Abstract
Trypanosomatids are protozoan parasites and the causative agent of infamous infectious diseases. These organisms regulate their gene expression mainly at the post-transcriptional level and possess characteristic RNA processing mechanisms. In this study, we analyzed the complete repertoire of Leishmania major small nucleolar (snoRNA) RNAs by performing RNA-seq analysis on RNAs that were affinity-purified using the C/D snoRNA core protein, SNU13, and the H/ACA core protein, NHP2. This study revealed a large collection of C/D and H/ACA snoRNAs, organized in gene clusters generally containing both snoRNA types. Abundant snoRNAs were identified and predicted to guide trypanosome-specific rRNA cleavages. The repertoire of snoRNAs was compared to that of the closely related Trypanosoma brucei, and 80% of both C/D and H/ACA molecules were found to have functional homologues. The comparative analyses elucidated how snoRNAs evolved to generate molecules with analogous functions in both species. Interestingly, H/ACA RNAs have great flexibility in their ability to guide modifications, and several of the RNA species can guide more than one modification, compensating for the presence of single hairpin H/ACA snoRNA in these organisms. Placing the predicted modifications on the rRNA secondary structure revealed hypermodification regions mostly in domains which are modified in other eukaryotes, in addition to trypanosome-specific modifications.
Keywords: snoRNA, Leishmania, rRNA processing, H/ACA, C/D, methylation, pseudouridylation
Introduction
RNA modification has drawn the attention of many scientists, because modifications are not only enriched on stable RNAs such as rRNA, tRNA and snRNAs but also in coding and other non-coding RNAs. The transcriptome-wide mapping of such modifications suggests that folding, stability and activity are modulated and regulated by RNA modifications.1,2
The two major rRNA modifications, 2′-O-methylation (Nm) and pseudouridine (Ψ) formation, are prevalent on rRNA and small nuclear RNAs (snRNAs) in eukaryotes and are guided by small nucleolar RNAs (snoRNAs). The 2′-O-methylations are guided by C/D box snoRNAs. These boxes, together with the short sequences near the 5′ and 3′ ends of the RNA, are essential for processing, localization, and stabilization of these molecules. Most of the guide RNAs carry internal boxes related to the C and D boxes, known as C′ and D′ boxes. The recognition of the target is based on complementarity of 10 to 21 nucleotides (nt) between these 2 molecules, located upstream of the D and D′ sequences (reviewed in3,4). The methylation site is situated 5 nt upstream from the D and D′ boxes, within the domain of interaction between the snoRNA and the substrate.5
In most eukaryotes, the snoRNAs that guide pseudouridylation consist of 2 hairpin domains connected by a single-stranded hinge, the H domain, and by a tail, the ACA box. A short RNA recognition motif on the snoRNA base-pairs with the target and directs the conversion of uridine to pseudouridine. The pseudouridine is usually located 14 to 16 nt upstream from the H box or the ACA box of the snoRNA.6 Most of the well-studied C/D and H/ACA box RNAs characterized to date at both structural and functional levels are from humans, or from the yeast Saccharomyces cerevisiae.7 However, studies in last decade also characterized snoRNAs in other organisms, such as the amoeba diplomonad Dictyostelium discoideum,8 the parasitic protozoan Giardia lamblia9 and Entameoba histolytica,10 and the malaria parasite Plasmodium falciparum.11 snoRNAs were also studied in model organisms such as Drosophila12 and the nematode Caenorhabditus elegans.13
Trypanosomatids are parasitic protozoa that are the causative agents of several infamous diseases, such as African trypanosomiasis, caused by Trypanosoma brucei, Chagas' disease, caused by Trypanosoma cruzi; and Leishmaniasis, caused by Leishmania species. Leishmania spp. are obligatory intracellular parasites that cause a spectrum of human diseases, with an annual incidence of 2 million cases in 88 countries. The parasite cycles between 2 hosts, namely, the insect host, where Leishmania parasites grow as flagellated extracellular promastigotes; and the mammalian host, where they proliferate as aflagellated intracellular amastigotes.14
rRNA processing events in Trypanosomatids are unique. The large subunit rRNA undergoes trypanosome-specific cleavages during rRNA maturation, yielding 2 large rRNA molecules and 4 small RNAs, ranging in size from 76 to 226 nt.15
Several specific features were found in snoRNAs of trypanosomatids. Most, if not all H/ACA RNAs are composed of a single hairpin RNA and carry an AGA box instead of an ACA box.16,17 The first discovered trypanosome H/ACA-like RNA, the spliced leader-associated RNA 1 (SLA1), guides modification of a unique short-lived RNA,18 the spliced leader RNA (SL RNA). This RNA is the donor of the spliced leader sequence to all trypanosome mRNAs.19 Silencing of the pseudouridine synthase (CBF5) by RNA interference in T. brucei provided evidence for the role of SLA1 in trans-splicing.20 We proposed that SLA1 has a chaperone function and escorts the SL RNA early in its biogenesis until it is assembled with Sm proteins.21
Using bioinformatics and experimental tools, we recently performed a genome-scale analysis of snoRNAs that guide methylations and pseudouridylations on rRNAs in both T. brucei and L. major. Our data suggested that most snoRNAs are clustered in reiterated repeats that carry a mixed population of C/D and H/ACA-like RNAs. Predicting the modifications guided by these RNAs and using partial mapping data, allowed us to identify 57 C/D snoRNAs that potentially guide 84 Nm modifications, and 34 H/ACA like RNAs that target rRNA, suggesting a high occurrence of Nms compared to pseudouridines on T. brucei rRNA.16 Based on T. brucei snoRNAs, we identified 23 gene clusters in L. major that encode 62 C/D snoRNAs that potentially guide 79 methylations, and 37 H/ACAs that can guide 30 pseudouridylation reactions. In general, the pattern of Nm modifications is highly conserved between L. major and T. brucei.17
Using RNA-seq of small RNPs we expanded the repertoire of T. brucei snoRNAs and identified 79 C/D and 63 H/ACA-like snoRNA, suggesting that these organisms also harbor a large number of pseudouridines.22 Many H/ACA were shown to exist in clusters containing only H/ACA RNAs, and these escaped our previous screens, which identified H/ACA based on their presence in clusters with C/D snoRNAs. Abundant snoRNAs, mostly of the C/D type, were shown to function in rRNA processing.22,23
The analysis of modifications guided by T. brucei snoRNAs revealed the existence of additional species specific and increased overall modification levels at domains that are already modification-rich in other eukaryotes.16 About 40% of the trypanosome-specific modifications are situated in unique positions outside the highly conserved domains of the rRNA.16,17
In this study, the L. major repertoire of snoRNAs was determined by RNA-seq analysis of RNA affinity selected with the C/D and H/ACA specific proteins SNU13 and NHP2, respectively. The study identified 81 H/ACA and 80 C/D; among these are newly identified 13 C/D and 44 H/ACA snoRNAs. The snoRNAs vary in their abundance as can be observed by the RNA-seq reads and Northern analyses. Among the abundant snoRNAs, we identified 13 snoRNAs predicted to function in trypanosome-specific rRNA processing. The putative role of 2 such snoRNAs in rRNA processing was studied by in vivo psoralen cross-linking and fractionation on RNP complexes. The predicted rRNA modifications guided by the identified snoRNAs were placed on the secondary structure of rRNA. Our data suggest the presence of hyper-modifications in domains that are also modification-rich in other eukaryotes. The repertoire of L. major snoRNAs is highly related to that of T. brucei. However, species-specific snoRNAs and modifications were also identified. The relatedness of H/ACA RNAs in T. brucei and L. major was studied, suggesting the mechanism by which snoRNAs may have been generated during evolution. Flexibility in the generation of a pseudouridylation pocket was detected, which potentially enables a single hairpin H/ACA RNA to guide more than one target, thus compensating for the presence of single-hairpin RNAs in trypanosomes compared to double-hairpin RNAs in other eukaryotes.
Materials and Methods
Oligonucleotides
The list of oligonucleotides used in this study is given in Table S-1.
RNA preparation and primer extension analysis of RNAs
RNA was prepared using TRI Reagent (Sigma). Primer extension analysis was performed as described previously,24 using 5′-end-labeled oligonucleotides specific to target RNAs, as indicated in the figure legends. The extension products were analyzed on 6% polyacrylamide-7 M urea gels.
RT-PCR
RNA was treated with the “DNase-free” reagent (Ambion) according to the manufacturer's protocol for 30 minutes to remove DNA contamination. Reverse transcription was performed by random priming (Reverse transcription system, Promega). The samples were heated for 5 min at 70°C, followed by chilling on ice for 5 minutes. Next, 1 unit of AMV-reverse transcriptase (Promega) was added, together with 1 unit RNase inhibitor (Promega) and the elongation reaction was performed according to the manufacturer's instructions at 25°C for 10 min, and then at 50°C for 60 min (Promega kit). The resulting cDNA was used for PCR amplification using primers as specified in Table S-1.
Purification of the SNU13 and NHP2 RNPs
Tandem affinity purification was performed from whole cell extracts. The cell pellet from L. major (2×1011 cells) was washed twice with PBS and once with buffer I (20 mM Tris-HCl (pH 7.7), 150 mM KCl, and 3 mM MgCl2). The cells were resuspended in 15 ml of buffer II (buffer I with 1 mM DTT and 10 μg/ml leupeptin), equilibrated in a nitrogen cavitation bomb (Parr Instruments Co.) at 750 p.s.i. N2 for 1h at 4°C, and disrupted by release from the bomb. After release of the pressure, protease inhibitor mixture (Roche Applied Science) was added, and the extract was treated with 0.5% Triton X-100. The extract was incubated at 4°C for 15 min and cleared by centrifugation (15,000 ×g), and the supernatant was incubated while rotating for 2 h with rabbit IgG-agarose beads (200 μl) (Sigma). The beads were washed 5 times with TEV buffer (buffer I with 0.5 mM DTT, 0.5 mM EDTA) and incubated overnight in 1.5 ml of TEV buffer with 200 units of tobacco etch virus protease (Promega). After centrifugation, the supernatant was incubated with 50 μl of Strep-T actin-Sepharose beads (IBA) for 1h. The beads were washed with buffer III (TEV buffer with 2 mM CHAPS (GE Healthcare)), and the complexes were eluted with elution buffer (100 mM Tris-Cl (pH 8), 150 mM NaCl, 1 mM EDTA) containing 2.5 mM d-desthiobiotin (Sigma). After removing the proteins by phenol-extraction, the RNA (1–2 μg) was fragmented using the Ambion RNA fragmentation kit (AM8740). The RNA was dephosphorylated at the 3′ end using T4 Polynucleotide kinase (PNK) (in the absence of ATP). The 5′ end of the RNA was repaired using PNK in the presence of tracer radioactive [γ32P]ATP. The material was separated on 15% polyacrylamide denaturing gel and the radioactive bands at a size of ∼ (25–40) nt were excised from the gel. RNA was eluted and the 3′ adaptor was ligated (3′-RAppCTGTAGGCACCATCAAT/3′DDG) with T4 RNA ligase 2, (New England Biolabs). The reaction was loaded on a 15% polyacrylamide denaturing gel, the radioactive higher molecular weight bands(40–60 nt) were excised, and the 5′ RNA adaptor (5′-ACACGACGCUCUUCCGAUCU-3′) was ligated using T4 RNA ligase. RNA was extracted and cDNA was synthesized in the presence of radiolabelled dCTP as tracer. The cDNA was purified from a 15% polyacrylamide gel and subjected to PCR using “Platinum” DNA polymerase (Invitrogen). The PCR was sequenced by Illumina sequencing, as previously described.22
“RNA walk”
Cross-linking was performed essentially as described in.25 Briefly, L. major cells were harvested and resuspended at 5×107 cells/ml and washed twice with PBS. Cells (∼109) were concentrated and incubated on ice. 4-Aminomethyl-trioxsalen hydrochloride (AMT) was added to the cells at a concentration of 0.2 mg/ml. Cells treated with AMT were kept on ice and irradiated using a UV lamp at 365 nm at a light intensity of 10 μW/cm2 for 30 minutes. Next, the cells were washed once with PBS and deproteinized by digestion with proteinase K (200 μg/ml in 1% SDS for 60 minutes). RNA was prepared using TRIzol reagent. Approximately 250 μg of RNA was used for affinity selection, essentially as described in.23 After affinity selection, the RNA was subjected to RT-PCR as described26
Mapping RNA-seq reads to the genome
The 36 nts sequence reads obtained from the Illumina Genome Analyzer were first trimmed of Illumina adapters using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), and reads of 15 bases or less were discarded from subsequent analysis. The remaining reads were mapped to the L. major draft genome (TriTrypDB-2.5; http://tritrypdb.org/common/downloads/release-2.5/Lmajor/) using SMALT v0.7.5 (http://www.sanger.ac.uk/resources/software/smalt/) with the default parameters, allowing non-unique reads to be mapped randomly to their best match in the genome. Next, the reads were imported and visualized in the IGV viewer.27,28
Sucrose gradient fractionation of RNP complexes
Whole cell extracts were prepared from 2×1010 L. major cells as previously described,29 and fractionated on a 10–30% sucrose gradient. Gradients were centrifuged at 4°C for 3h at 35,000 rpm in a Beckman SW41 rotor. Then, 1 ml fractions were collected using the ISCO gradient fractionation system. The absorbance profile at 245 nm was determined to locate the positions of the 80S monosome and the polysomes.
