Abstract
Predominantly identified in pathogenic Gram-positive bacteria, sortase-dependent pili are also found in commensal species, such as the probiotic-marketed strain Lactobacillus rhamnosus strain GG. Pili are typically associated with host colonization, immune signalling and biofilm formation. Comparative analysis of the N-terminal domains of pilin-specific sortases from various piliated Gram-positive bacteria identified a conserved motif, called GYPSY, within the signal sequence. We investigated the function and role of the GYPSY residues by directed mutagenesis in homologous (rod-shaped) and heterologous (coccoid-shaped) expression systems for pilus formation. Substitutions of some of the GYPSY residues, and more specifically the proline residue, were found to have a direct impact on the degree of piliation of Lb. rhamnosus GG. The present findings uncover a new signalling element involved in the functionality of pilin-specific sortases controlling the pilus biogenesis of Lb. rhamnosus GG and related piliated Gram-positive species.
Introduction
In Gram-positive bacteria, sortase enzymes play an essential role in displaying a large variety of effector proteins to the cell surface, allowing the bacterial cells to interact with their host and environment. Typically, sortases are either involved in anchoring proteins to the cell wall (the so-called housekeeping sortases) or in polymerizing pilin proteins (pilin-specific sortases) [1–3]. Due to their ability to display colonization or virulence factors, sortases have been widely investigated and constitute an alternative and promising target for antimicrobial treatment [4]. Sortases are not essential for bacterial cell survival but do significantly impact on the binding to host tissues, signaling to the host, or escaping the host immune response [5,6]. More recently, sortases have also been used to modify proteins in vitro, illustrating their use in synthetic biology [7].
Sortases are cysteine transpeptidases that attach proteins harboring a conserved pentapeptide motif (LPXTG) motif to another cell surface protein (polymerization reaction) or to peptidoglycan lipid II (anchoring reaction) [3]. Following the characterization of the first sortase enzyme Sa-SrtA and its gene in Staphylococcus aureus [8,9], genomic analysis revealed a remarkable diversity of sorting pathways, sequence motifs and sortase subfamilies [10]. In all studied systems, protein secretion and anchoring is a finely coordinated and regulated process that relies on multiple mechanisms and signaling pathways, the details of which are still emerging [11]. It has been recognized that the distribution of surface proteins on their cell walls differs between rod-shaped and coccoid cells. In Streptococcus pyogenes, proteins anchored to the cell wall are localized in separate foci in the vicinity of the septum [12], whereas in Bacillus subtilis proteins are secreted on the cell wall in a spiral shape along the cell axis [13]. In addition, the co-localization of the secretory machinery and sortases allow an efficient protein anchoring or assembly [14,15]. Sortase enzymes themselves harbor a number of conserved motifs essential for their specific localization. Thus, the lack of the positively charged C-terminal domain of Enterococcus faecalis sortases alter their distribution on the cell wall, resulting in the lack of pili [14]. Similarly, sortase-anchored proteins are retained in specific focal sites in the absence of sortases, suggesting that both sortases and their respective substrates are addressed to dedicated assembly sites, called ‘pilusosome’ [16]. It has been suggested that these foci are localized in the cell envelope regions abundant in lipid II, to which LPXTG proteins are ultimately cross-linked [17]. In S. aureus, the spatial distribution of the Sa-SrtA-anchored proteins is dependent on the presence of a conserved motif (YSIRK/GS) in their signal sequence and those with this motif are directed near the septum, whereas others lacking this motif are retained at peripheral foci [18]. Still, the complete secretory and anchoring machinery has been not fully comprehended and, more specifically, sortase trafficking and signaling systems have been poorly examined. The present study aims at identifying novel signal sequence elements within sortases involved in the translocation and secretion of the pilin-specific sortase enzymes in Lactobacillus rhamnosus GG. By comparing the pilus-specific sortases of well-characterized pilus systems in Gram-positive bacteria, we found a distinct N-terminal domain, termed here the GYPSY motif, located in the signal sequence. Hence, we identified the critical residues in the GYPSY motif in an experimental system based on pilus gene cassettes that were either expressed in Lactococcus lactis or a pilus-deficient Lb. rhamnosus derivative. The latter were examined by immunogold staining transmission electron microscopy, immunoblotting and mucus binding assays. The results show that the proline residue in the GYPSY motif is essential for the function of pilin-specific sortases and therefore pilus assembly in not only L. rhamnosus GG but potentially also in other piliated Gram-positive bacteria, which harbor this conserved GYPSY motif within the peptide signal of their pilin-specific sortases.
Experimental Procedures
Bioinformatics sequence analysis
The protein sequences of twenty-four pilin-specific sortases identified in 16 pilus gene clusters from 9 different Gram-positive bacterial species were obtained from public depository databases, including i.e. Lactobacillus rhamnosus GG [19], Corynebacterium glutamicum R [20], Enterococcus faecium TX0016 [21–23], Bacillus cereus ATCC 14579 [24,25], Streptococcus agalactiae 2603V/R [26–30], Streptococcus pneumoniae TIGR4 [31–33], Enterococcus faecalis V583 [34,35], Corynebacterium diphtheriae NCTC 13129 [36–38] and Lactococcus lactis IL1403 [39,40]. The sequence alignment of pilin-specific sortases was generated using MUSCLE [41]. The LOGO motif was generated using the MEME suite [42]. Further GYPSY motif search using KEGG database (http://www.genome.jp/tools/motif/) was performed. The input motif used for the motif search was GX5YPX2SX2Y, which corresponds to the motif initially identified in Lb. rhamnosus GG SrtC1. Search was restricted to prokaryotic KEGG database and proteins, where the motif start position was at most 30 amino acids from the start methionine. The secondary structure of GG SrtC1 was also analysed with PSIPRED [43,44] which showed the residues of the GYPSY motif to be part of a non-continuous helix. This motif was further modelled ab initio and the structural illustrations were generated using PyMOL software [45].
Bacterial strains, plasmids, growth media and conditions
Bacterial strains and expression vectors used in the present study are shown in Table 1. Lb. rhamnosus strains GG and GG-PB12 (termed PB12 in the present work) were cultured in MRS broth in anaerobic conditions at 37°C. Lb. rhamnosus PB12 transformants were selected in MRS agar medium with 5 μg/ml chloramphenicol (Sigma Aldrich, MO, USA). L. lactis subsp. cremoris strain NZ9000 [46] and MG1363 derivatives were propagated anaerobically at 30°C in M17 broth (Oxoid, UK) with 0.5% (w/v) D-glucose (Sigma). L. lactis subsp. cremoris transformants were selected in M17 agar medium with 0.5% (w/v) D-glucose and 5 μg/ml chloramphenicol.
