Abstract
The spatial organization of chromatin is critical in establishing cell-type dependent gene expression programs. The inner nuclear membrane protein emerin has been implicated in regulating global chromatin architecture. We show emerin associates with genomic loci of muscle differentiation promoting factors in murine myogenic progenitors, including Myf5 and MyoD. Prior to their transcriptional activation Myf5 and MyoD loci localized to the nuclear lamina in proliferating progenitors and moved to the nucleoplasm upon transcriptional activation during differentiation. The Pax7 locus, which is transcribed in proliferating progenitors, localized to the nucleoplasm and Pax7 moved to the nuclear lamina upon repression during differentiation. Localization of Myf5, MyoD, and Pax7 to the nuclear lamina and proper temporal expression of these genes required emerin and HDAC3. Interestingly, activation of HDAC3 catalytic activity rescued both Myf5 localization to the nuclear lamina and its expression. Collectively, these data support a model whereby emerin facilitates repressive chromatin formation at the nuclear lamina by activating the catalytic activity of HDAC3 to regulate the coordinated spatiotemporal expression of myogenic differentiation genes.
Keywords: Emerin, HDAC3, Chromatin remodeling, Myogenesis, Nuclear organization nuclear, Lamina, Nucleoskeleton
Introduction
Three-dimensional organization of chromatin in the interphase nucleus plays a critical role in determining cell fate and coordinating gene expression programs in multicellular organisms. Defective chromatin architecture has emerged as an underlying cause of many genetic diseases, cancer, and aging (Misteli 2010). The nuclear envelope, encompassing the nuclear lamina, the inner and outer nuclear membranes, and nuclear pore complexes (NPCs), is fundamental to maintaining chromatin architecture and tissue-specific gene expression (Van De Vosse et al. 2010). Dynamic association of chromatin with the nuclear lamina is essential for the coordinated temporal regulation of gene expression during development and differentiation to maintain tissue homeostasis (Peric-Hupkes and van Steensel 2011). Currently, the molecular mechanisms regulating establishment, maintenance, and dynamics of repressive chromatin at the nuclear envelope are unclear.
The nuclear envelope is composed of two lipid bilayers: the outer nuclear membrane (ONM), which is contiguous with the endoplasmic reticulum, and the inner nuclear membrane (INM; Wilson and Berk 2010). Emerin, one of more than 100 putative integral membrane proteins of the INM, interacts with A- and B-type lamins, type V intermediate filament proteins underlying the inner nuclear membrane (Berk et al. 2013). Together with other integral membrane proteins, emerin and lamins form a nuclear envelope-associated protein mesh-work named the nuclear lamina (Simon and Wilson 2013). Emerin binds to a wide range of chromatin-associated proteins, transcription regulators, and nucleoskeletal components (Holaska et al. 2003; 2004; 2006; Holaska and Wilson 2007; Lee et al. 2001; Salpingidou et al. 2007). Emerin and lamins together form many diverse complexes with roles ranging from mRNA transcription and splicing to DNA replication, mechanotransduction, chromatin remodeling, and nuclear architecture (Berk et al. 2013; Holaska 2008).
Mutations in emerin cause X-linked Emery–Dreifuss Muscular Dystrophy (X-EDMD), a disease characterized by tendon contracture, skeletal muscle wasting, cardiac conduction defects, and fatal irregular heart rhythms (Muchir and Worman 2007). While the molecular pathogenesis of X-EDMD is still unknown, recent evidence suggests the skeletal muscle defects are caused by the inability to properly regenerate skeletal muscle. For example, skeletal muscle from both X-EDMD patients and mice lacking emerin show increased expression of muscle regeneration pathway components, suggesting repression of these genes normally requires emerin (Bakay et al. 2006; Melcon et al. 2006). Emerin-downregulated myoblasts also exhibit impaired differentiation (Frock et al. 2006; Huber et al. 2009). Further, signaling pathways important for skeletal muscle regeneration were disrupted in emerin-null myogenic progenitors, including Wnt, IGF-1, TGF-β, and Notch (Koch and Holaska 2012).
Muscle regeneration is a multi-step process that repairs damaged muscle (Karalaki et al. 2009). Upon injury, satellite cells, also called muscle stem cells or myogenic progenitors, are activated and begin proliferating (Zammit et al. 2006a). A small fraction of these cells retain the satellite cell gene expression program and replenish their niche while activated myogenic progenitors initiate the myogenic differentiation program. Activated satellite cells will differentiate further and fuse to the injured myofiber to repair the skeletal muscle damage. The fate of myogenic progenitors is controlled by a complex interplay between the paired-box transcription factors Paired Box 3 (Pax3) and Paired Box 7 (Pax7), which regulate muscle stem cell specification and activation, and the muscle regulatory transcription factors Myogenic Determination Factor 1 (MyoD) and Myogenic Regulatory Factor 5 (Myf5), which regulate later stages of myogenic differentiation (Rudnicki et al. 2008). Pax3 and Pax7 regulate skeletal muscle formation in the embryo, and pools of Pax7+/Pax3+ cells replenish injured skeletal muscle tissue throughout life, activating MyoD and Myf5 expression by binding directly to their promoters and cis-regulatory elements (Lagha et al. 2008). MyoD and Myf5 bind to E-box sequences in promoters of downstream myogenic differentiation genes such as myogenin and myosin heavy chain, there-by committing the cell to terminal differentiation into a skeletal muscle fiber (Cao et al. 2010; Kuang et al. 2008). While the complete transcriptional network controlling muscle differentiation and regeneration is complex, the paradigm of Pax3 and Pax7 maintaining a proliferative undifferentiated state and subsequent activation of MyoD and Myf5 committing myogenic progenitors to terminal differentiation is well established (Wang and Rudnicki 2011; Zammit et al. 2006b).
The nuclear lamina is an essential regulator of genomic organization and chromatin architecture and is important for ensuring spatiotemporally coordinated gene expression during differentiation. Whole-genome studies indicate that nearly 40 % of the genome associates with the nuclear lamina across many cell types and that those regions, called lamina-associated domains (LADs), are generally transcribed at lower rates than non-LADs (Guelen et al. 2008; Pickersgill et al. 2006). These large regions (0.1–10 Mb) of the genome are characterized by repressive chromatin marks such as H3K27me3 and H3K9me2, high repeat density, and low gene density (Ikegami et al. 2010; Peric-Hupkes et al. 2010). Components of the nuclear lamina mediate the establishment of repressive chromatin, as the expression of genes repositioned to the lamina is repressed (Finlan et al. 2008; Reddy et al. 2008). The association of genomic regions with the nuclear lamina is dynamic in response to differentiation cues and external stimuli (Mehta et al. 2010; Peric-Hupkes et al. 2010). These dynamics are accompanied by coordinate changes in chromatin modifications consistent with transcriptional activation upon dissociation from the lamina (Kind et al. 2013; Peric-Hupkes and van Steensel 2011).