SnoRNA quantification and annotation
Raw read counts for each snoRNA were obtained using Multicov from the Bedtools suite (v 2.17.0). For each snoRNA that appears multiple times in the genome, the counts for each genomic location were combined. Reads Per Kilobase per Million (RPKM) was utilized as the quantification method to obtain a measure of each gene's expression.30 Reads mapping to unannotated loci were chosen as potential novel snoRNAs. In order to determine if the reads mapping to unannotated loci were derived from known annotated sequences, they were merged, extracted and further analyzed by BLAST31 against the L. major known coding sequences. Those reads that were not similar to any known coding sequence were then run as input to a variety of programs to identify putative snoRNAs. snoScan version 0.9b32 was used to identify C/D snoRNAs. snoGPS33 and PsiScan34 were used to test if the ncRNA candidates were likely to be H/ACA snoRNAs. In the final stage, manual examination of the remaining sequences in each library was performed to look for the classical C/D and H/ACA motifs (TGAUGA/CUGA and AGA) in the SNU13 and NHP2 libraries, respectively.
Prediction of targets on rRNA
For C/D snoRNAs, the potential targets (for 2′-O-methylation) in rRNA and on snRNAs were determined using BLAST31 to search for a complementary match to an rRNA or U snRNA target. For this study, the program was used to search for complementarity to rRNA that complies with the +5 guiding rule. Additionally, the targets were also predicted based on the data available from their T. brucei homologues. For H/ACAs, which have a well-defined structure, sequence folding by MFOLD35 was performed. The resulting sequences from the internal loop were extracted. A PERL script was used to scan the rRNA and UsnRNAs for a compatible target for the potential pocket based on the guiding rules established for yeast, mammals, and plants (http://www.bio.umass.edu/biochem/rna-sequence/Yeast_snoRNA_Database/snoRNA_DataBase.html; http://bioinf.scri.sari.ac.uk/cgi-bin/plant_snorna/conservation.
The annotations of the newly identified snoRNA in this study were submitted to GeneDB.
Identifying L. major H/ACA and C/D homologues in T. brucei
The repertoire of H/ACA snoRNAs in L. major and T. brucei was compared to establish homologues between the 2 species. Homologues were found for each H/ACA as detailed in Doniger et al. 2009.36 The H/ACAs were “split,” and the pseudouridylation pockets and the rest of the H/ACA were aligned independently for each homologous pair using the Needle program from the EMBOSS 6.1.0 package.37 The repertoire of C/D snoRNAs in L. major and T. brucei was compared to establish homologues between the 2 species. BLAST31 was used to find matching guide regions.
Results
Preparation of snoRNA-specific libraries
The recent genome wide-search for small ncRNAs in T. brucei doubled the number of identified H/ACA-like RNAs,22 suggesting that the previous bioinformatic studies performed in both T. brucei and L. major16,17 had missed a large fraction of the H/ACA-like repertoire. Previous studies had identified these RNAs based on their chromosomal location in the vicinity of C/D snoRNAs. However, the study in T. brucei indicated that H/ACA-like RNAs are found not only in clusters with C/D snoRNA, but also in clusters containing only H/ACA RNAs or as solitary genes.22 The T. brucei RNA-seq was performed on the small RNome, and may have missed snoRNAs, especially non-abundant molecules.22 To identify the complete repertoire of these RNAs, we established a system to specifically sequence snoRNAs by affinity selecting these RNA via their association with their cognate RNA binding proteins. To this end, the L. major snoRNP core SNU13 (C/D) and the NHP2 (H/ACA) were cloned into the expression vector, pSAP1, carrying a C-terminal tag composed of protein A binding domain, the tobacco virus protease cleavage site and streptavidin-binding peptide.38 L. major transgenic cell lines were prepared and used to affinity purify the snoRNAs. After affinity-selection, the levels of the selected snoRNAs were compared to different small RNAs by primer extension. The results (Fig. 1A) demonstrated the specific selection of LM26Cs1H4 snoRNA, with no background from SL RNA or the C/D snoRNA LM26C1C1. The same experiment was performed with an SNU13 tagged cell line, and the results indicated efficient selection of LM26C1C1 with no background from the SL RNA and LM26Cs1H4 (Fig. 1A). Next, we scaled up the affinity purification as described in Materials and Methods, and the RNA of the last purification step was de-proteinized and fractionated on a denaturing gel and stained with silver (Fig. 1B). The silver stain of the NHP2-selected RNA indicated the presence of RNA longer than the majority of the selected RNAs, ranging in size from 70 to 100 nt. Most of the SNU13 selected RNAs were in the size range of 60–70 nts. Next, the affinity selected RNA was fragmentized by mild alkaline hydrolysis and libraries were prepared as described in Materials and Methods. cDNA was prepared and amplified. The fragments were sequenced and 21 and 33 million reads from the NHP2 and SNU13 libraries, respectively, were mapped to the L. major genome.
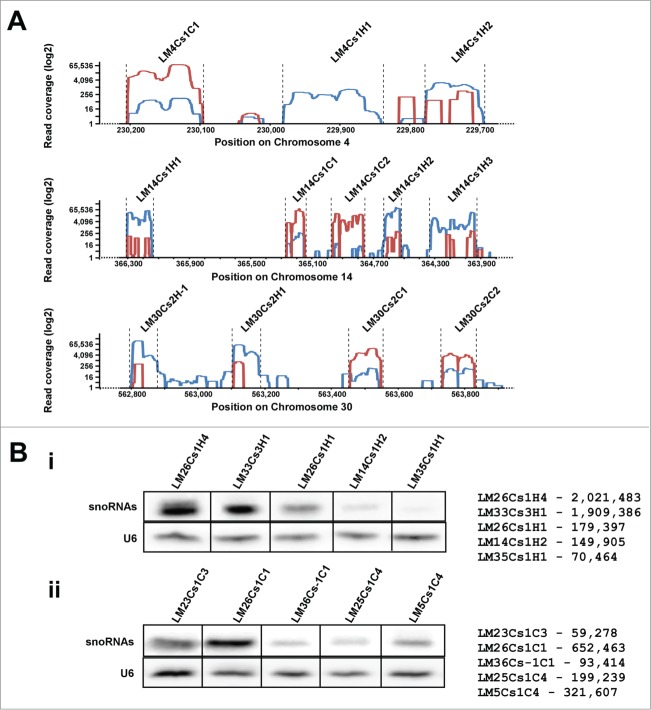
Figure 1.
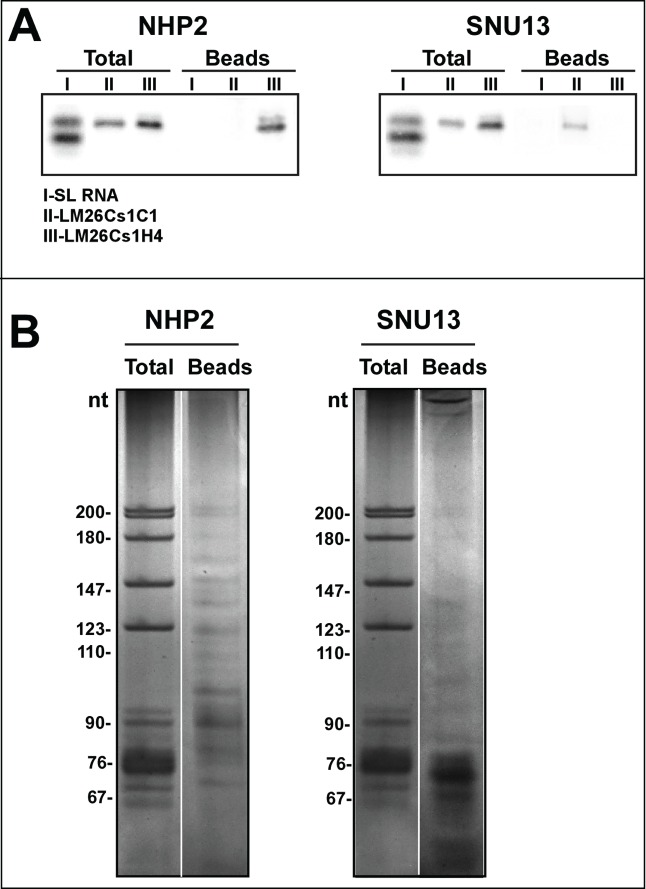
RNA Affinity purified with NHP2 and SNU13 tagged proteins. (A)The specificity of the affinity selection by NHP2 and SUN13 tagged proteins. RNA from the beads (1/10 of the amount loaded was subjected to primer extension with a sequence-specific probe (Table S1), and separated on a 6% polyacrylamide denaturing gel. The identities of the tagged protein are indicated and the tested RNAs are listed below the image. (B) Purification of RNAs associated with NHP2 and SNU13. Purification was performed using 2×1011 cells, as described in Materials and Methods. The purified RNPs were de-proteinized, and the RNA was separated on a 10% polyacrylamide-denaturing gel and stained with silver. The size of the pBR322 DNA-Msp I digest is indicated. Total RNA (1 μg) was used as a marker. Beads, the RNA extracted from the beads after the last step of purification.
Almost 90% of the reads belong to the expected snoRNA class with almost no contamination from the other snoRNA family (i.e. no C/D in the NHP2 library and no H/ACA in the SNU13 library) (Fig. 2A). Additionally, contamination with the most abundant RNAs (rRNA and tRNA) was minimal (Fig. 2B). Based on the number of reads that matched the genome (between 20 to 30 million reads), we believe that we identified the complete repertoire of snoRNAs in the cell.
Figure 2.
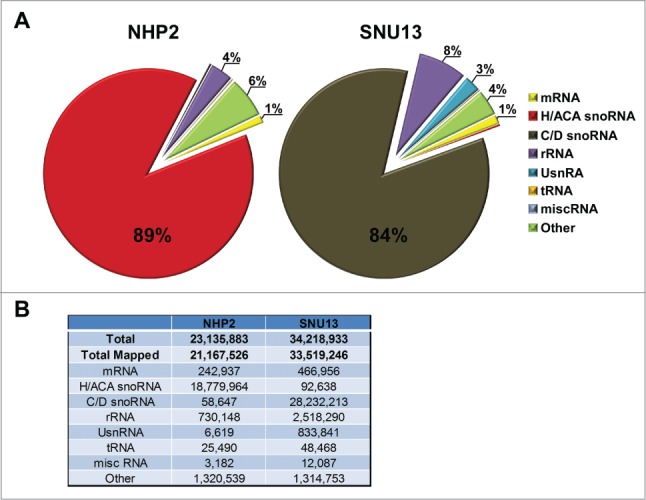
The quantities and identities of snoRNAs selected by NHP2 and SNU13. (A) The percentage of different RNAs molecules among the reads; 91% and 98% of the reads were mapped to the genome from NHP2 and SNU13 libraries. (B) The number of reads for each of the RNAs presented in panel A.
Next, the reads were imported and visualized in the IGV genome browser (Fig. 3A). To observe the distribution of reads within a given cluster, 3 different genomic clusters were plotted, and read distribution along the respective coding regions is presented. The results indicated that although all the snoRNA coding regions are represented, the distribution of reads was not uniform along the coding regions and certain portions of the molecules were severely underrepresented. The results clearly demonstrated that whereas the reads from the H/ACA molecules covered the entire coding region of the molecule, the reads for C/D snoRNAs were fragmented. This abnormality might be the result of the alkaline hydrolysis that was performed prior to the generation of the library. We observed that the fragmentation increased the number of reads per molecule, but the distribution of the reads was distorted. The bias observed for the C/D molecules may result from the fact the C/D snoRNA are composed mainly of single-stranded RNA, and only a very short stem is present at the termini. As a result, these snoRNAs are more sensitive to the fragmentation. In contrast, H/ACA RNA forms stem-loop structures, and thus, most of the RNA is present in dsRNA configuration except the pseudouridylation pocket and the apical loop. H/ACA molecules therefore seem to be less susceptible to degradation. Interestingly, this bias was not observed in our previous study22 when we prepared a small RNP library, most probably because the protocol did not include the alkaline hydrolysis step. However, the fragmentation resulted in a greater number of reads per molecule (our unpublished results).
Figure 3.
(A) Schematic presentation of the reads in distinct snoRNA clusters. The reads from the NHP2 and SNU13 are shown in red and blue, respectively. (B) Primer extension to validate the amount of snoRNA. Total RNA (10 μg) was subjected to primer extension. Numbers of reads corresponding to each of the snoRNA species tested are presented.
A very striking finding was the variation in the number of reads for the different snoRNAs ranging from 2 million counts to thousands of counts per molecule (Table S2). To examine the correlation between the number of reads and the abundance of the RNA, a primer extension experiment was performed. The results indicate a correlation for the H/ACA but not for C/D snoRNAs, reflecting the bias introduced due to severe degradation of the C/D during alkaline hydrolysis, as discussed above (Fig. 3B).
