ABSTRACT
Transcription termination delineates the 3′ ends of transcripts, prevents otherwise runaway RNA polymerase (RNAP) from intruding into downstream genes and regulatory elements, and enables release of the RNAP for recycling. While other eukaryotic RNAPs require complex cis-signals and/or accessory factors to achieve these activities, RNAP III does so autonomously with high efficiency and precision at a simple oligo(dT) stretch of 5–6 bp. A basis for this high density cis-information is that both template and nontemplate strands of the RNAP III terminator carry distinct signals for different stages of termination. High-density cis-information is a feature of the RNAP III system that is also reflected by dual functionalities of the tRNA promoters as both DNA and RNA elements. We review emerging developments in RNAP III termination and single strand nontemplate DNA use by other RNAPs. Use of nontemplate signals by RNAPs and associated transcription factors may be prevalent in gene regulation.
KEYWORDS: Nontemplate strand, RNA Polymerase, RNA Polymerase III, RPC11, RPC37, RPC53, transcription termination
Abbreviations
- RNAP
RNA polymerase
- C37
RPC37
- PTC
pre-termination complex
Background and context
All of the multisubunit RNAPs are evolutionarily related, comprised of 5 (bacterial), 12 (Archaeal RNAP and RNAP II), 14 (RNAP I) or 17 (RNAP III) subunits.1 After initiation and promoter clearance, RNAPs form very stable elongation complexes (ECs) which promote synthesis of long RNAs and prevent costly premature or spontaneous termination.2 Accordingly, programmed termination of transcription by RNAPs proceed through graded mechanical adjustments that slow and destabilize the elongating enzyme and ultimately release it from the DNA template and the nascent RNA.3 Bacterial RNAP requires a 2-part cis-acting termination recognition signal comprised of an A-rich tract in the template DNA strand closely preceded by a G+C-rich inverted dyad repeat (Fig. 1A) that forms a hairpin in the nascent transcript, and a compound mechanism by the polymerase that serves to destabilize the EC at multiple contact points, for intrinsic, i.e., factor-independent, termination.3 Although key details of the molecular mechanisms involved in this process as well as for factor-dependent termination by bacterial RNAP remain uncertain, an excellent review is available.3 Briefly, the RNAP pauses upon incorporation of a run of U's corresponding to the A-rich tract, and concurrent sensing of formation of the upstream RNA hairpin in the RNA exit channel of the RNAP causes alterations that contribute to disruption of the oligo(rU:dA). Factor-mediated termination by bacterial RNAP uses Rho-helicase, a RNA-binding protein that associates with a C-rich region of the nascent transcript and then tracks along the transcript like a torpedo until it contacts the elongating polymerase and mediates effects that destabilizes the EC that has stopped at a pause site and thereby promotes termination.3
Figure 1.
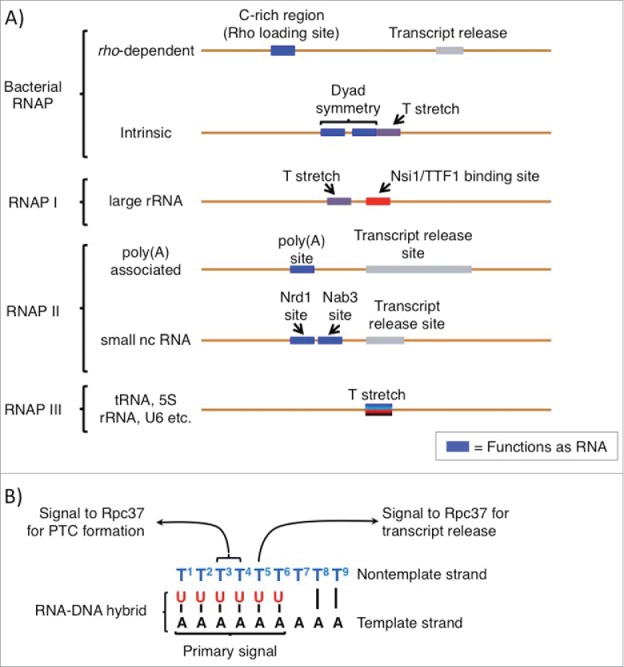
Cis-acting transcription termination signals for different multisubunit RNA polymerases. (A) Cis-termination signals in DNA for the different RNA polymerases indicated on the left are shown. Characteristics of the elements are labeled. Those shown as blue boxes function as RNA, as sequence-specific protein binding sites or as secondary structure elements. For descriptions of trans-acting factors, Nrd1 and Nab3, and TTF-1, see Porrua and Libri2 and Richard and Manley.4 In general, for the T stretches indicated as such, they are the transcript release sites; the others are less well defined and indicated as gray rectangles and can be spread over some distance. (B) Different components of the RNAP III oligo(dT) terminator and the sequence-specific signals and determinants referred to in the text are indicated.
