Abstract
The human genome contains some thousands of long non coding RNAs (lncRNAs). Many of these transcripts are presently considered crucial regulators of gene expression and functionally implicated in developmental processes in Eukaryotes. Notably, despite a huge number of lncRNAs are expressed in the Central Nervous System (CNS), only a few of them have been characterized in terms of molecular structure, gene expression regulation and function. In the present study, we identify linc-NeD125 as a novel cytoplasmic, neuronal-induced long intergenic non coding RNA (lincRNA). Linc-NeD125 represents the host gene for miR-125b-1, a microRNA with an established role as negative regulator of human neuroblastoma cell proliferation. Here, we demonstrate that these two overlapping non coding RNAs are coordinately induced during in vitro neuronal differentiation, and that their expression is regulated by different mechanisms. While the production of miR-125b-1 relies on transcriptional regulation, linc-NeD125 is controlled at the post-transcriptional level, through modulation of its stability. We also demonstrate that linc-NeD125 functions independently of the hosted microRNA, by reducing cell proliferation and activating the antiapoptotic factor BCL-2.
Keywords: apoptosis, cell proliferation, long non coding RNA, microRNA, miR-125, neuroblastoma, neuronal differentiation, post-transcriptional regulation, transcriptional regulation
Introduction
The largest variety of non coding RNAs (ncRNAs), both small and long, are abundantly expressed in mammalian CNS where they participate in addressing stem cells toward specific differentiation fates, and contribute in survival, maturation and neuronal activities.1-5 Even though small and long ncRNAs (lncRNAs) represent the two halves of the ncRNA world, they intermingle with one another both functionally, when long RNAs control the availability and activity of small RNAs, such as microRNAs (miRNAs),6 and structurally, when small RNAs are embedded inside lncRNA genes. In this latter case, a unique primary transcript can generate two mature RNA molecules whose expression, as well as function, may be coordinated or independent.
Due to their poor sequence conservation among even closely related organisms and the lacking of shared biochemical and structural features, the study of lncRNAs is still at a preliminary stage 7. Despite their abundance in the CNS, only a handful of lncRNAs have been structurally and functionally characterized thus far.8-11 Diversely, several miRNAs have been thoroughly described as molecules controlling cell proliferation and/or differentiation programs in different organisms. Specifically, miR-124 and miR-9 are considered the most abundant brain-enriched miRNAs.12-15
Another miRNA that is abundantly expressed in animal brain and upregulated during neurogenesis is miR-125, the ortholog of the heterocronic lin-4.16-18 The function of miR-125 has been extensively studied. It promotes neuronal19 and astrocyte20 differentiation and is implicated in synaptic plasticity.21 We contributed to the study of miR-125 function defining its participation in the control of neural precursor cell proliferation 22 and its involvement in cell growth arrest and apoptosis of human Neuroblastoma (NB)- and Medulloblastoma (MB)-derived cell lines 12, 23. Accordingly, miR-125 is downregulated in the corresponding primary tumors suggesting it may have a tumor-suppressor function 12, 23.
The relevant role of miR-125 in cell fate decision prompted us to study its biogenesis. Among the three miR-125 coding genes, we focused on miR-125b-1 that is organized in cluster with miR-100 and let-7, representing the only cluster of miRNAs that is conserved from flies to humans 24.
The study of miR-125b-1 host gene led us to the discovery of a novel lincRNA, that we named linc-NeD125. Here, we report the molecular and functional characterization of linc-NeD125, which is up-regulated during in vitro neuronal differentiation of NB and MB cells. We identify the minimal promoter driving its constitutive expression in proliferating conditions and unveil a post-transcriptional regulatory mechanism responsible for its induction upon the differentiation stimulus. We also demonstrate here that linc-NeD125 may act autonomously from the hosted miRNA by negatively regulating cell proliferation and apoptosis.
Results
Identification of a novel, neuronal-induced lincRNA as the host gene for miR-125b-1
UCSC genome browser (assembly 2009)25 shows that miR-125b-1 is located on chromosome 11q23 and embedded inside the third intron of a RefSeq26 annotated non-protein coding RNA, named MIR100HG (hereafter, RefSeq MIR100HG; NCBI Reference Sequence: NR_024430.1). In the same intron, at a distance of about 45 kb from miR-125b-1, miR-100 and let-7a-2 are also positioned (Fig. 1A upper scheme).
Figure 1.
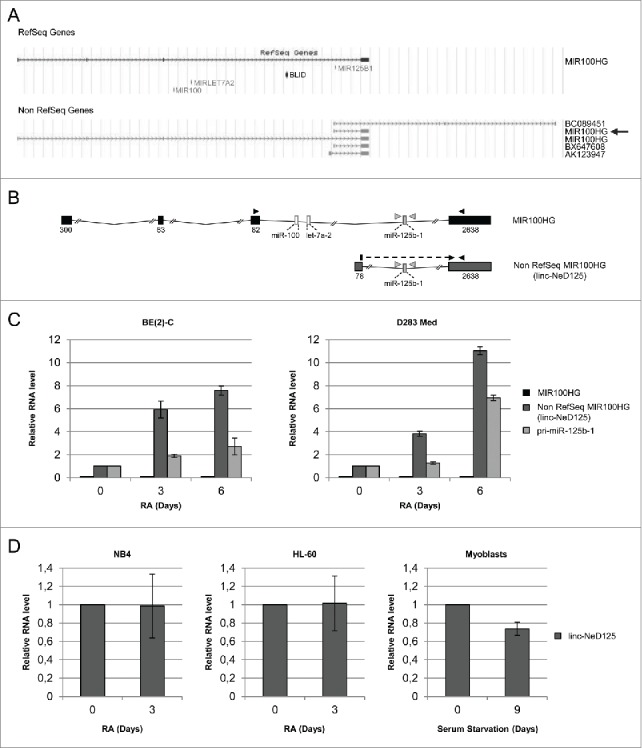
Structure and expression profile of miR-125b-1 containing transcripts. (A) Genomic organization of MIR125B1 locus, according to UCSC genome browser. RefSeq (upper panel) and Non RefSeq (lower panel) genes are depicted. The arrow points to non RefSeq MIR100HG short isoform corresponding to linc-NeD125. (B) Schematic representation of RefSeq MIR100HG and Non RefSeq MIR100HG/linc-NeD125. Boxes (black for MIR100HG or gray for linc-NeD125) represent exons, lines are introns; exon sizes are also reported. miRNA coding regions are depicted as thin boxes. Arrowheads indicate primers used for qRT-PCR analyses. (C) Expression profiles of miR-125b-1 containing transcripts. Histograms show the relative amount of RefSeq MIR100HG (black bars), Non RefSeq MIR100HG/linc-NeD125 (dark gray bars) and pri-miR-125b-1 (light gray bars) in BE(2)-C (left panel) and in D283 Med (right panel) cells, as determined by qRT-PCR analyses. In both panels, the expression levels were evaluated at specific time-points of RA treatment (days, as indicated below). Day 0 was set as 1 only for linc-NeD125 and pri-miR-125b-1. RefSeq MIR100HG expression was almost undetectable both in proliferating and in differentiating cells; however, for facilitating data visualization, its expression was represented as a black line in the histogram. Gapdh mRNA was used as a loading control. Data are presented as mean values ± SEM from at least 3 independent experiments (BE(2)-C) or from 3 technical replicates (D283 Med). (D) Expression profiles of linc-NeD125 in NB4, HL-60 and human myoblasts in conditions of cell proliferation (day 0) or upon specific treatments (indicated below each histogram) triggering differentiation. Day 0 was set as 1. Gapdh mRNA was used as a loading control.
To correlate pri-miR-125b-1 expression with that of its putative host gene MIR100HG, we profiled their expression in proliferating versus differentiating cells by qRT-PCR. As model system, we used the BE(2)-C cell line deriving from human Neuroblastoma, a pediatric tumor of the sympathetic nervous system. Treatment of BE(2)-C cells with Retinoic Acid (RA) inhibits proliferation and triggers neuronal differentiation.12 The main advantage of this model system is the production in 6 days of a homogeneous population of cells, displaying neuronal morphology (Fig. S1A) and showing modulation of several neuronal differentiation markers.27
We treated BE(2)-C cells with RA for specific time points (0, 3 and 6 days) and verified by qRT-PCR: i) the increased expression of neuronal differentiation markers, as the neuropeptide VGF, the cytoscheletal component bIII-tubulin (TUBB3) and the neuronal-specific microRNA miR-124; ii) the reduced expression of the neural repressor ID2 (Inhibitor of DNA binding-2) and the pro-proliferative factor N-MYC (Fig. S1B). In parallel, the expression of the putative miR-125b-1 host gene, RefSeq MIR100HG, was analyzed. Its expression was evaluated using specific combinations of oligonucleotides designed to amplify the exonic sequences of interest (Fig. 1B and Fig. S2A). We found that RefSeq MIR100HG was not significantly expressed either in proliferating (0 days) or in differentiating (3 and 6 days) cells (Fig. 1C, left panel and Fig. S2B). Differently, pri-miR-125b-1 was induced upon RA treatment, reaching a peak of expression at 6 days (Fig. 1C). These results indicate that RefSeq MIR100HG is not the host gene for miR-125b-1 in NB cell lines induced to neuronal differentiation.
