Abstract
Virulence of the most deadly malaria parasite Plasmodium falciparum is linked to the variant surface antigen PfEMP1, which is encoded by about 60 var genes per parasite genome. Although the expression of particular variants has been associated with different clinical outcomes, little is known about var gene expression at the onset of infection. By analyzing controlled human malaria infections via quantitative real-time PCR, we show that parasite populations from 18 volunteers expressed virtually identical transcript patterns that were dominated by the subtelomeric var gene group B and, to a lesser extent, group A. Furthermore, major changes in composition and frequency of var gene transcripts were detected between the parental parasite culture that was used to infect mosquitoes and Plasmodia recovered from infected volunteers, suggesting that P. falciparum resets its var gene expression during mosquito passage and starts with the broad expression of a specific subset of var genes when entering the human blood phase.
Author Summary
Parasites of the species Plasmodium falciparum, which are responsible for the most severe forms of malaria, escape from the human immune response by antigenic variation. A repertoire of 60 var genes codes for a broad range of different variant antigens presented on the surface of infected erythrocytes. These antigens mediate adhesion to blood vessels, thereby disturbing the blood microcirculation and causing life-threatening organ dysfunctions. To better understand antigenic variation in vivo we analyzed the var gene expression profiles in blood samples from 18 malaria naïve volunteers, who were experimentally infected with cryopreserved sporozoites isolated from Anopheles mosquitoes. Our in-depth analysis revealed a broad, but remarkably uniform expression pattern of a specific set of var gene variants. Moreover, the results clearly show that this var gene expression program is specifically activated after mosquito transmission of the parasite. These findings are of particular importance for our understanding of the strategy of malaria parasites to establish and maintain infections in the human host.
Introduction
Malaria is one of the most frequently occurring parasitic diseases worldwide with an estimated 198 million clinical cases in 2013 and a death toll of more than 0.5 million [1]. The virulence of the most deadly species of human malaria parasites, Plasmodium falciparum, is directly linked to the variable surface protein PfEMP1 (P. falciparum erythrocyte membrane protein 1) [2,3]. Members of the PfEMP1 family enable the parasite to adhere to a large variety of surface receptors on microvasculature linings or in case of pregnancy to the maternal side of the placenta in order to avoid spleen passage and subsequent clearance (reviewed in [4]). Expression switching between different PfEMP1 variants correlates with changes in the antigenic and adhesion phenotype of the parasite [5]. Each parasite possesses about 60 var genes coding for different PfEMP1 variants, which are expressed in a mutually exclusive manner meaning that generally only a single PfEMP1 variant is exposed on the surface of the infected erythrocyte at a time while all other gene copies are silenced [6]. Historically, the global var gene repertoire present in the parasite population was assumed to be highly diverse and the number of variants almost unlimited. But in the recent past, evidence is arising that every parasite genotype is organized similarly and exhibits roughly the same numbers of var gene variants of each subgroup (A, B, C and E) defined by PfEMP1 protein domain composition as well as by chromosomal localization, direction of transcription and particular 5’-UTR sequences of their encoding var genes [7–9]. Moreover, the comparison of seven P. falciparum genomes revealed 23 PfEMP1 domain cassettes (DCs), which seem to form conserved recombination and receptor-binding units [10]. Both observations point to a more conserved var gene repertoire than previously assumed.
The reference strain NF54 possesses 10 group A var gene copies including the interstrain conserved subfamilies var1 and var3, which are all located near the end of chromosomes and have a transcriptional direction towards the telomeres. With exception of the short var3 PfEMP1 variants, A-type PfEMP1 proteins have an extended, multiple domain composition and a non CD36-binding head structure consisting of a DBLα1 and a CIDRα1/β/γ/δ domain. Their pattern of expression has been linked to severe disease outcome [11–20]. The 37 member group B var genes are located closest to the telomere and transcribed towards the centromere and their expression has been associated with both severe and mild malaria [13,15,19–21]. Recently, expression of A- and B-type var genes encoding the interstrain conserved PfEMP1 DCs 8 and 13, which bind the endothelial receptor EPCR, as well as the DC number 5, known to mediate PECAM1 binding, were linked to severe malaria in young children [18,22,23]. Interestingly, some genes are chimeras of group B and A or C var genes and are thought to represent intermediate groups between the major groupings [8]. In the NF54 genome, 4 and 9 members form these intermediate groups B/A and B/C, respectively, which all have a 5’-UTR characteristic for B-type var genes. In contrast to B/A genes, which are very similar in location and transcriptional orientation to group B genes, the chromosomal characteristics of group B/C genes are in common with group C genes. The 13 group C genes are located at chromosome internal clusters, transcribed towards the telomeres and possess a C-type 5’-UTR. C-type PfEMP1 variants are known to be expressed in parasites causing asymptomatic infection and in long-term in vitro cultivated parasites [11,13,24–27]. Most B- and C-type PfEMP1 proteins have a 4-domain extracellular structure including a CD36-binding head structure consisting of a DBLα0 and a CIDRα2–6 domain plus another DBL and CIDR domain [7,10].
Despite this large repertoire of variant surface proteins, the vast majority of P. falciparum infections do not lead to severe disease, suggesting that parasite sequestration is a well-adapted process and increased parasite transmission to mosquitoes outweighs losses due to host death. Hence, PfEMP1 facilitates repeated and long-lasting infections of the human host even after repeated exposures. But, remarkably, particular PfEMP1 subtypes appear to be specialized for infection of malaria naïve hosts where the interaction of PfEMP1 with endothelial and circulating cells directly causes obstruction of blood circulation, which contributes along with immunopathology to organ failure. In malaria endemic areas, severe malaria mainly affects young children under the age of five lacking a sufficient immune response from previous Plasmodium infections [28]. Therefore, a better understanding of the var gene expression in malaria naïve individuals and of the mechanisms that control the expression of particular PfEMP1 types is of particular importance. Three previous studies analyzed var gene expression in three experimentally infected naive individuals [29–31]. Peters et al. found a single dominant B-type var transcript in two volunteers, whereas Lavstsen et al. and Wang et al. detected a more broad activation of most var genes at the early onset of the blood infection. Based on these results two different strategies used by the malaria parasite to initiate an infection in the human host are currently discussed in the scientific community. The first model suggests that the parasites may use an ordered hierarchical var gene expression program, meaning that most of the parasites express a single var type in the first generation after egress from the liver. In the following replication cycles the parasite switches to other var gene variants determined by the intrinsic rate for each gene to be turned on or off. This would provide an efficient mechanism to evade the host's immune system in concordance with protection of the remaining PfEMP1 variants from unnecessary exposure to the immune system [29,32]. The second concept postulates the early exploration of the suitability of the available host sequestration receptors. Accordingly, the parasite population released from the liver expresses all var genes and later on selective forces favor the survival of parasites expressing certain PfEMP1 variants with the best adhesion properties and for which the human host has no pre-existing variant-specific immunity [30].
To clarify these contradicting concepts the study presented here made use of a controlled human malaria infection (CHMI) trial in which volunteers were injected with aseptic, purified, cryopreserved P. falciparum NF54 sporozoites (PfSPZ Challenge) of a single production lot produced under cGMP [33] and var gene expression was analyzed in samples from 18 malaria naïve hosts at the early onset of blood infection. The data presented here reveal a strategy, which favors the expression of a broad subset of var gene variants from subtelomeric locations while repressing var variants that are conserved between strains or that are located at chromosome internal sites. Moreover, we clearly show for the first time that ex vivo var gene expression patterns differ significantly from the expression profile of the parasite culture that was used to produce the PfSPZ Challenge lot used for CHMI, indicating that the expression of subtelomeric var genes is specifically turned on in vivo. Therefore our data support aspects of both postulated models: the parasite population seems to activate a whole subset of var genes allowing for exploration of the situation in the host which is shaped by available host receptors (e.g. CSA in pregnant women) and pre-existing variant specific immunity, whereas a different subset of var genes remains infrequently expressed.
