Abstract
Background
The search for novel chemical entities targeting essential and parasite-specific pathways is considered a priority for neglected diseases such as trypanosomiasis and leishmaniasis. The thiol-dependent redox metabolism of trypanosomatids relies on bis-glutathionylspermidine [trypanothione, T(SH)2], a low molecular mass cosubstrate absent in the host. In pathogenic trypanosomatids, a single enzyme, trypanothione synthetase (TryS), catalyzes trypanothione biosynthesis, which is indispensable for parasite survival. Thus, TryS qualifies as an attractive drug target candidate.
Methodology/Principal Finding
A library composed of 144 compounds from 7 different families and several singletons was screened against TryS from three major pathogen species (Trypanosoma brucei, Trypanosoma cruzi and Leishmania infantum). The screening conditions were adjusted to the TryS´ kinetic parameters and intracellular concentration of substrates corresponding to each trypanosomatid species, and/or to avoid assay interference. The screening assay yielded suitable Z’ and signal to noise values (≥0.85 and ~3.5, respectively), and high intra-assay reproducibility. Several novel chemical scaffolds were identified as low μM and selective tri-tryp TryS inhibitors. Compounds displaying multi-TryS inhibition (N,N'-bis(3,4-substituted-benzyl) diamine derivatives) and an N5-substituted paullone (MOL2008) halted the proliferation of infective Trypanosoma brucei (EC50 in the nM range) and Leishmania infantum promastigotes (EC50 = 12 μM), respectively. A bis-benzyl diamine derivative and MOL2008 depleted intracellular trypanothione in treated parasites, which confirmed the on-target activity of these compounds.
Conclusions/Significance
Novel molecular scaffolds with on-target mode of action were identified as hit candidates for TryS inhibition. Due to the remarkable species-specificity exhibited by tri-tryp TryS towards the compounds, future optimization and screening campaigns should aim at designing and detecting, respectively, more potent and broad-range TryS inhibitors.
Author Summary
Parasites from the genus Trypanosoma and Leishmania are etiologic agents for a group of neglected diseases with high morbidity and mortality rates in the developing world. Inasmuch as vaccine development is hampered by the successful mechanisms employed by the pathogens to evade the host immune response, chemotherapy remains as a safe option to fight these diseases. However, new drugs with better pharmacological performance (i.e. safety, efficacy and ease of administration) than those in current use are urgently needed. The thiol-redox metabolism of trypanosomatids offers an excellent opportunity for the development of more selective and efficacious medicines because it depends on a molecule, trypanothione (a bis-glutathionyl derivative of spermidine), unique and indispensable to the pathogens. Here we report the identification of novel inhibitors of trypanothione synthetase from three major trypanosomatid species of medical and veterinary relevance. Although highly conserved in sequence, trypanothione synthetases display significant species-specifity towards compounds, pointing to structural differences as determinants of ligand selectivity. Most of the active compounds presented two-digit μM inhibitory activity and serve as primary scaffolds to develop more potent inhibitors. Among them, N,N'-bis(benzyl)-substituted diamine and paullone derivatives are interesting candidates because of their potent and/or selective anti-trypanosomal and anti-trypanothione synthetase activity.
Introduction
Protozoan parasites from the genus Trypanosoma and Leishmania are responsible for diseases affecting humans and their livestock. The zoonotic character of these diseases, which involve different insect species as vectors and wild animals as reservoirs, hamper the implementation of successful control strategies [1]. Immuno-prophylaxis is not yet available and for some species, such as T. brucei spp. and T. cruzi, appears unfeasible due to complex immune-evasion mechanisms [2, 3]. So far, and probably for several decades ahead, chemotherapy remains as the sole choice of treatment. Only a handful of drugs are available to fight Chagas’s disease (T. cruzi), sleeping sickness (T. brucei gambiense and T. b. rhodesiense) and the different forms of leishmaniasis (Leishmania spp.). Unfortunately, they suffer from several drawbacks encompassing low efficacy, resistance and route of administration [4–7]. Moreover, several of these drugs (e.g. nifurtimox, benznidazole and melarsoprol) present a non-specific mode of action that accounts for their high toxicity [8, 9]. Thus, the discovery of new chemical entities targeting specific and indispensable components of parasite metabolism is a priority for trypanosomiasis and leishmaniasis.
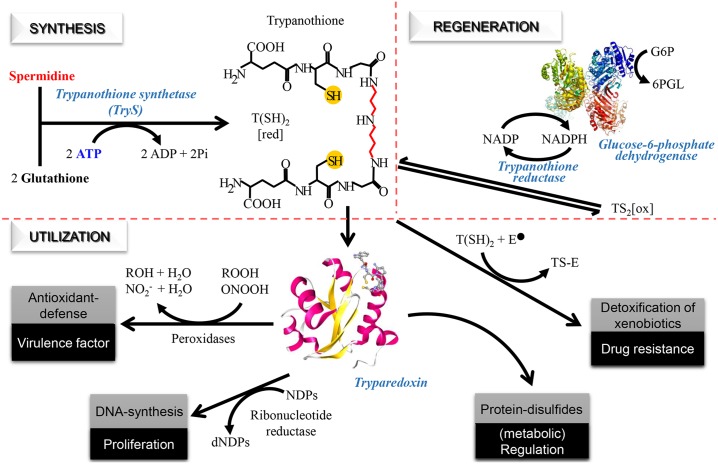
The thiol-dependent redox metabolism is one of the unique metabolic features that distinguish trypanosomatids from humans and offer reliable molecular targets for selective drug development [10]. An example of genetic hallmarks of trypanosomatids is the lack of genes coding for glutathione reductase and thioredoxin reductase [11–13], which fuel the major redox systems of most living organisms (i.e. the glutathione/glutaredoxin system and the thioredoxin system) with reducing equivalents. Instead, trypanosomatids rely on the low molecular mass thiol N1, N8-bis(glutathionyl)spermidine [trypanothione, T(SH)2] and the flavoenzyme trypanothione reductase for sustaining the intracellular redox homeostasis (Fig 1). T(SH)2 delivers reducing potential to different redoxin proteins, which, by acting on different targets, control vital functions (for a review see [10]; Fig 1). Proposed functions of T(SH)2 further comprise neutralization of xeno- and endobiotics (e.g. methylglyoxal, iron and nitric oxide), coordination of iron-sulfur complexes and reduction of ascorbate [14, 15].
Fig 1. Trypanothione dependent redox metabolism.
The chemical structure of trypanothione (N1,N8-bis(glutathionyl)spermidine; T(SH)2) is depicted at the center. Synthesis: trypanothione synthetase catalyzes the ligation of two molecules of gluthatione to one of spermidine using the energy provided by two ATP molecules. Regeneration: trypanothione reductase maintains trypanothione in the reduced state at expenses of NADPH, which can be supplied by the oxidative phase of the pentose phosphate pathway via glucose 6-phosphate dehydrogenase. Utilization: reduced trypanothione is involved in multiple functions such as the detoxification of xenobiotics, cell proliferation, defense against oxidants and protein thiol-redox homeostasis. The multipurpose oxidoreductase tryparedoxin plays an important role catalyzing electron transfer from T(SH)2 to different molecular targets (e.g. peroxidases, ribonucleotide reductase and protein disulfides). G6P: glucose-6-phosphate, 6PGL: 6-phosphogluconolactone, T(SH)2: reduced trypanothione, TS2[ox]: oxidized trypanothione, NDPs: nucleosides diphosphate, dNDP: deoxinucleosides diphosphate, E-: electrophilic species, TS-E: trypanothione-electrophile adduct, ROOH: hydroperoxide, ONOOH: peroxynitrite, NO2-: nitrite.
The biosynthesis of T(SH)2 is achieved in two consecutive steps each involving the ligation of a glutathione (GSH) molecule by its glycine carboxyl group to the free N1 and N8 amine groups of spermidine (SP). Both reactions are catalysed by the C-terminal ligase domain of trypanothione synthetase (TryS; EC 6.3.1.9) at the expense of ATP (Fig 1). Some trypanosomatids species harbour (L. infantum, L. donovani, L. mexicana and T. cruzi) or express (Crithidia fasciculata) a gene coding for glutathionylspermidine synthetase (GspS; EC 6.3.1.8), which synthesizes the reaction intermediate N8 mono-glutathionylspermidine.
The importance of TryS activity for parasite viability has been demonstrated in vitro and in vivo for T. b. brucei [16–19] and L. infantum [20] by means of genetic and pharmacological approaches. In addition, TryS presents several advantages as a drug target candidate: (i) it is encoded by a single copy gene [11–13], (ii) the structure of TryS from L. major has been elucidated [21], (iii) TryS has been shown to provide metabolic control to the trypanothione pathway in T. cruzi [22], and (iv) kinetic information is available for several TryS [18, 22–27].
At an early state of knowledge, the rational inhibitor design was undertaken using GspS of C. fasciculata (CfGspS) and Escherichia coli (EcGspS) as test enzymes and compounds isosteric with GSH or related transition state analogues as chemical scaffolds [28–34]. Preliminary studies with GSH analogues identified the γ-glutamyl moiety as critical for molecular recognition [28]. Further work revealed that addition of acidic groups to the L-γ-Glu-L-Leu dipeptide resulted in CfGspS inhibitors of reasonable potency such as the phosphonic (Ki ~ 60 μM) [29], the boronic (Ki ~ 81 μM and Ki* ~ 18 μM) [34] or the diaminopropionic acid derivative (Ki ~ 7.2 μM and Ki* ~ 21 μM) [35].
Transition state mimics, previously identified as potent inhibitors of EcGspS [30–32], proved to be equally active against recombinant CfGspS. Notably, a Gsp-phosphinate derivative was capable to inhibit recombinant TryS from L. major, T. cruzi and T. b. brucei albeit with apparent Ki values 16–40-fold higher than that obtained for CfGspS (Ki of 18.6 nM) [36]. Unfortunately, these compounds displayed null biological activity against pathogenic trypanosomatids at 100 μM. Nevertheless, this phosphinate remained the only compound able to target TryS from three different pathogenic trypanosomatids.
The anti-proliferative activity of GSH derivates (N,S-blocked GSH diesters, S-2,4-dinitrophenyl-GSH) [37–39] and GSH-related phosphinopeptides [40] ranked from 35 to 0.2 μM in different trypanomatids. The poor biological activity exhibited by these inhibitors has been ascribed to their peptidic nature susceptible to hydrolysis by esterases and amidases [37].
Based on the ATP-dependency of TryS, a compound library of protein kinase inhibitors was screened against recombinant CfTryS. This led to the identification of N5-acetamide paullones (benzo[2,3]azepino[4,5-b]indol-6-ones) as potent inhibitors of CfTryS [10, 41]. Recently, we reported the finding of a related 10-trifluormethylated acetamide derivative of this paullone as nM inhibitor of LiTryS (IC50 350 nM) [20]. Information on the activity of these paullones against TryS and parasites from other pathogenic species is lacking.
More recently, a HTS campaign against TbTryS with a library of ~ 62000 compounds identified several hits that upon optimization yielded leaders with inhibitory activity against TbTryS in the nM range (IC50 values of 45, 95 and 140 nM for DDU86439, DDD85811 and DDD86243, respectively) and EC50 values towards infective T. b. brucei that ranked from 5 to 10 μM. The compounds allowed the chemical validation of TbTryS as drug target [17, 18].
We here describe the setting-up of a screening technique to detect TryS inhibitors and report the identification and chemical validation of inhibitors from TryS from three major trypanosomatid species (Trypanosoma brucei brucei: T. b. brucei, Trypanosoma cruzi: T. cruzi and Leishmania infantum: L. infantum).
Materials and Methods
Organisms and Reagents
Unless otherwise stated all reagents were of analytical grade and purchased from Sigma-Aldrich, J.T. Baker, Carlo Erba Reagents SA, Gibco, Invitrogen, Life Technologies, Enzo Life Sciencies, Roche.
Heterologous Expression and Purification of Recombinant TryS
TryS from different trypanosomatids was produced in recombinant form with an N-terminal His-tag. The constructs pET-15b TbTryS [27], pRSET-B TcTryS and pET-28c(+)LiTryS [20] were kind gifts of Alan Fairlamb (Dundee University, Dundee, Scotland), Sergio Guerrero (Universidad Nacional del Litoral, Santa Fe, Argentina) and Helena Castro (Institute for Molecular and Cell Biology, Porto, Portugal), respectively. They were used to express TryS of T. b. brucei 427 (MITat1.4, GenBank accession protein id CAC87573.1), T. cruzi strain Tulahuen 0 (GenBank accession protein id AAO00722.1) and L. infantum JPCM5 (GenBank accession protein id CAM69145.1). E. coli strain BL21 (DE3) or Tuner (DE3) (Novagen) served as expression host. For a detailed description of the expression and purification protocols see S1 Text.
Protein concentration was determined using the Bicinconinic Acid assay with bovine serum albumin as standard. The protocols described above yielded 4–8 mg of recombinant TryS per liter of culture medium with ≥ 95% purity and homogeneous specific activity.