We also analyzed the expression profile of lncRNA_N2 (AK0191713 transcript), previously reported in a human transcriptome analysis28 and described as the miR-125b-1, miR-100 and let-7a-2 host gene with a crucial function in neuronal differentiation of human neural stem cells.29 We found that this transcript was almost undetectable both in undifferentiated and in RA-treated BE(2)-C cells (Fig. 2SC).
Other predicted RNA species that might host miR-125b-1 were then searched in the UCSC genome browser. The non RefSeq annotated genes, depicted in the lower scheme of Fig. 1A, were analyzed. Only one of them, also named MIR100HG (pointed by an arrow in Fig. 1A lower scheme), was found to be expressed and upregulated during in vitro neuronal differentiation (Fig. S2D). This transcript showed the same expression profile as pri-miR-125b-1, indicating it may be the miR-125b-1 host gene in our cellular system (Fig. 1B lower scheme andFig. 1C left panel). Therefore, it was renamed linc-NeD125 (Neuronal Differentiation lincRNA hosting miR-125).
Non RefSeq MIR100HG, here renamed linc-NeD125, was reported in UCSC genome browser as a non coding RNA, since it does not harbour putative Open Reading Frames (ORFs) encoding polypeptides longer than 100 amino acids.30 To further verify its non coding nature, we analyzed the occurrence of small ORFs by the sORF finder program (http://evolver.psc.riken.jp/). We found that linc-NeD125 could potentially encode three short peptides of 16, 17 and 27 amino acids (Fig. S3A). However, in vitro transcription/translation assay of the mature transcript indicated that it does not display any coding capacity (Fig. S3B), and can therefore be referred to as a bona fide long non coding RNA.
To verify whether linc-NeD125 is a neuronal-induced transcript, we profiled its expression in other in vitro differentiation models. As an additional neural system, we used the D283 Med cell line, that can also be induced toward neuronal differentiation by RA-treatment31. However, differently from BE(2)-C cells, the D283 Med cell line originates from human Medulloblastoma, a tumor of the CNS. Moreover, we used two human Acute Promyelocytic Leukemia (APL) cell lines, the NB4 and HL-60 cells, induced to granulocytic differentiation by RA-treatment32. Further, we tested linc-NeD125 expression in human primary myoblasts differentiated to myotubes.33
qRT-PCR analyses carried out on total RNA from proliferating and differentiating cells indicated that, in agreement with what observed in BE(2)-C cells, RefSeq MIR100HG was barely detectable in RA-treated D283 Med cells, whereas linc-NeD125 and pri-miR-125b-1 were induced (Fig. 1C right panel and Fig. S4).
Differently, linc-NeD125 was expressed, but not up-regulated, during both haematopoietic and muscle in vitro differentiation (Fig. 1D). Altogether, these results reject the possibility that linc-NeD125 induction may be a side effect of RA administration, and indicate that the transcript is specifically induced during in vitro neuronal differentiation.
Investigation of linc-NeD125 evolutionary conservation was carried out by a phylogenetic analysis across 18 placental mammal species (Fig. 2A). This analysis showed separate branching of Primates from more evolutionarily distant Mammals, such as mouse and rat. Among non-human Primates, the Hominidae (chimpanzee, gorilla and orangutan) showed the highest sequence conservation (99.20, 98.68 and 97.76 percent identity, respectively). Notably, the same phylogenetic pattern was obtained when the mature or the primary linc-NeD125 transcripts were considered (data not shown). However, even though sequence conservation drops off to about 70% in mouse (Fig. 2B), linc-NeD125 gene structure, consisting of 2 exons and a miR-125b-1-containing intron, is conserved (Fig. 2C).
Figure 2.
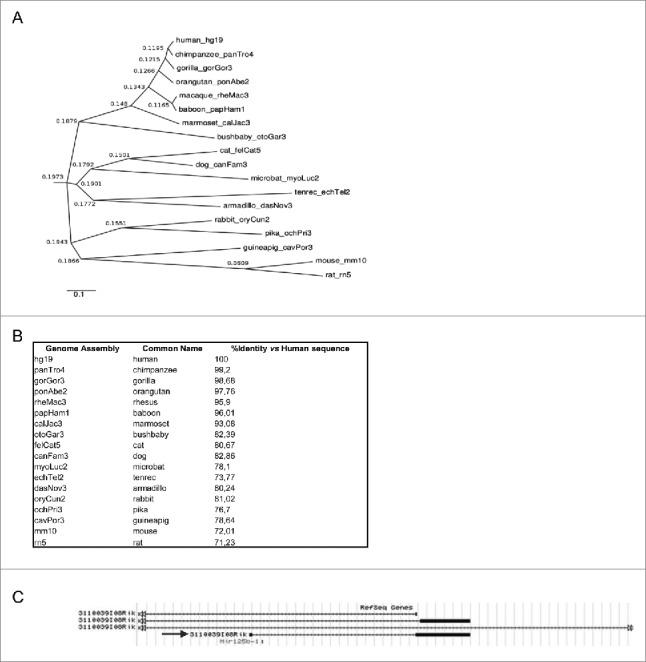
Phylogenetic analysis inferred from multiple sequence alignment of mature linc-NeD125. (A) The dendrogram shows grouping of 18 placental mammal species based on the Neighbor Joining (NJ) agglomeration method applied to the distance matrix of percent divergence computed between each pair of sequences. Leaf nodes in the dendrogram are labeled with the name of the species and its specific genome assembly included in the multiple sequence alignment. Labels at the connection point between 2 branches show their distance score. The scale bar indicates rate of residue substitution per site. (B) Linc-NeD125 sequence identity between human and each of the 18 species reported in the dendogram, expressed as percentage relatively to the human sequence. (C) Genomic organization of the murine Mir125b-1 locus, according to UCSC genome browser. The arrow points to the transcript showing the same organization of human linc-NeD125.
Molecular characterization of linc-NeD125
The increased levels of the mature linc-NeD125 during in vitro neuronal differentiation led us to further investigate its structure and function.
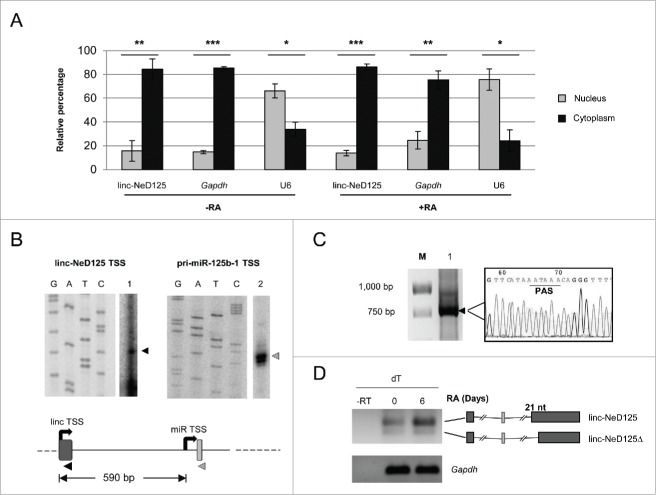
The linc-NeD125 locus spans 11,349 kb and comprises 2 exons of 78 bp and 2,638 bp, and a 8,633 bp long intron containing miR-125b-1, but not including miR-100 and let-7a-2. We started linc-NeD125 characterization by determining its sub-cellular localization. Nucleus/cytoplasm fractionation was carried out on BE(2)-C cells both in growing (−RA) and in differentiating (+RA) conditions. qRT-PCR analysis performed on linc-NeD125 and, as controls, on Gapdh mRNA and U6 snRNA , indicated that it is predominantly localized into the cytoplasmic compartment in both conditions (Fig. 3A).
Figure 3.
Molecular characterization of linc-NeD125. (A) Linc-NeD125 sub-cellular localization. Proliferating (-RA) or 6-days RA-treated (+RA) BE(2)-C cells were fractionated into nuclear (gray bars) and cytoplasmic (black bars) fractions and linc-NeD125 transcript was evaluated by qRT-PCR analysis. Gapdh mRNA and U6 snRNA were used as cytoplasmic and nuclear controls, respectively. Data are represented as percentage of the values relative to an exogenous internal control and presented as the means ± SEM from 3 independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001. (B) Linc-NeD125 (left panel) and pri-miR-125b-1 (right panel) TSS identification. The primer extension reactions (lanes 1 and 2) were run in parallel with the sequencing reactions (lanes G, A, T, and C). The arrowheads indicate the extended products. In the beneath schematic representation, linc-NeD125 TSS (linc TSS) and pri-miR-125b-1 TSS (miR TSS) are represented by black curved arrows. The distance between the 2 TSSs is reported as nucleotide length. Primers used for the primer extension reactions are also depicted as arrowheads. (C) Identification of linc-NeD125 3′ end by 3′ RACE. The reaction product (lane 1) was gel-fractionated along with a DNA ladder (lane M). The arrowhead points to the amplified product, whose sequence is also reported; PAS stands for Poly-Adenylation Signal. (D) RT-PCR analysis of linc-NeD125 carried out with oligo-dT primers from untreated (day 0) or RA-treated (day 6) BE(2)-C cells. Control reaction lacking Reverse Transcriptase is loaded in lane –RT. Gapdh mRNA was used as a loading control. The two linc-NeD125 splicing variants are schematized on the right and their difference in length (21 nucleotides) is reported. Boxes represent exons, lines are introns, MIR125B1 coding regions is depicted as a light gray thin box.