Results
Infection kinetics of the volunteers
Our study was carried out in the frame of a dose-finding CHMI trial, in which malaria-naïve volunteers were infected with increasing doses of PfSPZ Challenge [33]. All individuals underwent CHMI with PfSPZ from the same lot of PfSPZ Challenge either by intravenous (iv, n = 24) or intradermal (id, n = 6) injections of 50 to 3,200 sporozoites. Samples from a subset of the volunteers inoculated with 200 to 3,200 sporozoites by intravenous injection (n = 15) or 2,500 sporozoites by intradermal injection (n = 3) were assessed (Table 1). All volunteers were examined every 12 hours from day 5 after PfSPZ injection and were treated immediately once their thick blood smear became parasite positive by microscopy. On average, the subset of volunteers included for var transcript analysis became parasite positive by thick blood smear ~12 days after infection (mean = 12.0; standard deviation (SD): ±1.2 days post infection) with parasitemias ranging between 2.5 and 54 parasites per μl of blood (mean = 9.1; SD: ±10.1 parasites/μl) (Table 1). In concordance with previous CHMI studies liver stage development took about 6 days and parasite kinetics measured by qPCR in this study suggest that parasites of the third generation after liver release were generally detected at day 11 and 12 post infection [30,33,34]. Accordingly, samples from volunteers infected intravenously either with 800 or 3,200 PfSPZ contained third generation blood phase parasites, whereas volunteer 02.1, the only one infected with 200 PfSPZ, and volunteers infected intradermally with 2,500 PfSPZ had fourth and fifth generation blood phase parasites (day 13–15) (Table 1). The blood from 18 infected volunteers was preserved for transcript profiling of the entire NF54 var gene repertoire at the day of patent infection defined as presence of ring stage parasites in the thick blood smear.
Table 1. Overview of volunteer characteristics infected with PfSPZ Challenge.
| Volunteer ID | Gender | No. ofPfSPZ | Route of administration | Time* | Parasites/μl** |
|---|---|---|---|---|---|
| 02.1 | m | 200 | iv | 13.94 | 54 |
| 08.1 | m | 800 | iv | 10.88 | 2.5 |
| 08.2 | f | 800 | iv | 11.29 | 6 |
| 08.3 | m | 800 | iv | 11.99 | 6 |
| 08.4 | f | 800 | iv | 12.26 | 8.5 |
| 08.5 | m | 800 | iv | 12.08 | 9 |
| 25.1 | f | 2,500 | id | 15.34 | 11 |
| 25.2 | m | 2,500 | id | 13.87 | 8 |
| 25.3 | m | 2,500 | id | 12.89 | 3.5 |
| 32.1 | m | 3,200 | iv | 10.50 | 6.5 |
| 32.2 | m | 3,200 | iv | 11.05 | 7.5 |
| 32.3 | m | 3,200 | iv | 11.52 | 5.5 |
| 32.4 | f | 3,200 | iv | 11.11 | 6.5 |
| 32.5 | m | 3,200 | iv | 10.99 | 7.5 |
| 32.6 | m | 3,200 | iv | 11.00 | 10 |
| 32.7 | m | 3,200 | iv | 10.45 | 7 |
| 32.8 | m | 3,200 | iv | 11.55 | 9.5 |
| 32.9 | f | 3,200 | iv | 12.48 | 9 |
* Days between volunteer infection and positive thick blood smear (day malaria)
** Parasite density at diagnosis determined by thick blood smear
f: female; id: intradermal; iv: intravenous; m: male
Var transcript levels in NF54 sporozoite infected volunteers at the day of patent infection
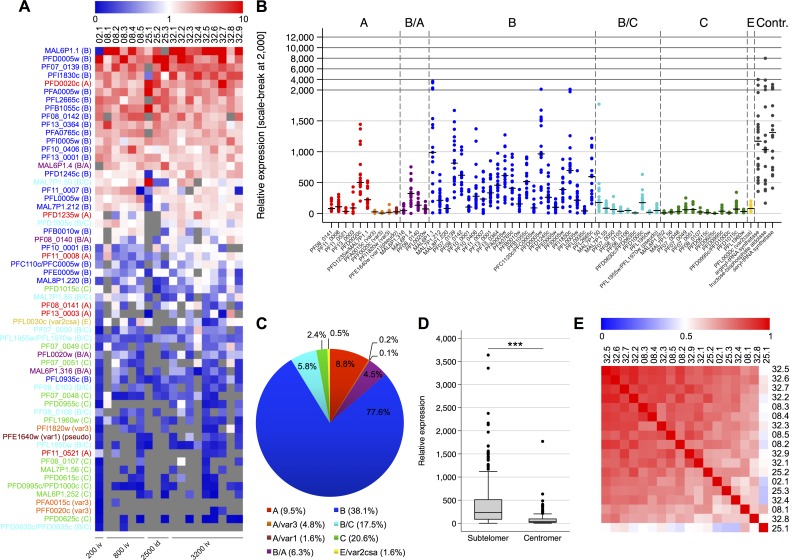
Analysis of var gene expression profiles in individual volunteers showed that transcripts of all var gene variants were detectable during the early blood stage NF54 infection. Interestingly, the individual gene expression profiles seemed to be very similar between the volunteer samples and the variability of the expression values for each var gene was mainly caused by differences in total var expression levels between the samples (Figs 1A and S1). In all samples a B-type variant was expressed at the highest level, mostly the variants MAL6P1.1/PF3D7_0632800 (IDs 08.1, 08.2, 08.4, 25.1, 32.2, 32.5 and 32.9) or PFD0005w/PF3D7_0400100 (IDs 02.1, 08.3, 25.2, 25.3, 32.1, 32.3 and 32.7) (Figs 1A and 1B and S2). Across all patients, the highest transcript levels were detected for MAL6P1.1/PF3D7_0632800 (median = 988.4; interquartile range (IQR): 479.2–1,535.9), followed by PFD0005w/PF3D7_0400100 (median = 964.5; IQR: 430.0–1,523.7), PF07_0139/PF3D7_0733000 (median = 814.9; IQR: 558.2–1,173.1), PFI1830c/PF3D7_0937800 (median = 695.8; IQR: 496.6–883.7) and PF08_0142/PF3D7_0800100 (median = 618.8; IQR: 464.0–667.7) (S2 Fig). All these genes are categorized as group B var genes, reflecting the dominant expression of group B var genes in vivo (Fig 1C). Conversely, transcript abundance of the var gene groups C and E showed some of the lowest expression levels detected. Within group A, the highest relative expression value was always detected for the EPCR-binding variant PFD0020c/PF3D7_0400400 (median = 500.5; IQR: 351.1–793.4), while the conserved group A var1 pseudogene PFE1640w/PF3D7_0533100 and the three var3 genes PFA0015c/PF3D7_0100300, PFI1820w/PF3D7_0937600 and PFF0020c/PF3D7_0600400 are among the 12 genes with lowest expression values (Figs 1A–1C and S2). A comparison of the expression levels between subtelomerically located var genes with those located on internal chromosome clusters revealed a significant difference between both gene subsets (Wilcoxon rank-sum test; p = <0.0001) (Fig 1D). Overall, transcript profiles showed high positive correlations between all patients, irrespective of infection route and dose (Figs 1E and S1). Furthermore, the observed expression profiles highly correlated with the expression profile obtained from a single volunteer in a previous study by Wang et al. [30] (Spearman’s Rank Correlation coefficient = 0.77, p < 0.001) (S3 Fig). In contrast, the observed var gene expression patterns showed no correlation with the data obtained from a previous study in which var gene activation from a null-var background induced by promoter-titration was monitored in an attempt to mimic the early onset of infection in vitro [35] (S3 Fig). In summary, the var gene expression pattern in NF54 parasites at the early onset of blood infection seems to be remarkably uniform at the level of both var gene groups and single var genes. In contrast to the high abundance of transcripts of a restricted subset of var genes which we previously observed in established clinical malaria cases [32], the expression profiles in parasites from all volunteers at this early stage of infection are rather broad without a single gene clearly dominating the infection (Fig 1A and 1B).