The extended repertoire L. major C/D and H/ACA RNAs
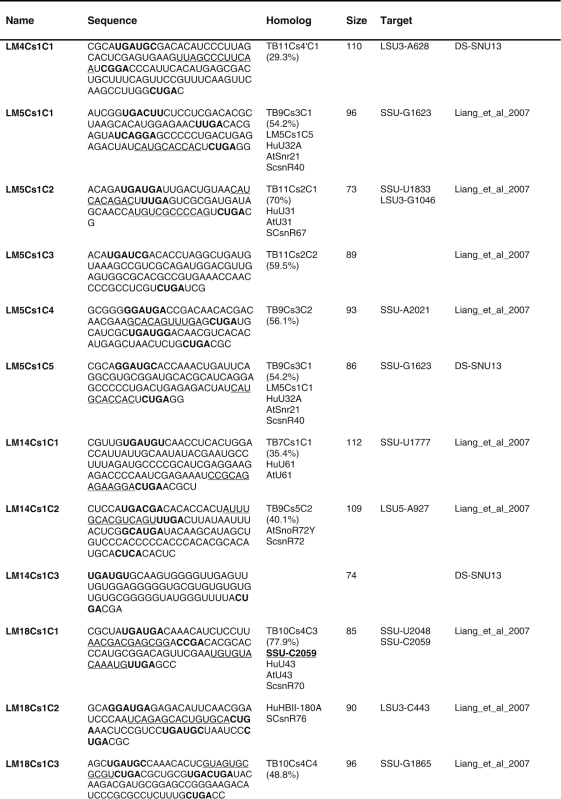
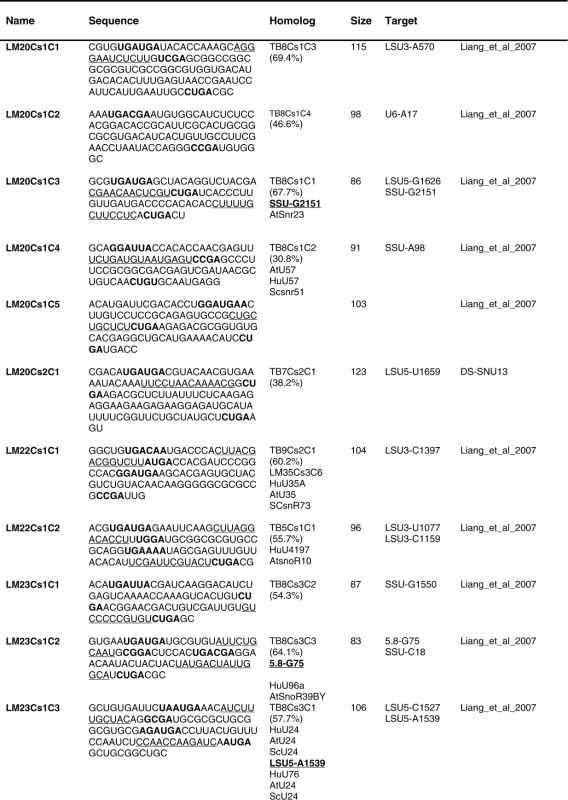
Next, the RNA-seq information was used to describe the repertoire of L. major C/D snoRNAs. 80 C/Ds snoRNAs were identified ranging in size from 70 to 150 nt (Table 1). The 5′ end of the molecule is indicated 3–5 nt upstream of the C box, and the 3′ end is located 2–5 nt downstream of the D box. Several long C/D molecules were revealed, and among these is LM20Cs1C2, which is predicted to guide Nm on U6. This molecule has no target on rRNA and may target other RNA classes (possibly other snRNAs or tRNAs).
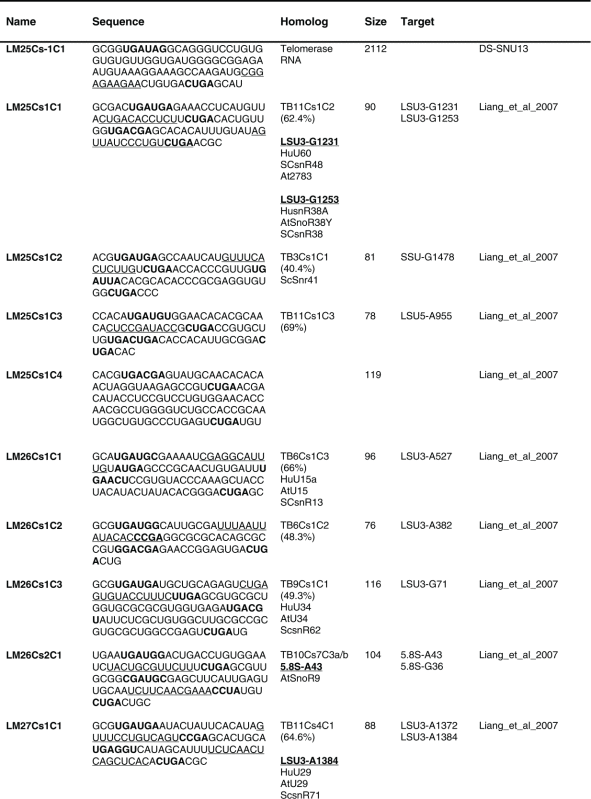
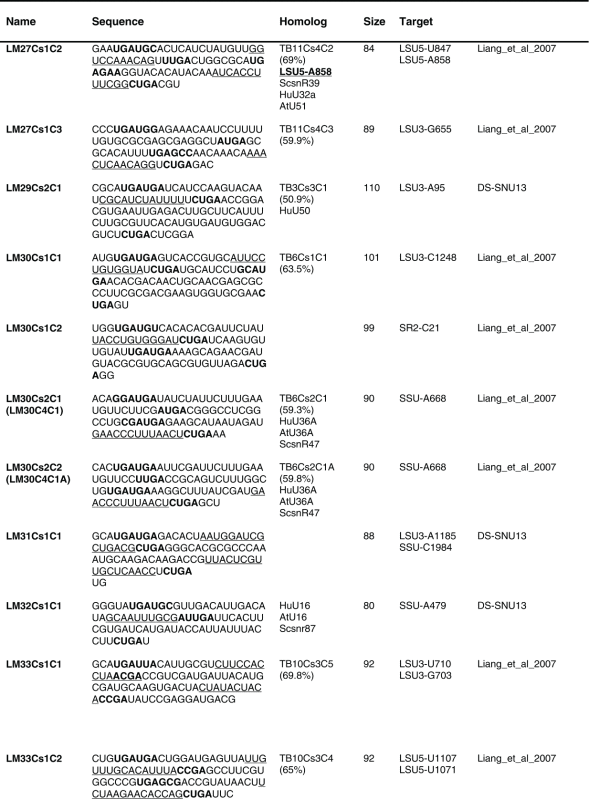
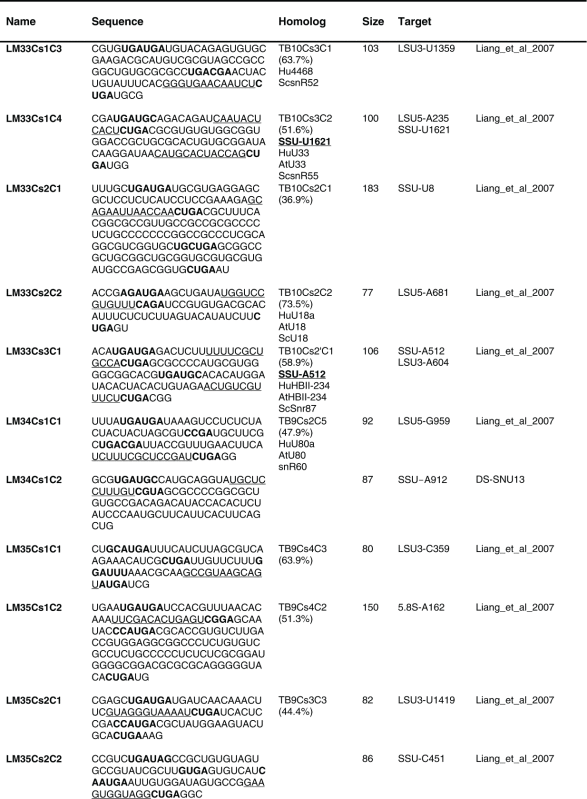
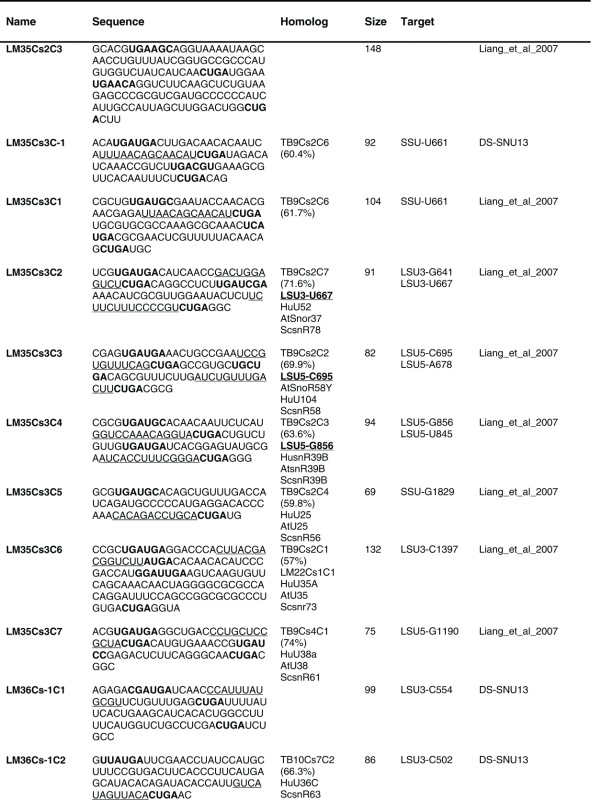
Table 1.
The sequence of the L. major C/D snoR N As. The boundaries of the molecules are based on the R NA-seq data of the R NA selected with S NU13 The boxes are indicated in bold and the domain of complementarity to the target is underlined. The homolog in T. brucei and the targets are indicated, and the source for the identification of the molecule is given. D S, deep sequencing from the current study.
 (Continued on next page) (Continued on next page) |
 |
 (Continued on next page) (Continued on next page) |
 |
 (Continued on next page) (Continued on next page) |
 |
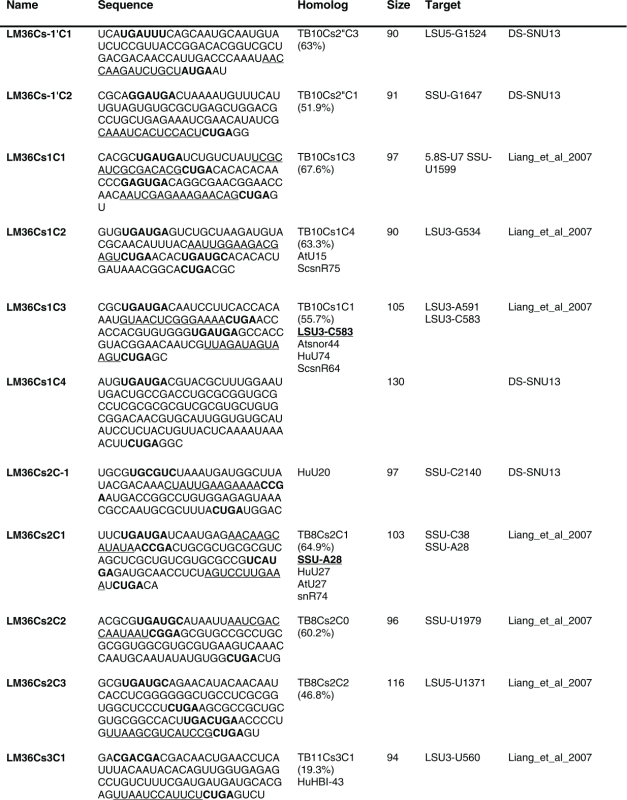 (Continued on next page) (Continued on next page) |
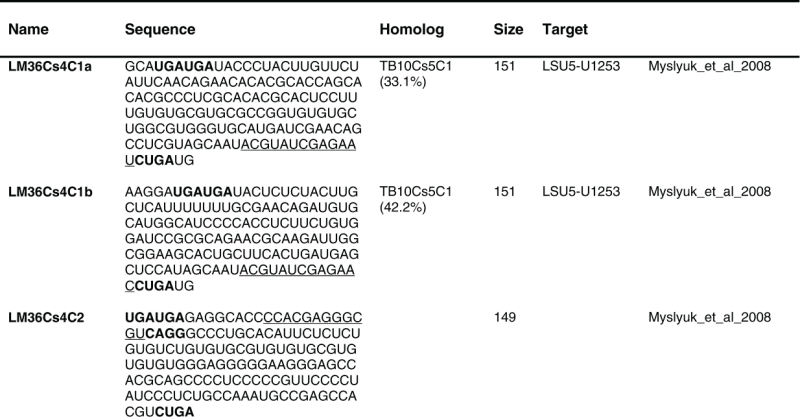 |
Previously, we described 64 C/D molecules in L.major, and 30 T. brucei homologues were assigned.16 Among the 80 C/D molecules identified here, 65 have homologues in T. brucei. Homology was determined solely on the target sites, since the sequence of the C/D outside the target site is very variable.
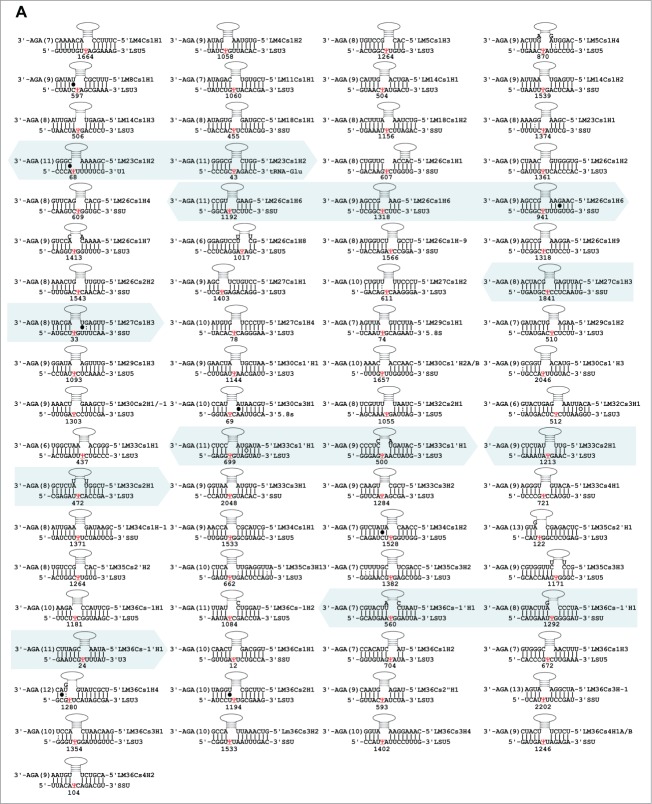
The proposed targets of the C/D snoRNAs are presented in Figure 4. Of these, 92 Nms 88 sites were identified only on rRNAs. For 8 molecules, we could not predict a target. Our target search was stringent, requiring at least a 10 bp duplex.