Identification of linc-NeD125 5′- and 3′-ends was carried out on total RNA from RA-treated BE(2)-C cells by primer extension and 3′RACE, respectively. By using an oligonucleotide annealing in the linc-NeD125 first exon (Fig. 3B, lower scheme), we identified its Transcriptional Start Site (TSS) (Fig. 3B, left panel). We found that it matches the 5′-end of the short MIR100HG transcript annotated among the non RefSeq genes in the UCSC genome browser view (pointed by an arrow in Fig. 1A). Notably, a further primer extension analysis with an oligonucleotide annealing into the pri-miRNA stem-loop structure allowed the identification of an additional TSS, located 30 nt upstream of the pre-miR-125b-1 (Fig. 3B, right panel).
The 3′-end of linc-NeD125 was found to overlap with a canonical polyadenylation signal, indicating that at least a fraction of this transcript is polyadenylated (Fig. 3C). An RT-PCR analysis carried out on total RNA using oligo dTs as primers confirmed the occurrence of linc-NeD125 polyadenylation, and highlighted the existence of two isoforms deriving from alternative splicing. These differ in length (21 nt at the 5′-end of exon II) and in the expression profile. In particular, the longer isoform (linc-NeD125) is expressed at higher levels both in proliferating and differentiating cells and is the only one induced upon the differentiation stimulus (Fig. 3D). Differently, the lower band (linc-NeD125Δ), matching the predicted transcript BX647608 (Fig. 1A lower scheme), was expressed at very low level in our model system.
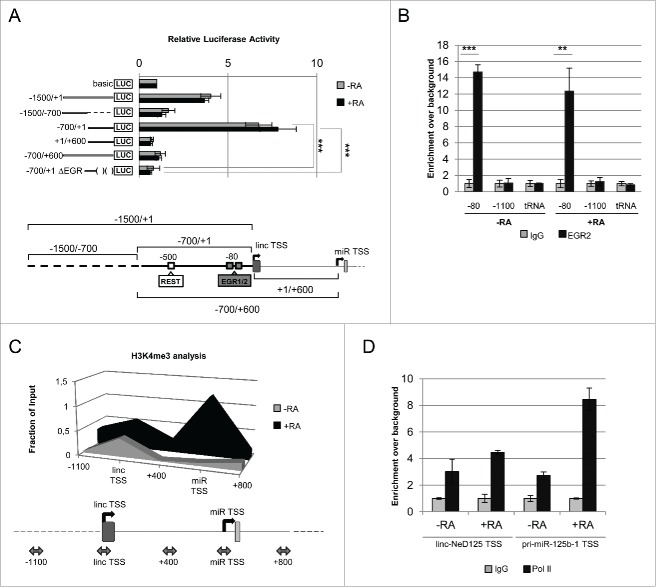
The identification of two distinct TSSs prompted us to identify the cis-acting elements responsible for the transcription of the two ncRNAs. To this end, we focused on the 1.5 kb region upstream of linc-NeD125 TSS and exploited the Luciferase (LUC) reporter gene approach. As depicted in Figure 4A, the whole region (−1500/+1) as well as two sub-modules of this region (−1500/−700 and −700/+1) were fused to the LUC ORF in a promoter-less plasmid (basic), and the derived constructs were transfected into untreated and RA-treated BE(2)-C cells. Evaluation of the luciferase activity indicated that, in both cell conditions, the longest fragment (−1500/+1) was able to induce transcription of the reporter gene by about 4-folds compared to the control construct (basic). The region encompassing −1500/−700 was unable to promote luciferase activity, whereas the 700 bp upstream of the TSS (−700/+1) provided the strongest activity, independently of the differentiation stimulus (Fig. 4A).
Figure 4.
Linc-NeD125 transcriptional regulation. (A) Identification of linc-NeD125 core promoter by luciferase assay. Upper panel: the luciferase-based reporter construct, containing the 1.5-kb region upstream of the linc-NeD125 TSS (−1500/+1), and its deletion mutant derivatives (−1500/−700), (−700/+1) and (−700/+1ΔEGR) were tested in both proliferating (–RA, gray bars) and 6-days RA-treated (+RA, black bars) BE(2)-C cells. The additional constructs containing the intronic region between the 2 TSSs (+1/+600) and the same region extended to include linc-NeD125 minimal promoter (−700/+600) were also analyzed in the same conditions. The promoter-less reporter construct (basic) was used as a control. Data are presented as mean values ± SEM from at least 3 independent experiments. ***p < 0.001. Lower panel: schematic representation of the sequence modules spanning the linc-NeD125 TSS that were cloned upstream of the luciferase reporter gene. The core promoter is depicted as a thick line; the putative binding sites for REST and EGR1/2 factors (according to ChIP-Mapper Web Server) are indicated by white and gray boxes, respectively. Their positions relative to linc-NeD125 TSS are also reported. (B) ChIP-qPCR experiments of EGR2 occupancy on linc-NeD125 promoter. Chromatin samples were obtained from untreated (-RA) or 6 days RA-treated (+RA) BE(2)-C cells and incubated with anti-EGR2 (black bars) or rabbit IgG as a control (gray bars). Immunoprecipitation levels are expressed as enrichment over background (IgG). A region lacking of putative EGR binding sites (−1100) and the tRNAGlu (tRNA) were PCR-amplified as negative controls. Data are represented as mean values ± SEM from 3 independent experiments.**p < 0.01, ***p < 0.001. (C) Analysis of H3K4 trimethylation (H3K4me3) on linc-NeD125 and pri-miR-125b-1 TSSs. ChIP assays were performed on chromatin samples from untreated (-RA, gray area) or 6 days RA-treated (+RA, black area) BE(2)-C cells, incubated with antibodies against H3K4me3 or IgG. In the scheme below, PCR-amplified regions are positioned with respect to linc-NeD125 TSS and indicated by double-headed arrows. Immunoprecipitation levels are expressed as fraction of the Input. (D) ChIP analysis of RNA Pol II on linc-NeD125 and pri-miR-125b-1 TSSs. ChIP assays were performed on chromatin samples from untreated (-RA) or 6 days RA-treated (+RA) BE(2)-C cells, incubated with anti-Pol II antibody (black bars) or IgG (gray bars). Immunoprecipitation levels are expressed as enrichment over background (IgG).
In addition, to investigate whether pri-miR-125b-1 transcription could be directed by an independent promoter, a further construct that included the intronic region between the two TSSs (+1/+600) was generated. Moreover, to test whether this intronic sequence may confer RA responsiveness to the upstream region, it was fused to the −700/+1 sequence, generating the −700/+600 construct (Fig. 4A). Transfection of these constructs in untreated and RA-treated cells indicated that they were not able either to promote or to regulate transcription of the reporter gene (Fig. 4A).
In conclusion, our results identified the −700/+1 region as the gene locus core promoter and highlighted its constitutive activity in NB cells.
Regulation of linc-NeD125 gene expression
To identify the transcription factors driving linc-NeD125 expression, a Chip-Mapper (mapper.chip.org)34 in silico analysis of the region encompassing the core promoter (−700/+1 region) was carried out. This analysis predicted putative binding sites for the neuronal repressor REST and the neuronal activators Early Growth Response (EGR) 1 and 2 (Fig. 4A, lower scheme). EGR1 and 2 factors share a highly homologous DNA-binding domain that recognizes an identical DNA response element.35 ChIP assays were performed to verify the involvement of such factors, which are modulated during in vitro neuronal differentiation (Fig. S5A), in linc-NeD125 expression. Figure 4B shows that EGR2 is already bound to the core promoter in proliferating cells (-RA) and that its association to the promoter persists in differentiating cells (+RA). Conversely, no binding for REST and EGR1 was detected (Fig. S5B-C). Since EGR2 knockdown by RNAi was inefficient, we assessed its contribution to linc-NeD125 expression by deriving, from the reporter construct containing the core promoter (−700/+1 region), a deletion mutant lacking the EGR binding sites (−700/+1 ΔEGR construct of Fig. 4A). Notably, in the absence of the two putative EGR binding sites, the luciferase activity was abolished both in untreated and in RA-treated cells (Fig. 4A), indicating a role for EGR2 in driving the transcription of the reporter gene. This result, together with the occurrence of EGR2 on the minimal promoter both in proliferating and in differentiating conditions, indicates that this factor is responsible for the constitutive transcription of the gene locus.
To assess whether the induction of linc-NeD125 and/or pri-miR-125b-1 along differentiation was transcriptionally regulated, we investigated the chromatin structure and the RNA polymerase II (RNA pol II) recruitment in the regions encompassing the two TSSs, in proliferating vs. differentiating conditions.
Analysis by ChIP assay of the chromatin mark H3K4me3, which specifically correlates with 5′ regions of active genes 36, 37, indicated that its level did not change around linc-NeD125 TSS upon the differentiation stimulus, whereas it strongly increased around the pri-miR-125b-1 TSS (Fig. 4C). Parallel RNA pol II ChIP assay showed that its recruitment on the linc-NeD125 TSS was not significantly affected by RA-treatment, whereas its association with the pri-miR-125b-1 TSS strongly increased upon the differentiation stimulus (Fig. 4D). These results indicate that the two RNA molecules are differentially transcribed in response to RA-treatment, with the pri-miR-125b-1 specifically induced.
