Abbreviations
- Cm
2′-O-methyl-cytidine
- dm5C
5-methyl-2-deoxyribocytidine
- dU
2′-deoxyuridine
- Gm
2′-O-methyl-guanosine
- LC-MS/MS
liquid chromatography-mass spectrometry/mass spectrometry
- LOD
limit of detection
- LOQ
limit of quantification
- m5C, mC
5-methylcytidine
- rm5C
5-methyl-ribocytidine
- Um
2′-O-methyl-uridine.
Introduction
Nucleoside methylations and other nucleic acid modifications have recently encountered a surge in interest, prompted, among other things, by the detection of methylation and active demethylation of DNA and mRNA by similar mechanisms. In DNA, deoxycytidine methylation by Dnmt enzymes generates 5-methyldeoxycytidine,1 an important epigenetic mark that typically causes inactivation of transcription of the methylated promoter region. Recent exciting developments have shown that these marks are not concrete-cast, but can be actively removed by the oxidative action of TET enzymes,2 which generate, through a series of 2-electron oxidations, first hydroxymethylcytidine (hm5C), then formyldeoxycytidine (f5C),3 and finally carboxydeoxycytidine (ca5C), which may eventually regenerate deoxycytidine by decarboxylation. The apparent functional homolog in mRNA is m6A, which appears to reduce translation efficiency. Here, too, the methylation can be removed by TET-related enzymes generating first hydroxymethyladenosine (hm6A), then formyladenosine (f6A).4,5,6 Also, the presence of 5-methylcytidine in mRNA has been reported early on, and has recently raised renewed interest, although its function is as yet unclear and putative conversion into hydroxymethylcytidine is yet to be demonstrated.32 These developments are enhanced by the development of highly sensitive detection methods7,8 including the adaptation of the so-called bisulfite sequencing from DNA, where it is well established,9 to RNA, where its application has significantly contributed to the present high level of interest.10,11,12 However, bisulfite sequencing alone does not yield unassailable results13,14 and we have thus looked to expand the limits of detection of m5C in both DNA and RNA by LC-MS/MS. Here we report a straightforward regimen that provides values for the limit of quantification (LOQ) in the triple digit attomol range. Its application to presumed negative controls, namely synthetic oligonucleotides, surprisingly detected significant amounts of m5C in both types of synthetic nucleic acids.
Results
Development and characteristics of a highly sensitive LC-MS/MS method
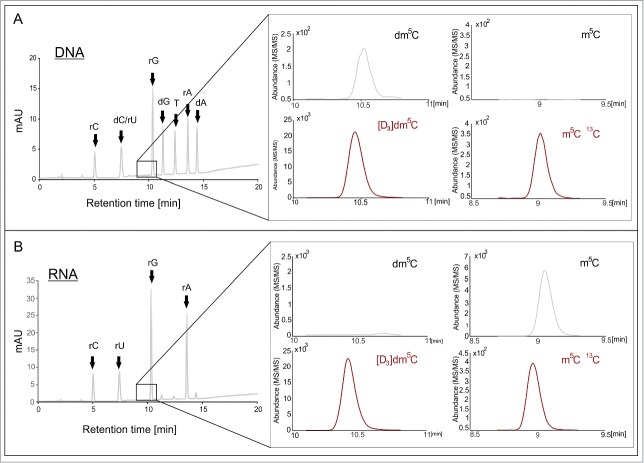
For the detection and quantification of m5C in both DNA and RNA, 2 related protocols were derived from a highly sensitive LC-MS/MS method15 which was based on conventional RP-18 chromatography and a triple quadrupole detector. A typical elution gradient elutes the nucleoside mixture obtained upon digestion of DNA or RNA by a mix of nucleases and phosphatase, as is shown in Figure 2. Optimized instrument parameters including in particular fragmentor voltage, collision energy and cell accelerator voltage are detailed in the methods section (Table 1). An exploratory run with this protocol, which had originally been designed to detect potential DNA methylation by Dnmt2,13,14 was conducted on commercial synthetic DNA oligonucleotides, which we had meant to be negative controls. Surprisingly, we detected significant amounts of m5C, which prompted us to conduct a more systematic quantification of m5C in oligonucleotides of commercial origin, including also RNA oligonucleotides. Figure 1A and 1B show the corresponding mass transitions. The elution profiles for RNA and DNA are presented in Figure 2.
Figure 2.
