelin biogenesis. In this process, discrete sets of proteins and lipids are specifically assembled to generate and maintain distinct structural and functional domains necessary for nerve function [1–5]. During myelination, positive and negative regulators must be tightly controlled so that myelin thickness is strictly proportional to axonal diameters. However, the molecular mechanisms that promote and regulate myelination as well as the molecular machineries responsible for the transport and targeting of vesicles during myelin biogenesis are largely unknown. For example, Kif1b is the only motor protein identified thus far implicated in central nervous system (CNS) myelination in Danio rerio (zebrafish) [6].
We previously reported that in Schwann cells the Kif13b motor protein (also known as guanylate kinase-associated kinesin [GAKIN] in humans) is part of a complex that titrates membrane formation during Schwann cell myelination [7]. We found that Kif13b interacts with the Discs large 1 (Dlg1) scaffold in Schwann cells and that the downregulation of either Kif13b or Dlg1 expression in Schwann cell/dorsal root ganglia (DRG) neuron co-cultures decreases myelination in vitro [7]. Another study independently reported that Dlg1-silenced Schwann cells in vitro showed migration defects and reduced expression of the polarity protein Par3 [8]. Occasionally, silenced cells overcame their migration defect and myelinated, but the resulting myelin segments were thicker than those of controls, which indicated Dlg1 as a negative regulator of myelin sheath thickness [8]. This role was further assessed in vivo, as we and others subsequently reported that mouse nerves lacking Dlg1 expression specifically in Schwann cells have hypermyelination, myelin outfoldings, and demyelination as a consequence of myelin instability [8,9]. Dlg1 is thought to act in complex with phosphatase and tensin homolog (PTEN) to reduce AKT (v-AKT murine thymoma viral oncogene homolog) activation; thus, it is a brake on myelination [8].
Kif13b kinesin is a plus end motor protein that mediates the transport of several cargos in polarized cells [10–16]. In PC12 cells, Kif13b negatively regulates centaurin-α1/PIP3BP (phosphatidylinositol-3,4,5-trisphosphate binding protein), a GTPase activating protein (GAP) for Arf6 (ADP-ribosylation factor) GTPase and promotes Arf6 plasma membrane activation [16]. In neurons, Kif13b transports centaurin-α1/PIP3BP and PIP3 to the tip of neurites to promote neuronal polarity [11].
To further investigate the function of the Kif13b/Dlg1 complex in myelination in vivo, we generated a novel Kif13b floxed allele and conditional knock-out mouse models with specific ablation of Kif13b or Dlg1 in either Schwann cells or oligodendrocytes. Here, we report that Kif13b has opposite roles in the control of myelination in the peripheral nervous system (PNS) and CNS. Our data indicate that in Schwann cells, Kif13b interacts with p38γ mitogen-activated protein kinase (MAPK) to promote phosphorylation and ubiquitination of Dlg1. Consistent with this observation, loss of Kif13b results in reduced levels of p38γ MAPK, increased Dlg1 expression, and reduced myelin thickness.
Finally, we report that Kif13b also controls Dlg1 function in oligodendrocytes by promoting its negative regulation. However, our data indicate that, in contrast to Schwann cells, Dlg1 does not reduce but rather enhances AKT activation in oligodendrocytes. Thus, Kif13b is a novel negative regulator of CNS myelination.
Results
Kif13b Regulates Myelin Thickness in Schwann Cells
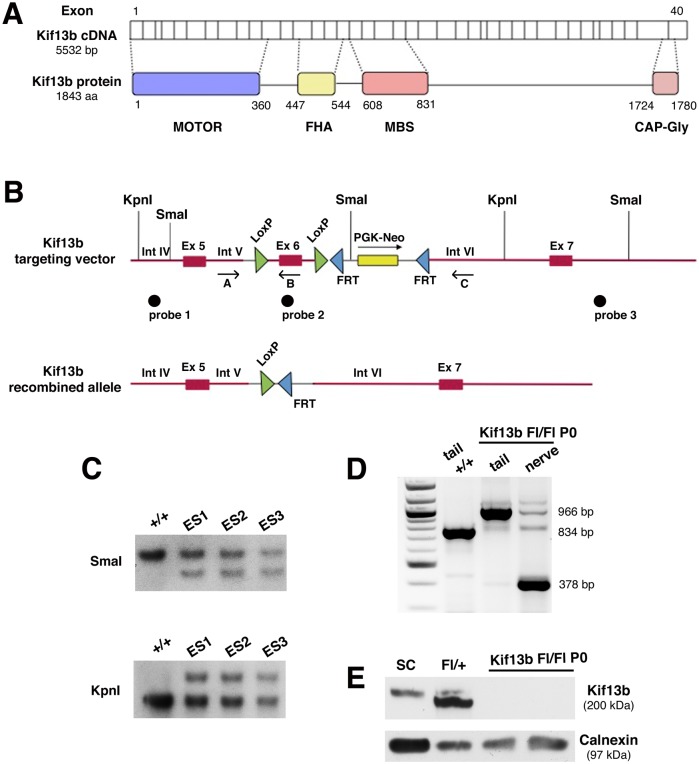
We previously reported that, in the peripheral nerve, Kif13b is mainly detected in cytosolic compartments of myelin-forming and non-myelin-forming Schwann cells [7]. To investigate the role of Kif13b in Schwann cells in vivo, we generated a Kif13b Floxed (hereafter, Kif13b Fl) allele in which exon 6 was flanked by lox-P sites. Using the Cre/loxP technology, excision of exon 6 produces a frameshift leading to the introduction of a premature stop codon and to nonsense-mediated mRNA decay (Fig 1A–1C). To ablate Kif13b specifically in Schwann cells, we generated Kif13b Fl/Fl P0-Cre mice, in which the myelin protein zero (MPZ) promoter drives Cre recombinase expression specifically in Schwann cells, starting from E13.5 [17,18]. Deletion of exon 6 was documented by PCR analysis on DNA from the sciatic nerve, where a recombination band of 378 bp was specifically detected (Fig 1D). Kif13b protein expression was ablated in sciatic nerve lysates from Kif13b Fl/Fl P0-Cre mice, thus also confirming that Kif13b is mainly expressed by Schwann cells in the nerve (Fig 1E).
Fig 1. Generation of the Kif13b-null mouse in Schwann cells.
(A) Kif13b protein domains. (B) The Kif13b targeting vector and the recombined allele following deletion of exon 6. Thick red lines represent genomic sequences, and thin black lines correspond to the pFlrt1 vector used for homologous recombination. (C) Southern blot analysis performed to identify ES cell recombinant clones. (D) PCR analysis using primers A and C performed on DNA from tail and sciatic nerve of wild type (+/+) and Kif13b Fl/Fl P0-Cre mice. (E) Western blot analysis on lysates from mouse sciatic nerves and isolated rat Schwann cells shows reduction of Kif13b expression in the mutant.
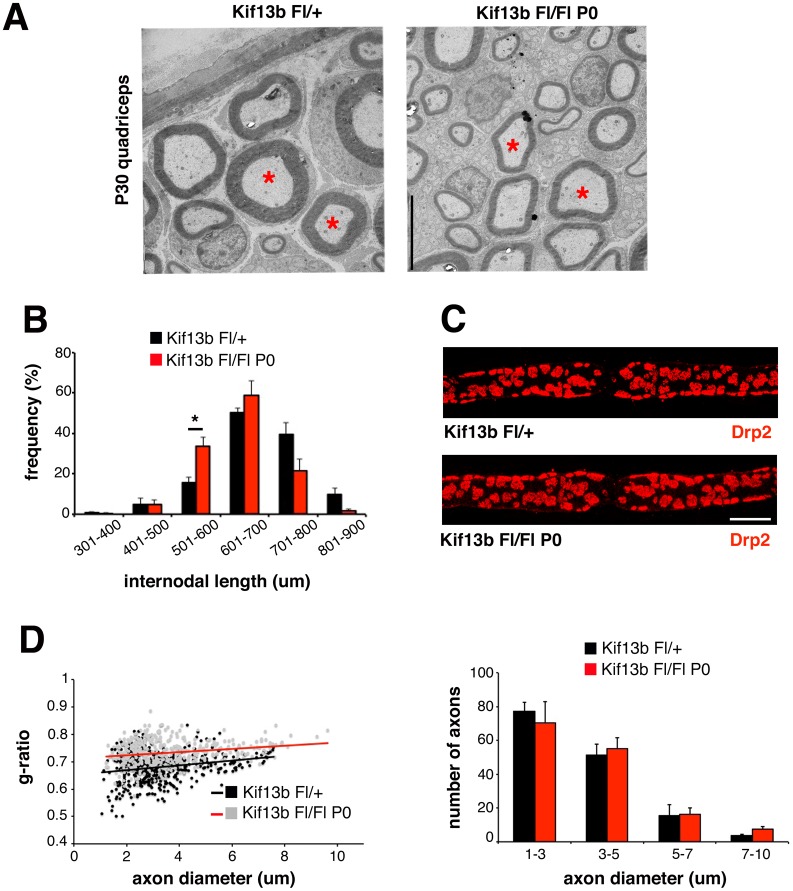
We then analyzed Kif13b Fl/Fl P0-Cre sciatic nerves starting at P30 by performing semithin section and ultrastructural analyses. In mutant nerves, we noted a higher number of fibers displaying Schwann cell nuclei and the surrounding cytoplasm, suggesting a shorter internodal length (Fig 2A). Consistent with this, we found that Kif13b Fl/Fl P0-Cre quadriceps nerves had indeed a higher percentage of fibers with shorter internodes, particularly in the range between 500 and 600 μm (Fig 2B). Cajal bands are cytoplasmic channels located at the abaxonal surface of myelinated fibers and are involved in the biosynthesis and assembly of myelin [3]. Ablation of the Schwann cell protein Periaxin disrupts Cajal bands and is also associated with reduced longitudinal growth of Schwann cells [19]. However, subsequent work from the same group has shown that loss of Cajal bands as a result of Drp2 ablation causes focal hypermyelination and concomitant demyelination [20]. We analyzed Kif13b Fl/Fl P0-Cre quadriceps nerves, but we did not detect major differences in Cajal band structures between mutant and control nerves (Fig 2C). Our findings are consistent with the view that the longitudinal growth of Schwann cells does not correlate with Cajal band integrity [20]. As myelin thickness is proportional to axonal diameter and internodal length [3], we evaluated myelin thickness in Kif13b Fl/Fl P0-Cre nerves by performing ultrastructural analysis. By measuring the g-ratio—the ratio between axonal diameter and fiber diameter—we observed reduced myelin thickness in mutant quadriceps nerves at P30, which displayed increased g-ratio values as compared to controls (ultrastructural analysis, Kif13b Fl/Fl P0-Cre, 0.75 ± 0.008, 575 fibers; Kif13b Fl/+, 0.70 ± 0.009, 516 fibers, n = 3 animals per genotype, p = 0.03). At P20, myelin thickness was normal in Kif13b Fl/Fl P0-Cre nerves, suggesting that myelination is not delayed in this mutant (ultrastructural analysis, g-ratio values: Kif13b Fl/Fl P0-Cre, 0.715 ± 0.007, 400 fibers; Kif13b Fl/+, 0.71 ± 0.004, 403 fibers, n = 3 animals per genotype, p = 0.49). Reduced myelin thickness was still present in nerves of older mice at 8 mo, as g-ratio values were increased in mutant nerves (Fig 2D).
Fig 2. Kif13b regulates myelin thickness in Schwann cells.