In eukaryotes, the 3 major classes of genes that produce, large rRNA, mRNA and tRNA are transcribed by RNAPs I, II and III respectively.4,5 RNAPs I and II also use multipartite cis-termination signals, RNA-binding factors and 5′–3′ exonucleases that track the polymerases according to the torpedo model.2,4 Moreover, depending on the type of RNAP II-transcribed gene, poly(A)-associated or small ncRNA (Fig. 1A), different classes of cis-elements trigger termination (for extensive review see ref 2). Briefly, RNAP II termination of poly(A) pre-mRNA involves multiple ancillary factors, and the termination mechanism is tightly linked to cleavage of the nascent RNA ∼50 nucleosides downstream of the AATAAA poly(A) addition site (Fig. 1A).2 Note that by this mechanism the actual termination site at which the RNAP releases from the template can be variable distances downstream from the poly(A) site (Fig. 1A).2 The small ncRNA RNAP II termination pathway utilizes different cis-signals as well as the Nrd1 and Nab3 trans-acting factors (Fig. 1A, for extensive review see ref 2). By comparison to the other RNAPs, RNAP III has a most simple cis-termination signal, a short oligo(T) stretch4 that functions without need for an upstream hairpin6 (Fig. 1A), and the mechanism provides for decisive, precise and efficient termination.
Similar to bacteria, the archaea also contain a single RNAP that must transcribe all gene classes although with 12 subunits its composition is much more like RNAP II.1 Archaeal RNAP and RNAP III share a similar feature of termination, the cis-element of an oligo(T) stretch, although their mechanisms of termination differ in certain other regards.7 Significantly unlike RNAP III, termination by Archaeal RNAP is confounded by the fact that it must suppress termination at the large number of oligo(T) stretches found within Archaeal genes.7 Among other possibilities this suggests that additional cis-information can modulate the termination function of oligo(T) sequences in Archaea, although the mechanisms involved remain unknown.8 We are left with the conclusion that of all the multisubunit RNAPs, RNAP III indeed appears to have the most simple and direct link between its cis-termination signal and its termination mechanism.
What characteristics distinguish the RNAP III system toward such relative efficiency and apparent simplicity of its termination mechanism? One clue is that it is the only eukaryotic RNAP that exclusively synthesizes short RNAs. As the hundreds of tRNA genes that occupy most eukaryotic genomes are its most abundant substrates, its major products are limited to about 100 nt in length, and the small number of longer substrate RNAs are no more than a few hundred nt. One feature of a short length RNA that allows a simple oligo(dT) motif to serve as a cis-element for termination, is avoidance of that sequence motif in the body of the transcript, as is the case for the tRNAs and other RNAP III products (this becomes less likely as transcript length and complexity increases). Thus, one can imagine that as part of the evolutionary specialization of RNAP III for short transcripts, the risk of premature truncation was low relative to the benefit of a facile termination mechanism, especially if this can be linked to an efficient recycling or reinitiation mechanism, as is the case for RNAP III.9 The tRNAs are required in high levels to fuel protein synthesis in rapidly growing cells. An estimation from tRNA levels in yeast indicates that RNAP III generates ∼2 transcripts per gene per second, making it the most efficient transcription reinitiation machine.10 The strategies used by RNAP III for such high efficiency appear to have included great stability of DNA-bound transcription factors to sustain innumerable rounds of transcription reinitiation (reviewed in 9) and acquisition of integral RNAP subunits that perform functions carried out by dissociable transcription factors for other RNAPs.11,12 Although these are some of features that streamline the RNAP III transcription cycle for high efficiency (reviewed in 9), it seems likely that others, including specialization of the active center (see below), remain to be elucidated.