Kinetic Characterization
The kinetic characterization of His-tagged TryS was performed using the LDH/PK assay which couples ATP regeneration to NADH oxidation. The end-point assay based on detection of inorganic phosphate (Pi) by the BIOMOL GREEN reagent was used to estimate the apparent Ki for ADP. All reactions were performed at room temperature (RT, 20–25°C) and a detailed description of both assays is provided in S1 Text. The apparent kinetic parameters (KM and Vmax) were calculated by fitting plots of initial velocity (v) vs. substrate concentracion ([S]), determined at saturating concentration of co-substrates, to the Michaelis Menten equation assisted by the software OriginPro 8. For GSH, the KM and Ki values were determined using the following equation v = Vmax / (1 + KM / [GSH] + [GSH] / Ki), which considers the nonproductive binding of GSH to the substituted enzyme [42]. The apparent Ki for ADP was estimated from linear fitting of the plots [E]/v vs. [ADP] at different concentrations of ATP, where [E] is enzyme concentration.
Compound Library
The compound library involves 144 chemical entities that are clustered by chemical scaffold as follow: (A) 6-arylpyrido[2,3-d]pyrimidine-2,7-diamine derivatives (APPDA; S1 Table) developed as ATP-competitive inhibitors of bacterial D-Alanine:D-Alanine ligase [43] and biotin carboxylase [44]; (B) 1-(benzo[d]thiazol-2-yl)-4-benzoyl-3-hydroxy-5-phenyl-1H-pyrrol-2(5H)-one derivatives (BBHPP; S2 Table); (C) N,N'-bis(3,4-substituted-benzyl) diamine derivatives (BDA; S3 Table) that display potent anti-malarial or -trypanosomal/leishmanial activity [45, 46] and are simplified derivatives of compounds interfering with the parasite’s polyamine metabolism [45, 47–50]; (D) benzofuroxan (BZ; S4 Table) [51–55]; (E) 4,5-dihydroazepino[4,5-b]indol-2(1H,3H,6H)-one derivatives (AI; S5 Table), some of which with reported anti-TryS or -T. b. brucei activity [56]; (F) 1H-purine-2,6(3H,7H)-dione (PD), including the 3-butyl-7-methyl-8-((3-(trifluoromethyl) phenylthio)methyl)3,4,5,7-tetrahydro-1H-2,6-dione) (kindly provided by Dr. Luise Krauth-Siegel, Heidelberg University, Germany; S6 Table) that was reported as inhibitor of the T(SH)2-dependent oxidoreductase tryparedoxin [57]; (G) 2-aminooxazole-5-carboxamide derivatives (AOCA; S7 Table); (H) and several singletons: N,N-dibenzyl-1-ethyl-3-methyl-1H-pyrazole-5-carboxamide; 2-amino-N,N-dibenzyl-4-methyl thiazole-5-carboxamide; prochlorperazine; tert-butyl 8-aminooctylcarbamate; tert-butyl 12-aminododecylcarbamate; N-(8-aminooctyl)acetamide.
TryS Screening Assay Development
General considerations
The BIOMOL GREEN assay was adapted for the screening of potential TryS inhibitors. All assays (screening and interference test) were performed manually (non-automated) in quadruplicates, at RT and using a total reaction volume of 50 μL in 96-well plates. The colorimetric reaction was initiated by adding 200 μL BIOMOL GREEN reagent and after 20 min incubation at RT the A650 was measured with a MultiScan EX plate reader (Thermo SCIENTIFIC). The interference of reagents (DMSO, NaCl, glycerol, DTT, SP and GSH) with the colorimetric reaction was evaluated individually using 20 μM K2HPO4 as Pi standard. The assay conditions (e.g. reagents, TryS units, time) were adjusted to yield optimal signal detection (20 μM Pi) taking into account the intracellular concentration reported or estimated for each substrate and/or to avoid interference with the colorimetric reaction.
The linearity range, detection limit (e.g. [Pi] corresponding to 3σn-1 the A650nm of the blank) and interference was estimated from plots of blank-corrected A650nm vs. [Pi] or the reagent concentration for each condition tested. The plots were prepared and analyzed with OriginPro 8 (OriginLab Corporation). The errors are expressed as 2σn-1.
Screening assay
For all TryS, ATP was used at 150 μM because of high background signal at higher concentrations and SP fixed at 2 mM, in agreement with the intracellular levels calculated from data reported in the literature for trypanosomatids [58–60]. GSH was adjusted to 0.05, 0.57 and 0.25 mM for TbTryS, TcTryS and LiTryS, respectively, to avoid substrate inhibition or to approach physiological concentrations [14].
A master mix (MM) solution containing all the substrates at 1.25-fold their end concentration in assay was prepared in screening reaction buffer (5 mM DTT, 10 mM MgSO4, 0.5 mM EDTA, 100 mM HEPES pH 7.4, 9 mM NaCl, 10% v/v DMSO) and kept on ice until use. Microtiter plate wells were loaded with 5 μL of test compound, DMSO (reaction control) or TryS-specific inhibitor (inhibition control) and 40 μL of MM. The reactions were then started by adding 5 μL of TryS (1, 2 and 3 x 10−5 μmol.min-1.mL-1 for TcTryS, LiTryS and TbTryS, respectively) and stopped after 15 min with 200 μL BIOMOL GREEN reagent.
The compounds used as inhibition control were added at concentrations that inhibited 50% TryS activity: 30 μM and 150 nM MOL2008 for TbTryS and LiTryS, respectively, and 30 μM J18 for TcTryS. Blanks were prepared for each condition by adding 5 μL of screening reaction buffer with 150 mM NaCl instead of enzyme.
The TryS activity is calculated as follow: % TryS activity = {[(A650nm Ci−A650 nm CiB) / (A650nm E−A650nm EB)] x 100}, where A650 nm Ci is the mean absorbance at 650 nm for the reaction test with compound i, A650 nm CiB is the mean absorbance at 650 nm for the blank control with compound i, A650 nm E is the mean absorbance at A650 nm for the reaction control with DMSO and A650 nm EB is the mean absorbance at A650 nm for the blank control with DMSO. The assays yielded an intra-assay coefficient variation (CV = σn-1E / E) ≤ 2.5%, a signal to background coefficient (E/EB) of ~ 3.5 and a Z’ factor [61] ≥ 0.85.
Screening work-flow
The solubility in DMSO of each compound was evaluated immediately prior to assay. Compounds insoluble at 3 mM in DMSO were excluded from the screening. Soluble compounds were stepwise diluted in DMSO up to 0.3 mM. For the primary screening all compounds were tested (inhibition and blank) at a fixed concentration of 30 μM. Compounds that decrease or increase TryS activity ≤ 55% or ≥ 105%, respectively, were tested for interference as described in S1 Text.
For compounds that at 30 μM decreased TryS activity ≤ 55%, the IC50 values were obtained from 10-point dose-response curves (% TryS activity ± 2σn-1 vs. Log nM compound) fitted to a four-parameter sigmoidal equation (Boltzmann model). The reaction conditions were identical to those described for the screening assay and, whenever corresponding, the TryS activity values were corrected by the interference at each compound’s concentration.
Biology
Proliferation assays for parasites and macrophages
The biological activity of selected compounds was evaluated in the infective form of T. b. brucei strain 427, cell line 514–1313 (WT) and cell line RNAi-TryS [16], as well as Leishmania infantum promastigotes (MHOM MA67ITMAP263; kindly provided by Dr. Helena Castro and Dr. Ana Tomas, IBMC, Portugal). The assay conditions to assess the cytotoxicity of compounds towards parasites and murine macrophages (cell line J774) are described in S1 Text. All assays were performed in duplicates or triplicates.
Western blot analysis of TryS expression
Total cell extracts from the WT and RNAi-TryS cell lines of T. b. brucei grown in the presence or absence of oxytetracycline were separated under reducing conditions on an SDS / 10% PAGE. The blotted membranes were incubated with mouse polyclonal antibodies against TbTryS (dilution 1:1000) followed by a horseradish peroxidase-conjugated anti-mouse IgG (dilution 1:10000). Reactive bands were detected by chemiluminescence using ECL plus (GE Healthcare) and quantified by densitometric analysis using the free software ImageJ (http://imagej.nih.gov/ij/).
Analysis of thiols
The method is a modified version of that described in [62]. Briefly, L. infantum promastigotes or bloodstream T. b. brucei (WT and RNAi-TryS cell lines) treated or not with compounds (MOL2008 or EAP1-47, respectively) for 24 h were harvested by centrifugation (2000g, 10 min at 4°C), washed twice with PBS and resuspended at a density of 108 cells/0.1 mL of 40 mM Hepes pH 8.0 and 2 mM mono-bromobimane in methanol. After incubation at 70°C for 3 minutes, the samples were cooled on ice, then added of 100 μL 4 M methanesulfonic acid (adjusted to pH 1.5 with LiOH) and incubated overnight at 4°C. The supernatant containing the derivatized low molecular mass thiols was centrifuged at 13000g 40 min at 0°C. Labeled thiols were separated using solvent A: 0.25% camphorsulfonic acid pH 2.64 and solvent B: 25% 1-propanol in solvent A and the following conditions: 100 μL of the sample, corresponding to 5x107 cells, was injected onto a C18, 5 μm, 4.6 x 150 mm column connected to an HPLC (both from Agilent) pre-equilibrated in 12% solvent B followed by a linear gradient from 12–50% solvent B over 36 minutes at a flow of 1 mL/min. Specific thiol concentrations were determined from peak areas relative to different masses of derivatized standards (i.e. 100, 75, 50, 25, 10, 5 and 0.5 nmol) of GSH and T(SH)2, the last produced according to [63].
Accesion numbers
Results
Kinetic parameters of TryS and its implicancies for enzyme regulation and inhibition
The kinetic studies of TryS employed in this work were performed with three major aims, first to test the quality of the recombinant enzymes and establish optimal assay conditions, second, to disclose kinetic data previously not addressed for TryS from T. cruzi strain Tulahuen 0 and L. infantum (strain JPCM5) and a reaction product (ADP), and third, to compare the kinetic behavior of different TryS and its implication for enzyme regulation and inhibition. The kinetic data available for TryS from earlier [18; 22–27] and the present work are presented in Table 1 and S1 Fig.
Table 1. Kinetic parameters for recombinant tritryp trypanothione synthetase (TryS).
| Parasite | T. cruzi | T. b. brucei | L. major [26] | L. infantum [20] | C. fasciculate [23] | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Silvio X10 clone 7 [25] | Tulahuen 0 | Ninoa [22] | 427 (MITat1.4) [27] | 427a [24] | Friedlin | JPCM5 | USb | ||
| KM GSHc | 570 ± 50 | 140 ± 13 | 760 ± 210 | 56.2 ± 10.7d | 23.8 ± 2.3e | 142 ± 20 | 34 ± 4 | 89 ± 7 | 170 ± 23 | 1175 |
| Ki GSH | 1200 ± 100 | 1690 ± 172 | Nof | 36.5 ± 6.7d | 55 ± 6e | 254 ± 45 | 143 ± 20 | 1000 ± 80 | 699 ± 75 | Yes |
| KM SPg | 625 ± 39 | 702 ± 70 | 860 ± 95 | 37.8 ± 5.0d | 45.4 ± 2.0e | 238 ± 25 | 92 ± 25 | 940 ± 140 | 1434 ± 135 | 7424 |
| KM ATP | 53 ± 3 | 41 ± 6 | 70 ± 40 | 7.1 ± 0.4d | 8.6 ± 0.6e | 19 ± 5 | 6.6 ± 0.5 | 63 ± 2 | 43 ± 7 | 52 |
| Ki ADP | NDh | 60 ± 6 | NDh | NDh | NDh | 40 ± 5 | NDh | NDh | 90 ± 8 | NDh |
The apparent KM and Ki values (μM) were determined using the lactate dehydrogenase (LDH)/pyruvate kinase (PK) assay and the Ki ADP was assessed using the end-point assay. Whenever available, the values are expressed as the mean ± S.D.. The parameters determined for the N-terminal His-tagged tritryp TryS studied in this work are shown in bold letters and the references to data published earlier in brackets. Please, refer to S8 Table for details about the conditions employed in the kinetic characterization of each enzyme.
a Trypanosoma brucei brucei, strain 427, cell line 449.
b US, unknown strain.
c GSH, glutathione
d Kinetic parameters determined for untagged Trypanosoma brucei TryS (TbTryS) using the PK/LDH coupled assay [27].
e Kinetic parameters determined for untagged TbTryS using the end-point assay [18].
f No, denotes lack of GSH inhibition at 4.5 mM.
g SP, spermidine.
h ND, not determined.
LiTryS presented apparent KM values of 166 ± 62 μM for GSH, 1335 ± 167 μM for SP and 42 ± 10 μM for ATP, and a apparent Ki for GSH of 680 ± 160 μM, all of which are in the same order of magnitude as those reported for non-tagged TryS from the related species L. major [26].