(A) Ultrastructural analysis of quadriceps nerves at P30. Red asterisks indicate thinner myelinated fibers in the mutants as compared to control fibers with similar axonal diameter. (B) Analysis of internodal length in quadriceps nerves at P30. The percentage (%) of fibers in the range 501–600 μm is 15.7 ± 2.61 for Kif13b Fl/+ and 33.7 ± 4.26 for Kif13b Fl/Fl P0-Cre. n = 3 animals per genotype, p = 0.031. (C) Immunohistochemistry using anti-Drp2 antibody on teased fibers shows that Cajal bands structure is preserved in Kif13b Fl/Fl P0-Cre quadriceps nerves. (D) Quantification of the g-ratio as a function of axonal diameter (Kif13b Fl/Fl P0-Cre, 0.73 ± 0.006, 447 fibers; Kif13b Fl/+, 0.68 ± 0.005, 443 fibers, n = 3 animals per genotype, p = 0.003) and axonal diameter distribution at 8 mo, ultrastructural analysis. The total number of fibers was normal. Bar in (A) and (C) is 5 μm.
Finally, following crush nerve injury, remyelinating Kif13b Fl/Fl P0-Cre nerves also displayed thinner myelin (S1 Fig).
In conclusion, our data indicate that loss of Kif13b specifically in Schwann cells affects longitudinal and radial myelin growth during development and remyelination after injury. Of note, myelination is not delayed in Kif13b mutant nerves at early stages of development, suggesting that Kif13b-mediated regulation occurs only during active myelination after P20.
Kif13b Loss in Schwann Cells Is Associated with Increased Dlg1 Expression and Stability
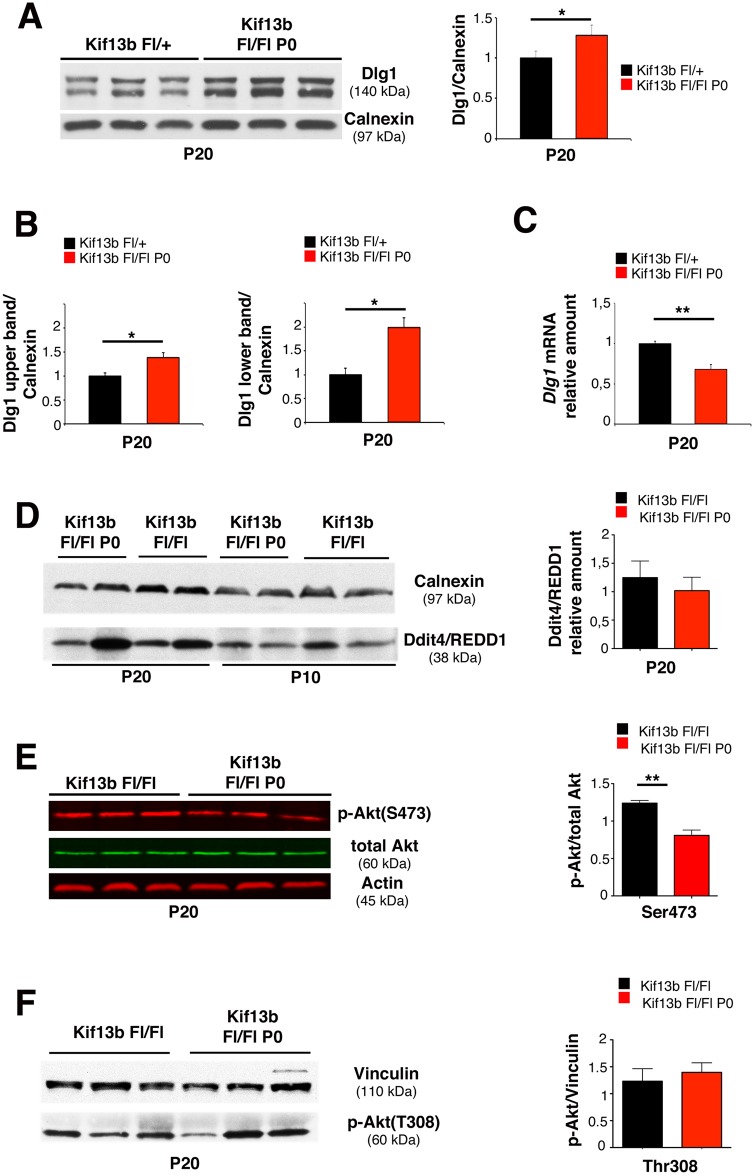
To investigate the molecular basis of the observed myelin phenotype, we looked at the expression level of Dlg1, a known interactor of Kif13b and a negative regulator of Schwann cell myelination in vivo [7–9]. Interestingly, we found that Dlg1 expression level was increased in Kif13b Fl/Fl P0-Cre nerves at P20 (Fig 3A). Note that the increase is particularly evident in the lower band (Fig 3B), which corresponds to a hypo-phosphorylated isoform of Dlg1 [21,22]. In contrast, Dlg1 mRNA levels were downregulated in mutant nerves (Fig 3C), which suggested that Dlg1 protein was more stable in the absence of Kif13b. To assess whether other negative regulators could contribute to the observed effect on myelination, we also looked at Ddit4/REDD1 expression levels. Ddit4/REDD1 is a known negative regulator of myelination, which downregulates the mechanistic target of rapamycin (mTOR) pathway by activating the tuberous sclerosis complex TSC1/2 [9]. We found that Ddit4 was similarly expressed between wild-type and mutant nerves at P10 and P20 (Fig 3D).
Fig 3. Increased Dlg1 expression in Kif13b Fl/Fl P0-Cre nerves.
(A) Dlg1 expression is increased in Kif13b Fl/Fl P0-Cre nerve lysates, with quantification, n = 3, p = 0.042, respectively. (B) Quantification of the Dlg1 upper and lower isoform bands as shown in panel (A) n = 3, p = 0.041, and p = 0.022, three independent experiments. (C) qPCR analysis shows that Dlg1 mRNA is decreased in Kif13b Fl/Fl P0-Cre nerves, n = 3, p = 0.008. (D) Expression levels of Ddit4/REDD1 in Kif13b Fl/Fl P0-Cre nerves at P10 and P20, with quantification at P20, n = 4, p = 0.564, representative of three independent experiments. (E) Phosphorylation of AKT at S473 is reduced in Kif13b Fl/Fl P0-Cre nerves, with quantification n = 3, p = 0.006, two independent experiments. (F) Phosphorylation of AKT at T308 in Kif13b Fl/Fl P0-Cre nerves, with quantification n = 3, p = 0.6, representative of three independent experiments.
Dlg1 interacts with Kif13b in Schwann cells and is known to potentiate PTEN phosphatase activity on PIP3, thus downregulating AKT activation [7–9]. Consistent with this, phosphorylation of AKT at S473 was decreased in Kif13b Fl/Fl P0-Cre nerves as compared to controls at P20, when AKT phosphorylation starts to decline during postnatal nerve development (Fig 3E) [9]. On the contrary, in Kif13b Fl/Fl P0-Cre nerves, phosphorylation of AKT at T308 was not significantly different from controls (Fig 3F). This finding may indicate activation of the feedback loop involving mTOR and molecules upstream of PI3K, as also already observed in other mutants [9,23–26].
Finally, we found normal expression levels of NRG1 type III (and the phosphorylation of its receptor ErbB2), Krox20, and Oct6, known regulators of myelin initiation, further supporting that reduced myelin thickness of Kif13b Fl/Fl P0-Cre nerves is associated with enhanced negative regulation of postnatal myelination and not with a delay in myelin program initiation (S2 Fig).
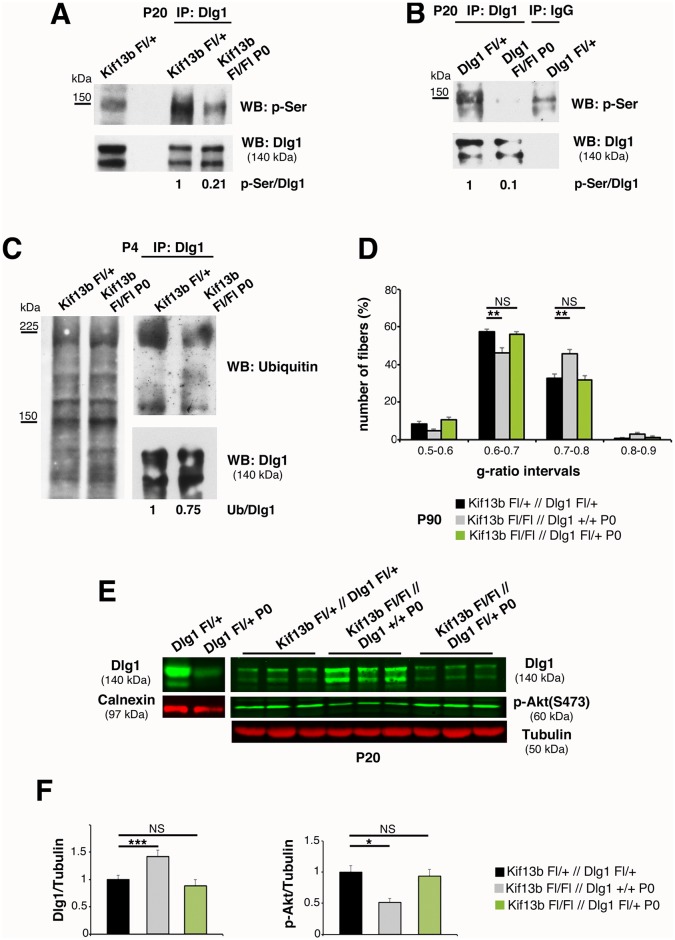
Phosphorylation is known to modulate protein–protein interactions necessary for the cytoskeletal localization of Dlg1 [27,28]. In particular, serine phosphorylation correlates with Dlg1 inactivation, and hyperphosphorylated Dlg1 interacts with ubiquitin ligases, which mediate its ubiquitination and degradation [8,21,22,27–30]. Thus, we hypothesized that increased Dlg1 protein levels in Kif13b Fl/Fl P0-Cre nerves could result from reduced serine phosphorylation and/or ubiquitination. By immunoprecipitating Dlg1 from sciatic nerve lysates at P20, we observed a decrease of Dlg1-serine phosphorylation in Kif13b Fl/Fl P0-Cre nerves compared to controls (Fig 4A). As expected, in Dlg1 Fl/Fl P0-Cre sciatic nerve lysates, the phosphorylated band was not detected (Fig 4B). Then, to evaluate whether the decrease in serine-phosphorylation correlated with increased stability, we determined the pattern of Dlg1 ubiquitination. Consistent with our hypothesis, by immunoprecipitating Dlg1 from Kif13b Fl/Fl P0-Cre nerves at P4 and P10, we found that Dlg1 was less ubiquitinated in mutant nerve lysates when compared to controls (Fig 4C).
Fig 4. Kif13b negatively regulates Dlg1 expression and activity.
(A) Immunoprecipitation of Dlg1 from Kif13b Fl/Fl P0-Cre nerves and controls at P20 followed by western blot analysis using an anti-phospho-serine antibody shows that Dlg1 is less phosphorylated in the mutant. Two independent experiments. (B) Immunoprecipitation of Dlg1 from Dlg1 Fl/Fl P0-Cre nerves indicates that the band at 140–150 KDa is Dlg1. The residual Dlg1 protein in mutant nerves could result from non-recombined fibroblasts within the nerve as described in [9]. (C) Immunoprecipitation of Dlg1 at P4 followed by western blot analysis shows that Dlg1 is less ubiquitinated in mutant nerves. Two independent experiments. (D) G-ratio value distribution shows that haploinsufficiency of Dlg1 in the Kif13b Fl/Fl P0-Cre background rescues the hypomyelination, p = 0.0093 and p = 0.0077, n = 4 animals per genotype, semithin section analysis. (E) Western blot analysis shows restored Dlg1 expression and AKT phosphorylation levels in Kif13b Fl/Fl//Dlg1 Fl/+; P0-Cre nerve lysates, with quantification in (F), p = 0.0004 (Dlg1) and p = 0.020 (p-Akt), n = 3. The genetic reduction of 50% of Dlg1 is sufficient to reduce Dlg1 expression level in lysates from Dlg1 Fl/+ P0-Cre.