The high density of cis-information in the RNAP III system is not only found in the terminator but also extends to the tRNA gene promoter elements. Most notably, these A and B box elements are gene-internal13 (Fig. 2A) and as such minimize the size of the transcription unit as compared to genes transcribed by other RNAPs whose control regions typically extend far upstream. Second, as internal sequence motifs they are of dual use, both as DNA elements to which different subunits of transcription factor IIIC bind14 (Fig. 2B), and as conserved structural elements of the tRNAs themselves, the D loop and T loop (green and yellow respectively in Figs. 2A-C). Of somewhat similar duality is the oligo(dT) terminator which demarcates the 3′ border of the TFIIIC complex in chromatin15 and produces the nascent RNA 3′ oligo(U) ends (blue color circles in Fig. 2C) to which the UUU 3′-OH-sequence-specific binding protein known as La binds and serves as chaperone during RNA processing.16
Figure 2.
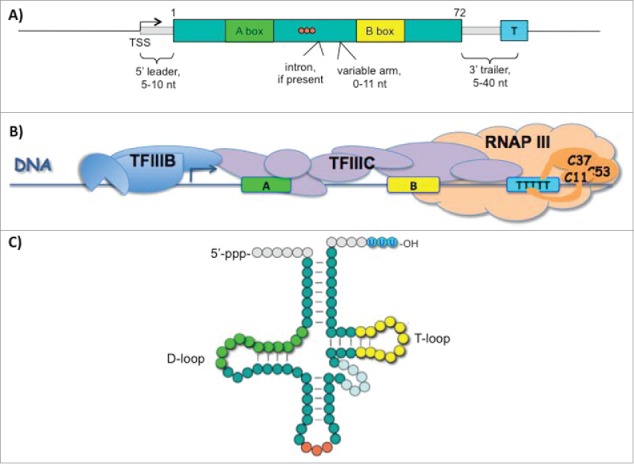
High-density of cis-information in the RNAP III transcription promoter and terminator control system; dual functions as tRNA elements. (A) Typical tRNA gene with internal A and B box promoter and terminator control elements, color-coded to match B & C, is shown.13 (B) Highly schematic representation of transcription factors (TF) IIIB, TFIIIC and RNAP III bound to a tRNA gene (for more high resolution see14). RNAP III is depicted as terminating transcription at the oligo(dT) stretch, with the termination subcomplex (C53/C37/C11) portrayed in darker color. (C) Schematic representation of unprocessed nascent precursor-tRNA with control elements color-coded as in A; orange circles represent the anticodon (see text).
However, recent data indicate that the cis-information density of the RNAP III terminator is greater than expected and perhaps more so than for any other RNAP system, as its template and nontemplate strands contain distinct signals to direct termination.17 Below we focus on recent analyses that show that the oligo(dT) terminator contains 3 signals to direct RNAP III termination; while the template strand oligo(dA) acts as a principal destabilizing signal, specific nontemplate strand dT residues direct 2 phases of termination, pausing of a metastable pretermination complex (PTC), and transcript release. First however, we will briefly review some principles of transcription termination that have been learned mostly from studies of bacterial RNAP (see 3).
Basics of transcription termination
Transcription elongation complexes are highly dynamic yet stable as they move along the DNA adding nucleotides at a fast rate (e.g., ∼50 nt/second), and very resistant to premature release of the transcripts or the template DNA. Several features enable this: grip on the downstream DNA in a cleft of the RNAP by its mobile clamp domain, interaction between the growing RNA and amino acid side chains in the RNA exit channel of the RNAP, and a grasp on the 8–9 bp RNA:DNA hybrid that is maintained in the active center of the RNAP.18,19 However, very important to the stability of the complex is the collective thermodynamic strength of the base pairs of the RNA:DNA hybrid itself.3 Upon encountering oligo(dT), i.e., oligo(dA) on the template strand, an elongating RNAP forms an oligo(rU:dA) hybrid in the active center (e.g., see Fig. 1B). Since dT-rich or oligo(dT) sequences comprise bacterial RNAP and RNAP III terminators, it was suggested that the unique thermodynamic instability of the oligo(rU:dA) hybrid, which far exceeds that of any other sequence combination, would be an important determinant of termination,20 consistent with current models for termination by RNAP III and other RNAPs.