Fig 1. Var transcription profiles of parasites recovered from infected volunteers at the day of first microscopically detectable parasitemia.
(A) Heat map showing the individual var gene expression for all volunteer samples taken when parasites were present in the thick blood smear ranked by mean expression. To correct for individual differences in the overall var expression levels, the expression for each var gene was normalized against the total var expression in each sample. The color scale indicates the relative expression levels with red representing values above the median, blue representing values below the median, and white representing median. Grey means not detected. The number of sporozoites (200, 800, 2500 and 3200) and mode of injection (iv = intravenous, id = intradermal) used for each volunteer are indicated below. (B) The distribution of the relative gene expression per var gene and control genes is shown in a dot plot for all volunteer samples at the day of patent infection defined as parasites present in the thick blood smear (n = 18). Each point represents a var gene expression value relative to the normalizing gene sbp1 observed per volunteer sample and the median expression per var gene is marked. Housekeeping genes used as controls, var gene names and groups are indicated. (C) Proportion of var gene expression by group across all volunteers at the day of patent infection. For comparison, genomic proportion of each var gene group is indicated after the color code. (D) Comparison of the expression levels between subtelomeric and centromeric var gene variants. The box plot shows the distribution of transcript levels for each individual var gene relative to sbp1 according to the chromosomal localization of the genes for all 18 volunteer samples at the day of patent infection. Gene expression varied significantly between both gene sets (Wilcoxon rank-sum test, p<0.0001) with median expression of 232 (IQR: 87–510) for telomeric var genes and 40 (IQR: 18–94) for centromeric var genes. (E) The heat map of pairwise Spearman’s rank correlation coefficients (R) between expression profiles illustrates the positive correlation between all 18 volunteer samples at the day of patent infection. Volunteer samples were ranked by the sum of their correlation coefficients. The color scale indicates the correlation coefficient in the range from 0 to 1. With exception of isolate 25.1 versus the isolates 02.1 (p = 0.0015), 25.3 (p = 0.0029) and 32.4 (p = 0.0030) all expressions correlated at a significance level below 0.001.
One interesting outlier from the general expression profile is the pattern detected in volunteer 02.1, in which the intermediate group B/A gene MAL6P1.4/PF3D7_0632500 was not expressed and transcript abundance of the neighboring B-type var gene MAL6P1.1/PF3D7_0632800 was significantly reduced in comparison to all other volunteer samples. Analysis of the genomic DNA by qPCR showed that this was due to a loss of the MAL6P1.4/PF3D7_0632500 gene from the entire parasite population and a partial loss of the MAL6P1.1/PF3D7_0632800 variant, which was only present in the genome of about 2.5% of the parasites in volunteer 02.1 (S4 Fig).
Var transcript levels in PfSPZ Challenge-infected volunteers over time
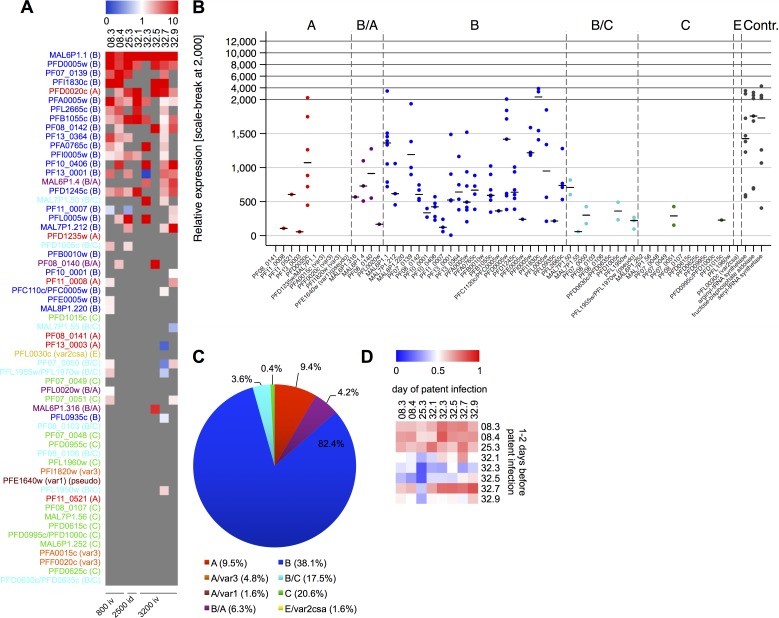
In addition to patient samples obtained at the day of first positive thick blood smear, samples were collected from 8 volunteers at time points with sub-microscopic parasitemia, prior to patent infection. Var transcript profiles were assessed on blood samples obtained either at day 9 (volunteers 32.1, 32.3, 32.5, 32.7) or day 11 post infection (volunteers 08.3, 08.4, 25.3, 32.9), thus, 1 or 2 days before the infected volunteer became microscopically positive (Fig 2A). Hence, the expression profiling only detected highly expressed variants because parasite load in these samples was extremely low (Fig 2A and 2B). qPCR confirmed high expression of B-type var genes and the A-type var gene PFD0020c/PF3D7_0400400 in these early samples (Fig 2A–2C). The expression profiles showed positive correlations between the two parasite generations in vivo (Fig 2D, Spearman’s Rank correlation) suggesting a stable var gene expression pattern during the first few blood phase parasite replication cycles at least in this group of malaria naïve individuals.
Fig 2. Var transcription profiles of parasites from infected volunteers 1–2 days before parasites were microscopically detectable.
(A) Heat map showing the individual var gene expression profiles for samples obtained from volunteers one or two days before parasites were present in the thick blood smear. To correct for individual differences in the overall var expression levels, the expression for each var gene was normalized against the total var expression in each sample. Expression is ranked by mean expression obtained from “day of patent infection” samples (see Fig 1A). The color scale indicates the relative expression levels with red representing values above the median, blue representing values below the median, and white representing median. Grey means not detected. (B) Relative gene expression is shown in a dot plot for the volunteer samples one time point before thick blood smear positivity (n = 8). Gene IDs of var genes and controls are indicated on the x-axis, var gene groups are indicated above the graph. (C) The distribution of var transcripts according to var group affiliation in the volunteer samples 1–2 days before parasites were detected in the thick blood smear is displayed by summarization of the total var gene expression and calculation of the proportion for each var group. The genomic proportion of each var gene group is indicated after the color code. (D) The pairwise Spearman’s rank correlation heat map demonstrates a positive correlation between the expression profiles on the day of first microscopically detectable parasitemia and 1–2 days before in the same volunteer and between volunteer samples.