Our data suggest that the hypomyelination in Kif13b Fl/Fl P0-Cre nerves results from increased Dlg1 stability/activity and enhanced negative regulation of AKT. Hence, we hypothesized that 50% reduction of Dlg1 gene expression in the Kif13b Fl/Fl P0-Cre background might rebalance Dlg1 levels and rescue the phenotype. Thus, we generated Kif13b Fl/Fl//Dlg1 Fl/+; P0-Cre mice, and we compared these mutants with Kif13b Fl/Fl//Dlg1 +/+; P0-Cre mouse nerves. By performing western blot analysis, we observed that Dlg1 expression and AKT phosphorylation levels in Kif13b Fl/Fl//Dlg1 Fl/+; P0-Cre sciatic nerve lysates were rescued at a level similar to controls (Fig 4E and 4F). Accordingly, myelin thickness in Kif13b Fl/Fl//Dlg1 Fl/+; P0-Cre nerves was also restored (Fig 4D). Overall, our data suggest that Kif13b negatively regulates Dlg1 stability and activity in Schwann cells. Thus, in kif13b Fl/Fl P0-Cre nerves, increased Dlg1 activity reduces AKT signaling and myelination.
Kif13b Regulates Myelin Thickness in Oligodendrocytes
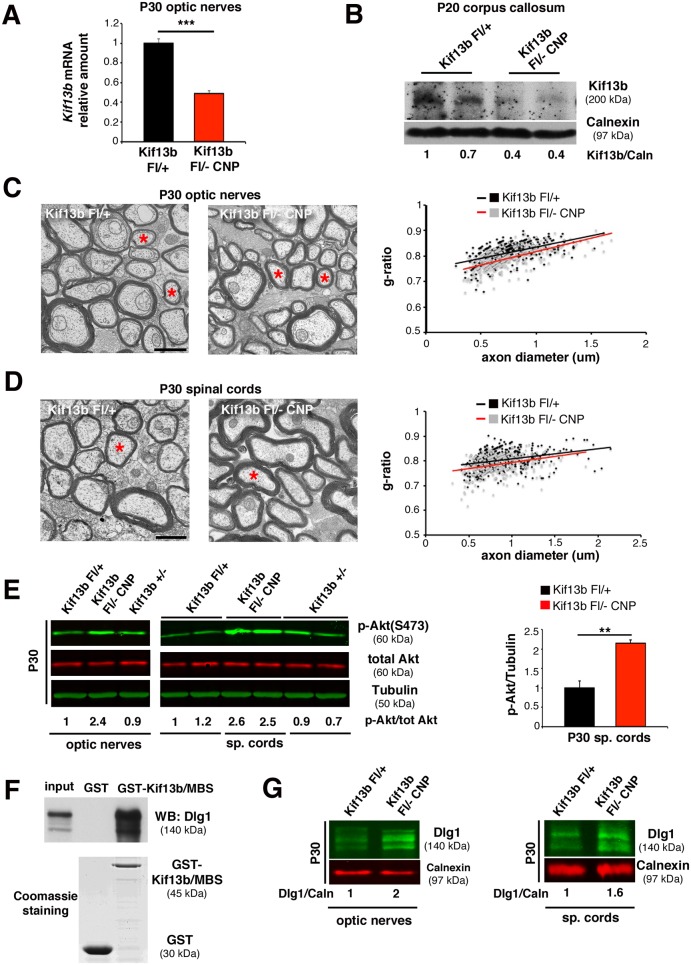
Since Kif13b regulates PNS myelination, we sought to assess whether Kif13b has a similar role in the CNS. First, we confirmed Kif13b mRNA expression in optic nerves and in myelinated tracts of the corpus callosum (Fig 5A and 5B). Then, we generated a Kif13b Fl/- CNP-Cre mouse with conditional inactivation of Kif13b in newly generated oligodendrocytes [31]. To achieve maximum efficiency of CNP-Cre mediated recombination, we generated a compound heterozygous mouse for a Kif13b Fl allele and a Kif13b - (null) allele. We first assessed downregulation of Kif13b mRNA expression in Kif13b Fl/- CNP-Cre optic nerves by performing quantitative RT-PCR analysis (Fig 5A). Western blot analysis confirmed reduction of Kif13b protein expression in lysates from corpus callosum of Kif13b Fl/- CNP-Cre mice (Fig 5B). We then performed morphological analysis of optic nerves and spinal cords at P30. Surprisingly, we observed increased myelin thickness with decreased g-ratios in both Kif13b Fl/- CNP-Cre optic nerves and spinal cords as compared to either Kif13b Fl/+ or Kif13b -/+ controls (Fig 5C and 5D). However, at P90, myelin thickness in either Kif13b Fl/- CNP-Cre optic nerves or spinal cords was normal, suggesting a transient effect of Kif13b loss (S3 Fig).
Fig 5. Kif13b regulates myelin thickness in oligodendrocytes.
(A) RT-PCR analysis on optic nerves shows reduction of Kif13b mRNA in the mutant (n = 5, p = 0.0004). (B) Western blot analysis on lysates from corpus callosum at P20 shows reduction of Kif13b protein expression in the mutant. (C) Ultrathin analysis and quantification of the g-ratio as a function of axonal diameter in Kif13b Fl/- CNP-Cre and control optic nerves at P30. Red asterisks indicate fibers with similar diameter to be compared in the two genotypes (Kif13b Fl/- CNP-Cre, 0.792 ± 0.005, 296 fibers; Kif13b Fl/+, 0.814 ± 0.006, 345 fibers, n = 4 animals per genotype, p = 0.030; Kif13b +/- 0.813 ± 0.004, 435 fibers, n = 5 animals per genotype). (D) Ultrathin analysis and quantification of the g-ratio as a function of axonal diameter in Kif13b Fl/- CNP-Cre and control spinal cords at P30 (Kif13b Fl/- CNP-Cre, 0.785 ± 0.003, 364 fibers; Kif13b Fl/+, 0.803 ± 0.007, 403 fibers, n = 5 animals per genotype, p = 0.016). (E) Western blot analysis on lysates from optic nerves and spinal cords at P30 shows increased AKT phosphorylation at S473 in the mutant. Three independent experiments. Quantification of AKT phosphorylation levels from spinal cord lysates, p = 0.005, n = 3. (F) Pull down assay from P11 rat optic nerves using GST-Kif13b/MBS as a bait indicates Dlg1 and Kif13b interaction. Three independent experiments. (G) Western blot analysis on optic nerve and spinal cord lysates shows increased Dlg1 expression in Kif13b mutant oligodendrocytes. Three independent experiments. Bar in (C) and (D) is 1 μm.
At the molecular level, AKT phosphorylation at S473 was enhanced in both Kif13b Fl/- CNP-Cre optic nerves and spinal cords at P30 (Fig 5E), consistent with the observed hypermyelination and the role of AKT in promoting CNS myelination [32]. We then explored whether, as in the PNS, Kif13b regulates myelination by controlling Dlg1 expression levels. First, we assessed whether Kif13b interacts with Dlg1 in oligodendrocytes in vivo. By performing GST pull down assays from rat optic nerve lysates using GST-Kif13b/MBS as a bait, we identified Dlg1, suggesting the existence of a Kif13b/Dlg1 complex (Fig 5F). Interestingly, we noted that in spinal cord and optic nerve lysates Dlg1 isoforms were expressed in the range 140–150 KDa, as already observed in sciatic nerves (Fig 3A), where Dlg1 is not expressed in the axon [7,8]. This finding suggests that the Kif13b/Dlg1 interaction likely occurs in oligodendrocytes and not in axons/neurons, where the main Dlg1/SAP97 isoform runs at a different molecular weight (97KDa). Next, we evaluated Dlg1 protein expression in Kif13b Fl/- CNP-Cre mice and we found increased Dlg1 levels in both Kif13b Fl/- CNP-Cre optic nerves and spinal cords at P30 (Fig 5G). This result is consistent with the hypothesis that Kif13b negatively regulates Dlg1 expression also in the CNS.
Overall, our findings indicate that Kif13b is a transient negative regulator of myelination in the CNS as its downregulation in oligodendrocytes increases myelin thickness and enhances AKT activation. Moreover, we suggest that also in the CNS Kif13b interacts with Dlg1 and negatively regulates its stability.
Dlg1 Enhances AKT Activation in Oligodendrocytes
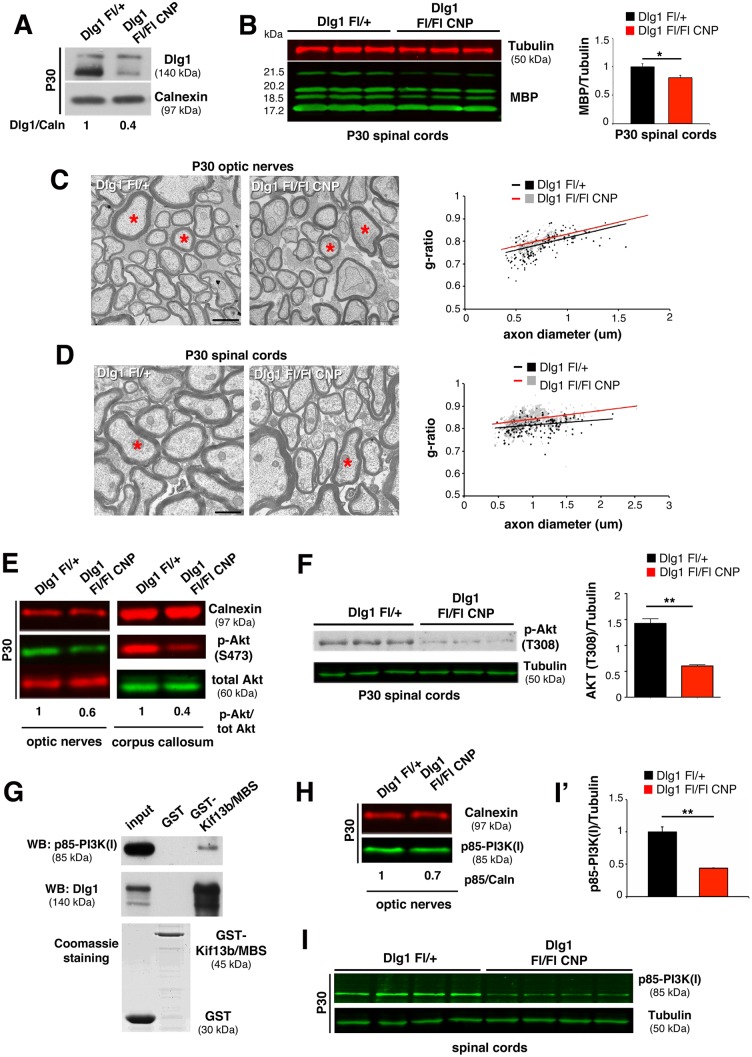
In Kif13b Fl/- CNP-Cre mice, increased myelin thickness is associated with enhanced Dlg1 expression. However, if Dlg1 acts as a negative regulator of myelination in oligodendrocytes as well, we would have expected to observe hypomyelination and not hypermyelination. Thus, we hypothesized that in oligodendrocytes Dlg1 might have the opposite role in the control of myelination, being a promoter rather than an inhibitor. To test this hypothesis, we generated Dlg1 Fl/Fl CNP-Cre conditional knockout mice in which Dlg1 was ablated in oligodendrocytes. We first demonstrated a reduction of Dlg1 protein expression in Dlg1 Fl/Fl CNP-Cre optic nerves at P30 (Fig 6A). Then, we performed morphological analyses of optic nerves and spinal cords starting at P30. Consistent with our hypothesis, mutant optic nerves and spinal cords displayed reduced myelin thickness and increased g-ratios (Fig 6C and 6D). Hypomyelination was also evidenced by decreased myelin basic protein (MBP) expression levels in spinal cord lysates from Dlg1 Fl/Fl CNP-Cre mice (Fig 6B). As in the case of Kif13b Fl/- CNP-Cre mutants, myelin thickness of Dlg1 Fl/Fl CNP-Cre optic nerves and spinal cords was normal at P90, suggesting a transient role of Dlg1 in the control of myelination (S3 Fig).
