Abstract
Long noncoding RNAs (lncRNAs) are pivotal regulators of genome structure and gene expression. LncRNAs can directly interact with chromatin-modifying enzymes and nucleosome-remodeling factors to control chromatin structure and accessibility of genetic information. Moreover, lncRNA expression can be controlled by chromatin-remodeling factors, suggesting a feedback circuit of regulation. Here, we discuss the recent advances of lncRNA studies, focusing on the function and mechanism of lncRNA–chromatin interactions.
Keywords: chromatin modification, chromatin remodeling, human diseases, lncRNA promoter, lncRNA regulation, long non-coding RNA, RNA therapeutics
Long noncoding RNAs (lncRNAs), which are defined as non-protein coding transcripts longer than 200 nucleotides, have emerged as important regulators of cell physiology and pathology. Transcriptome profiling has identified an increasing number of lncRNAs with tissue-specific expression; however, only dozens of lncRNAs have been characterized in vivo with regard to the mechanisms of action.1-7 The majority of lncRNAs remains unknown for their biological functions, roles in diseases, and precise mechanisms. One unique feature of lncRNAs is their biochemical abilities to interact with a wide range of molecules, and through specific RNA functional domains lncRNAs can form a variety of RNA–RNA, RNA–DNA, or RNA–protein complexes,8 conferring functional diversities to lncRNAs. Recent discoveries of lncRNAs reveal a broad association of lncRNAs with the epigenetic machinery to control chromatin structure and gene expression. LncRNAs can directly interact with many histone- and DNA-modifying enzymes to participate in covalent modifications of histones or DNA. Furthermore, an lncRNA was recently found to be capable of modulating the non-covalent, ATP-dependent chromatin remodeling process,9 indicating an extensive role of lncRNAs in chromatin regulation. The mechanisms of how lncRNAs control chromatin by covalent modifications are previously reviewed.10-12 Here, we discuss the recent progress of lncRNA studies, with an emphasis on the mechanism and function of lncRNA that modulates chromatin remodeling. We also discuss how lncRNA expression is regulated and how lncRNA–chromatin interactions can be used to design new therapeutic strategies for human diseases.
Ongoing characterization of long non-coding RNAs
The genome that encodes lncRNAs contains sequences that feature promoter, initiation codons, termination sites, and splice sites.13,14 A recent computational biology study concluded that human cells contain at least 91,013 expressed transcripts, 68% of which (58,648) were classified as lncRNAs.15 The putative lncRNA promoters are enriched with histone 3 lysine 4 trimethylation (H3K4me3), RNA polymerase II (Pol II), and DNase I hypersensitivity sites,15 suggesting that lncRNA expression is actively regulated. This is consistent with the tissue- or developmental stage-specific expression of many lncRNAs. Like the mRNAs (mRNAs), lncRNAs are produced by independent transcription unit predominantly through the polymerase II complex, and most lncRNAs are polyadenylated.16 The lncRNA genomic loci harbor histone modifications, and the RNA transcripts exhibit alternative splicing that uses splicing signals (GT/AG) with exon and intron lengths comparable to those of mRNAs.16 However, in terms of exons number, a large fraction (42%) of lncRNAs consists of 2 exons in contrast to that (6%) of protein-coding genes.16 Although lncRNA transcripts and promoters are more conserved evolutionarily relative to random control regions,15 the degree of conservation of lncRNAs is less than that of mRNAs, and lncRNAs usually do not contain open reading frames that have cross-species mutations.17,18 This generally low conservation of lncRNA primary sequence, however, doesn't preclude potential conservation of secondary or tertiary structure or the presence of conserved functional domains.19 Indeed, approximately 1% of lncRNA genomic loci harbor ultra-conserved elements, defined as DNA longer than 200 nucleotides that have nearly identical sequences across multiple species,15 suggesting the presence of lncRNA functional domains. Despite the modest overall conservation, 7% of the lncRNAs are found to overlap with disease-associated SNPs, indicating an association of these lncRNAs with diseases.15
LncRNAs are found in all subcellular fractions, with a subset of lncRNAs enriched in the nuclei, while the majority of lncRNAs located in the cytosol or in the ribosomal fractions.20 Given that lncRNAs can functionally interact with many proteins, the association of lncRNAs with ribosomes could not be simply perceived as representing their protein-coding tendency. However, many lncRNAs do contain small open reading frames, and an emerging trend in the field is to explore the peptide- or micropeptide-coding potential of the RNA transcripts that were annotated as lncRNAs.21,22 This aspect of lncRNAs is largely obscure, given the difficulty of identifying open reading frames that encode small peptides. Recent genome-wide and peptideomic studies revealed hundreds of small peptides that are encoded by annotated lncRNAs in vertebrates.21,22 Functions of some small peptides encoded by “lncRNAs” are known. For example, a skeletal muscle-specific lncRNA encodes a 46-amino acid micropeptide Myoregulin (MLN), which is critical for inhibiting the membrane pump SERCA to regulate calcium uptake into the sarcoplasmic reticulum for muscle relaxation.23 Another example is ELABELA, a 32-amino acid peptide hormone encoded by a previously annotated lncRNA.24 During early cardiovascular development ELABELA transmits developmental signals from ectoderm to endoderm, and then to mesoderm through the putative Apelin receptor APLNR and Gata5- and Sox7-mediated pathways.24 Therefore, for annotated lncRNAs that contain one or more open reading frames, efforts should be made to distinguish their functions resulting from the RNA molecules or from the translated small peptides.