Comparison of var transcript levels between pre-mosquito parasites and parasites recovered from infected volunteers at the day of patent infection
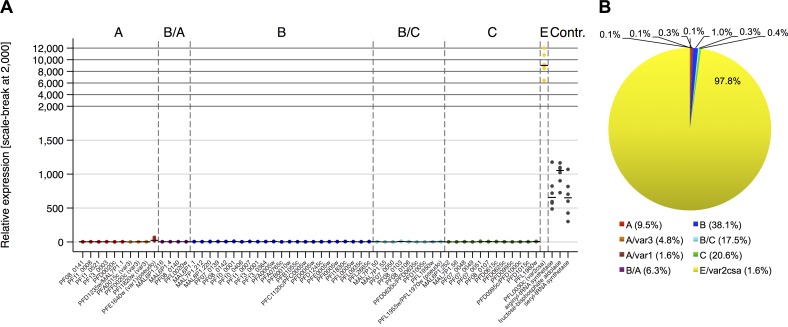
In order to investigate whether mosquito passage results in reprogramming of the var gene transcription pattern, we compared ex vivo var gene expression profiles in the volunteers with the var gene expression profiles in the parental parasite line used to produce the PfSPZ Challenge injected into the subjects. For this purpose, two vials of frozen NF54 parasites from the Sanaria Master Cell Bank RKV01-092505 (MCB) were independently thawed and cultured and var gene expression levels were analyzed after 6, 8 and 21 parasite replication cycles, respectively. The results indicate that the in vitro-adapted pre-mosquito NF54 line stably expressed exclusively the var2csa gene PFL0030c/PF3D7_1200600 (median = 9,012.7; IQR: 8,525.2–10,804.4) (Fig 3A and 3B).
Fig 3. Var transcription profiles of parental parasite lines used for volunteer infection.
(A) The gene expression of each var gene and control genes relative to sbp1 expression is shown in dot plots for the pre-mosquito Master Cell Bank (MCB) parasite line. Each point represents values observed per test generation for the MCB samples taken from two independently thawed parasite stocks after 6, 8 and 21 parasite generations, respectively (n = 6). The median expression per var gene is shown, housekeeping genes used as controls, var gene names and groups are indicated. (B) The MCB cell line exclusively expresses the group E var2csa gene as shown by the proportion of var transcripts according to var group affiliation. The genomic proportion of each var gene group is indicated after the color code.
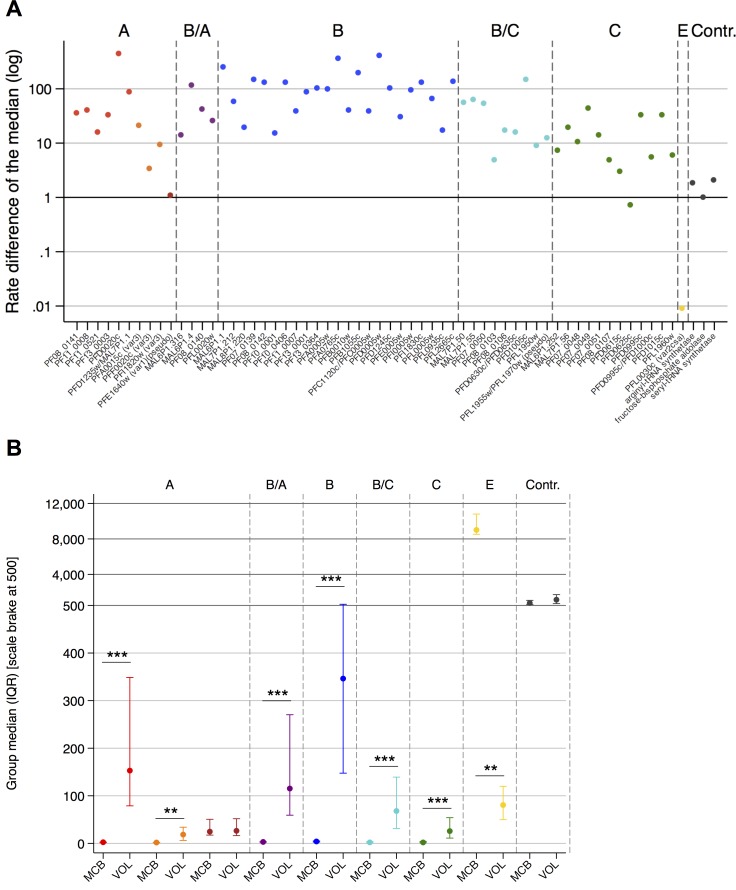
Given the abundant expression of var2csa in the parental NF54 parasites and the monoallelic expression of var genes [6,36] it was unsurprising that direct comparisons of transcript levels between the parental NF54 parasite line MCB and parasites recovered from the infected volunteers revealed increased expression in vivo of the entire gene family except var2csa (Fig 4A). The elevated expression of the var groups in the infected volunteer samples was significant; A (p <0.0001) including the var3 subfamily (p = 0.0028), B/A (p <0.0001), B (p <0.0001), B/C (p <0.0001) and C (p <0.0001). The only exception was the decreased expression of the group E var2csa gene (p = 0.0045) and the var1 subfamily pseudogene PFE1640w/PF3D7_0533100 was expressed at very low, unchanging levels in both NF54 and volunteers (Fig 4B). Thus, the var gene expression profiles of the ex vivo patient samples revealed substantial changes in comparison to the pre-mosquito NF54 parasites, which indicates epigenetic reprogramming of var genes during mosquito and/or liver passage.
Fig 4. Modification of var transcription by mosquito transmission.
(A) The rate difference of the median expression for each individual var variant and the housekeeping controls shows the overexpression of the entire var gene family in vivo with exception of the var2csa gene PFL0030c in comparison to the parental Master Cell Bank (MCB) parasite line. Each point reflects the median for the volunteer samples at the day of patent infection (n = 18) divided by the median observed for the parasite generations 6, 8 and 21 from two vials of the MCB cell line (n = 6). Housekeeping genes used as controls, var gene names and groups are indicated. (B) Differences in gene expression on group level between the pre-mosquito MCB parasite lines and parasites recovered from the infected volunteers (VOL) at the day of patent infection determined by thick blood smear are displayed as group median with interquartile range (IQR). Significant differences in distributions between MCB and VOL series were tested via a Wilcoxon rank-sum test using a Bonferroni corrected significance level. The graph contains a scale brake at the y-axis to account for the huge variability in the gene expressions. Group affiliations are indicated above the graph. Red (A), orange (A, subfamily var3), dark red (A, subfamily var1), purple (B/A), blue (B), turquoise (B/C), green (C) and yellow (E).
Discussion
Analyses of the role of PfEMP1 in malaria pathogenesis and protection are hampered by the high sequence variability of the encoding var genes. Hence, quantitative expression data of the entire var gene repertoire from P. falciparum patient isolates are not well documented. The most common strategy for analyzing var gene expression in parasite strains with variable genomic background uses degenerate primer pairs targeting conserved sequence blocks for semi-quantitative RT-PCR or qPCR approaches. Using this strategy a higher frequency of A- and B-type PfEMP1 transcripts was detected in malaria patients suffering from severe disease in comparison to mild or asymptomatic malaria [11–21]. In contrast to malaria patients who have already developed clinical symptoms of the disease and parasites ran through an unknown number of cycles, parasites from CHMI studies are only allowed to progress through a few, well-characterized replication cycles in vivo before the volunteers have to be treated. Accordingly, patients infected in CHMI studies have very low levels of parasitemia and do not develop complications (reviewed in [34]). Using samples from such a CHMI study we performed the first in depth quantitative analysis of var gene expression on 18 volunteer samples at the early onset of blood infection. All volunteers were infected with the NF54 strain and the same cryopreserved PfSPZ lot was used for intravenous or intradermal injection. In agreement with the expression of all or most of the NF54 var gene repertoire in parasites from recently infected volunteers express, there is also evidence that parasites tend to express many var genes rather than a single dominant variant in individuals with low levels of naturally acquired immunity [12,32,37]. Interestingly, genes possessing A- and B-type 5’-UTRs revealed a significant higher expression level in comparison to genes with C- or E-type 5’-UTR. Moreover, the expression level also seems to be influenced by the chromosomal position of the gene since the subtelomerically located A-, B/A- and B-type genes show higher expression levels than centromerically located genes of the B/C and C groups. This observation is in line with previously described differences in on- and off-rates for subtelomerically versus centromerically located var gene variants indicating that subtelomerically located variants have a much higher expression dynamic [25,36,38,39]. Lowest transcript abundances in vivo were detected for centromeric genes with C-type 5'-UTR. The only exceptions are the var gene variants of the var1 and var3 subfamilies, which show a very low expression level despite their A-type promoter sequence and their subtelomeric location. Together with the E-type var2csa (PFL0030c/PF3D7_1200600) gene all interstrain conserved var gene variants reveal only minor transcript abundances in all volunteers analyzed.