The apparent kinetic parameters of TcTryS strain Tulahuen 0 (KM GSH = 123 ± 23 μM, KM SP = 685 ± 105 μM, KM ATP = 41 ± 6 μM and Ki GSH = 1600 ± 230 μM) are similar to those published for TcTryS strain Silvio X10 clone 7 [25] and strain Ninoa [22], except that inhibition by GSH was not observed for TryS from the last strain [22].
The apparent KM values for substrates and Ki for GSH obtained for His-tagged TbTryS (KM GSH = 135 ± 43 μM, KM SP = 238 ± 51 μM, KM ATP = 18 ± 6 μM and Ki GSH = 242 ± 102 μM) were almost 3- to 7-fold higher than those reported for the untagged version of this protein (KM GSH = 23.8–56.2 μM, KM SP = 37.8–92 μM, KM ATP = 6.6–8.6 μM and Ki GSH = 36.5–143 μM) by different laboratories [24, 27], which may be ascribed to different assay conditions (summarized in S8 Table) as highlighted in previous publications [23, 24]. Nonetheless, all values determined here for His-tagged tritryp TryS differed in less than one order of magnitude from those reported for tag-free versions from identical or homologue proteins, hence the recombinant form of the enzymes were rated as suitable for the screening assay.
Inhibition by ADP has been reported to occur for CfGspS with a Ki of 80 μM [64] but information is lacking for related enzymes from pathogenic trypanosomatids. As shown here, ADP competed for the ATP-binding site of TbTryS, LiTryS and TcTryS (S1 Fig) with apparent Ki values of 40 ± 5 μM, 90 ± 8 μM and 60 ± 6 μM, respectively (Table 1). It is worth to note that these Ki values represent overall estimates of product inhibititon because the assay conditions used do not allow distinguishing a preferential inhibition of the first or second biosynthetic step catalyzed by TryS.
Comparison of the kinetic parameters for TryS from three major trypanosomatid species obtained under similar experimental conditions (this study), shows that the enzyme from African trypanosomes presents KM values for ATP and SP that are remarkable lower (2.3‒5.6-fold) than those for TryS from L. infantum and T. cruzi (Table 1). A similar conclusion can be drawn from studies carried out for related TryS in other laboratories [18, 22, 24–27]. At variance with Leishmania spp. and T. cruzi, T. brucei spp. is an extracellular pathogen that fully relies on de novo synthesis of polyamines and glycolysis to fulfil its metabolic and energetic needs [58, 65]. Thus, TryS from African trypanosomes presents kinetic parameters that guarantee the production of the indispensable metabolite trypanothione [16, 17] under conditions of restricted supply of ATP and SP that the parasite may face during its complex life cycle (e.g. differentiation, which entails drastic reprogramming of energetic metabolism, and different nutrient availability in vector and host). All three tritryp TryS displayed similar KM values for GSH, which contrast with earlier studies reporting a higher affinity of T. b. brucei TryS for this substrate [18, 22, 24, 27]. As previously reported [18, 23–27], tritryp TryS were susceptible to substrate inhibition by GSH, although the inhibitory efficiency varied between species as follows: T. b. brucei (apparent Ki/KM GSH = 1.8) > L. infantum (apparent Ki/KM GSH = 4.1) > T. cruzi (apparent Ki/KM GSH = 13.0). This suggests that the T. b. brucei enzyme is particularly sensititive to inhibition by the substrate GSH. However, the physiological role of this inhibition mechanism, if any, remains questionable because substrate accumulation will further enhance TryS inhibition and GSH cannot surrogate T(SH)2 functions in T. brucei [16]. For the leishmanial and T. cruzi TryS, substrate inhibition will become relevant only at high concentrations of GSH (e.g. > 1.5 mM), which, except for a few examples of L. donovani parasites grown to mid-log phase (e.g. 2.27 mM GSH for axenic amastigotes from the strain BOB and 1.68 mM for promastigotes from the strain LV9) [14], appears to be a non-physiological condition. Indeed, L. infantum parasites harbouring a single trys allele and with a TryS content about 50% lower than that of wildtype cells show no phenotype in vitro and in infected animals [20].
The Ki ADP / KM ATP ratios for tri-tryp TryS range from1.5 to 2.2-fold, suggesting that ADP may be a physiological modulator of TryS activity. However, taking into account that the intracellular concentration of ATP for trypanosomatids (e.g. 2–4 mM and 0.58 mM for infective T. b. brucei [65, 66] and T. cruzi [67], respectively, and 0.87 mM for L. donovani promastigotes [68]) is > 14-fold in excess with respect to the respective KM values for tritryp TryS (e.g. 18–60 μM; Table 1) and that the ATP/ADP ratio reported or estimated by us for trypanosomatids is between 3 to 10 [65, 66, 69–71], such regulatory role of ADP in T(SH)2 biosynthesis may be questioned. A recent kinetic analysis of TbTryS assisted by computational modeling of the enzymatic mechanism [24] highlighted that the activated enzyme is particularly sensitive to inhibition by GSH and T(SH)2. Because the experimental setup employed in this study did not consider inhibition by ADP and excluded ADP as an integral component of the enzyme activated complex, the real contribution of this product to TryS inhibition remains to be addressed.
Screening
Assay development and validation
Under reaction conditions resembling the screening assay and using KH2PO4 as Pi standard, the detection limit and linearity range of the colorimetric reaction with BIOMOL GREEN was ~ 1.9 μM Pi and 2‒50 μM Pi, respectively (S2 Fig). DTT and NaCl (salt present in enzyme buffer) added up to 5 mM and 30 mM, respectively, do not affect assay’s readout (S3A and S3D Fig). DMSO (compound solvent) and glycerol (cryoprotective agent added to TryS) interfered with BIOMOL GREEN signal in an additive fashion. For instance, 10% v/v DMSO (e.g. concentration used in the assay) and 4% v/v glycerol reduced assay sensitivity 8.4 ± 1.5% (n = 8) and ~ 12% (S3C Fig), respectively, whereas addition of both reagents (DMSO 10% v/v and glycerol 4% v/v) decreased ~ 25% assay sensitivity (S3D Fig). In order to reduce the signal interference, assays were performed at glycerol concentrations < 1% v/v.
Assay linearity is affected at ATP concentrations > 150 μM due to increase background signal and ADP-mediated inhibition of TryS (S1 Fig; see previous section). Thus, for practical reasons ATP was adjusted to 150 μM, a concentration that is 4‒27-fold lower than the estimated physiological concentration of the substrate in different trypanosomatids [66–70] but ~3.5 and 8-fold higher than the KM values for TcTryS and LiTryS, and TbTryS, respectively (Table 1).
GSH added up to 2.5 mM does not interfere with the colorimetric signal. For assays with TcTryS, GSH was used at 570 μM, a value within the physiological concentrations reported for infective forms of T. cruzi strains (i.e. 340 to 1180 μM) [14]. For the screening involving LiTryS, GSH was employed at a concentration of 250 μM, which is near the intracellular content estimated for amastigote forms of L. donovani (250 μM) and L. major (310 μM) isolated from hamsters and mice, respectively [14]. In order to avoid substrate inhibition of TbTryS, GSH was adjusted to 50 μM a value close to the corresponding KM (Table 1) and 5- fold lower than the intracellular concentration (i.e. 230–260 μM) reported for this substrate in trypanosomes isolated from infected rats [14], respectively.
Irrespective of the TryS species, SP, a substrate not interfering with the colorimetric reagent (S3B Fig), was fixed to 2 mM for the screening assays. This concentration is 8.4-, 2.9- and 1.5-fold higher than the KM values for TbTryS, TcTryS and LiTryS, respectively and close to that we estimated for the clinically relevant forms of T. b. brucei (SP ~ 2 mM for bloodstream stage), T. cruzi (~ 3 mM for amastigotes and trypomastigotes) and L. mexicana (~ 7 mM for amastigotes) according to information available on polyamine content [58–60], cell volumes and/or total protein content per cell for each parasite species [14, 71, 72].
The linearity A650nm corr vs. time was evaluated for each TryS at substrates concentrations used in the screening assay (see Experimental Procedures, S4 Fig). For LiTryS (2.3 x 10−5 μmol.min-1.mL-1), the assay was linear for at least 30 min whereas the production of Pi by TcTryS (3.5 x 10−6 μmol.min-1.mL-1) and TbTryS (1.5 x 10−5 μmol.min-1.mL-1) displayed a linear relation up to 15 min. As indicated above, the trypanosomal TryS were more sensitive than LiTryS to inhibition by ADP, which may explain the lower turnover of the former at longer reaction times. Therefore, a reaction end-point of 15 min was selected for the screening assays, which corresponds to the formation of 20 μM Pi and ADP for all three enzymes.
The assay conditions reported above for each enzyme yielded a Z’ factor [61] of ≥0.85, a signal background coefficient (E/EB) of ~ 3.5 and a CV (σn-1E/E) intra-assay ≤ 2.5%, which validate the screening test as highly robust and reproducible.
Compound screening at near physiological concentrations of substrates
The screening work flow is shown in Fig 2 and all data here reported correspond to assays that fulfilled the quality control parameters indicated above.
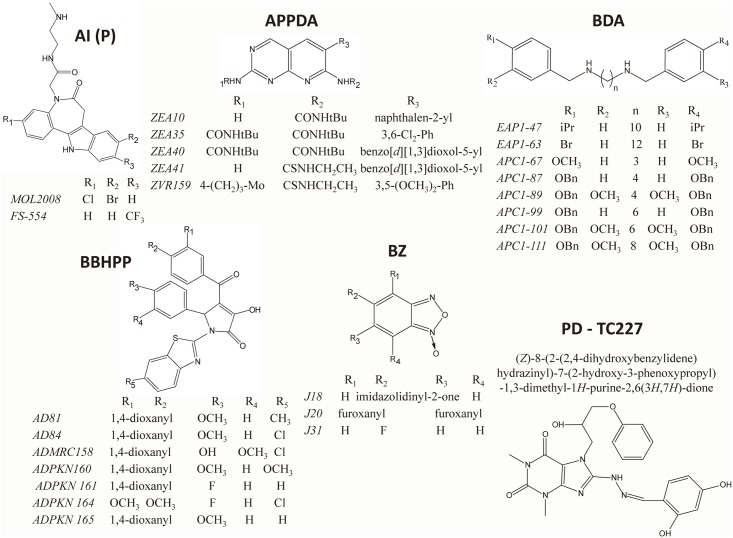
Fig 2. Structure of compounds affecting tritryp trypanothione synthetase activity.
AI (P), 4,5-dihydroazepino[4,5-b]indol-2(1H,3H,6H)-one derivatives, paullones derivatives, (FS-554 and MOL2008), five APPDA, 6-arylpyrido[2,3-d]pyrimidine-2,7-diamine derivatives (ZEA10, ZEA35, ZEA40, ZEA41 and ZVR159), eight BDA, N,N'-bis(3,4-substituted-benzyl) diamine derivatives (EAP1-47, EAP1-63, APC1-67, APC1-87, APC1-89, APC1-99, APC1-101 and APC1-111), seven BBHPP, 1-(benzo[d]thiazol-2-yl)-4-benzoyl-3-hydroxy-5-phenyl-1H-pyrrol-2(5H)-one derivatives (AD81, AD84, ADMRC158, ADKPN160, ADKPN161, ADKPN164 and ADKPN165), three BZ, benzofuroxan derivatives (J18, J20 and J31) and one PD, 1H-purine-2,6(3H,7H)-dione derivatives [(Z)-8-(2-(2,4-dihydroxybenzylidene)hydrazinyl)-7-(2-hydroxy-3-phenoxy propyl)-1,3-dimethyl-1H-purine-2,6(3H,7H)-dione, TC227]. iPr, tBu, OBn, Mo and Ph, correspond to an isopropyl, tert-butyl, O-benzyl, 4 -morpholinyl and phenyl substitution, respectively.
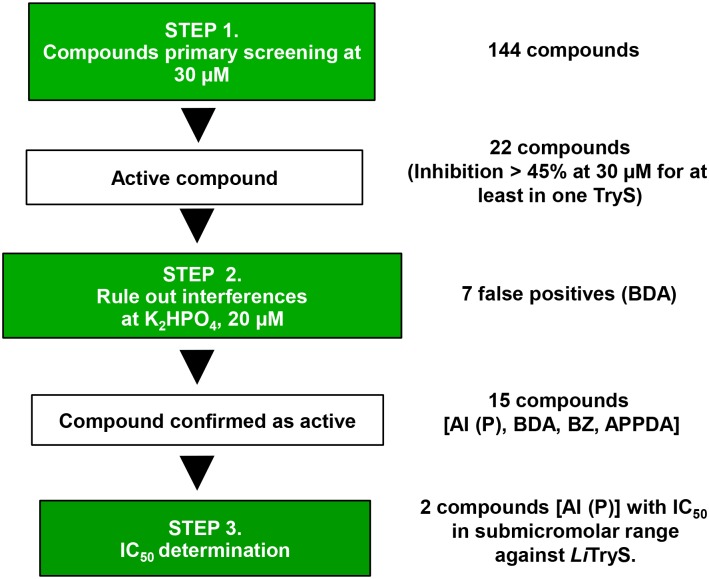
The initial screening performed with 144 compounds belonging to 7 different families (Fig 3 and S1–S7 Tables) and tested at 30 μM against tritryp TryS, allowed the identification of 22 compounds that lowered Pi signal to ≤ 55%. An assay interference counter-screen confirmed 15 of them as true TryS inhibitors (Table 2) and the remaining 7, all being BDA, as false positive readouts (S3 Table).