Method development for the detection and quantification of dm5C and rm5C by LC-MS/MS. (A) Elution profiles of DNA oligonucleotide hydrolysate, spiked with S. cerevisiae RNA SIL-IS and [D3]dm5C monitored by UV-absorption and mass chromatograms from LC- MS/MS. (B) Shows the elution profiles of a hydrolyzed RNA sample, spiked with both internal standards as well as the corresponding MS/MS data. (A and B) show distinct contamination of the samples with the matching m5C derivative.
Table 1.
QQQ parameters of the dynamic MRM method
| Modified nucleoside | Molecular Weight [Da] | Precursor ion [m/z] | Product ion [m/z] | Fragmentor Voltage [V] | Collision Energy [eV] | Cell accelerator voltage [V] | Retention time [min] |
|---|---|---|---|---|---|---|---|
| rm5C | 257 | 258 | 126 | 40 | 9 | 2 | 9.0 |
| rm5C 13C | 267 | 268 | 131 | 40 | 9 | 2 | 9.0 |
| dm5C | 241 | 242 | 126 | 60 | 5 | 2 | 10.4 |
| [D3]dm5C | 244 | 245 | 129 | 60 | 5 | 2 | 10.7 |
Figure 1.
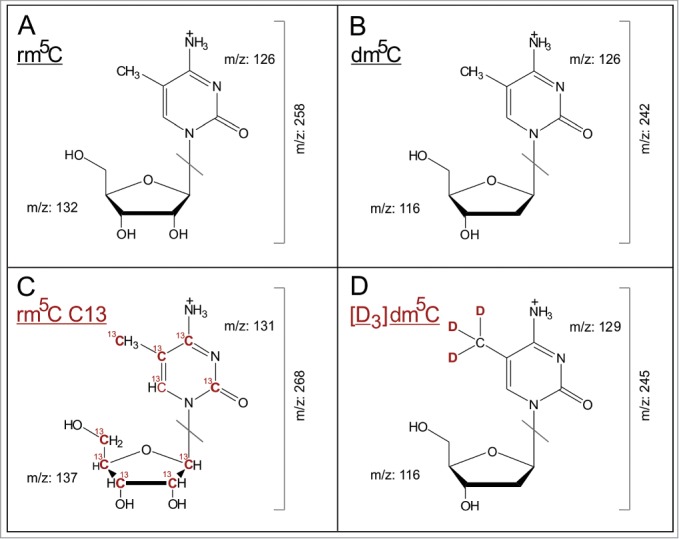
Mass transitions of the analyzed modified nucleosides. (A–D) show the [M+H]+ structures of analyzed nucleosides and their stable isotope labeled internal standards, as well as the collision induced dissociation fragmentation during MS/MS analysis.
To ascertain the limit of quantification (LOQ) and limit of detection (LOD), quantification of genuine 5-methyl-2-deoxyribocytidine (dm5C) in DNA and of 5-methyl-ribocytidine in RNA (rm5C), authentic samples of these substances were weighed and dissolved to obtain a dilution series for external calibration. Supplemental Figure 1 shows double logarithmic plots of amount of substance versus MS signal for both compounds. Insets on the left show the peak for 200 amol dm5C, the lowest amount used for calibration, with a signal-to-noise ratio (S/N) of 13.1, which is well above the definition of the limit of quantification (LOQ) of S/N>10. For rm5C, S/N at 500 amol was 45.2, showing that subsequent quantifications were accurate with LOQs in the triple digit attomol range, which is keeping with latest reports.16 For all calibrations, linear regression coefficients (R2) were better than 0.98.
Detection and quantification of m5C in synthetic oligonucleotides of commercial origin
From a collection of about several hundred oligonucleotides acquired from various commercial sources for different unrelated projects, one to 3 oligonucleotides per commercial supplier and nucleic acid species were randomly chosen. These oligonucleotides, both RNA and DNA, were of synthetic origin, i.e. synthesized by conventional solid phase phosphoramidite chemistry,17 except for one RNA oligonucleotide from Thermo/Dharmacon, which, according to the manufacturer's information, had been synthesized by ACE chemistry.18,19 For each oligonucleotide, an aliquot was digested to mononucleosides by sequential treatments with nucleases and phosphatase, according to established protocols.20,21 Upon injecting amounts corresponding to 20 pmol cytosine per injection, contained quantities of m5C were measured as an LC-MS/MS signal and converted to absolute amounts of substance. To perform quantification according to the state-of-the-art, we implemented the use of stable isotope labeled-internal standards (SIL-IS) as depicted in Figure 1C and 1D. For dm5C, the corresponding SIL-IS had been made by chemical synthesis,31 while that for rm5C was obtained from yeast cultures according to a recently published method.15 The UV signal of (deoxy-) cytidine in conjunction with an analogous calibration (R2 = 1, Fig. S2) was used to determine the precise injected amount of substance of this nucleoside, such that the ratio of m5C/C could be determined in each sample.