The first study on var gene expression in ex vivo samples of 3D7-infected volunteers applied a semi-quantitative RT-PCR strategy followed by sequencing of a large number of clones. On day 12.5 or 13.5 post infection most of the detected transcripts belonged to subtelomeric var genes of group A and B, consistent with the results presented here [29]. In contrast to the broad expression of most var genes detected in our study, a single B-type PfEMP1 transcript, PF11_0007/PF3D7_1100100, was found most frequently and clearly dominated the expression profile in both volunteers analyzed by Peters and colleagues. One possible explanation for this divergent observation is the susceptibility of the semi-quantitative RT-PCR method to primer and cloning bias, which can result in overestimation of transcript frequencies. In another study from Lavstsen et al. parasites from six infected volunteers were analyzed by qPCR after approximately a month of in vitro-cultivation. In line with our data, all parasite lines established from first-generation parasites after liver release showed a similar var gene expression pattern and transcripts of all var genes could be detected [31]. In contrast to the data obtained in our study, 9 of the 10 lowest transcribed genes were classified as A or B/A var gene, which may be the result of the higher expression dynamic of these genes and/or the in vitro-cultivation of the parasites before analysis. More recently, NF54 parasites isolated from a Dutch volunteer in another CHMI study were analyzed using a more accurate qPCR approach [30]. Due to the very low parasitemia in CHMI studies the authors were able to analyze transcript abundances of the full NF54 var repertoire in a single blood sample only taken at day 11 after infection. Interestingly, apart from slight differences in frequencies, they observed exactly the same pattern of var gene transcription at the early onset of blood infection and, remarkably, the highest transcript levels were observed for the same group B var gene variant, MAL6P1.1/PF3D7_0632800 (synonym PFF1595c). Moreover, the three least polymorphic var gene subfamilies var1, var2csa and var3 were also among the 10 lowest transcript levels in the Dutch volunteer. The positive correlation between both in vivo data sets was highly significant (S3 Fig). These highly reproducible results indicate that an intrinsic var gene expression program of the NF54 parasite exists which seems to drive higher levels of transcription of A- and B-type 5'-UTR controlled genes with the exception of the conserved var1 and var3 genes at the onset of blood infection. Later on, growth advantages of parasites best adapted to their host’s adhesion surfaces may select for parasites expressing particular var gene variants, subsequently resulting in the typical, dominant expression of one or a few var genes as we observed in natural infections. But whether this var gene expression pattern is a general strategy used by all parasite strains to establish human blood infections waits to be confirmed. Further work is also needed to address the question whether var gene expression patterns differ between parasites from experimentally infected malaria-naïve humans and those who already underwent natural or experimental P. falciparum infections and, accordingly, possess a pre-formed immune response against PfEMP1 variants expressed in previous infection(s).
Another study reversibly silenced all endogenous var genes by promoter titration to mimic the moment when parasites enter the human blood phase [35]. In agreement with our data, two weeks after drug removal these “null-var” parasites with erased epigenetic memory from previous in vitro generations broadly activated all var genes. However, although A- and B-type variants are also activated, these parasites tend to express predominantly group C var genes, which becomes even more clearly after one and three month of cultivation. Accordingly, central var genes are the most highly activated genes in vitro, which is in clear contrast to the expression pattern we observe in vivo where subtelomerically located var genes dominate. Accordingly, Spearman’s rank correlation analysis showed no significant correlation between the data set from Fastman et al. and our in vivo data (S3 Fig). Maybe the observed pattern in vivo is the result of resetting plus in vivo selection while the resetting of the var gene repertoire by promoter titration is the result of the resetting alone. Additionally, because Fastman et al. found highly activated B and C-type var genes adjacent to rarely activated genes they concluded that the probability of var genes to be turned on or off are independent of their chromosomal location or promoter type per se and rather seems to be associated with intrinsic properties of each gene. Although we also found expression level differences between adjacent gene variants, e.g. PFI1820 and PFI1830, our ex vivo data show a clear association of the subtelomeric position as well as A- and, especially, B-type promoters with higher expression level.
The comparison between var gene expression in the pre-mosquito NF54 parasites and in the ex vivo patient samples revealed substantial changes indicating a kind of resetting of the var gene expression program during mosquito and/or liver passage of parasites. Pre-mosquito MCB parasites predominantly transcribed the var2csa gene PFL0030c/PF3D7_1200600 and this gene was also among the most abundant transcripts in Sanaria’s Working Cell Bank lot SAN02-073009, which was derived from the MCB and used to produce the PfSPZ Challenge lot for this particular volunteer infection (personal communication Matthias Frank), which is in clear contrast to the broad expression pattern of the subtelomeric var gene groups in the patient samples. Previous studies have indicated that mosquito passage leads to a shift in the transcribed var profile towards expression of subtelomeric genes [29] and promiscuous var transcription [30]. By using a parental parasite that appeared to exclusively express var2csa and robust and exhaustive methods to quantitate the entire var repertoire, we prove conclusively for the first time that the expressed var repertoire is dramatically reset following mosquito passage. Two mechanisms could explain these results: either the var epigenetic program is altered following passage through the mosquito leading to erasure of epigenetic memory and activation of subtelomeric var genes by first generation merozoites; or parental parasites expressing var2csa were selected against after exiting asexual replication in vitro. Although the latter explanation is possible it would mean that nearly all of the parental parasites would either fail to passage through the mosquito or to establish the infection in the human host. In fact, some parasites transcribing var2csa express no PfEMP1 on the surface of the infected erythrocyte, because var2csa is often repressed at the level of translation [40]. Those parasites fail to adhere to endothelial receptors and would be rapidly cleared by the spleen. Resetting of the epigenetic var program following mosquito passage is supported by the observation that release of artificial repression of the entire var repertoire in asexual parasites leads to promiscuous var activation within the population [35], similar to the var profiles following mosquito passage that we and Wang et al. observed [30]. Furthermore, in NF54 sporozoites var gene expression is largely silenced, which is in agreement with a resetting of the var expression program during mosquito passage [41]. Our findings synthesize these various studies into a complete picture of mosquito passage causing epigenetic reprogramming to allow expression of any member of the var repertoire but with an apparent bias towards the subtelomeric var genes. Whether this bias was the consequence of phenotype selection in the first rounds of asexual replication remains unknown.
Interestingly, the modification of variable antigen expression by vector transmission has recently also been shown for the rodent malaria parasite P. chabaudi [42]. Spence and colleagues showed that the high virulence of serially blood-passaged parasites is attenuated by parasite transmission through mosquitos and this correlates with altered expression of the cir (chabaudi interspersed repeats) multi-gene family. Moreover, the activation of 114 cir variants (57%) after mosquito passage of P. chabaudi parasites closely reflects the broad activation pattern of all or most subtelomerically located var gene variants at the onset of blood infection in malaria naïve volunteers. Although the pir (Plasmodium interspersed repeats) multi-gene family is unique to P. vivax, P. knowlesi and the rodent clade of malaria parasites without any significant sequence similarity in the P. falciparum genome [43], the reprogramming of antigenic variant gene expression by vector transmission seems to be universal in the Plasmodium genus [29,42,44,45]. In summary, our data from this study clearly show differences in var gene expression before mosquito passage of the NF54 strain and early onset of blood infections, providing novel evidence for an epigenetic reprogramming or resetting of virulence gene expression during parasite transmission as proposed previously [29,32,42].