Multiple lines of evidence support emerin’s role in regulating chromatin architecture at the nuclear lamina. Primary fibroblasts from X-EDMD patients have significantly less heterochromatin at the nuclear lamina and have a markedly relaxed chromatin architecture (Meaburn et al. 2007; Mewborn et al. 2010; Ognibene et al. 1999). We recently showed that emerin binds to histone deacetylase 3 (HDAC3) and that emerin directly activates its histone deacetylase activity (Demmerle et al. 2012). Emerin-null myogenic progenitors exhibit higher levels of active histone marks such as H4K5 acetylation, and lower levels of repressive marks such as H3K27 trimethylation, indicating loss of emerin causes a more relaxed chromatin architecture (Demmerle et al. 2012). Given the changes in chromatin architecture in emerinnull myogenic progenitors, we hypothesized that localization of myogenic regulatory loci to the nuclear lamina would be disrupted in emerin-null myogenic progenitors throughout differentiation.
3D DNA-ImmunoFISH was used to interrogate the sub-nuclear positioning of myogenic regulatory loci in proliferating and differentiating wildtype or emerin-null myogenic progenitors to address the role of emerin in genomic organization during myogenic differentiation. Both emerin and HDAC3 were required for the coordinated temporal localization of MyoD, Myf5, and Pax7 to the nuclear periphery during differentiation; failure to establish proper localization of these loci resulted in aberrant expression of these critical myogenic genes. Localization of Myf5 to the nuclear lamina and repression of Myf5 was rescued in emerin-null myogenic progenitors by targeted activation of HDAC3 catalytic activity. These results show that emerin and HDAC3 promote repressive chromatin architecture at the nuclear periphery, which is required for coordinating the spatial dynamics and temporal expression of myogenic regulatory loci during differentiation.
Materials and methods
Cell culture
Wildtype and emerin-null H2K mouse myogenic progenitors were a generous gift from Tatiana Cohen and Terry Partridge (Children’s National Medical Center, Washington, DC) and were isolated as previously described (Koch and Holaska 2012). Proliferating myogenic progenitors were grown in proliferative media consisting of complete high-glucose DMEM vitrogen) supplemented with 20 % heat-inactivated FBS (Invitrogen), 2 % chick embryo extract (Accurate Chemical), 2 % l-glutamine (Invitrogen), 1 % penicillin–streptomycin (Invitrogen) and 20 units/ml γ-interferon (Millipore). Proliferating cells were plated at a density of ~650 cells/cm2 and grown at 33 °C and 10 % CO2. Differentiating cells were plated at a density of 25,000 cells/cm2 in proliferative conditions for 24 h (Day 0), then switched to differentiation media consisting of DMEM supplemented with 5 % horse serum (Invitrogen) and 2 % l-glutamine, and grown at 37 °C and 5 % CO2. Cells between passages 5 and 8 were used for all analyses. GFP-emerin- and GFP-S54F-emerin-containing constructs were electroporated into cells as previously described (Koch and Holaska 2012).
Antibodies
For a complete list of the antibodies used in FISH, ChIP, immunofluorescence, and Western Blot experiments, see Supplemental Table S4.
Chromatin Immunoprecipitation
Ten million proliferating H2K myogenic progenitors were cultured, trypsinized, centrifuged at 160×g for 5 min and resuspended in 10 ml proliferative media. Protein was cross-linked with 1 % formaldehyde at room temperature for 10 min, and then quenched with glycine for 5 min at room temperature. Cross-linked cells were centrifuged at 160×g for 5 min, media was aspirated and the pellet was resuspended in 1 mL PBS containing protease inhibitors (Pierce). Cells were centrifuged at 800×g for 5 min at 4°C, PBS was aspirated, the pellet was resuspended in 500 μl SDS lysis buffer (1 % SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.0) with protease inhibitors, and cells were incubated on ice for 10 min. Lysed cells were sonicated on ice for 40 pulses of 20 s, with a 30 s pause between pulses using a FB 120 sonicator (Fisher). Lysates were then centrifuged at 13,000×g for 10 min at 4°C to pellet insoluble material, and the supernatant was separated into 50 μL aliquots. One aliquot was combined with 450 μL ChIP dilution buffer (0.01 % SDS 1.1 % Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl, 167 mM NaCl) for each ChIP. 5 μl of the diluted sample was removed as input. For each ChIP, 10 μg emerin, 2–3 μg RNAPII, or 1 μg IgG antibody was added to the sample with 20 μl of Magna ChIP protein A+G magnetic beads (Millipore). Samples were rotated overnight at 4 °C. Beads were collected using a magnet, the supernatant was aspirated, and each sample was washed four times with RIPA buffer (150 mM NaCl, 1 % NP40, 0.5 % sodium deoxycholate, 0.1 % SDS, 50 mM Tris–HCl) for 3 min at 4 °C with rotation and once with TE (10 mM Tris–HCl, 1 mM EDTA, pH 8.0). Washed pellets and 1 % input samples were resuspended in 100 μL ChIP elution buffer (50 mM NaHCO3, 1 % SDS, 200 mM NaCl) with 10 ng proteinase K (5 Prime) and incubated with shaking at 62 °C and 1,000 rpm for 4 h. Samples were incubated at 95 °C for 10 min to inactivate proteinase K and cooled to room temperature. Beads were separated using a magnet and the supernatant transferred to a new tube for purification. DNA was recovered from the ChIP and input samples using the QIAquick PCR purification kit (Qiagen) per manufacturer instructions. qPCR was performed as described on selected gene promoters using primers listed in Supplemental Table S1.
shRNA knockdown
Myogenic progenitors were transduced with lentiviral particles of copGFP control (sc-108084, Santa Cruz) and either scrambled shRNA (sc-108080, Santa Cruz) or HDAC3 shRNA (sc-35539-V, Santa Cruz) at an MOI of 80 per manufacturer’s instructions in the presence of 5 μg/ml Polybrene (Santa Cruz). Cells were expanded for 5 days under proliferative conditions prior to fixation. Cells for imaging were selected based on their expression of GFP.
Western Blots
Equal numbers of cells was separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked for 1 h in 3 % BSA in PBS containing 0.1 % Tween 20 (PBS-T) followed by incubation for 2 h at room temperature or overnight at 4 °C with the indicated primary antibodies in 3 % BSA in PBS-T. Membranes were washed 5 times in PBS-T and placed in the appropriate secondary antibody (Thermo Scientific #31430 or #31460) at a concentration of 1:10,000 for 2 h at room temperature. Antibody binding was assayed using enhanced chemiluminescence detection reagent (GE Healthcare). Images were scanned and band intensities were determined using Quantity One software (BioRad).
Quantitative Real-Time PCR
RNA was isolated from proliferating or terminally differentiated wildtype or emerin-null myogenic progenitors using the RNeasy Kit (Qiagen) per manufacturer instructions. cDNA was generated from mRNA using MMLV Reverse Transcriptase (Invitrogen) per manufacturer instructions. qPCR was performed using a combination of individual primer sets for analysis of mRNA expression or ChIP enrichment (Supplementary Tables S1 and S2, respectively) using SYBR GreenER qPCR SuperMix (Invitrogen) on a BioRad iQ5 machine in triplicate. Three biological replicates were analyzed for all experiments.