Most surprisingly, our method, which was set to probe for both, dm5C and rm5C, detected dm5C in all DNA samples, and rm5C in all RNA samples, albeit in strongly varying amounts. No rm5C was detected in DNA and inversely, no dm5C was detected in RNA. Supplemental Figure 1 illustrates that the quantification range for m5C in both types of oligonucleotides was largely superior to the LOQ, so the sensitivity of method and equipment were more than sufficient for accurate quantification. Figure 4A shows that the dm5C content varied from ppm to percent among 6 commercial suppliers, where DNA oligonucleotides from suppliers 1, 2 and 3 showed detectable, but rather low dm5C amounts in the double to triple digit ppm range. Dm5C amounts in oligonucleotides from suppliers 4, 5 and 6 ranged from 0.1 up to 3%. Quantification of rm5C in RNA turned up variations from double/ triple digit ppm range (oligonucleotides from sources 1 and 4) over low percentile range (oligonucleotides from suppliers 1, 2, 3, 7, 8) up to single digit percentiles for suppliers 4 and 9.
Figure 3.
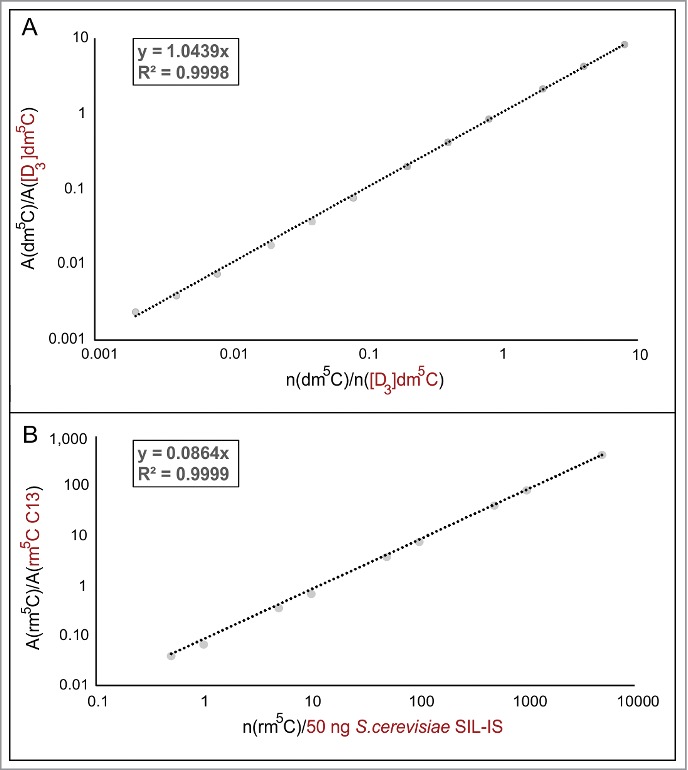
Calibration of stable isotope labeled-internal standards (SIL-IS). (A) Various amounts of pure dm5C nucleoside were spiked with a constant molar amount (250 fmol) of chemically synthesized [D3]dm5C. The slope of the double logarithmic plot of the ratio of the areas and the amount of nucleoside can be used to calculate the unknown amount of dm5C in the sample. (B) Calibration of S. cerevisiae SIL-IS, containing 13C labeled rm5C, was performed in the same manner. Instead of a constant molar amount of SIL-IS a constant mass of 50 ng was used.
Figure 4.