Materials and Methods
Ethics statement
The ethics committee of the University Clinic and the Medical Faculty of the University of Tübingen approved the study and the U.S. Food and Drug Administration Agency (FDA) provided regulatory oversight. Investigation of the var gene expression pattern in early blood stage infection was an exploratory objective of the trial. The study was conducted according to the principles of the Declaration of Helsinki in its 6th revision as well as International Conference on Harmonization–Good Clinical Practice (ICH-GCP) guidelines. The study registration code with ClinicalTrials.gov is NCT01624961. All volunteers, aged 18 to 45 years, provided written informed consent and understanding of the study and procedures was assessed with a quiz [33].
CHMI trial and blood sampling
At the Institute of Tropical Medicine in Tübingen, Germany, healthy, malaria-naïve volunteers were infected with aseptic, purified, cryopreserved NF54 sporozoites (PfSPZ Challenge) from a single lot manufactured by Sanaria Inc., USA [33].
Blood samples for thick blood smears were taken daily from the onset of merozoite release at 5 days after sporozoite injection. Blood samples for var transcription profiling were taken at a 48 hours interval. Sampling continued until parasites were detected in thick blood smears when anti-malaria treatment was initiated. Then, erythrocytes from all infected volunteers were separated from leukocytes by Lymphoprep (Axis-Shield) gradient centrifugation followed by filtration through Plasmodipur filter (EuroProxima). In total, 4.5–9.0 ml of packed red blood cells were obtained from 18 volunteers at the day of first positive P. falciparum parasitemia by thick blood smear and from 8 volunteers at one time point before the thick blood smear was positive (1–2 days).
Parasite cultivation
Two frozen vials (A and B) of NF54 parasites from Sanaria’s MCB lot RKV01-092505 were separately thawed and maintained in culture using a protocol adopted from Trager and Jensen [46]. A hematocrit of 5% was adjusted using human O+ red blood cells and the parasite culture medium was supplemented with 10% heat-inactivated human serum.
DNA purification
For DNA purification, MCB parasites were cultivated for 6 parasite replication cycles to obtain a higher yield. Genomic DNA was prepared from ring stage parasites established from both cell stocks using the QIAamp DNA Blood Mini Kit (Qiagen).
RNA purification and cDNA synthesis
For RNA purification ring stage parasites from MCB vials A and B were taken after 6, 8 and 21 parasite replication cycles after thawing, respectively. One cycle of invasion before harvesting, parasite growth was synchronized twice at an interval of 6 hours using sorbitol [47]. Cell pellets of MCB and of the leukocyte depleted patient blood samples were rapidly lysed in 10 volumes pre-warmed TRIzol (Invitrogen) and stored at -80°C. RNA purification was performed according to the manufacturers instruction of the PureLink RNA Kit (Life Technologies) including DNase treatment on the column. Absence of DNA was checked using 50 ng RNA as template for a qPCR run with the sbp1 primer pair. If necessary, DNasing was repeated until the sample was free of any DNA contamination by qPCR. Afterwards, cDNA was synthetized with the SuperScript III Reverse Transcriptase (Invitrogen) primed with random hexamers (Invitrogen) at 50°C for 1 hour. As possible, a cDNA synthesis reaction without enzyme was performed and analyzed in parallel by qPCR.
Quantitative real-time PCR
Quantitative amplification was conducted in the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) using the SDS software version 2.3 (Applied Biosystems). Template was mixed with the SYBR Green PCR Master Mix (Applied Biosystems) and 0.3 μM forward and reverse primer in a final volume of 10 μl. Reactions were incubated at 50°C for 2 min, at 95°C for 10 min, then subjected to 40 cycles of 95°C for 15 s and 60°C for 1 min and a subsequent melting step (60–95°C). The specificity of each primer pair was confirmed after each qPCR run by dissociation curve analysis.
Analysis was performed using sbp1 (PFE0065w/PF3D7_0501300) to normalize and gDNA from MCB parasites to calibrate the individual var gene expression data. Relative quantification of the NF54 var repertoire by 2-ΔΔCt analysis was performed using a previously described primer set [48] supplemented with new primer pairs for PF07_0049/PF3D7_0712000, PFL1950w/PF3D7_1240300, PFE0005w/PF3D7_0500100 and PF07_0051/PF3D7_0712600. Furthermore, primer pairs targeting the housekeeping genes fructose-bisphosphate aldolase (PF14_0425/PF3D7_1444800), seryl-tRNA synthetase (PF07_0073/PF3D7_0717700) and arginyl-tRNA synthetase (PFL0900c/PF3D7_1218600) were included (S1 Table). Relative expression data were corrected for amplification efficiency of each primer pair, which was determined by dilution of a single gDNA from 3D7 over 5 logs of concentration (S1 Table). Expression and correlation heat maps were generated using Multiple Experiment Viewer (MeV). The expression plots were programmed with Stata version 14. In all expression dot plots a scale break of the y-axis was introduced at 2,000 due to the huge difference of the relative expression values between gene groups.
Statistical analysis
Spearman’s rank correlation was applied to assess whether gene expression patterns between parasites isolated from different infected volunteers or between different parasite generations from the same infected volunteer were comparable. Correlation coefficients (R-values) were displayed in a heat-map, where color codes define the correlation levels (Figs 1E and 2D). Furthermore, Spearman’s rank correlation was used to compare the gene expression patterns observed in this study with the results obtained by Wang et al. [30] and Fastman et al. [35] (S3C Fig).
To compare var gene expression levels between infected volunteers, and the pre-mosquito cell line MCB differences in the median gene expressions were calculated for each individual var gene variant. Therefore, the median ratio was calculated. A value of 1 indicates no difference, >1 higher gene expression in the infected volunteers and a value <1 higher gene expression in the MCB cell line (Fig 4A).
To describe the expression of genes with different chromosomal localizations or per var gene group observations within respective groups were pooled and the median expression along with interquartile rage (IQR) was calculated. Differences in gene expressions between the respective groups were tested via a Wilcoxon rank sum test. This analysis was applied to assess differences in gene expression levels of subtelomerically versus centromerically located var gene variants in the volunteer samples taken at the day of first microscopically detectable parasitemia (Fig 1D). Furthermore, expression level differences of var gene groups (i.e., A, A var3 and var1 subfamilies, B/A, B, B/C, C, E) and control genes between volunteer samples at the day of patent infection versus the pre-mosquito parasite line MCB were also tested via Wilcoxon rank sum test (Fig 4B). Bonferroni corrected significance level was used to account for multiple comparisons. Since comparison was done among 9 var gene groups, the p-value was corrected by the multiplication with 9.
All analyses were done using STATA 14 (College Station, TX: StataCorp LP) or GraphPad Prism 4.
Supporting Information
(PDF)
Spearman’s rank correlation coefficient (R) and significances (p-values) are indicated above each graph.
(PDF)
The median var transcript level relative to the sbp1 transcript level with IQR is shown for all 18 volunteer samples. Expression values for genes of the different var groups are presented in red (A), orange (subfamily var3), dark red (subfamily var1), purple (B/A), blue (B), turquoise (B/C), green (C) and yellow (E).