Fig 6. Dlg1 enhances AKT activation and regulates CNS myelination.
(A) Dlg1 expression is reduced in Dlg1 Fl/Fl CNP-Cre optic nerve lysates at P30. (B) Decreased MBP expression in mutant spinal cords, with quantification, p = 0.05, n = 3. (C) Ultrathin analysis and quantification of the g-ratio as a function of axonal diameter in optic nerves at P30 (Dlg1 Fl/Fl CNP-Cre, 0.824 ± 0.006, 294 fibers; Dlg1 Fl/+ 0.804 ± 0.002, 310 fibers, n = 5 animals per genotype, p = 0.017). The red asterisks in the mutant indicate fibers with thinner myelin. (D) Ultrathin analysis and the g-ratio as a function of axonal diameter in spinal cords at P30 (Dlg1 Fl/Fl CNP-Cre, 0.842 ± 0.004, 271 fibers, Dlg1 Fl/+, 0.815 ± 0.002, 213 fibers, n = 4 animals per genotype, p = 0.003). (E) Reduced S473 AKT phosphorylation in Dlg1 Fl/Fl CNP-Cre mice, two independent experiments. (F) Phosphorylation of AKT at T308 is also reduced in Dlg1 Fl/Fl CNP-Cre mice spinal cords, with quantification, n = 3, p = 0.001. (G) GST pull down assay from P11 rat optic nerves using GST-Kif13b/MBS as a bait indicates Dlg1 and p85 interaction. Two independent experiments. (H–I') p85 expression is reduced in lysates from Dlg1 Fl/Fl CNP-Cre optic nerves and spinal cords (I), with quantification from spinal cord lysates, p = 0.003, n = 4 animals per genotype, five independent experiments. Bar in (C) and (D) is 1 μm.
To investigate the mechanism by which Dlg1 promotes myelination in oligodendrocytes, we examined the phosphorylation state of AKT in lysates from optic nerves and corpus callosum of Dlg1 Fl/Fl CNP-Cre mutants. We found that AKT phosphorylation at both S473 and T308 was reduced in both Dlg1 Fl/Fl CNP-Cre optic nerves and corpus callosum as compared to controls, consistent with the decreased myelination (Fig 6E and 6F).
Since (1) AKT phosphorylation depends on PIP3 levels and on the activity of the PI3K class I and (2) Dlg1 has been described to interact with the regulatory subunit of PI3K class I, p85, in epithelial cells [29], we hypothesized that also in oligodendrocytes Dlg1 may interact with p85, influencing PI3K activity upstream of AKT.
To address this point, we first explored p85 expression levels in optic nerves and spinal cords at P30 and found that p85 protein levels were reduced in Dlg1 Fl/Fl CNP-Cre mice (Fig 6H–6I'). Next, GST pull down experiments from P11 rat optic nerve lysates demonstrated that Dlg1 and p85 are interactors of GST-Kif13b/MBS (Fig 6G), thus providing evidence for the existence of a complex involving Kif13b, Dlg1, and p85. Interestingly, by performing co-immunoprecipitation and pull down experiments, we did not observe interaction between p85 and the Kif13b/Dlg1 complex in the PNS in sciatic nerves. Consistent with this, p85 was similarly expressed in Kif13b Fl/Fl P0-Cre and Dlg1 Fl/Fl P0-Cre mutant sciatic nerves as compared to controls (S4 Fig). These findings suggest that in the PNS, in contrast to the CNS, the Kif13b/Dlg1 complex does not involve p85.
In conclusion, similarly to Schwann cells, downregulation of Kif13b expression in oligodendrocytes is associated with increased Dlg1 levels. However, in the CNS, Dlg1 promotes myelination. Thus, downregulation of Kif13b expression in oligodendrocytes causes hypermyelination.
p38γ MAPK Regulates Dlg1 Stability in Both PNS and CNS
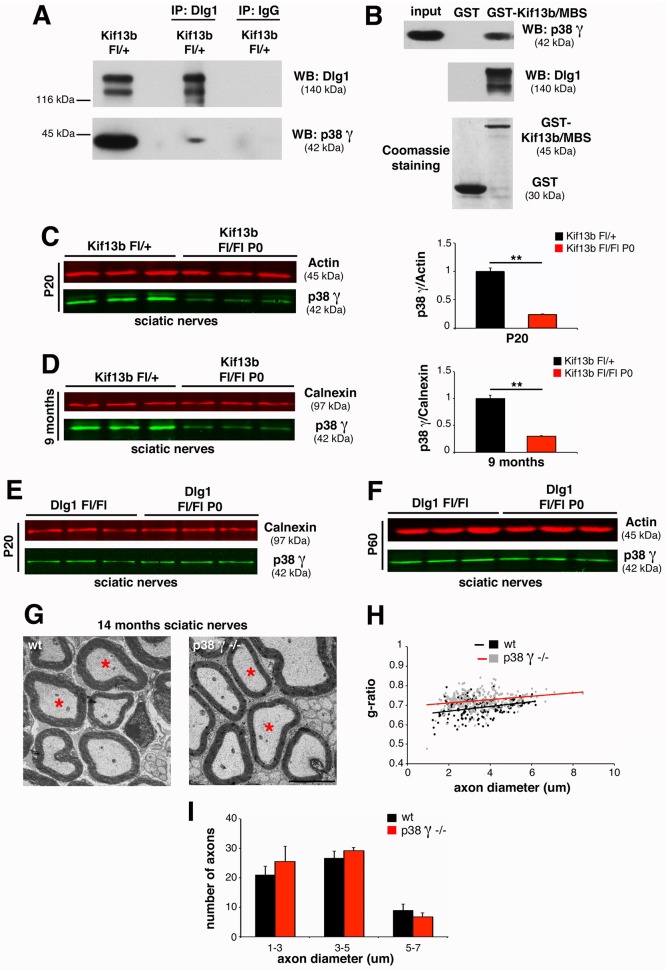
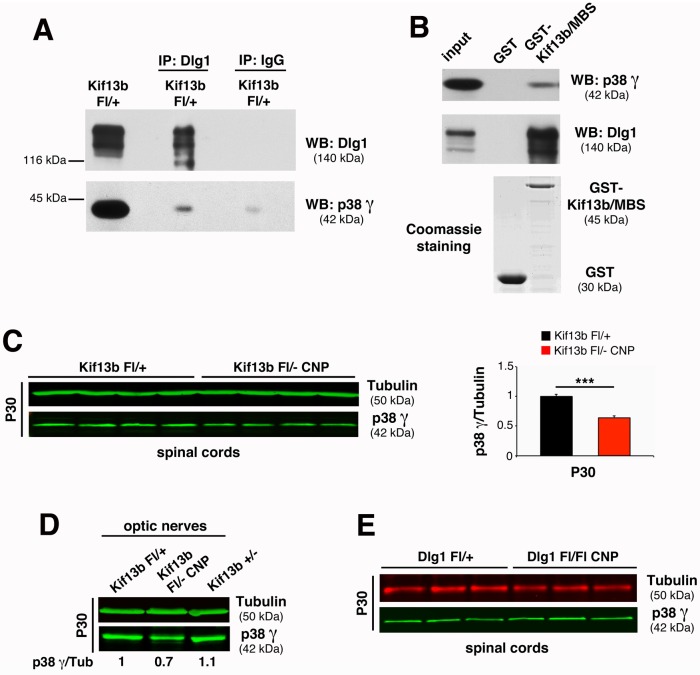
Finally, we asked how downregulation of Kif13b expression results in increased Dlg1 stability in both PNS and CNS. In previous yeast two-hybrid screening analyses, we had found that the PDZ2+3 domain of Dlg1 directly interacts with the p38γ MAPK isoform [7,33], as also previously reported for HEK293 cells [28]. Since p38γ can phosphorylate and negatively regulate the interaction of Dlg1 with cytoskeletal protein partners, we further investigated the interaction of Kif13b, p38γ, and Dlg1 in the nerve in vivo. We first confirmed Dlg1 and p38γ interaction by performing co-immunoprecipitation experiments from sciatic nerve lysates (Fig 7A). Next, we observed that Kif13b/MBS-GST was able to pull down both Dlg1 and p38γ from nerve lysates, suggesting that Kif13b, p38γ, and Dlg1 may be part of the same complex (Fig 7B). To provide further evidence for this hypothesis, we investigated p38γ expression levels in mutants with conditional ablation of either Kif13b or Dlg1 in Schwann cells. Interestingly, p38γ expression levels were decreased in Kif13b Fl/Fl P0-Cre sciatic nerves at both P20 and 9 mo (Fig 7C and 7D) but not in Dlg1 Fl/Fl P0-Cre nerves (Fig 7E and 7F), suggesting that p38γ acts downstream of Kif13b and upstream of Dlg1. To confirm these results, we analyzed the sciatic nerves of p38γ knock-out mutants. As expected, nerves from p38γ-null mice were hypomyelinated (Fig 7G–7I), supporting the hypothesis that p38γ is a novel promoter of Schwann cell myelination.
Fig 7. p38γ MAPK acts in complex with Kif13b and Dlg1 in the PNS.
(A) Dlg1 and p38γ co-immunoprecipitate from mouse sciatic nerve lysates at P20. Two independent experiments. (B) Kif13b/MBS-GST used to pull down Dlg1 and p38γ from rat sciatic nerve lysates at P11. Two independent experiments. (C) p38γ expression levels are reduced in Kif13b Fl/Fl P0-Cre nerve lysates at both P20 and (D) at 9 mo, with quantification, p = 0.005 at P20 and p = 0.007 at 9 mo, n = 3. (E, F) p38γ expression is not reduced in lysates from Dlg1 Fl/Fl P0-Cre nerves at P20 and P60. (G, H) G-ratio as a function of axonal diameter in p38γ-null sciatic nerves at 14 mo, performed by ultrastructural analysis (wild-type, 0.688 ± 0.008, 170 fibers, n = 3 animals, p38γ-null 0.726 ± 0.009, 249 fibers, n = 4 animals, p = 0.023). Total number of axons does not change in the two genotypes. (I) Fiber diameter distribution in p38γ-null nerves at 14 mo of age. Bar in (G) is 2.5 μm.
Finally, since our data suggest that Kif13b may similarly regulate Dlg1 also in the CNS, we assessed whether a Kif13b, p38γ, and Dlg1 complex could be detected in oligodendrocytes. As expected, Dlg1 and p38γ co-immunoprecipitate from optic nerve lysates (Fig 8A) and Kif13b/MBS-GST is able to pull down both Dlg1 and p38γ (Fig 8B). Even if not as striking as in Schwann cells, p38γ expression levels were decreased in Kif13b Fl/- CNP-Cre optic nerve lysates but not in Dlg1 Fl/Fl CNP-Cre, suggesting that p38γ acts downstream of Kif13b and upstream of Dlg1 (Fig 8C–8E).
Fig 8. p38γ MAPK acts in complex with Kif13b and Dlg1 in the CNS.
(A) Dlg1 and p38γ co-immunoprecipitate from rat optic nerve lysates at P11. Two independent experiments. (B) Kif13b/MBS-GST pulls down Dlg1 and p38γ from rat optic nerve lysates at P11. Two independent experiments. (C,D) p38γ expression levels are reduced in Kif13b Fl/- CNP-Cre optic nerve and spinal cord lysates at P30, with quantification, p = 0.0005, n = 4 animals per genotype, representative of four independent experiments. (E) p38γ expression is not reduced in Dlg1 Fl/Fl CNP-Cre spinal cords at P30.
As p38α is the MAPK isoform known to regulate myelination in both PNS and CNS [34–41], we assessed whether Kif13b/Dlg1 may also form a complex with p38α. Interestingly, by performing pull down experiments, we found that Dlg1 does not interact with p38α in either optic or sciatic nerve lysates. Consistent with this, expression levels of p38α in either sciatic nerves or spinal cords of Kif13b conditional knock-out mutants were similar to controls (S5 Fig).