Fig 3. Screening work flow.
The different steps, the most relevant assay conditions and the go/no-go criteria of the screening campaign are indicated in boxes. The figures on the right refer to the number of compounds screened and that subsequently advanced during the campaign. From 144 compounds, 22 compounds lowered assay signal ≥ 45% for at least one TryS. From these 22, 7 BDA were false positive and the remaining 15 compounds were confirmed as enzyme inhibitors. Two of them are AI with potency in the submicromolar range against LiTryS. AI (P), 4,5-dihydroazepino[4,5-b]indol-2(1H,3H,6H)-one derivatives (P, paullone); APPDA, 6-arylpyrido[2,3-d]pyrimidine-2,7-diamine derivatives; BZ, benzofuroxan derivatives; BDA, N,N'-bis(3,4-substituted-benzyl) diamine derivatives.
Table 2. Inhibitors of tritryp trypanothione synthetase (TryS) identified in this work and their biological activity.
| Compound | Structurea | TcTryS | LiTryS | TbTryS | T. b. Brucei EC50 (μM)b | L. infantum EC50 (μM)c | SId |
|---|---|---|---|---|---|---|---|
| MOL2008 | AI (P) | 40.5 ± 5.9%; (1.00) | 0.15 ± 0.06 μM e; (0.99); 0.35 ± 0.03 f | 59.0 ± 6.0%; (0.97) | 4.3 ± 0.7 | 12.6 ± 1.6 | 2.4 (0.8) |
| FS-554 | AI (P) | 55.5 ± 3.8%; (1.02) | 0.35 ± 0.05 μM e; (0.99); 0.36 ± 0.04 f | ~75 μM; (0.94 at 300 μM) | ND | 112.3 ± 1.1 g | ND (0.6) |
| ZEA10 | APPDA | 32.3 ± 4.0% | 32.1 ± 6.4% | 48.7 ± 2.8%; (1.41) | 0.28 ± 0.08 | ND | >4 |
| ZEA35 | APPDA | 61.3 ± 7.6%; (1.01) | 13.8 ± 9.7% | 24.3 ± 6.1% | ND | ND | ND |
| ZEA40 | APPDA | 52.2 ± 1.4%; (1.02) | 15.3 ± 8.3% | 25.3 ± 5.2% | ND | ND | ND |
| ZEA41 | APPDA | 65.4 ± 1.5%; (0.99) | 13.6 ± 3.4% | 20.9 ± 4.4% | ND | ND | ND |
| EAP1-47 | BDA | 53.5 ± 1.3%; (1.48) | 47.8 ± 1.8%; (1.34) | 51.1 ± 4.2%; (1.49) | 0.20 ± 0.02 | ND | 15 |
| EAP1-63 | BDA | 30.8 ± 3.8% | 25.9 ± 9.3% | 47.5 ± 2.8%; (1.30) | 0.090 ± 0.007 | ND | 124 |
| EAP1-67 | BDA | 23.8 ± 6.6%; (1.34) | 51.5 ± 5.3%; (1.35) | 42.4 ± 1.6%; (1.33) | ND | ND | ND |
| APC1-87 | BDA | 26.6 ± 4.1% | 56.7 ± 7.9%; (1.26) | 37.9 ± 4.0% | ND | ND | ND |
| APC1-89 | BDA | 25.6 ± 1.4% | 23.4 ± 2.7%; (1.32) | 50.9 ± 1.5%; (1.31) | 0.061 ± 0.001 | ND | 164 |
| APC1-99 | BDA | 33.4 ± 4.1% | 39.1 ± 3.4% | 48.9 ± 1.7%; (1.05) | 0.015 ± 0.001 | ND | 67 |
| APC1-101 | BDA | 17.0 ± 2.1%; (1.40) | 26.3 ± 10.1%; (1.36) | 61.2 ± 1.8%; (1.33) | 0.28 ± 0.09 | ND | 10 |
| APC1-111 | BDA | 28.9 ± 4.1%; (1.57) | 49.6 ± 5.7%; (1.51) | 55.6 ± 3.2%; (1.52) | 0.040 ± 0.001 | ND | 67 |
| J18 | BZ | 63.5 ± 1.5%; (1.00) | 41.3 ± 3.6%; (1.00) | 7.3 ± 5.9% | ND | ND | ND |
In vitro and in vivo (cell) activity profile of most promising TryS inhibitors. Enzyme inhibition is expressed as % TryS inhibition ± 2σn-1 and IC50 values in μM concentration ± 2σn-1. For compounds affecting BIOMOL GREEN signal, the interference factor used to correct TryS activity is provided in brackets (see S1 Text). All values reported stem from at least duplicates. ND, not determined.
a the chemical scaffolds are: AI (P), 4,5-dihydroazepino[4,5-b]indol-2(1H,3H,6H)-one derivatives, paullone derivatives; APPDA, 6-arylpyrido[2,3-d]pyrimidine-2,7-diamine derivatives; BDA, N,N'-bis(3,4-substituted-benzyl) diamine derivatives; BZ, benzofuroxan derivatives.
b Cytotoxicity against bloodstream Trypanosoma brucei brucei expressed as effective concentration 50: EC50.
c Cytotoxicity against Leishmania infantum promastigotes is expressed as effective concentration 50: EC50.
d Selectivity index calculated as the ratio: EC50 for murine macrophages (cell line J774) vs. EC50 for T. b. brucei or EC50 for L. infantum (value in brackets).
e IC50 value calculated from 3 independent experiments and expressed as mean ± 2 S.D.
f Slope from IC50 plots calculated from 3 independent experiments and expressed as mean ± 2 S.D.
g EC50 value reported by [20]
There was no correlation between the rate of hit identification for each enzyme (i.e. 5.6% for TbTryS and 4.2% for TcTryS and LiTryS), and the potency of the compounds (i.e. 2 out 6 were nM inhibitors of LiTryS whereas all compounds targeting the trypanosomal TryS were two digit μM inhibitors, see Table 2). Supporting the existence of species-specific differences between TryS, the individual enzymes were preferentially targeted: LiTryS by AI; TbTryS by BDA and TcTryS by APPDA. Only 4 compounds out of 15, each two BDA and AI, were able to target multiple TryS, though to different extent. EAP1-47 [N1,N10-bis(4-isopropylbenzyl)decane-1,10-diamine] and APC1-111 [N1,N8-bis(4-(benzyloxy)-3-methoxybenzyl)octane-1,8-diamine] at 30 μM halved activity of all three TryS or TbTryS and LiTryS, respectively; MOL2008 and FS-554 were two digits nM inhibitors of leishmanial TryS (Fig 4) that, to a remarkable minor degree (~ 50% inhibition at 30 μM), also targeted both trypanosomal enzymes or TcTryS, respectively.
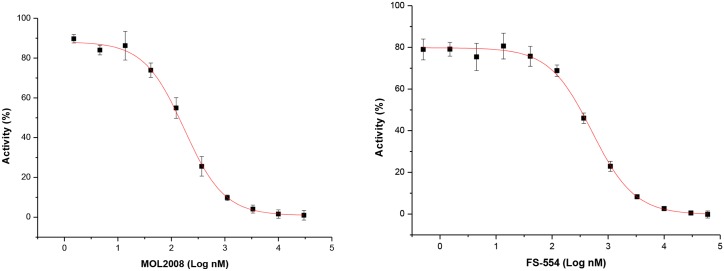
Fig 4. Inhibition plot of Leishmania infantum trypanothione synthetase (TryS) by paullones.
The data are presented as mean TryS activity (%) ± 2 S.D. (n = 4) as function of Log10 concentration (nM) of compound adjusted to a four-parameter Boltzmann sigmoidal equation. Representative dose-response plot obtained for MOL2008 (IC50 = 0.14 ± 0.03 μM; slope plot 0.38 ± 0.03) and for FS-554 (IC50 = 0.32 ± 0.73 μM; slope plot 0.37 ± 0.03).
Early investigations had shown that the N5-substituted paullone derivatives MOL2008 and FS-554 are potent inhibitors of TryS from C. fasciculata (IC50 = 30 nM) [10] and L. infantum (IC50 = 350 nM) [20], respectively, but information on their activity towards the homologue enzyme from related trypanosomatids was missing. MOL2008 was the most potent inhibitor of LiTryS (IC50 = 0.15 ± 0.06 μM) identified in the screening that displayed a moderate activity against TbTryS (IC50 ~ 30 μM) and TcTryS (40% TryS inhibition at 30 μM; see Table 2). On the other hand, FS-554 showed moderate activity against TcTryS (~50% inhibition at 30 μM) and a weak activity against TbTryS (~50% inhibition at 75 μM) (Table 2). Substitution of position N5 appears key to confer anti-LiTryS activity to AI since the inhibitory activity of unsubstituted derivatives was almost abrogated (compound 11a and 11b in S5 Table). Given the amine nature of the N5 substitutent, a potential SP-competitive mechanism of MOL2008 on LiTryS was investigated. Assays performed at fixed concentration of co-substrates (250 μM GSH and 150 μM ATP) and variable concentrations of SP (0.2 to 16 mM) show that enzyme inhibition was inversely proportional to polyamine concentration (S9 Table) and confirmed the proposed inhibition mechanism. A collection of 4-azapaullones substituted in position 9 and 11 was recently assayed for their anti-T. b. brucei and TbTryS activity [56]. Only derivatives containing in position 9 an α,β-unsaturated carbonyl substituted with a phenyl, phenyl(amino) or benzylamino group showed a minor anti-TbTryS activity (30–40% inhibition at 30 μM). Given the species-specificity displayed by tri-tryp TryS, the complete set of 4-azapaullones was tested against the L. infantum and T. cruzi enzyme (S5 Table). None of the compounds inhibited TcTryS to a significant level (enzyme inhibition ≤ 17% at 30 μM compound) whereas LiTryS showed again to be more susceptible to inhibition by the paullone scaffold, with 4-chloro-phenyl > furan-2-yl > 4-methoxy-phenyl analogues in 9-position reducing 32–44% enzyme activity at 30 μM. Identical substitutions (i.e. 4-methoxy-phenyl and 4-chloro-phenyl) in 11-position of 4-azapaullones rendered analogues with similar inhibitory activity (30–35%) towards LiTryS.
The BDA (S5 Fig, S3 Table) exerted a low to moderate inhibition of TryS that allows performing a SAR analysis. For all TryS, the inclusion of bulky substitutions (R1 = R4 = OBn: APC1-87, APC1-99 and APC1-109, or in addition R2 = R3 = OMe: APC1-111, APC1-101 and APC1-89) at the benzyl rings yields derivatives with higher activity (40–60% inhibition) than the unsubstituted congeners and analogues containing less bulky groups (R2 = R3 = OMe, R1 = R4 = OH) (S5A Fig). In contrast to LiTryS, the trypanosomal enzymes were particularly sensitive to the length of the linker, with derivatives bridged by 3- or 10-carbon being less active. This suggests a substituent additive effect to the length of the carbon chain with a possible steric fulfillment demand of the interacting pocket. For the halogenated BDA, the length of the linker appears to be a common determinant of activity against all TryS, whereas each enzyme displayed certain specificity for the nature of the halogen atom (S5B Fig). Thus, derivatives with a 12-carbon linker are more potent TryS inhibitors than those consisting of shorter chains and the halogen substitution yielding the highest activity (~50% inhibition at 30 μM) for this scaffold is F (EAP1-69), Cl (EAP1-67) and Br (EAP1-63) for TcTryS, LiTryS and TbTryS, respectively. On the other hand, the inclusion of an isopropyl group at position R1 = R4 of BDA with a 10-carbon linker (EAP1-47) improved up to 40% tri-tryp TryS inhibition when compared to the unmodified compound EAP1-9 and produced the best multi-TryS inhibitor here identified (50% inhibition at 30 μM).
From a group of 35 APPDA tested, three derivatives containing a substituted phenyl or benzyl group in position R3 (i.e. ZEA35, ZEA 40 and ZEA 41) showed selectivity and a moderate potency towards TcTryS (50% inhibition at 30 μM; Table 3). Replacing the R3 benzyl of ZEA41 by a naphthalen group (ZEA10) increased the activity against TbTryS (50% inhibition at 30 μM) and LiTryS (17% increased inhibition) but was detrimental to TcTryS inhibition (18% lower inhibitory activity). TcTryS was also target of selective inhibition by several BZ with J18 being the most active (IC50 ~30 μM), and the derivatives J20 and J21 reducing about 40% enzyme activity when tested at 30 μM. J18 produced a similar degree of inhibition of LiTryS, whereas TbTryS was almost refractory to inhibition by the BZ (S4 Table).