Table 1. Bacterial strains and plasmids used in the present study.
| Strains or plasmids | Relevant properties and characteristics | References |
|---|---|---|
| Strains | ||
| E. coli BL21-DE3 | Expression host | NEB, MA, USA |
| Lb. rhamnosus GG | type strain (ATCC 53103) | Valio Ltd. culture collection |
| Lb. rhamnosus PB12 | GG derivative strain, truncated srtC1 gene | [47] |
| L. lactis NZ9000 | MG1363 strain, pepN::nisRnisK genes | [46] |
| L. lactis ppiA | ppiA mutant of MG1363 by single crossing over, EmR | [48] |
| L. lactis Ctl (ppiA+) | control ppiA+ strain, EmR | [48] |
| Plasmids | ||
| pET-SpaA | pET-52b(+) encoding a 798–bp spaA fragment, AmpR | This study |
| pNZ44 | L. lactis expression vector, CmR | [49] |
| pAC44 | pNZ44 encoding spaA and srtC1 | [11] |
| pGG44 | pAC44 derivative, AA replacement SrtC1Y29G, P30G | This study |
| pYG44 | pAC44 derivative, AA replacement SrtC1P30G | This study |
| pGP44 | pAC44 derivative, AA replacement SrtC1Y29G | This study |
| pNZ44-SrtC1 | pNZ44 encoding srtC1 gene | This study |
| pNZ44-SrtC1Y29G, P30G | pNZ44-SrtC1, AA replacement SrtC1Y29G, P30G | This study |
| pNZ44-SrtC1P30G | pNZ44-SrtC1, AA replacement SrtC1P30G | This study |
| pNZ44-SrtC1Y29G | pNZ44-SrtC1, AA replacement SrtC1Y29G | This study |
CmR, chloramphenicol resistance marker, AmpR, ampicillin resistance marker, EmR, erythromycin resistance
Site-directed mutagenesis of the SpaA-SrtC1 pilus cassette
Oligonucleotide PCR primers were synthesized by Oligomer Oy (Helsinki, Finland). Expression vectors are shown in Table 1. To perform the site-directed mutagenesis, we amplified the plasmid pAC44 using back-to-back phosphorylated PCR oligonucleotides, as previously described [11]. The resulting PCR amplicon was then self-ligated using T4 DNA ligase (Promega). The ligation product was dialyzed and then electroporated into L. lactis electrocompetent cells [50]. Transformants were selected on M17 agar plates supplemented with 0.5% (w/v) D-Glucose and chloramphenicol (final concentration 5 μg/ml), screened and sequenced prior phenotypic analysis. Similar procedure was performed for pGG44, pYG44 and pGP44.
Complementation of pilus-less Lb. rhamnosus PB12
The full-length srtC1 gene was cloned into pNZ44 [49] resulting in the construct pNZ44-SrtC1. Briefly, the srtC1 gene was amplified by PCR using Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Fischer Scientific, PA, USA) and flanked with NcoI and SpeI restriction sites and a ribosome binding site (RBS). Both insert (srtC1 gene amplicon) and pNZ44 plasmid were digested by NcoI and SpeI (FastDigest restriction enzymes, Thermo Fischer Scientific) for 30 min at 37°C and then ligated overnight at 4°C with T4 DNA ligase (Promega). The ligation product was then cloned into L. lactis NZ900 as detailed above. Three additional derivatives pNZ44-SrtC1Y29G,P30G, pNZ44-SrtC1P30G and pNZ44-SrtC1Y29G were generated using the same mutagenesis approach as described above. The correct construction of all four vectors was verified by DNA sequencing analysis. Next, electrocompetent Lb. rhamnosus PB12 were prepared and transformed with pNZ44-SrtC1, pNZ44-SrtC1Y29G,P30G, pNZ44-SrtC1P30G and pNZ44-SrtC1Y29G following the same protocol as in a previous study [51].
Production of polyclonal antibodies directed against SpaA pilin protein
A 798 bp-long fragment of the spaA gene was codon-optimized, in vitro synthesized and cloned into pET-52b(+) (Merck Millipore, Germany) by GenScript USA Inc. (NJ, USA). The plasmid was then introduced into E. coli BL21-DE3 (NEB, MA, USA) and the SpaA fragment was produced as described in the QIAexpressionist manual (Qiagen, Germany). SpaA protein was purified using PrepEase Histidine-tagged Protein Purification Midi Kit (Affimetrix, CA, USA) and desalted using Econo-Pac 10DG Desalting Columns (Biorad, CA, USA). Following concentration using Amicon Ultra-4 Centrifugal Filter Unit with Ultracel-10 membrane (Merck Millipore, Germany), the SpaA protein was analysed by protein gel electrophoresis and quantified by NanoDrop (Thermo Fischer Scientific). Rabbit immunization procedures were performed at the Laboratory Animal Centre (Large Animal Unit) of the University of Helsinki (Finland) [52]. Terminal bleeds were collected and the SpaA antiserum was aliquoted and stored at -80°C for further use.
Cell-wall extraction and Western blotting analysis
Lb. rhamnosus cell wall extracts were prepared as previously described [11,53] and then examined by Western-blotting analysis [54] using polyclonal anti-SpaA rabbit antibodies (dilution 1/ 25,000) and goat anti-rabbit IgG (H + L)-horseradish peroxidase conjugate (dilution 1/100,000, Biorad) as primary and secondary antibodies, respectively.
Immunogold staining transmission electron microscopy
Bacterial cells harvested from overnight cultures were immunogold-stained using rabbit polyclonal anti-SpaA antibodies (dilution 1/100) and gold particule (10 nm) conjugated protein A (dilution 1/55) and then immobilized on copper grids as described before [55]. Sample grids were observed using JEM-1400 transmission electron microscope (JEOL Ltd., Japan).
Mucus binding assays
Mucus adhesion assays of the different Lb. rhamnosus strains were performed, as described previously [47,56]. The ratios between the bound bacterial cells and the total bacterial cells resulted in percentages of binding to pig stomach type II mucus (Sigma Aldrich, MO, USA) and were based on twelve technical replicates of three biological repeats.
Results