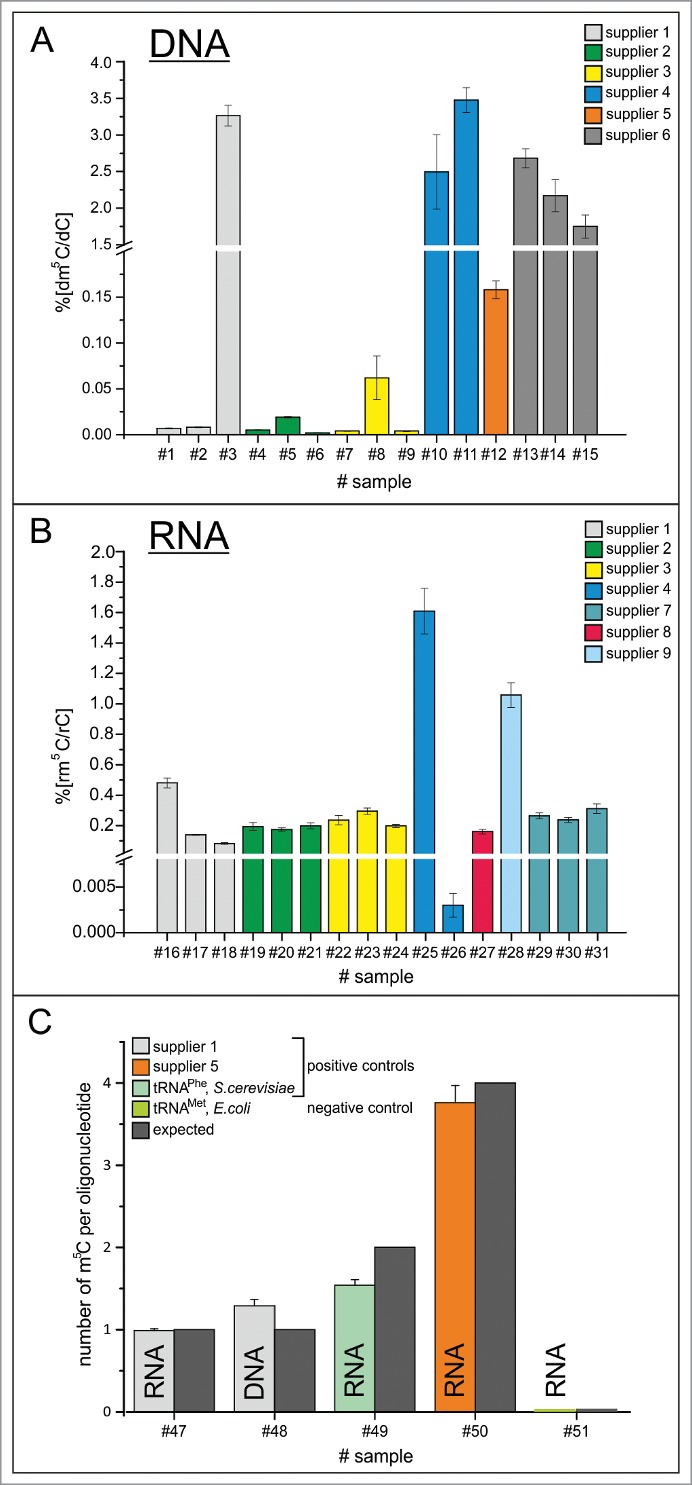
Quantification results of dm5C and rm5C. (A) Ratios of dm5C/dC in commercial oligonucleotides from 6 different commercial suppliers. (B) Ratios of rm5C/rC in commercial oligonucleotides from 6 different commercial suppliers and one academic lab. (C) Absolute quantification of m5C per oligonucleotide on RNA and DNA of supplier 1, commercially supplied tRNAPhe of S.cerevisae and RNA of supplier 5, as well as isolated tRNAMet, E. coli as negative control. Oligonucleotides #47–50 contain m5C. The expected amount is indicated by the dark gray bars.
Controls
The quantifications presented show standard deviations from 3 technical replicates of the same oligonucleotide digest. Further guards against experimental pitfalls included blank runs between samples, which proved the absence of any nucleoside carryover between runs. Measurements on a subset of oligonucleotides have been conducted at 3 distinct time points over a period of 18 months, and by 3 different people. The first 2 sets of measurements were conducted with external calibration, but the most recent used our SIL-IS approach. The m5C contents varied within a range expected according to our published experience with both calibration techniques.15 Of note, however, the relative content among the various oligonucleotides was reproduced faithfully over the timespan of a year.
As positive controls, bona fide m5C-containing nucleic acids were analyzed both of synthetic and of biological origin (Fig. 4C). For synthetic oligonucleotides, quantification of m5C was found to be in good agreement with the m5C content predicted based on nucleoside composition of the sequence. Furthermore, tRNAPhe from Saccharomyces cerevisiae, which is known to contain 2 m5C sites, showed roughly 1.5 mol m5C per mol tRNA, suggesting incomplete modification in the physiological setting of the original yeast culture.
Furthermore several negative controls have been included. Interestingly, both CTP and dCTP triphosphates showed m5C contents barely above LOD, but below LOQ. Consequently, an in vitro transcribed RNA showed a similarly low rm5C content (below 0.1 per mille). Since Escherichia coli tRNA is not known to contain any m5C residues, we isolated tRNAMet from commercially available total tRNA of E. coli culture, using a biotinylated cDNA. Significantly, this tRNA did not contain any detectable rm5C.