Figure 4.
Potential base-pair interactions between the C/D snoRNAs and their targets. For each snoRNA and target pair, the name of the snoRNA, the domain of complementarity, and the nucleotide to be modified on its target are indicated in red. SSU, small subunit; LSU 5 or 3 indicate the 5′ and 3′ ends of the large rRNA subunit. The snoRNAs that direct the 2 Nms are shaded by blue background and if these exist in 2 lanes the shaded boxes are marked with arrowheads. The D or D’ boxes are underlined.
Eight molecules appeared to be Leishmania-specific, since they lack homologues in T. brucei, and they do have predicted targets in Leishmania. We cannot, however, exclude the possibility that the T. brucei homologues for these molecules exist but have yet to be identified. Only 21 of the L. major C/D snoRNAs are proposed to have 2 targets based on at least 10 nt complementarity to each of the target RNAs. Note that most of the predicted target sites are on rRNA. Searching for snoRNAs that can target Nm on snRNAs revealed a single target (see Discussion).
Among the C/D molecules, we identified highly abundant ones such as LM5Cs1C2, LM5Cs1C3, LM5Cs1C4, LM18Cs1C3, LM22Cs1C2, LM23Cs1C2, LM25Cs1C4, LM26Cs1C1, LM33Cs3C1, LM35Cs2C1, LM35Cs3C4, LM35Cs3C6, LM35Cs3C5 (Table S2). Since the T. brucei abundant snoRNAs were implicated in rRNA processing, we anticipated that their L. major homologues have similar functions (see below).
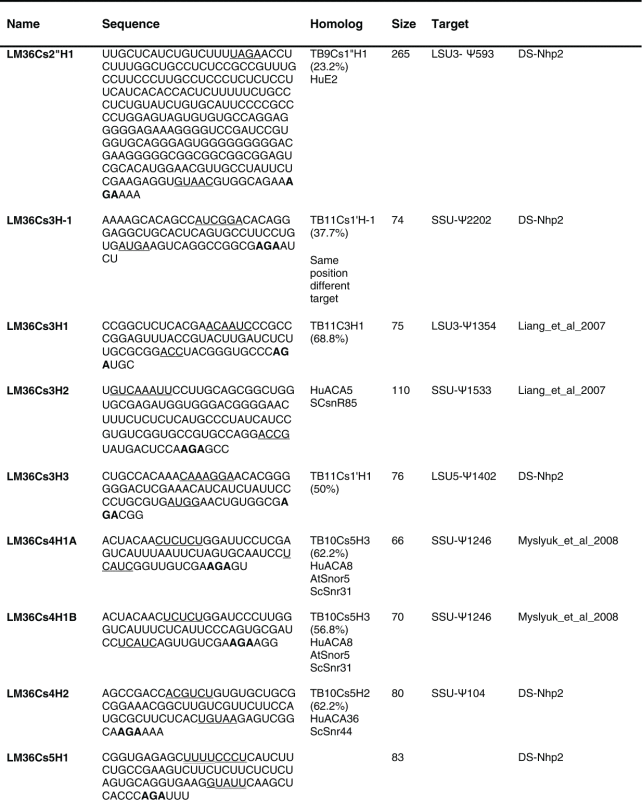
Next we analyzed the H/ACA RNAs that are associated with NHP2 and identified 81 molecules (Table 2). Our previous study17 identified only 37 H/ACA RNAs. The RNAs ranged in size from 70 to ∼300 nt. Of the H/ACA RNA only 67 have homologues in T. brucei or other organisms such as human, yeast and plants. 15 of the molecules are Leishmania-specific. Interestingly, the size of the H/ACA is much larger than C/D snoRNAs and molecules of sizes as large as 192 and 265 nt were also detected.
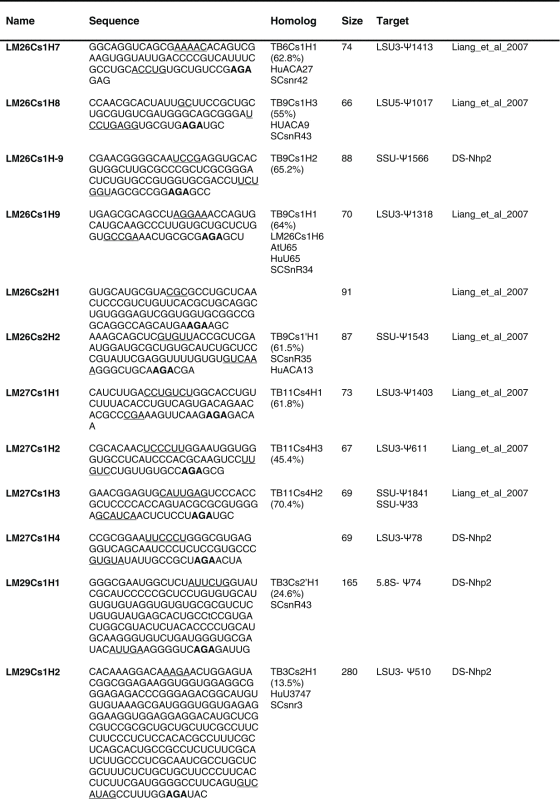
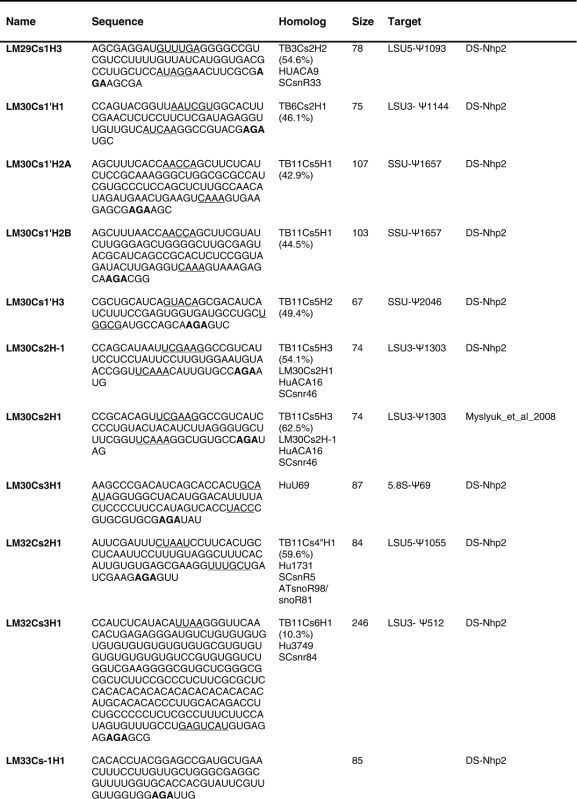
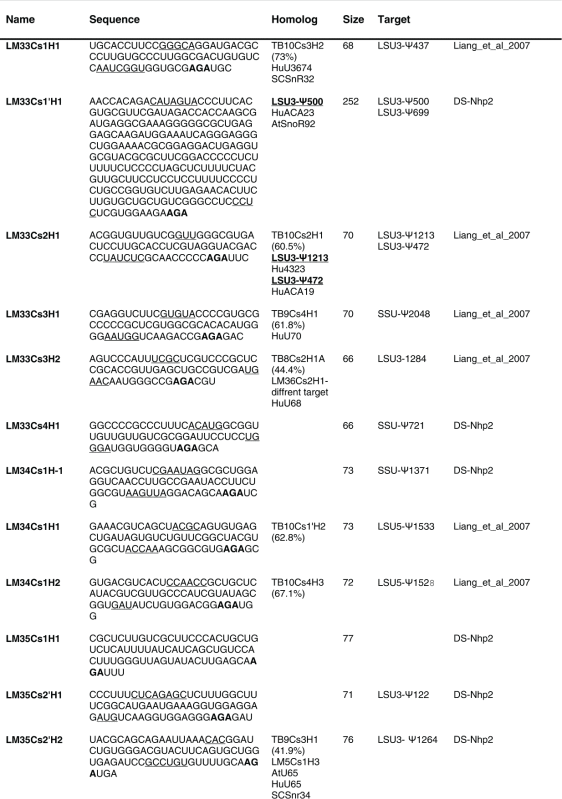
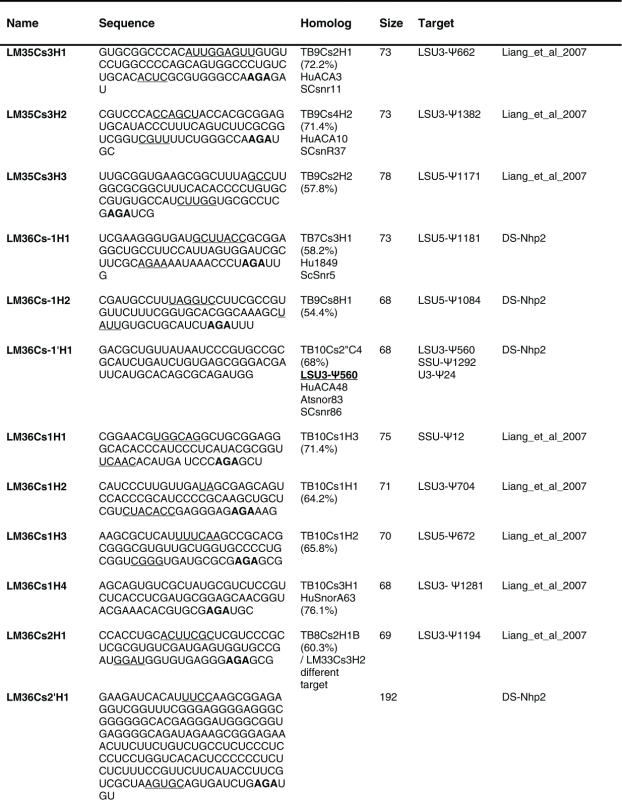
Table 2.
The sequence of the L. major H/ACA snoRNAs. The boundaries of the molecules are based on the RNA-seq data of the RNA selected with NHP2 The AGA is shown in bold. The homolog in T. brucei and the targets are indicated, and the source of the identification of the molecule is given. DS, deep sequencing from the current study.
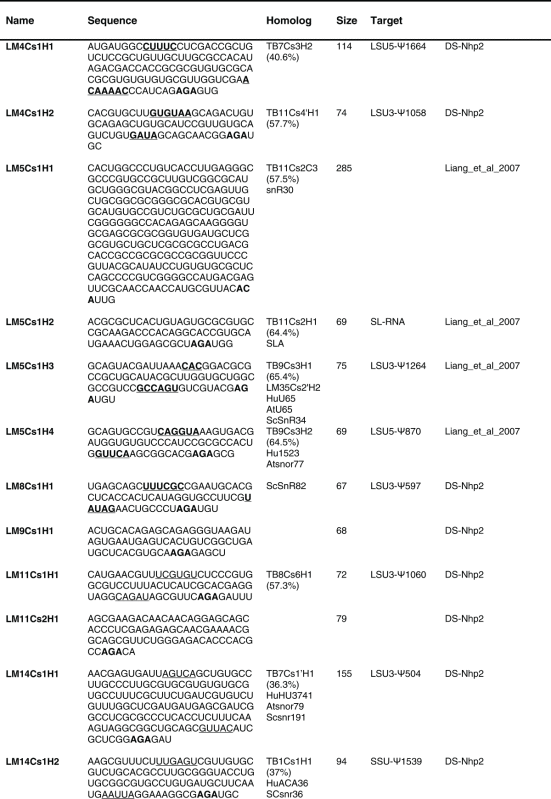 (Continued on next page) (Continued on next page) |
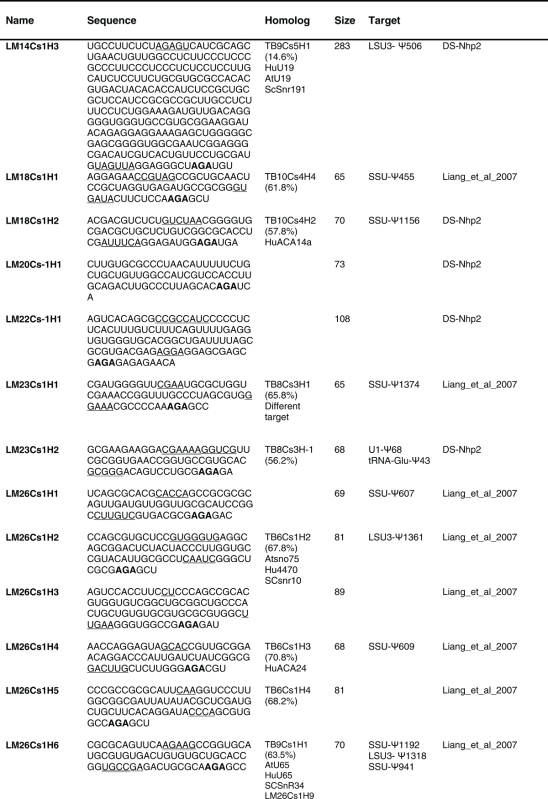 |
 (Continued on next page) (Continued on next page) |
 |
 (Continued on next page) (Continued on next page) |
 |
 |
In most cases, an A is present 1 nt upstream of stem I (94% in T. brucei and 67% in L. major). In L. major, C can also appear in this position (about 30% of the time), while G and U are rarely found. Stem I is usually perfect and can range from 4 to 8 nt in length. In most cases, stem I is 6 to 7 bp long, and compensatory changes are often found to support the integrity of the stem. The pseudouridylation pocket varies in size from 12 to 17 nt. Stem II also varies in size, but a perfect stem of 4 to 7 nt is present immediately adjacent to the pseudouridylation pocket.