Theophylline Treatment
Proliferating myogenic progenitors were treated with H2O alone or 10 μM theophylline (PHR1023, Sigma) in H2O for 4 h prior to fixation. Experiments were blinded and cells were selected for acquisition with the random selection algorithm RanCoo, a generous gift from Nadine Schroden (Memorial Sloan-Kettering Cancer Center, New York, NY).
3D DNA-ImmunoFISH
Immunofluorescence coupled with fluorescent DNA in situ hybridization on preserved nuclei (3D DNA-ImmunoFISH) was performed as described previously (Reddy et al. 2008; Solovei et al. 2002) with some modifications specific for myogenic precursors (See Supplemental Material—Extended Methods). All BACs (Supplementary Table S3) were obtained from the BACPAC Resource Center at Children’s Hospital Oakland Research Institute (Oakland, CA).
Imaging and Analysis
Images were obtained using 1.40 NA objectives (×63.0 or ×100) with Nyquist sampling and pixel sizes <50 nm on a TCS SP5-II laser-scanning confocal microscopy system (Leica Microsystems, GmbH). For 3D acquisition, Z-steps did not exceed 0.2 μm. Randomly selected nuclei were analyzed only when clear lamin staining was visible, and two foci in the 488 or 549 nm channels corresponding to biotin- or digoxigenin-labeled probes, respectively, were observed within the nucleus. Distances between foci and the nuclear lamina were calculated by creating a histogram of fluorescence intensities over a line orthogonal to the lamin signal using the region-of-interest tool in Leica’s ASF software. The distance between the intensity peaks, rounded to the nearest 0.1 μm, was recorded. Distributions of loci were compared using the Kolgomorov–Smirnov Test calculator at http://www.physics.csbsju.edu/stats/KS-test.html. Distributions were significant to p≤0.05 unless otherwise noted. Images were processed in FIJI and Adobe Illustrator.
Results
Association of myogenic regulatory loci with emerin
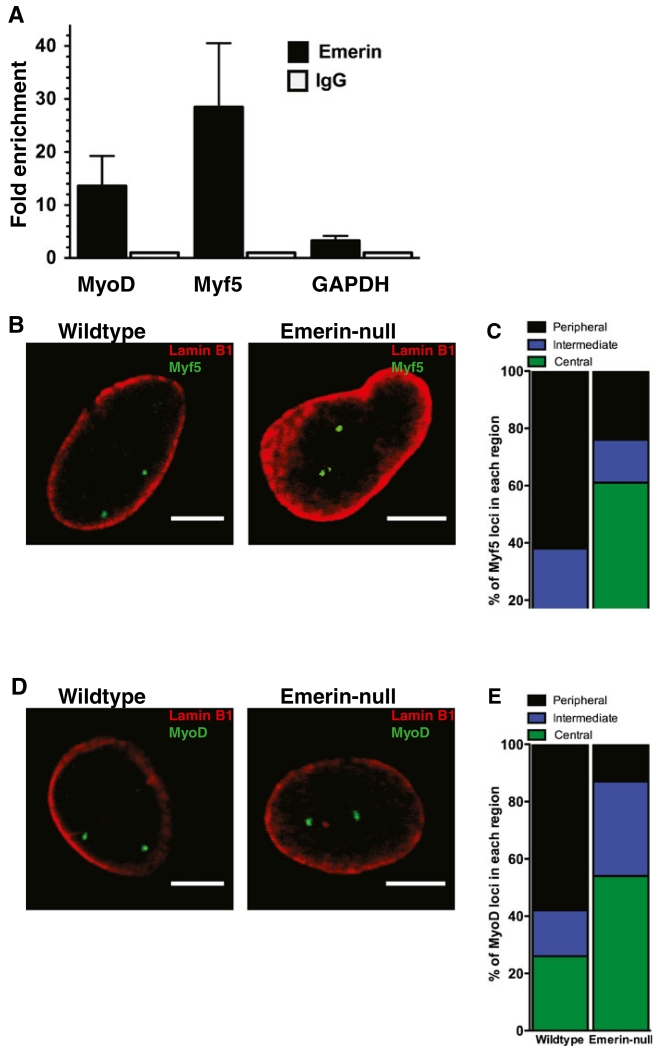
Previous studies have suggested the myogenic regulatory loci Myf5 and MyoD localize to the nuclear periphery in proliferating C2C12 myoblasts concomitant with their repression (Lee et al. 2006; Yao et al. 2011). Whether these loci interacted with emerin to ensure their nuclear envelope localization was tested, as previous evidence suggests emerin contributes to maintenance of repressive chromatin architecture. Chromatin immunoprecipitation (ChIP) with antibodies against emerin in wildtype primary mouse myogenic progenitors was done to test if MyoD and Myf5 loci associate with emerin. Emerin was enriched 4.5- and 11.5-fold relative to IgG at the promoters of MyoD and Myf5, respectively (Fig. 1a). Visualization of the Myf5 locus by 3D DNA-ImmunoFISH showed 63 and 84 % of Myf5 loci were within 1 and 2 μm of the nuclear lamina, respectively, in wildtype myogenic progenitors (Fig. 1b, c). Staining for lamin B1 was used to define the nuclear lamina. The MyoD locus also preferentially localized to the nuclear periphery in wildtype myogenic progenitors, with 58 and 74 % of MyoD loci within 1 and 2 μm of the lamina, respectively (Fig. 1d, e), consistent with previous results (Lee et al. 2006; Yao et al. 2011). Emerin-null myogenic progenitors had only 25 and 39 % of Myf5 loci within 1 and 2 μm of the lamina, respectively (Fig. 1b, c). Only 13 and 46 % of MyoD loci localized within 1 and 2 μm of the nuclear lamina, respectively (Fig. 1d, e). These findings indicate emerin is required for the nuclear lamina positioning of these myogenic regulatory genes, since based on the size of myogenic progenitor nuclei random localization to the nuclear periphery would result in 28 % of loci within 1 μm of the nuclear periphery.
Fig. 1.
Emerin binds to myogenic regulatory factor loci and regulates their association with the nuclear lamina. a ChIP-qPCR of MyoD and Myf5 promoters with antibodies against emerin or IgG in wildtype proliferating myogenic progenitors. Fold-enrichment was determined by comparing emerin to IgG. Error bars represent S.E.M. n=3. b 3D-ImmunoFISH showing the localization of Myf5 in proliferating wildtype and emerin-null myogenic progenitors. Lamin B1 staining is red and Myf5 is green. c Quantification of localization data for Myf5 in proliferating myogenic progenitors. n=50. Peripheral is ≤1 μm from the nuclear lamina. Intermediate is between 1 μm and 2 μm from the nuclear lamina. Central is ≥2 μm from the nuclear lamina. d 3D-ImmunoFISH showing the localization of MyoD in proliferating wildtype and emerin-null myogenic progenitors. Lamin B1 staining is red and MyoD is green. e Quantification of localization data for MyoD in proliferating myogenic progenitors. n=50. Peripheral is ≤1 μm from the nuclear lamina. Intermediate is between 1 μm and 2 μm from the nuclear lamina. Central is ≥2 μm from the nuclear lamina. Scale bars are 5 μm.