Table 3. Compounds enhancing tritryp trypanothione synthetase (TryS) activity.
| Compound | Core structurea | TcTryS | LiTryS | TbTryS |
|---|---|---|---|---|
| ZVR159 | APPDA | 168.6 ± 4.1; (1.10) | 100.6 ± 5.8 | 81.3 ± 1.6 |
| AD81 | BBHPP | 109.7 ± 8.9 | 83.9 ± 4.2 | 128.2 ± 3.6; (0.93) |
| AD84 | BBHPP | 98.7 ± 5.1 | 82.1 ± 4.0 | 119.0 ± 6.3; (0.95) |
| ADMRC158 | BBHPP | 97.4 ± 6.1 | 85.3 ± 8.7 | 119.1 ± 4.6; (0.88) |
| ADPKN160 | BBHPP | 97.6 ± 3.7 | 77.9 ± 3.6 | 116.4 ± 4.2; (0.87) |
| ADPKN161 | BBHPP | 90.8 ± 5.8 | 83.5 ± 9.2 | 126.7 ± 0.8; (0.86) |
| ADPKN164 | BBHPP | 99.6 ± 5.7 | 66.0 ± 3.4 | 123.2 ± 7.0; (0.83) |
| ADPKN165 | BBHPP | 107.1 ± 4.5 | 83.5 ± 6.0 | 166.3 ± 5.5; (0.79) |
| J20 | BZ | 64.4 ± 3.8 | 111.5 ± 14.5; (0.97) | 118.8 ± 2.4; (0.97) |
| J31 | BZ | 118.8 ± 6.1; (1.00) | 84.1 ± 0.001 | 96.2 ± 3.7 |
| TC227 | PD | 161.6 ± 5.3; (0.98) | 88.5 ± 7.1 | 102.0 ± 2.3 |
Enzyme activity is expressed as % TryS activity ± 2σn-1 and in brackets is provided the interference factor used to correct TryS activity of compounds affecting BIOMOL GREEN signal (see S1 Text). All values reported stem from at least triplicates. TcTryS, TbTryS and LiTryS are the trypanothione synthetase of Trypanosoma cruzi, Trypanosoma brucei and Leishmania infantum, respectively. ND, not determined.
a the chemical scaffolds are: APPDA, 6-arylpyrido[2,3-d]pyrimidine-2,7-diamine derivative; BBHPP, 1-(benzo[d]thiazol-2-yl)-4-benzoyl-3-hydroxy-5-phenyl-1H-pyrrol-2(5H)-one derivatives; BZ, benzofuroxan derivatives; PD, 1H-purine-2,6(3H,7H)-dione derivative.
Certain BBHPP (ADMRC150, ADMRC154, ADMRC159 and ADPKN164) showed specificity for targeting LiTryS with an average inhibitory activity of 35% at 30 μM and marginal or, even, enhancing activity of trypanosomal TryS (Table 3 and S2 Table).
Under the assay conditions used for the screening, none of the PD, AOCA and singletons inhibited tri-tryp TryS to a significant level (S6 and S7 Tables). In this respect, prochlorperazine has been early reported as a SP-competitive inhibitor of TbTryS with an IC50 of 19 μM, when assayed at a subsaturating concentration of the competing substrate (i.e. 25 μM SP) [18]. To investigate the potential of prochlorperazine to target TryS from other species and validate their mode of inhibition, we tested the activity of this compound against different TryS and at different SP concentrations. At 2 mM SP, the estimated intracellular concentration of this polyamine in trypanosomatids [58–60], none of the TryS was inhibited by prochlorperazine added at 30 and 300 μM. Lowering the SP concentration in the assay to 0.24 mM SP, 300 μM prochlorperazine reduces the activity of TbTryS, TcTryS and LiTryS by 58%, 34% and 0%, respectively. On the one hand, this result show that high intracellular concentrations of prochlorperazine have to be reached to impair T(SH)2 synthesis to a significant level and, on the other hand, highlights that more stringent assay conditions, as those representing nearly physiological substrate concentrations, will avoid the detection of hits with low pharmacological potential.
The screening also allowed the identification of compounds that increased 16–69% the activity of the trypanosomal TryS (i.e. for TbTryS: 7 BBHPP and 2 BZ and for TcTryS the APPDA ZVR159 and the PD TC227) (Table 3). Strikingly, none of the compounds tested here produced a similar stimulatory effect on the activity of the leishmanial enzyme. In order to shed light on the mechanism underlying the activation of TryS, the most active compounds were tested against trypanosomal TryS in the presence of different combinations of co-substrates. As shown in S10 Table, enhacement of TryS ATPase activity by non-chemically related compounds (ZVR159, TC227 and ADPKN165) was detected only when both co-substrates, GSH and SP, were present in the reaction mixture. These results are suggestive of an allosteric mechanism of TryS activation.
Biological activity of compounds inhibiting TryS from African trypanosomes and L. infantum
The biological activity of the most active inhibitors of TbTryS (i.e. IC50 ≤ 30 μM; MOL2008, ZEA10, EAP1-47, EAP1-63, APC1-89, APC1-99, APC1-101, APC1-111) and LiTryS (i.e. IC50 < 1 μM; MOL2008 and FS-554) was evaluated against the infective form of T. b. brucei or promastigotes of L. infantum and murine macrophages. Except for the paullone MOL2008 (EC50 4.3 ± 0.7 μM), all other TbTryS inhibitors presented anti-T. b. brucei activity in the nM range. The most active being the BDA APC1-99 (EC50 15 ± 1 nM), APC1-111 (EC50 40 ± 1 nM), APC1-89 (EC50 61 ± 1 nM) and EAP1-63 (EC50 90 ± 7 nM), followed by ZEA10, EAP1-47 and APC1-101 with EC50 between 200–280 nM (Table 2). In comparison to paullone FS-554 (EC50 112.3 ± 1.1 μM) [20], MOL2008 displayed a 10-fold higher anti-leishmanial activity (EC50 12.6 ± 1.6 μM).
The selectivity of the anti-parasitic effect was assessed using murine macrophages. Most compounds, except for MOL2008, FS-554 and ZEA 10, presented a selectivity index (SI) ≥ 10 with EAP1-63 (SI = 124) and APC1-89 (SI = 164) having the highest selectivity towards bloodstream T. b. brucei (Table 2).
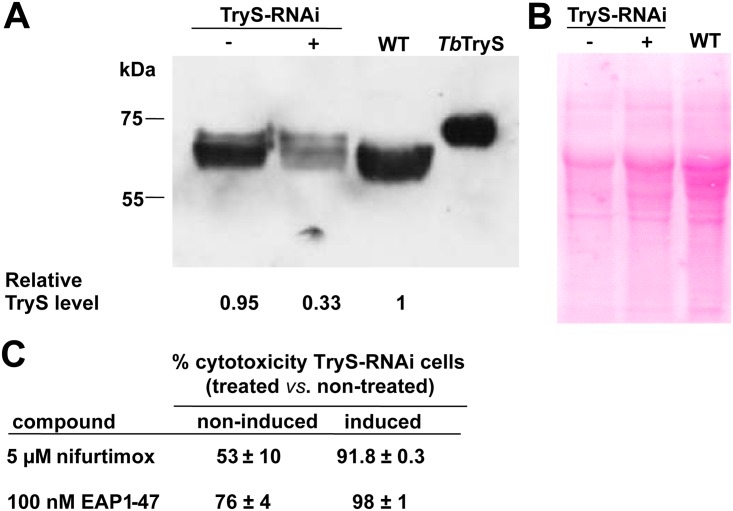
In order to get an insight into the on-target activity of promising compounds, the intracellular thiol content of WT parasites exposed for 24 h to EAP1-47 and MOL2008 was determined. For bloodstream T. b. brucei treated with 50 nM EAP1-47, T(SH)2 content decreases by 28% while GSH level increases by 39% with respect to untreated cells, which overall resembles the metabolite changes observed for parasites with 48 h RNAi-downregulated expression of TryS (S11 Table and [16]). A compound acting on T(SH)2 metabolism is expected to display an increased cytotoxicity against parasites depleted in TryS. The cytotoxic effect of 100 nM EAP1-47 towards trypanosomes with a TryS content that is 1/3 of that corresponding to non-induced RNAi or wild-type cells was slightly increased (1.3-fold; Fig 5). For comparison, nifurtimox, a drug inducing thiol depletion [73], added at 5 μM (EC50 determined for WT cells) displayed a 1.8-fold increased potency towards TryS-depleted parasites. Strikingly, identical assays performed with the chemically-related and trypanosome-selective compounds EAP1-63 and APC1-99 did not show differences in the potency of these compounds towards WT or TryS-depleted parasites. Taking together, these results shows that EAP1-47 is interfering with T(SH)2 biosynthesis, although the almost two order of magnitude difference between TbTryS inhibition and EC50 indicates that the compound has also other molecular targets in vivo. In contrast, the lack of enhanced cytotoxicity displayed by EAP1-63 and APC1-99 towards the TryS-RNAi induced cell line, is an strong indication that their potent trypanocidal activity is unrelated to interference with T(SH)2 metabolism.
Fig 5. Biological activity of compounds against infective Trypanosoma brucei brucei with downregulated expression of trypanothione synthetase (TryS).
A) Western blot analysis of cell extracts from 2x107 T. b. brucei from the wildtype (WT), 48 h tetracycline-induced (+) and non-induced (-) TryS-RNAi cell line. Two hundred ng of recombinant TbTryS was loaded as control. Bands from the molecular weight marker are indicated on left. The picture at the bottom shows the abundance of TryS for each condition as estimated by densitometry and expressed relative to the level of the WT cell line. B) Ponceau staining of the Western blot membrane that served as normalization control of protein load for each condition. C) Cytotoxicity (%) ± S.D. (n = 2) for tetracycline-induced (+) and non-induced (-) TryS-RNAi T. b. brucei treated with 5 μM nifurtimox or 100 nM EAP1-47.
On the other hand, L. infantum promastigotes in the log growth phase exposed to MOL2008 at its EC50 (12 μM) for 24 h presented a marked decrease in the intracellular pool of thiols. From two independent experiments, T(SH)2 and GSH content decreases ≥ 90% and > 30%, respectively, upon MOL2008 treatment (S11 Table). Interestingly and at variance with the metabolic changes observed for the genetic (RNAi) or chemical (EAP1-47) silencing of TryS in T. b. brucei, inhibition of TryS by MOL2008 did not lead to accumulation of the substrate GSH in L. infantum. Further experiments are required to establish whether the reduction on GSH level is caused by paullone-mediated inhibition of any of the two ATP-dependent enzymes in charge of glutathione synthesis or is a consequence of a species-specific regulatory mechanism triggered by T(SH)2 depletion.
Discussion
The screening of a compound library consisting of 7 major chemical scaffolds and several singletons against TryS from three major pathogenic species of trypanosomatids at near physiological concentration of substrates led to the identification of 15 inhibitor molecules with μM (APPDA, BDA, BZ, AI) and sub-μM (AI) potency and a remarkable species-specificity for the molecular target. Despite the high sequence identity between tri-tryp TryS (e.g. the amino acid sequence identity is 71.6% for TbTryS/TcTryS, 64.7% for TbTryS/LiTryS and 63.4% for TcTryS/LiTryS) and the almost strict conservation of residues involved in substrate binding [20, 21], only 4 compounds were able to target TryS from different species, with only one of them (a BDA) inhibiting all three TryS with moderate potency (50% at 30 μM). This behavior highlights the existence of structural differences between tritryp TryS that determine their specificity for ligands. In support of this observation, single aminoacid substitutions in the homologue enzyme from C. fasciculata were previously reported to produce drastic changes in enzyme activity [23, 36] and our comparative analysis of the kinetic parameters of tritryp TryS obtained under similar assay conditions reveals considerable differences between species.
Early investigations have shown that substitutions at different positions of the paullone scaffold yield nM inhibitors of TryS from C. fasciculata or L. infantum (N5-substituted AI) [10, 20] and potent anti-T. b. brucei agents with moderate inhibition towards TbTryS (11-substituted 4-azapaullone) [56]. By extending the analysis of AI activity towards tritryp TryS, we have confirmed that the inclusion of a N-[2-(methylamino)ethyl] acetamide in position N5 confers SP-competitive inhibition of LiTryS activity, since compounds lacking this substitution were almost inactive. From the tritryp TryS evaluated here, LiTryS presented the higher KM for SP, which indicates the enzyme binds less efficiently this substrate probably due to an unfavorable conformation of the polyamine binding site. However, the shape adopted by the SP-binding site of LiTryS appears suitable for accommodating the N5-substituent present in MOL2008 and FS554. In support of this hypothesis, the trypanosomal TryS displayed a 2- to 6-fold lower KM values for SP and a marked refractoriness to inhibition by N5-substituted paullones (> 200-fold higher IC50). The opposite behavior was observed for prochlorperazine, a SP-competitive inhibitor of TbTryS [18]. In this case, the degree of enzyme inhibition was inversely proportional to the SP KM value for each TryS, with TbTryS being the most sensitive to inactivation followed by TcTryS, whereas LiTryS was refractory to inactivation (300 μM prochlorperazine) even at a sub-KM concentration of SP (240 μM vs. KM of 1335 μM). Altogether, these data strongly point to the existence of remarkable differences in the polyamine binding site of tritryp TryS that should be carefully considered for the design of new AI with multi-species activity. In this respect, our study also demonstrated that in spite of the potent anti-T. b. brucei activity [56], AI substituted with α,β-unsaturated carbonyl chains in position 9 or 11 should be disregarded as ligands for future optimization because of their null to marginal anti-TryS activity.