LncRNAs as partners of chromatin-modifying enzymes
As previously reviewed,10-12 lncRNAs can regulate gene expression through their interactions with chromatin-modifying enzymes, which catalyze covalent changes of histones or DNA on the chromatin to affect the expression of genetic information. LncRNAs are known to associate with many histone- or DNA-modifying enzymes, including Polycomb Repressive Complex (PRC),25 MLL/TrxG complex,26 histone demethylase LSD1,27 DNA methyltransferase DNMT1,28 and DNA demethylation regulator GADD45a.29 Although the detailed biochemical mechanisms remain to be determined, lncRNAs may act through 2 different mechanisms to covalently modulate chromatin. First, lncRNAs can directly bind to chromatin-modifying enzymes, thus serving as a guide to anchor chromatin modifiers to targeted genomic regions or functioning as a decoy to sequester chromatin modifiers from specific genomic sites. A recent study showed that the lncRNA TARID (TCF21 antisense RNA inducing demethylation) acts by coupling with GADD45A to direct the DNA demethylation machinery to specific gene loci in cancer cells to regulate gene expression.29 Second, lncRNAs can be incorporated into the chromatin-modifying complex and function as part of the complex or as a scaffold to assemble the complex for chromatin modification. In the fruit fly, for example, the lncRNA roX2, after being structurally remodeled by an RNA helicase MLE1,30,31 becomes incorporated into the male-specific lethal (MSL) protein complex, which then recruits a histone acetyltransferase (MOF) to induce histone acetylation and activate gene transcription in the male X chromosome.32,33 The lncRNA Xist provides another interesting example. Xist is bound by ATRX protein to promote PRC2 loading onto Xist and subsequent spreading of PRC2 along the X chromosome.34 PRC2 then causes widespread H3K27 trimethylation and gene inactivation in the X chromosome.
LncRNAs as partners of ATP-dependent chromatin-remodeling factors
Besides covalent modifications of DNA and histones, active remodeling of the nucleosome composition and position provides another route for regulating chromatin and gene expression. However, it remains largely unaddressed whether lncRNAs are involved in the nucleosome remodeling process. Our recent study shed some light on how lncRNA may regulate nucleosome remodeling and gene expression through the ATP-dependent chromatin-remodeling complex.9
We identified a cluster of alternatively spliced, cardiac-specific, and nuclei-enriched lncRNAs that are associated with myosin heavy chain gene Myh7 in mice. The myosin heavy chain associated RNA transcript was named Myheart (Mhrt).9 Mhrt can protect the heart from stress-induced cardiac hypertrophy and failure through its direct inhibition of Brg1 chromatin function. Brg1 is an ATPase catalytic subunit of the SWI/SNF-like BAF chromatin-remodeling complex, and it is essential for the development of pathological cardiac hypertrophy35 (Figs. 1A and 2). Brg1 contains a SF2 RNA helicase domain, which can bind to chromatinized DNA and tether Brg1 to its genomic targets for nucleosome remodeling and gene regulation. The Brg1 helicase domain can also bind to Mhrt RNA with high affinity, thus enabling