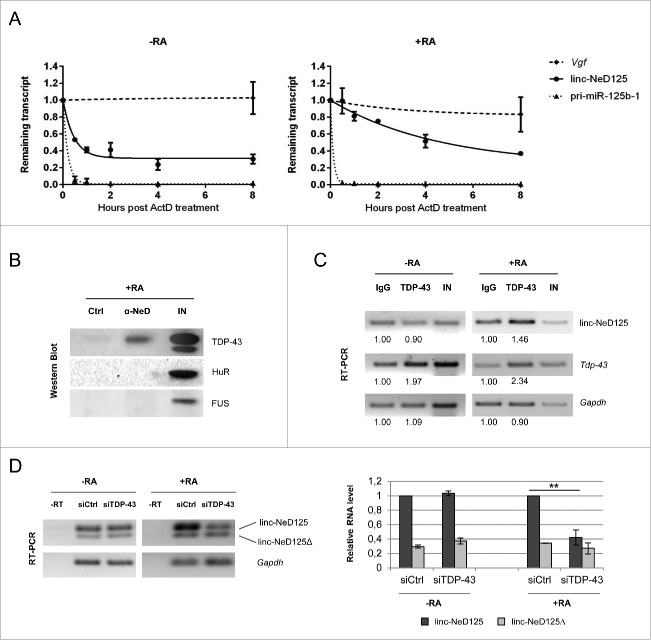
The finding that linc-NeD125 is constitutively transcribed in untreated and RA-treated cells, suggests that its induction during differentiation can rely on post-transcriptional gene regulation. To test this possibility, linc-NeD125 half-life was evaluated by Actinomycin D (ActD) administration to untreated and RA-treated BE(2)-C cells. qRT-PCR quantification of the remaining amount of linc-NeD125 transcript upon ActD administration revealed a 10 fold-longer half-life in differentiating (t1/2 = 3.1 hr) vs proliferating cells (t1/2=18 min) (Fig. 5A). This result indicates that the increased levels of linc-NeD125 during differentiation are mainly due to its increased stability.
Figure 5.
Linc-NeD125 post-transcriptional regulation. (A) Untreated (-RA, left panel) or 4 days RA-treated (+RA, right panel) BE(2)-C cells were maintained in the presence of Actinomycin D (ActD). Linc-NeD125 transcript was analyzed by qRT-PCR at the indicated time points (hours) after ActD addition. Vgf mRNA64 and pri-miR-125b-1 levels were also analyzed as controls. Results were normalized against Gapdh mRNA and expressed with respect to ActD-untreated cells. Fitted curves were modeled using one-phase exponential decay. (B) Linc-NeD125 RNA pull-down in RA-treated BE(2)-C cells: Western blot analysis of TDP-43, HuR and FUS from Input (lane IN), specific probe-bound fraction (lane α-NeD) or unspecific RNA probe fraction (lane Ctrl). (C) RT-PCR analysis of RNA from TDP-43 CLIP assays in untreated (-RA) or RA-treated (+RA) BE(2)-C cells. Linc-NeD125, Tdp-43 (positive control) 65 or Gapdh (negative control) transcripts were PCR-amplified from Input (lane IN) and extracts immunoprecipitated for TDP-43 (lane TDP-43) or IgG (lane IgG). Relative quantities from one representative experiment are expressed respect to control samples (IgG), set as 1. (D) Left panel: RT-PCR analyses of linc-NeD125 in proliferating (-RA) or differentiating (+RA) BE(2)-C cells transfected with control non-targeting siRNAs (lane siCtrl) or siRNAs against TDP-43 (lane siTDP-43). Control reactions lacking Reverse Transcriptase are loaded in lane -RT. Gapdh mRNA was used as a loading control. Right panel: quantification of linc-NeD125 (dark gray bars) and linc-NeD125Δ (light gray bars) in proliferating (-RA) or differentiating (+RA) BE(2)-C cells depleted for TDP- 43. linc-NeD125 level in control cells (siCtrl) was set as 1, in both conditions. Histogram shows mean values ± SEM from three independent experiments. **p < 0.01.
Next, we looked for protein partners that might interact with linc-NeD125 in RA-treated cells. A subset of RNA binding proteins (RBPs), whose activity has been related to lncRNA biogenesis and function, has been initially considered. Specifically, Hu antigen R (HuR), that contributes to pri-miRNA 38 and lncRNA 39, 40 processing and stability, and FUS/TLS and TDP-43, which share some common lncRNA targets 41 were taken into account. Computational inspection of the linc-NeD125 sequence revealed putative binding sites for the three RBPs (Fig. S6). RNA-protein interactions were then validated in RA-treated cells by pull-down experiments, which allowed to selectively isolate the endogenous full-length isoform of linc-NeD125 (Fig. S7A). Western blot analyses showed that only TDP-43 was specifically associated with linc-NeD125 (Fig. 5B). Parallel CLIP analysis carried out both in proliferating and differentiating BE(2)-C cells confirmed such interaction, and indicated that it takes place only upon the differentiation stimulus (Fig. 5C and Fig. S7B).
To investigate the effect of TDP-43 binding, we analyzed linc-NeD125 levels in TDP-43 depleted cells. We found that TDP-43 knockdown (Fig. S7C) exerted not effect in growing conditions, whereas it caused a significant decrease of the full-length transcript in differentiating cells (Fig. 5D), when linc-NeD125 stability increases and it is physically associated with TDP-43.
Linc-NeD125 negatively controls NB cell proliferation and apoptosis
To verify whether linc-NeD125 may function independently from the hosted miRNA, its mature transcript lacking the intronic miRNA-coding sequence was expressed in an inducible BE(2)-C cell line upon doxycycline administration (Fig. 6A).
Figure 6.
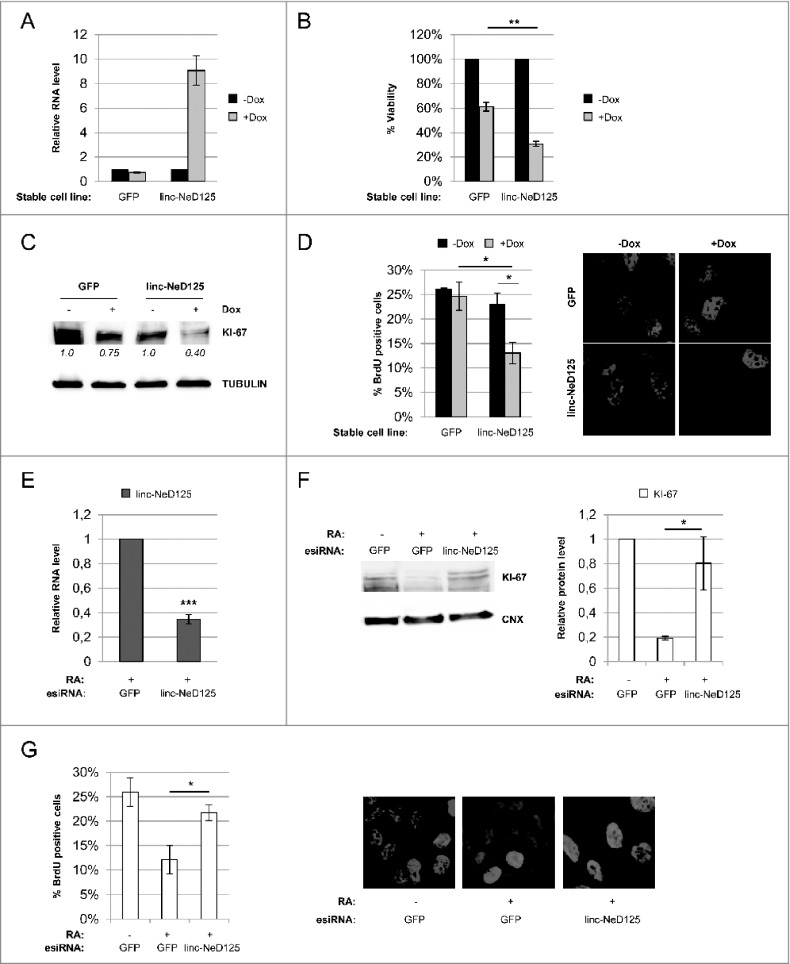
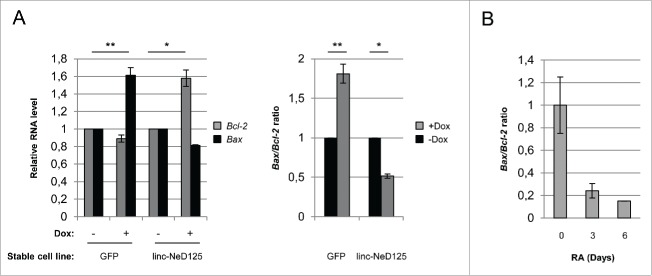
Role of linc-NeD125 in apoptosis. (A) Left panel: qRT-PCR analysis of Bcl-2 and Bax mRNA levels in linc-NeD125 and GFP overexpressing BE(2)-C cell lines upon 3 days of doxycycline-treatment. –Dox samples were set as 1. Gapdh mRNA was used as a loading control. Right panel: Bax/Bcl-2 mRNA ratio as evaluated from values reported in left panel. Histograms show mean values ± SEM from three independent experiments. *p < 0.05; **p < 0.01. (B) Bax/Bcl-2 ratio derived from qRT-PCR profiling of Bax and Bcl-2 mRNA (shown is Fig. S8) during in vitro BE(2)-C differentiation. Time points (days) of RA treatment are indicated below.