RNAP III is hypersensitive to termination
The core subunits of the multisubunit RNAPs that comprise their active centers (the 2 largest subunits) that are sensitive to termination share sequence and structural homology with RNAP III (reviewed in 5). These RNAPs also share similar mechanisms of transcription initiation.11 While this suggests that similar principles should apply to RNAP III termination as have been elucidated for bacterial RNAP, the data on RNAP III suggests that it has been sensitized for facile and efficient termination, including an active center structure that is less binding and more sensitive to the oligo(rU:dA) hybrid than the other RNAPs.5,21 In addition, the C37 subunit contributes to termination apparently by recognizing the terminator sequence element (below).22
Termination by RNAP III
Biochemical studies revealed 2 distinct mechanisms of RNAP III termination: core and holo-enzyme mechanisms.22 The distinguishing features of these are the dependence or not on 2 components, the 3 subunit RPC53/RPC37/RPC11 (C53/37/11) “termination-reinitiation” subcomplex and the length of the oligo(dT) stretch. The 17-subunit RNAP III holoenzyme can terminate on as few as 5 or 6Ts; while a 14 subunit core version of RNAP III that lacks these 3 subunits (previously referred to as pol IIIΔ)23 that can be isolated from a rpc11-mutant yeast, requires 9Ts for efficient termination.22 Even on a 9T tract, RNAP III-holoenzyme terminates almost exclusively in its proximal region and RNAP III-core terminates almost exclusively in its distal region.22 As more than 80% of tRNA gene terminators in S. cerevisiae have a 6 or 7T terminator,24,25 the holo-enzyme mechanism would appear to be essential for RNAP III function.
This distinction in dT length suggests that the core mechanism reflects sensitivity of the RNAP III core to the thermodynamic instability of the oligo(rU:dA) hybrid, independent of the C53/37/11 subcomplex. Accordingly, because the holoenzyme mechanism differs in that it can terminate on a shorter oligo(rU:dA) hybrid and requires C53/37/11, it might be hypothesized that these subunits increase the destabilizing effects of a short oligo(rU:dA) hybrid. Although this is attractive as a simple hypothesis, the data suggest a more complex mechanism of action.17 Earlier biochemical analysis indicated that the C53/37/11 subcomplex reduces the elongation rate of RNAP III,23 increasing residence time on the terminator and thereby increasing the opportunity for the kinetic coupling model of termination.26 Recent experiments indicate a more intricate mechanism than simple reduction in elongation rate.
Biochemical analysis of the kinetics of the RNAP III holoenzyme mechanism of termination revealed that the transition from elongation to termination occurs via a metastable but catalytically active intermediate.17 The data suggest that C37 is the key facilitator of this transformation, while C11 and C53 are required during other phases of the termination process or contribute supporting roles, respectively.17 Dissection of the mechanism revealed 2 steps: formation of a metastable pretermination complex (PTC) followed by transcript release. In further support of this, deletion of 5 amino acids from a C-terminal region of C37 that were previously found as hotspots in decreased termination mutants in vivo,27 uncoupled these steps in the biochemical assays.17 Surprisingly however, although the model that C53/37/11 may increase oligo(rU:dA) hybrid instability might predict interaction between these 2 components, the data instead suggested that the target of C37 (Rpc37) action in the RNAP III termination pathway is the nontemplate, i.e., oligo(dT) strand of the terminator (Fig. 1B).17
Terminating RNAP III on a 2-rail track: The template and nontemplate strands
Termination can occur by either RNAP III-holo or RNAP III-core on single-stranded oligo(dA) in the absence of a nontemplate strand.17 On the other hand, if the template strand does not have an oligo(dA) stretch, no termination occurs even if oligo(dT) exists in the nontemplate strand of the heteroduplex (17, unpublished). These observations argue that a principal signal for RNAP III termination derives from the oligo(dA) stretch of the template strand. However, total absence of a nontemplate strand or changes in its base identity of specific Ts altered termination qualitatively as well as quantitatively17 indicating that the nontemplate strand plays an active role in transcription termination (also see 28).