Overall, these findings suggest a similar mechanism of Kif13b and p38γ-mediated regulation of Dlg1 in both PNS and CNS, with opposite outcomes on the control of myelination, as Dlg1 is a brake on myelination in the PNS and a positive regulator in the CNS.
Discussion
Microtubule-based kinesin motors have many cellular functions, including the transport of a variety of cargos to different parts of the cell [42]. Motors can also be used to place cargos on a long distance, such as signaling complexes or developmental determinants in neurons or embryos, respectively. However, unconventional functions have recently emerged, and kinesins have also been reported to act as scaffolding proteins and signaling molecules [43]. In particular, Kif13b has been recently shown in hepatocytes to work as a scaffold and to enhance caveolin-1 dependent internalization of LRP11 receptor [12]. In T cells, Kif13b acts as a signaling molecule that controls CARD11 scaffold localization at the synapse and downregulates TCR signaling [13]. In this work, we further extend these findings on unconventional roles of kinesins and propose a novel mechanism by which the Kif13b motor protein regulates Dlg1 scaffold activity and titrates the PI3K/AKT signaling with two opposite outcomes in PNS and CNS myelination (Fig 9).
Fig 9. Kif13b regulates myelination in PNS and CNS through the Dlg1 scaffold.
The PI3K-AKT-mTOR signaling axis is one of the signaling pathways regulating myelination in both PNS and CNS, as recently reviewed [49–56,64]. Our results suggest that in Schwann cells, Kif13b is a positive regulator of myelination. Kif13b promotes p38γ MAPK-mediated Dlg1 phosphorylation and ubiquitination. Dlg1, in complex with PTEN, is known to reduce AKT activation and thus negatively regulates myelination. Dlg1 loss is associated with increased myelin thickness and myelin outfoldings, as a result of increased PIP3 and AKT phosphorylation levels [8,9,47]. On the contrary, in oligodendrocytes, loss of Kif13b-mediated negative regulation of Dlg1 and the consequent increase in Dlg1 levels are associated with transient hypermyelination. Of note, we found that in the CNS Dlg1 is a promoter and not an inhibitor of myelination, and it likely modulates PI3K class I activity.
Kif13b Regulates PNS Myelination through the Dlg1 Scaffold
Here we report that downregulation of Kif13b expression in Schwann cells is associated with reduced myelin thickness, decreased AKT activation, and increased levels of Dlg1, a known brake on PNS myelination acting on the PIP3-AKT-mTOR pathway [8,9]. As Kif13b and Dlg1 interact in Schwann cells [7], we hypothesized that Kif13b may control myelination through the Dlg1 scaffold itself, by regulating its stability and function. Indeed, in support of our hypothesis, in Kif13b Fl/Fl //Dlg1 Fl/+; P0-Cre double mutant sciatic nerves, Dlg1 expression levels and myelin thickness are similar to wild type.
Interestingly, we report here that in Kif13b Fl/Fl P0-Cre nerves, in which Dlg1 expression levels are increased, myelination is not delayed at very early stages of postnatal nerve development, and reduced myelin thickness is evident when AKT activation starts to physiologically decline, after P20 [8,9]. This observation is consistent with the phenotype of mutant mice lacking Dlg1, specifically in Schwann cells [9]. We previously reported a transient increase in myelin thickness and occasional myelin outfoldings in Dlg1 Fl/Fl P0-Cre nerves starting from P10 [9]. However, even if enhanced, myelination was not accelerated in Dlg1 Fl/Fl P0-Cre nerves, and, at very early stages of postnatal nerve development, the number of myelinated fibers and myelin thickness were similar to control nerves. Thus, Dlg1 may act as a brake on myelination to downregulate AKT activation at the peak of myelination, when AKT phosphorylation starts to decline. In support to this hypothesis, myelin outfoldings, a focal form of hypermyelination that is thought to be linked to AKT overactivation and loss of Dlg1-mediated negative control on myelination, are observed in the nerve after 3 w of postnatal development [44].
Dlg1 stability is controlled by phosphorylation and ubiquitination [8,21,22,27,28,30,45,46]. In Drosophila, the PAR1 kinase directly phosphorylates Dlg1 at conserved sites and negatively regulates its mobility and targeting at postsynaptic membranes of neuromuscular junctions [27]. Osmotic stress-induced serine phosphorylation of Dlg1 by p38γ MAP kinase can induce Dlg1 dissociation from the glucokinase-associated dual specificity phosphatase (GKAP) and the cytoskeleton, negatively regulating Dlg1 [28]. Finally, phosphorylated DLG1 interacts with the β-TrCP ubiquitin ligase receptor, which mediates ubiquitination of the protein [30]. Thus, we investigated whether enhanced Dlg1 protein expression levels in Kif13b Fl/Fl P0-Cre nerves correlated with a decrease in serine phosphorylation and/or ubiquitination. Consistent with our hypothesis, we found that in Kif13b Fl/Fl P0-Cre nerves Dlg1 is less phosphorylated and less ubiquitinated, suggesting that Kif13b promotes radial myelin growth by directly or indirectly influencing Dlg1 stability and expression.
We also suggest that p38γ MAPK could be the kinase that, downstream of Kif13b, phosphorylates Dlg1 to regulate its activity. Indeed, p38γ MAPK is known to interact with and to phosphorylate serine residues of Dlg1 in other cells [28]. We identified p38γ in a yeast two-hybrid screening analysis using a nerve cDNA library and Dlg1 as a bait [7,33]. Moreover, we show that p38γ, Dlg1, and Kif13b form a complex in the nerve. More importantly, sciatic nerves of p38γ-null mice are hypomyelinated, thus confirming the hypothesis that p38γ, by phosphorylating and negatively regulating Dlg1, acts as a promoter of myelination downstream of Kif13b. Unfortunately, antibodies that can specifically recognise phosphorylated p38γ are not available to assess whether activated p38γ could interact with Kif13b and Dlg1.
Interestingly, the role of p38γ MAPK in the regulation of PNS myelination has not yet been assessed. Previous studies suggested that p38 MAPK mediates laminin signaling in vitro to promote Schwann cell elongation and alignment at the very first stages of differentiation [34]. Hossain et al., suggested that p38 directs Schwann cell differentiation by regulating Krox-20 expression, thus further supporting the role of p38 MAPK as a positive regulator of PNS myelination [35]. However, on the basis of the MAPK inhibitors used, the observed effect was likely to be mediated by the p38α or p38β [35]. A more recent study reported that in vitro p38 MAPK promotes the de-differentiated state of Schwann cells during Wallerian degeneration, by inducing c-Jun expression and by inhibiting myelin gene expression, and also suggested that p38 MAPK is a negative regulator of Schwann cell differentiation and myelination during development [36]. On the basis of the antibodies used recognizing the phosphorylated state of MAPK as well as the MAPK inhibitor used (SB203580), other isoforms rather than p38γ are more likely to mediate this function [36].
How can both Kif13b and p38γ control Dlg1 phosphorylation, ubiquitination, and stability? Kif13b could transport and localize the kinase at membranes where Dlg1 is enriched to downregulate, in complex with PTEN, PIP3 levels, and AKT activation [47]. Indeed, in Kif13b-null but not in Dlg1-null nerves p38γ expression levels are reduced, thus suggesting that p38γ is downstream of Kif13b and upstream of Dlg1. Alternatively, the binding of Kif13b with Dlg1, which is mediated by the membrane-associated guanylate kinase homologue binding stalk (MBS) and guanylate kinase homologue (GUK) domains, respectively, may relieve intramolecular inhibition in either Kif13b or Dlg1, as already reported [48]. For example, following Kif13b binding, a conformational change in Dlg1 (open state) can be induced so that target residues for serine phosphorylation can be exposed and accessible to p38γ kinase-mediated phosphorylation. Unfortunately, p38γ-specific inhibitors are not available to further investigate these mechanisms.
Our data convey a novel function for Kif13b/p38γ as negative regulators of Dlg1 in the PI3K/AKT signalling pathway. Interestingly, Kif13b has already been proposed as a negative regulator in other studies. For example, in PC12 cells, KIF13B negatively regulates centaurin-α1/PIP3BP (PIP3 binding protein), a GAP for Arf6, thus promoting Arf6 GTPase plasma membrane activation [16]. Further, in T cells, KIF13B negatively regulates TCR signaling to NF-kB, by redistributing the CARD11 scaffold from the center of the synapse to a more distal region [13].
Kif13b Regulates CNS Myelination through the Dlg1 Scaffold
We also show that Kif13b is a negative regulator of CNS myelination. Indeed, we observed that downregulation of Kif13b expression in oligodendrocytes results in increased myelin thickness and AKT activation, consistently with the role of AKT in promoting CNS myelination [32]. Similar to PNS, we found that Kif13b interacts with Dlg1 and that loss of Kif13b is associated with increased Dlg1 levels, thus suggesting a negative regulation mediated by Kif13b on Dlg1. Given these similarities, we investigated whether the increased Dlg1 level and stability in oligodendrocytes could also result from a decrease in p38γ-mediated phosphorylation. Indeed, we found that Kif13b, Dlg1, and p38γ MAPK interact in optic nerves and that p38γ expression is decreased in Kif13b but not in Dlg1 mutants, as already observed in the PNS. These findings suggest that p38γ may act downstream of Kif13b and upstream of Dlg1 to negatively regulate Dlg1 activity.
The role of the p38γ isoform in the regulation of CNS myelination has not been yet assessed. As for PNS, only p38α has been investigated in the CNS. Inhibition of p38α activity or expression in vitro in a co-culture system has been reported to prevent oligodendrocyte progenitor differentiation and myelination [37–39]. Another study suggested that p38α MAPK supports myelin gene expression in the brain through several mechanisms acting on both positive and negative regulators of differentiation [40]. More recently, myelination was found to be impaired in mice with conditional inactivation of p38α MAPK in oligodendrocyte progenitor cells [41]. Interestingly, the same authors observed an opposite effect of p38α MAPK in remyelination, as mutant mice exhibited a more efficient remyelination as compared to controls following demyelination [41]. These studies further support the notion that the regulation of myelination is a very complicated process, in which different signals arising from the extracellular matrix, axons, and astrocytes in the CNS must be correctly integrated in time and space within the same cell to achieve homeostasis [49–56].
If Dlg1 is a brake on myelination in the CNS as in the PNS, how can loss of Kif13b and elevation of Dlg1 result in increased CNS myelin thickness? Surprisingly, our data indicate that in oligodendrocytes Dlg1 is a positive and not a negative regulator of myelination, as its loss is associated with reduced myelin thickness and AKT activation. Interestingly, in addition to Dlg1, other molecules have been found to control myelination with opposite roles in PNS and CNS [57–60]. For example, myosin light chain II phosphorylation promotes myelination in the PNS and inhibits myelination in the CNS [57].
To determine the mechanism by which Dlg1 could promote CNS myelination acting on the PI3K-AKT pathway, we sought to investigate the regulatory subunit of PI3K class I, p85, a known interactor of Dlg1 in epithelial cells [29]. Consistent with this, we found that Dlg1 interacts with p85 in the optic nerve, likely to modulate PI3K class I activity, PIP3 levels, and ultimately AKT activation. Interestingly, phosphorylation of DLG1 on serine and threonine is known to prevent DLG1 interaction with SH2 domains of p85/PI3K [29]. Thus, we could speculate that Dlg1, when hypophosphorylated, may display a higher affinity for the SH2 domains of p85, whose activation is necessary for PI3K activity regulation [61].
Whether in oligodendrocytes Dlg1 also promotes myelination by other mechanisms, which can converge on AKT activation, remains to be determined.