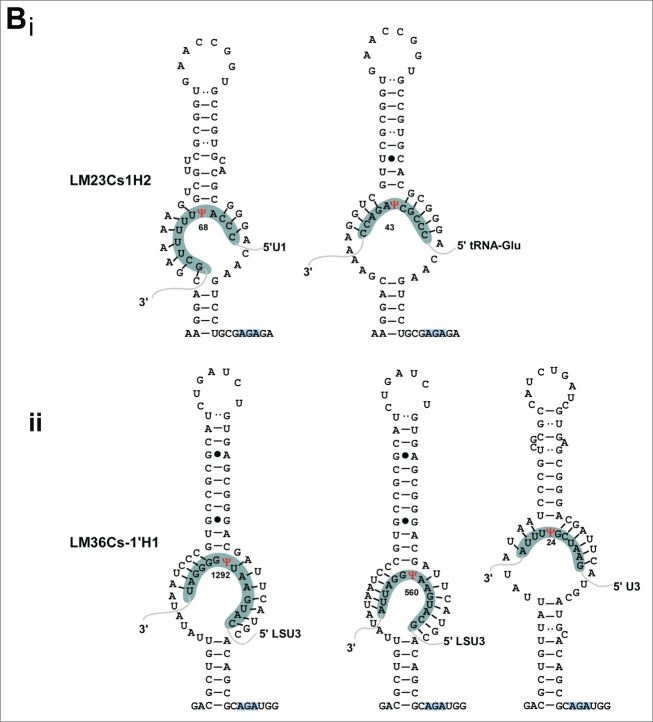
The targets for the L. major H/ACA are given in Figure 5A. For 12 molecules we were unable to suggest any target. For LM23Cs1H2, we could suggest a target on U1, but this molecule also has a predicted target on tRNA, suggesting that trypanosomatid H/ACA molecules may have a flexible pocket, and that a single molecule can guide the pseudouridylation on more than one substrate (Fig. 5Bi). Another example is LM36Cs-1′H1which can potentially guide a Ψ on U3 and on rRNA. And the molecules, LM27Cs1H3, LM33Cs1′H1, LM33Cs2H1 that can potentially target Ψ on 2 different rRNA domains (Table 2). An H/ACA (LM36Cs-1′H1) can guide Ψ on 3 different targets (Fig. 5Bii). An additional such RNA is LM26Cs1H6 which can potentially target 3 different Ψs on rRNA.
Figure 5.
For figure legend, see page 1245.
Figure 5.
(A) Potential base-pair interaction between the H/ACA snoRNAs and their targets. The target sites are indicated in red and the positions are given. The designation of the targets is as detailed in Figure 4. The snoRNAs shaded with blue background and arrow heads can potentially guide more than a single modification. (B) H/ACA snoRNAs which can potentially guide pseudouridylation on more than a single site; i, LM23Cs1H2; ii, LM36Cs-1′H1.
Interestingly, we identified 14 molecules which are longer than average (100 nt and more). Of these, 11 have a target on rRNA, but because of their size these may target modifications on additional RNAs (see below). Interestingly, long H/ACA molecules were not identified in T. brucei. However, each L. major long H/ACA molecule carries a region that is homologous to short T. brucei molecules. For instance, a portion of LM14Cs1H1 is homologous to TB7Cs1′H1, which targets a Ψ site that is conserved in evolution. In addition, a portion of LM14Cs1H3 is homologous to TB9Cs5H1 and the modification proposed to be guided by this RNA is also conserved in evolution. It is possible that these long molecules maybe the outcome of a fusion of more than one H/ACA molecule, and hence, may target more than one type of RNA. However, we could not find evidence that the long snoRNAs resulted from the fusion of snoRNAs located adjacent to the homologous T. brucei snoRNA.
The presence of 19 H/ACA snoRNAs which are found in Leishmania but not in T. brucei was surprising. Although, we have recently expanded the repertoire of T. brucei snoRNAs, this study identified 81 H/ACA in L. major, whereas only 63 H/ACA are known in T. brucei,22 suggesting that the published T. brucei repertoire is not complete.
Genomic organization of snoRNA clusters and differences between T. brucei and L. major
Functional information can at times be derived from the genomic organization of the snoRNAs. One such an example is the SLA1 locus that carries SLA1, snR30,20 as well as TB11Cs1C1 and TB11Cs1C2, which were demonstrated to be trypanosome-specific snoRNAs involved in rRNA processing.22,23 We therefore examined the organization of the snoRNA clusters to identify clusters that may carry snoRNAs with special function such as rRNA processing. The L. major snoRNAs are organized in 49 chromosomal loci (Fig. S3) compared to the 23 loci previously described.17 The majority (32) are mixed clusters with both types of snoRNAs. However, we also detected solitary snoRNAs (19 H/ACA and 10 C/D). In contrast to T. brucei, where the majority of the snoRNA clusters are repeated multiple times, in L. major only 22 loci are reiterated (Fig. S3). Interestingly, in some chromosomal loci, only certain portions of the cluster are repeated (such repeats are indicated by yellow squares in Fig. S3). For instance, only part of LM18Cs1 is repeated 10 times (the A repeat). Another complex genomic arrangement is seen in LM27Cs1, which is composed of 2 clusters, C and D, which appear in alternate order and are repeated different numbers of times in each position.
Trypanosome snoRNAs implicated in rRNA processing
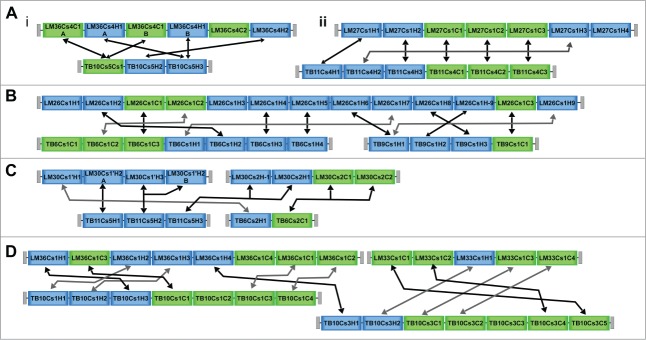
The synteny analysis of T. brucei and L. major snoRNA clusters (Fig. 6) revealed clusters that contain the same set of homologous snoRNAs despite the fact that their order within the cluster differs; such a case is LM36Cs4 and its homolog TB10Cs5. However, the LM cluster contains a unique C/D snoRNA, LM36Cs4C2 (L.m specific snoRNA) (Figs. 6Ai). The same relatedness applies to LM27Cs1 and TB11Cs4, and also in this case, the cluster contains an LM specific snoRNA, LM27Cs1H4 (Figs. 6Aii). In other cases, it seems that an L. major cluster emerged from fusion of 2 T. brucei clusters; this is the case for LM26Cs1, which is related to 2 T. brucei clusters, TB6Cs1 and TB9Cs1 (Fig. 6B). However, more complex relationships also exist, such as 2 LM clusters (LM30Cs1', LM30Cs2) which are related to 2 TB clusters (TB6Cs1, TB11Cs5); the clusters in each species are related to 2 clusters in the other species (Fig. 6C). Interestingly, the Leishmania, related clusters are found next to each other, while they are encoded on 2 different T. brucei chromosomes. The most complex relationship was found for LM33Cs1, which shares 5 out its 6 snoRNAs with TB10Cs3; this cluster has one snoRNA that has an homolog in LM36Cs1, but the latter cluster is most closely related to TB10Cs1. Interestingly the clusters related to the LM cluster are localized on the same T. brucei chromosome (Fig. 6D).
Figure 6.
The relatedness between genome organization of snoRNA in L. major and T. brucei. (A) i - Genomic organization of the related clusters LM36Cs4 and TB10Cs5; ii - LM27Cs1 and TB11Cs4. (B) Relatedness of LM26Cs1 to the TB6Cs1 and T9Cs1 clusters. (C) Relatedness of LM30Cs1′ LM30Cs2 to TB6Cs2 and TB11Cs5. (D) Complex relationship of LM36Cs1 and LM33Cs1 to the T. brucei clusters TB10Cs1 and TB10Cs3.
Interestingly, snoRNAs involved in rRNA processing are found in distinct clusters. The SLA1 locus in T. brucei (TB11Cs2) is homologous to LM5Cs1 and contains abundant snoRNAs with special functions including rRNA processing (Figs. 7Ai). An additional cluster of this type is LM35Cs3, which is homologous to TB9Cs2, which contains the abundant snoRNA TB9Cs2C1, implicated in rRNA processing22 and is homologous to LM35Cs3C6. In addition, the LM35Cs3 cluster contains LM35Cs3C4, which is homologous to the abundant TB9Cs2C3 snoRNA. This cluster carries another abundant C/D snoRNA, LM35Cs3C5, which is implicated to direct cleavage of 5′ the ETS but seems to be a Leishmania-specific snoRNA (see below) (Figs. 7Aii). LM36Cs1 is another such cluster (Fig. 7A-iii) that carries 2 C/D snoRNAs implicated in rRNA processing, LM36Cs1C3, which is homologous to TB10Cs1C1, and LM36Cs1C2, which is homologous to TB10Cs1C4 and was shown to be involved in trypanosome-specific rRNA processing.22 Additional snoRNAs implicated in rRNA processing are present in LM22Cs1, LM23Cs1, and LM26Cs1 but these are not organized in the same way as in T. brucei.
Figure 7.
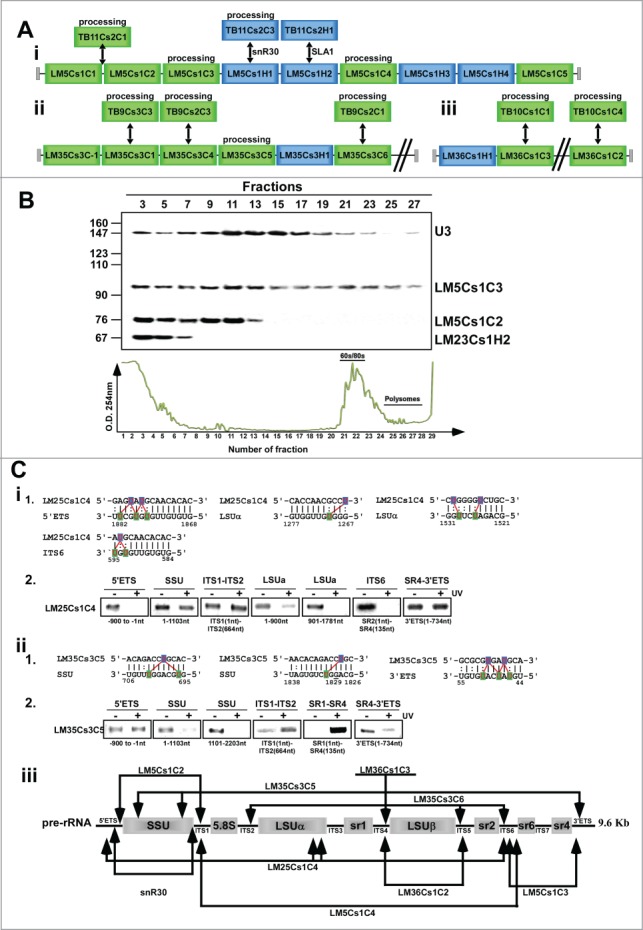
L. major snoRNAs implicated in rRNA processing. (A) L. major homologues to T. brucei snoRNAs implicated in rRNA processing. (B) Genomic relatedness between clusters encoding snoRNAs involved in rRNA processing. i. LM5Cs1 and TB11Cs2; ii. LM35Cs3/TB9Cs3; iii. LM36Cs1/TB10Cs1. B - Migration of the snoRNAs on sucrose gradients. RNPs were prepared and fractionated as described in Materials and Methods. RNA was extracted from the fractions and separated on 10% polyacrylamide-denaturing gel, and subjected to Northern analysis with the indicated probes. The sizes of pBR322 DNA-MspI digest marker are indicated on the left, and the identity of the RNAs on the right. (C) “RNA walk” analysis to validate snoRNA-rRNA interactions. i-1- The proposed cleavage sites of LM35csC5 and LM25Cs1C4 and proposed interaction domain of LM25Cs1C4 and pre-rRNA with rRNA. i-2- “RNA walk” analyses of the interaction site of the 2 snoRNAs. RT-PCR of rRNA domains interacting with TB25Cs1C4. cDNA was prepared from RNA affinity selected with LM25Cs1C4 from total RNA prepared after AMT cross-linking. The domains and the size of the amplified fragments are indicated. ii (1 and 2). The same as in (i) but for LM35CSC5. iii. Schematic representation of the interaction site and potential cleavages by snoRNA implicated in rRNA processing. The proposed cleavage site of pre-rRNA is indicated by arrows.