a competitive inhibition mechanism by which Mhrt sequesters Brg1 from the genomic DNA loci and inhibits Brg1's gene regulation. Mhrt provides the first example of lncRNA capable of inhibiting the function of an ATP-dependent chromatin-remodeling factor in vivo. This Mhrt–Brg1 interaction thus showcases a direct link between lncRNA and nucleosome remodeling. Besides restructuring nucleosomes, Brg1 is a central player in recruiting other epigenetic factors to modify chromatin at specific genomic loci. Brg1 recruits at least 4 other classes of chromatin-modifying enzymes—Parp, Hdac, G9a/Glp, and Dnmt3—to converge on the promoter of Myh6, a cardiac-specific molecular motor gene. Once recruited to the target Myh6 promoter, these chromatin-modifying enzymes act in concert to generate repressive chromatin that mediates stress-induced Myh6 gene silencing and contributes to the development of cardiomyopathy (35 and Chang lab unpublished Data). Mhrt, by inhibiting Brg1 from targeting Myh6 promoter,9 is likely capable of inhibiting the Brg1-centered covalent modifications of histones and DNA, which include poly-ADP-ribosylation (catalyzed by Parp), histone deacetylation (by Hdac), H3K9 methylation (by G9a/Glp), and DNA methylation (by Dnmt3). Further testing of such additional epigenetic actions of Mhrt will provide new insights into how lncRNA can lead to a cascade of epigenetic changes through a key chromatin remodeler.
Figure 1.
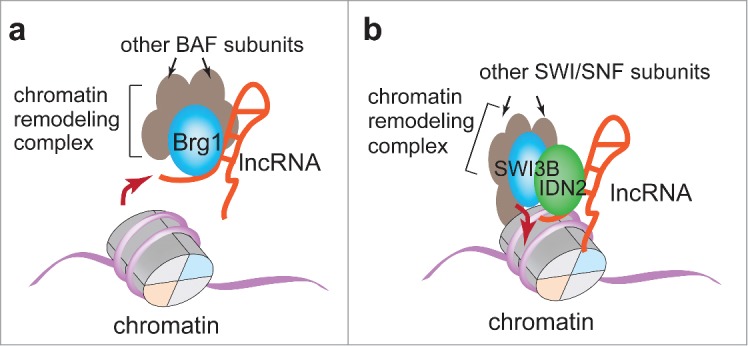
Two models of how lncRNAs regulate chromatin remodeling. (A), The lncRNA Mhrt can sequester Brg1/BAF chromatin-remodeling complex from their genomic targets to inhibit nucleosome remodeling.9 The action is mediated by direct interactions between Mhrt and Brg1. (B), Pol V-transcribed lncRNA tethers SWI3B-containing SWI/SNF chromatin-remodeling complex to genomic loci to repress gene expression in Arabidopsis.36 Such action is accomplished through a RNA-binding protein IDN2, which binds both lncRNA and SWI3B.
Figure 2.
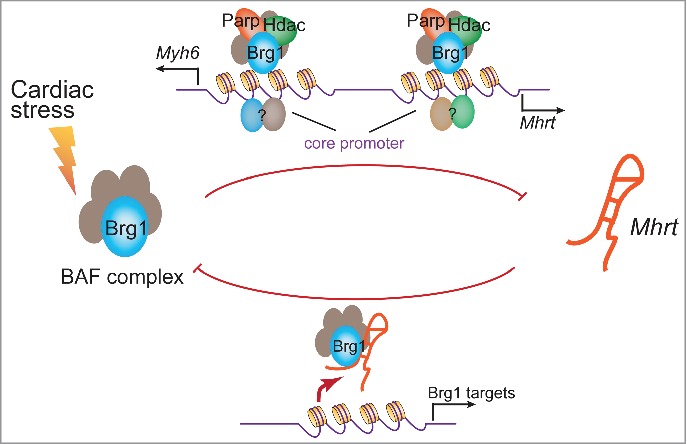
Reciprocal regulation of Brg1 and Mhrt. Cardiac stress activates Brg1 to complex with HDAC and PARP to repress Myh6 and Mhrt transcription through 2 separate promoter elements (purple).9,12,35 Mhrt, on the other hand, can inhibit Brg1's chromatin targeting.9,12
Studies of Arabidopsis suggested a model in which lncRNAs recruit Swi/Snf ATP-dependent chromatin-remodeling complex to genomic sites to position nucleosomes and silence gene expression36 (Fig. 1B). In Arabidopsis thaliana, lncRNAs that are transcribed by RNA polymerase V (Pol V) from genomic sites containing retrotransposons and repetitive DNA elements are usually associated with gene silencing.37 Transcription of these lncRNAs facilitates local heterochromatin formation to repress gene expression.37 The Pol V-transcribed lncRNAs can bind to IDN2 (an RNA-binding protein essential for transcription silencing),38,39 which then binds to the SWI3B subunit of Swi/Snf complex to recruit the remodeling complex to the sites of lncRNA transcription to establish repressive chromatin environment.36 Therefore, lncRNAs can indirectly recruit chromatin-remodeling complex to local genomic sites to reposition nucleosomes for gene repression.