The effect of linc-NeD125 ectopic expression on cell viability was initially evaluated. Resazurin assay, which measures cell metabolic activity42, revealed a 50% reduction of cell viability following linc-NeD125 overexpression, compared to the control (Fig. 6B).
To verify whether such reduction could be contributed by altered cell proliferation and/or apoptosis, the involvement of linc–NeD125 in the control of these cellular processes was investigated.
Cell proliferation was analyzed by two distinct approaches. Analysis of the growth marker KI-67, which is expressed during all active phases of cell cycle43, showed a 60% reduction of the protein levels upon linc-NeD125 forced expression, compared to untreated cells (Fig. 6C). BrdU assay, which measures the rate of DNA replication44 , highlighted a 50% decrease of BrdU incorporation following linc-NeD125 ectopic expression (Fig. 6D). These results that demonstrate a role for linc-NeD125 in the control of cell growth were corroborated through mirror experiments. Linc-NeD125 depletion was performed through transfection of endoribonuclease-prepared siRNAs (esiRNAs) into RA-treated BE(2)-C cells, which caused a 70% decrease of the transcript (Fig. 6E). The following dosage of KI-67 protein and BrdU assay showed an almost complete recovery of KI-67 protein levels (Fig. 6F), and an 80% rescue of BrdU incorporation (Fig. 6G), with respect to control cells.
Overall, these data indicate that linc-NeD125 depletion counteracts the effect of RA on cell proliferation and clearly establish a relevant role for linc-NeD125 in the control of NB cell growth.
Then, we evaluated the effect of linc-NeD125 on apoptosis by measuring the expression levels of the pro-apoptotic BAX and anti-apoptotic BCL-2 genes, which constitute control checkpoints in the programmed cell death pathway. Intriguingly, as detected by qRT-PCR, the levels of Bcl-2 increased upon linc-NeD125 overexpression, whereas those of Bax were almost unaffected (Fig. 7A, left panel). Evaluation of Bax/Bcl-2 ratio, which determines cell susceptibility to apoptosis,45,46 revealed a 50% decrease upon linc-NeD125 ectopic expression with respect to the control cells (Fig. 7A right panel).
Figure 7.
For figure legend, see page . Figure 7 (see previous page). Role of Linc-NeD125 in cell growth. (A) Assessment of linc-NeD125 overexpression in BE(2)-C stable cell lines upon doxycycline treatment. qRT-PCR analysis of linc-NeD125 levels in untreated (-Dox) or 3 days doxycycline-treated (+Dox) BE(2)-C-linc-NeD125 cells (or BE(2)-C-GFP cells as a control). –Dox samples were set as 1. Gapdh mRNA was used as a loading control. Histogram shows mean values ± SEM from 3 independent experiments. (B) Resazurin-based viability assay performed in untreated (-Dox) or doxycycline-treated (+Dox) BE(2)-C-linc-NeD125 cells (or BE(2)-C-GFP cells as a control) in proliferating conditions. Cell viability is expressed as percentage relative to untreated (-Dox); values are the means ± SEM from 3 independent experiments. **p < 0.01. (C) Protein lysates from the samples in (B) were probed for the proliferation marker KI-67. TUBULIN was used as a loading control. KI-67 levels from densitometric analysis are relative to –Dox samples and shown below each lane. (D) BrdU- based proliferation assay was performed in untreated (-Dox) or doxycycline-treated (+Dox) BE(2)-C-linc-NeD125 cells (or BE(2)-C-GFP cells as a control) in proliferating conditions Left panel: quantification of BrdU incorporation; values are the means ± SEM from 3 independent experiments. *p < 0.05. Right panels: representative images of untreated (-Dox) or doxycycline-treated (+Dox) BE(2)-C-GFP or BE(2)-C-linc-NeD125 cells immuno-stained for BrdU (red); nuclei were counter-stained with DAPI (blue). (E) linc-NeD125 knockdown efficiency was monitored by qRT-PCR in RA-treated BE(2)-C cells transfected with specific esiRNA. Values are relative to control cells transfected with esiRNA against GFP and shown as the means ± SEM from 3 independent experiments. ***p < 0.001. (F) KI-67 protein levels were assessed in RA-treated cells depleted of linc-NeD125, by Western blot. As controls, proliferating and differentiating BE(2)-C cells transfected with esiRNA against GFP were also analyzed. The histogram on the right shows KI-67 densitometric analysis relative to –RA sample. Values are the means ± SEM from 3 independent experiments. Statistics as in (D). (G) BrdU assay was performed in RA-treated cells depleted of linc-NeD125. As controls, proliferating and differentiating BE(2)-C cells transfected with esiRNA against GFP were also analyzed. Histogram on the left shows quantification of BrdU incorporation. Values are the means ± SEM from 3 independent experiments. Statistics as in (D). Representative images of the assay are shown on the right. BrdU (red); DAPI (blue).
Interestingly, an increase of BCL-2 levels was previously observed upon RA treatment of BE(2)-C cells.47 Besides confirming this finding by evaluating Bcl-2 RNA levels at specific time points during in vitro neuronal differentiation (Fig. S8 left panel), we also found unaltered levels of Bax mRNA (Fig. S8 right panel).
Overall, these results indicate that linc-NeD125 enforced expression mimics the effects of RA administration on the control of Bax/Bcl-2 ratio (Fig. 7B).
We finally tested whether linc-NeD125 may be involved in cell differentiation. Reverse genetics approach allowed us to conclude that altered expression of the mature transcript did not affect the levels of neuronal markers (Fig. S9A-B).
Based on these data, we conclude that linc-NeD125 may be considered an anti-proliferative molecule, which also counteracts cell apoptosis during in vitro neuronal differentiation.
Discussion
In this work, we uncovered linc-NeD125 as a novel neuronal-induced, long intergenic non coding RNA that hosts miR-125b-1 into its intron. The GENCODE v7 catalog of human long non coding RNAs revealed that their introns represent a minor source of small RNAs: in particular, miRNA coding sequences are 3.5 fold-enriched in lncRNA exons with respect to introns.48 While the exonic arrangement may account for the many lncRNAs that solely function as precursors for small RNAs, intronic organization may be compatible with an intrinsic lncRNA function. Several linc-NeD125 features suggest it could be intrinsically functional, beyond merely serving as a miRNA precursor. Among them, we considered: i) its specific induction along neuronal differentiation, ii) its accumulation as a spliced, stable molecule upon differentiation stimulus, iii) its high level of evolutionary conservation in Primates.
We underscored a relevant role for linc-NeD125 in the control of cell metabolism, which resulted in 50% decrease of cell viability upon its ectopic expression in NB proliferating cells. Notably, this outcome was produced by the mature linc-NeD125 that can not generate the microRNA, indicating an autonomous function for the transcript. Importantly, cell viability reduction was accompanied by a 50% decrease of cell proliferation, evaluated both at the molecular (KI67 dosage) and at DNA activity (BrdU incorporation) levels, leading us to conclude that linc-NeD125 controls cell vitality mainly by arresting cell cycle progression.
An unexpected role for linc-NeD125 as an antiapoptotic factor came out from the analysis of BAX and BCL-2 genes, which represent critical intracellular checkpoints in the apoptotic pathway. The ectopic expression of linc-NeD125 in growing cells increased the expression levels of the antiapoptotic gene BCL-2, leading to a decrease of Bax/Bcl-2 ratio, which is characteristic for apoptosis-resistant cells.49 Since Bax/Bcl-2 ratio determines cell susceptibility to apoptosis, we propose that linc-NeD125 may function as a regulator of critical apoptosis control points.
It is noteworthy that RA-treatment of BE(2)-C cells results in conversion to an N-type morphology and extension of longer processes, in association with a 3 fold-increase in BCL-2 protein levels.47 Since it has been proposed that, during differentiation, BCL-2 confers a selective survival advantage to cells that up-regulate its expression,47 and since high levels of bcl-2 mRNA 50,51 and BCL-2 protein are found in the developing nervous system where it plays important functions in the regulation of neuron survival,52 one possibility is that linc-NeD125 may enforce the survival of cells that undergo neuronal differentiation.
The finding that linc-NeD125 functions independently of the hosted miR-125b-1, raises the question whether the two overlapping RNA species are also autonomously regulated. We identified a minimal locus promoter that displays a constitutive activity and found that transcription of linc-NeD125 and miR-125b-1 may initiate from distinct TSSs. We highlighted that the differential TSS usage was strictly dependent on the cell conditions. In particular, in proliferating cells both the TSSs are utilized at basal levels, as assessed by H3K4me3 occupancy and RNA Pol II recruitment, whereas the transition from proliferation to differentiation favors the selection of intronic TSS. Therefore, while the TSS upstream of linc-NeD125 may be considered as a constitutively active TSS, the intronic one is a differentiation-regulated TSS, which allows the transcriptional control of pri-miR-125b-1 expression. Further studies will be necessary to identify the cis- and trans-acting factors involved in such regulation.
A post-transcriptional regulatory mechanism is, instead, responsible for the accumulation of linc-NeD125, upon the differentiation stimulus. In this condition, linc-NeD125 levels are rapidly regulated by increasing the stability of its transcript, rather than the transcription rate of its gene.