(PDF)
(A) Heat map showing the individual var gene expression profiles for all volunteer samples on the day of patent infection and for the volunteer sample obtained by Wang et al. [30]. Furthermore, var gene expression data is shown for two 3D7 cell lines (E1 and G6), in which a “null-var” phenotype was created in vitro, var gene expression was reactivated and analyzed two weeks, one and three months after drug removal [35]. Var genes are ranked by var gene group and mean expression. To correct for individual differences in the overall var expression levels and for the use of different normalizing genes in the three studies, the expression of each var gene was normalized against the total var expression in each sample. Samples were hierarchically clustered using average linkage clustering with Spearman’s Rank Correlation as distance metric. The color scale indicates the relative expression levels with red representing values above the median, blue representing values below the median, and white representing median. Grey means not detected. (B) Scatter plots of normalized var gene expression data obtained by Wang et al. [30] and Fastman et al. [35] against mean var gene expression from all ex vivo volunteer samples at the day of first microscopically detectable parasitemia. Spearman’s rank correlation coefficient (R) and significances (p-values) are indicated above each graph. (C) Heat map of pairwise Spearman’s rank correlation coefficients (R) of var gene expression profiles from Wang et al. and Fastman et al. [30,35] against the mean ex vivo var gene expression of all volunteers at the day of patent infection. The color scale indicates the correlation coefficient with red indicating positive correlation, blue indicating negative correlation and white indicating no correlation.
(PDF)
(A) var gene expression in parasites recovered from volunteer 02.1, who received 200 sporozoites intravenously 14 days before. Shown are transcript levels of all NF54 var genes and three housekeeping controls as indicated. For comparison, the median expression value calculated from all volunteer samples is displayed for each gene as a black line. Both genes with remarkably low expression values, MAL6P1.4/PF3D7_0632500 and MAL6P1.1/PF3D7_0632800, are highlighted in red boxes. (B) var qPCR analysis with genomic DNA extracted from two vials of Master Cell Bank parasites (MCB-A and -B), Working Cell Bank parasites (WCB) and parasites recovered from volunteers 02.1 (VOL 02.1) and 32.5 (VOL 32.5). Shown are Ct (cycle of treshold) values for each individual var gene primer pair. The primer pair for the variant MAL6P1.4 failed to detect an amplification product in the sample from volunteer 02.1 within 40 PCR cycles (Ct ≥ 40). An increased Ct value for the variant MAL6P1.1 of 5.3 PCR cycles reflects a reduction of 38.9 fold on genomic DNA level (2ΔCt).
(PDF)
Acknowledgments
We would like to thank all volunteers who participated in the clinical trial at the Institute of Tropical Medicine in Tübingen, Germany, and Sandra Dimonte, Ellen Bruske and Matthias Frank for providing genomic DNA from Sanarias WCB and parasites recovered from volunteers 02.1 and 32.5. Special thanks go to Heidrun von Thien for her technical support and Tim-Wolf Gilberger for critical reading of the manuscript. Furthermore, we thank Christian W. Wang and Yair Fastman for sharing their data.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The clinical trial and the var gene expression analysis were supported by the Federal Ministry of Education and Research in the framework of the German Centre for Infection Research (DZIF) (WP4). RK, BM and PGK received funding by the German Center for Infection Research (DZIF) (WP4). The publication of this article was funded by the Open Access Fund of the Leibniz Association. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organisation (2014) World Malaria Report 2014.
- 2. Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, et al. (1995) Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82: 77–87. [DOI] [PubMed] [Google Scholar]
- 3. Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, et al. (1995) The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82: 89–100. [DOI] [PubMed] [Google Scholar]
- 4. Rowe JA, Claessens A, Corrigan RA, Arman M (2009) Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: molecular mechanisms and therapeutic implications. Expert Rev Mol Med 11: e16 10.1017/S1462399409001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, et al. (1995) Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82: 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, et al. (2006) A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 439: 1004–1008. [DOI] [PubMed] [Google Scholar]
- 7. Kraemer SM, Smith JD (2003) Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol Microbiol 50: 1527–1538. [DOI] [PubMed] [Google Scholar]
- 8. Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG (2003) Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar J 2: 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kraemer SM, Kyes SA, Aggarwal G, Springer AL, Nelson SO, et al. (2007) Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics 8: 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T (2010) Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes-divide and conquer. PLoS Comput Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, et al. (2004) Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med 199: 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bull PC, Berriman M, Kyes S, Quail MA, Hall N, et al. (2005) Plasmodium falciparum variant surface antigen expression patterns during malaria. PLoS Pathog 1: e26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaestli M, Cockburn IA, Cortes A, Baea K, Rowe JA, et al. (2006) Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J Infect Dis 193: 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, et al. (2006) Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol 150: 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rottmann M, Lavstsen T, Mugasa JP, Kaestli M, Jensen AT, et al. (2006) Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun 74: 3904–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Warimwe GM, Keane TM, Fegan G, Musyoki JN, Newton CR, et al. (2009) Plasmodium falciparum var gene expression is modified by host immunity. Proc Natl Acad Sci U S A 106: 21801–21806. 10.1073/pnas.0907590106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lavstsen T, Turner L, Saguti F, Magistrado P, Rask TS, et al. (2012) Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci U S A 109: E1791–1800. 10.1073/pnas.1120455109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bertin GI, Lavstsen T, Guillonneau F, Doritchamou J, Wang CW, et al. (2013) Expression of the domain cassette 8 Plasmodium falciparum erythrocyte membrane protein 1 is associated with cerebral malaria in Benin. PLoS One 8: e68368 10.1371/journal.pone.0068368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Almelli T, Ndam NT, Ezimegnon S, Alao MJ, Ahouansou C, et al. (2014) Cytoadherence phenotype of Plasmodium falciparum-infected erythrocytes is associated with specific pfemp-1 expression in parasites from children with cerebral malaria. Malar J 13: 333 10.1186/1475-2875-13-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Almelli T, Nuel G, Bischoff E, Aubouy A, Elati M, et al. (2014) Differences in Gene Transcriptomic Pattern of Plasmodium falciparum in Children with Cerebral Malaria and Asymptomatic Carriers. PLoS One 9: e114401 10.1371/journal.pone.0114401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Merrick CJ, Huttenhower C, Buckee C, Amambua-Ngwa A, Gomez-Escobar N, et al. (2012) Epigenetic dysregulation of virulence gene expression in severe Plasmodium falciparum malaria. J Infect Dis 205: 1593–1600. 10.1093/infdis/jis239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, et al. (2013) Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature 498: 502–505. 10.1038/nature12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berger SS, Turner L, Wang CW, Petersen JE, Kraft M, et al. (2013) Plasmodium falciparum expressing domain cassette 5 type PfEMP1 (DC5-PfEMP1) bind PECAM1. PLoS One 8: e69117 10.1371/journal.pone.0069117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sharp S, Lavstsen T, Fivelman QL, Saeed M, McRobert L, et al. (2006) Programmed transcription of the var gene family, but not of stevor, in Plasmodium falciparum gametocytes. Eukaryot Cell 5: 1206–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frank M, Dzikowski R, Amulic B, Deitsch K (2007) Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol 64: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Falk N, Kaestli M, Qi W, Ott M, Baea K, et al. (2009) Analysis of Plasmodium falciparum var genes expressed in children from Papua New Guinea. J Infect Dis 200: 347–356. 10.1086/600071 [DOI] [PubMed] [Google Scholar]