Material and Methods
Antibodies
The following primary antibodies were used: mouse anti-KIF13B (provided by Dr. A. Chishti); mouse anti-Dlg1 (Stressgen); mouse anti-phosphoserine (Alexis Biochemicals); rabbit anti-ubiquitin (Santa Cruz Biotechnology); rabbit anti-DRP2 (provided by Dr. D. Sherman); rabbit anti-phospho-Akt (Ser473) (Cell Signaling); rabbit anti-phospho-Akt (Thr308) (Cell Signaling); rabbit anti-Akt (pan) (Cell Signaling); rabbit anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (Cell Signaling); rabbit anti-p44/42 MAP Kinase (Cell Signaling); rabbit anti-Neuregulin-1α/β1/2 (C20) (Santa Cruz Biotechnology); rabbit anti-p-Neu (Tyr 1248)-R (i.e., p-ErbB-2) (Santa Cruz Biotechnology); rabbit anti-Neu (C-18) (i.e., ErbB-2) (Santa Cruz Biotechnology); rabbit anti-PI3 Kinase p85 (Cell Signaling); rat anti-MBP (Millipore); rabbit anti-p38α (Santa Cruz); rabbit anti-p38γ (R&D Systems); rabbit anti-calnexin (Sigma-Aldrich); mouse anti-β-tubulin (Sigma-Aldrich); rabbit anti-actin (Sigma-Aldrich). For immunofluorescence, secondary antibodies included fluorescein (FITC)-conjugated and rhodamine (TRITC)-conjugated donkey anti-mouse or rabbit IgG (Jackson ImmunoResearch). For western blotting, secondary antibodies included horseradish peroxidase (HRP)-conjugated goat anti-rabbit and rabbit anti-mouse immunoglobulins (Dako), and IRDye 800- and 680-conjugated goat anti-mouse, goat anti-rabbit, and goat anti-rat IgG (Li-Cor Biosciences). As negative control in immunoprecipitation experiments, ChromPure mouse IgG whole molecules were used (Jackson ImmunoResearch).
Animals
All experiments involving animals were performed in accordance with Italian national regulations and covered by experimental protocols reviewed by local Institutional Animal Care and Use Committees.
The pFlrt-1 vector, including lox-P sites, FRT-flanked neomycin resistance gene (neo), and PGK-TK, was used to target the Kif13b gene. The selected Kif13b mouse genomic regions to be inserted in the targeting vector were amplified from a BAC clone spanning the Kif13b gene and obtained from The Center for Applied Genomics (The Hospital for Sick Children, Ontario, Canada).
To generate the targeting vector for homologous recombination, a 503 bp BamHI fragment including exon 6 and flanking intronic regions was first inserted between lox-P sites in pFlrt-1. In a second step, a 4,606 bp fragment containing exon 7 was inserted into the BstBI site downstream of the PGK-neo cassette to constitute the long arm for homologous recombination. Finally, a fragment of 2,000 bp containing exon 5 was cloned into the SalI site upstream to the first lox-P and represented the short arm for homologous recombination.
After electroporation of TBV2 embyonic stem cells (129S2/SvPas), recombinant clones were screened by Southern blot analysis. Digestion with KpnI and hybridization with two probes designed on exon 6 (inside the recombination) and upstream of exon 5 (outside the 5′ end of the recombination) revealed two bands of 7,671 bp (wild type) and of 9,671 bp (containing the neo cassette).
Similarly, SmaI digestion of genomic DNA and hybridization using a probe designed at the 3′ end of the targeted region, outside the recombination boundaries, detected two bands of 7,694 bp (the targeted allele, since one SmaI restriction site is present within the neo cassette) and of 9,151 bp (the wild-type allele).
Two different correctly targeted clones were injected into C57BL6 blastocysts (Core Facility for Conditional Mutagenesis San Raffaele/Telethon Transgenic Service) to obtain transmission of the Floxed allele through the germline.
The neo cassette was removed in vivo by crossing heterozygous Kif13b Fl (neo)/+ with Flpe transgenic mice. Heterozygous Kif13b Fl/+ animals were crossed with P0-Cre [17,18] transgenic mice to excise exon 6 specifically in Schwann cells. To generate Kif13b Fl/Fl P0-Cre conditional knockout mice, Kif13b Fl/+ P0-Cre animals were crossed with homozygous Kif13b Fl/Fl. Kif13b Fl/Fl mouse nerves had normal myelin thickness and mean g-ratio values similar to wild-type mice, thus suggesting that Kif13b Fl/+ does not represent a hypomorphic allele.
To obtain Kif13b Fl/- CNP-Cre [31] mice with conditional inactivation of Kif13b in oligodendrocytes, Kif13b Fl/+ mice were first crossed with CMV-Cre transgenic mice. Then, after germline segregation of the CMV-Cre transgene, Kif13b -/+ (without CMV-Cre) were crossed with Kif13b Fl/+ CNP-Cre mice to obtain Kif13b Fl/- CNP-Cre conditional null. In this way, we increased CNP-Cre mediated recombination efficiency on the Floxed allele in the Kif13b Fl/- CNP-Cre genotype.
The Dlg1 Fl (C57/BL6 strain) allele has been already reported (Zhou et al., 2008). To generate Dlg1 conditional knockout mice in oligodendrocytes, homozygous Dlg1 Fl/Fl mice were crossed with heterozygous Dlg1 Fl/+ mice carrying the CNP-Cre transgene.
To obtain 50% reduction of Dlg1 specifically in Schwann cells in a Kif13b Fl/Fl P0-Cre background, Kif13b Fl/Fl P0-Cre mice were first crossed with Dlg1 Fl/Fl P0-Cre mice. Then, Kif13b Fl/+//Dlg1 Fl/+; P0-Cre double heterozygous mice were crossed to obtain Kif13b Fl/Fl//Dlg1 Fl/+; P0-Cre mice. These latter were compared with Kif13b Fl/Fl//Dlg1 +/+; P0-Cre mice and controls (only floxed alleles without Cre) within the same litters.
The generation of p38γ-null mice has been already reported [28].
For all the experiments involving animals, n ≥ 5 animals per genotype of either sex were analysed.
PCR Analysis
Genotype analysis on Kif13b mutant mice was carried out on tail genomic DNA using primer pairs A plus B (415 bp floxed band and 342 bp wild type band) or A plus C (966 bp floxed band, 834 bp wild type band, and 378 bp recombined band). Genotype analysis of the Dlg1 floxed allele and of the p38γ-null locus has already been reported [9,28].
RT-PCR was performed as described previously [7,9]. Designed probes were used to amplify mouse Kif13b and the endogenous reference transcript calnexin. The comparative Ct method was used. As calibrator, a control sample ΔCt was chosen for each selected transcript. The ΔΔCt (ΔCt of each normalized selected transcript minus ΔCt of the calibrator) was calculated. Expression levels of Kif13b mRNA are indicated as 2-ΔΔCt values. For statistical analysis, SD was calculated for triplicate samples of each reaction and SEM is indicated on the average of the determinations from different animals.
Morphological Analysis
Three to five animals per genotype for each time point were analysed. Semithin analysis of quadriceps and sciatic nerves and ultrastructural analysis of optic nerves and spinal cords were performed as described previously [62].
To perform morphometric analysis, digitalized images of fiber cross sections were obtained from corresponding levels of the quadriceps or sciatic nerves with a 100x objective and Leica DFC300F digital camera (Milan, Italy). Five images per animal were analysed with the Leica QWin software (Leica Microsystem) and the g-ratio calculated as the ratio between the mean diameter of an axon (without myelin) and the mean diameter of the same axon including the myelin sheath.
For morphometric analysis on ultrastructural sections, 20 images per animal were taken at 4000x (LEO 912AB Transmission Electron Microscope, Milan, Italy) and the g-ratio values determined by measuring axon and fiber diameters.
Internodal lengths were measured as described using Openlab (PerkinElmer) [19], and 100 internodes of two quadriceps nerves were evaluated for each animal (n = 3).
Sciatic Nerve Crush
Adult mice were anesthetized with avertin (trichloroethanol, 0.02 ml/g of body weight), and crush injury was performed as previously described [63]. After skin incision, the sciatic nerve was exposed and crushed distal to the sciatic notch for 20 s with fine forceps previously cooled in dry ice. To identify the site of injury, forceps were previously dropped into vital carbon. The nerve was replaced under the muscle and the incision sutured.
Preparation of Tissue Lysates
Protein lysates from mouse sciatic nerves, corpus callosum, optic nerves, and spinal cords for western blot analysis were prepared using a lysis buffer containing 2% SDS, 50 mM Tris buffer pH 8.0, 150 mM NaCl, 10 mM NaF, 1 mM NaVO3, and complete protease and phosphatase inhibitors (Roche). For the detection of phosphorylated antigens, samples were lysed with a buffer containing 1%TX-100. Protein quantification was performed using BCA assay (Pierce, Thermo Scientific).
Co-Immunoprecipitation
Mouse and rat sciatic nerves were lysed in a buffer containing 1% NP-40, 150 mM NaCl, 50 mM Tris buffer pH 8.0, 10 mM NaF, 1 mM NaVO3, and complete protease and phosphatase inhibitors (Roche). Following centrifugation at 13,000 rpm for 15 min at 4°C, equal amounts of protein lysates were incubated with 6–8 ug of mouse anti-Dlg1 antibody (Stressgen) or mouse IgG for control (Jackson ImmunoResearch). After 3 h of incubation with the antibody at 4°C, 35 μl of protein G agarose (settled) (Sigma-Aldrich) was added to immunocomplexes within the lysates and incubated for 1 h and 30 min at 4°C. The agarose beads were washed two times with cold PBS-Tween 0.1% and once with cold PBS. The immunoprecipitated product was denatured in Laemmly buffer (Biorad) with β-mercaptoethanol and resolved by SDS-PAGE.
Glutathione S-transferase- (GST) Binding Assays
Kif13b/MBS cDNA was cloned into pGEX-4T2 expression vector and expressed together with GST alone in Escherichia coli BL21(DE3) cells [7]. Recombinant proteins were purified directly from bacterial extract on glutathione-Sepharose 4 Fast Flow beads.
Rat sciatic and optic nerves were lysed in a buffer containing 1% NP-40, 50 mM Tris buffer pH 7.4, 10% glycerol, 100 mM NaCl, 10 mM NaF, and 1 mM NaVO3. Equal amounts of protein lysates were incubated for 4 h at 4°C with immobilized GST-Kif13b/MBS proteins and GST as control. After three washes with a buffer containing 0.5% NP-40, 50 mM Tris buffer pH 7.4, 10% glycerol, 100 mM NaCl, 10 mM NaF, and 1 mM NaVO3, the bead pellets were dissolved in Laemmly buffer with β-mercaptoethanol, resolved by SDS-PAGE, and analyzed by immunoblotting. To show the relative amount used of GST-Kif13B/MBS and GST, beads were dissolved again in Laemmly buffer with β-mercaptoethanol, resolved by SDS-PAGE, and the gels stained with Coomassie.
Western Blotting
SDS-PAGE gels were transferred to PVDF membranes (Millipore) or to nitrocellulose (Millipore) at 4°C in 20% methanol blotting buffer. Filters were blocked in 5% dry milk in PBS-0.1% Tween 20 overnight at 4°C and immunoblotted with primary antibodies diluted in 3% dry milk in PBS-0.1% Tween. For phosphorylated antigens, an additional blocking was performed for 30 min at RT in 3% bovine serum albumin (BSA) (Sigma-Aldrich), 0.5% gelatin, 0.1% Tween, 1 mM EDTA pH 8.0, 0.15 M NaCl, 10 mM Tris buffer pH 7.5, followed by incubation with primary antibodies diluted in the same blocking solution. Secondary antibodies, either horseradish peroxidase-conjugated (Dako) or IRDye 800- and 680-conjugated (Li-Cor Biosciences), were used and immunoblots revealed by using either ECL/ECL-prime developing systems and films for chemiluminescent detection (Amersham) or by Odyssey CLx Infrared Imaging System (Li-Cor Biosciences).
Statistical Analysis
Statistical analysis was performed using the Student t test, two tails, unequal variants, and α = 0.005 were considered. All results are shown as mean ± SEM.