We highlighted a role for the RNA binding protein TDP-43 as a positive regulator of linc-NeD125 gene expression. Overall, the evidence that: i) TDP-43 directly interacts with the mature form of linc-NeD125, ii) such interaction takes place only in differentiation conditions, when linc-NeD125 is stabilised, and iii) TDP-43 depletion caused a drastic reduction of linc-NeD125, suggest that TDP-43 may contribute to the control of linc-NeD125 stability. This activity played by TDP-43 in RA-treated cells is in line with its role as a neuronal activity-responsive factor53 and as a protein directly involved in neuronal differentiation.27, 54
In conclusion, we unveiled different layers of gene expression regulation that converge on the same gene locus to satisfy the demand of increased amounts of 2 ncRNAs, miR-125b-1 and linc-NeD125, during neuronal differentiation.
Notably, we characterized the function of the newly identified linc-NeD125 by demonstrating that its activity contributes to establish the conditions required for cells undergoing differentiation: as an antiproliferative factor it ensures decreasing of cell growth, whereas as an antiapoptotic factor it supports cell survival.
Materials and Methods
Oligonucleotides
Oligonucleotide sequences used in this study are listed in Table S1.
Cell cultures and treatments
Human neuroblastoma-derived BE(2)-C (ATCC® CRL-2268™) cells were cultured in RPMI medium 1640, supplemented with 10% fetal bovine serum (EU Standard, South American Origin), 1% sodium pyruvate, 1% L-glutamine and 1% penicillin/streptomycin (Gibco). Human medulloblastoma-derived D283 Med (ATCC® HTB-185™) cells were grown in MEM medium, supplemented with 20% fetal bovine serum (EU Standard, South American Origin), 1% sodium pyruvate, 1% L-glutamine and 1% penicillin/streptomycin, 1% non-essential aminoacid solution (EuroClone). BE(2)-C and D283 Med cell lines were induced to differentiate by 10 μM and 2.5 μM respectively all-trans-Retinoic Acid (RA, Sigma-Aldrich). D283 Med cells were cultured on poly-L-lysine (Sigma-Aldrich)-coated plates before RA-treatment. HL-60 (ATCC® CCL−240™) and NB4 cell lines were cultured and differentiated with RA as described in Salvatori et al., 2011.32 Myoblasts were treated as indicated in Ballarino et al., 2015.33
Plasmids were transiently transfected into untreated or RA-treated BE(2)-C cells by Lipofectamine and Plus Reagent (Invitrogen) in OPTI-MEM I medium (Gibco), according to manufacturer's directions. For linc-NeD125 stability studies, untreated or 3 days RA-treated BE(2)-C cells were cultured in the presence of 5 μg/ml of actinomycin D (Sigma-Aldrich), for the time points indicated in Figure 5A.
TDP-43 knockdown was performed as described in Di Carlo et al., 2013.27
For linc-NeD125 loss-of-function studies, esiRNA against linc-NeD125 (or GFP as a control) were purchased from Sigma. Differentiating BE(2)-C cells were transiently transfected for 3 days with 500 ng of each esiRNA. Linc-NeD125 esiRNA sequence is available upon request.
RNA extraction and analysis
Total RNA was extracted from confluent plates of untreated or RA-treated BE(2)-C or D283 Med cells using QIAzol (Qiagen), as recommended by the manufacturer's protocol. Prior to cDNA synthesis, 1 μg of RNA samples was treated with 1 U of DNase (Thermo Scientific), according to the manufacturer's directions.
For quantitative RT-PCR (qRT-PCR) assays, cDNA was generated using miScript II Reverse Transcription Kit (Qiagen) or SuperScript® III First-Strand Synthesis SuperMix (Invitrogen) for miR-124 or lncRNAs and mRNAs, respectively. The qRT-PCR detection was performed using quantiTect SYBR® Green PCR Master Mix (Qiagen) on a 7500 Fast Real-Time PCR (Applied Biosystem). Relative expression levels of the selected targets were normalized over Gapdh mRNA levels. miR-124 levels were normalized over U6 snRNA.
For miR-125b-1 putative host gene expression analyses and for linc-NeD125 isoforms determination, cDNA was obtained using SuperScript® III First-Strand Synthesis System for RT-PCR (Invitrogen) in the presence of oligo dT or random hexamers, and analyzed through semi-quantitative reverse transcriptase-PCR (RT-PCR). Amplifications were fractionated along a 2% agarose gel and revealed by ChemiDoc XRS+ Molecular Imager (Bio-Rad). Gapdh mRNA was used as loading control.
In vitro translation
Linc-NeD125 cDNA was cloned into pcDNA 3.1+ (Life Technologies), under the T7 promoter control. The resulting construct was subjected to coupled in vitro transcription and translation, in the presence of 35S-labeled methionine (Perkin Elmer), according to the TNT® Coupled Reticulocyte Lysate Systems protocol (Promega). A plasmid expressing the Firefly luciferase, provided by the kit, was used as a positive control. Translation products were fractionated at 150 V for 20 min or 1h along denaturing SDS-PAGE and labeled protein bands were detected by autoradiography.
Phylogenetic analysis
Genomic coordinates for the linc-NeD125 were retrieved through the UCSC genome browser (http://genome.ucsc.edu/) based on human genome assembly version hg19. Multiple alignment blocks relative to the two linc-NeD125 exons were obtained through Galaxy55 based on PhastCons56 multiple alignments data of vertebrate species for 18 species, including primates (human, chimpanzee, gorilla, orangutan, macaque, marmoset, baboon, bushbaby) and other placental mammals (armadillo, cat, dog, guineapig, microbat, mouse, pika, rabbit, rat, and tenrec).
Evolutionary relationships between sequences were inferred by using ClustalW 57 and graphically rendered in a phylogenetic tree by using the web tool Phylodendron (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html).
Cell fractionation
Untreated and 6 days RA-treated BE(2)-C cells, were fractionated according to the Ambion® PARIS™ system (Life Technologies). An in vitro-transcribed exogenous RNA (mmu-linc-MD1) 58 was added to each fraction, and used as internal control. RNA was extracted using miRNeasy Mini Kit (Qiagen). Equal volumes of cytoplasmic and nuclear DNA-free RNA were retro-transcribed using miScript II Reverse Transcription Kit (Qiagen) and analyzed by qRT-PCR. Values are normalized against the internal control and expressed as percentage of total RNA (cytoplasm + nucleus).
Primer extension analysis
The positions of linc-NeD125 and miR-125b-1 TSSs were mapped by primer extension. Ten μg of DNA-free RNA were reverse-transcribed with specific γ-32P-labeled oligonucleotides listed in Table S1. Reaction products were fractionated by urea-PAGE together with the sequencing reactions, obtained from M13 single strand DNA using the Sequenase Version 2.0 DNA Sequencing Kit (GE Healthcare).
Rapid amplification of 3′ cDNA ends (3′ RACE)
Linc-NeD125 3′ end was mapped by 3′ RACE System (Invitrogen). Two μg of total DNA-free RNA from BE(2)-C cells were retro-transcribed according to the manufacturer's protocol. The generated cDNA was amplified using the oligonucleotide 3′RACE_linc-NeD125 and sequenced.
Promoter analysis
DNA fragments mapping upstream of linc-NeD125 TSS were PCR-amplified from BE(2)-C genomic DNA, employing the oligonucleotides listed in Table S1, and cloned into pGL4.10 vector (Promega), upstream of the firefly Luc2 cDNA. Each plasmid was co-transfected into BE(2)-C cells together with a control plasmid expressing the RLuc gene (pRL-TK, Promega). After 6 days of incubation in the presence or absence of RA, cells were assayed with the Dual-Luciferase® Reporter Assay System (Promega).
Protein extraction and western blot assay
Whole-cell protein extracts were prepared from BE(2)-C cells lysed in RIPA buffer.27 Samples were separated by electrophoresis on 10% poly-acrylamide gel (Invitrogen), and electroblotted onto nitrocellulose membrane (Whatman™ Protran™ Nitrocellulose Blotting Membrane, Fisher Scientific). Immunoblots were incubated overnight with anti-REST antibody (Millipore, 07–579), anti-EGR1 antibody (Cell Signaling, #4154), anti-EGR2, anti-HuR, anti-FUS, anti-CNX and anti-GAPDH antibodies (Santa Cruz Biotechnology, sc-20690X, sc-5261, sc-47711, sc-11397, and sc-32233, respectively), anti-TDP-43 antibody (Proteintech, 10782–2-AP), anti-α-Tubulin antibody (Sigma-Aldrich, T5168) and anti-KI-67 antibody (Abcam, ab16667). Staining was performed by SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). Each immunoblot is representative of at least 3 biological replicates. All the experiments were quantified as described in Di Carlo et al., 2013.27
Chromatin Immunoprecipitation (ChIP) Assays
For the analysis of H3K4me3 levels, REST and EGR1 binding on linc-NeD125 promoter, chromatin extracts from proliferating or 6 days RA-treated BE(2)-C cells were immunoprecipitated with anti-trimethyl Histone H3 (Lys4) (Millipore 07–473), anti-REST , or anti-EGR1 according to Laneve et al., 201059 with minor modifications. Normal rabbit IgG, or mouse-IgG (Santa Cruz Biotechnology, sc-2027 and sc-2025, respectively) were used as negative controls. The immunoprecipitation levels were expressed as fraction of Input after background (IgG) subtraction. For EGR2 transcription factor and RNA Pol II, ChIP analyses were performed on chromatin extracts from proliferating or 6 days RA-treated BE(2)-C cells according to manufacturer's specifications of MAGnify Chromatin Immunoprecipitation System kit (Invitrogen). Sheared chromatin was immunoprecipitated with anti-Egr2 or anti-Pol II (Santa Cruz Biotechnology, sc-899). Normal rabbit IgG, provided by the kit, was used as negative control.