Both N5-substituted paullones identified as potent LiTryS inhibitors displayed an IC50 slope (Hill coefficient) below the unity (~0.35), which suggests binding of the inhibitor to non-equivalent binding pockets or to partitioning of the compounds into an inactive, less potent or inaccessible form at higher concentrations [74]. Since both paullones displayed good solubility in the concentration range tested (~1 nM-50 μM) and produced 100% LiTryS inhibition, we ruled out insolubility or aggregation phenomena as responsible of this behavior. On the other hand, the dose-response plots do not show the presence of a second inflection point that would be indicative of a second binding pocket for paullones. TryS are monomeric but mechanistically complex (trisubstrate) enzymes [23, 24] whose substrate binding sites display large conformational changes during catalysis [21, 75, 76], hence, it is possible that the low Hill slopes reflect an equilibrium between two or more forms of the enzyme that interact differentially with the paullones. Further mechanistic studies are needed to shed light on this issue.
From this screening, MOL2008 was the most potent inhibitor identified for LiTryS that, in addition, shows potential for further optimization of its anti-trypanosomal TryS activity (IC50 ~30 μM). The related paullone FS554 was chemically validated in a previous study using transgenic cell lines of L. infantum [20]. Here we show that MOL2008 is almost one order of magnitude more potent towards L. infantum promastigotes than FS554 and, more importantly, that it targets in vivo trypanothione biosynthesis. The future design of MOL2008 analogues should also aim at improving its biological properties, which according to our data are far from optimal (EC50 in the μM range and SI ≤ 2) and suggest off-target effects in host cells.
Precedent studies demonstrated that disustituted polyamines are potent antiproliferative agents that interfere with the polyamine metabolism of Plasmodium and African trypanosomes [47–50]. On the basis that the substituted diamine moiety may eventually be recognized as ligand by TryS, a collection of simplified derivatives was screened for their anti-synthetase activity. Two BDA (EAP1-47 and APC1-111) displayed inhibitory activity, albeit moderate (IC50 ~ 30 μM), against multiple TryS. SAR analysis revealed the need to fulfill a specific steric demand on the enzyme interacting pocket. A rational optimization of these compounds seems difficult, since the screening performed here included a wide diversity of derivatives (mono- and di-substituted, with bulky groups or halogen atoms, and with linker of different length) that yielded only weak TryS inhibitors. Nevertheless, as in a fragment-based approach, certain moieties of the most active BDA can be selected as substituents of novel or known TryS inhibitors.
With few exceptions, most APC derivatives tested here were previously shown to display low to sub-μM potency towards infective T. b. brucei [46]. Our study confirmed and disclosed the potent (EC50 15–280 nM) and selective (SI = 10–164) anti-parasitic activity of APC analogues and of two halogenated BDA from the EAP series, respectively, all of which exherted a moderate inhibition of TbTryS (IC50 ~ 30 μM). Interestingly, EAP1-47 induced changes in the intracellular pool of low molecular thiols that overall resembled those triggered by RNAi-mediated silencing of TryS [16] and displayed a moderate increase in its cytotoxicity towards TryS-defficient cells that was not paralleled by the more potent and selective molecules APC1-99 and EAP1-63. Nonetheless, as pointed out before, the higher in vivo (anti-trypanosomal activity) vs. in vitro (TbTryS inhibition) activity of EAP1-47 suggests that this compound is targeting other essential molecular target(s) in addition to TryS.
The positive loop of substrate inhibition of TryS by GSH has a direct impact on the design of inhibitors. On the one hand, it suggests that groups competing with GSH can be excluded from the ligand scaffold to avoid unnecessary substrate competition and to reduce the molecular mass of the inhibitor. On the other hand, it shows that full inhibition of TryS is not required because the phenomenon of substrate inhibition will amplify in vivo the inactivation of the enzyme, which represents a pharmacological advantage of this molecular target.
The four APPDA identified as moderate inhibitors of, preferentially, the T. cruzi TryS, are structuraly related to compounds originally designed as inhibitors of bacterial D-Ala:D-Ala ligase. Interestingly, these derivatives presented a 10-fold lower IC50 towards the trypanosomal enzyme than the analogue compounds against the corresponding bacterial target (best IC50 = 260 μM) [43]. The APPDA may serve as primary scaffolds to develope analogues with improved anti-TryS activity and, consequently, superior selectivity index. TcTryS, and to minor extent LiTryS, was target of inhibition by a BZ substituted with an imidazolone. A BZ with an additional imidazolone group retained activity against TcTryS but enhanced ATP hydrolysis by LiTryS and TbTryS, indicating that these ligands present a molecular pattern recognized by the active site of the enzymes that should be further investigated.
Our work also disclosed several compounds enhancing ATP consumption by trypanosomal TryS. Adding value to the existence of distinctive structural features between tritryp TryS, TbTryS was targeted specifically by large heterocyclic compounds from the BBHPP whereas TcTryS was the most promiscuous enzyme, being activated by a PD, an APPDA and the BZ mentioned above. A single compound, a bis-imidazolone BZ, increased LiTryS activity. According to our results, the activation of ATPase activity in trypanosomal TryS by these compounds takes place only when both co-subtrates, namely GSH and SP, are present in the reaction. More detailed studies are required to establish whether such allosteric activation of TryS is paralleled by an increased production of T(SH)2. Compounds promoting the non-productive consumption of ATP are attractive candidates for biological evaluation, because the metabolic outcome of their action should be similar to that of true enzyme inhibitors.
In summary, this study: (i) led to the identification and on-target validation of a potent inhibitor of leishmanial TryS and of chemical scaffolds that can be further developed into inhibitors of wide spectrum against TryS from different trypanosomatid species, (ii) highlights the existence of remarkable kinetic and structural differences between tritryp TryS that prompt to obtain 3D structures of TryS from different trypanosomatids in order to guide a structure-based rationale design of multi-TryS inhibitors; iii) demonstrates the relevance of running a HTS under near physiological concentration of subtrates to minimize the possibility to detect weak competitive inhibitors with low potential to act in vivo
Supporting Information
The plots of velocity (v) / [E](s-1) vs. different substrate or product concentrations (μM) are shown for: A) Trypanosoma cruzi trypanothione synthetase (TcTryS) with a.1) spermidine (SP), a 2) ATP, a.3) glutathione (GSH) and a.4) ADP; B) Trypanosoma brucei trypanothione synthetase (TbTryS) with b.1) SP; b.2) ATP; b.3) GSH, and b.4) ADP; C) Leishmania infantum trypanothione synthetase (LiTryS) with c.1) SP, c.2) ATP, c.3) GSH, and c.4) ADP. See Materials and Methods, S1 Text and S8 Table for details about assay conditions.
(TIF)
In a total volume of 50 μL containing all assay reagents [150 μM ATP, 2 mM spermidine (SP), 250 μM glutathione (GSH), 5 mM DTT, 10 mM MgSO4, 0.5 mM EDTA, 100 mM HEPES pH 7.4, 9 mM NaCl and 10% v/v DMSO] except TryS, K2HPO4 was added at concentrations ranging from 1.25 to 140 μM. Two hundred μL BIOMOL GREEN reagent were added per well and the colorimetric reaction was allowed to develop for 20 min. The absorbance at 650 nm was measured in each well using a MultiScan EX plate reader (Thermo SCIENTIFIC). The mean A650 nm corr values ± 2 S.D. BIOMOL GREEN signal is plotted against [K2HPO4]. A linear regression model based on the least square method was fitted for K2HPO4 concentrations from 0 to 50 μM.
(TIF)
The corrected absorbance of BIOMOL GREEN at 650 nm (A650 nm corr) is ploted against different concentrations of: A) DTT (mM) for a reaction containing 20 μM K2HPO4 in 150 μM ATP, 2 mM spermidine (SP), 250 μM glutathione (GSH), 10% v/v DMSO and 9 mM NaCl. The A650 nm corr is expressed as percentage relative to A650 nm corr at 5 mM DTT; B) SP (mM) for a reaction containing 20 μM K2HPO4, 150 μM ATP, 2 mM SP, 250 μM GSH, 5 mM DTT, 10% v/v DMSO and 9 mM NaCl. The A650 nm corr is expressed as percentage relative to A650 nm corr at SP 2 mM; C) glycerol (%) for a reaction containing 20 μM K2HPO4, 150 μM ATP, 2 mM SP, GSH 250 μM, 5 mM DTT and 9 mM NaCl. The A650 nm corr is expressed as percentage relative to A650 nm corr at 0% v/v glycerol; D) GSH (mM) for a reaction containing 20 μM K2HPO4, 150 μM ATP, DTT 5 mM, 10% v/v DMSO, 9 or 30 mM NaCl, and 0 or 4% v/v glycerol. The A650 nm corr is expressed as percentage relative to A650 nm corr at 0% v/v glycerol, 0 mM NaCl and 2.5 mM GSH (black squares), 4% v/v glycerol and 0 mM NaCl (red circles), 4% v/v glycerol and 30 mM NaCl (blue triangle) and 4% v/v glycerol and 9 mM NaCl (green triangle).
(TIF)
Pi production (A650 nm corr) was monitored at different time points (min) for recombinant Leishmania infantum trypanothione synthetase, LiTryS (a, b), Trypanosoma cruzi trypanothione synthetase, TcTryS (c, d) and Trypanosoma brucei trypanothione synthetase, TbTryS (f, g). The assays were conducted at RT in a total reaction volume of 50 μL containing 150 μM ATP, 2 mM SP, 5 mM DTT, 10 mM MgSO4, 0.5 mM EDTA, 100 mM HEPES pH 7.4, 10% v/v DMSO and 9 mM NaCl, with variable concentrations of gluthatione (GSH) and enzyme units according to the TryS species: 250 μM GSH and 2.3 x 10−5 μmol.min-1.mL-1 for LiTryS, 570 μM GSH and 3.5 x 10−6 μmol.min-1.mL-1 for TcTryS, and 50 μM GSH and 1.5 x 10−5 μmol.min-1.mL-1 for TbTryS. Blanks lacking enzyme were prepared for each condition. The TryS reaction was stopped at different time points by adding 200 μL BIOMOL GREEN reagent. The plates were incubated for 20 min at RT and A650 nm measured using a MultiScan EX plate reader (Thermo SCIENTIFIC). The mean A650 nm corr ± 2 S.D. of BIOMOL GREEN signal is plotted vs. time. A linear regression model based on the least square method was applied to each plot to estimate the linearity range.