Among the snoRNAs involved in rRNA processing/maturation are snoRNAs that are suggested to function in processing of the small subunit rRNA (SSU) and the large subunit rRNA (LSU) such as LM5Cs1C3 (homologous to TB11Cs2C2), and those that are implicated in SSU processing (LM5Cs1C1). To examine whether 2 distinct large RNP complexes exist for LSU and SSU processing, whole cell extracts were prepared from L. major, fractionated on 10–30% sucrose gradients, and the fractions were analyzed by Northern analysis using the probes specified. The results (Fig. 7B) indicated that U3, which is implicated in SSU processing, peaked in factions 11–17, and LM5Cs1C2 which is also implicated is SSU processing peaks in fractions 9–13, but LM5Cs1C3 which is the homolog of TB11Cs2C2 and is implicated in LSU processing23 was also found in the higher S value complexes (a peak in fractions 9–13 and a second peak in fractions 19–23). In contrast, LM23Cs1H2 which is implicated in tRNA modification was found only on small RNPs and at the top of the gradient (fractions 3–7). The results implied that fractionation of RNPs may be used to suggest the function of a snoRNA and its involvement processing of SSU, LSU or both or even other targets.
Of special interest are those snoRNAs which are Leishmania-specific, such as LM25Cs1C4, and LM35Cs3C5. The bioinformatic predictions suggested that LM25Cs1C4 potentially interacts with the 5′ETS, LSUα, and ITS6 (Figs. 7Ci-1). We therefore examined whether this snoRNA interacts with these domains by “RNA-walk” a method that we used previously to map the interaction of snoRNAs with their target site.22,23 In brief, cells are treated with AMT-psoralen, which intercalates between the duplex, and upon UV treatment a covalent linkage is introduced. The in vivo cross-linking enables capture of interactions that take place in cells. To select for the small RNA-target duplexes, they are purified by affinity selection using an anti-sense oligonucleotide complementary to the small RNA. The site of interaction between the small RNA and its target does not allow the reverse transcriptase to copy this domain, and as a result cDNA prepared with random-primers cannot be amplified by PCR using specific primers covering the cross-linked adduct.
The four possible target interactions of LM25Cs1C4 were examined using RT-PCR on different domains along the pre-rRNA. The results (Figs. 7Ci-2) indicated specific reduction in the level of 5′ETS, LSUα and ITS6, but not in other domains on the rRNA, thereby supporting the bioinformatic prediction that this snoRNA interacts with pre-rRNA, possibly for processing. Next, we examined the interaction of LM35Cs3C5 with its targets. The bioinformatics predictions suggested that LM35Cs3C5 interacts with SSU, and potentially guides the methylation at position Gm1829. In addition, this snoRNA potentially interacts with the 3′ ETS (Fig. 7Cii-1). ‘RNA walk’ was used to verify these interactions by examining the amplification of different domains along the pre-rRNA. The results supported the interactions with both SSU and the 3′ETS (Fig. 7Cii-2). Interestingly, this snoRNA may direct cleavages to liberate both SSU and LSU (Fig. 7C-iii). Our results (Fig. 7) suggested that the LSU processing is mediated by a complex that is distinct from the SSU processome, extensively studied in other eukaryotes (see Discussion). Although most of the snoRNAs implicated in rRNA were predicted to either process SSU or LSU, several snoRNA were predicted to be involved in processing both subunits (Fig. 7C-iii).
The rich repertoire of both Ψs and Nms in trypanosomatids compared to other eukaryotes
The rich repertoire of snoRNAs suggested the existence of an unusually high level of modifications that maybe related to parasites cycling between the 2 hosts.39 Although the repertoire of H/ACA identified in this study almost doubled the amount of predicted Ψs from 37 to 69, the number of predicted Nms is 93, hence higher than the number of Ψs.
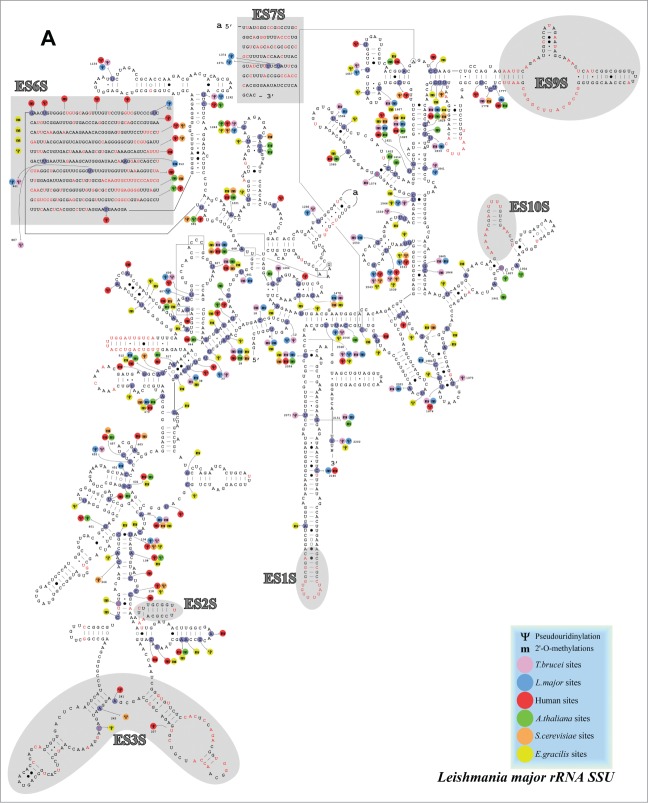
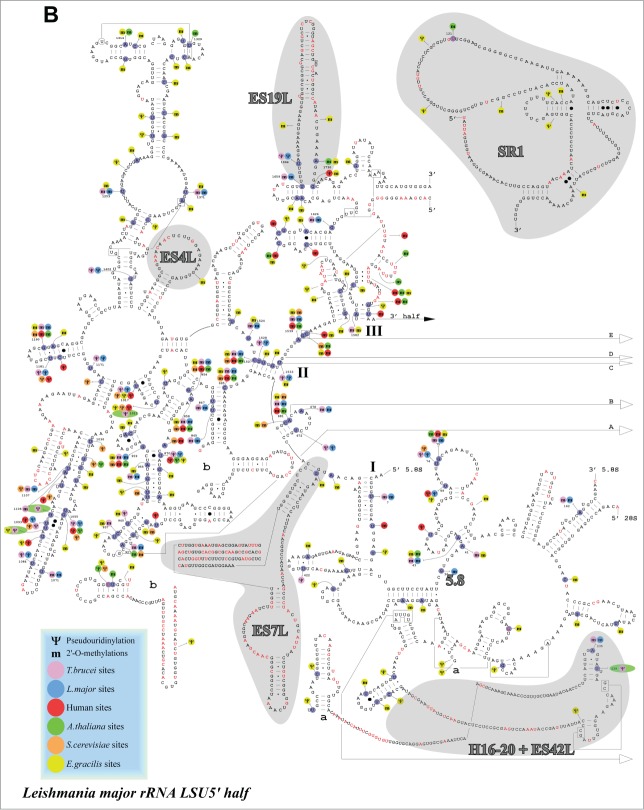
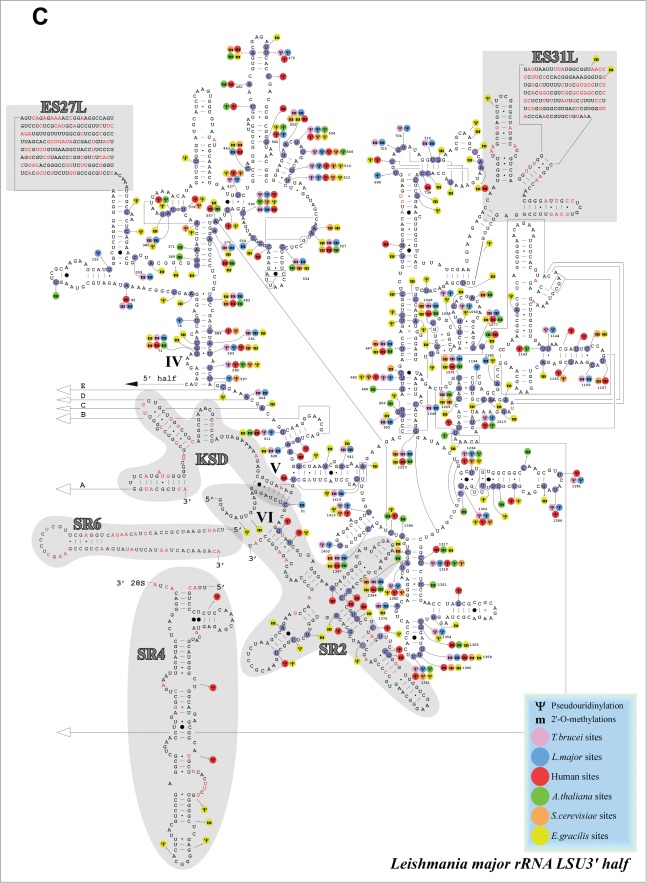
The repertoire of Ψs and Nms on SSU and LSU compared to the modifications present in yeast, human, plants, and Euglena are presented (Fig. 8). Among the 88 predicted Nms sites, 47 were found to be modified in either yeast, mammals or plants. 47 Nm sites are shared with Euglena. 75 Nms are shared between T. brucei and L. major. Among the 67 Ψs, 35 are found in yeast, mammals or plants, 55 are shared between L. major and T. brucei, and 28 Ψs are shared between L. major and Euglena. Interestingly, trypanosomatid-specific modifications were not evenly distributed among the rRNA: SSU (11Nm, 9Ψ), LSU 5′ (12 Nm, 2Ψ), LSU 3′ (19 Nm, 14Ψ) and included more specific Nms than Ψs. In several domains, modifications were found exclusively in trypanosomatids and Euglena; LSU 5′ positions 1253 −1373 contained 2 modifications shared with trypanosomatids, and 14 Euglena-specific ones, and positions 1659–1725 (ES19L) contained 2 trypanosomatid-specific and 2 Euglena modifications. In the LSU3′, most of the trypanosomatid and Euglena specific modifications existed in highly modified domains. However, position 699–740 contains 4 trypanosomatid-specific but 7 Euglena-specific modifications. Thus, the data suggested that the hypermodification in trypanosomatids and Euglena is found mostly in modification-rich domains. Interestingly, species-specific modifications also exist and may play a role in ribosome function in these organisms (see Discussion).
Figure 8.
For figure legend, see page 1250.
Figure 8.
For figure legend, see page 1250.
Figure 8.
Localization of modified nucleotides on the secondary structure of rRNA. (A) Modification of SSU. Location of modified nucleotides on the structure of rRNA. The Nm are marked as m, and the pseudouridines as Ψ. The secondary structure was predicted based on the structure presented for T. brucei at htttp://www.icmb.utexas.edu, adjusting it to the L. major rRNA sequence. The identity of the small rRNA fragments and distinct domains is indicated and shaded. The modifications in different eukaryotes are designated by different colors, as indicated to the right. (B) As in A but for the 5′ half of LSU. (C) The same as in A and B, but for the 3′ part of LSU. The domains of the rRNA are indicated.
How could different snoRNAs arise: A lesson from comparing L. major and T. brucei homologues
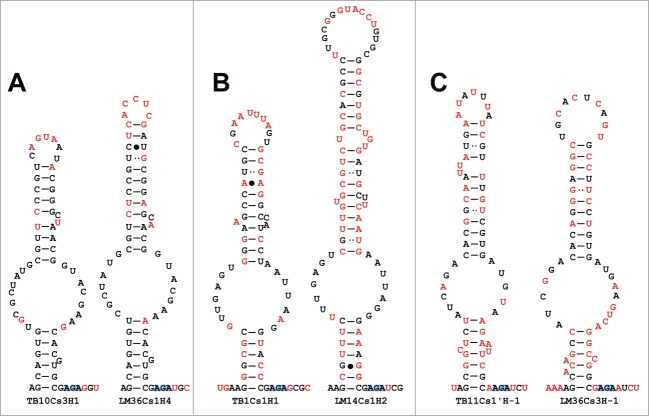
When comparing homologous H/ACA snoRNAs in L. major and T. brucei, we noticed that the pseudouridylation pocket, the functional heart of the molecule, and the rest of the molecule, the body, evolved at different rates, yielding different types of snoRNA homologues. The first group is composed of homologous pairs that share overall identity and guide the same Ψ sites (for instance TB10Cs3H1 and LM36Cs1H4) (Fig. 9A). These homologues share 65% identity or greater in the pseudouridylation pocket and 55–65% identity over the rest the H/ACA. These account for 29 of the homolog pairs. The second group includes snoRNAs that share low similarity in their body region (≤ 55% identity) but share almost identical pockets (90 – 100%) i.e., the sequence and even the structure (length of the second stem) are different between the homologues, but these are predicted to guide the same modification i.e. carry out the same function (e.g. TB1Cs1H1 and LM14Cs1H2) (Fig. 9B). Eighteen snoRNAs pairs are associated with this group. Interestingly, the last group is composed of snoRNAs that share low similarity in their body region (≤ 55% identity), with differences in the pseudouridylation pocket (55–85% identity) but guide the same Ψ site (LM36Cs3H-1 and TB11Cs1pH-1). There are 14 snoRNAs pairs associated with this group (Fig. 9C). Kruskal-Wallis tests revealed significant differences in pocket identity (P < 0.001) and body identity (P < 0.001) as well as the interaction between them (P < 0.001) among these 3 groups.
Figure 9.
The relatedness between the L. major and T. brucei snoRNAs. (A–C) Secondary structure of the snoRNA in each of the 3 groups. The nts that differ in sequence are shown in red. The identifiers of each group are detailed in the text.