A standard curve for each primer pair was generated by qPCR, with different dilutions of Input chromatin, and used to quantify the immunoprecipitates. Data are presented as enrichment over background.
Biotin RNA pull-down assay
Linc-NeD125 pull-down assay was performed by using a short antisense probe, namely α-NeD RNA bait, to capture the endogenous linc-NeD125 and its protein partners. More in depth, a 173 nt-long antisense probe, specifically matching the splice junction of linc-NeD125 full length isoform, was produced by PCR reaction on the plasmid ePB-Puro-TT-linc-NeD125 as a substrate, with the primers reported in Table S1. As a control, pcDNA3.1+ Multiple Cloning Site (MCS) was PCR amplified, with the oligos reported in Table S1, generating an unspecific RNA bait of 177 nt, after in vitro transcription in the presence of Biotin-rUTP.
Four days RA-treated BE(2)-C cells were incubated in Lysis buffer (20 mM HEPES pH 7, 150 mM KCl, 5 mM MgCl2, 0.5% NP-40, 5% glycerol, 2 mM EDTA, 0.5 mM DTT), and 2.5 mg of total extract was incubated with 1 µg of αNeD125 biotinylated RNA bait or with 1 µg of a biotinylated control (pCDNA3.1+ MCS).40 Baits were previously heated for 10 min at 80°C and blocked on ice. Then, 100 ml of Streptavidin MagneSphere paramagnetic particles (Promega) saturated with 150 µg of tRNA were added to the extract for 15 min at room temperature. After three washes with EMSA buffer (20 mM HEPES-KOH, 150 mM KCl, 2 mM MgCl2, 1 mM DTT, 5% glycerol, 0,01% Triton), the bound complexes were eluted and analyzed by protein gel blot. Biotinylated baits were obtained from PCR-generated templates by using the T7 RiboMAX™ Express Large Scale RNA Production System (Promega) in the presence of 10 mM Biotin-rUTP (Roche).
Cross-linking Immunoprecipitation (CLIP) assay
BE(2)-C cells were grown with or without RA as described above, UV-crosslinked at 4000×100 µj/cm2 energy, and lysed in 100 mM NaCl, 20 mM TRIS-HCl pH 8, 0.5 mM EDTA, 0.5% NP-40 for 20 minutes on ice. Total extracts were incubated with 5 μg of TDP-43 antibody (isotype matched IgG were used as a negative control) and immunoprecipitated with ProteinA/G-sepharose resin (Pierce). After extraction by miRNeasy Mini Kit (Qiagen), immunoprecipitated RNA was retro-transcribed and analyzed by RT-PCR using specific primers for linc-NeD125, Tdp-43 or Gapdh mRNAs.
Generation of stable and inducible BE(2)-C cell line expressing full-length linc-NeD125
Linc-NeD125 exon I and exon II were PCR-amplified from BE(2)-C genomic DNA and ligated (in a ratio 1:1) to obtain the full-length linc-NeD125 isoform. The resulting DNA fragment was cloned between the BamHI and NotI restriction sites of the enhanced piggyBac transposable vector epB-Puro-TT.60 Helper and transposon plasmids (epB-Puro-TT-linc-NeD125, or epB-Puro-TT-GFP as a control) were transfected into BE(2)-C cells in a ratio 1:4, as described above. Selection with puromycin (0.5 μg/ml, Life Technologies) was initiated 48 hours after transfection and maintained until resistant colonies became visible, thus generating stably transfected BE(2)-C cells. When indicated (+Dox) cells were treated with doxycycline (100 ng/ml, Sigma-Aldrich) for 3 days.
Cell viability and proliferation assays
Cell viability was assessed by resazurin-based assay (Sigma-Aldrich) as previously reported61 with minor modifications. Five hundred BE(2)-C cells, bearing the inducible version of linc-NeD125 or a GFP-expressing construct as a control, were seeded in triplicate on a 96 well clear plates and grown with or without doxycycline for 3 days Fluorescence was measured with EnVision® Multilabel Reader (PerkinElmer) at 595 nm wavelength. To determine cell proliferation, the same cells were grown in parallel on coverslips and subjected to a 3-hour pulse of BrdU (Sigma-Aldrich) after doxycycline treatment. Cells were then fixed and stained for BrdU as in Ogrunc et al., 2014.62 BrdU positive cells were counted with CellProfiler software (cellprofiler.org).63 At least 150 cells were analyzed for each sample.
Statistical analyses
Statistical significance was determined by 2-tailed Student's t-test. A p value <0.05 was considered statistically significant. Data shown here are the mean ± SEM from at least 3 biological replicates, unless otherwise indicated. Statistical analyses of RNA stability were performed using GraphPadPrism (GraphPad Software).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank M. Arceci and M. Marchioni for technical support. We thank A. Rosa, J. Lenzi and M. Ballarino for helpful discussion. We also thank A. Rosa for gently providing the piggyBac transposable vector epB-Puro-TT.
Funding
This work was partially supported by grants from: Epigen-Epigenomics Flagship Project, ERC-2013-AdG 340172–MUNCODD, HFSP (RGP0009/2014), AriSLA full grant 2014 “ARCI.” VDC was supported by a fellowship from the Institute Pasteur, Fondazione Cenci-Bolognetti.
References
- 1.Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci 2006; 29:77-103; PMID:16776580; http://dx.doi.org/ 10.1146/annurev.neuro.29.051605.112839 [DOI] [PubMed] [Google Scholar]
- 2.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev 2007; 87:799-823; PMID:17615389; http://dx.doi.org/ 10.1152/physrev.00036.2006 [DOI] [PubMed] [Google Scholar]
- 3.Mercer TR, Dinger ME, Mariani J, Kosik KS, Mehler MF, Mattick JS. Noncoding RNAs in Long-Term Memory Formation. Neuroscientist 2008; 14:434-45; PMID:18997122; http://dx.doi.org/ 10.1177/1073858408319187 [DOI] [PubMed] [Google Scholar]
- 4.Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience 2013; 235:200-14; PMID:23337534; http://dx.doi.org/ 10.1016/j.neuroscience.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet 2013; 29:461-8; PMID:23562612; http://dx.doi.org/ 10.1016/j.tig.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 6.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014; 15:7-21; PMID:24296535; http://dx.doi.org/ 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 7.Sabin LR, Delas MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Mol Cell 2013; 49:783-94; PMID:23473599; http://dx.doi.org/ 10.1016/j.molcel.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al.. The transcriptional landscape of the mammalian genome. Science 2005; 309:1559-63; PMID:16141072; http://dx.doi.org/ 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- 9.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al.. Antisense transcription in the mammalian transcriptome. Science 2005; 309:1564-6; PMID:16141073; http://dx.doi.org/ 10.1126/science.1112009 [DOI] [PubMed] [Google Scholar]
- 10.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al.. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007; 447:799-816; PMID:17571346; http://dx.doi.org/ 10.1038/nature05874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al.. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012; 22:1760-74; PMID:22955987; http://dx.doi.org/ 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, Bozzoni I, Caffarelli E. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc Natl Acad Sci U S A 2007; 104:7957-62; PMID:17483472; http://dx.doi.org/ 10.1073/pnas.0700071104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Annibali D, Gioia U, Savino M, Laneve P, Caffarelli E, Nasi S. A new module in neural differentiation control: two microRNAs upregulated by retinoic acid, miR-9 and -103, target the differentiation inhibitor ID2. PLoS One 2012; 7:e40269; PMID:22848373; http://dx.doi.org/ 10.1371/journal.pone.0040269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun AX, Crabtree GR, Yoo AS. MicroRNAs: regulators of neuronal fate. Curr Opin Cell Biol 2013; 25:215-21; PMID:23374323; http://dx.doi.org/ 10.1016/j.ceb.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 2011; 9:113-8; PMID:21802386; http://dx.doi.org/ 10.1016/j.stem.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002; 12:735-9; PMID:12007417; http://dx.doi.org/ 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- 17.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843-54; PMID:8252621; http://dx.doi.org/ 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 18.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993; 75:855-62; PMID:8252622; http://dx.doi.org/ 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- 19.Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol 2009; 29:5290-305; PMID:19635812; http://dx.doi.org/ 10.1128/MCB.01694-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shenoy A, Danial M, Blelloch RH. Let-7 and miR-125 cooperate to prime progenitors for astrogliogenesis. EMBO J 2015; 34:1180-94; PMID:25715649; http://dx.doi.org/ 10.15252/embj.201489504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akerblom M, Petri R, Sachdeva R, Klussendorf T, Mattsson B, Gentner B, Jakobsson J. microRNA-125 distinguishes developmentally generated and adult-born olfactory bulb interneurons. Development 2014; 141:1580-8; PMID:24598163; http://dx.doi.org/ 10.1242/dev.101659 [DOI] [PubMed] [Google Scholar]