(TIF)
All compounds were evaluated at 30 μM against Trypanosoma cruzi trypanothione synthetase, TcTryS (Tc), Leishmania infantum trypanothione synthetase, LiTryS (Li) and Trypanosoma brucei trypanothione synthetase, TbTryS (Tb). TryS activity is expressed as percentage (see S3 Table). “n” denotes the number of carbons in the linker between both nitrogens. A) Oxygen substituted (OMe: methoxy, OBn: O-benzyl, OH: hydroxyl) BDA with the number indicating the position of the substitution in the phenyl ring. B) Halogenated (F, Cl and Br) and alkylated (iPr: isopropyl) BDA.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Dr. Sergio Guerrero (Universidad Nacional del Litoral, Argentina), Dr. Ana Tomas, Dr. Helena Castro (Institute for Molecular and Cell Biology, Portugal) and Dr. Alan Fairlamb (Dundee University, Scotland) for providing expression plasmids for different TryS or L. infantum promastigotes. The Recombinant Protein Unit from the Institut Pasteur de Montevideo is acknowledged for providing facilities and assistance during protein purification. Dr. Lucía Piacenza, Dr. Rafael Radi and Damian Estrada from the CEINBIO (Centro de Investigaciones Biomédicas, Universidad de la República, Uruguay) are gratefully acknowledged for providing facilities and assistance for the HPLC analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The financial support of Agencia Nacional de Investigación e Innovación (ANII, Uruguay) is gratefully acknowledged by DB (POS_NAC_2013_1_114477), LFi (POS_NAC_2011_1_3284) and MAC (project DCI-ALA/2007/19.040). MAC acknowledges the support of MERCOSUR Structural Convergence Fund (FOCEM) and Institut Pasteur ACIP call 2015 (project ACIP 17-2015). AM acknowledges the support of Comisión Sectorial de Investigación Científica, Universidad de la Republica Uruguay (project nr. 3404). TC, AD, JŠ, LPM, CK, LFl and MAC acknowledge the support of the European Cooperation in the field of Technical and Scientific Research (COST), Action CM0801. FM and CK acknowledge the support from the German Bundesministerium für Bildung und Forschung (KMU-innovativ 5, Förderkennzeichen 0315814). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Barrett MP, Croft SL. Management of trypanosomiasis and leishmaniasis. Br Med Bull. 2012; 104: 175–96. 10.1093/bmb/lds031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gadelha C, Holden JM, Allison HC, Field MC. Specializations in a successful parasite: What makes the bloodstream-form African trypanosome so deadly?. Mol Biochem Parasitol. 2011; 179: 51–58. 10.1016/j.molbiopara.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 3.Nagajyothi F, Machado FS, Burleigh BA, Jelicks LA, Scherer PE, Mukherjee S, et al. Mechanisms of Trypanosoma cruzi persistence in Chagas disease. Cell Microbiol. 2012; 14: 634–643. 10.1111/j.1462-5822.2012.01764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filardi LS, Brener Z. Susceptibility and natural resistance of Trypanosoma cruzi strains to drugs used clinically in Chagas disease. Trans R Soc Trop Med Hyg. 1987; 81: 755–759. [DOI] [PubMed] [Google Scholar]
- 5.Croft SL, Sundar S, Fairlamb AH. Drug Resistance in Leishmaniasis. Clin Microbiol Rev. 2006; 19: 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graf FE, Ludin P, Wenzler T, Kaiser M, Brun R, Pyana PP, et al. Aquaporin 2 mutations in Trypanosoma brucei gambiense field isolates correlate with decreased susceptibility to pentamidine and melarsoprol. PLoS Negl Trop Dis. 2013; 7(10): e2475 10.1371/journal.pntd.0002475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart ML, Burchmore RJS, Clucas C, Hertz-Fowler C, Brooks K. et al. Multiple genetic mechanisms lead to loss of functional TbAT1 expression in drug-resistant trypanosomes. Eukaryot Cell. 2010; 9: 336–343. 10.1128/EC.00200-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson S, Wyllie S. Nitro drugs for the treatment of trypanosomatid diseases: past, present, and future prospects. Trends Parasitol. 2014; 30: 289–298. 10.1016/j.pt.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balasegaram M, Harris S, Checchi F, Ghorashian S, Hamel C, Karunakara U. Melarsoprol versus eflornithine for treating late-stage Gambian trypanosomiasis in the Republic of the Congo. Bull World Health Organ. 2006; 84: 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jäger T (Editor), Koch O (Editor), Flohé L. (Editor), Selzer PM (Series Editor). Trypanosomatid Diseases: Molecular Routes to Drug Discovery (Drug Discovery in Infectious Diseases). Oxford, UK: Wiley-Blackwell; 2013. [Google Scholar]
- 11.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005; 309: 416–422. [DOI] [PubMed] [Google Scholar]
- 12.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005; 309: 409–415. [DOI] [PubMed] [Google Scholar]
- 13.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005; 309: 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krauth-Siegel RL, Comini M. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim Biophys Acta. 2008; 1780: 1236–1248. 10.1016/j.bbagen.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 15.Manta B, Comini M, Medeiros A, Hugo M, Trujillo M, Radi R. Trypanothione: A unique bis-glutathionyl derivative in trypanosomatids. Biochim Biophys Acta. 2013; 1830: 3199–3216. 10.1016/j.bbagen.2013.01.013 [DOI] [PubMed] [Google Scholar]
- 16.Comini M, Guerrero SA, Haile S, Menge U, Lünsdorf H, Flohé L. Validation of Trypanosoma brucei trypanothione synthetase as drug target. Free Radic Biol Med. 2004; 36: 1289–1302. [DOI] [PubMed] [Google Scholar]
- 17.Wyllie S, Oza SL, Patterson S, Spinks D, Thompson S, Fairlamb AH. Dissecting the essentiality of the bifunctional trypanothione synthetase-amidase in Trypanosoma brucei using chemical and genetic methods. Mol Microbiol. 2009; 74: 529–540. 10.1111/j.1365-2958.2009.06761.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torrie LS, Wyllie S, Spinks D, Oza SL, Thompson S, Harrison JR, et al. Chemical validation of trypanothione synthetase a potential drug target for human trypanosomiasis. J Biol Chem. 2009; 284: 36137–36145. 10.1074/jbc.M109.045336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spinks D, Torrie LS, Thompson S, Harrison JR, Frearson JA, Read KD, et al. Design, synthesis and biological evaluation of Trypanosoma brucei trypanothione synthetase inhibitors. Chem Med Chem. 2012; 7: 95–106. 10.1002/cmdc.201100420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sousa AF, Gomes-Alves AG, Benítez D, Comini M, Flohé L, Jaeger T, et al. Genetic and chemical analysis of trypanothione biosynthesis in the glutathionylspermidine synthetase-containing parasite Leishmania infantum reveals that only trypanothione synthetase is essential for survival. Free Radic Biol Med. 2014; 73: 229–238. [DOI] [PubMed] [Google Scholar]
- 21.Fyfe PK, Oza SL, Fairlamb AH, Hunter WN. Leishmania trypanothione synthetase-amidase structure reveals a basis for regulation of conflicting synthetic and hydrolytic activities. J Biol Chem. 2008; 283: 17672–17680. 10.1074/jbc.M801850200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olin-Sandoval V, González-Chávez Z, Berzunza-Cruz M, Martínez I, Jasso-Chávez R Becker I, et al. Drug target validation of the trypanothione pathway enzymes through metabolic modeling. FEBS J. 2012; 279: 1811–1833. 10.1111/j.1742-4658.2012.08557.x [DOI] [PubMed] [Google Scholar]
- 23.Comini M, Menge U, Wissing J, Flohé L. Trypanothione synthesis in Crithidia revisited. J Biol Chem. 2005; 280: 6850–6860. [DOI] [PubMed] [Google Scholar]
- 24.Leroux AE, Haanstra JR, Bakker BM, Krauth-Siegel RL. Dissecting the catalytic mechanism of Trypanosoma brucei trypanothione synthetase by kinetic analysis and computational modelling. J Biol Chem. 2013; 288: 23751–23764. 10.1074/jbc.M113.483289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oza SL, Tetaud E, Ariyanayagam MR, Warnon SS, Fairlamb AH. A single enzyme catalyses formation of trypanothione from glutathione and spermidine in Trypanosoma cruzi. J Biol Chem. 2002; 277: 35853–35861. [DOI] [PubMed] [Google Scholar]
- 26.Oza SL, Shaw MP, Wyllie S, Fairlamb AH. Trypanothione biosynthesis in Leishmania major. Mol Biochem Parasitol. 2005; 139: 107–116. [DOI] [PubMed] [Google Scholar]
- 27.Oza SL, Ariyanayagam MR, Aitcheson N, Fairlamb AH. Properties of trypanothione synthetase from Trypanosoma brucei. Mol Biochem Parasitol. 2003; 131: 25–33. [DOI] [PubMed] [Google Scholar]
- 28.De Craecker S, Verbruggen C, Rajan PK, Smith K, Haemers A, Fairlamb AH. Characterization of the peptide substrate specificity of glutathionylspermidine synthetase from Crithidia fasciculata. Mol Biochem Parasitol. 1997; 84: 25–32. [DOI] [PubMed] [Google Scholar]
- 29.Verbruggen C, De Craecker S, Rajan PK, Jiago XY, Borloo M, Smith K, et al. Phosphonic acid and phosphinic acid tripeptides as inhibitors of glutathionylspermidine synthetase. Bioorg Med Chem Lett. 1996; 6: 253–258. [Google Scholar]
- 30.Kwon DS, Lin CH, Chen S, Coward JK, Walsh CT, Bollinger JM Jr. Dissection of Glutathionylspermidine synthetase/ amidase from Escherichia coli into autonomously folding and functional synthetase and amidase domains. J Biol Chem. 1997; 272: 2429–2436. [DOI] [PubMed] [Google Scholar]
- 31.Lin C, Chen S, Kwon DS, Coward JK, Walsh CT. Aldehyde and phosphinate analogs of glutathione and glutathionylspermidine: potent, selective binding inhibitors of the E. coli bifunctional glutathionylspermidine synthetase/amidase. Chem Biol. 1997; 4: 859–866. [DOI] [PubMed] [Google Scholar]
- 32.Chen S, Lin CH, Walsh CT, Coward JK. Novel inhibitors of trypanothione biosynthesis: synthesis and evaluation of a phosphinate analog of glutathionyl spermidine (GSP), a potent, slow binding inhibitor of GSP synthetase. Bioorg Med Chem Lett. 1997; 7: 505–510. [Google Scholar]
- 33.Chen S, Lin CH, Kwon DS, Walsh CT, Coward JK. Design, Synthesis, and Biochemical Evaluation of phosphonate and phosphonamidate analogues of glutathionylspermidine as inhibitors of glutathionylspermidine synthetase/amidase from Escherichia coli. J Med Chem. 1997; 40: 3842–3850. [DOI] [PubMed] [Google Scholar]
- 34.Amssoms K, Oza SL, Ravaschino E, Yamani A, Lambeir A, Rajan P, et al. Glutathione-like tripeptides as inhibitors of glutathionylspermidine synthetase. Part 1: Substitution of the glycine carboxylic acid group. Bioorg Med Chem Lett. 2002: 12: 2553–2556. [DOI] [PubMed] [Google Scholar]
- 35.Amssoms K, Oza S.L, Augustyns K, Yamani A, Lambeir A.M, Bal G, et al. Glutathione-like tripeptides as inhibitors of glutathionylspermidine synthetase. Part 2: substitution of the glycine part. Bioorg Med Chem Lett. 2002; 12: 2703–2705. [DOI] [PubMed] [Google Scholar]
- 36.Oza SL, Chen S, Wyllie S, Coward JK, Fairlamb AH. ATP-dependent ligases in trypanothione biosynthesis—kinetics of catalysis and inhibition by phosphinic acid pseudopeptides. FEBS J. 2008; 275: 5408–5421. 10.1111/j.1742-4658.2008.06670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Silva C, Daunes S, Rock P, Yardley V, Croft SL. Structure-activity study on the in vitro antiprotozoal activity of glutathione derivatives. J Med Chem. 2000; 43: 2072–2078. [DOI] [PubMed] [Google Scholar]
- 38.Daunes S, D’Silva C, Kendrick H, Yardley V, Croft SL. QSAR study on the contribution of Log P and Es to the in vitro antiprotozoal. Activity of glutathione derivatives. J Med Chem. 2001; 44: 2976–2983. [DOI] [PubMed] [Google Scholar]
- 39.Daunes S, D’Silva C. Glutathione derivatives active against Trypanosoma brucei rhodesiense and T. brucei brucei in vitro. Antimicrob Agents Chemother. 2002; 46: 434–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravaschino EL, Docampo R, Rodriguez JB. Design, synthesis, and biological evaluation of phosphinopeptides against Trypanosoma cruzi targeting trypanothione biosynthesis. J Med Chem. 2006; 49: 426–435. [DOI] [PubMed] [Google Scholar]
- 41.Stuhlmann F, Jäger T, Flohé L, Schinzer D. N5-substituted 2-(6-oxo-6,7-dihydro-5H-benzo[2,3]azepino[4,5-b]indol-5-yl)-acetamides for treating tropical diseases. EP 1 757 607 A1. 2007. Feb 28. http://www.google.com/patents/EP1757607A1?cl=en
- 42.Haldane JBS. Enzymes. New York: Longmans, Green; 1930. [Google Scholar]
- 43.Škedelj V, Arsovska E, Tomašić T, Kroflič A, Hodnik V, Hrast M, et al. 6-Arylpyrido[2,3-d]pyrimidines as novel ATP-competitive inhibitors of bacterial D-Alanine:D-Alanine ligase. PloS One 2012; 7(8): e39922 10.1371/journal.pone.0039922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller JR, Dunham S, Mochalkin I, Banotai C, Bowman M, Buist S, et al. A class of selective antibacterials derived from a protein kinase inhibitor pharmacophore. Proc Natl Acad Sci. U S A. 2009; 106: 1737–1742. 10.1073/pnas.0811275106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Labadie GR, Choi SR, Avery MA. Diamine derivatives with antiparasitic activities. Bioorg Med Chem Lett. 2004; 14: 615–619. [DOI] [PubMed] [Google Scholar]
- 46.Caminos AP, Panozzo-Zenere EA, Wilkinson SR, Tekwani BL, Labadie GR. Synthesis and antikinetoplastid activity of a series of N,N’-substituted diamines. Bioorg Med Chem Lett. 2012; 22: 1712–1715. 10.1016/j.bmcl.2011.12.101 [DOI] [PubMed] [Google Scholar]
- 47.Bitonti AJ, McCann PP, Sjoerdsma A. The effects of polyamine analogs on malaria parasites in vitro and in vivo. Adv Exp Med Biol. 1998; 250: 717–726. [DOI] [PubMed] [Google Scholar]
- 48.Bitonti AJ, Dumont JA, Bush TL, Edwards ML, Stemerick DM, McCann PP, et al. Bis(benzyl)polyamine analogs inhibit the growth of chloroquine-resistant human malaria parasites (Plasmodium falciparum) in vitro and in combination with a-difluoromethylornithine cure murine malaria. Proc Natl Acad Sci USA. 1989; 86: 651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bitonti AJ, Dumont JA, Bush TL, Stemerick DM, Edwards ML, McCann PP. Bis(benzyl)polyamine analogs as novel substrates for polyamine oxidase. J Biol Chem. 1990; 265: 382–388. [PubMed] [Google Scholar]
- 50.Byers TL, Bush TL, McCann PP, Bitonti AJ. Antitrypanosomal effects of polyamine biosynthesis inhibitors correlate with increases in Trypanosoma brucei brucei S-adenosyl-L-methionine. Biochem J. 1991; 274: 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Šarlauskas J, Miliukienė V, Anusevičius Ž, Misevičienė L, Krikštopaitis K, Nemeikaitė-Čėnienė A., et al. Redox properties and prooxidant cytotoxicity of benzofuroxans: a comparison with nitrobenzenes, Chemija. 2009; 20: 109–115. [Google Scholar]
- 52.Šarlauskas J, Anusevičius Ž, Misiūnas A. Benzofuroxan (Benzo[1,2-c]1,2,5-oxadiazole N-oxide) derivatives as potential energetic materials: studies on their synthesis and properties. Central European Journal of Energetic Materials. 2012; 9: 365–386. [Google Scholar]
- 53.Šarlauskas J.Synthesis of energetic materials containing benzimidazole core. Proceedings of Seminar on New Trends in Research of Energetic Materials 11(part 2); 2010. p. 730–737. [Google Scholar]
- 54.Šarlauskas J. Synthesis of some new heterocyclic derivatives of benzofuroxan. in Proceedings of 7th Natl. Lithuanian Conference “Chemistry” 7; 2005. p. 104–109.