A series of oligoribonucleotides without any cytidines was designed to contain only 2 nucleoside species. Three permutations thus yielded oligoribonucleotides composed of (i) rA and rU (ii)rA and rG, and (iii) rG and rU, respectively. The analog series of oligodeoxyribonucleotides consequently containing compositions (i) dA/dT (ii) dA/dG and (iii) dG/dT. Importantly, none of these oligonucleotides contained detectable amounts of rm5C nor of dm5C, clearly associating m5C to the presence of (deoxy-)cytidine in the oligonucleotides both for DNA and for RNA.
Discussion
Appropriate analytics
Detection and concurrent quantification of nucleosides with high sensitivity by LC-MS/MS techniques typically relies on a selection step of the ionized nucleoside, followed by a collision induced fragmentation step, and finally on a detection step of the ionized nucleobase fragment. In the gold standard equipment configuration for this purpose, each of these steps is performed in a quadrupole element.33,34 Of note, the mass spectra for method development were recorded in a scanning mode to determine optimal collision parameters in the second quadrupole to maximize the so called product ion. In quantification mode, the detector is exclusively focused on the continuous detection of the latter. Coincidence of chromatography retention time and mass spectrometric fragmentation patterns are commonly accepted proof of identity, which we confirmed with commercially available samples of C, dC, dm5C, and rm5C. Embodiment of these principles differs strongly for each given instrument; even between to triple-quads of the same company. Hence, parameters such as e.g. collision energy must be adapted de novo in every lab. However, several labs consistently report LOD and LOQ values comparable to the ones we obtained.33,34 We conclude that we have performed state-of-the art detection and quantification of m5C in DNA and RNA by internal calibration.15
Potential origin
A number of controls were conducted to ascertain that the source of detected m5C was indeed the synthetic oligonucleotides under investigation. Most significantly, oligoribonucleotides that did not contain cytosines in their sequence showed neither rm5C nor dm5C, and neither did oligodeoxyribonucleotides of similar composition. This associates the occurrence of m5C to the use of their respective chemical building blocks during solid phase synthesis, and essentially rules out the possibility that low levels of m5C might originate from thymidine amination during a deprotection step of the exocyclic amine functions using ammonia. We have analyzed an oligoribonucleotide synthesized in a renowned academic laboratory, and find low, but detectable levels of rm5C, that are similar to those of several commercial suppliers. This suggests that the principle problem is not in proficiency and state-of-the art solid phase oligonucleotide synthesis, but might rather be related to the source of building blocks for the latter. Note that for most suppliers similar values were obtained with different oligonucleotides, expect in one case (Fig. 4B), where the significant difference between 2 oligonucleotides might conceivably originate from different phosphoramidite batches.
Potential impact
Although the above is rather standard in nucleoside quantification, we are the first to detect trace amounts of 5-methylcytosine in all the investigated oligonucleotides. Considering that this modified nucleoside is liable to decrease or entirely suppress gene expression, this may be of significance in experiments using synthetic genes. While dm5C contents in the ppm range are unlikely to have any effect, contents of up to 3%, as detected in DNA oligonucleotides from 3 suppliers, may well have a significant impact on gene expression e.g., in transformation and transfection experiments. Of potential interest might also be an impact of dm5C in recognition by the mammalian innate immune system. For example, recognition by TLR9 of so-called CpG motifs and subsequent signaling are reported to be suppressed by the presence of dm5C.22 For RNA, the presence of randomly incorporated rm5C residues in mRNA has been similarly reported to suppress immune stimulation and to enhance translation.23,24 Further effects of m5C in RNA25 include contribution to metabolic stability26,27 and protection against degradation by nucleases.28 Since, in general, 5-methylated pyrimidines stabilize helical structures in nucleic acids, presumably by increased base-stacking25,29,30 high content of randomly incorporated m5C in the single digit percentile range might potentially bias duplex melting experiments, e.g. broadening hyperchromatic transitions in UV-melting experiments.