- 22.Gioia U, Di Carlo V, Caramanica P, Toselli C, Cinquino A, Marchioni M, Laneve P, Biagioni S, Bozzoni I, Cacci E, et al.. Mir-23a and mir-125b regulate neural stem/progenitor cell proliferation by targeting Musashi1. RNA Biol 2014; 11:1105-12; PMID:25483045; http://dx.doi.org/ 10.4161/rna.35508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R, Giangaspero F, Farcomeni A, et al.. MicroRNA profiling in human medulloblastoma. Int J Cancer 2009; 124:568-77; PMID:18973228; http://dx.doi.org/ 10.1002/ijc.23948 [DOI] [PubMed] [Google Scholar]
- 24.Sokol NS. Small temporal RNAs in animal development. Curr Opin Genet Dev 2012; 22:368-73; PMID:22578317; http://dx.doi.org/ 10.1016/j.gde.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12:996-1006; PMID:12045153; http://dx.doi.org/ 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 2012; 40:D130-5; PMID:22121212; http://dx.doi.org/ 10.1093/nar/gkr1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Carlo V, Grossi E, Laneve P, Morlando M, Dini Modigliani S, Ballarino M, Bozzoni I, Caffarelli E. TDP-43 regulates the microprocessor complex activity during in vitro neuronal differentiation. Mol Neurobiol 2013; 48:952-63; PMID:24113842; http://dx.doi.org/ 10.1007/s12035-013-8564-x [DOI] [PubMed] [Google Scholar]
- 28.Imanishi T, Itoh T, Suzuki Y, O'Donovan C, Fukuchi S, Koyanagi KO, Barrero RA, Tamura T, Yamaguchi-Kabata Y, Tanino M, et al.. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol 2004; 2:e162; PMID:15103394; http://dx.doi.org/ 10.1371/journal.pbio.0020162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J 2012; 31:522-33; PMID:22193719; http://dx.doi.org/ 10.1038/emboj.2011.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 2008; 4:e1000176; PMID:19043537; http://dx.doi.org/ 10.1371/journal.pcbi.1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E, Screpanti I, et al.. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J 2008; 27:2616-27; PMID:18756266; http://dx.doi.org/ 10.1038/emboj.2008.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvatori B, Iosue I, Djodji Damas N, Mangiavacchi A, Chiaretti S, Messina M, Padula F, Guarini A, Bozzoni I, Fazi F, et al.. Critical Role of c-Myc in Acute Myeloid Leukemia Involving Direct Regulation of miR-26a and Histone Methyltransferase EZH2. Genes Cancer 2011; 2:585-92; PMID:21901171; http://dx.doi.org/ 10.1177/1947601911416357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballarino M, Cazzella V, D'Andrea D, Grassi L, Bisceglie L, Cipriano A, Santini T, Pinnar∫ C, Morlando M, Tramontano A, et al.. Novel long noncoding RNAs (lncRNAs) in myogenesis: a miR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol Cell Biol 2015; 35:728-36; PMID:25512605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marinescu VD, Kohane IS, Riva A. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res 2005; 33:D91-7; PMID:15608292; http://dx.doi.org/ 10.1093/nar/gki103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckmann AM, Wilce PA. Egr transcription factors in the nervous system. Neurochem Int 1997; 31:477-510; discussion 7-6; http://dx.doi.org/ 10.1016/S0197-0186(96)00136-2 [DOI] [PubMed] [Google Scholar]
- 36.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ 3rd, Gingeras TR, et al.. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005; 120:169-81; PMID:15680324; http://dx.doi.org/ 10.1016/j.cell.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 37.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 2013; 152:1021-36; PMID:23452851; http://dx.doi.org/ 10.1016/j.cell.2013.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhury NR, de Lima Alves F, de Andres-Aguayo L, Graf T, Caceres JF, Rappsilber J, Michlewski G. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev 2013; 27:24-38; PMID:23307866; http://dx.doi.org/ 10.1101/gad.199190.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47:648-55; PMID:22841487; http://dx.doi.org/ 10.1016/j.molcel.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell 2014; 53:506-14; PMID:24440503; http://dx.doi.org/ 10.1016/j.molcel.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, et al.. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci 2012; 15:1488-97; PMID:23023293; http://dx.doi.org/ 10.1038/nn.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Czekanska EM. Assessment of cell proliferation with resazurin-based fluorescent dye. Methods Mol Biol 2011; 740:27-32; PMID:21468965; http://dx.doi.org/ 10.1007/978-1-61779-108-6_5 [DOI] [PubMed] [Google Scholar]
- 43.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol 2000; 182:311-22; PMID:10653597; http://dx.doi.org/ 10.1002/(SICI)1097-4652(200003)182:3%3c311::AID-JCP1%3e3.0.CO;2-9 [DOI] [PubMed] [Google Scholar]
- 44.Darzynkiewicz Z, Juan G. Analysis of DNA content and BrdU incorporation. Curr Protoc Cytom 2001; Chapter 7:Unit 7. [DOI] [PubMed] [Google Scholar]
- 45.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl−2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 1993; 74:609-19; PMID:8358790; http://dx.doi.org/ 10.1016/0092-8674(93)90509-O [DOI] [PubMed] [Google Scholar]
- 46.Yang E, Korsmeyer SJ. Molecular thanatopsis: a discourse on the BCL2 family and cell death. Blood 1996; 88:386-401; PMID:8695785 [PubMed] [Google Scholar]
- 47.Hanada M, Krajewski S, Tanaka S, Cazals-Hatem D, Spengler BA, Ross RA, Biedler JL, Reed JC. Regulation of Bcl−2 oncoprotein levels with differentiation of human neuroblastoma cells. Cancer Res 1993; 53:4978-86; PMID:8402688 [PubMed] [Google Scholar]
- 48.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al.. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012; 22:1775-89; PMID:22955988; http://dx.doi.org/ 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams JM, Cory S. The Bcl−2 protein family: arbiters of cell survival. Science 1998; 281:1322-6; PMID:9735050; http://dx.doi.org/ 10.1126/science.281.5381.1322 [DOI] [PubMed] [Google Scholar]
- 50.Abe-Dohmae S, Harada N, Yamada K, Tanaka R. Bcl−2 gene is highly expressed during neurogenesis in the central nervous system. Biochem Biophys Res Commun 1993; 191:915-21; PMID:8466531; http://dx.doi.org/ 10.1006/bbrc.1993.1304 [DOI] [PubMed] [Google Scholar]
- 51.Merry DE, Veis DJ, Hickey WF, Korsmeyer SJ. bcl−2 protein expression is widespread in the developing nervous system and retained in the adult PNS. Development 1994; 120:301-11; PMID:8149910 [DOI] [PubMed] [Google Scholar]
- 52.Anilkumar U, Prehn JH. Anti-apoptotic BCL−2 family proteins in acute neural injury. Front Cell Neurosci 2014; 8:281; PMID:25324720; http://dx.doi.org/ 10.3389/fncel.2014.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem 2008; 105:797-806; PMID:18088371; http://dx.doi.org/ 10.1111/j.1471-4159.2007.05190.x [DOI] [PubMed] [Google Scholar]
- 54.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A 2012; 109:3347-52; PMID:22323604; http://dx.doi.org/ 10.1073/pnas.1112427109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blankenberg D, Taylor J, Nekrutenko A. Making whole genome multiple alignments usable for biologists. Bioinformatics 2011; 27:2426-8; PMID:21775304; http://dx.doi.org/ 10.1093/bioinformatics/btr398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 2005; 15:1034-50; PMID:16024819; http://dx.doi.org/ 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2002; Chapter 2:Unit 2 3; PMID:18792934 [DOI] [PubMed] [Google Scholar]
- 58.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011; 147:358-69; PMID:22000014; http://dx.doi.org/ 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laneve P, Gioia U, Andriotto A, Moretti F, Bozzoni I, Caffarelli E. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res 2010; 38:6895-905; PMID:20624818; http://dx.doi.org/ 10.1093/nar/gkq604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J 2011; 30:237-48; PMID:21151097; http://dx.doi.org/ 10.1038/emboj.2010.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song H, Fares M, Maguire KR, Siden A, Potacova Z. Cytotoxic effects of tetracycline analogues (doxycycline, minocycline and COL-3) in acute myeloid leukemia HL-60 cells. PLoS One 2014; 9:e114457; PMID:25502932; http://dx.doi.org/ 10.1371/journal.pone.0114457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogrunc M, Di Micco R, Liontos M, Bombardelli L, Mione M, Fumagalli M, Gorgoulis VG, d'Adda di Fagagna F. Oncogene-induced reactive oxygen species fuel hyperproliferation and DNA damage response activation. Cell Death Differ 2014; 21:998-1012; PMID:24583638; http://dx.doi.org/ 10.1038/cdd.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al.. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006; 7:R100; PMID:17076895; http://dx.doi.org/ 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagasaki K, Sasaki K, Maass N, Tsukada T, Hanzawa H, Yamaguchi K. Staurosporine enhances cAMP-induced expression of neural-specific gene VGF and tyrosine hydroxylase. Neurosci Lett 1999; 267:177-80; PMID:10381005; http://dx.doi.org/ 10.1016/S0304-3940(99)00359-6 [DOI] [PubMed] [Google Scholar]
- 65.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al.. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 2011; 14:459-68; PMID:21358643; http://dx.doi.org/ 10.1038/nn.2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.