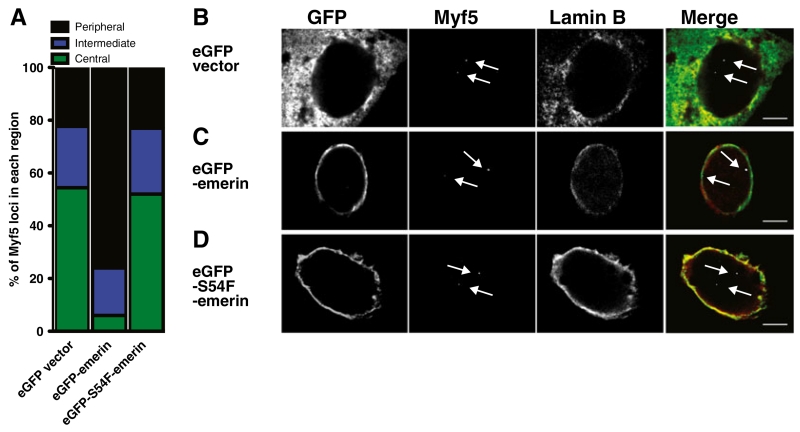
Emerin-null myogenic progenitors were electroporated with wildtype emerin-containing constructs to confirm emerin mediates the localization of Myf5 to the nuclear periphery. In eGFP-emerin-expressing cells, 77 % of Myf5 loci were within 1 μm of the nuclear lamina (Fig. 2a, c). eGFP-expressing emerin-null cells had only 27 % of Myf5 loci within 1 μm of the nuclear lamina (Fig. 2a, b), a similar distribution to untransfected emerin-null myogenic progenitors (Fig 1b). Emerin mutant S54F, an emerin mutant that fails to bind HDAC3 and activate HDAC3 activity (Demmerle et al. 2012), failed to rescue Myf5 localization (Fig. 2a, d). The inability of the emerin S54F mutant to rescue Myf5 localization suggests emerin binding to or activation of HDAC3, or both, mediates the association of myogenic loci with the nuclear lamina.
Fig. 2.
Association of Myf5 with the nuclear lamina is dependent on emerin. a Quantification of FISH data from emerin-null proliferating myogenic progenitors transfected with either eGFP, an N-terminal eGFP-emerin fusion or eGFP-S54F emerin mutant. n=50 for each class. p<0.05 when comparing eGFP-emerin to either vector or S54F-emerin. Peripheral is ≤1 μm from the nuclear lamina. Intermediate is between 1 μm and 2 μm from the nuclear lamina. Central is ≥2 μm from the nuclear lamina. b 3D-ImmunoFISH of eGFP transfected myogenic progenitors. c 3D-ImmunoFISH of eGFP-emerin transfected myogenic progenitors. d 3D-ImmunoFISH of eGFP-S54F-emerin transfected myogenic progenitors. b–d Lamin B1 is red, eGFP is green, Myf5 is white. Arrows indicate Myf5 loci. Scale bars are 5 μm
Sub-nuclear positioning of myogenic regulatory loci during differentiation is emerin-dependent
Pax7 localization was examined by 3D DNA-ImmunoFISH to test if the Pax7 locus is recruited to the nuclear periphery upon transcriptional repression during differentiation. In proliferating wildtype myogenic progenitors, 57 % of Pax7 loci were more than 2 μm away from the nuclear periphery, while only 9 % of loci localized to within 1 μm of the nuclear periphery (Fig 3a, b). The nucleoplasmic distribution of Pax7 in proliferating myogenic progenitors was not altered in emerin-null cells, as expected (Fig. 3a, b). After day 1 of differentiation (48 h after plating at high density and 24 h after serum withdrawal), when Pax7 is repressed, 53 % of Pax7 loci localized within 1 μm of the nuclear periphery (Fig. 3a, b). In contrast, emerin-null myogenic progenitors had only 11 % of Pax7 loci within 1 μm of the nuclear periphery after 1 day of differentiation (Fig. 3a, b). Emerin-null cells differentiated for 1 day showed a 12-fold increase in Pax7 mRNA expression (Fig. 3e). Additionally, Myf5 mRNA was upregulated 3.1-fold in proliferating emerin-null myogenic progenitors (Fig. 3e). Thus, aberrant temporal nuclear lamina localization of Myf5 and Pax7 are concomitant with transcriptional activation of Myf5 and Pax7.
Fig. 3.
Emerin regulates changes in genomic architecture during myogenic differentiation. a 3D-ImmunoFISH of Pax7 localization in proliferating and differentiating wildtype and emerin-null myogenic progenitors. Lamin B1 is red, Pax7 is green. Scale bars are 5 μm. b Quantification of localization data for Pax7. p<0.05 when comparing wildtype localization at differentiation day 1 to proliferating cells or emerin-null differentiation day 1; n=50. Peripheral is ≤1 μm from the nuclear lamina. Intermediate is between 1 μm and 2 μm from the nuclear lamina. Central is ≥2 μm from the nuclear lamina. (c) 3D-ImmunoFISH of MyoD and Myf5 localization in wildtype and emerin-null myogenic progenitors at differentiation day 1. Lamin B1 is red and MyoD or Myf5 is green. Scale bars are 5 μm. d Quantification of localization data for MyoD and Myf5 in wildtype and emerin-null myogenic progenitors at day 1 of differentiation. Peripheral is ≤1 μm from the nuclear lamina. Intermediate is between 1 μm and 2 μm from the nuclear lamina. Central is ≥2 μm from the nuclear lamina. e qPCR of Myf5 and Pax7 mRNA in emerin-null proliferating or differentiation day 1 myogenic progenitors relative to wildtype proliferating or differentiation day 1 myogenic progenitors, respectively, normalized to GAPDH. Error bars represent S. E. M. n=3. f Percentage of MyoD, Myf5, Pax3, and Pax7 loci in wildtype myogenic progenitors within 1 μm of the nuclear lamina throughout differentiation. Dotted line represents the percentage of loci expected within 1 μm of the nuclear lamina by chance. g Percentage of MyoD, Myf5, Pax3, and Pax7 loci in emerin-null myogenic progenitors within 1 μm of nuclear lamina throughout differentiation. Dotted line represents percentage of loci expected within 1 μm of the nuclear lamina by chance
To further define the temporal localization of myogenic regulatory loci, movement of Myf5, MyoD, Pax7, and Pax3 loci was analyzed every day during differentiation until terminally differentiated myotubes were formed (day 3). Myf5 and MyoD moved into the nuclear interior after day 0 of differentiation (Fig. 3c, d), concomitant with activation of Myf5 and MyoD expression (Wang and Rudnicki 2011). While day 0 is 24 h after plating at high density (2.5×104 cells/cm2 compared to 6.5×102 cells/cm2 while proliferating), it is prior to serum withdrawal to induce differentiation. Normal culture conditions are used during day 0 and cells are considered at day 1 of differentiation once they have spent 24 h in differentiation conditions and 48 h at high density. MyoD and Myf5 stayed in the nucleoplasm throughout differentiation, as expected (Fig. 3f). The absence of emerin does not affect the sub-nuclear positioning of MyoD and Myf5 loci once they are in the nuclear interior (Fig. 3c, d, g). Pax7 and Pax3 moved from the nuclear interior to the nuclear periphery upon differentiation, concurrent with their downregulation (Wang and Rudnicki 2011). MyoD, Myf5, Pax3 and Pax7 loci were randomly distributed within the nucleus throughout differentiation of emerin-null progenitors, as denoted by the dashed line indicating the random localization of these loci based on the size of these nuclei (28 %; Fig. 3g).