Material and Methods
Isolation of tRNAMet species from E. coli
tRNAMet was isolated from E. coli total tRNA (Roche, Basel, Switzerland) by hybridization with a complementary, biotinylated DNA-oligonucleotide (sequence: 5′-biotin-AAATGGTGGCTACGACGGGATTCGAACCTGTGACCCCATCATTATGAGTGATGTGCTCTAACCAACGAGCTACGTAGCC-Atto 488–3′, IBA, Goettingen, Germany) followed by immobilization on streptavidin-coated magnetic beads (Dynabeads® MyOne™ Streptavidin T1, Life Technologies, Darmstadt, Germany). The hybridization step was performed in 5x SSC buffer (20x: 3 M NaCl, 300 mM trisodium citrate, pH 7.0) using 100 pmol biotinylated oligonucleotide and 150 µg total tRNA per 25 µL beads. Samples were denatured at 90°C for 3 min, subsequently hybridized at 65°C for 10 min and cooled to room temperature. Dynabeads® were washed 3 times using Binding & Washing buffer (5 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 1 M NaCl) according to the manual and then equilibrated once in 5x SSC buffer before adding the hybridized samples. Immobilization of the hybrid was performed at 25°C under shaking for 30 min. Subsequently, the supernatant containing non-target tRNAs was removed and the beads were washed once in 1x SSC buffer and 3 times in 0.1x SSC buffer. Finally, the beads were resuspended in MilliQ water and heated to 75°C for 3 minutes to elute the target tRNA. In order to remove any remaining DNA-oligonucleotide from the eluted tRNA, a DNase I (Fermentas, St. Leon-Roth, Germany) digestion step was performed at 37°C in DNase I buffer (Fermentas, St. Leon-Roth, Germany) for 2 hours followed by 10 % denaturing polyacrylamide gel electrophoresis and ethanol precipitation.
Preparation of oligonucleotides for LC-MS/MS analysis
DNA- and RNA-oligonucleotides synthesized by the following suppliers were tested: Biomers (Ulm, Germany), BioTeZ (Berlin, Germany), Eurofins Genomics (Ebersberg, Germany), IBA (Goettingen, Germany), MWG Biotech (Ebersberg, Germany), Sigma Aldrich (Munich, Germany), Thermo Scientific (Schwerte, Germany). One non-commercial RNA oligonucleotide was synthesized and provided by the Micura lab (Innsbruck, Austria). Native tRNAs (methionine specific from E. coli, phenylalanine specific from S. cerevisiae) as well as deoxy- and ribocytidinetriphosphates were purchased from Sigma-Aldrich (Munich, Germany) respectively Thermo Scientific (Schwerte, Germany). More detailed information for all analyzed nucleic acids are shown in Supplemental Table 1. As a control CTP/ dCTP (Fermentas, St. Leon-Roth, Germany) were treated the same way as the oligonucleotides.
Prior to LC-MS/MS analysis, the DNA- and RNA-Oligonucleotides were digested into nucleosides using the following protocol: 100 pmol of each oligonucleotide were incubated at 37°C for 2 h in the presence of 0.3 U nuclease P1 (Roche Diagnostics, Mannheim, Germany), 1/10 vol. of 10 x nuclease P1 buffer (0.2 M NH4OAc pH 5.0, ZnCl2 0.2 mM) and 0.1 U snake venom phosphodiesterase (Worthington, Lakewood, USA). Afterwards 1/10 vol. of 10 x fast alkaline phosphatase buffer (Fermentas, St. Leon-Roth, Germany) and 1 U fast alkaline phosphatase (Fermentas, St. Leon-Roth, Germany) were added, followed by an incubation at 37°C for 1 h. Of each sample, aliquots corresponding to 20 pmol rC/dC (calculated for the intact oligonucleotide according to the manufacturer's quantification) were submitted to LC-MS/MS analysis.
Procedure for LC-MS/MS measurements
The prepared nucleoside samples were analyzed on an Agilent 1260 series equipped with a diode array detector (DAD) and a Triple Quadrupole mass spectrometer (Agilent 6460). A Synergy Fusion RP column (4 μm particle size, 80 Å pore size, 250 mm length, and 2 mm inner diameter) from Phenomenex (Aschaffenburg, Germany) was used at 35°C. The solvents consisted of 5 mM ammonium acetate buffer adjusted to pH 5.3 using acetic acid (solvent A) and pure acetonitrile (solvent B). The elution (flow rate 0.35 mL/min) started with 100 % solvent A followed by a linear gradient to 8 % solvent B at 10 min. Solvent B was increased further to 40% over 10 minutes. Over the next 3 minutes Solvent B was decreased back to 0%. Initial conditions were regenerated by rinsing with 100 % solvent A for additional 7 minutes. For detection of the major nucleosides, the effluent from the column was first measured photometrically at 254 nm by the DAD, and modified nucleosides were then detected by the mass spectrometer equipped with an electrospray ion source (Agilent Jet Stream). ESI parameters were as follows: gas temperature 350°C, gas flow 8 L/min, nebulizer pressure 50 psi, sheath gas temperature 350°C, sheath gas flow 12 L/min, capillary voltage 3500 V. The MS was operated in the positive ion mode using Agilent Mass Hunter software in the DMRM (dynamic multiple reaction monitoring) mode. The monitored mass transitions, instrument settings and retention time windows can be seen in Table 1. The peak areas were determined by employing Agilent MassHunter Qualitative Analysis Software. In the case of the major nucleosides, peak areas were extracted from the recorded UV chromatograms in order to avoid saturation of the mass signals. The peak areas of the modified nucleosides were measured by LC-MS/MS. All samples were analyzed in technical triplicates.