In silico analysis of the N-terminal domain of pilin-specific sortases from different piliated Gram-positive bacteria
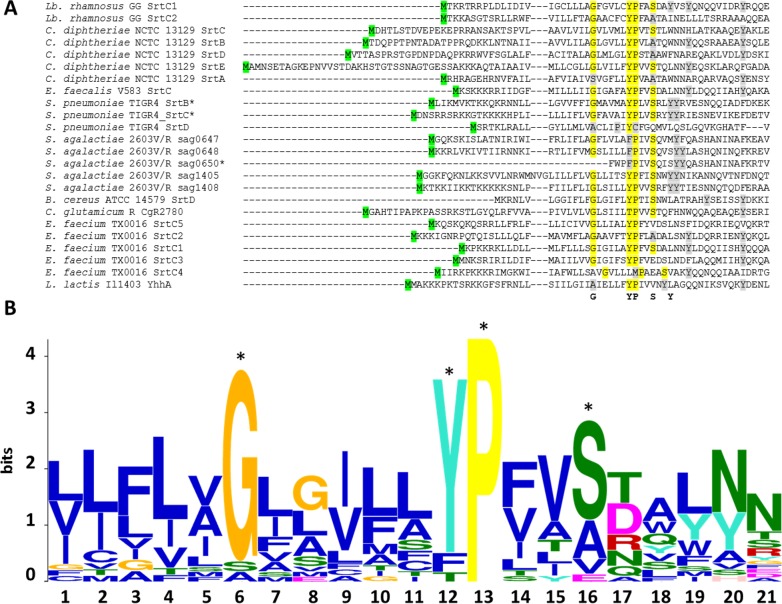
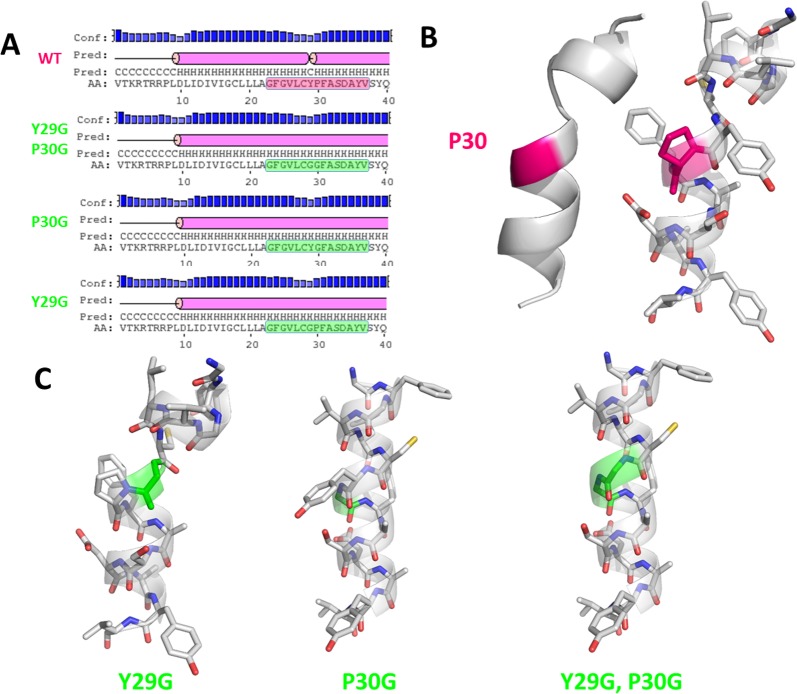
The sequences of twenty-four pilin-specific sortases from 16 different pilus gene clusters were analysed and revealed the conservation of residues located within the peptide signal sequence, termed the GYPSY motif (Fig 1). It generally consists of the following consensus sequence GX5YPX2SXnY, n⊰[2;6]. More specifically, the juxtaposition of the tyrosine (Y29) residue and the proline (P30) residue was found in all analysed sequences, except in S. pneumoniae TIGR4 SrtD and E. faecium TX0016 SrtC4. In the latter, a methionine (M) residue was present instead of the first tyrosine (Y) residue. TX0016 SrtC4 has been reported to belong to a pilus gene cluster located on a megaplasmid [57], possibly resulting in a high degree of sequence polymorphism due to the mobility potential of such DNA element. In S. agalactiae strain 2603V/R, one of its two pilus gene clusters also harboured a set of three pilin-specific sortases all having a phenylalanine (F) residue replacing the first tyrosine (Y) residue. In all 16 pilus gene clusters, there is a high diversity in terms of gene order. However, the GYPSY motif was widely conserved, indicating that it may play a similar role in these species, possibly in the translocation and secretion of pilin-specific sortases to the cell wall. Additional sequence searches further identified the GYPSY motif in a larger number of pilin-specific sortase from strains belonging to Enterococcus, Lactobacillus and Streptococcus spp. Remarkably, the analyzed housekeeping sortases did not have such a GYPSY motif within their N-terminal domains, as observed for example in Lb. rhamnosus GG, L. lactis MG1363, E. faecalis V583 or S. agalactiae 2603V/R genomes (data not shown). Additional bioinformatics analysis using Motif Search for the GX5YPX2SX2Y query sequence revealed that proteins harboring the latter motif within the first 30 amino acid of the N-terminal domain were prevalently annotated as sortases (S1 Fig), further illustrating that such motif within the signal peptide is somehow characteristic to sortases. Secondary structure prediction of the Lb. rhamnosus SrtC1 GYPSY motif and ab initio modelling revealed that the GYPSY motif forms a broken α-helix (Fig 2). Proline has the particularity to introduce kink [58], impacting on the geometry and therefore biological functions [59–61]. Substitution of the proline (P30) residue by a glycine (G) residue is predicted to remove the kink induced by the presence of the proline residue thus forming an uninterrupted helix, as illustrated in SrtC1Y29G,P30G and also in SrtC1P30G (Fig 2). Although the secondary structure of SrtC1Y29G was predicted to be an uninterrupted helix by PSIPRED (Fig 2), we concluded that the helix in the GYPSY motif may be also broken owing to the presence of the proline (P30) as in the wild-type SrtC1 sequence and as suggested by the mutagenesis experiments detailed below. We hypothesized that mutant variants SrtC1Y29G,P30G and SrtC1P30G may have an impact on pilus formation. Experimental models used in the present study aimed at determining the functional roles of each these residues in regard with our hypothesis and predicted models.
Fig 1. Sequence alignment of the N-terminal domains of pilin-specific sortases from various piliated Gram-positive species and LOGO motif analysis.
(A) The alignment was generated using MUSCLE [41]. The so-called GYPSY motif is highlighted in yellow, with the tyrosine residue and the proline residue constituting the functional core of the motif. Residues with a partial degree of conservation within the GYPSY motif are shaded in light grey. The start methionine is highlighted in green. Protein sequences were obtained from public databases, as indicated in the text. In the table, the start of the three protein sequences marked with an asterisk (*) were re-analyzed in the present study. In addition, all sortase sequences excluding E. faecium SrtC4, S. pneumoniae TIGR4 SrtD and S. agalactiae 2603V/R sag0650 were analyzed using the MEME suite [42], resulting in the LOGO motif depicted in (B). Legend: blue, hydrophobic residue; green, polar, non-charged residue; magenta, acidic residue and red, positively charged residue.
Fig 2. Modelling of the region containing the GYPSY motif of the Lb. rhamnosus GG SrtC1 with kinked proline residue.
(A) Output prediction results using PSIPRED server [43,44] indicated that the GYPSY motif introduces a breakage within an α-helix as opposed to some mutant variants; (B) and (C), ab initio model of the GYPSY motif. The kinked proline residue is shaded in fuchsia. The relevant AA modification(s) introduced in the present study are shown in vivid green.
With regard to the distinct functions of pilin-specific sortases (polymerization) and housekeeping sortases (anchoring), we hypothesized that the GYPSY motif, and intrinsically its conformation, may be functionally important for the stability, translocation and/or secretion of the pilin-specific sortases. This would then constitute an additional regulatory pathway, controlling the pilus assembly in Gram-positive species. We used site-directed mutagenesis together with biochemical, microscopic and complementary microbiological analysis methods to test this hypothesis using the rod-shaped and coccoid model systems derived from Lb. rhamnosus GG and Lactococcus lactis subsp. cremoris strains MG1363 and NZ9000.