HDAC3 is required for Myf5 localization to the nuclear periphery
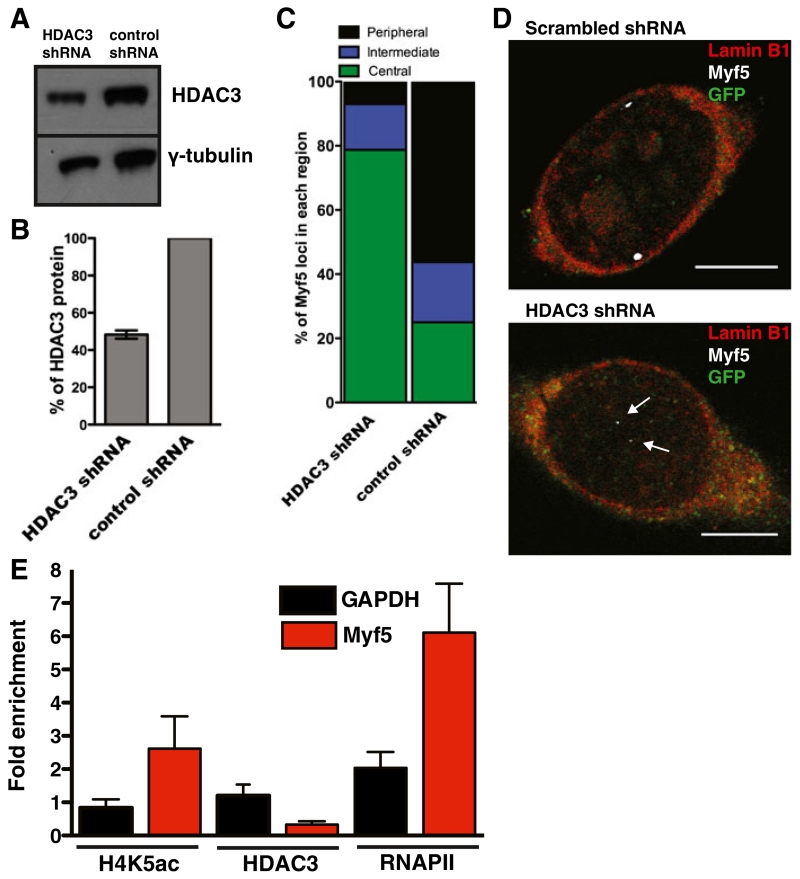
HDAC3 was downregulated in wildtype myogenic progenitors to test if HDAC3 was required for localization of these loci to the nuclear envelope, since emerin activates HDAC3 activity (Demmerle et al. 2012) and HDAC3 downregulation decreased association of LADs with the nuclear periphery (Zullo et al. 2012). HDAC3 was downregulated by 50 % in myogenic progenitors (Fig. 4a, b). 3D DNA-ImmunoFISH of Myf5 was performed in cells expressing HDAC3 shRNA or scrambled shRNA co-transduced with control constructs expressing GFP; 58 and 77 % of Myf5 loci localized within 1 and 2 μm of the nuclear lamina, respectively, in progenitors treated with scrambled shRNA (Fig. 4c, d). Only 8 and 22 % of Myf5 localized within 1 and 2 μm of the nuclear lamina, respectively, in HDAC3-downregulated progenitors (Fig. 4c, d).
Fig. 4.
HDAC3 knockdown reduces localization of Myf5 with the nuclear lamina. a Wildtype myogenic progenitors treated with control or HDAC3 shRNA were separated by SDS-PAGE and western blotted with antibodies against HDAC3 and γ-tubulin. b Quantification of HDAC3 levels in HDAC-downregulated progenitors was normalized to γ-tubulin and control shRNA-treated cells. Error bars are S.E.M. n=3. c Quantification of Myf5 localization in control and HDAC3-downregulated myogenic progenitors. n=50. p<0.05. Peripheral is ≤1 μm from the nuclear lamina. Intermediate is between 1 μm and 2 μm from the nuclear lamina. Central is ≥2 μm from the nuclear lamina. d 3D-ImmunoFISH of Myf5 localization in control and HDAC3-downregulated myogenic progenitors co-transfected with GFP. GFP is green, Lamin B1 is red, Myf5 is white. Arrows indicate Myf5 loci. Scale bars are 5 μm. e ChIP-qPCR of Myf5 and GAPDH promoters with antibodies against RNA Polymerase II (RNAPII), H4K5ac, and HDAC3 in wildtype and emerin-null proliferating myogenic progenitors. Fold-enrichment was determined by comparing emerin-null cells to wildtype. p≤0.05 between wildtype and emerin-null for all three antibodies. Error bars represent S.E.M. n=3
Based on these results, we predicted that occupancy of HDAC3 at the Myf5 promoter would be decreased. HDAC3 is the catalytic subunit of the Nuclear Co-Repressor Complex (NCoR), which preferentially deacetylates lysine 5 of histone H4 (H4K5ac). Thus, we predicted H4K5ac levels at the Myf5 promoter would be increased in emerin-null cells. ChIP of the Myf5 and GAPDH promoters was done using antibodies against HDAC3 and H4K5ac in wildtype and emerin-null proliferating myogenic progenitors. HDAC3 binding to the Myf5 promoter decreased 70 % and H4K5ac and RNA Polymerase II showed 3.5- and 5.5-fold increases, respectively, in emerin-null progenitors (Fig. 4e). Enrichment of HDAC3, H4K5ac, and RNA Polymerase II were unchanged at the GAPDH promoter in emerin-null cells (Fig. 4e).