- 27. Enderes C, Kombila D, Dal-Bianco M, Dzikowski R, Kremsner P, et al. (2011) Var Gene promoter activation in clonal Plasmodium falciparum isolates follows a hierarchy and suggests a conserved switching program that is independent of genetic background. J Infect Dis 204: 1620–1631. 10.1093/infdis/jir594 [DOI] [PubMed] [Google Scholar]
- 28. Gupta S, Snow RW, Donnelly CA, Marsh K, Newbold C (1999) Immunity to non-cerebral severe malaria is acquired after one or two infections. Nat Med 5: 340–343. [DOI] [PubMed] [Google Scholar]
- 29. Peters J, Fowler E, Gatton M, Chen N, Saul A, et al. (2002) High diversity and rapid changeover of expressed var genes during the acute phase of Plasmodium falciparum infections in human volunteers. Proc Natl Acad Sci U S A 99: 10689–10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang CW, Hermsen CC, Sauerwein RW, Arnot DE, Theander TG, et al. (2009) The Plasmodium falciparum var gene transcription strategy at the onset of blood stage infection in a human volunteer. Parasitol Int 58: 478–480. 10.1016/j.parint.2009.07.004 [DOI] [PubMed] [Google Scholar]
- 31. Lavstsen T, Magistrado P, Hermsen CC, Salanti A, Jensen AT, et al. (2005) Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malar J 4: 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bachmann A, Predehl S, May J, Harder S, Burchard GD, et al. (2011) Highly co-ordinated var gene expression and switching in clinical Plasmodium falciparum isolates from non-immune malaria patients. Cell Microbiol 13: 1397–1409. 10.1111/j.1462-5822.2011.01629.x [DOI] [PubMed] [Google Scholar]
- 33. Mordmüller B, Supan C, Sim KL, Gómez-Pérez GP, Salazar CLO, et al. (2015) Direct venous inoculation of Plasmodium falciparum sporzoites for controlled human malaria infection: a dose-finding trial in two centres. Malar J 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sauerwein RW, Roestenberg M, Moorthy VS (2011) Experimental human challenge infections can accelerate clinical malaria vaccine development. Nat Rev Immunol 11: 57–64. 10.1038/nri2902 [DOI] [PubMed] [Google Scholar]
- 35. Fastman Y, Noble R, Recker M, Dzikowski R (2012) Erasing the epigenetic memory and beginning to switch-the onset of antigenic switching of var genes in Plasmodium falciparum . PLoS One 7: e34168 10.1371/journal.pone.0034168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dzikowski R, Frank M, Deitsch K (2006) Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog 2: e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Warimwe GM, Recker M, Kiragu EW, Buckee CO, Wambua J, et al. (2013) Plasmodium falciparum var gene expression homogeneity as a marker of the host-parasite relationship under different levels of naturally acquired immunity to malaria. PLoS One 8: e70467 10.1371/journal.pone.0070467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horrocks P, Pinches R, Christodoulou Z, Kyes SA, Newbold CI (2004) Variable var transition rates underlie antigenic variation in malaria. Proc Natl Acad Sci U S A 101: 11129–11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blomqvist K, Normark J, Nilsson D, Ribacke U, Orikiriza J, et al. (2010) var gene transcription dynamics in Plasmodium falciparum patient isolates. Mol Biochem Parasitol 170: 74–83. 10.1016/j.molbiopara.2009.12.002 [DOI] [PubMed] [Google Scholar]
- 40. Amulic B, Salanti A, Lavstsen T, Nielsen MA, Deitsch KW (2009) An upstream open reading frame controls translation of var2csa, a gene implicated in placental malaria. PLoS Pathog 5: e1000256 10.1371/journal.ppat.1000256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang CW, Mwakalinga SB, Sutherland CJ, Schwank S, Sharp S, et al. (2010) Identification of a major rif transcript common to gametocytes and sporozoites of Plasmodium falciparum . Malar J 9: 147 10.1186/1475-2875-9-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spence PJ, Jarra W, Levy P, Reid AJ, Chappell L, et al. (2013) Vector transmission regulates immune control of Plasmodium virulence. Nature 498: 228–231. 10.1038/nature12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frech C, Chen N (2013) Variant surface antigens of malaria parasites: functional and evolutionary insights from comparative gene family classification and analysis. BMC Genomics 14: 427 10.1186/1471-2164-14-427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brannan LR, McLean SA, Phillips RS (1993) Antigenic variants of Plasmodium chabaudi chabaudi AS and the effects of mosquito transmission. Parasite Immunol 15: 135–141. [DOI] [PubMed] [Google Scholar]
- 45. McLean SA, Phillips RS, Pearson CD, Walliker D (1987) The effect of mosquito transmission of antigenic variants of Plasmodium chabaudi . Parasitology 94 (Pt 3): 443–449. [DOI] [PubMed] [Google Scholar]
- 46. Trager W, Jensen JB (1976) Human malaria parasites in continuous culture. Science 193: 673–675. [DOI] [PubMed] [Google Scholar]
- 47. Lambros C, Vanderberg JP (1979) Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol 65: 418–420. [PubMed] [Google Scholar]
- 48. Duffy MF, Byrne TJ, Carret C, Ivens A, Brown GV (2009) Ectopic recombination of a malaria var gene during mitosis associated with an altered var switch rate. J Mol Biol 389: 453–469. 10.1016/j.jmb.2009.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Spearman’s rank correlation coefficient (R) and significances (p-values) are indicated above each graph.
(PDF)
The median var transcript level relative to the sbp1 transcript level with IQR is shown for all 18 volunteer samples. Expression values for genes of the different var groups are presented in red (A), orange (subfamily var3), dark red (subfamily var1), purple (B/A), blue (B), turquoise (B/C), green (C) and yellow (E).
(PDF)
(A) Heat map showing the individual var gene expression profiles for all volunteer samples on the day of patent infection and for the volunteer sample obtained by Wang et al. [30]. Furthermore, var gene expression data is shown for two 3D7 cell lines (E1 and G6), in which a “null-var” phenotype was created in vitro, var gene expression was reactivated and analyzed two weeks, one and three months after drug removal [35]. Var genes are ranked by var gene group and mean expression. To correct for individual differences in the overall var expression levels and for the use of different normalizing genes in the three studies, the expression of each var gene was normalized against the total var expression in each sample. Samples were hierarchically clustered using average linkage clustering with Spearman’s Rank Correlation as distance metric. The color scale indicates the relative expression levels with red representing values above the median, blue representing values below the median, and white representing median. Grey means not detected. (B) Scatter plots of normalized var gene expression data obtained by Wang et al. [30] and Fastman et al. [35] against mean var gene expression from all ex vivo volunteer samples at the day of first microscopically detectable parasitemia. Spearman’s rank correlation coefficient (R) and significances (p-values) are indicated above each graph. (C) Heat map of pairwise Spearman’s rank correlation coefficients (R) of var gene expression profiles from Wang et al. and Fastman et al. [30,35] against the mean ex vivo var gene expression of all volunteers at the day of patent infection. The color scale indicates the correlation coefficient with red indicating positive correlation, blue indicating negative correlation and white indicating no correlation.
(PDF)
(A) var gene expression in parasites recovered from volunteer 02.1, who received 200 sporozoites intravenously 14 days before. Shown are transcript levels of all NF54 var genes and three housekeeping controls as indicated. For comparison, the median expression value calculated from all volunteer samples is displayed for each gene as a black line. Both genes with remarkably low expression values, MAL6P1.4/PF3D7_0632500 and MAL6P1.1/PF3D7_0632800, are highlighted in red boxes. (B) var qPCR analysis with genomic DNA extracted from two vials of Master Cell Bank parasites (MCB-A and -B), Working Cell Bank parasites (WCB) and parasites recovered from volunteers 02.1 (VOL 02.1) and 32.5 (VOL 32.5). Shown are Ct (cycle of treshold) values for each individual var gene primer pair. The primer pair for the variant MAL6P1.4 failed to detect an amplification product in the sample from volunteer 02.1 within 40 PCR cycles (Ct ≥ 40). An increased Ct value for the variant MAL6P1.1 of 5.3 PCR cycles reflects a reduction of 38.9 fold on genomic DNA level (2ΔCt).
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.