Alteration of the peptide signal impacts on piliation in a lactococcal model
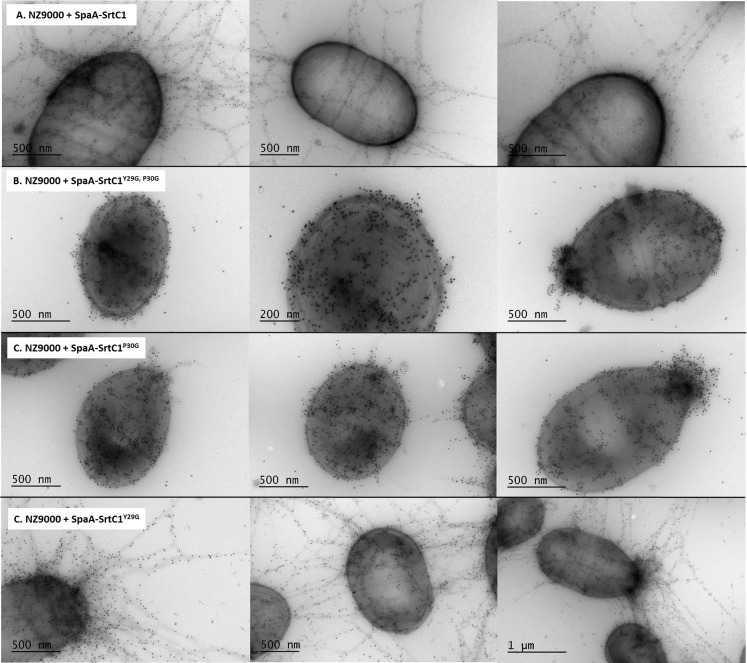
In previous work, we expressed the Lb. rhamnosus GG spaA-srtC1 in L. lactis NZ9000 to investigate the signalling pathway involved in sortase specificity during pilus biogenesis [11]. Using the same lactococcal model system complemented with a plasmid encoding the SpaA major pilin subunit and the pilin-specific sortase SrtC1, we specifically mutated the tyrosine (Y29) and proline (P30) residues of the GYPSY motif of SrtC1. The GYPSY motif is highly conserved in pilin-specific sortases, including the ones found in L. lactis (Fig 1) and absent in its housekeeping sortases analyzed, possibly indicating that it may constitute an original signalling in pilin-specific sortase translocation. In contrast with NZ9000 producing SpaA and SrtC1 (positive control), the replacement of both tyrosine (Y29) and proline (P30) residues by glycine (G) residues within the GYPSY motif of SrtC1 reduced the degree of SpaA piliation in Lactococcus lactis cells. Detailed analysis by immune-electron microscopy showed SpaA pilin subunits to appear predominantly on the cell wall in a monomeric or oligomeric form (Figs 3 and 4). Interestingly, in a small fraction of the L. lactis NZ9000 population producing SpaA-SrtC1Y29G,P30G, some SpaA aggregates also accumulated at the poles of the cells (Fig 3C). This peculiar phenotype was, however, observed in only few cells. The SrtC1 Y29G substitution did not significantly impact on pilus assembly in L. lactis NZ9000, whereas the SrtC1 P30G substitution impaired pilus assembly. Few cells displaying short pili and SpaA aggregates could also be observed at the cell poles. Immuno-electron microscopy observations in L. lactis mutant derivatives indicated that the proline (P30) residue had a significant impact on the functionality of SrtC1 and therefore negatively affects the assembly of pili. We did not address whether the alteration of the peptide signal (especially the GYPSY motif) has a deleterious effect on the stability of SrtC1 and/or the translocation/secretion of SrtC1 to the cell wall of L. lactis NZ9000. Nevertheless, it seems reasonable to believe that the substitution of 1 or 2 amino-acid residues is not likely to impact on the overall stability of the whole protein SrtC1 but rather on some associated mechanisms that required specific and well-conserved motifs.
Fig 3. Electron microscopy observations of L. lactis NZ9000 producing various SpaA pilus structures.
Bacterial cells were immunogold-labeled with anti-SpaA serum and gold particles (10 nm). Legend: A, NZ9000 + SpaA-SrtC1; B, NZ9000 + SpaA-SrtC1Y29G, P30G; C, NZ9000 + SpaA-SrtC1P30G; D, NZ9000 + SpaA-SrtC1Y29G.
Fig 4. Western blotting analysis of L. lactis NZ9000 expressing different spaA-srtC1 gene variants, where the GYPSY motif has been altered by AA substitution.
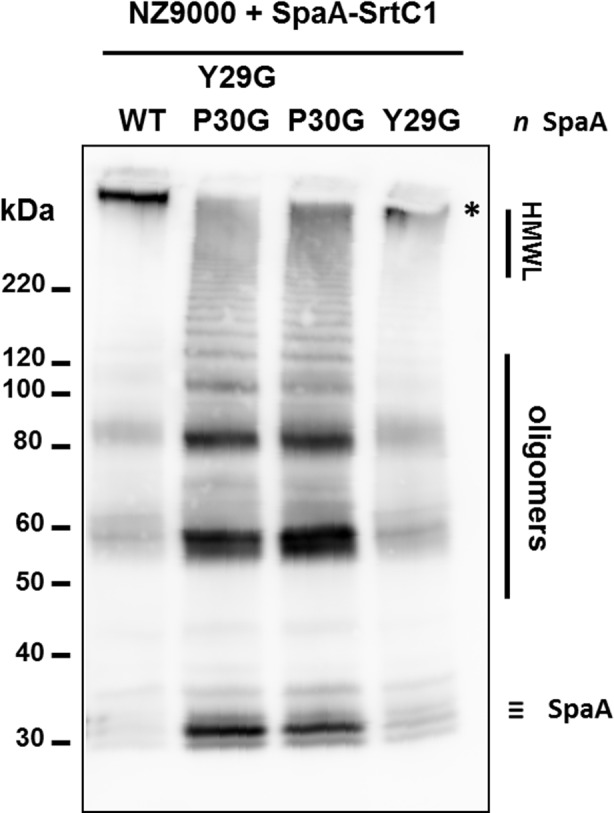
SpaA pilin proteins were detected using anti-SpaA polyclonal antibodies. Legend: #, protein weight marker; HMWL, high molecular weight ladder; *, band corresponding to elongated pilus structures.
Complementation of pilus-less Lb. rhamnosus PB12 and restoration piliation
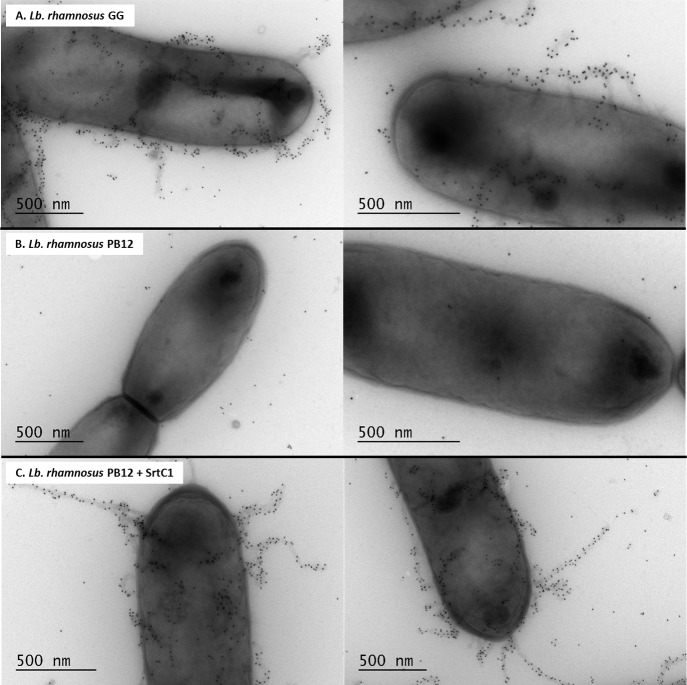
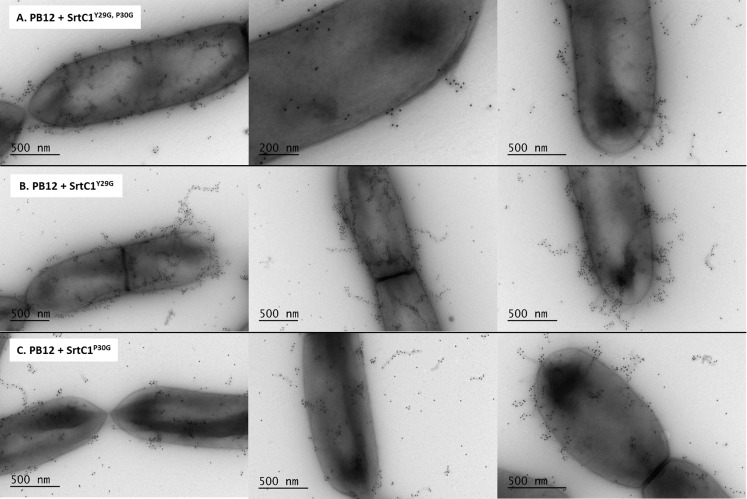
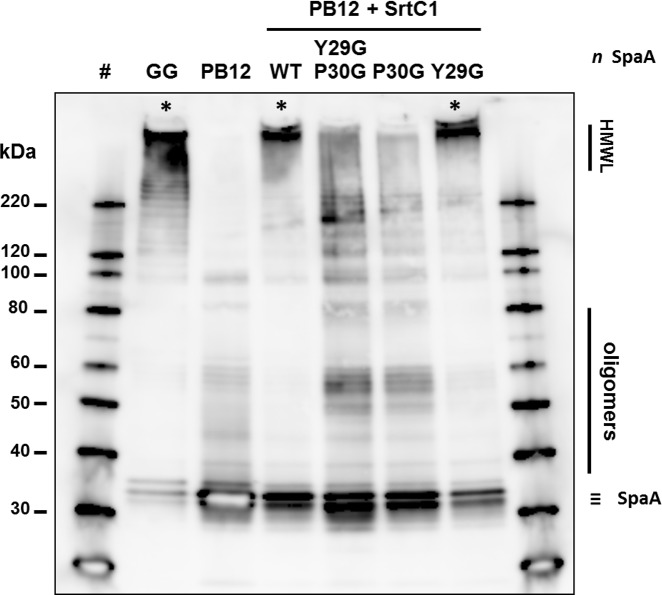
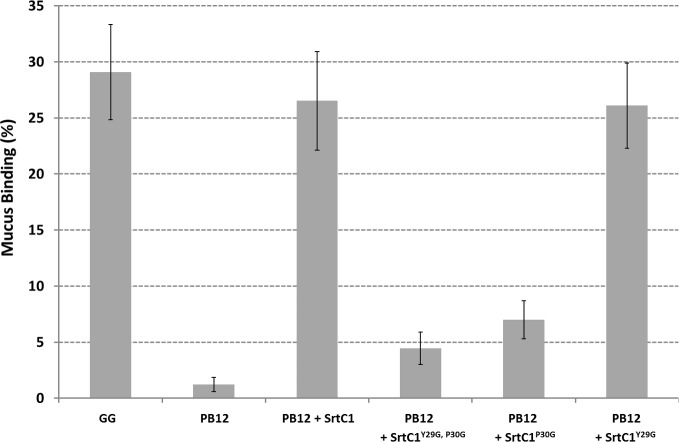
The pilus-less Lb. rhamnosus PB12 is a well-characterized, non-GMO derivative of strain GG that harbors a truncated srtC1 gene due to the presence a stop codon within the srtC1 open reading frame [47]. We used this strain to further investigate the GYPSY motif by complementing it with different SrtC1 variants. The introduction of the full-length srtC1 gene into PB12 restored the piliation, as observed by electron microscopy (Fig 5). This confirmed that the pilus-less phenotype observed in PB12 was only due to the truncation of SrtC1 and not any of the other collateral chromosomal mutations reported [47]. Numerous and elongated pilus structures could be