Though the nontemplate strand of the terminator under study was a 9T stretch, substitutions of individual nucleotides revealed specific effects on different stages of termination.17 Substitution of the 3rd and 4th Ts impaired slowing of the complex and formation of the PTC while substitution of the 5th T produced a distinct effect, deficiency for transcript release by the PTC. The results suggest that C37 recognizes the nontemplate strand in a sequence-specific manner, and can distinguish nucleotide positions at different phases of termination (also see 28 for insightful discussion).
The nontemplate strand in the transcription bubble: exposed for regulatory factors
During transcription, the template strand is buried within the catalytic center and is otherwise not accessible.29 On the other hand, DNA-protein cross linking as well as crystal structures of elongation complexes formed with RNAP II reveal that the path of the nontemplate strand of the transcription bubble is along the lobe and protrusion of the Rpb2 subunit, exposed at the surface of the complex.30,31 This accessibility of the nontemplate strand may indicate it as an active player in modulation of RNAP activity during different stages of transcription.
The heterodimer RNAP III subunits C53/37 share homology with RNAP II transcription initiation factor subunits TFIIF and RNAP I integral subunits RPA49/RPA34.5.12 Given that all 3 of these homologous complexes occupy analogous positions on their respective RNAPs,32,33 it should be suspected that they may interact with the nontemplate strand albeit possibly to aid different outcomes.
The nontemplate strand as a target of regulatory factors
The bacterial initiation factor σ binds to the nontemplate strand of the −10 promoter element in a sequence specific manner during transcription initiation34 and stabilizes initial promoter opening.35 Moreover, the β subunit of RNAP exhibits sequence specific interaction with the dG nucleotide at +2 of the nontemplate strand.36 Crystal structures of backtrack-arrested RNAP II suggests that such a binding pocket for a +2 nucleotide of the nontemplate strand may also exist for eukaryotic RNAPs.36,37 Sequence specific preference for nontemplate +2 dG residue modulates elongation of transcription as it counteracts pausing by stabilizing the post-translocated state.38
A universally conserved family of transcription elongation factors binds the nontemplate strand of the transcription bubble.39 NusG binds to the bacterial RNAP elongation complex after the dissociation of σ factor and interacts with the T rich, single stranded nontemplate DNA.40,41 NusG paralog RfaH is recruited to the elongation complex by the sequence specific binding to the ops sequence on single stranded nontemplate strand of the transcription bubble.42 Interestingly, the eukaryotic member of the NusG family of factors, Spt5, also localized in close proximity to the nontemplate strand of transcription bubble of RNAP II.43,28 All of these observations suggest that the role of the nontemplate strand as a sequence-specific regulator of transcription initiation, elongation and/or termination are more widespread than previously thought.
Finally, apart from transcription bubbles, more extended R loops also expose single stranded DNA as potential targets of regulatory mechanisms. Displacemet of single stranded DNA at tRNA loci has been documented on a genome-wide scale.44 Another example is the DNA methylation-resistant CG island-containing promoters that are characterized by R loops.45 These suggest that the individual single strands of the DNA may play definitive roles in regulation of gene expression.
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Funding
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
References
- 1.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 2011; 9:85-98; PMID:21233849; http://dx.doi.org/ 10.1038/nrmicro2507 [DOI] [PubMed] [Google Scholar]
- 2.Porrua O, Libri D. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol 2015; 16:190-202; PMID:25650800; http://dx.doi.org/ 10.1038/nrm3943 [DOI] [PubMed] [Google Scholar]