In most cases, we can detect a clear ortholog in which the body and/or pocket are maintained. However, there are several cases in which we suspect a more complicated evolutionary history. We have identified 15 H/ACAs in L. major as likely paralogues (identity in body ≥ 60%). This may complicate identifying a single T. brucei homolog for each L. major H/ACA (Fig. S4A). Nonetheless, most of these paralogues seem to have arisen as a result of a duplication event in L. major, and thus map to a single T. brucei snoRNA. Such a case is TB10Cs5H3, which has 2 paralogues, LM36Cs4H1a that has an identical pseudouridylation pocket and 34.6% identity across the body, as well as LM36Cs4H1b, which has the same pocket but shares an overall identity of 56.8%. A few additional examples are presented in (Fig. S4B). Interestingly, the TB9Cs3H1 has 2 homologues LM5Cs1H3 and LM35Cs2′H2, which have almost the same pocket, suggesting that 2 different snoRNAs can be modified to target the same nt and this may be a mechanism whereby new snoRNAs are generated i.e., by only modifying the pocket after duplication of an existing snoRNA. In fact, it was possible to find 2 Leishmania paralogues for a single T. brucei snoRNA. Generation of paralogues from different snoRNA was also found in T. brucei. Such a case is LM18Cs1H2 and LM33Cs3H1 (61.5% identity), which we implicated as paralogues yet appear to have different homologues in T. brucei (TB10Cs4H2, TB9Cs4H1). This may present an example of a duplication event in a common ancestor (Fig. S4C).
Another interesting case involves a snoRNA in T. brucei, TB8Cs2H1A and B. It appears in the T. brucei genome in 2 forms, which are almost exactly identical copies with a 2 base pair change in the pocket area. Each pocket matches a different LM homolog (LM33Cs3H2 and LM36Cs2H1). Several of the paralogues (LM5Cs1H3 and LM35Cs2′H2, LM30Cs2H-1 and LM30Cs2H1, LM26Cs1H9 and LM26Cs1H6) guide the same modification on rRNA. Although the paralogues may guide the same modification, we cannot rule out the possibility that these may undergo conformational changes and direct the modification on another target. Indeed, we have found the example of LM27Cs1H3, in which a single snoRNA can potentially guide 2 different modifications on rRNA.
Discussion
This study describes comprehensively the repertoire of L. major H/ACA and C/D snoRNAs, identifying potential targets of 92 Nm and 73 Ψ sites on rRNA. Based on these studies, the number of predicted modifications is much higher compared to yeast, which has approximately 50 modifications of each type, despite having the same genome size. The high number of modifications in trypanosomes may suggest a role in maintaining ribosome function when cycling between the insect and the mammalian host.40 One striking finding is that except for 2 cases, all the snoRNAs described in this study have the potential to guide modifications on rRNA. Interestingly, the single-hairpin H/ACA RNA have flexibility in generating more than a single pseudouridylation pocket, and hence, a single hairpin RNA can potentially direct modifications on more than one site. The comparison between the repertoire of snoRNAs in T. brucei and L. major demonstrated how related snoRNA were engineered during evolution to direct modifications on the same or different sites. Duplication of snoRNA genes was needed to give rise to novel and species-specific snoRNAs. Abundant snoRNA emerged in both species to carry out trypanosome-specific rRNA processing events. The flexibility in the structure and function of trypanosome snoRNAs enables a relatively small repertoire of RNAs to guide a rich repertoire of modifications and carry out rRNA processing activities that are trypanosome-specific.
snoRNAs involved in trypanosome-specific rRNA processing: the LSU processome and snoRNAs with dual functions
One of the unique and striking properties of trypanosome rRNA is the fact that it is generated by cleavage of the LSU, resulting in small rRNA fragments that are held in the ribosome by base pairing. It was not clear for a long time why this fragmentation takes place, and how it is mediated. A recent study using high resolution cryo-electron microscopy solved the structure of the T. brucei ribosome.41 The results revealed that the rRNA expansion regions that are highly variable in sequence and in size are extended in T. brucei. This is also true for L. major (Fig. 8). It was suggested that the cleavages of the rRNA fragments is necessary to accommodate for the increase in size of rRNA due to an increase in rRNA expansion domains. Our previous study supported the notion that trypanosome-specific snoRNAs evolved to direct these cleavages. Indeed, we have previously demonstrated the role of TB11Cs2C1 and TB11Cs2C2 in rRNA processing23 and also the role of TB10Cs4C4, TB6Cs1C3, TB0Cs2C1, in trypanosome-specific rRNA fragmentation.22 We suggested that TB11Cs2C1 may be the homolog of U14. Such special snoRNAs may also exist in Euglena, which also undergoes extensive rRNA processing.42 However, despite the description of C/D and H/ACA RNAs in this organism, none of these have yet been implicated in rRNA processing. Interestingly, although the Euglena snoRNA repertoire resembles that of trypanosomes, it does possess a U14 homolog.43
snoRNAs involved in rRNA processing are clustered together in both T. brucei and L. major (Fig. 7). Of special interest is the finding that not all of the snoRNAs species implicated in rRNA processing are homologous in the 2 organisms. Among the 13 molecules implicated in rRNA processing44 in T. brucei, only 9 have homologues in Leishmania while the rest are T. brucei specific, suggesting that different snoRNAs in these 2 species were selected to carry out these special processing functions. Indeed, all the snoRNAs implicated in rRNA processing also guide modifications on rRNA and thus these special snoRNAs may have emerged from snoRNAs involved in modification. The question remains how snoRNAs were selected for this additional function. It is possible that these snoRNAs were selected because of their chromosomal location and abundance. snoRNAs involved in rRNA processing were shown to be abundant in both T. brucei and L. major.
It is not currently known how the trypanosome differentially regulates the level of snoRNAs. The level of a snoRNA might be influenced by a post-transcriptional modification such as polyadenylation (our unpublished results).
The data presented in this study suggest that the SSU processome may be distinct from the LSU processome. However, relatively little is known about the biochemistry of the LSU in other organisms.45 Recent bioinformatics analysis identified many factors involved in 40S processome and 60S processome function in T. brucei.44 Tagging of factors from these complexes, and affinity purification and mass-spectrometry analyses should shed light on whether these 2 processomes function in a coordinated manner. Interestingly, we identified snoRNAs (Fig. 7) which are implicated in both SSU and LSU processing. It will therefore be interesting to identify possible cross-talk between the SSU and LSU processome, especially before the separation of the pre-40S complex from the pre-60S complexes.
The rich repertoire of trypanosome rRNA modifications
Recent studies suggest that Ψ modification on rRNA is important for the translation of a distinct subset of mRNAs such as mRNA harboring an internal ribosome entry site in mammals.46 We have recently begun to analyze the Ψ at the whole-transcriptome genome level using a methodology similar to that used in recent studies that performed whole transcriptome mapping of this modification.47 It will be of great interest to compare the pattern during the 2 life stages of the parasite and examine if these changes are correlated with growth at different temperatures, and/or are essential for preferential translation of distinct mRNAs which are developmentally regulated.
Of special interest are the specific modifications in the expansion regions that are highly expanded in the trypanosome ribosome. It is not currently known why these domains are extensively expanded in trypanosomes. One possibility is that these domains bind factors involved in mRNA stability and translation, and thus regulate the stability and translation of mRNAs that are developmentally regulated in these parasites. Indeed, modifications were also found in these expanded domains in other eukaryotes but distinct positions are modified in the trypanosome rRNA (Fig. 8).
We suggested that the number of Nms is much higher than the number of Ψs in these organisms.17 Even after we revealed the high number of H/ACA RNAs, there are still more expected Nms guided by snoRNAs compared to Ψs on rRNA. The Nms have been suggested to confer stability to rRNA and are found in thermophylic Archaea.48 Indeed, studies in T. brucei showed that certain Nm positions are more extensively modified in the bloodstream form of the parasite than in the procyclic stage, which propagates in the fly.40 Transcriptome-wide mapping of Nm is required to examine whether the changes in Nm are typical only of rRNA, or of other RNAs such as snRNAs or even mRNAs. The recent whole transcriptome mapping of Ψ suggests that this modification is also prevalent on mRNAs and is not only present on stable RNAs.47 The Ψs were also shown to be induced under heat-shock.47 Studies are underway to map the transcriptome-wide Ψ and Nms in the 2 life stages of the parasite, since both modifications are known to confer structural rigidity on localized RNA structure.49 Indeed, these 2 modifications influence the RNA structure by favoring the C3′-endoribose conformation, diminishing the distance between the bases and enhancing stacking, which contributes to RNA stability.50
In Euglena, an organism that is evolutionarily related to trypanosomes, the LSU is fragmented to 14 pieces51 and the degree of modification (350 modified nt on rRNA) is correlated with the level of rRNA fragmentation. In addition, the mismatch level in helical regions is 3-fold higher than in the same domain in human rRNA, and indeed, these domains are highly modified in Euglena.51 Thus, breaking of helical stem structures in rRNA during evolution may have forced the generation of species-specific modification in trypanosomatids, and may explain the need for the development of species-specific snoRNA to direct these compensating modifications. Interestingly, trypanosomatids exhibit modifications which are common only to Euglena and in domains outside the domains enriched in modifications in other eukaryotes. It is plausible that both gain and loss of modifications during evolution shaped the repertoire found in Euglena and trypanosomes. The loss is represented by the absence of modifications which are conserved in other eukaryotes (Fig. 8). However, each of these organisms has acquired large number of species-specific modifications which are present in variable regions and may represent gain over evolutionary time.51 However, it seems that the enhanced modifications found in both trypanosomes and Euglena may have evolved to cope with the fragmentation of the LSU. It will be of great interest to examine which of these modifications are constitutive or are induced under certain conditions.
The evolution of snoRNAs, a lesson from T. brucei and L. major
The comparison between L. major and T. brucei snoRNAs revealed that in 48% of the cases, H/ACA snoRNAs are true homologues, since these guide the same modifications in both species and the snoRNA share a high level of sequence similarity. Included in this family are the snoRNAs which share high similarity, but the sequence of the pseudouridylation pocket was changed to accommodate for differences in the rRNA sequence between the 2 species. However, a larger group of snoRNAs belong to a family of 43 (52%) snoRNAs which guide the same Ψ but share little sequence similarity. This may suggest how new snoRNAs can be generated by copying an existing snoRNA and “changing” the pseudouridylation pocket. This scenario resulted in different snoRNAs that guide the same modification. Indeed, we revealed cases in which 3 different snoRNAs are implicated in guiding the same modification. It will be interesting to determine if these modifications are critical for the function of the ribosome, and if they are guided by snoRNAs that are themselves developmentally regulated. Indeed, recent studies of snoRNAs in cancer demonstrated that certain snoRNAs are changed during the neoplastic process to guide a different set of modifications by minor sequence changes in the pseudouridylation pocket. It was suggested that the translation of distinct mRNA species requires different types of modification and that a particular pattern is favored to translate mRNAs that are essential for the survival and metastasis of cancer cells.46 Thus, novel snoRNAs with different function may arise in all organisms from Leishmania to man by just changing the pocket. Indeed, in Euglena as well, recent studies suggest that frequent gene duplication is a common mechanism driving snoRNA emergence leading to both a large number of snoRNAs and clustered patterning of rRNA modification.51
One of the most striking observations made in our snoRNA studies16,17,22 is that the vast majority of snoRNAs described are predicted to guide modifications on rRNA but not on snRNAs. Our recent mapping of Ψs on rRNA (unpublished) indicates that all the sites suggested to be guided by the H/ACA are valid, since these modifications exist on rRNA. We have also mapped Ψ and Nms on U snRNAs by primer extension sequencing and showed that at least 22 of the Ψs are guided by snoRNAs (our unpublished data). The question remains which snoRNAs guide these modifications and whether small Cajal body RNA (scaRNA) may be involved. The only scaRNA-like molecule identified so far is SLA1, which guides modification on the SL RNA.18 This RNA is localized in a distinct site in the nucleus and outside the nucleolus.21 In addition, SLA1 is bound by the protein MTAP52 which exhibits homology to WD40 protein bound by scaRNA in mammals.53 Thus, MTAP may specify the scaRNA-like RNAs in trypanosomes as well.
snoRNAs in pathogenic protozoan parasites
Are the specific features of snoRNAs and their guided modifications related to their parasitic life especially cycling between 2 hosts. To answer this question we compared the features of trypanosome snoRNA to those found Plasmodium (malaria). In malaria, as in humans, many snoRNAs are encoded in introns and are associated with genes involved in ribosome metabolism.11 It was suggested that malaria snoRNAs were duplicated in evolution via retroposition. However, no information is available on differences between snoRNA expression and modifications in the parasite propagating in the 2 hosts and this aspect will be interesting to study.
Since many of the protozoan parasites diverged early in evolution from the eukaryotic lineage it was interesting to compare their snoRNA properties to those of trypanosomes. As in trypanosomes, in Entameoba histolytica snoRNAs are single hairpin RNAs, but these possess an ACA rather than an AGA box. The E. histolytica snoRNAs are more related in sequence to yeast and human than to Plasmodium and trypanosomes.10 Studies from Giradia lamblia, one of the most ancient eukaryotes, suggest that, as opposed to trypanosomes, their C/D snoRNAs are only single guiders and all H/ACAs are composed of double-stem-loop hairpins in contrast to trypanosomes. Thus, Giardia snoRNAs are more related to the snoRNA found in fungi and metazoans9 and the single hairpin snoRNA is not typical to ancient eukaryotes and is specific only to certain sub-groups.