LncRNA transcription can be controlled by ATP-dependent chromatin-remodeling factors
The expression of lncRNAs is under tight regulatory control, with many lncRNAs showing tissue- or developmental stage-specific expression.15,40,41,42 Such specific expression of lncRNAs raises a critical question regarding how lncRNA transcription is regulated in a precise temporal and spatial manner. The regulation of Mhrt expression may shed some light on the control mechanism.9 In mice Mhrt is specifically expressed in the heart: it is present at low level in fetal cardiomyocytes, and its transcription increases as the heart matures from the fetal stage to the neonatal period and to adulthood. This expression pattern coincides with that of Myh6, a cardiac-specific molecular motor gene essential for heart function. Mhrt and Myh6 are down-regulated when the hearts of adult mice or humans are pathologically stressed, and such down-regulation of Mhrt and Myh6 is crucial for the development of hypertrophy and heart failure.9,35 Mhrt and Myh6 share the same promoter, whose activity is cardiac-specific and bidirectional—transcribing Myh6 in one direction and Mhrt in the other (Fig. 2).9,35 This bidirectional promoter harbors 2 distinct promoter elements to separately control the transcription of Myh6 and Mhrt,9,35 and both promoter elements are repressed by the chromatin-remodeler Brg19,35 (Fig. 2). Brg1 has an expression pattern opposite to that of Mhrt and Myh6. Brg1 expression is activated in fetal hearts, silenced in adult hearts, but reactivated by cardiac stress in adult hearts.35 Once reactivated in adult hearts, Brg1 forms a chromatin repressor complex with Hdac and Parp on the bidirectional promoter to simultaneously inhibit Myh6 and Mhrt, resulting in cardiomyopathy.9,35 In the heart, Mhrt transcription is repressed by Brg1, whereas Mhrt RNA itself can inhibit Brg1's chromatin targeting.9 This reciprocal inhibition indicates the presence of a Brg1–Mhrt feedback circuit critical for controlling cardiac physiology and pathology.9,35 Moreover, the Brg1–Mhrt interaction illustrates that the ATP-dependent chromatin-remodeling factor can control the expression of lncRNA under different pathophysiological conditions.
A recent yeast genetic screening identified a large number of putative lncRNA repressors, which include 8 genes encoding subunits of 4 different ATP-dependent chromatin-remodeling complexes Rsc (RSC1, RSC2, and HTL1), Isw2 (ITC1), Ino80 (IES2), and Swr1 (SWR1, ARP6, and YAF9).43 These chromatin-remodeling factors repress over 250 antisense lncRNAs (chromatin remodeling-repressed antisense transcripts or CRRATs) to maintain normal expression of mRNA transcripts that overlap with those antisense lncRNAs.43 Therefore, the highly conserved ATP-dependent chromatin-remodeling factors may serve as global lncRNA repressors.43 The repression of antisense lncRNAs may be a common mechanism of mRNA regulation. This view is consistent with the observation that Brg1 represses the expression of Mhrt, which is antisense to Myh7, to promote the expression of Myh7 mRNA in pathologically stressed hearts.9,35 Future investigations are needed to pinpoint how chromatin-remodeling complex coordinates with other epigenetic regulators and transcriptional machinery to modulate transcription of lncRNAs, which may form a foundation to develop novel therapeutics by manipulating the expression of disease-associated lncRNAs.