Activation of HDAC3 by theophylline restores lamina localization of Myf5
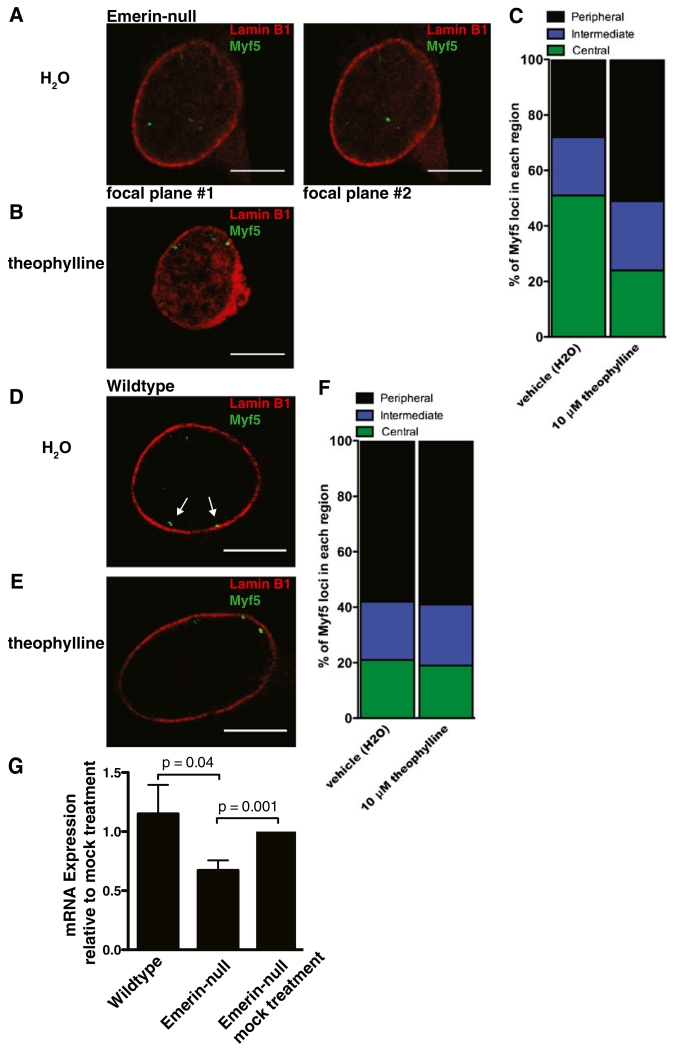
It was unclear from these studies if localization of myogenic regulatory genes to the nuclear lamina was mediated by emerin binding to HDAC3 at the nuclear lamina or by emerin stimulating HDAC3 catalytic activity, or both. Theophylline was used to test this hypothesis, as it is a small molecule activator of HDAC3 catalytic activity (Barnes 2003). Theophylline was previously shown to act as a phosphodiesterase inhibitor and adenosine-receptor antagonist at high concentrations (100 μM), but its ability to activate HDAC3 activity at low concentrations (10 μM) was discovered only in the last decade (Ito et al. 2002). 3D DNA-ImmunoFISH of the Myf5 locus was performed on emerin-null myogenic progenitors treated with a vehicle control (H2O) or 10 μM theophylline. In the vehicle control, 27 and 44 % of Myf5 loci localized within 1 and 2 μm of the nuclear lamina, respectively (Fig. 5a, c). In emerin-null myogenic progenitors treated with 10 μM theophylline for 4 h, 46 and 76 % of Myf5 loci were localized within 1 and 2 μm of the nuclear lamina, respectively (Fig. 5b, c). In wildtype myogenic progenitors treated with the vehicle control 58 and 79 % of Myf5 loci localized within 1 and 2 μm of the nuclear lamina, respectively (Fig. 5d, f). There was no significant difference in wildtype myogenic progenitors treated with 10 μM theophylline for 4 h, as 59 and 81 % of Myf5 loci localized within 1 and 2 μm of the nuclear lamina, respectively (Fig. 5e, f). Theophylline treatment repositioned the Myf5 locus to the nuclear lamina in emerin-null myogenic progenitors with a concomitant 0.33-fold reduction in Myf5 mRNA expression (Fig 5g); wildtype myogenic progenitors were unaffected.
Fig. 5.
Stimulation of HDAC3 activity restores association of Myf5 with the nuclear lamina and reduces Myf5 mRNA expression. a 3D-ImmunoFISH analysis of emerin-null proliferating myogenic progenitors treated with vehicle (H2O). Lamin B1 is red, Myf5 is green. b 3D-ImmunoFISH of emerin-null proliferating myogenic progenitors treated with 10 μM theophylline. Lamin B1 is red, Myf5 is green. c Quantification of Myf5 localization in vehicle and theophylline-treated emerin-null proliferating myogenic progenitors. n=50. p<0.05. Peripheral is ≤1 μm from the nuclear lamina. Intermediate is between 1 μm and 2 μm from the nuclear lamina. Central is ≥2 μm from the nuclear lamina. d 3D-ImmunoFISH of wildtype proliferating myogenic progenitors treated with vehicle (H2O). Lamin B1 is red, Myf5 is green. e 3D-ImmunoFISH of wildtype proliferating myogenic progenitors treated with 10 μM theophylline. Lamin B1 is red, Myf5 is green. f Quantification of Myf5 localization in vehicle and theophylline-treated wildtype proliferating myogenic progenitors. n=50. Scale bars are 5 μm. g qPCR of Myf5 mRNA in wildtype and emerin-null cells treated with 10 μM theophylline for 4 h relative to mock treatment. Expression was normalized to GAPDH. Error bars represent S.E.M. n=3
Discussion
Emerin Regulates Dynamic Genomic Organization During Myogenesis
This study established a novel mechanism for the dynamic localization of myogenic regulatory loci at the nuclear lamina. Our studies were aimed at determining how emerin regulates the spatial organization of myogenic regulatory loci during differentiation and how the functional interaction between emerin and HDAC3 contributes to dynamic genome organization during differentiation. Here, we show myogenic regulatory loci MyoD, Myf5, Pax3 and Pax7 dynamically occupy the nuclear periphery, concomitant with repression during differentiation. Both emerin and HDAC3 are required for Myf5 localization to the nuclear lamina. Further, emerin binding to HDAC3 was shown to be important for Myf5 localization. Failure to establish proper coordinated temporal localization of these loci at the nuclear periphery during myogenic differentiation accompanies aberrant expression of these critical myogenic regulatory genes. Collectively, these data support the hypothesis that the functional interaction between emerin and HDAC3 is required for establishment of proper genomic architecture during myogenic differentiation.
Model for Regulation of Chromatin Architecture at the Nuclear Periphery by Emerin and HDAC3
We propose a mechanism whereby genomic regions containing the MyoD, Myf5, and Pax7 loci are rapidly deacetylated by HDAC3 when in contact with the nuclear periphery due to increased activity of HDAC3 when bound to emerin. These regions would then be further repressed by other chromatin remodeling complexes, including, but not limited to the G9a and PRC2 complexes. Our data support a model whereby deacetylation of H4K5 by the NCoR complex, facilitated by the catalytic activation of HDAC3 by emerin, directs the deposition of repressive epigenetic marks at the Myf5 locus, as well as the Pax3/7 and MyoD loci, to repress their transcription during myogenic differentiation. In this model, loss of emerin shifts the acetylation/deacetylation kinetics at the nuclear lamina to favor acetylation of the Myf5 locus, leading to developmentally inappropriate localization of this locus to the nuclear interior and aberrant transcription. This may further disrupt the dynamic deposition and removal of active and repressive marks that allow myogenic loci to be stably repressed at the nuclear lamina. To fully and rigorously test this model, it will be necessary to establish whether deacetylation by HDAC3 is the initiating factor in this cascade and to determine the temporal dynamics for the deposition of repressive epigenetic modifications.