Images
Figures were prepared using Adobe Photoshop version 11.0 (Adobe Systems).
Supporting Information
(XLSX)
(A) Nerve regeneration 1 mo after crush injury shows that Kif13b Fl/Fl P0-Cre nerves regenerate properly, as assessed by semithin section analysis. Only the number of big caliber axons was reduced in crushed Kif13b Fl/Fl P0-Cre nerves (as a percentage, Kif13b Floxed/+, 3.404 ± 0.865; Kif13b Fl/Fl P0-Cre, 0.883 ± 0.352, n = 5 animals per genotype, p = 0.027). (B) Quantification of the g-ratio as a function of the axonal diameter in crushed sciatic nerves shows that Kif13b Fl/Fl P0-Cre nerves are hypomyelinated 2 mo after injury (Kif13b Fl/Fl P0-Cre, 0.713 ± 0.009, 1924 fibers; Kif13b Fl/+, 0.681 ± 0.007,1944 fibers, n = 3 animals per genotype, p = 0.005).
(TIF)
(A) Expression levels with quantification of Nrg1-III, of ErbB2 (B), and of Erk phosphorylation (C) in Kif13b Fl/Fl P0-Cre sciatic nerves. (D) Expression levels with quantification of Krox20 and Oct6 (E) in Kif13b Fl/Fl P0-Cre sciatic nerves.
(TIF)
(A,B) The g-ratio as a function of axonal diameter in Kif13b Fl/- CNP-Cre optic nerves and spinal cords shows normal myelin thickness at P90 (optic nerve: Kif13b Fl/- CNP-Cre, 0.792 ± 0.007, 454 fibers; Kif13b Fl/+, 0.811 ± 0.006, 408 fibers, n = 5 animals per genotype, p = 0.077. Spinal cord: Kif13b Fl/- CNP-Cre, 0.787 ± 0.014, 228 fibers; Kif13b Fl/+, 0.789 ± 0.013, 218 fibers, n = 3 animals per genotype, p = 0.897). (C,D) The g-ratio as a function of axonal diameter in Dlg1 Fl/Fl CNP-Cre optic nerves and spinal cords shows normal myelin thickness at P90 (optic nerve: Dlg1 Fl/Fl CNP-Cre, 0.811 ± 0.008, 696 fibers; Dlg1 Fl/+, 0.815 ± 0.004, 734 fibers, n = 4 animals per genotype, p = 0.647. Spinal cord: Dlg1 Fl/Fl CNP-Cre, 0.792 ± 0.013, 403 fibers; Dlg1 Fl/+, 0.789 ± 0.002, 390 fibers, n = 3 animals per genotype, p = 0.855).
(TIF)
Immunoprecipitation of Dlg1 from mouse sciatic nerves at P20 followed by western blot analysis using an anti-p85 antibody shows that Dlg1 does not interact with p85 in Schwann cells. (B) GST pull down assay from P11 rat optic nerves using GST-Kif13b/MBS as a bait indicates that Dlg1 does not interact with p85 in Schwann cells, two independent experiments. (C) Expression levels of p85 in sciatic nerves of Kif13b Fl/Fl P0-Cre mutants and controls at P20, with quantification. (D) Expression levels of p85 in sciatic nerves of Dlg1 Fl/Fl P0-Cre mutants and controls at P20, with quantification. (E) Expression levels of p85 in sciatic nerves of both Kif13b Fl/Fl P0-Cre and Dlg1 Fl/Fl P0-Cre mutants at 5 mo, with quantification.
(TIF)
(A) GST pull down assay from P11 rat optic nerves performed using GST-Kif13b/MBS as a bait indicates that Dlg1 does not interact with p38α in rat optic nerves at P11 or (B) in sciatic nerves at P11. (C) Expression levels of p38α in both Kif13b Fl/Fl P0-Cre sciatic nerves and Kif13b Fl/Fl CNP-Cre spinal cords is similar to controls.
(TIF)
Acknowledgments
We thank Dr L. Wrabetz for the P0-Cre transgenic line and Dr. A. Christhi for antibodies. We are grateful to Dr. C. Taveggia for useful discussion and the critical reading of the manuscript.
Abbreviations
- AKT
v-AKT murine thymoma viral oncogene homolog
- Arf6
ADP-ribosylation factor
- CNS
central nervous system
- Dlg1
Discs large 1
- DRG
dorsal root ganglia
- GAKIN
guanylate kinase-associated kinesin
- GAP
GTPase activating protein
- GKAP
glucokinase-associated dual specificity phosphatase
- GUK
guanylate kinase homologue
- MAPK
mitogen-activated protein kinase
- MBP
myelin basic protein
- MBS
membrane-associated guanylate kinase homologue binding stalk
- MPZ
the myelin protein zero gene
- mTOR
mechanistic target of rapamycin
- PIP3
phosphatidylinositol-3,4,5-trisphosphate
- PI3K
phosphatidylinositol 3-kinase
- PNS
peripheral nervous system
- PTEN
phosphatase and tensin homolog
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
AB was supported by Telethon-Italy (GPP10007D and GPP12017), http://www.telethon.it; Association Française contre les Myopathies (AFM)-France, http://www.afm-telethon.com; the ERA-Net for research programs on rare diseases E-Rare 2 (E-RARE 11-040), http://www.erare.eu; and FISM, Fondazione Italiana Sclerosi Multipla (2010/R/8), http://www.aism.it/home.aspx. DLS and PJB were supported by the Wellcome Trust, http://www.wellcome.ac.uk. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Simons M, Trotter J (2007) Wrapping it up: the cell biology of myelination. Curr Opin Neurobiol 17: 533–540. [DOI] [PubMed] [Google Scholar]
- 2. Nave KA (2010) Myelination and support of axonal integrity by glia. Nature 468: 244–252. 10.1038/nature09614 [DOI] [PubMed] [Google Scholar]
- 3. Sherman DL, Brophy PJ (2005) Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci 6: 683–690. [DOI] [PubMed] [Google Scholar]
- 4. Eshed-Eisenbach Y, Peles E (2013) The making of a node: a co-production of neurons and glia. Current opinion in neurobiology 23: 1049–1056. 10.1016/j.conb.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zoupi L, Savvaki M, Karagogeos D (2011) Axons and myelinating glia: An intimate contact. IUBMB life 63: 730–735. 10.1002/iub.513 [DOI] [PubMed] [Google Scholar]
- 6. Lyons DA, Naylor SG, Scholze A, Talbot WS (2009) Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nature genetics 41: 854–858. 10.1038/ng.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bolis A, Coviello S, Visigalli I, Taveggia C, Bachi A, et al. (2009) Dlg1, Sec8, and Mtmr2 regulate membrane homeostasis in Schwann cell myelination. J Neurosci 29: 8858–8870. 10.1523/JNEUROSCI.1423-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cotter L, Ozcelik M, Jacob C, Pereira JA, Locher V, et al. (2010) Dlg1-PTEN interaction regulates myelin thickness to prevent damaging peripheral nerve overmyelination. Science 328: 1415–1418. 10.1126/science.1187735 [DOI] [PubMed] [Google Scholar]
- 9. Noseda R, Belin S, Piguet F, Vaccari I, Scarlino S, et al. (2013) DDIT4/REDD1/RTP801 is a novel negative regulator of Schwann cell myelination. The Journal of neuroscience: the official journal of the Society for Neuroscience 33: 15295–15305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Asaba N, Hanada T, Takeuchi A, Chishti AH (2003) Direct interaction with a kinesin-related motor mediates transport of mammalian discs large tumor suppressor homologue in epithelial cells. J Biol Chem 278: 8395–8400. [DOI] [PubMed] [Google Scholar]
- 11. Horiguchi K, Hanada T, Fukui Y, Chishti AH (2006) Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. The Journal of cell biology 174: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanai Y, Wang D, Hirokawa N (2014) KIF13B enhances the endocytosis of LRP1 by recruiting LRP1 to caveolae. The Journal of cell biology 204: 395–408. 10.1083/jcb.201309066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lamason RL, Kupfer A, Pomerantz JL (2010) The dynamic distribution of CARD11 at the immunological synapse is regulated by the inhibitory kinesin GAKIN. Molecular cell 40: 798–809. 10.1016/j.molcel.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xing BM, Yang YR, Du JX, Chen HJ, Qi C, et al. (2012) Cyclin-dependent kinase 5 controls TRPV1 membrane trafficking and the heat sensitivity of nociceptors through KIF13B. The Journal of neuroscience: the official journal of the Society for Neuroscience 32: 14709–14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanada T, Lin L, Tibaldi EV, Reinherz EL, Chishti AH (2000) GAKIN, a novel kinesin-like protein associates with the human homologue of the Drosophila discs large tumor suppressor in T lymphocytes. The Journal of biological chemistry 275: 28774–28784. [DOI] [PubMed] [Google Scholar]
- 16. Venkateswarlu K, Hanada T, Chishti AH (2005) Centaurin-alpha1 interacts directly with kinesin motor protein KIF13B. Journal of cell science 118: 2471–2484. [DOI] [PubMed] [Google Scholar]
- 17. Feltri ML, D'Antonio M, Previtali S, Fasolini M, Messing A, et al. (1999) P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann N Y Acad Sci 883: 116–123. [PubMed] [Google Scholar]
- 18. Feltri ML, D'Antonio M, Quattrini A, Numerato R, Arona M, et al. (1999) A novel P0 glycoprotein transgene activates expression of lacZ in myelin-forming Schwann cells. Eur J Neurosci 11: 1577–1586. [DOI] [PubMed] [Google Scholar]
- 19. Court FA, Sherman DL, Pratt T, Garry EM, Ribchester RR, et al. (2004) Restricted growth of Schwann cells lacking Cajal bands slows conduction in myelinated nerves. Nature 431: 191–195. [DOI] [PubMed] [Google Scholar]
- 20. Sherman DL, Wu LM, Grove M, Gillespie CS, Brophy PJ (2012) Drp2 and periaxin form Cajal bands with dystroglycan but have distinct roles in Schwann cell growth. The Journal of neuroscience: the official journal of the Society for Neuroscience 32: 9419–9428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Narayan N, Massimi P, Banks L (2009) CDK phosphorylation of the discs large tumour suppressor controls its localisation and stability. Journal of cell science 122: 65–74. 10.1242/jcs.024554 [DOI] [PubMed] [Google Scholar]
- 22. Massimi P, Narayan N, Cuenda A, Banks L (2006) Phosphorylation of the discs large tumour suppressor protein controls its membrane localisation and enhances its susceptibility to HPV E6-induced degradation. Oncogene 25: 4276–4285. [DOI] [PubMed] [Google Scholar]
- 23. Sherman DL, Krols M, Wu LM, Grove M, Nave KA, et al. (2012) Arrest of myelination and reduced axon growth when Schwann cells lack mTOR. The Journal of neuroscience: the official journal of the Society for Neuroscience 32: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Norrmen C, Figlia G, Lebrun-Julien F, Pereira JA, Trotzmuller M, et al. (2014) mTORC1 controls PNS myelination along the mTORC1-RXRgamma-SREBP-lipid biosynthesis axis in Schwann cells. Cell reports 9: 646–660. 10.1016/j.celrep.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 25. Norrmen C, Suter U (2013) Akt/mTOR signalling in myelination. Biochemical Society transactions 41: 944–950. 10.1042/BST20130046 [DOI] [PubMed] [Google Scholar]
- 26. Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 149: 274–293. 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y, Guo H, Kwan H, Wang JW, Kosek J, et al. (2007) PAR-1 kinase phosphorylates Dlg and regulates its postsynaptic targeting at the Drosophila neuromuscular junction. Neuron 53: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sabio G, Arthur JS, Kuma Y, Peggie M, Carr J, et al. (2005) p38gamma regulates the localisation of SAP97 in the cytoskeleton by modulating its interaction with GKAP. The EMBO journal 24: 1134–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laprise P, Viel A, Rivard N (2004) Human homolog of disc-large is required for adherens junction assembly and differentiation of human intestinal epithelial cells. The Journal of biological chemistry 279: 10157–10166. [DOI] [PubMed] [Google Scholar]