Determination of dC/rC amount
Deoxycytidine (dC) and cytidine (C) (Sigma-Aldrich, Munich, Germany) were dissolved in pure water to a final concentration of 10 mM. These stock solutions were then used to prepare 4 calibration solutions of 100, 10, 1 and 0.1 µM. By varying the injection volume a calibration curve from 2–700 pmol dC, respectively rC could be obtained. Using the linear equation of this curves, the dC/rC amount in the samples could be assigned via the area under the curve (AUC) of the (deoxy-)cytidine UV-peaks. The spiked amount of 13C RNA was measured separately. Due to co-elution of the 13C cytidine and 12C cytidine (Fig. 1) in the samples the AUC of 13C cytidine has to be subtracted to obtain the AUC of 12C cytidine in the samples. The same procedure was applied to rU and dC (Fig. 1).
Quantification of rm5C using stable isotope labeled internal standard of S. cerevisiae
In order to quantify the rm5C content of the RNA samples, 13C-labeled total RNA from S. cerevisiae was used as a stable isotope-labeled internal standard (SIL-IS) as described for total RNA from E. coli previously.15 Briefly, 9 calibration solutions containing 0.01–500 fmol/µL rm5C (Sigma Aldrich, Munich, Germany) and 50 ng/µL SIL-IS were prepared and analyzed by LC-MS/MS (injection volume 10 µL/sample). For determination of a nucleoside-isotope response factor for rm5C, the ratio of the extracted areas of the 12C-rm5C and 13C-rm5C peaks was calculated for each calibration solution. The resulting response factor from the linear equation was then used for rm5C quantification in the RNA samples.
Quantification of dm5C, using chemically synthesized [D3]dm5C internal standard
Twelve different ratios of the unlabeled dm5C (purchased from Sigma-Aldrich, Steinheim, Germany) and stable isotope labeled [D3]dm5C (generous gift from Carell lab, Munich) have been measured, while the amount of the deuterated nucleoside was constant with 2.5 fmol/ injection. The results were plotted as a linear calibration curve where the ratio of the AUC of the LC-MS/MS chromatograms (unlabeled/labeled) was applied to the y-axis and the ratios of the molar amounts to the y-axis. The gradient of these curve was used for the calculation of the modified nucleoside's amount in the sample.31
The obtained results were then utilized to calculate the percentage of dm5C per dC.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ronald Micura (Innsbruck) for providing a reference oligoribonucleotide, and Thomas Carell (Munich for [D3]dm5C.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
Funding
This work was supported by the DFG in the framework of the SPP 1784 (HE 3397/14 to MH).