observed in the complemented PB12 in contrast to PB12, which was devoid of any pili. The complementation of PB12 with SrtC1 mutant variants further supported the initial observations done in the lactococcal model. The introduction of either SrtC1Y29G,P30G or SrtC1P30G into Lb. rhamnosus PB12 did not result in wild-type piliation but rather in SpaA oligomers (Fig 6). The SrtC1 Y29G substitution, however, did not prevent the pilus assembly and resulted in a phenotype similar to Lb. rhamnosus GG, as observed by EM. Immunoblotting analysis using polyclonal SpaA antibodies also showed that PB12 complemented with SrtC1 or SrtC1Y29G harboured long pilus structures (high molecular weight ladder) similarly to the piliated type strain Lb. rhamnosus GG (Fig 7). In complemented strains PB12 with SrtC1Y29G,P30G or SrtC1P30G, SpaA oligomers could be observed and the degree of high molecular weight SpaA multimers was lower. In contrast, in GG and also PB12 with SrtC1 and PB12 with SrtC1Y29G, we were able to observe a higher degree of piliation, as indicated with an asterisk (*). It is noteworthy that the different SrtC1 protein variants are clearly expressed, since immunoblotting data (Fig 7) do show pilin polymerization or at least some degree of pilin oligomerization in the different mutants, as opposed to the negative control PB12, which is devoid of functional sortase SrtC1. In the latter (PB12), only SpaA monomers are produced. To further support our data and provide more quantitative measures of the degree of piliation, we measured the mucus binding ability of the different strains using an in vitro binding assay, where we measured the fraction of mucus-bound radiolabelled cells from the total amount of cells applied to the mucus-coated wells (Fig 8). Lb. rhamnosus PB12 complemented with either SrtC1 or SrtC1Y29G displayed a high mucus binding capacity, which was almost at comparable levels with that of the wild-type Lb. rhamnosus GG. In contrast, the introduction of SrtC1Y29G,P30G and SrtC1P30G into strain PB12 did not significantly increase the mucus binding ability, indicating that SrtCY29G,P30G and SrtC1P30G are not fully functional. Immuno-electron microscopy data, immunoblotting analysis and mucus binding assays were all concordant and showed that the substitution of the proline (P30) residue within the GYPSY motif impaired directly or indirectly the pilus assembly. As also observed in the results obtained in our lactococcal model, the reduction of SrtC1 activity remains to be identified, but we hypothesized that the substitution of 1 or 2 amino acid residues did not impact on the stability and expression of the different variants SrtC1, but rather on some specific mechanisms relating to the translocation of SrtC1 to the cell membrane.
Fig 5. Electron microscopy observations of Lb. rhamnosus PB12 and its SrtC1-complemented derivatives.
Bacterial cells were immunogold-labeled with anti-SpaA serum and gold particles (10 nm). Legend: A, Lb. rhamnosus GG; B, Lb. rhamnosus PB12; C, Lb. rhamnosus PB12 + SrtC1.
Fig 6. Electron microscopy observations of Lb. rhamnosus PB12 complemented with SrtC1 variants.
Bacterial cells were immunogold-labeled with anti-SpaA serum and gold particles (10 nm). Legend: A, Lb. rhamnosus PB12 complemented with SrtC1 Y29G, P30G; B, Lb. rhamnosus PB12 complemented SrtC1 Y29G; C, L. rhamnosus PB12 complemented SrtC1 P30G.
Fig 7. Western blotting analysis of Lb. rhamnosus GG and PB12 expressing different srtC1 gene variants, where the GYPSY motif has been altered by AA substitution.
SpaA pilin proteins were detected using anti-SpaA polyclonal antibodies. Legend: #, protein weight marker; HMWL, high molecular weight ladder; *, band corresponding to elongated pilus structures.
Fig 8. Mucus binding assays of Lb. rhamnosus GG and PB12 expressing different srtC1 gene variants, where the GYPSY motif has been altered by AA substitution.
The averages and standard deviations were obtained from a total of twelve technical replicates from three biological replicates.