Future perspective: the translational value of lncRNA–chromatin biology
LncRNAs have emerged as an important class of molecules implicated in human diseases, including cancers, cardiovascular diseases, neural degenerative disease, as well as metabolic disorders.2,4-7,12 The profound lncRNA–chromatin interface provides new opportunities to unlock the potential of this mechanism for diagnostic or therapeutic applications. First, lncRNAs are more tissue-specific in contrast to the chromatin-regulating or epigenetic machinery that tends to operate widely in many tissues. Targeting lncRNAs may therefore result in better tissue specificity with lower general toxicity. Current technology holds great promise for therapeutic manipulation of lncRNAs through siRNA interfering, antisense oligonucleotides, aptamers, ribozymes, or CRISPR-mediated RNA cleavage.44-46 In addition, direct interactions between lncRNAs and chromatin-regulating factors can be used as an avenue for developing assays to screen for small molecules and large molecule biologics that augment or dampen the lncRNA–chromatin interactions. Finally, more and more lncRNAs have been discovered as disease-causal or as the hub of new pathways that contribute to disease development.2,4-7,12 Such increasingly fast discoveries of disease-associated lncRNAs are offering us unprecedented opportunities to develop novel therapeutics through the lncRNA mechanisms. The lessons we have learned from targeting microRNAs and delivering RNA-related reagents over the last decade can be modified and transferred to the lncRNA field to realize the therapeutic potential of lncRNAs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
C.-P.C. is the Charles Fisch Scholar of Cardiology and was supported by National Institutes of Health (NIH; HL118087, HL121197, HL085345), the American Heart Association (AHA; National Scientist Development Award; Established Investigator Award 12EIA8960018), Lucile Packard Heart Center Research Program, Oak Foundation, Baxter Foundation, Children's Heart Foundation, March of Dimes Foundation (#6-FY11-260), Indiana University (IU) School of Medicine—IU Health Strategic Research Initiative, and the IU Physician-Scientist Initiative, endowed by Lilly Endowment.
References
- 1.Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, et al.. Considerations when investigating lncRNA function in vivo. eLife 2014; 3:e03058; PMID:25124674; http://dx.doi.org/ 10.7554/eLife.03058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861-74; PMID:22094949; http://dx.doi.org/ 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 3.Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trend Cell Biol 2014; 24:594-602; PMID:25022466; http://dx.doi.org/ 10.1016/j.tcb.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maass PG, Luft FC, Bahring S. Long non-coding RNA in health and disease. J Mol Med (Berl) 2014; 92:337-46; PMID:24531795; http://dx.doi.org/ 10.1007/s00109-014-1131-8 [DOI] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell 2009; 136:629-41; PMID:19239885; http://dx.doi.org/ 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 6.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trend Cell Biol 2011; 21:354-61; PMID:21550244; http://dx.doi.org/ 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10:155-9; PMID:19188922; http://dx.doi.org/ 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 8.Quinn JJ, Ilik IA, Qu K, Georgiev P, Chu C, Akhtar A, Chang HY. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol 2014; 32:933-40; PMID:24997788; http://dx.doi.org/ 10.1038/nbt.2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al.. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014; 514:102-6; PMID:25119045; http://dx.doi.org/ 10.1038/nature13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinn JL. lncRNAs: linking RNA to chromatin. Cold Spring Harb Perspect Biol 2014; 6:pii: a018614; PMID:25085913; http://dx.doi.org/ 10.1101/cshperspect.a018614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81:145-66; PMID:22663078; http://dx.doi.org/ 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang CP, Dorn GW 2nd, Thum T, Heymans S; Cardiolinc network . Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol 2015; PMID:25855606 [DOI] [PubMed] [Google Scholar]
- 13.Brent MR, Guigo R. Recent advances in gene structure prediction. Curr Opin Struct Biol 2004; 14:264-72; PMID:15193305; http://dx.doi.org/ 10.1016/j.sbi.2004.05.007 [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Li S, Zhang Y, Zheng H, Xu Z, Ye J, Yu J, Wong GK. Vertebrate gene predictions and the problem of large genes. Nat Rev Genet 2003; 4:741-9; PMID:12951575; http://dx.doi.org/ 10.1038/nrg1160 [DOI] [PubMed] [Google Scholar]