Interactions between HDAC3 and Emerin mediate the interaction of myogenic regulatory loci with the nuclear lamina
HDAC3 was recently shown to be required for the interaction of some lamina-associated sequences (LASs) with the nuclear lamina through a proposed interaction with the inner nuclear membrane protein LAP2β (Zullo et al. 2012). Localization of some of these LASs was mediated by cKrox, the mammalian ortholog of Drosophila GAGA-associated factor, through its association with GAGA sequences. This is unlikely to be the only mechanism guiding association of repressed chromatin with the nuclear periphery, since not all LASs contain GAGA repeat sequences and cKrox downregulation failed to affect all LASs. Further, downregulation of LAP2β did not significantly reduce LAS localization to the lamina (Zullo et al. 2012). Our findings suggest emerin binding to HDAC3 and activation of its catalytic activity as an additional, and possibly predominant, mechanism for recruitment and maintenance of lamina-associated genomic regions.
Competing Models of Gene Positioning at the Nuclear Periphery
There are two non-mutually exclusive models to explain how the interaction between emerin and HDAC3 regulate chromatin architecture of developmentally regulated genes. One model is that binding of HDAC3 to emerin mediates the localization of targeted loci to the nuclear envelope with emerin as a molecular tether, analogous to models for the interaction between LAP2β, cKrox and HDAC3 (Somech et al. 2005; Zullo et al. 2012). An alternative model is that emerin binding to HDAC3 not only anchors these regions to the periphery, but also stimulates HDAC3 catalytic activity to initiate or maintain repressive chromatin architecture at the nuclear lamina leading to transcriptional repression. The evidence presented here shows the catalytic activity of HDAC3 is critical for localization of loci to the nuclear periphery, since activation of HDAC3 catalytic activity increases the peripheral localization of Myf5 in the absence of emerin. Thus, emerin binding to HDAC3 does not act solely to physically anchor HDAC3-associated repressed loci to the nuclear lamina, but also acts to repress these loci through stimulation of HDAC3 catalytic activity. It should be noted that 28 % of HDAC3 remains associated with the lamina in the absence of emerin (Demmerle et al. 2012) and the rescue of Myf5 localization to the lamina might result from increased activity of this pool of lamina-associated HDAC3.
Our evidence that activation of HDAC3 catalytic activity by theophylline, a drug used for over 70 years to treat asthma and chronic obstructive pulmonary disease, restores sub-nuclear positioning and repression of Myf5 suggests this may be a promising treatment for ameliorating the skeletal muscle wasting seen in X-EDMD. While broad HDAC inhibitors are widely used (Tang et al. 2013), to our knowledge this is the first instance of a HDAC activator, rather than a repressor, being used to attenuate gene expression. Experiments decoupling HDAC3 binding and catalytic activity will be needed to rigorously determine the mechanism underlying the functional interaction between emerin and HDAC3 and how this interaction regulates association of distinct loci with the nuclear lamina to transcriptionally repress genes within these loci.
Supplementary Material
Acknowledgments
We thank Harinder Singh, Ignacio Demarco, Joseph Zullo, and Eric Bertolino for assistance with 3D DNA-ImmunoFISH and reagents; Christine Labno at the University of Chicago Light Microscopy Core for valuable guidance with image acquisition and processing; and Aaron Mull of the Holaska lab for experimental help. This work was supported by the Ellison Medical Foundation (J.M.H. and J.D.) and the National Institutes of Health (T32 GM007197 and T32 HL007381, A.J.K.).
Abbreviations
- ChIP
Chromatin immunoprecipitation
- ESCs
Embryonic stem cells
- FISH
Fluorescent in situ hybridization
- HDAC3
Histone deacetylase 3
- INM
Inner nuclear membrane
- LAD
Lamina-associated domain
- LASs
Lamina-associated sequences
- LBR
Lamin B receptor
- Myf5
Myogenic factor 5
- MyoD
Myoblast determination protein 1
- NCoR
Nuclear corepressor complex
- NPC
Nuclear pore complex
- ONM
Outer nuclear membrane
- Pax3
Paired box 3
- Pax7
Paired box 7
- PRC2
Polycomb repressive complex 2
- X-
X-linked Emery–Dreifuss muscular
- EDMD
dystrophy
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10577-013-9381-9) contains supplementary material, which is available to authorized users.
Ethical Standards Statement These experiments comply with the current laws for ethical conduct in the USA. This article does not contain any studies with human or animal subjects performed by any of the authors.
Competing Interests Justin Demmerle, Adam J Koch, and James M Holaska declare no competing interests.
Contributor Information
Justin Demmerle, Department of Medicine, Section of Cardiology, The University of Chicago, 5841 S. Maryland Ave, MC6088, Rm A607, Chicago, IL 60637, USA.