- 30. Mantovani F, Banks L (2003) Regulation of the discs large tumor suppressor by a phosphorylation-dependent interaction with the beta-TrCP ubiquitin ligase receptor. The Journal of biological chemistry 278: 42477–42486. [DOI] [PubMed] [Google Scholar]
- 31. Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, et al. (2003) Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet 33: 366–374. [DOI] [PubMed] [Google Scholar]
- 32. Flores AI, Narayanan SP, Morse EN, Shick HE, Yin X, et al. (2008) Constitutively active Akt induces enhanced myelination in the CNS. The Journal of neuroscience: the official journal of the Society for Neuroscience 28: 7174–7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bolis A, Coviello S, Bussini S, Dina G, Pardini C, et al. (2005) Loss of Mtmr2 phosphatase in Schwann cells but not in motor neurons causes Charcot-Marie-Tooth type 4B1 neuropathy with myelin outfoldings. J Neurosci 25: 8567–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fragoso G, Robertson J, Athlan E, Tam E, Almazan G, et al. (2003) Inhibition of p38 mitogen-activated protein kinase interferes with cell shape changes and gene expression associated with Schwann cell myelination. Experimental neurology 183: 34–46. [DOI] [PubMed] [Google Scholar]
- 35. Hossain S, de la Cruz-Morcillo MA, Sanchez-Prieto R, Almazan G (2012) Mitogen-activated protein kinase p38 regulates Krox-20 to direct Schwann cell differentiation and peripheral myelination. Glia 60: 1130–1144. 10.1002/glia.22340 [DOI] [PubMed] [Google Scholar]
- 36. Yang DP, Kim J, Syed N, Tung YJ, Bhaskaran A, et al. (2012) p38 MAPK activation promotes denervated Schwann cell phenotype and functions as a negative regulator of Schwann cell differentiation and myelination. The Journal of neuroscience: the official journal of the Society for Neuroscience 32: 7158–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haines JD, Fang J, Mushynski WE, Almazan G (2010) Mitogen-activated protein kinase activated protein kinase 2 (MK2) participates in p38 MAPK regulated control of oligodendrocyte differentiation. Glia 58: 1384–1393. 10.1002/glia.21014 [DOI] [PubMed] [Google Scholar]
- 38. Fragoso G, Haines JD, Roberston J, Pedraza L, Mushynski WE, et al. (2007) p38 mitogen-activated protein kinase is required for central nervous system myelination. Glia 55: 1531–1541. [DOI] [PubMed] [Google Scholar]
- 39. Bhat NR, Zhang P, Mohanty SB (2007) p38 MAP kinase regulation of oligodendrocyte differentiation with CREB as a potential target. Neurochemical research 32: 293–302. [DOI] [PubMed] [Google Scholar]
- 40. Chew LJ, Coley W, Cheng Y, Gallo V (2010) Mechanisms of regulation of oligodendrocyte development by p38 mitogen-activated protein kinase. The Journal of neuroscience: the official journal of the Society for Neuroscience 30: 11011–11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chung SH, Biswas S, Selvaraj V, Liu XB, Sohn J, et al. (2015) The p38alpha mitogen-activated protein kinase is a key regulator of myelination and remyelination in the CNS. Cell death & disease 6: e1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goldstein LS (2001) Molecular motors: from one motor many tails to one motor many tales. Trends in cell biology 11: 477–482. [DOI] [PubMed] [Google Scholar]
- 43. Hirokawa N, Niwa S, Tanaka Y (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68: 610–638. 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- 44. Bolino A, Bolis A, Previtali SC, Dina G, Bussini S, et al. (2004) Disruption of Mtmr2 produces CMT4B1-like neuropathy with myelin outfolding and impaired spermatogenesis. J Cell Biol 167: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hanada T, Lin L, Chandy KG, Oh SS, Chishti AH (1997) Human homologue of the Drosophila discs large tumor suppressor binds to p56lck tyrosine kinase and Shaker type Kv1.3 potassium channel in T lymphocytes. The Journal of biological chemistry 272: 26899–26904. [DOI] [PubMed] [Google Scholar]
- 46. Koh YH, Popova E, Thomas U, Griffith LC, Budnik V (1999) Regulation of DLG localization at synapses by CaMKII-dependent phosphorylation. Cell 98: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goebbels S, Oltrogge JH, Kemper R, Heilmann I, Bormuth I, et al. (2010) Elevated phosphatidylinositol 3,4,5-trisphosphate in glia triggers cell-autonomous membrane wrapping and myelination. The Journal of neuroscience: the official journal of the Society for Neuroscience 30: 8953–8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamada KH, Hanada T, Chishti AH (2007) The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry 46: 10039–10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mitew S, Hay CM, Peckham H, Xiao J, Koenning M, et al. (2014) Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience 276: 29–47. 10.1016/j.neuroscience.2013.11.029 [DOI] [PubMed] [Google Scholar]
- 50. Emery B (2010) Regulation of oligodendrocyte differentiation and myelination. Science 330: 779–782. 10.1126/science.1190927 [DOI] [PubMed] [Google Scholar]
- 51. Emery B (2010) Transcriptional and post-transcriptional control of CNS myelination. Curr Opin Neurobiol 20: 601–607. 10.1016/j.conb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 52. Nave KA (2010) Oligodendrocytes and the "micro brake" of progenitor cell proliferation. Neuron 65: 577–579. 10.1016/j.neuron.2010.02.026 [DOI] [PubMed] [Google Scholar]
- 53. Nave KA (2010) Myelination and the trophic support of long axons. Nature reviews Neuroscience 11: 275–283. 10.1038/nrn2797 [DOI] [PubMed] [Google Scholar]
- 54. Fancy SP, Chan JR, Baranzini SE, Franklin RJ, Rowitch DH (2011) Myelin regeneration: a recapitulation of development? Annual review of neuroscience 34: 21–43. 10.1146/annurev-neuro-061010-113629 [DOI] [PubMed] [Google Scholar]
- 55. Zuchero JB, Barres BA (2013) Intrinsic and extrinsic control of oligodendrocyte development. Curr Opin Neurobiol 23: 914–920. 10.1016/j.conb.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pereira JA, Lebrun-Julien F, Suter U (2012) Molecular mechanisms regulating myelination in the peripheral nervous system. Trends in neurosciences 35: 123–134. 10.1016/j.tins.2011.11.006 [DOI] [PubMed] [Google Scholar]
- 57. Wang H, Tewari A, Einheber S, Salzer JL, Melendez-Vasquez CV (2008) Myosin II has distinct functions in PNS and CNS myelin sheath formation. The Journal of cell biology 182: 1171–1184. 10.1083/jcb.200802091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chan JR, Watkins TA, Cosgaya JM, Zhang C, Chen L, et al. (2004) NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron 43: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palazuelos J, Crawford HC, Klingener M, Sun B, Karelis J, et al. (2014) TACE/ADAM17 is essential for oligodendrocyte development and CNS myelination. The Journal of neuroscience: the official journal of the Society for Neuroscience 34: 11884–11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. La Marca R, Cerri F, Horiuchi K, Bachi A, Feltri ML, et al. (2011) TACE (ADAM17) inhibits Schwann cell myelination. Nat Neurosci 14: 857–865. 10.1038/nn.2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B (2010) The emerging mechanisms of isoform-specific PI3K signalling. Nature reviews Molecular cell biology 11: 329–341. 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- 62. Wrabetz L, Feltri ML, Quattrini A, Imperiale D, Previtali S, et al. (2000) P(0) glycoprotein overexpression causes congenital hypomyelination of peripheral nerves. J Cell Biol 148: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Triolo D, Dina G, Lorenzetti I, Malaguti M, Morana P, et al. (2006) Loss of glial fibrillary acidic protein (GFAP) impairs Schwann cell proliferation and delays nerve regeneration after damage. Journal of cell science 119: 3981–3993. [DOI] [PubMed] [Google Scholar]
- 64. Nave KA, Salzer JL (2006) Axonal regulation of myelination by neuregulin 1. Curr Opin Neurobiol 16: 492–500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(A) Nerve regeneration 1 mo after crush injury shows that Kif13b Fl/Fl P0-Cre nerves regenerate properly, as assessed by semithin section analysis. Only the number of big caliber axons was reduced in crushed Kif13b Fl/Fl P0-Cre nerves (as a percentage, Kif13b Floxed/+, 3.404 ± 0.865; Kif13b Fl/Fl P0-Cre, 0.883 ± 0.352, n = 5 animals per genotype, p = 0.027). (B) Quantification of the g-ratio as a function of the axonal diameter in crushed sciatic nerves shows that Kif13b Fl/Fl P0-Cre nerves are hypomyelinated 2 mo after injury (Kif13b Fl/Fl P0-Cre, 0.713 ± 0.009, 1924 fibers; Kif13b Fl/+, 0.681 ± 0.007,1944 fibers, n = 3 animals per genotype, p = 0.005).
(TIF)
(A) Expression levels with quantification of Nrg1-III, of ErbB2 (B), and of Erk phosphorylation (C) in Kif13b Fl/Fl P0-Cre sciatic nerves. (D) Expression levels with quantification of Krox20 and Oct6 (E) in Kif13b Fl/Fl P0-Cre sciatic nerves.
(TIF)
(A,B) The g-ratio as a function of axonal diameter in Kif13b Fl/- CNP-Cre optic nerves and spinal cords shows normal myelin thickness at P90 (optic nerve: Kif13b Fl/- CNP-Cre, 0.792 ± 0.007, 454 fibers; Kif13b Fl/+, 0.811 ± 0.006, 408 fibers, n = 5 animals per genotype, p = 0.077. Spinal cord: Kif13b Fl/- CNP-Cre, 0.787 ± 0.014, 228 fibers; Kif13b Fl/+, 0.789 ± 0.013, 218 fibers, n = 3 animals per genotype, p = 0.897). (C,D) The g-ratio as a function of axonal diameter in Dlg1 Fl/Fl CNP-Cre optic nerves and spinal cords shows normal myelin thickness at P90 (optic nerve: Dlg1 Fl/Fl CNP-Cre, 0.811 ± 0.008, 696 fibers; Dlg1 Fl/+, 0.815 ± 0.004, 734 fibers, n = 4 animals per genotype, p = 0.647. Spinal cord: Dlg1 Fl/Fl CNP-Cre, 0.792 ± 0.013, 403 fibers; Dlg1 Fl/+, 0.789 ± 0.002, 390 fibers, n = 3 animals per genotype, p = 0.855).
(TIF)
Immunoprecipitation of Dlg1 from mouse sciatic nerves at P20 followed by western blot analysis using an anti-p85 antibody shows that Dlg1 does not interact with p85 in Schwann cells. (B) GST pull down assay from P11 rat optic nerves using GST-Kif13b/MBS as a bait indicates that Dlg1 does not interact with p85 in Schwann cells, two independent experiments. (C) Expression levels of p85 in sciatic nerves of Kif13b Fl/Fl P0-Cre mutants and controls at P20, with quantification. (D) Expression levels of p85 in sciatic nerves of Dlg1 Fl/Fl P0-Cre mutants and controls at P20, with quantification. (E) Expression levels of p85 in sciatic nerves of both Kif13b Fl/Fl P0-Cre and Dlg1 Fl/Fl P0-Cre mutants at 5 mo, with quantification.
(TIF)
(A) GST pull down assay from P11 rat optic nerves performed using GST-Kif13b/MBS as a bait indicates that Dlg1 does not interact with p38α in rat optic nerves at P11 or (B) in sciatic nerves at P11. (C) Expression levels of p38α in both Kif13b Fl/Fl P0-Cre sciatic nerves and Kif13b Fl/Fl CNP-Cre spinal cords is similar to controls.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.