References
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 2005; 74:481-514; PMID:15952895; http://dx.doi.org/ 10.1146/annurev.biochem.74.010904.153721 [DOI] [PubMed] [Google Scholar]
- 2.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 2013; 14:341-56; PMID:23698584; http://dx.doi.org/ 10.1038/nrm3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O, et al.. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol 2014; 10:574-81; PMID:24838012; http://dx.doi.org/ 10.1038/nchembio.1532 [DOI] [PubMed] [Google Scholar]
- 4.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genetics 2014; 15:293-306; PMID:24662220; http://dx.doi.org/ 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 5.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet 2013; 29:108-15; PMID:23218460; http://dx.doi.org/ 10.1016/j.tig.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al.. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun 2013; 4:1798; PMID:23653210; http://dx.doi.org/ 10.1038/ncomms2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201-6; PMID:22575960; http://dx.doi.org/ 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 8.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012; 149:1635-46; PMID:22608085; http://dx.doi.org/ 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinders J, Paszkowski J. Bisulfite methylation profiling of large genomes. Epigenomics 2010; 2:209-20; PMID:22121871; http://dx.doi.org/ 10.2217/epi.10.6 [DOI] [PubMed] [Google Scholar]
- 10.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 2009; 37:e12; PMID:19059995; http://dx.doi.org/ 10.1093/nar/gkn954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 2013; 9:e1003602; PMID:23825970; http://dx.doi.org/ 10.1371/journal.pgen.1003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 2012; 40:5023-33; PMID:22344696; http://dx.doi.org/ 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer M, Lyko F. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nat Genet 2010; 42:920-1; author reply 1; PMID:20980983; http://dx.doi.org/ 10.1038/ng1110-920 [DOI] [PubMed] [Google Scholar]
- 14.Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat Genet 2009; 41:696-702; PMID:19412177; http://dx.doi.org/ 10.1038/ng.360 [DOI] [PubMed] [Google Scholar]
- 15.Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res 2014; 42(18):e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capuano F, Mulleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem 2014; 86:3697-702; PMID:24640988; http://dx.doi.org/ 10.1021/ac500447w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy S, Caruthers M. Synthesis of DNA/RNA and their analogs via phosphoramidite and H-phosphonate chemistries. Molecules 2013; 18:14268-84; PMID:24252996; http://dx.doi.org/ 10.3390/molecules181114268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartsel SA, Kitchen DE, Scaringe SA, Marshall WS. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods Mol Biol 2005; 288:33-50; PMID:15333896 [DOI] [PubMed] [Google Scholar]
- 19.Scaringe SA, Kitchen D, Kaiser RJ, Marshall WS. Preparation of 5′-silyl-2′-orthoester ribonucleosides for use in oligoribonucleotide synthesis. Methods Mol Biol 2005; 288:33-50; PMID:18428924 [DOI] [PubMed] [Google Scholar]
- 20.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol 1990; 193:782-90; PMID:1706062; http://dx.doi.org/ 10.1016/0076-6879(90)93450-Y [DOI] [PubMed] [Google Scholar]
- 21.Kellner S, Kollar LB, Ochel A, Ghate M, Helm M. Structure-function relationship of substituted bromomethylcoumarins in nucleoside specificity of RNA alkylation. PloS one 2013; 8:e67945; PMID:23844135; http://dx.doi.org/ 10.1371/journal.pone.0067945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al.. A Toll-like receptor recognizes bacterial DNA. Nature 2000; 408:740-5; PMID:11130078; http://dx.doi.org/ 10.1038/35047123 [DOI] [PubMed] [Google Scholar]
- 23.Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 2008; 16:1833-40; PMID:18797453; http://dx.doi.org/ 10.1038/mt.2008.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalpke A, Helm M. RNA mediated Toll-like receptor stimulation in health and disease. RNA Biol 2012; 9:828-42; PMID:22617878; http://dx.doi.org/ 10.4161/rna.20206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motorin Y, Lyko F, Helm M. Five-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res 2010; 38:1415-30; PMID:20007150; http://dx.doi.org/ 10.1093/nar/gkp1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 2012; 19:900-5; PMID:22885326; http://dx.doi.org/ 10.1038/nsmb.2357 [DOI] [PubMed] [Google Scholar]
- 27.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al.. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 2014; PMID:25063673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010; 24:1590-5; PMID:20679393; http://dx.doi.org/ 10.1101/gad.586710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayrapetyan A, Seidu-Larry S, Helm M. Function of modified nucleosides in RNA stabilization. In Grosjean H, Ed., DNA and RNA Modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution. Austin, TX: Landes Bioscience, 2006; pp. 550-9. [Google Scholar]
- 30.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2011; 2:611-31; PMID:21823225; http://dx.doi.org/ 10.1002/wrna.79 [DOI] [PubMed] [Google Scholar]
- 31.Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T, Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 2010 Dec 23; 5(12):e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, Pfeifer GP, Xu GL, Wang Y. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc 2014 Aug 20; 136(33):11582-5; http://dx.doi.org/ 10.1021/ja505305z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capuano F, Mülleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem. 2014 Apr 15; 86(8):3697-702; http://dx.doi.org/ 10.1021/ac500447w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber SM, van Delft P, Mendil L, Bachman M, Smollett K, Werner F, Miska EA, Balasubramanian S. Formation and abundance of 5-hydroxymethylcytosine in RNA. Chembiochem 2015 Mar 23; 16(5):752-5; http://dx.doi.org/ 10.1002/cbic.201500013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domingo O, Hellmuth I, Jäschke A, Kreutz C, Helm M. Intermolecular ‘cross-torque’: the N4-cytosine propargyl residue is rotated to the ‘CH’-edge as a result of Watson-Crick interaction. Nucleic Acids Res 2015 Jun 23; 43(11):5275-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.