- 3.Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol 2011; 412:793-813; PMID:21439297; http://dx.doi.org/ 10.1016/j.jmb.2011.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Gen Dev 2009; 23:1247-69; PMID:19487567; http://dx.doi.org/ 10.1101/gad.1792809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arimbasseri AG, Rijal K, Maraia RJ. Transcription termination by the eukaryotic RNA polymerase III. Biochimica et biophysica acta 2013; 1829:318-30; PMID:23099421; http://dx.doi.org/ 10.1016/j.bbagrm.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arimbasseri AG, Kassavetis GA, Maraia RJ. Comment on “Mechanism of eukaryotic RNA polymerase III transcription termination”. Science 2014; 345:524; PMID:25082694; http://dx.doi.org/ 10.1126/science.1253783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santangelo TJ, Cubonova L, Skinner KM, Reeve JN. Archaeal intrinsic transcription termination in vivo. J Bacteriol 2009; 191:7102-8; PMID:19749050; http://dx.doi.org/ 10.1128/JB.00982-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grohmann D, Werner F. Recent advances in the understanding of archaeal transcription. Curr Opin Microbiol 2011; 14:328-34; PMID:21596617; http://dx.doi.org/ 10.1016/j.mib.2011.04.012 [DOI] [PubMed] [Google Scholar]
- 9.Arimbasseri AG, Rijal K, Maraia RJ. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription 2014; 5:e27639; PMID:25764110; http://dx.doi.org/ 10.4161/trns.27369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moir RD, Willis IM. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochimica et biophysica acta 2013; 1829:361-75; PMID:23165150; http://dx.doi.org/ 10.1016/j.bbagrm.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 2012; 45:439-46; PMID:22365827; http://dx.doi.org/ 10.1016/j.molcel.2012.01.023 [DOI] [PubMed] [Google Scholar]
- 12.Carter R, Drouin G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evolution 2010; 27:1035-43; PMID:20026480; http://dx.doi.org/ 10.1093/molbev/msp316 [DOI] [PubMed] [Google Scholar]
- 13.Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase III transcription control elements: themes and variations. Gene 2012; 493:185-94; PMID:21712079; http://dx.doi.org/ 10.1016/j.gene.2011.06.015 [DOI] [PubMed] [Google Scholar]
- 14.Male G, von Appen A, Glatt S, Taylor NM, Cristovao M, Groetsch H, Beck M, Müller CW. Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat Commun 2015; 6:7387; PMID:26060179; http://dx.doi.org/ 10.1038/ncomms8387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagarajavel V, Iben JR, Howard BH, Maraia RJ, Clark DJ. Global ‘bootprinting’ reveals the elastic architecture of the yeast TFIIIB-TFIIIC transcription complex in vivo. Nucleic Acids Res 2013; 41:8135-43; PMID:23856458; http://dx.doi.org/ 10.1093/nar/gkt611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip Rev RNA 2011; 2:362-75; PMID:21572561; http://dx.doi.org/ 10.1002/wrna.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arimbasseri Aneeshkumar G, Maraia Richard J. Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element. Mol Cell 2015; 58:1124-32; PMID:25959395; http://dx.doi.org/ 10.1016/j.molcel.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem 2000; 275:6530-6; PMID:10692458; http://dx.doi.org/ 10.1074/jbc.275.9.6530 [DOI] [PubMed] [Google Scholar]
- 19.Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu Rev Biochem 2009; 78:335-61; PMID:19489723; http://dx.doi.org/ 10.1146/annurev.biochem.76.052705.164655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin FH, Tinoco I. DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res 1980; 8:2295-9; PMID:6159577; http://dx.doi.org/ 10.1093/nar/8.10.2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman N, Jakobi A, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen W, et al.. Molecular structures of unbound and transcribing RNA polymerase III. Nature 2015; PMID:26605533; http://dx.doi.org/ 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.22. Maraia RJ, Rijal K. Structural biology: A transcriptional specialist resolved. Nature. 2015; PMID:26605522; http://dx.doi.org/23401852 10.1038/nature16317. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arimbasseri AG, Maraia RJ. Distinguishing core and holoenzyme mechanisms of transcription termination by RNA polymerase III. Mol Cell Biol 2013; 33:1571-81; PMID:23401852; http://dx.doi.org/ 10.1128/MCB.01733-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J 2006; 25:118-28; PMID:16362040; http://dx.doi.org/ 10.1038/sj.emboj.7600915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dT) termination signal recognition by S. cerevisiae RNA polymerase III. J Biol Chem 2005; 280:19551-62; PMID:15788403; http://dx.doi.org/ 10.1074/jbc.M412238200 [DOI] [PubMed] [Google Scholar]