The study presented here describes the comprehensive repertoire of snoRNAs in L. major, and suggests how snoRNAs may have evolved to carry out different functions. Whereas the snoRNAs identified here are proposed to carry out rRNA modification, we have yet to identify the snoRNAs guiding the modifications on snRNAs. The simple rules that were applied to identify modifications on rRNA do not seem to apply to snRNAs, and as suggested above, trypanosomes may have the ability to utilize single hairpin H/ACA snoRNAs to guide more than a single modification. This study also identified the snoRNAs implicated in trypanosome-specific rRNA processing. The peculiar properties of trypanosome snoRNAs are not related to their parasitic life, but rather to the need for rRNA fragmentation, as in Euglena. However, we are only at the beginnings of understanding rRNA processing in these organisms, as well as the extent by which modification mediated by snoRNAs or enzymes regulate gene expression and ribosome function. The trypanosome genome encodes for numerous pseudouridine synthases and methyltransferases, which should also contribute to the complexity of RNA modifications in these important parasites.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors dedicate this study to the memory of Elisabetta Ullu, an excellent, inspiring scientist, a leader of our field and above all a dear friend and generous colleague.
Funding
This work was supported by a grant from the Israel-US Binational Science Foundation (BSF), and the I-core Center of Excellence grant no 1796/12 from the Israel Science Foundation and by NIH grant [RO1 AI 056333] to EU. S.M. holds the David and Inez Myers Chair in RNA silencing of diseases.
References
- 1.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2011; 2:611-31; PMID:21823225; http://dx.doi.org/ 10.1002/wrna.79 [DOI] [PubMed] [Google Scholar]
- 2.Yu YT, Meier UT. Sf mod: Rna-guided isomerization of uridine to pseudouridine - pseudouridylation. RNA Biol 2015; 11:1483-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 1996; 86:823-34; PMID:8797828; http://dx.doi.org/ 10.1016/S0092-8674(00)80156-7 [DOI] [PubMed] [Google Scholar]
- 4.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 2011; 3:397-414; PMID:22065625; http://dx.doi.org/ 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- 5.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 1996; 85:1077-88; PMID:8674114; http://dx.doi.org/ 10.1016/S0092-8674(00)81308-2 [DOI] [PubMed] [Google Scholar]
- 6.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997; 89:799-809; PMID:9182768; http://dx.doi.org/ 10.1016/S0092-8674(00)80263-9 [DOI] [PubMed] [Google Scholar]
- 7.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol 1997; 9:337-42; PMID:9159079; http://dx.doi.org/ 10.1016/S0955-0674(97)80005-1 [DOI] [PubMed] [Google Scholar]
- 8.Aspegren A, Hinas A, Larsson P, Larsson A, Soderbom F. Novel non-coding RNAs in Dictyostelium discoideum and their expression during development. Nucleic Acids Res 2004; 32:4646-56; PMID:15333696; http://dx.doi.org/ 10.1093/nar/gkh804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo J, Teng M, Zhang GP, Lun ZR, Zhou H, Qu LH. Evaluating the evolution of G. lamblia based on the small nucleolar RNAs identified from Archaea and unicellular eukaryotes. Parasitol Res 2009; 104:1543-6; PMID:19326145; http://dx.doi.org/ 10.1007/s00436-009-1403-3 [DOI] [PubMed] [Google Scholar]
- 10.Kaur D, Gupta AK, Kumari V, Sharma R, Bhattacharya A, Bhattacharya S. Computational prediction and validation of C/D, H/ACA and Eh_U3 snoRNAs of Entamoeba histolytica. BMC Genomics 2012; 13:390; PMID:22892049; http://dx.doi.org/ 10.1186/1471-2164-13-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra PC, Kumar A, Sharma A. Analysis of small nucleolar RNAs reveals unique genetic features in malaria parasites. BMC Genomics 2009; 10:68; PMID:19200392; http://dx.doi.org/ 10.1186/1471-2164-10-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang ZP, Zhou H, He HL, Chen CL, Liang D, Qu LH. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. RNA 2005; 11:1303-16; PMID:15987805; http://dx.doi.org/ 10.1261/rna.2380905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morita K, Saito Y, Sato K, Oka K, Hotta K, Sakakibara Y. Genome-wide searching with base-pairing kernel functions for noncoding RNAs: computational and expression analysis of snoRNA families in Caenorhabditis elegans. Nucleic Acids Res 2009; 37:999-1009; PMID:19129214; http://dx.doi.org/ 10.1093/nar/gkn1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al.. The genome of the kinetoplastid parasite, Leishmania major. Science 2005; 309:436-42; PMID:16020728; http://dx.doi.org/ 10.1126/science.1112680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White TC, Rudenko G, Borst P. Three small RNAs within the 10 kb trypanosome rRNA transcription unit are analogous to domain VII of other eukaryotic 28S rRNAs. Nucleic Acids Res 1986; 14:9471-89; PMID:3797245; http://dx.doi.org/ 10.1093/nar/14.23.9471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang XH, Uliel S, Hury A, Barth S, Doniger T, Unger R, Michaeli S. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA 2005; 11:619-45; PMID:15840815; http://dx.doi.org/ 10.1261/rna.7174805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang XH, Hury A, Hoze E, Uliel S, Myslyuk I, Apatoff A, Unger R, Michaeli S. Genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Leishmania major indicates conservation among trypanosomatids in the repertoire and in their rRNA targets. Eukaryot Cell 2007; 6:361-77; PMID:17189491; http://dx.doi.org/ 10.1128/EC.00296-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang XH, Xu YX, Michaeli S. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA 2002; 8:237-46; PMID:11911368; http://dx.doi.org/ 10.1017/S1355838202018290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol 2011; 6:459-74; PMID:21526946; http://dx.doi.org/ 10.2217/fmb.11.20 [DOI] [PubMed] [Google Scholar]
- 20.Barth S, Hury A, Liang XH, Michaeli S. Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J Biol Chem 2005; 280:34558-68; PMID:16107339; http://dx.doi.org/ 10.1074/jbc.M503465200 [DOI] [PubMed] [Google Scholar]
- 21.Hury A, Goldshmidt H, Tkacz ID, Michaeli S. Trypanosome spliced-leader-associated RNA (SLA1) localization and implications for spliced-leader RNA biogenesis. Eukaryot Cell 2009; 8:56-68; PMID:19028994; http://dx.doi.org/ 10.1128/EC.00322-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaeli S, Doniger T, Gupta SK, Wurtzel O, Romano M, Visnovezky D, Sorek R, Unger R, Ullu E. RNA-seq analysis of small RNPs in Trypanosoma brucei reveals a rich repertoire of non-coding RNAs. Nucleic Acids Res 2012; 40:1282-98; PMID:21976736; http://dx.doi.org/ 10.1093/nar/gkr786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta SK, Hury A, Ziporen Y, Shi H, Ullu E, Michaeli S. Small nucleolar RNA interference in Trypanosoma brucei: mechanism and utilization for elucidating the function of snoRNAs. Nucleic Acids Res 2010; 38:7236-47; PMID:20601683; http://dx.doi.org/ 10.1093/nar/gkq599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandelboim M, Barth S, Biton M, Liang XH, Michaeli S. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J Biol Chem 2003; 278:51469-78; PMID:14532264; http://dx.doi.org/ 10.1074/jbc.M308997200 [DOI] [PubMed] [Google Scholar]
- 25.Liu L, Ben-Shlomo H, Xu YX, Stern MZ, Goncharov I, Zhang Y, Michaeli S. The trypanosomatid signal recognition particle consists of two RNA molecules, a 7SL RNA homologue and a novel tRNA-like molecule. J Biol Chem 2003; 278:18271-80; PMID:12606550; http://dx.doi.org/ 10.1074/jbc.M209215200 [DOI] [PubMed] [Google Scholar]
- 26.Lustig Y, Wachtel C, Safro M, Liu L, Michaeli S. 'RNA walk' a novel approach to study RNA-RNA interactions between a small RNA and its target. Nucleic Acids Res 2010; 38:e5; PMID:19854950; http://dx.doi.org/ 10.1093/nar/gkp872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol 2011; 29:24-6; PMID:21221095; http://dx.doi.org/ 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2012; 14:178-92; PMID:22517427; http://dx.doi.org/ 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tkacz ID, Gupta SK, Volkov V, Romano M, Haham T, Tulinski P, Lebenthal I, Michaeli S. Analysis of spliceosomal proteins in Trypanosomatids reveals novel functions in mRNA processing. J Biol Chem 2010; 285:27982-99; PMID:20592024; http://dx.doi.org/ 10.1074/jbc.M109.095349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5:621-8; PMID:18516045; http://dx.doi.org/ 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-10; PMID:2231712; http://dx.doi.org/ 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 32.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science 1999; 283:1168-71; PMID:10024243; http://dx.doi.org/ 10.1126/science.283.5405.1168 [DOI] [PubMed] [Google Scholar]
- 33.Schattner P, Decatur WA, Davis CA, Ares M Jr., Fournier MJ, Lowe TM. Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res 2004; 32:4281-96; PMID:15306656; http://dx.doi.org/ 10.1093/nar/gkh768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myslyuk I, Doniger T, Horesh Y, Hury A, Hoffer R, Ziporen Y, Michaeli S, Unger R. Psiscan: a computational approach to identify H/ACA-like and AGA-like non-coding RNA in trypanosomatid genomes. BMC Bioinformatics 2008; 9:471; PMID:18986541; http://dx.doi.org/ 10.1186/1471-2105-9-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 1999; 288:911-40; PMID:10329189; http://dx.doi.org/ 10.1006/jmbi.1999.2700 [DOI] [PubMed] [Google Scholar]
- 36.Doniger T, Michaeli S, Unger R. Families of H/ACA ncRNA molecules in trypanosomatids. RNA Biol 2009; 6; PMID:19652533; http://dx.doi.org/ 10.4161/rna.6.4.9270 [DOI] [PubMed] [Google Scholar]
- 37.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 2000; 16:276-7; PMID:10827456; http://dx.doi.org/ 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- 38.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J 2003; 22:913-24; PMID:12574127; http://dx.doi.org/ 10.1093/emboj/cdg083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uliel S, Liang XH, Unger R, Michaeli S. Small nucleolar RNAs that guide modification in trypanosomatids: repertoire, targets, genome organisation, and unique functions. Int J Parasitol 2004; 34:445-54; PMID:15013734; http://dx.doi.org/ 10.1016/j.ijpara.2003.10.014 [DOI] [PubMed] [Google Scholar]
- 40.Barth S, Shalem B, Hury A, Tkacz ID, Liang XH, Uliel S, Myslyuk I, Doniger T, Salmon-Divon M, Unger R, et al.. Elucidating the role of C/D snoRNA in rRNA processing and modification in Trypanosoma brucei. Eukaryot Cell 2008; 7:86-101; PMID:17981991; http://dx.doi.org/ 10.1128/EC.00215-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 2013; 503:539-43; PMID:24185006; http://dx.doi.org/ 10.1038/nature12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnare MN, Gray MW. Sixteen discrete RNA components in the cytoplasmic ribosome of Euglena gracilis. J Mol Biol 1990; 215:73-83; PMID:2118960; http://dx.doi.org/ 10.1016/S0022-2836(05)80096-8 [DOI] [PubMed] [Google Scholar]
- 43.Moore AN, Russell AG. Clustered organization, polycistronic transcription, and evolution of modification-guide snoRNA genes in Euglena gracilis. Mol Genet Genomics 2011; 287:55-66; PMID:22134850; http://dx.doi.org/ 10.1007/s00438-011-0662-8 [DOI] [PubMed] [Google Scholar]
- 44.Michaeli S. rRNA Biogenesis in Trypanosomes In: Bindereif A, ed. RNA Metabolism in Trypanosomes. Belin, Heidelberg: Springer-Verlag, 2012:123-48. [Google Scholar]
- 45.Turowski TW, Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev RNA 2014; 6:129-39; PMID:25176256; http://dx.doi.org/ 10.1002/wrna.1263 [DOI] [PubMed] [Google Scholar]
- 46.McMahon M, Contreras A, Ruggero D. Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip Rev RNA 2014; 6:173-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al.. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014; 159:148-62; PMID:25219674; http://dx.doi.org/ 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang TH, Rozhdestvensky TS, d'Orval BC, Bortolin ML, Huber H, Charpentier B, Branlant C, Bachellerie JP, Brosius J, Huttenhofer A. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res 2002; 30:921-30; PMID:11842103; http://dx.doi.org/ 10.1093/nar/30.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 2000; 49:341-51; PMID:10902565; http://dx.doi.org/ 10.1080/152165400410182 [DOI] [PubMed] [Google Scholar]
- 50.Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry 1992; 31:1040-6; PMID:1310418; http://dx.doi.org/ 10.1021/bi00119a012 [DOI] [PubMed] [Google Scholar]
- 51.Schnare MN, Gray MW. Complete modification maps for the cytosolic small and large subunit rRNAs of Euglena gracilis: functional and evolutionary implications of contrasting patterns between the two rRNA components. J Mol Biol 2011; 413:66-83; PMID:21875598; http://dx.doi.org/ 10.1016/j.jmb.2011.08.037 [DOI] [PubMed] [Google Scholar]
- 52.Zamudio JR, Mittra B, Chattopadhyay A, Wohlschlegel JA, Sturm NR, Campbell DA. Trypanosoma brucei spliced leader RNA maturation by the cap 1 2′-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex. Mol Cell Biol 2009; 29:1202-11; PMID:19103757; http://dx.doi.org/ 10.1128/MCB.01496-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 2009; 34:47-57; PMID:19285445; http://dx.doi.org/ 10.1016/j.molcel.2009.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.