Expression of SpaA-SrtC1 in L. lactis MG1363 ppiA mutant
The identification of the GYPSY motif in the SrtC1 peptide signal and its effect on pilus biogenesis highlighted an important role of the proline residue (P30) in the functionality of pilin-specific sortase SrtC1. In Fig 2, we showed that the proline (P) residue introduce a breakage of the left-hand α-helix and hypothesized that the proline kink is conformationally required to ensure stability and/or translocation of SrtC1 to the cell wall. It is known that cis-trans isomerization of the proline (P) by prolyl peptidyl isomerase (PPI) may also play a role in protein folding kinetics, geometry and biological functions. To further verify this, the pilus cassette encoding SpaA-SrtC1 was then electroporated into L. lactis MG1363 ppiA mutant and its control derivative [48] (Table 1). The L. lactis MG1363 ppiA mutant has no functional prolyl peptidyl isomerase, preventing any cis-trans isomerization transition. Pilus phenotype was analyzed by immunoblotting blotting (Fig 9) and revealed that the L. lactis ppiA mutant did produce SpaA pili, as also observed in the control, suggesting that the prolyl peptidyl isomerase ppiA is not involved in the stability and/or translocation of SrtC1 and therefore not required for pilus biogenesis as tested in a lactococcal model.
Fig 9. Western blotting analysis of L. lactis MG1363 PPiA KO mutant and its complemented mutant, where the SpaA-SrtC1 cassette was introduced.
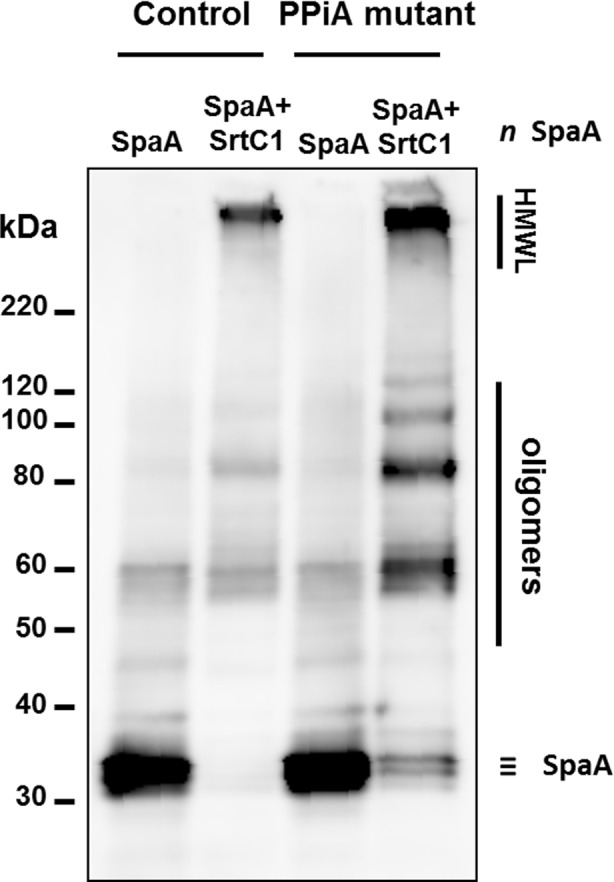
SpaA pilin proteins were detected using anti-SpaA polyclonal antibodies. Legend: #, protein weight marker; HMWL, high molecular weight ladder; *, band corresponding to elongated pilus structures.
Discussion
In contrast with other piliated Gram-positive species studied so far, Lb. rhamnosus GG has been shown to confer health promoting properties for the host and therefore has been widely used in food industry [62–65]. The identification of mucus-binding pili in GG [19] has opened the way to address their role in the probiotic and host signalling response [45]. Much of the research that has been done on the Lb. rhamnosus pilus biogenesis [11,54,55], showed some similarities in terms of function, assembly or gene order with other pilus clusters from Gram-positive piliated species and more specifically lactobacilli [66]. The present study expanded on this and provided new insight into the signalling pathways involved in the pilus assembly in Lb. rhamnosus GG. We showed that the GYPSY motif is highly conserved in pilin-specific sortases in Gram-positive piliated species.
We, therefore, proposed that the presence of the GYPSY motif in these bacterial species is likely to play a role in regard with the functionality of pilin-specific sortases that control the biogenesis of pili. Specifically, the proline residue is found in most sortase protein sequences analysed, possibly indicating its role in the conformation of the peptide signal of pilin-specific sortases in Lb. rhamnosus but also C. diphtheriae, E. faecium or S. pneumoniae. Other residues of the GYPSY motif did show a higher degree of diversity, such the tyrosine (Y) residue, which was replaced by a phenylalanine (F) residue in all sortases of one pilus gene cluster from S. agalactiae 2603V/R or by a methionine (M) residue in the SrtC4 from E. faecium TX0016. The tyrosine (Y) residue juxtaposing the proline (P) residue may be therefore not functionally important. Using mutagenesis, this was confirmed in both L. lactis and Lb. rhamnosus, where the P30G substitution had the highest impact on the degree of piliation, whereas the Y29G substitution did not. Other small residues constituting the GYPSY motif (G23 and S33) were not investigated but may have a limited role since they seem to be less conserved.
Phenotype analysis indicated that the loss of the proline (P30G) residue reduced the pilus assembly and only SpaA monomers and aggregates could be visualized by EM and immunoblotting in L. lactis (Table 2). Remarkably, a subpopulation of L. lactis NZ9000 cells (expressing SpaA-SrtC1Y29G,P30G or SpaA-SrtC1P30G) display an accumulation of SpaA-mers and few short pili at the poles of the cells. We hypothesized that such phenotype may be possibly attributed to the poor stability/translocation of SrtC1, resulting in the assembly of few short and aborted pili, which are then moved away from the septum upon growth and division of the bacterial cells. In Lb. rhamnosus, we also demonstrated that the GYPSY motif and more specifically the proline (P30) residue was important for SrtC1 functionality and indirectly for pilus assembly (Table 2). Its location within the peptide signal further suggests that the proline may determine a specific conformation of the peptide signal, directly relating to the secretion-competent state or stability of the pilin-specific sortase SrtC1. The close vicinity of the GYPSY motif with the predicted peptide signal cleavage (S33) suggests a possible link between peptide signal, stability and translocation of SrtC1. Proline residue confers unique properties to protein structure and we proposed that the presence of a proline kink is functionally important, although we do not currently know if it has an effect on SrtC1 stability or translocation. We also ruled out the possibility of a prolyl peptidyl isomerase-mediated regulatory pathway (cis-trans isomerization), since we showed that prolyl peptidyl isomerase was not required for the production of pili. It, therefore, remains to be determined what are the regulatory controls and mechanisms associated with the GYPSY motif, i.e. protein translocation, stability or chaperone activity. Peptide signals are expected to be functionally important for the specific and coordinated addressing and translocation of pilin subunits and sortases to the pilusosomes (assembly sites). The recent description of a signal peptidase involved in processing secreted proteins suggests that a number of players remains to be identified with regard to their respective role in pilin and sortase targeting [67]. The present study is the first report that described the presence of proline within the peptide signal, as a critical signal element for the functionality of pilin-specific sortases.
Table 2. Summary of pilus phenotypes observed in derivatives of Lactococcus lactis NZ9000, Lactobacillus rhamnosus GG and PB12.
The AA replacement is shaded in light grey within the GYPSY motif (bold AA residues).
| Bacterial Derivatives | GYPSY Motif | Pili Assembly | Description of Pilus Phenotype |
|---|---|---|---|
| Lactobacillus rhamnosus GG | GFGVLCYPFASDAY | + | High molecular weight pili |
| Lactococcus lactis NZ9000+pAC44 | |||
| - wild-type SrtC1 | GFGVLCYPFASDAY | + | High molecular weight pili |
| - SrtC1Y29G, P30G | GFGVLCGGFASDAY | +/- | SpaA multimers |
| - SrtC1Y29G | GFGVLCGPFASDAY | + | High molecular weight pili |
| - SrtC1P30G | GFGVLCYGFASDAY | +/- | SpaA multimers |
| Lactobacillus rhamnosus PB12 | |||
| - negative control | n/a | - | No pili, SpaA monomers |
| - complemented with wild-type SrtC1 | GFGVLCYPFASDAY | + | High molecular weight pili |
| - complemented with SrtC1Y29G, P30G | GFGVLCGGFASDAY | +/- | SpaA multimers |
| - complemented with SrtC1Y29G | GFGVLCGPFASDAY | + | High molecular weight pili |
| - complemented with SrtC1P30G | GFGVLCYGFASDAY | +/- | SpaA multimers |
Legend: +, presence of pili; -, absence of pili; +/-, presence of short and/or rare pili; n/a, not applicable.