- 26.Iben JR, Maraia RJ. Yeast tRNAomics: tRNA gene copy number variation and codon use provide bioinformatics evidence of a new wobble pair in a eukaryote. RNA 2012; 18:1358-72; PMID:22586155; http://dx.doi.org/ 10.1261/rna.032151.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin DJ, Burgess RR, Richardson JP, Gross CA. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci USA 1992; 89:1453-7; PMID:1741399; http://dx.doi.org/ 10.1073/pnas.89.4.1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rijal K, Maraia RJ. RNA polymerase III mutants in TFIIFα-like C37 cause terminator readthrough with no decrease in transcription output. Nucleic Acids Res 2013; 41:139-55; PMID:23093604; http://dx.doi.org/ 10.1093/nar/gks985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artsimovitch I, Belogurov Georgi A. Creative Math of RNA Polymerase III Termination: Sense Plus Antisense Makes More Sense. Mol Cell 2015; 58:974-6; PMID:26091347; http://dx.doi.org/ 10.1016/j.molcel.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 30.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 2001; 292:1876-82; PMID:11313499; http://dx.doi.org/ 10.1126/science.1059495 [DOI] [PubMed] [Google Scholar]
- 31.Barnes Christopher O, Calero M, Malik I, Graham Brian W, Spahr H, Lin G, Cohen AE, Brown IS, Zhang Q, Pullara F, et al.. Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble. Mol Cell 2015; 59:258-69; PMID:26186291; http://dx.doi.org/ 10.1016/j.molcel.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrecka J, Treutlein B, Arcusa MA, Muschielok A, Lewis R, Cheung AC, Cramer P, Michaelis J. Nano positioning system reveals the course of upstream and nontemplate DNA within the RNA polymerase II elongation complex. Nucleic Acids Res 2009; 37:5803-9; PMID:19620213; http://dx.doi.org/ 10.1093/nar/gkp601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu CC, Lin YC, Chen HT. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol 2011; 31:2715-28; PMID:21536656; http://dx.doi.org/ 10.1128/MCB.05151-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez-Tornero C, Moreno-Morcillo M, Rashid UJ, Taylor NMI, Ruiz FM, Gruene T, Legrand P, Steuerwald U, Müller CW. Crystal structure of the 14-subunit RNA polymerase I. Nature 2013; 502:644-9; PMID:24153184; http://dx.doi.org/ 10.1038/nature12636 [DOI] [PubMed] [Google Scholar]
- 35.Feklistov A, Darst Seth A. Structural Basis for Promoter −10 Element Recognition by the Bacterial RNA Polymerase σ Subunit. Cell 2011; 147:1257-69; PMID:22136875; http://dx.doi.org/ 10.1016/j.cell.2011.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Bushnell David A, Kornberg Roger D. Lock and Key to Transcription: σ-DNA Interaction. Cell 2011; 147:1218-9; PMID:22153066; http://dx.doi.org/ 10.1016/j.cell.2011.11.033 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural Basis of Transcription Initiation. Science 2012; 338:1076-80; PMID:23086998; http://dx.doi.org/ 10.1126/science.1227786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 2011; 471:249-53; PMID:21346759; http://dx.doi.org/ 10.1038/nature09785 [DOI] [PubMed] [Google Scholar]
- 39.Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 2014; 344:1285-9; PMID:24926020; http://dx.doi.org/ 10.1126/science.1253458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.NandyMazumdar M, Artsimovitch I. Ubiquitous transcription factors display structural plasticity and diverse functions: NusG proteins - Shifting shapes and paradigms. BioEssays 2015; 37:324-34; PMID:25640595; http://dx.doi.org/ 10.1002/bies.201400177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomar SK, Artsimovitch I. NusG-Spt5 Proteins—Universal Tools for Transcription Modification and Communication. Chem Rev 2013; 113:8604-19; PMID:23638618; http://dx.doi.org/ 10.1021/cr400064k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yakhnin AV, Babitzke P. NusG/Spt5: are there common functions of this ubiquitous transcription elongation factor? Curr Opin Microbiol 2014; 18:68-71; PMID:24632072; http://dx.doi.org/ 10.1016/j.mib.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 2002; 109:193-203; PMID:12007406; http://dx.doi.org/ 10.1016/S0092-8674(02)00724-9 [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Rucobo FW, Sainsbury S, Cheung ACM, Cramer P. Architecture of the RNA polymerase–Spt4/5 complex and basis of universal transcription processivity. EMBO J 2011; 30:1302-10; PMID:21386817; http://dx.doi.org/ 10.1038/emboj.2011.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet 2014; 10:e1004716; PMID:25357144; http://dx.doi.org/ 10.1371/journal.pgen.1004716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 2012; 45:814-25; PMID:22387027; http://dx.doi.org/ 10.1016/j.molcel.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]