- 55.Šarlauskas J, Misevičienė L, Marozienė A, Karvelis L, Stankevičiūtė J, Krikštopaitis K, et al. The study of NADPH-dependent flavoenzyme-catalyzed reduction of benzo[1,2-c]1,2,5-oxadiazole N-oxides (Benzofuroxans). Int J Mol Sci 2014; 15: 23307–31. 10.3390/ijms151223307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maiwald F, Benítez D, Charquero D, Abad Dar M, Erdmann H, Preu L, et al. 9- and 11-substituted 4-azapaullones are potent and selective inhibitors of African trypanosome. Eur J Med Chem. 2014; 18: 274–283. [DOI] [PubMed] [Google Scholar]
- 57.Fueller F, Jehle B, Putzker K, Lewis JD, Krauth-Siegel RL. High Throughput Screening against the peroxidase cascade of African trypanosomes identifies antiparasitic compounds that inactivate tryparedoxin. J Biol Chem. 2012; 12: 8792–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor MC, Kaur H, Blessington B, Kelly JM, Wilkinson SR. Validation of spermidine synthase as a drug target in African Trypanosomes. Biochem J. 2008; 409: 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coombs GH, Sanderson BE. Amine production by Leishmania mexicana. Ann Trop Med Parasitol. 1985; 79: 409–415. [DOI] [PubMed] [Google Scholar]
- 60.Ariyanayagam MR, Oza SL, Mehlert A, Fairlamb AH. Bis(glutathionyl)spermine and other novel trypanothione analogues in Trypanosoma cruzi. J Biol Chem. 2003; 278: 27612–27619. [DOI] [PubMed] [Google Scholar]
- 61.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen.1999; 4: 67–73. [DOI] [PubMed] [Google Scholar]
- 62.Fairlamb AH, Henderson GB, Bacchi CJ, Cerami A. In vivo effects of difluoromethylornithine on trypanothione and polyamine levels in bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol. 1987; 24: 185–191. [DOI] [PubMed] [Google Scholar]
- 63.Comini MA, Dirdjaja N, Kaschel M, Krauth-Siegel RL. Preparative enzymatic synthesis of trypanothione and trypanothione analogues. Int J Parasitol. 2009; 39: 1059–1062. 10.1016/j.ijpara.2009.05.002 [DOI] [PubMed] [Google Scholar]
- 64.Koenig K, Menge U, Kiess M, Wray V, Flohé L. Convenient isolation and kinetic mechanism of glutathionylspermidine synthetase from Crithidia fasciculata. J Biol Chem. 1997; 272: 11908–11915. [DOI] [PubMed] [Google Scholar]
- 65.Haanstra JR, van Tuijl A, van Dam J, van Winden W, Tielens AG, van Hellemond JJ, et al. Proliferating bloodstream-form Trypanosoma brucei use a negligible part of consumed glucose for anabolic processes. Int J Parasitol. 2012; 42: 667–673. 10.1016/j.ijpara.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 66.Visser N, Opperdoes FR. (1980) Glycolysis in Trypanosoma brucei. Eur J Biochem. 1980; 103: 623–632. [DOI] [PubMed] [Google Scholar]
- 67.Martins RM, Covarrubias C, Rojas RG, Silber AM, Yoshida N. Use of L-Proline and ATP production by Trypanosoma cruzi metacyclic forms as requirements for host cell invasion. Infect Immun. 2009; 77: 3023–3032. 10.1128/IAI.00138-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zilberstein D, Dwyer DM. Proton motive force-driven active transport of D-glucose and L-proline in the protozoan parasite Leishmania donovani. Proc Natl Acad Sci USA. 1985; 82:1716–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bakker BM, Michels PAM, Opperdoes FR, Westerhoff HV. Glycolysis in bloodstream form Trypanosoma brucei can be understood in terms of the kinetics of the glycolytic enzymes. J Biol Chem. 1997; 272: 3207–3215. [DOI] [PubMed] [Google Scholar]
- 70.Graven P, Tambalo M, Scapozza L, Perozzo R. Purine metabolite and energy charge analysis of Trypanosoma brucei cells in different growth phases using an optimized ion-pair RP-HPLC/UV for the quantification of adenine and guanine pools. Exp Parasitol. 2014; 141: 28–38. 10.1016/j.exppara.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 71.Stoppani AO, Docampo R, de Boiso JF, Frasch AC. Effect of inhibitors of electron transport and oxidative phosphorylation on Trypanosoma cruzi respiration and growth. Mol Biochem Parasitol. 1980; 2: 3–21. [DOI] [PubMed] [Google Scholar]
- 72.Hart DT, Vickerman K, Coombs GH. A quick, simple method for purifying Leishmania mexicana amastigotes in large mumbers. Parasitol. 1981; 82: 345–355. [DOI] [PubMed] [Google Scholar]
- 73.Boiani M, Piacenza L, Hernández P, Boiani L, Cerecetto H, González M, Denicola A. Mode of action of nifurtimox and N-oxide-containing heterocycles against Trypanosoma cruzi: is oxidative stress involved? Biochem Pharmacol. 2010; 79: 1736–1745. 10.1016/j.bcp.2010.02.009 [DOI] [PubMed] [Google Scholar]
- 74.Coopeland R A (Editor). Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists. New Jersey, USA: John Wiley & Sons, Inc.; 2005. [PubMed] [Google Scholar]
- 75.Koch O, Cappel D, Nocker M, Jager T, Flohé L, Sotriffer CA, et al. Molecular Dynamics Reveal Binding Mode of Glutathionylspermidine by Trypanothione Synthetase. PloS One. 2013; 8(2): e56788 10.1371/journal.pone.0056788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pai CH, Chiang BY, Ko TP, Chou CC, Chong CM, Yen FJ, et al. Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase. EMBO J. 2006; 25: 5970–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The plots of velocity (v) / [E](s-1) vs. different substrate or product concentrations (μM) are shown for: A) Trypanosoma cruzi trypanothione synthetase (TcTryS) with a.1) spermidine (SP), a 2) ATP, a.3) glutathione (GSH) and a.4) ADP; B) Trypanosoma brucei trypanothione synthetase (TbTryS) with b.1) SP; b.2) ATP; b.3) GSH, and b.4) ADP; C) Leishmania infantum trypanothione synthetase (LiTryS) with c.1) SP, c.2) ATP, c.3) GSH, and c.4) ADP. See Materials and Methods, S1 Text and S8 Table for details about assay conditions.
(TIF)
In a total volume of 50 μL containing all assay reagents [150 μM ATP, 2 mM spermidine (SP), 250 μM glutathione (GSH), 5 mM DTT, 10 mM MgSO4, 0.5 mM EDTA, 100 mM HEPES pH 7.4, 9 mM NaCl and 10% v/v DMSO] except TryS, K2HPO4 was added at concentrations ranging from 1.25 to 140 μM. Two hundred μL BIOMOL GREEN reagent were added per well and the colorimetric reaction was allowed to develop for 20 min. The absorbance at 650 nm was measured in each well using a MultiScan EX plate reader (Thermo SCIENTIFIC). The mean A650 nm corr values ± 2 S.D. BIOMOL GREEN signal is plotted against [K2HPO4]. A linear regression model based on the least square method was fitted for K2HPO4 concentrations from 0 to 50 μM.
(TIF)
The corrected absorbance of BIOMOL GREEN at 650 nm (A650 nm corr) is ploted against different concentrations of: A) DTT (mM) for a reaction containing 20 μM K2HPO4 in 150 μM ATP, 2 mM spermidine (SP), 250 μM glutathione (GSH), 10% v/v DMSO and 9 mM NaCl. The A650 nm corr is expressed as percentage relative to A650 nm corr at 5 mM DTT; B) SP (mM) for a reaction containing 20 μM K2HPO4, 150 μM ATP, 2 mM SP, 250 μM GSH, 5 mM DTT, 10% v/v DMSO and 9 mM NaCl. The A650 nm corr is expressed as percentage relative to A650 nm corr at SP 2 mM; C) glycerol (%) for a reaction containing 20 μM K2HPO4, 150 μM ATP, 2 mM SP, GSH 250 μM, 5 mM DTT and 9 mM NaCl. The A650 nm corr is expressed as percentage relative to A650 nm corr at 0% v/v glycerol; D) GSH (mM) for a reaction containing 20 μM K2HPO4, 150 μM ATP, DTT 5 mM, 10% v/v DMSO, 9 or 30 mM NaCl, and 0 or 4% v/v glycerol. The A650 nm corr is expressed as percentage relative to A650 nm corr at 0% v/v glycerol, 0 mM NaCl and 2.5 mM GSH (black squares), 4% v/v glycerol and 0 mM NaCl (red circles), 4% v/v glycerol and 30 mM NaCl (blue triangle) and 4% v/v glycerol and 9 mM NaCl (green triangle).
(TIF)
Pi production (A650 nm corr) was monitored at different time points (min) for recombinant Leishmania infantum trypanothione synthetase, LiTryS (a, b), Trypanosoma cruzi trypanothione synthetase, TcTryS (c, d) and Trypanosoma brucei trypanothione synthetase, TbTryS (f, g). The assays were conducted at RT in a total reaction volume of 50 μL containing 150 μM ATP, 2 mM SP, 5 mM DTT, 10 mM MgSO4, 0.5 mM EDTA, 100 mM HEPES pH 7.4, 10% v/v DMSO and 9 mM NaCl, with variable concentrations of gluthatione (GSH) and enzyme units according to the TryS species: 250 μM GSH and 2.3 x 10−5 μmol.min-1.mL-1 for LiTryS, 570 μM GSH and 3.5 x 10−6 μmol.min-1.mL-1 for TcTryS, and 50 μM GSH and 1.5 x 10−5 μmol.min-1.mL-1 for TbTryS. Blanks lacking enzyme were prepared for each condition. The TryS reaction was stopped at different time points by adding 200 μL BIOMOL GREEN reagent. The plates were incubated for 20 min at RT and A650 nm measured using a MultiScan EX plate reader (Thermo SCIENTIFIC). The mean A650 nm corr ± 2 S.D. of BIOMOL GREEN signal is plotted vs. time. A linear regression model based on the least square method was applied to each plot to estimate the linearity range.
(TIF)
All compounds were evaluated at 30 μM against Trypanosoma cruzi trypanothione synthetase, TcTryS (Tc), Leishmania infantum trypanothione synthetase, LiTryS (Li) and Trypanosoma brucei trypanothione synthetase, TbTryS (Tb). TryS activity is expressed as percentage (see S3 Table). “n” denotes the number of carbons in the linker between both nitrogens. A) Oxygen substituted (OMe: methoxy, OBn: O-benzyl, OH: hydroxyl) BDA with the number indicating the position of the substitution in the phenyl ring. B) Halogenated (F, Cl and Br) and alkylated (iPr: isopropyl) BDA.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.