Adam J. Koch, Committee on Development, Regeneration and Stem Cell Biology, The University of Chicago, Chicago, IL, USA
James M. Holaska, Department of Medicine, Section of Cardiology, The University of Chicago, 5841 S. Maryland Ave, MC6088, Rm A607, Chicago, IL 60637, USA; Committee on Genetics, Genomics and Systems Biology, The University of Chicago, Chicago, IL, USA; Committee on Development, Regeneration and Stem Cell Biology, The University of Chicago, Chicago, IL, USA
References
- Bakay M, Wang Z, Melcon G, et al. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 2006;129:996–1013. doi: 10.1093/brain/awl023. doi:10.1093/brain/awl023. [DOI] [PubMed] [Google Scholar]
- Barnes PJ. Theophylline: new perspectives for an old drug. Am J Respir Crit Care Med. 2003;167:813–818. doi: 10.1164/rccm.200210-1142PP. doi:10.1164/rccm.200210-1142PP. [DOI] [PubMed] [Google Scholar]
- Berk JM, Tifft KE, Wilson KL. The nuclear envelope LEM-domain protein emerin. Nucleus. 2013;4(4):298–314. doi: 10.4161/nucl.25751. doi:10.4161/nucl.25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. doi:10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmerle J, Koch AJ, Holaska JM. The Nuclear envelope protein emerin binds directly to histone deacetylase 3 (HDAC3) and activates HDAC3 activity. J Biol Chem. 2012;287:22080–22088. doi: 10.1074/jbc.M111.325308. doi:10.1074/jbc.M111.325308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4(3):e1000039. doi: 10.1371/journal.pgen.1000039. doi:10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock RL, Kudlow BA, Evans AM, et al. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev. 2006;20:486–500. doi: 10.1101/gad.1364906. doi:10.1101/gad.1364906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Holaska JM. Emerin and the nuclear lamina in muscle and cardiac disease. Circ Res. 2008;103:16–23. doi: 10.1161/CIRCRESAHA.108.172197. doi:10.1161/CIRCRESAHA.108.172197. [DOI] [PubMed] [Google Scholar]
- Holaska JM, Wilson KL. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry. 2007;46:8897–8908. doi: 10.1021/bi602636m. doi:10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Lee KK, Kowalski AK, Wilson KL. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J Biol Chem. 2003;278:6969–6975. doi: 10.1074/jbc.M208811200. doi:10.1074/jbc.M208811200. [DOI] [PubMed] [Google Scholar]
- Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2:E231. doi: 10.1371/journal.pbio.0020231. doi:10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Rais-Bahrami S, Wilson KL. Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum Mol Genet. 2006;15:3459–3472. doi: 10.1093/hmg/ddl423. doi:10.1093/hmg/ddl423. [DOI] [PubMed] [Google Scholar]
- Huber MD, Guan T, Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol Cell Biol. 2009;29:5718–5728. doi: 10.1128/MCB.00270-09. doi:10.1128/MCB.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Egelhofer TA, Strome S, Lieb JD. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010;11:R120. doi: 10.1186/gb-2010-11-12-r120. doi:10.1186/gb-2010-11-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Lim S, Caramori G, et al. A molecular mechanism of action of theophylline: Induction of histone deacetylase activity to decrease inflammatory gene expression. Proc Natl Acad Sci U S A. 2002;99:8921–8926. doi: 10.1073/pnas.132556899. doi:10.1073/pnas.132556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalaki M, Fili S, Philippou A, Koutsilieris M. Muscle regeneration: cellular and molecular events. In Vivo. 2009;23:779–796. [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. doi:10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Koch AJ, Holaska JM. Loss of emerin alters myogenic signaling and miRNA Expression in mouse myogenic progenitors. PLoS ONE. 2012;7:e37262. doi: 10.1371/journal.pone.0037262. doi:10.1371/journal.pone.0037262.t002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Gillespie MA, Rudnicki MA. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell. 2008;2:22–31. doi: 10.1016/j.stem.2007.12.012. doi:10.1016/j.stem.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Lagha M, Sato T, Bajard L, et al. Regulation of skeletal muscle stem cell behavior by Pax3 and Pax7. Cold Spring Harb Symp Quant Biol. 2008;73:307–315. doi: 10.1101/sqb.2008.73.006. doi:10.1101/sqb.2008.73.006. [DOI] [PubMed] [Google Scholar]
- Lee KK, Haraguchi T, Lee RS, et al. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J Cell Sci. 2001;114:4567–4573. doi: 10.1242/jcs.114.24.4567. [DOI] [PubMed] [Google Scholar]
- Lee H, Quinn JC, Prasanth KV, et al. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 2006;20:784–794. doi: 10.1101/gad.1392006. doi:10.1101/gad.1392006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Cabuy E, Bonne G, et al. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell. 2007;6:139–153. doi: 10.1111/j.1474-9726.2007.00270.x. doi:10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11:R5. doi: 10.1186/gb-2010-11-1-r5. doi:10.1186/gb-2010-11-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcon G, Kozlov S, Cutler DA, et al. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum Mol Genet. 2006;15:637–651. doi: 10.1093/hmg/ddi479. doi:10.1093/hmg/ddi479. [DOI] [PubMed] [Google Scholar]
- Mewborn SK, Puckelwartz MJ, Abuisneineh F, et al. Altered chromosomal positioning, compaction, and gene expression with a Lamin A/C gene mutation. PLoS ONE. 2010;5:e14342. doi: 10.1371/journal.pone.0014342. doi:10.1371/journal.pone.0014342.t002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. doi:10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Worman HJ. Emery–Dreifuss muscular dystrophy. Curr Neurol Neurosci Rep. 2007;7:78–83. doi: 10.1007/s11910-007-0025-3. doi:10.1007/s11910-007-0025-3. [DOI] [PubMed] [Google Scholar]
- Ognibene A, Sabatelli P, Petrini S, et al. Nuclear changes in a case of X-linked Emery–Dreifuss muscular dystrophy. Muscle Nerve. 1999;22:864–869. doi: 10.1002/(sici)1097-4598(199907)22:7<864::aid-mus8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, van Steensel B. Role of the nuclear lamina in genome organization and gene expression. Cold Spring Harb Symp Quant Biol. 2011;75:517–524. doi: 10.1101/sqb.2010.75.014. doi:10.1101/sqb.2010.75.014. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, et al. Molecular maps of the reorganization of genome–Nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. doi:10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. doi:10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. doi:10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Le Grand F, McKinnell I, Kuang S. The molecular regulation of muscle stem cell function. Cold Spring Harb Symp Quant Biol. 2008;73:323–331. doi: 10.1101/sqb.2008.73.064. doi:10.1101/sqb.2008.73.064. [DOI] [PubMed] [Google Scholar]
- Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, et al. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. doi:10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Wilson KL. Partners and post-translational modifications of nuclear lamins. Chromosoma. 2013;122:13–31. doi: 10.1007/s00412-013-0399-8. doi:10.1007/s00412-013-0399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Cavallo A, Schermelleh L, et al. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH) Exp Cell Res. 2002;276:10–23. doi: 10.1006/excr.2002.5513. doi:10.1006/excr.2002.5513. [DOI] [PubMed] [Google Scholar]
- Somech R, Shaklai S, Geller O, et al. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci. 2005;118:4017–4025. doi: 10.1242/jcs.02521. doi:10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- Tang J, Yan H, Zhuang S. Histone deacetylases as targets for treatment of multiple diseases. Clin Sci. 2013;124:651–662. doi: 10.1042/CS20120504. doi:10.1042/CS20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Vosse DW, Wan Y, Wozniak RW, Aitchison JD. Role of the nuclear envelope in genome organization and gene expression. WIREs Syst Biol Med. 2010;3:147–166. doi: 10.1002/wsbm.101. doi:10.1002/wsbm.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Rudnicki MA. Satellite cells, the engines of muscle repair. Nat Rev Mol Cell Biol. 2011;13:127–133. doi: 10.1038/nrm3265. doi:10.1038/nrm3265. [DOI] [PubMed] [Google Scholar]
- Wilson KL, Berk JM. The nuclear envelope at a glance. J Cell Sci. 2010;123:1973–1978. doi: 10.1242/jcs.019042. doi:10.1242/jcs.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Fetter RD, Hu P, et al. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 2011;25:569–580. doi: 10.1101/gad.2021411. doi:10.1101/gad.2021411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem. 2006a;54:1177–1191. doi: 10.1369/jhc.6R6995.2006. doi:10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]
- Zammit PS, Relaix F, Nagata Y, et al. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci. 2006b;119:1824–1832. doi: 10.1242/jcs.02908. doi:10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Pique-Regi R, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. doi:10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.