The structures of housekeeping and pilin-specific sortases have been solved in a number of species [68]. Unfortunately, very little is known about the detailed conformation of peptide signals of these as these have not been included in the solved crystal structures. Their hydrophobic propensity prevents their structural studies, leaving us with in silico prediction to understand their structure and function. Due to the pivotal role of sortases in displaying interaction factors in pathogenic and non-pathogenic bacteria, a further understanding of the mechanisms governing the translocation and maturation of sortases would offer new approaches in the development of anti-microbial inhibitors blocking the interaction capabilities of bacteria towards the host.
Supporting Information
The input motif used for the search was the following one: G-x-x-x-x-x-Y-P-x-x-S-x-x-Y, which corresponds to the motif identified in L. rhamnosus GG SrtC1. We restricted our search to prokaryotic KEGG gene database and only considered proteins, where the motif start position was at most 30 amino acids from the start methionine. Out of 685 hits found in the prokaryotic KEGG gene database, 87 of them did have the GX5YPX2SX2Y motif located at the N-terminal domain. Interestingly, 69 out of 87 were proteins annotated as sortase enzymes. (A) The list below shows the complete list of proteins found through this search. (B) Using these 69 sequences, we also generated the LOGO motif using the MEME Suite.
(PDF)
Acknowledgments
We are grateful to the personnel from the Animal Laboratory Centre (University of Helsinki), from the Electron Microscopy Unit (Institute of Biotechnology, Helsinki) and to the DNA Sequencing and Genomics lab (Institute of Biotechnology, Helsinki) for providing high quality technical assistance. We are also thanking Prof. Douwe van Sinderen (Alimentary Pharmabiotic Centre, University College Cork) for providing the pNZ44 expression vector. We are grateful to Dr. Isabelle Poquet (Micalis, INRA, France) for kindly providing the PPIase mutants. Finally, we would like to thank Dr. Matti Kankainen (Institute for Molecular Medicine Finland, University of Helsinki) for his input at the start of the project.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The present study was sponsored by the European Research Council (grant ERC 250172 - Microbes Inside, https://erc.europa.eu/), the University of Helsinki and the Academy of Finland (grants 141140 to AP, 252123 to FPD and 141130 to WMdV, http://www.aka.fi/en/).
References
- 1.Mandlik A, Swierczynski A, Das A, Ton-That H (2008) Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol 16: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proft T, Baker EN (2009) Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Life Sci 66: 613–635. 10.1007/s00018-008-8477-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clancy KW, Melvin JA, McCafferty DG (2010) Sortase transpeptidases: Insights into mechanism, substrate specificity, and inhibition. Peptide Sci 94: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maresso AW, Schneewind O (2008) Sortase as a target of anti-infective therapy. Pharmacol Rev 60: 128–141. 10.1124/pr.107.07110 [DOI] [PubMed] [Google Scholar]
- 5.Vanier G, Sekizaki T, Dominguez-Punaro MC, Esgleas M, Osaki M, Takamatsu D, et al. (2008) Disruption of srtA gene in Streptococcus suis results in decreased interactions with endothelial cells and extracellular matrix proteins. Vet Microbiol 127: 417–424. [DOI] [PubMed] [Google Scholar]
- 6.Kemp KD, Singh KV, Nallapareddy SR, Murray BE (2007) Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect Immun 75: 5399–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popp MW, Dougan SK, Chuang TY, Spooner E, Ploegh HL (2011) Sortase-catalyzed transformations that improve the properties of cytokines. Proc Natl Acad Sci U S A 108: 3169–3174. 10.1073/pnas.1016863108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O (1999) Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci U S A 96: 12424–12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazmanian SK, Liu G, Ton-That H, Schneewind O (1999) Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285: 760–763. [DOI] [PubMed] [Google Scholar]
- 10.Comfort D, Clubb RT (2004) A comparative genome analysis identifies distinct sorting pathways in Gram-positive bacteria. Infect Immun 72: 2710–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douillard FP, Rasinkangas P, von Ossowski I, Reunanen J, Palva A, de Vos WM (2014) Functional identification of conserved residues involved in Lactobacillus rhamnosus strain GG sortase specificity and pilus biogenesis. J Biol Chem 289: 15764–15775. 10.1074/jbc.M113.542332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlsson F, Stålhammar-Carlemalm M, Flärdh K, Sandin C, Carlemalm E, Lindahl G (2006) Signal sequence directs localized secretion of bacterial surface proteins. Nature 442: 943–946. [DOI] [PubMed] [Google Scholar]
- 13.Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, et al. (2004) Subcellular sites for bacterial protein export. Mol Microbiol 53: 1583–1599. [DOI] [PubMed] [Google Scholar]
- 14.Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, et al. (2009) Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol 191: 3237–3247. 10.1128/JB.01837-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu P, Bian Z, Fan M, Huang M, Zhang P (2008) Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch Oral Biol 53: 150–154. [DOI] [PubMed] [Google Scholar]
- 16.Guttilla IK, Gaspar AH, Swierczynski A, Swaminathan A, Dwivedi P, Das A, et al. (2009) Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in Gram-positive bacteria. J Bacteriol 191: 5603–5612. 10.1128/JB.00627-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raz A, Fischetti VA (2008) Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc Natl Acad Sci U S A 105: 18549–18554. 10.1073/pnas.0808301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeDent A, Bae T, Missiakas DM, Schneewind O (2008) Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. Embo J 27: 2656–2668. 10.1038/emboj.2008.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, et al. (2009) Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc Ntl Acad Sci U S A 106: 17193–17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yukawa H, Omumasaba CA, Nonaka H, Kós P, Okai N, Suzuki N, et al. (2007) Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiol 153: 1042–1058. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickx APA, Bonten MJM, van Luit-Asbroek M, Schapendonk CME, Kragten AHM, Willems RJL (2008) Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiol 154: 3212–3223. [DOI] [PubMed] [Google Scholar]
- 22.Qin X, Galloway-Peña JR, Sillanpää J, Roh JH, Nallapareddy SR, Chowdhury S, et al. (2012) Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol 12: 1471–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrickx APA, Schapendonk CME, van Luit-Asbroek M, Bonten MJM, van Schaik W, Willems RJL (2010) Differential PilA pilus assembly by a hospital-acquired and a community-derived Enterococcus faecium isolate. Microbiol 156: 2649–2659. [DOI] [PubMed] [Google Scholar]
- 24.Budzik JM, Marraffini LA, Schneewind O (2007) Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol 66: 495–510. [DOI] [PubMed] [Google Scholar]
- 25.Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, et al. (2003) Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423: 87–91. [DOI] [PubMed] [Google Scholar]
- 26.Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, et al. (2005) Genome analysis reveals pili in Group B Streptococcus. Science 309: 105 [DOI] [PubMed] [Google Scholar]
- 27.Telford JL, Barocchi M, Margarit I, Rappuoli R, Grandi G (2006) Pili in Gram-positive pathogens. Nat Rev Microbiol 4: 509–519. [DOI] [PubMed] [Google Scholar]
- 28.Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, Maione D, et al. (2006) Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol 61: 126–141. [DOI] [PubMed] [Google Scholar]