- 15.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al.. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015; 47(3):199-208; PMID:25599403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al.. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Gen Res 2012; 22:1775-89; PMID:22955988; http://dx.doi.org/ 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al.. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458:223-7; PMID:19182780; http://dx.doi.org/ 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al.. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 2010; 28:503-10; PMID:20436462; http://dx.doi.org/ 10.1038/nbt.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta 2014; 1840:1063-71; PMID:24184936; http://dx.doi.org/ 10.1016/j.bbagen.2013.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E, Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Gen Biol 2014; 15:R6; PMID:24393600; http://dx.doi.org/ 10.1186/gb-2014-15-1-r6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet 2014; 15:193-204; PMID:24514441; http://dx.doi.org/ 10.1038/nrg3520 [DOI] [PubMed] [Google Scholar]
- 22.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al.. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 2014; 33:981-93; PMID:24705786; http://dx.doi.org/ 10.1002/embj.201488411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, et al.. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015; 160(4):595-606; PMID:25640239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chng SC, Ho L, Tian J, Reversade B. ELABELA: a hormone essential for heart development signals via the apelin receptor. Dev Cell 2013; 27:672-80; PMID:24316148; http://dx.doi.org/ 10.1016/j.devcel.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 25.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA 2013; 19:429-42; PMID:23431328; http://dx.doi.org/ 10.1261/rna.037598.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al.. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011; 472:120-4; PMID:21423168; http://dx.doi.org/ 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010; 329:689-93; PMID:20616235; http://dx.doi.org/ 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, et al.. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 2013; 503:371-6; PMID:24107992; http://dx.doi.org/ 10.1038/nature12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et al.. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 2014; 55:604-14; PMID:25087872; http://dx.doi.org/ 10.1016/j.molcel.2014.06.031 [DOI] [PubMed] [Google Scholar]
- 30.Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, et al.. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell 2013; 51:156-73; PMID:23870142; http://dx.doi.org/ 10.1016/j.molcel.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maenner S, Muller M, Frohlich J, Langer D, Becker PB. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol Cell 2013; 51:174-84; PMID:23870143; http://dx.doi.org/ 10.1016/j.molcel.2013.06.011 [DOI] [PubMed] [Google Scholar]
- 32.Sass GL, Pannuti A, Lucchesi JC. Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc Natl Acad Sci U S A 2003; 100:8287-91; PMID:12829796; http://dx.doi.org/ 10.1073/pnas.1332749100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell 2000; 5:367-75; PMID:10882077; http://dx.doi.org/ 10.1016/S1097-2765(00)80431-1 [DOI] [PubMed] [Google Scholar]
- 34.Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell 2014; 159:869-83; PMID:25417162; http://dx.doi.org/ 10.1016/j.cell.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010; 466:62-7; PMID:20596014; http://dx.doi.org/ 10.1038/nature09130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Y, Rowley MJ, Bohmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell 2013; 49:298-309; PMID:23246435; http://dx.doi.org/ 10.1016/j.molcel.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008; 135:635-48; PMID:19013275; http://dx.doi.org/ 10.1016/j.cell.2008.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol 2009; 16:1325-7; PMID:19915591; http://dx.doi.org/ 10.1038/nsmb.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang CJ, Ning YQ, Zhang SW, Chen Q, Shao CR, Guo YW, Zhou JX, Li L, Chen S, He XJ. IDN2 and its paralogs form a complex required for RNA-directed DNA methylation. PLoS Genet 2012; 8:e1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Gen Dev 2011; 25:1915-27; PMID:21890647; http://dx.doi.org/ 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Li L, Xu H, Liu Y, Yang C, Cowley AW Jr., Wang N, Liu P, Liang M. Characteristics of long non-coding RNAs in the Brown Norway rat and alterations in the Dahl salt-sensitive rat. Scientific Rep 2014; 4:7146; PMID:25413633; http://dx.doi.org/ 10.1038/srep07146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014; 15:7-21; PMID:24296535; http://dx.doi.org/ 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 43.Alcid EA, Tsukiyama T. ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev 2014; 28:2348-60; PMID:25367034; http://dx.doi.org/ 10.1101/gad.250902.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol 2013; 45:1895-910; PMID:23748105; http://dx.doi.org/ 10.1016/j.biocel.2013.05.030 [DOI] [PubMed] [Google Scholar]
- 45.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol 2012; 19:60-71; PMID:22284355; http://dx.doi.org/ 10.1016/j.chembiol.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014; 516:263-6; PMID:25274302; http://dx.doi.org/ 10.1038/nature13769 [DOI] [PMC free article] [PubMed] [Google Scholar]