- 29.Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, Wessels MR, et al. (2002) Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc Natl Acad Sci U S A 99: 12391–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A 102: 13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, et al. (2006) A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A 103: 2857–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeMieux J, Hava DL, Basset A, Camilli A (2006) RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect Immun 74: 2453–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, et al. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293: 498–506. [DOI] [PubMed] [Google Scholar]
- 34.Nallapareddy SR, Sillanpää J, Mitchell J, Singh KV, Chowdhury SA, Weinstock GM, et al. (2011) Conservation of Ebp-type pilus genes among Enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets. Infect Immun 79: 2911–2920. 10.1128/IAI.00039-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, et al. (2003) Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299: 2071–2074. [DOI] [PubMed] [Google Scholar]
- 36.Ton-That H, Schneewind O (2003) Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol 50: 1429–1438. [DOI] [PubMed] [Google Scholar]
- 37.Cerdeño-Tárraga AM, Efstratiou A, Dover LG, Holden MTG, Pallen M, Bentley SD, et al. (2003) The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res 31: 6516–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swierczynski A, Ton-That H (2006) Type III Pilus of Corynebacteria: Pilus length is determined by the level of its major pilin subunit. J Bacteriol 188: 6318–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oxaran V, Ledue-Clier F, Dieye Y, Herry J- M, Péchoux C, Meylheuc T, et al. (2012) Pilus biogenesis in Lactococcus lactis: Molecular characterization and role in aggregation and biofilm formation. PLoS ONE 7: e50989 10.1371/journal.pone.0050989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, et al. (2001) The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11: 731–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41: W349–357. 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202. [DOI] [PubMed] [Google Scholar]
- 45.Lebeer S, Claes I, Tytgat HLP, Verhoeven TLA, Marien E, von Ossowski I, et al. (2012) Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol 78: 185–193. 10.1128/AEM.06192-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM (1998) Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64: 15–21. [Google Scholar]
- 47.Rasinkangas P, Reunanen J, Douillard FP, Ritari J, Uotinen V, Palva A, et al. (2014) Genomic characterization of non-mucus-adherent derivatives of Lactobacillus rhamnosus GG reveals genes affecting pilus biogenesis. Appl Environ Microbiol 80: 7001–7009. 10.1128/AEM.02006-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tremillon N, Morello E, Llull D, Mazmouz R, Gratadoux JJ, Guillot A, et al. (2012) PpiA, a surface PPIase of the cyclophilin family in Lactococcus lactis. PLoS One 7: e33516 10.1371/journal.pone.0033516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGrath S, Fitzgerald GF, van Sinderen D (2001) Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl Environ Microbiol 67: 608–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holo H, Nes IF (1989) High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55: 3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Keersmaecker SCJ, Braeken K, Verhoeven TLA, Perea Vélez M, Lebeer S, Vanderleyden J, et al. (2006) Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl Environ Microbiol 72: 4923–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanly WC, Artwohl JE, Bennett BT (1995) Review of polyclonal antibody production procedures in mammals and poultry. ILAR J 37: 93–118. [DOI] [PubMed] [Google Scholar]
- 53.Åvall-Jääskeläinen S, Hynönen U, Ilk N, Pum D, Sleytr UB, Palva A (2008) Identification and characterization of domains responsible for self-assembly and cell wall binding of the surface layer protein of Lactobacillus brevis ATCC 8287. BMC Microbiol 8: 165 10.1186/1471-2180-8-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Douillard FP, Ribbera A, Järvinen HM, Kant R, Pietilä TE, Randazzo C, et al. (2013) Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl Environ Microbiol 79: 1923–1933. 10.1128/AEM.03467-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reunanen J, von Ossowski I, Hendrickx APA, Palva A, de Vos WM (2012) Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol 78: 2337–2344. 10.1128/AEM.07047-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vesterlund S, Paltta J, Karp M, Ouwehand AC (2005) Measurement of bacterial adhesion—in vitro evaluation of different methods. J Microbiol Meth 60: 225–233. [DOI] [PubMed] [Google Scholar]
- 57.Kim DS, Singh KV, Nallapareddy SR, Qin X, Panesso D, Arias CA, et al. (2010) The fms21 (pilA)-fms20 locus encoding one of four distinct pili of Enterococcus faecium is harboured on a large transferable plasmid associated with gut colonization and virulence. J Med Microbiol 59: 505–507. 10.1099/jmm.0.016238-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanhoof G, Goossens F, De Meester I, Hendriks D, Scharpé S (1995) Proline motifs in peptides and their biological processing. FASEB J 9: 736–744. [PubMed] [Google Scholar]
- 59.Hardy JA, Nelson HCM (2000) Proline in alpha-helical kink is required for folding kinetics but not for kinked structure, function, or stability of heat shock transcription factor. Protein Sci 9: 2128–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Heijne G (1991) Proline kinks in transmembrane alpha-helices. J Mol Biol 218: 499–503. [DOI] [PubMed] [Google Scholar]
- 61.Wool GD, Vaisar T, Reardon CA, Getz GS (2009) An apoA-I mimetic peptide containing a proline residue has greater in vivo HDL binding and anti-inflammatory ability than the 4F peptide. J Lipid Res 50: 1889–1900. 10.1194/jlr.M900151-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernardeau M, Guguen M, Vernoux JP (2006) Beneficial lactobacilli in food and feed: long-term use, biodiversity and proposals for specific and realistic safety assessments. FEMS Microbiol Rev 30: 487–513. [DOI] [PubMed] [Google Scholar]
- 63.Saxelin M, Tynkkynen S, Mattila-Sandholm T, de Vos WM (2005) Probiotic and other functional microbes: from markets to mechanisms. Curr Opin Biotechnol 16: 204–211. [DOI] [PubMed] [Google Scholar]
- 64.de Vos WM (2011) Systems solutions by lactic acid bacteria: from paradigms to practice. Microb Cell Fact 10: S2 10.1186/1475-2859-10-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lebeer S, Vanderleyden J, De Keersmaecker SCJ (2010) Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol 8: 171–184. 10.1038/nrmicro2297 [DOI] [PubMed] [Google Scholar]
- 66.Sun Z, Harris HM, McCann A, Guo C, Argimón S, Zhang W, et al. (2015) Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6: 8322 10.1038/ncomms9322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young PG, Proft T, Harris PWR, Brimble MA, Baker EN (2014) Structure and activity of Streptococcus pyogenes SipA: a signal peptidase-like protein essential for pilus polymerisation. PLoS One 9: e99135 10.1371/journal.pone.0099135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neiers F, Madhurantakam C, Fälker S, Manzano C, Dessen A, Normark S, et al. (2009) Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity. J Mol Biol 393: 704–716. 10.1016/j.jmb.2009.08.058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The input motif used for the search was the following one: G-x-x-x-x-x-Y-P-x-x-S-x-x-Y, which corresponds to the motif identified in L. rhamnosus GG SrtC1. We restricted our search to prokaryotic KEGG gene database and only considered proteins, where the motif start position was at most 30 amino acids from the start methionine. Out of 685 hits found in the prokaryotic KEGG gene database, 87 of them did have the GX5YPX2SX2Y motif located at the N-terminal domain. Interestingly, 69 out of 87 were proteins annotated as sortase enzymes. (A) The list below shows the complete list of proteins found through this search. (B) Using these 69 sequences, we also generated the LOGO motif using the MEME Suite.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.